Significance
The Arctic is warming exceptionally rapidly, promoting an expansion of shrubs across the Arctic with global-scale climate implications. The Last Interglacial (∼125,000 y ago) was the most recent time the Arctic was warmer than present and thus serves as an analogue for Arctic greening in the near future. Ancient plant DNA in lake sediment from this time reveals major ecosystem changes in response to warmth, including an ∼400 km northward shift of dwarf birch relative to today. Enhanced shrub cover, corroborated by molecular and microfossil analyses, amplified warming during the Last Interglacial and will likely play a similar role in the future. This record constitutes the oldest authenticated plant DNA from lake sediment yet reported, increasing the technique’s temporal potential.
Keywords: paleoecology, Arctic greening, sedimentary ancient DNA, Last Interglacial
Abstract
Summer warming is driving a greening trend across the Arctic, with the potential for large-scale amplification of climate change due to vegetation-related feedbacks [Pearson et al., Nat. Clim. Chang. (3), 673–677 (2013)]. Because observational records are sparse and temporally limited, past episodes of Arctic warming can help elucidate the magnitude of vegetation response to temperature change. The Last Interglacial ([LIG], 129,000 to 116,000 y ago) was the most recent episode of Arctic warming on par with predicted 21st century temperature change [Otto-Bliesner et al., Philos. Trans. A Math. Phys. Eng. Sci. (371), 20130097 (2013) and Post et al., Sci. Adv. (5), eaaw9883 (2019)]. However, high-latitude terrestrial records from this period are rare, so LIG vegetation distributions are incompletely known. Pollen-based vegetation reconstructions can be biased by long-distance pollen transport, further obscuring the paleoenvironmental record. Here, we present a LIG vegetation record based on ancient DNA in lake sediment and compare it with fossil pollen. Comprehensive plant community reconstructions through the last and current interglacial (the Holocene) on Baffin Island, Arctic Canada, reveal coherent climate-driven community shifts across both interglacials. Peak LIG warmth featured a ∼400-km northward range shift of dwarf birch, a key woody shrub that is again expanding northward. Greening of the High Arctic—documented here by multiple proxies—likely represented a strong positive feedback on high-latitude LIG warming. Authenticated ancient DNA from this lake sediment also extends the useful preservation window for the technique and highlights the utility of combining traditional and molecular approaches for gleaning paleoenvironmental insights to better anticipate a warmer future.
The Arctic is greening as shrub biomass increases and vegetation ranges shift north in response to summer warming (1, 2). This process—one of the clearest terrestrial manifestations of climate change thus far—has major implications both for local ecosystems and for global energy balance and biogeochemical systems (3–5). In particular, taller shrubs darken otherwise snow-covered surfaces, contributing to the albedo feedback (6, 7), and enhanced evapotranspiration is expected to result in a positive greenhouse feedback (8). Shrub cover also impacts soil thermal regime, which may impact permafrost vulnerability (9–11). Because feedbacks related to Arctic greening are complex and potentially large in magnitude, estimating the extent and rate of northward shrub migration is a vital component of predicting future warming.
Past warm periods serve as valuable analogs for understanding the extent of Arctic greening under well-constrained climate conditions. The Last Interglacial (LIG; Marine Isotope Stage [MIS] 5e, 129 to 116 ka [thousands of years before present]) was ∼1 °C warmer than the preindustrial period globally, but the Arctic experienced amplified warming due to higher summer insolation anomalies and positive feedbacks at high latitudes (12, 13). The Eastern Canadian Arctic and Greenland, in particular, were likely ∼4 to 8 °C warmer in summer than present (Fig. 1) (14–18). LIG sediment records from this region thus provide an archive of the vegetation response to Arctic warming at levels comparable to predicted 21st-century climate change (19).
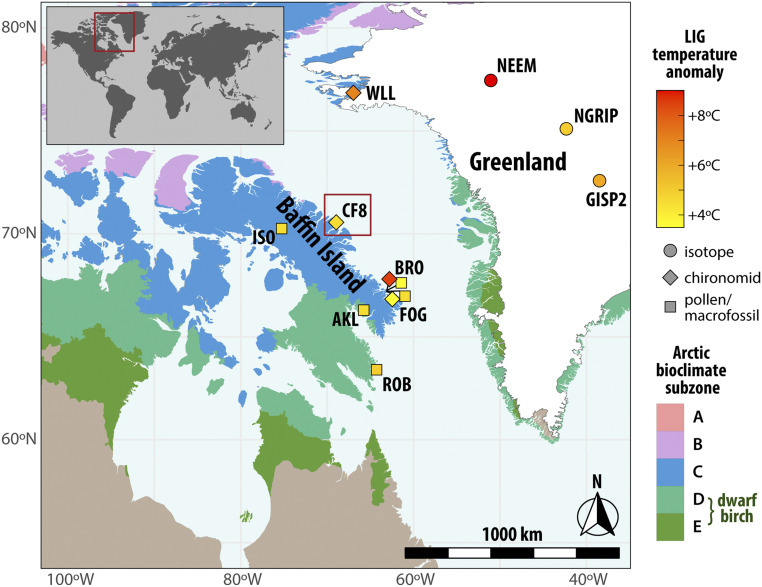
Fig. 1.
Map of Baffin Island and Lake CF8 study area. The symbols represent maximum LIG temperature anomalies based on terrestrial proxy records (shape indicates proxy type) from Baffin Island and Greenland (see SI Appendix, Table S1 for metadata). The shaded regions indicate Arctic bioclimate subzones delineations (29), including modern Betula range in subzones D and E. We note that a small outlier population of Betula occurs east of the D/E boundary on Baffin Island (not captured by vegetation map resolution) (38).
While most High Arctic lake basins were scraped clean by ice sheet erosion during the last glaciation and thus only contain postglacial sediments, lakes with small, low-relief catchments within regions of cold-based, slow-flowing ice were protected from erosion. Several such sites have been discovered on eastern Baffin Island, Arctic Canada and contain stratified records of multiple interglacials (20–22). Previous work from Lake CF8 on northeastern Baffin Island (Fig. 1 and SI Appendix, Fig. S1) demonstrates that its sediment record spans at least three interglacials (∼200 ka), including a substantially warmer-than-present LIG as indicated by chironomids, diatoms, and geochemical proxies (15, 23).
We targeted the multi-interglacial record from Lake CF8 to assess the vegetation response to pronounced warmth during the LIG and moderate warmth during the Holocene. Pollen produced by some key shrubs and trees, including Betula (birch), is efficiently wind-transported and thus present in lake sediments far north of their ranges (24, 25). We therefore analyzed both sedimentary ancient DNA (sedaDNA), which is sourced locally from within the lake catchment and does not include pollen-derived DNA (26), and fossil pollen to generate a robust vegetation record spanning the last ∼130 ka. Taken together, DNA-inferred plant communities and pollen-inferred July air temperatures provide insight into Arctic plant range shifts under strong summer warming.
Results
Lake CF8 (70.55818°N, 68.94968°W) is a small lake (surface area = 0.05 km2, max depth = ∼10 m; SI Appendix, Fig. S1) situated at 195 m above sea level on the Clyde Foreland, a broad coastal lowland of northeastern Baffin Island (Fig. 1). Its small catchment (∼0.2 km2) is typical of a High Arctic tundra (27, 28) or Arctic Bioclimate Subzone C (29). The modern plant communities include dry crustose and foliose lichens on rocks; wet graminoid and bryophyte communities on sorted-stripe slopes; and moist prostrate dwarf-shrub, sedge, and bryophyte communities on sorted polygons. Prostrate dwarf willows (Salix spp.) are rare (SI Appendix, Fig. S2). The upper slopes of the catchment feature frost-riven boulders, while glacial-fluvial sorting has deposited gravel and finer materials closer to the lake. Frost sorting has created stripes and polygons of finer materials that support the majority of soil development and vegetation cover.
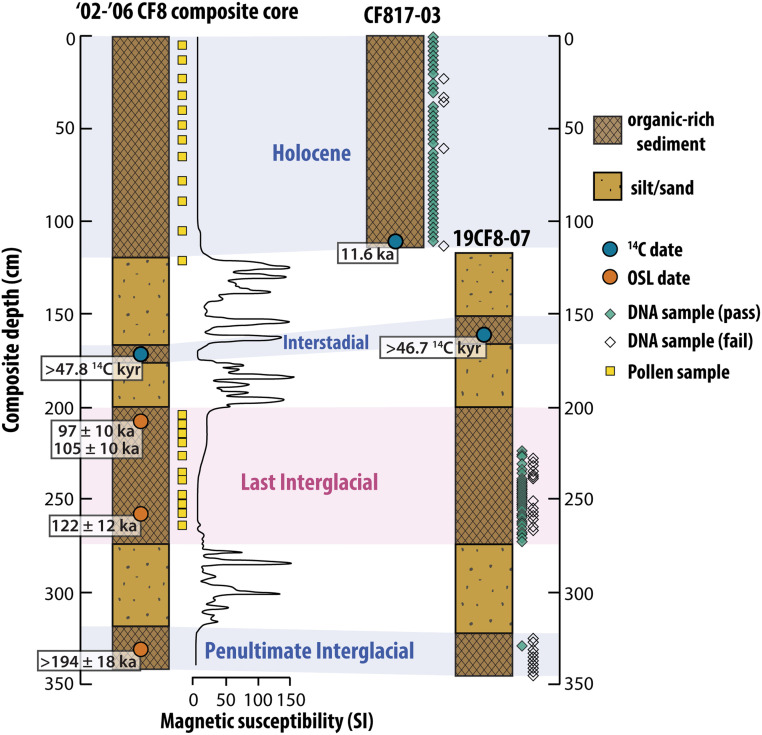
The CF8 sediment record is dominated by three organic-rich lacustrine sediment units that represent interglacial periods separated by sandy units that likely represent the waning stages of glacial periods following extensive depositional hiatuses (Fig. 2) (15, 20). The stratigraphy of cores collected in 2017 and 2019 correlate well with previously studied cores from CF8, lending confidence to interglacial assignments based on published age controls (SI Appendix, Fig. S3). In all cores, radiocarbon ages from plant macrofossils below the Holocene unit are >40 14C ka, indicating those units are beyond the radiocarbon dating window (Fig. 2 and SI Appendix, Table S2). The lowest interglacial unit in previously published cores yielded an optically stimulated luminescence (OSL) date of >194 ka and is thus assigned to the Penultimate Interglacial (PIG), MIS 7 (20). The next youngest interglacial is bounded by OSL dates of ∼122 and ∼100 ka (Fig. 2). Based on these dates and the lithostratigraphy of the core, we interpret this interval to represent the LIG but refrain from assigning absolute ages within the unit due to the large uncertainties (≥10 ka) associated with OSL dating. A thin nonglacial unit between the LIG and Holocene units is tentatively assigned to interstadial MIS 5a based on 14C dates >45 ka and the timing of maximum summer insolation across the Arctic (20, 30). We developed an age–depth model for the Holocene (12 ka to present) based on eight radiocarbon ages on plant macrofossils (SI Appendix, Fig. S4 and Table S2). Hereafter, we primarily focus on the LIG and Holocene units, where plant DNA preservation is adequate and interglacial assignment is most secure.
Fig. 2.
Lake CF8 core lithostratigraphy and sampling summary. Core diagrams show simplified sediment character and interglacial/interstadial assignments for previously published CF8 cores (2002 to 2006) (20, 23, 84) and new (2017 and 2019) CF8 cores. Magnetic susceptibility for 2002 to 2006 cores shows contrast between organic-rich (interglacial) units and minerogenic (deglacial) units. The key age control points (SI Appendix, Table S2) and pollen and DNA sample depths are shown with symbols defined in the legend.
We extracted and analyzed sedaDNA from the 2017 (Holocene) and 2019 (pre-Holocene) CF8 cores in dedicated ancient DNA facilities at Curtin University, Australia (see Materials and Methods). Targeting the P6 loop of the chloroplast trnL (UAA) intron (31), we amplified vascular plant DNA via PCR and sequenced the resulting amplicons following a standard metabarcoding approach (32). In the Holocene core, 41 of 46 samples passed our final quality filter (Fig. 2 and see Materials and Methods), indicating sufficient yields of endogenous plant DNA in 89% of analyzed levels. Following filtering, Holocene samples averaged 43,651 reads and 5.7 ± 2.8 (mean ±1 SD) taxa per sample, with no clear decrease in preservation over the last 12 ka (SI Appendix, Fig. S5). In the LIG core, 42 of 58 samples (72%) passed the final filter. Samples averaged 7,671 reads and 2.8 ± 1.0 taxa (SI Appendix, Fig. S5). Reduced yields and lower per-sample plant diversity in the LIG unit is an expected result of the >100-kyr age difference between LIG and Holocene samples, as DNA damage is known to accumulate through time in lake sediments (33). Of 30 total plant taxa identified, 12 are common to both interglacials, with 5 unique to the LIG and 13 occurring only in the Holocene (SI Appendix, Tables S3 and S4 and Fig. S2). Downcore trends in the dominant taxa (those occurring as >1% of reads in ≥2 samples) are summarized in Fig. 3. Pilot samples from the PIG largely failed to yield amplifiable DNA; only 1 of 12 samples passed final quality filtering, and it contained only one taxon (Salicaceae) (SI Appendix, Table S3). We thus consider the >190-ka unit from this site to be beyond the useful preservation window for the extraction and metabarcoding techniques employed here.
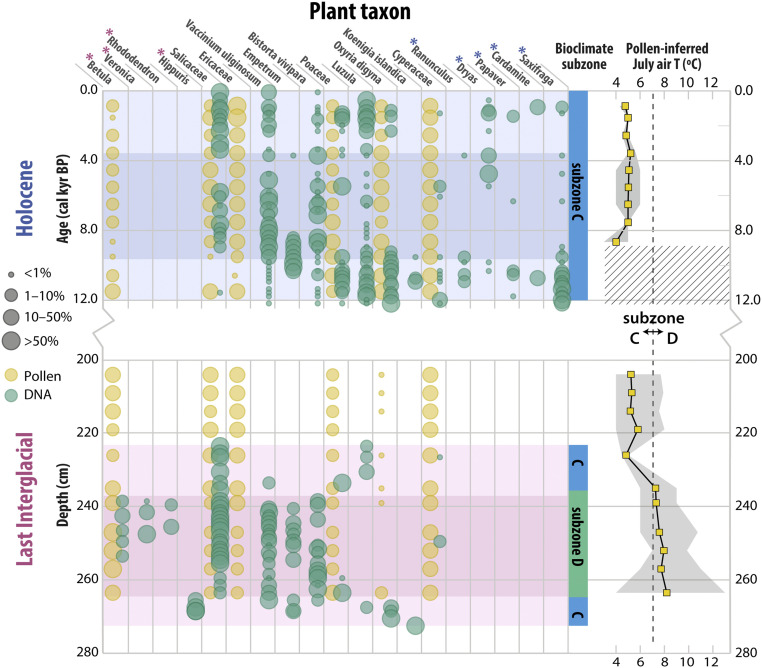
Fig. 3.
Vegetation and climate history at Lake CF8. Results from DNA metabarcoding (green circles; taxa occurring as >1% of reads in ≥2 samples included) and pollen (yellow circles; taxa averaging >5% included) are shown as circles scaled to relative abundance of sample reads or pollen grains (grouped into four percentage bins shown in legend). Note that in the LIG, no DNA analyses were completed above 223.5 cm, and no pollen analyses were completed below 263.5 cm. The pink and blue shading delineates biostratigraphic units (lower, middle, and upper for each interglacial) inferred from constrained hierarchical clustering of sedaDNA metabarcoding results. The pink and blue asterisks indicate taxa only occurring in the LIG and the Holocene, respectively, based on sedaDNA. Bioclimate subzone assignment (colors correspond to Fig. 1) is based on sedaDNA-inferred taxa presence within each unit. Pollen-inferred July air temperature estimate is based on the modern analog technique and includes the temperature range (gray band) from the five best modern analogs (see Materials and Methods). The dashed line indicates mean July temperature threshold (7 °C) defining the boundary between subzones C and D. The hatched area denotes interval with no adequate modern analog. Note the break in the y-axis and that Holocene samples are plotted on age scale while LIG samples are plotted on depth scale.
To verify the authenticity of the plant DNA within the LIG unit, we conducted shotgun sequencing on a subset of samples (see Materials and Methods and SI Appendix), one from the early Holocene (9.5 ka) and two from the LIG (at 242.5- and 246.5-cm depths). We investigated nucleotide misincorporation patterns by aligning each library to available complete plant reference genomes. For the two samples (both LIG) with a sufficient number of aligned reads aligned to the Salix brachista genome, we observed the increased rate of C to T (G to A) transitions at read starts (stops) known to characterize bona fide ancient DNA templates (SI Appendix, Figs. S6 and S7). These aligned reads were also short, with the LIG samples averaging 44 ± 11 base pairs (bp) in length (median ±1 SD) (SI Appendix, Figs. S8–S10).
We analyzed fossil pollen in sediment samples from previously published CF8 cores and aligned with the sedaDNA record using age–depth models (Holocene; SI Appendix, Fig. S4) and bulk geochemical trends (LIG; see SI Appendix, text and Fig. S3). Holocene sediments are dominated by pollen from herbs and heaths, while LIG sediments contain larger proportions of shrub taxa, including Betula (Fig. 3 and SI Appendix, Fig. S11 and Table S5). Pollen concentrations are on average higher in the LIG (72,600 grains per g dry sediment) compared to the Holocene (19,600 grains per g dry sediment) (Fig. 4 and SI Appendix, Table S5). We note that while concentrations in grains per gram of dry sediment account for differences in density between the two units, we cannot compare pollen fluxes because of a lack of age control within the LIG. An increase in pollen concentration in the late LIG is likely related to a reduction in background sedimentation rate rather than an increase in pollen flux. July air temperatures inferred from pollen assemblages (using the modern analog technique; see Materials and Methods) are highest in the LIG and ranged from ∼4 to 8 °C (Fig. 3).
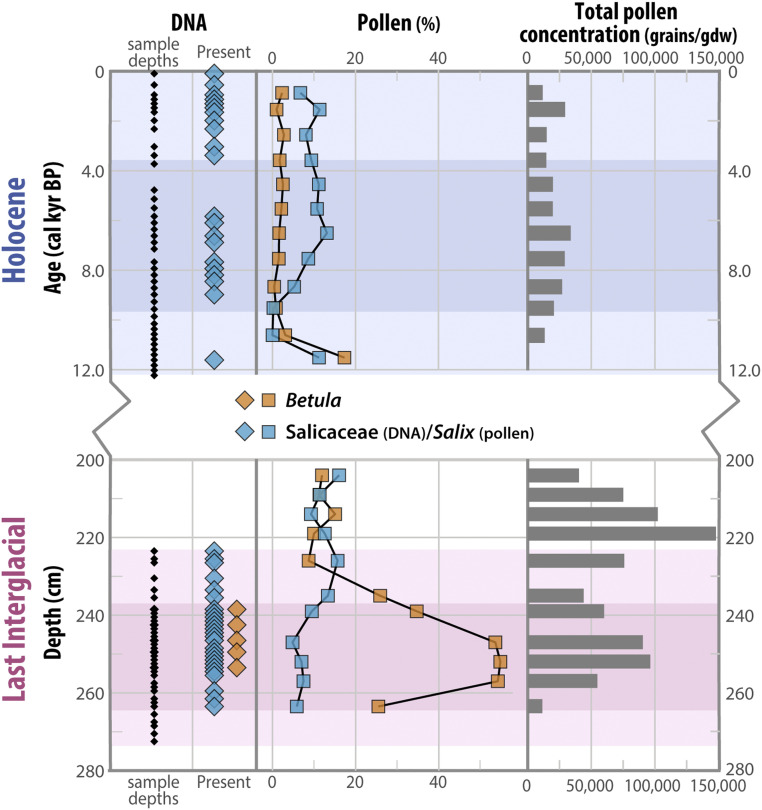
Fig. 4.
Multiproxy evidence for local Betula presence. The downcore trends for dominant woody shrubs Betula and Salicaceae (assumed Salix) based on DNA metabarcoding (diamonds; as presence) and pollen (squares; as percent of total pollen grains). Total pollen concentration (grains per g dry weight of sediment) highlights enhanced biomass in the LIG versus Holocene. The pink and blue shading delineates biostratigraphic units as in Fig. 3. Note the break in the y-axis and that Holocene samples are plotted on age scale while LIG samples are plotted on depth scale.
Discussion
Plant Community Changes across Interglacials.
The Holocene and LIG units contain genetic evidence for similar millennial-scale patterns of vegetation change, despite differences in peak plant communities. Constrained hierarchical clustering analysis resulted in the division of each interglacial into three biostratigraphic units based on the sedaDNA results (Fig. 3 and SI Appendix, Fig. S12) (34, 35). The distinct plant communities of the three units follow a broadly consistent interglacial pattern: 1) warming out of the glacial and early colonization of the landscape; 2) peak vegetation following peak summer warmth driven by high summer insolation; and 3) a shift back toward cold-tolerant taxa as summers cooled due to decreasing summer insolation. This consistent interglacial temporal pattern is captured by nonmetric multidimensional scaling ordination analysis (SI Appendix, Fig. S13) and is described below.
The lowest unit in both interglacials is characterized by early colonizers and/or cold-tolerant taxa, including Koenigia islandica, Oxyria digyna, and Luzula. The early Holocene also featured the relatively hardy taxa Saxifraga, Ranunculus, Papaver, and Carex, while the early LIG also featured the aquatic plant Hippuris. While the presence of Hippuris—which favors shallow water habitats (36)—could indicate a decrease in lake level during the early LIG, the current lake geometry includes some favorable shallow settings, and so a lake level change is not required.
Subsequently, a similar pattern of woody shrub establishment is apparent moving into the middle unit of both interglacials: High levels (>10%) of Empetrum occur first, followed by Vaccinium uliginosum, and then Salicaceae [(likely Salix spp (36)]. In the Holocene, Salicaceae is not consistently present until ∼8.9 ka, which was >3 ka after the catchment deglaciated. While absolute age control is unavailable for the LIG, a migration lag is apparent in the absence of Salicaceae from the lowest ∼10 cm (∼14%) of the interglacial unit. Betula [likely B. glandulosa or B. nana, dwarf birch (36)] is absent in the Holocene unit but appears in the LIG and remains present through much of its middle biostratigraphic unit. This result is consistent with warmer peak temperatures in the LIG than the Holocene, favoring northward migration of this subarctic shrub (17). The average pollen-inferred July temperature for the LIG middle unit is 8 °C (range: 6 to 11 °C), which is ∼3 °C above the modern mean July air temperature (SI Appendix) and the pollen-inferred Holocene average (5 °C) (Fig. 3). The absence of Betula DNA in the lowest ∼20 cm (∼27%) of the LIG despite favorable pollen-inferred temperatures suggests a pronounced migration lag—in concordance with the postglacial migration lag evident in a Holocene sedaDNA record from a southern Baffin Island lake core (32).
The upper unit of both interglacials is characterized by a shift back to relatively cold-tolerant taxa. This pattern is an expected manifestation of decreasing summer insolation through the interglacials (30), resulting in cooler summers—captured by a sharp decrease in pollen-inferred temperatures in the upper LIG unit and modest cooling in the late Holocene (Fig. 3). Betula disappears in the late LIG, and Empetrum is absent in the upper unit of both interglacials. The reappearance/expansion of Luzula and Poaceae occurs late in both interglacials. Saxifraga and Dryas also characterize the cooler late Holocene but have disappeared from the modern flora (SI Appendix, Fig. S2).
We note that while the pollen record largely corroborates the plant community at Lake CF8 through time, it indicates a less dynamic vegetation history than that inferred from sedaDNA (SI Appendix, Fig. S11). We suggest that this is due to reduced taxonomic resolution for some taxa and because pollen captures a more regional vegetation signal than sedaDNA (25, 37), including low to modest levels of wind-transported pollen from boreal trees, Alnus, and Betula when they were likely not growing near Lake CF8. Early in the interglacials following deglaciation, pollen may be sourced in part from older deposits on the landscape, whereas sedaDNA is unlikely to be preserved in reworked soils. We thus expect sedaDNA to more closely reflect local plant presence (and therefore timing of establishment/extirpation), whereas the pollen record represents a regionally smoothed vegetation history (25). Combining the two vegetation proxies—particularly given the greater preservation potential of pollen—enables a more robust approach to assessing ecosystem changes over the two interglacials compared to using either proxy alone.
Last Interglacial Shrub Range Shifts.
The DNA metabarcoding results provide compelling evidence that dwarf birch, a relatively cold-intolerant woody shrub that typifies the Low Arctic tundra and whose northern limit is currently ∼400 km south in the Eastern Canadian Arctic (38) (Fig. 1), was present at Lake CF8 during the LIG. Betula DNA was identified in five levels within the middle LIG unit, and it was not present (even prior to quality filtering) in any extraction controls or in any of the 46 Holocene samples (Fig. 3 and SI Appendix, Tables S3 and S4). Betula co-occurs with two other taxa, Veronica and Rhododendron, that were not present at CF8 during the Holocene. Veronica is currently found only at warmer, more southerly sites in the Eastern Canadian Arctic (36), while Rhododendron has been observed in Bioclimate Subzone C but is currently absent from the Clyde Foreland region. The interval with these three taxa defines the peak vegetation community at CF8 and is characteristic of Bioclimate Subzone D, which has a mean July temperature range of 7 to 9 °C (2 to 4 °C warmer than present; see SI Appendix) (29). This shift is consistent with peak LIG warmth of ∼3 °C above the late 20th century inferred from pollen (Fig. 3) and 4 to 5 °C above the late 20th century inferred from chironomid assemblages at CF8 (15) (Fig. 1).
A northward shift in the dwarf birch range is corroborated by a pronounced increase in Betula pollen during the middle LIG unit in CF8 sediment (Fig. 4). While low amounts of wind-transported Betula pollen are present throughout the CF8 Holocene record (<4% over the last 11 ka), the LIG sediment interval with DNA-inferred peak vegetation contains 25 to 55% Betula pollen. This peak in Betula pollen is consistent with local dwarf birch presence and an ∼400 km northward shift of Bioclimate Subzone D on Baffin Island. The average total pollen concentration for the LIG is nearly fourfold higher than the Holocene, further supporting an Arctic greening signal.
A northward migration of shrubs during the LIG has been inferred from other paleobotanical records from Baffin Island and elsewhere in the Arctic. A buried LIG soil section ∼20 km east of CF8 contains >50% Betula pollen, consistent with its local presence in the area (39). Pollen records from three lakes on southern Baffin Island (FOG, BRO, and AKL in Fig. 1) indicate a shrub tundra during the LIG, including the local establishment of Betula and possibly Alnus (alder) outside of their current limits, but range shift magnitudes are unclear due to the regional sourcing of pollen (17). Pollen in marine sediment southwest of Greenland also suggest enhanced shrub (alder and birch) cover on Greenland in the LIG compared to the Holocene (40). Moreover, substantial northward shifts of boreal treeline have been documented in paleorecords from northwest Alaska (41) and Siberia (42, 43). Widespread poleward range expansions during the LIG highlight important feedbacks between the climate system and biosphere during past warm periods.
Paleoclimate models have largely underestimated the magnitude of Arctic warming during the LIG (12, 44), and this discrepancy with proxy data may be related to models inadequately capturing vegetation-related feedbacks. Summer insolation at high northern latitudes during the early LIG was exceptionally high (45), with peak insolation at 127 ka occurring after sea level reached modern levels (46). This suggests that unlike during the Holocene, the penultimate continental ice sheets had largely disappeared prior to the insolation maximum of the LIG, enabling excess solar radiation to drive strong terrestrial warming (sensible heat) rather than ice ablation (latent heat). Thus, high-latitude landscapes were ice-free in time for more significant vegetation shifts compared to the Holocene. Models assessing LIG climate dynamics have identified vegetation changes, and especially the interplay between vegetation and snow cover (i.e., taller vegetation masking the high albedo of seasonal snow), as the most critical feedback amplifying high-latitude warming (47), including a near doubling of high-latitude temperature changes (48). Emerging climate model evidence suggests that Arctic vegetation feedbacks also play a large role in determining Greenland Ice Sheet mass balance and thus its contribution to sea level rise (49, 50). Such findings underscore the need for more spatially precise vegetation records from the LIG, especially as near-future climate change may be similarly amplified by as-yet unconstrained Arctic greening.
DNA Preservation in >100 ka Lake Sediment.
The Lake CF8 sediment record presents a valuable opportunity to assess the preservation potential of sedaDNA in favorable (cool) settings. Our DNA metabarcoding results indicate a systematic reduction in DNA preservation through progressively older interglacials (Fig. 2 and SI Appendix, Fig. S5), but sediment between ∼100 and 130 ka contains sufficiently well-preserved DNA for amplifying a 30- to 85-bp region of chloroplast DNA. This extends substantially the useful timeframe for authenticated lake sediment–based ancient DNA studies, which have so far focused on latest Pleistocene (<30 ka) and Holocene timescales (26, 51). We demonstrate through multiple lines of evidence that the LIG DNA is indeed endogenous and can provide ecologically meaningful information. The plant taxa inferred from sedimentary DNA within the CF8 LIG unit align well with ecological and paleoclimatic predictions, are replicable within the core, and are bioinformatically robust (see SI Appendix). Shotgun sequencing of LIG DNA extracts reveals clear damage patterns (nucleotide misincorporation and short fragment lengths) that confirm their ancient origin (SI Appendix, Figs. S6–S10).
As the greening of the Arctic proceeds due to human-caused climate change, precise and detailed paleoecological records from past warm periods are essential to anticipate the extent of future greening. Evidence for shrub range shifts in >100-ka sedimentary DNA underscore the potential to generate more taxonomically resolved, location-specific paleovegetation records from past interglacials than has been possible with traditional proxies alone. Given the potentially strong vegetation-related climate feedbacks initiated by northward shrub expansion, LIG sedaDNA records may elucidate the nature of environmental changes through warmer-than-present conditions and thereby help to improve the predictive power of climate models.
Materials and Methods
Core Collection, Description, and Age Control.
The Holocene sediment core (CF817-03) was collected in August 2017 using a Bolivia-type corer from an inflatable raft. The pre-Holocene sediment core (19CF8-07) was collected in May 2019 using a modified Nesje corer and aluminum core tube from the frozen lake surface. Both cores were taken from the central deep basin of the lake (SI Appendix, Fig. S1). Cores were transferred to cold room (4° C) storage prior to splitting and sampling within 9 mo of collection. Cores were split at the Trace and Environmental DNA (TrEnD) Lab at Curtin University (CF817-03) and the Sedimentology Lab at University of Colorado Boulder (19CF8-07) using bleach-cleaned tools and described immediately to note visual transitions between units.
A Holocene age–depth model was developed using eight radiocarbon ages on aquatic moss macrofossils, which are reliable in this region for radiocarbon dating (52). Macrofossils were sonicated in deionized water and freeze dried prior to preparation (acid–base–acid pretreatment and graphitization) in the Institute of Arctic and Alpine Research (INSTAAR) Laboratory for Accelerated Mass Spectrometry (AMS) Radiocarbon Preparation and Research. Radiocarbon measurements were made at the University of California Irvine AMS facility. The ages were used to create a Bayesian age–depth model in Bacon version 2.3.9.1 for R (53) (mem.strength: 4; mem.mean: 0.9; 116 1-cm sections; SI Appendix, Fig. S4), which relies on the IntCal20 calibration curve (54). Interpolated ages were then used to align Holocene DNA samples with pollen samples from a previously published age model (55).
Because internal age control was not possible for the pre-Holocene sediment, we aligned LIG units from core 19CF8-07 with previously published CF8 cores using bulk geochemistry (%C, C:N, and δ13C; see SI Appendix). Sediment subsamples were freeze dried, homogenized, and weighed into tin capsules for element analysis. Samples were run on a Flash 2000 Organic Elemental Analyzer interfaced with a Delta V Plus isotope ratio mass spectrometer via a Conflo IV in the University of Colorado Boulder Earth Systems Stable Isotope Lab following standard protocols.
DNA Metabarcoding.
Sediment sampling and DNA extractions were completed in an ultra-clean facility dedicated to ancient DNA work at the TrEnD Lab following established ancient DNA protocols for personal protective equipment and tool cleaning. Preparation for PCR was completed in a separate dedicated clean laboratory, and post-PCR work took place in a different building. Pristine sediment samples were taken from the center of the core (by scraping away uppermost sediment and avoiding core tube surfaces). A total of 2 500-mg aliquots were processed from each homogenized sediment subsample.
DNA digestion and extraction were completed following procedures described in Crump et al. (2019). Briefly, each aliquot was treated with 1 mL digest buffer following Grealy et al. (56), vortexed, and incubated in a hybridization oven at 55 °C with rotation for 24 h. An extraction control containing only digest buffer was prepared for each batch of 9 to 11 samples (SI Appendix, Table S4). Digests were concentrated to a volume of ∼50 µl using Vivaspin 500 centrifugal concentrators (Sigma Aldrich) and added to 1,300 µl of binding buffer following Dabney et al. (57). Samples were vortexed and then purified using a MinElute PCR Purification Kit (QIAGEN), eluted in a volume of 50 µl and stored at −20 °C.
An initial round of qPCR (StepOnePlus, Applied Biosystems) using full concentration, 1/10, and 1/100 dilution samples targeting the p6 loop of trnL–gh enabled assessment of suitable DNA templates and potential PCR inhibition in all samples. Based on preliminary qPCR results, optimal dilutions (typically 1/10 or 1/100 due to PCR inhibition) were then PCR amplified with unique (never previously used) multiplex identifiers (MID tags) for each metabarcoding assay via single-step amplification. Two PCR replicates were processed for each Holocene extract, and four PCR replicates were processed for LIG and PIG extracts to account for additional DNA degradation in older sediments. Two to four no-template controls and one to two positive controls were processed with each PCR plate. PCR reactions were as described in Crump et al. (2019), using 96-well plates and conditions set for 95 °C for 5 min, followed by 45 cycles of 95 °C for 30 s, 72 °C for 45 s, and 10 min at 72 °C.
The resulting amplicons were combined into “minipools” based on qPCR amplification curves and quantified via QIAxcel fragment analysis (QIAGEN), blended equimolarly into a sequencing library, purified with a QIAquick PCR Purification Kit (QIAGEN), and eluted in a volume of 32 µl. Illumina sequencing adapters were ligated onto the sequencing libraries using one of two PCR-free ligation methods: the Holocene library was end repaired, A tailed, and ligated using a method described in refs. 32 and 58, with the library purified between each step using a QIAquick PCR Purification Kit (QIAGEN) and eluted in a final volume of 30 µl; LIG libraries were ligated using the NxSeq AmpFREE Low DNA Library Kit (Lucigen) following manufacturer’s instructions, with an initial library concentration target of 500 ng and the final library purified using a QIAquick PCR Purification Kit (QIAGEN) and eluted in a volume of 60 µl.
Ligated libraries were size selected using a Pippin Prep (Sage Science) for 150 to 500 bp fragments, then cleaned via QIAquick PCR Purification Kit (QIAGEN). Libraries were quantified via Qubit Fluorometric Quantitation (ThermoFisher Scientific) and QIAxcel fragment analyzer (QIAGEN) to determine ligation efficiency. Dilutions of each library were denatured into single-stranded products and then prepared and loaded on an Illumina MiSeq Reagent Nano Kit version 2 (300 cycles) following manufacturer instruction.
Paired-end sequence data were demultiplexed using the insect package in R (59). Primers, adapters, and tags were then trimmed (exact matches only) and sequences shorter than 30 bp removed using cutadapt version 2.4 (60). Single-end sequences were first processed with OBITools (61): demultiplexed and primers/tags identified and removed (ngsfilter), filtered for length (minimum 30 bp; obigrep), and split into individual sample files (obisplit). All sequences were then dereplicated, denoised, merged (paired end only), and filtered for chimeras using the DADA2 pipeline (62). The resulting amplicon sequence variants were compared against both the National Center for Biotechnology Information (NCBI) GenBank database (63) and a curated Arctic–Boreal–Bryophyte database (64–66) using the BLASTn algorithm (67). Taxonomic assignments were made using a custom LCA algorithm in Python (68), requiring 100% of query coverage and 96% identity match with reference sequences.
Taxonomic assignments were further filtered for presumed contaminants/misassignments based on taxa present in the online version of the Flora of the Canadian Arctic Archipelago (36). Samples (with PCR replicates merged) were only retained if they passed the following final filtering criteria: 1) at least 1,000 reads assigned to ecologically plausible (“local”) taxa; 2) more local than contaminant sequences. We completed this filtering process with both the GenBank and curated Arctic–Boreal–Bryophyte database results and discarded a sample if it failed to pass the filter in either case. A minimum read cutoff of 10 reads was used for taxonomic observations, except in cases where a taxon was identified in >10 reads for any of the 12 negative controls, in which case we increased the read cutoff to 100 reads (Salicaceae, Empetrum, and O. Digyna) to account for low-level cross contamination (SI Appendix, Tables S3 and S4). Results derived from the Arctic–Boreal–Bryophyte database comparison were used for subsequent analyses.
We note that the LIG metabarcoding dataset includes two independent batches of sediment samples (overlapping depths from two halves of the same core), which we combined for main figures and interpretation; see SI Appendix, text and Fig. S14.
Shotgun Sequencing and Analysis.
Five extracts from select intervals of the CF8 core, along with a single library negative control, were prepared into shotgun libraries using a one-reaction, single-stranded library preparation developed to target degraded DNA (69). After adapter ligation, each library was purified using MinElute columns (Qiagen). A qPCR was carried out to determine the number cycles prior to amplifying with Illumina-compatible indices. Each library was subsequently amplified 11 to 18 cycles using 50 µL of Amplitaq 360 Gold Master Mix (ThermoFisher Scientific), 48 µL of DNA input, and 1 µL of 100-µM custom PCR primers containing custom 7-bp barcodes for downstream demultiplexing. Amplified PCR products were then purified using Sera-mag Speed Beads (GE Healthcare Sciences) with a 0.05% Tween concentration and eluted in 24 µL EBT buffer. Fragment size of each purified library was determined using a Fragment Analyzer (Agilent Technologies). Lastly, libraries were sequenced to 300 cycles using an Illumina NextSEq 500 instrument at UC, Santa Cruz Paleogenomics Lab.
Raw sequencing reads were trimmed and merged using SeqPrep (https://github.com/jstjohn/SeqPrep) and subsequently complexity filtered using PrinSEq (70). All complexity filtered reads were aligned to the NCBI full-nucleotide database using the BlastN command line tool and thereafter viewed with MEGAN (MEtaGenome ANalyzer) version 6 (71). Reads were then aligned to several species with readily available complete reference genomes on NCBI using Burrows-Wheeler Aligner (72), allowing each aligned read a minimum mapping quality score of 30. Nucleotide misincorporation plots were generated using MapDamage (73) with reads aligning to S. brachista. We note that we used the S. brachista genome because it was the highest quality, most complete Salix genome available and is genetically similar enough to the Salix species native to Arctic Canada to effectively identify Salix sequences in our samples. Complete aligned read-length plots accompanied with statistical analysis were generated with R (version 3.6).
Pollen Analysis and July Air Temperature Reconstruction.
Standard pollen preparation technique, including dispersion with KOH, digestion with HF and HCl, and acetolysis (74), were applied to 2.0 cm3 samples of fresh (wet) sediment. Pollen concentrations were determined by spiking with 3 Lycopodium spore tablets that contain 18,583 spores each (75). The basic sum used for relative frequency calculations included all spermatophyta taxa. The pollen sum averaged 450 grains in Holocene samples and 550 grains in LIG samples (SI Appendix, Table S5). Pollen concentrations were converted from volumetric (grains/cm3) to concentration by mass (grains/g sediment) using dry bulk density to account for differential compaction between the Holocene and LIG.
July air temperature reconstruction was performed using the modern analog technique (MAT). The reconstruction was made on the basis of a modern database including 828 pollen assemblages from lakes of the Boreal, Subarctic, and Arctic biomes of North America (north of 50°N) and Greenland, and corresponding temperature measurements (76, 77). The temperature data used are from the Climate Research Unit, University of East Anglia gridded climatology based on 1961 to 1990 meteorological averages (78). Pollen data include the 39 most common vascular taxa (18 woody plants and 21 herbs). For MAT, similarity between fossil and modern pollen assemblages is based on the squared chord distance (SCD) (79). In the present study, the adopted SCD threshold below which a modern sample is accepted as an analog is 0.26. To improve the statistical reliability, the July air temperature estimates were based on a set of five closest modern analogs. July air temperature reconstruction was performed with the “bioindic” package (ftp://ftp.cerege.fr/R/Package_bioindic) in R. The accuracy (RMSE) of the reconstruction is estimated to ±1.4 °C (r = 0.92). More details on the MAT approach used are given by ref. 77.
Data Analysis.
The Holocene and LIG units were subdivided into biostratigraphic units using constrained hierarchical clustering, computed via chclust (coniss method) in the R package Rioja (35). Holocene samples were analyzed using relative abundance of reads assigned to taxa, while LIG samples were analyzed with presence-absence data only; because LIG samples are highly degraded and yielded fewer taxa on average (∼2.8 versus 5.7 taxa in the Holocene), the relative abundance of reads is less meaningful and more reasonable results were obtained with presence-absence information only. The number of biostratigraphic units was determined by comparing the dispersion (as within-group sum of squares) between a broken stick model and hierarchical cluster grouping. The crossover point of the two models indicates the number of statistically significant groups based on hierarchical clustering—three groups for both interglacials (SI Appendix, Fig. S12) (34). To assess changes in vegetation at the community scale, we conducted nonmetric multidimensional scaling (NMDS) ordination analysis (80) on presence–absence metabarcoding data for the Holocene and LIG combined. NMDS was computed using the R package vegan (81) (metaMDS) using a Bray-Curtis dissimilarity matrix, yielding a minimum stress of 0.15.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Qikiqtaani Inuit and government of Nunavut for access to their land (Nunavut Research Institute Permits #0102217R-M and #0203819R-M). We thank CH2MHill Polar Services, Polar Continental Shelf Program, Joshua Akavak, Lasalie Joanasie, Zach Montes, and Devon Gorbey for field support and Mahsa Mousavi Mousaviderazmahalleh and Katrina West for bioinformatics expertise. We thank the INSTAAR Laboratory for AMS Radiocarbon Preparation and Analysis for sample preparation and insights. This research was funded by the US NSF (Office of Polar Programs Awards #1737712 to G.H.M. and J.S., #1737716 to E.K.T., and #1737750 to M.K.R.; NSF Graduate Research Fellowship Program Award #1144083 to S.E.C.; and NSF Doctoral Dissertation Research Improvement Award #1657743 to G.H.M. and S.E.C.), National Geographic Society, CU Center for the Study of Origins, CU Graduate School, and the Geological Society of America.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019069118/-/DCSupplemental.
Data Availability
Sequencing data have been deposited in the Dryad Digital Repository (https://doi.org/10.5061/dryad.pk0p2ngmq) (82) and pollen data have been deposited in the Neotoma Paleocology Database (https://data.neotomadb.org/48940) (83).
References
- 1.Elmendorf S. C., et al., Plot-scale evidence of tundra vegetation change and links to recent summer warming. Nat. Clim. Chang. 2, 8–12 (2012). [Google Scholar]
- 2.Myers-Smith I. H., et al., Complexity revealed in the greening of the Arctic. Nat. Clim. Chang. 10, 106–117 (2020). [Google Scholar]
- 3.Mekonnen Z. A., Riley W. J., Grant R. F., 21st century tundra shrubification could enhance net carbon uptake of North America Arctic tundra under an RCP8.5 climate trajectory. Environ. Res. Lett. 13, 054029 (2018). [Google Scholar]
- 4.Pearson R. G., et al., Shifts in Arctic vegetation and associated feedbacks under climate change. Nat. Clim. Chang. 3, 673–677 (2013). [Google Scholar]
- 5.Forkel M., et al., Enhanced seasonal CO2 exchange caused by amplified plant productivity in northern ecosystems. Science 351, 696–699 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Sturm M., Douglas T., Racine C., Liston G. E., Changing snow and shrub conditions affect albedo with global implications. J. Geophys. Res. 110, 1–13 (2005). [Google Scholar]
- 7.Chapin F. S. I. 3rd, et al., Role of land-surface changes in arctic summer warming. Science 310, 657–660 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Swann A. L., Fung I. Y., Levis S., Bonan G. B., Doney S. C., Changes in Arctic vegetation amplify high-latitude warming through the greenhouse effect. Proc. Natl. Acad. Sci. U.S.A. 107, 1295–1300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence D. M., Swenson S. C., Permafrost response to increasing Arctic shrub abundance depends on the relative influence of shrubs on local soil cooling versus large-scale climate warming. Environ. Res. Lett. 6, 045504 (2011). [Google Scholar]
- 10.Sturm M., et al., Snow–Shrub interactions in Arctic Tundra: A hypothesis with climatic implications. J. Clim. 14, 336–344 (2001). [Google Scholar]
- 11.Myers-Smith I. H., Hik D. S., Shrub canopies influence soil temperatures but not nutrient dynamics: An experimental test of tundra snow-shrub interactions. Ecol. Evol. 3, 3683–3700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto-Bliesner B. L., et al., How warm was the last interglacial? New model-data comparisons. Philos. Trans. A Math. Phys. Eng. Sci. 371, 20130097 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Members C-LIP , Last Interglacial Arctic warmth confirms polar amplification of climate change. Quat. Sci. Rev. 25, 1383–1400 (2006). [Google Scholar]
- 14.Dahl-Jensen D.et al.; NEEM community members , Eemian interglacial reconstructed from a Greenland folded ice core. Nature 493, 489–494 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Axford Y., et al., Chironomids record terrestrial temperature changes throughout Arctic interglacials of the past 200,000 yr. Geol. Soc. Am. Bull. 123, 1275–1287 (2011). [Google Scholar]
- 16.McFarlin J. M., et al., Pronounced summer warming in northwest Greenland during the Holocene and Last Interglacial. Proc. Natl. Acad. Sci. U.S.A. 115, 6537–6362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fréchette B., Wolfe A. P., Miller G. H., Richard P. J. H., de Vernal A., Vegetation and climate of the last interglacial on Baffin Island, arctic Canada. Palaeogeogr. Palaeoclimatol. Palaeoecol. 236, 91–106 (2006). [Google Scholar]
- 18.Yau A. M., Bender M. L., Robinson A., Brook E. J., Reconstructing the last interglacial at Summit, Greenland: Insights from GISP2. Proc. Natl. Acad. Sci. U.S.A. 113, 9710–9715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Post E., et al., The polar regions in a 2°C warmer world. Sci. Adv. 5, eaaw9883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briner J. P., Axford Y., Forman S. L., Miller G. H., Wolfe P., Multiple generations of interglacial lake sediment preserved beneath the Laurentide Ice Sheet. Geology 35, 887 (2007). [Google Scholar]
- 21.Steig E. J., Wolfe A. P., Miller G. H., Wisconsinan refugia and the glacial history of eastern Baffin Island, Arctic Canada: Coupled evidence from cosmogenic isotopes and lake sediments. Geology 26, 835 (1998). [Google Scholar]
- 22.Miller G. H., et al., Stratified interglacial lacustrine sediments from Baffin Island, arctic Canada: Chronology and paleoenvironmental implications. Quat. Sci. Rev. 18, 789–810 (1999). [Google Scholar]
- 23.Wilson C. R., et al., Arctic lake ontogeny across multiple interglaciations. Quat. Sci. Rev. 31, 112–126 (2012). [Google Scholar]
- 24.Birks H. H., The Late-Quaternary history of arctic and alpine plants. Plant Ecol. Divers. 1, 135–146 (2008). [Google Scholar]
- 25.Niemeyer B., Epp L. S., Stoof-Leichsenring K. R., Pestryakova L. A., Herzschuh U., A comparison of sedimentary DNA and pollen from lake sediments in recording vegetation composition at the Siberian treeline. Mol. Ecol. Resour. 17, e46–e62 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Parducci L., et al., Ancient plant DNA in lake sediments. New Phytol. 214, 924–942 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Kerwin M. W., Overpeck J. T., Webb R. S., Anderson K. H., Pollen-based summer temperature reconstructions for the eastern Canadian boreal forest, subarctic, and Arctic. Quat. Sci. Rev. 23, 1901–1924 (2004). [Google Scholar]
- 28.Polunin N., The real arctic: Suggestions for its delimitation, subdivision and characterization. J. Ecol. 39, 308–315 (1951). [Google Scholar]
- 29.Walker D. A., et al., The Circumpolar Arctic vegetation map. J. Veg. Sci. 16, 267–282 (2005). [Google Scholar]
- 30.Berger A., Loutre M. F., Insolation values for the climate of the last 10 million years. Quat. Sci. Rev. 10, 297–317 (1991). [Google Scholar]
- 31.Taberlet P., et al., Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crump S. E., et al., Arctic shrub colonization lagged peak postglacial warmth: Molecular evidence in lake sediment from Arctic Canada. Glob. Change Biol. 25, 4244–4256 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Pedersen M. W., et al., Postglacial viability and colonization in North America’s ice-free corridor. Nature 537, 45–49 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Bennett K. D., Determination of the number of zones in a biostratigraphical sequence. New Phytol. 132, 155–170 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Juggins S., rioja: Analysis of Quaternary Science Data (Version 0.9-21, R package, 2017).
- 36.Aiken S. G., et al., Flora of the Canadian Arctic Archipelago: Descriptions, Illustrations, Identification, and Information Retrieval (NRC Research Press, National Research Council of Canada, Ottawa, 2007). [Google Scholar]
- 37.Sjögren P., et al., Lake sedimentary DNA accurately records 20th Century introductions of exotic conifers in Scotland. New Phytol. 213, 929–941 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrews J. T., Mode W. N., Webber P. J., Miller G. H., Jacobs J. D., Report on the distribution of dwarf birches and present pollen rain, Baffin Island, N.W.T., Canada. Arctic 33, 50–58 (1980). [Google Scholar]
- 39.Miller G. H., Andrews J. T., Short S. K., The last interglacial-glacial cycle, Clyde foreland, Baffin Island, N.W.T.: Stratigraphy, biostratigraphy, and chronology. Can. J. Earth Sci. 14, 2824–2857 (1977). [Google Scholar]
- 40.de Vernal A., Hillaire-Marcel C., Natural variability of Greenland climate, vegetation, and ice volume during the past million years. Science 320, 1622–1625 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Edwards M., Hamilton T., Elias S., Bigelow N., Krumhardt A., Interglacial extension of the boreal forest limit in the Noatak valley, Northwest Alaska: Evidence from an exhumed river-cut bluff and debris apron. Arct. Antarct. Alp. Res. 35, 460–468 (2003). [Google Scholar]
- 42.Lozhkin A. V., Anderson P. M., The last interglaciation in Northeast Siberia. Quat. Res. 43, 147–158 (1995). [Google Scholar]
- 43.Zimmermann H. H., et al., The history of tree and shrub taxa on bol’shoy Lyakhovsky Island (new siberian Archipelago) since the last interglacial uncovered by sedimentary ancient DNA and pollen data. Genes (Basel) 8, E273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masson-Delmotte V., et al., “Information from paleoclimate archives in Climate change 2013: The physical science basis. Contribution of working group I to the fifth assessment report of the Intergovernmental panel on climate change” (Cambridge, UK, and New York, NY, 2013).
- 45.Yin Q. Z., Berger A., Individual contribution of insolation and CO2 to the interglacial climates of the past 800,000 years. Clim. Dyn. 38, 709–724 (2012). [Google Scholar]
- 46.Gallup C. D., Cheng H., Taylor F. W., Edwards R. L., Direct determination of the timing of sea level change during termination II. Science 295, 310–313 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Crucifix M., Loutre M. F., Transient simulations over the last interglacial period (126- 115 kyr BP): Feedback and forcing analysis. Clim. Dyn. 19, 417–433 (2002). [Google Scholar]
- 48.Schurgers G., Winguth A., The effect of land surface changes on Eemian climate. Clim. Dyn. 29, 357–373 (2007). [Google Scholar]
- 49.Stone E. J., Lunt D. J., The role of vegetation feedbacks on Greenland glaciation. Clim. Dyn. 40, 2671–2686 (2013). [Google Scholar]
- 50.Sommers A., et al., “Retreat of the Greenland ice sheet during the last interglacial” in American Geophysical Union Fall Meeting (2019).
- 51.Rawlence N. J., et al., Using palaeoenvironmental DNA to reconstruct past environments: Progress and prospects. J. Quaternary Sci. 29, 610–626 (2014). [Google Scholar]
- 52.Wolfe A. P., et al., “Geochronology of high latitude lake sediments” in Long-Term Environmental Changes in Arctic and Antarctic Lakes, Pienitz R., Douglas M. S. V., Eds. (Springer, Smol, JP, 2004), pp. 19–52. [Google Scholar]
- 53.Blaauw M., Christen J. A., Flexible paleoclimate age-depth models using an autoregressive gamma process. Bayesian Anal. 6, 457–474 (2011). [Google Scholar]
- 54.Reimer P. J., et al., The IntCal20 northern hemisphere radiocarbon age calibration curve (0-55 cal kBP). Radiocarbon 62, 725–757 (2020). [Google Scholar]
- 55.Axford Y., Briner J. P., Miller G. H., Francis D. R., Paleoecological evidence for abrupt cold reversals during peak Holocene warmth on Baffin Island, Arctic Canada. Quat. Res. 71, 142–149 (2009). [Google Scholar]
- 56.Grealy A. C., et al., A critical evaluation of how ancient DNA bulk bone metabarcoding complements traditional morphological analysis of fossil assemblages. Quat. Sci. Rev. 128, 37–47 (2015). [Google Scholar]
- 57.Dabney J., et al., Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozarewa I., Turner D. J., “Amplification-free library preparation for paired-end Illumina sequencing” in High-Throughput Next Generation Sequencing: Methods and Applications, Kwon Y. M., Ricke S. C., Eds. (Springer Science+Business Media, 2011), 733, pp. 257–266. [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson S. P., Davy S. K., Bunce M., Stat M., Taxonomic identification of environmental DNA with informatic sequence classification trees. Peer J Preprints [Preprint] (2018). 10.7287/peerj.preprints.26812v1. Accessed 31 July 2020. [DOI]
- 60.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011). [Google Scholar]
- 61.Boyer F., et al., obitools: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Callahan B. J., et al., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benson D. A., et al., GenBank. Nucleic Acids Res. 28, 15–18 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sønstebø J. H., et al., Using next-generation sequencing for molecular reconstruction of past Arctic vegetation and climate. Mol. Ecol. Resour. 10, 1009–1018 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Willerslev E., et al., Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 506, 47–51 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Soininen E. M., et al., Highly overlapping winter diet in two sympatric lemming species revealed by DNA metabarcoding. PLoS One 10, e0115335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 68.Mousavi-Derazmahalleh M., et al., eDNAFlow, an automated, reproducible and scalable workflow for analysis of environmental DNA (eDNA) sequences exploiting Nextflow and Singularity. Mol. Ecol. Resour. 10.1111/1755-0998.13356 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Troll C. J., et al., A ligation-based single-stranded library preparation method to analyze cell-free DNA and synthetic oligos. BMC Genomics 20, 1023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmieder R., Edwards R., Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huson D. H., et al., MEGAN community edition–Interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12, e1004957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jónsson H., Ginolhac A., Schubert M., Johnson P. L. F., Orlando L., mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Faegri K., Iversen J., Textbook of Pollen Analysis (Blackwell Scientific Publications, Oxford, ed. 3, 1975). [Google Scholar]
- 75.Benninghoff W. S., Calibration of pollen and spore density in sediments by addition of exotic pollen in known quantities. Pollen Spores 4, 232–233 (1962). [Google Scholar]
- 76.Whitmore J., et al., Modern pollen data from North America and Greenland for multi-scale paleoenvironmental applications. Quat. Sci. Rev. 24, 1828–1848 (2005). [Google Scholar]
- 77.Fréchette B., et al., Methodological basis for quantitative reconstruction of air temperature and sunshine from pollen assemblages in Arctic Canada and Greenland. Quat. Sci. Rev. 27, 1197–1216 (2008). [Google Scholar]
- 78.New M., Lister D., Hulme M., Makin I., A high-resolution data set of surface climate over global land areas. Clim. Res. 21, 1–25 (2002). [Google Scholar]
- 79.Overpeck J. T., Webb T. I., Prentice I. C., Quantitative interpretation of fossil pollen spectra: Dissimilarity coefficients and the method of modern analogs. Quat. Res. 23, 87–108 (1985). [Google Scholar]
- 80.Kruskal J. B., Nonmetric multidimensional scaling: A numerical method. Psychometrika 29, 115–129 (1964). [Google Scholar]
- 81.Oksanen J., et al., vegan: Community Ecology Package, R package version 2.5-6. https://cran.r-project.org/web/packages/vegan/index.html (2019).
- 82.Crump S. E., et al., Ancient plant DNA reveals High Arctic greening during the Last Interglacial. Dryad Digital Repository. 10.5061/dryad.pk0p2ngmq. Deposited 27 January 2021. [DOI] [PMC free article] [PubMed]
- 83.Fréchette B. and Crump S. E., Dataset 48940. Neotoma Paleocology Database. https://data.neotomadb.org/48940. Deposited 11 February 2021.
- 84.Axford Y., et al., Recent changes in a remote Arctic lake are unique within the past 200,000 years. Proc. Natl. Acad. Sci. U.S.A. 106, 18443–18446 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the Dryad Digital Repository (https://doi.org/10.5061/dryad.pk0p2ngmq) (82) and pollen data have been deposited in the Neotoma Paleocology Database (https://data.neotomadb.org/48940) (83).