Significance
The CD4 protein of primates has undergone rapid diversification, but the reasons for this remain unknown. Here we show that within-species diversity of the HIV/simian immunodeficiency virus (SIV) envelope (Env) binding (D1) domain is common among African primate species, and that these polymorphisms can inhibit SIV Env-mediated cell entry. Amino acid replacements in the D1 domain changed putative Env contact residues as well as potential N-linked glycosylation sites in many species, with evidence for parallel evolution and trans-specific polymorphism. These data suggest that the primate CD4 receptor is under long-term balancing selection and that this diversification has been the result of a coevolutionary arms race between primate lentiviruses and their hosts.
Keywords: CD4, primate lentiviruses, parallel evolution, trans-specific polymorphism, balancing selection
Abstract
Infection with human and simian immunodeficiency viruses (HIV/SIV) requires binding of the viral envelope glycoprotein (Env) to the host protein CD4 on the surface of immune cells. Although invariant in humans, the Env binding domain of the chimpanzee CD4 is highly polymorphic, with nine coding variants circulating in wild populations. Here, we show that within-species CD4 diversity is not unique to chimpanzees but found in many African primate species. Characterizing the outermost (D1) domain of the CD4 protein in over 500 monkeys and apes, we found polymorphic residues in 24 of 29 primate species, with as many as 11 different coding variants identified within a single species. D1 domain amino acid replacements affected SIV Env-mediated cell entry in a single-round infection assay, restricting infection in a strain- and allele-specific fashion. Several identical CD4 polymorphisms, including the addition of N-linked glycosylation sites, were found in primate species from different genera, providing striking examples of parallel evolution. Moreover, seven different guenons (Cercopithecus spp.) shared multiple distinct D1 domain variants, pointing to long-term trans-specific polymorphism. These data indicate that the HIV/SIV Env binding region of the primate CD4 protein is highly variable, both within and between species, and suggest that this diversity has been maintained by balancing selection for millions of years, at least in part to confer protection against primate lentiviruses. Although long-term SIV-infected species have evolved specific mechanisms to avoid disease progression, primate lentiviruses are intrinsically pathogenic and have left their mark on the host genome.
Simian immunodeficiency viruses (SIVs) comprise a large group of lentiviruses that infect over 45 African primate species, including numerous guenons (Cercopithecus spp.), African green monkeys (Chlorocebus spp.), mandrills and drills (Mandrillus spp.), mangabeys (Cercocebus spp.), colobus monkeys (Colobus spp., Piliocolobus spp.), as well as chimpanzees (Pan troglodytes) and western gorillas (Gorilla gorilla) (1). Although the prevalence rates and geographic distribution of these infections vary widely, most SIVs are host-specific (i.e., their genomes form species-specific clusters in phylogenetic trees) (2–5). This has enabled the identification of instances when SIVs have crossed species barriers, including from apes and monkeys to humans (6). Phylogenetic analyses have shown that both pandemic and nonpandemic forms of HIV type 1 (HIV-1) resulted from the cross-species transmission of SIVs infecting central chimpanzees (P. troglodytes troglodytes) and western lowland gorillas (G. gorilla gorilla), while the various groups of HIV type 2 (HIV-2) emerged following the transfer of SIVsmm strains naturally infecting sooty mangabeys (Cercocebus atys) (6–9).
SIVs have also jumped between nonhuman primate species, generating new SIV lineages. Cross-species transmission and recombination between ancestors of viruses today infecting greater spot-nosed (Cercopithecus nictitans), mustached (Cercopithecus cephus) and mona (Cercopithecus mona) monkeys (SIVgsn/SIVmus/SIVmon), and SIVrcm infecting red-capped mangabeys (Cercocebus torquatus) gave rise to SIVcpz in chimpanzees (10), and onward transmission of this virus to western lowland gorillas generated SIVgor (11). Additional cross-species transmissions and recombination events have generated mosaic SIV lineages in green monkeys (Chlorocebus sabaeus) and mandrills (Mandrillus sphinx) (12, 13). Finally, repeated introductions of diverse SIVs into the same primate species have resulted in cocirculating lineages, such as SIVkcol1 and SIVkcol2 in Kibale black-and-white colobus (Colobus guereza), and SIVmus-1, SIVmus-2, and SIVmus-3 in mustached monkeys (14, 15). Thus, primate lentiviruses have a high propensity to cross species barriers and have done so on numerous occasions throughout their evolutionary history.
Lentiviruses have existed for tens of millions of years as evidenced by the finding of endogenous viruses in the genomes of species from four orders of mammals, including lemurs (16, 17), colugos (18), rabbits (19, 20), and weasels (21, 22). Some SIVs, such as those infecting green monkeys (Chlorocebus spp.) (23, 24) and the lhoesti group of guenons (Allochrocebus spp.) (25, 26) are at least several million years old because they appear to have coevolved with their respective hosts since these diverged from a common ancestor. Although an upper limit of 6 to 10 million y has been suggested for SIVs based on the fact that they have so far been found only in African, but not Asian, lineages of Old World monkeys (6), certain features of antiviral defense genes suggest that monkeys may have been exposed to lentiviruses long before this (27). Cellular restriction factors, such as APOBEC3G and TRIM5, are exquisitely antiviral and are counteracted by dedicated SIV accessary proteins. These restriction factors have evolved under strong positive selection at sites specifically involved in the interaction with lentiviruses, in both African and Asian monkeys (28, 29). However, if SIV indeed infected the common ancestor of African and Asian monkeys, this would imply numerous subsequent infection losses from multiple host lineages. Thus, it remains unclear when lentiviruses first infected primates.
Among lentiviruses, those infecting primates are unique in their use of the CD4 receptor for entry into target cells. The viral envelope glycoprotein (Env) interacts with CD4 and subsequently undergoes conformational changes to expose the coreceptor binding site, which is required for viral–cell membrane fusion (30). CD4 is an immunoglobulin-like integral membrane protein that is expressed on multiple immune cells and stabilizes the interaction of the T cell receptor (TCR) with major histocompatibility complex class II (MHC II) molecules (31, 32). The most outward domain of CD4 (the D1 domain) binds a nonpolymorphic region on MHC II, which enhances TCR signaling (32). Importantly, the D1 domain is also the region that is bound by the HIV/SIV Env glycoprotein (33, 34). In HIV-infected humans and SIVmac-infected macaques, continuous high level viral replication leads to CD4+ T cell depletion, systemic immune activation, T cell exhaustion, and the development of AIDS (35, 36). Naturally occurring SIVs can also cause immunodeficiency and disease, as shown for chimpanzees and mandrills (37–40), indicating that these viruses are intrinsically pathogenic (41–43). However, a number of primate species with presumed longstanding SIV infections, such as African green monkeys, sooty mangabeys, and Ugandan red colobus monkeys (Piliocolobus tephrosceles), have evolved unique mechanisms that prevent disease progression despite continuous high viral replication (44–48). While the time required to evolve these adaptations is unknown, such protective mechanisms are absent from hosts that acquire new SIV infections.
Unlike restriction factors, which prevent or limit viral replication, CD4 is a dependency factor (i.e., a host protein that is required for successful infection). Since there are many examples of host receptors coevolving with pathogens (49–51), it has been assumed that pressures exerted by pathogenic SIVs are responsible for the rapid diversification of primate CD4 (52–54). However, direct evidence for this hypothesis has been lacking. Examining the functional consequences of CD4 diversity in chimpanzees, we recently found that naturally occurring amino acid replacements in the D1 domain were able to inhibit SIVcpz infection, both in vitro and in vivo (55). Protective mechanisms included charged residues at the CD4–Env interface and steric hindrance between CD4- and Env-encoded glycans, which were effective not only against SIVcpz but also other SIVs that chimpanzees frequently encounter. These results suggested that CD4 diversity protects wild chimpanzee populations from SIV infection, possibly by conferring a heterozygote advantage (55). Since humans lack polymorphisms and glycans in the D1 domain, we asked whether CD4 diversification was a unique adaptation of chimpanzees. Sequencing the D1 domain in members of 36 African primate species, we identified a remarkable degree of CD4 diversity, both within and between species. The observed polymorphisms altered the cell entry of a panel of diverse SIV Envs, with the level of restriction depending on the particular allele and virus strain analyzed. Thus, the diversification of the primate CD4 receptor appears to have resulted from an ancient arms race between primate lentiviruses and their hosts.
Results
Bonobos Exhibit Limited CD4 Diversity.
The CD4 receptor of chimpanzees contains amino acid substitutions at positions 25 (Q/R), 40 (Q/R), 52 (N/K), 55 (V/I), and 68 (P/T), all of which are the result of single nucleotide polymorphisms (SNPs) in exons 2 and 3 (55). These SNPs result in nine coding variants of the D1 domain, which appear to have evolved from an ancestral CD4 allele (QQNVP) by point mutation and recombination (55). Since bonobos (Pan paniscus) are the closest genetic relatives of chimpanzees (56), we asked whether they share CD4 polymorphisms. Bonobos live in the rain forests of the Democratic Republic of the Congo (DRC) south of the Congo River (SI Appendix, Fig. S1), and like chimpanzees, hunt and consume other primates (57); however, unlike chimpanzees, they do not appear to be SIV-infected (58). Extracting CD4 sequences from publicly available bonobo genome libraries (56), we obtained 14 full-length protein sequences, 13 of which were identical and differed from the ancestral chimpanzee QQNVP allele by a single amino acid replacement (Q429E) in the cytoplasmic domain (one sequence contained an additional R240W replacement in the D3 domain) (SI Appendix, Fig. S2). These results suggested that bonobos lack polymorphisms in the D1 domain.
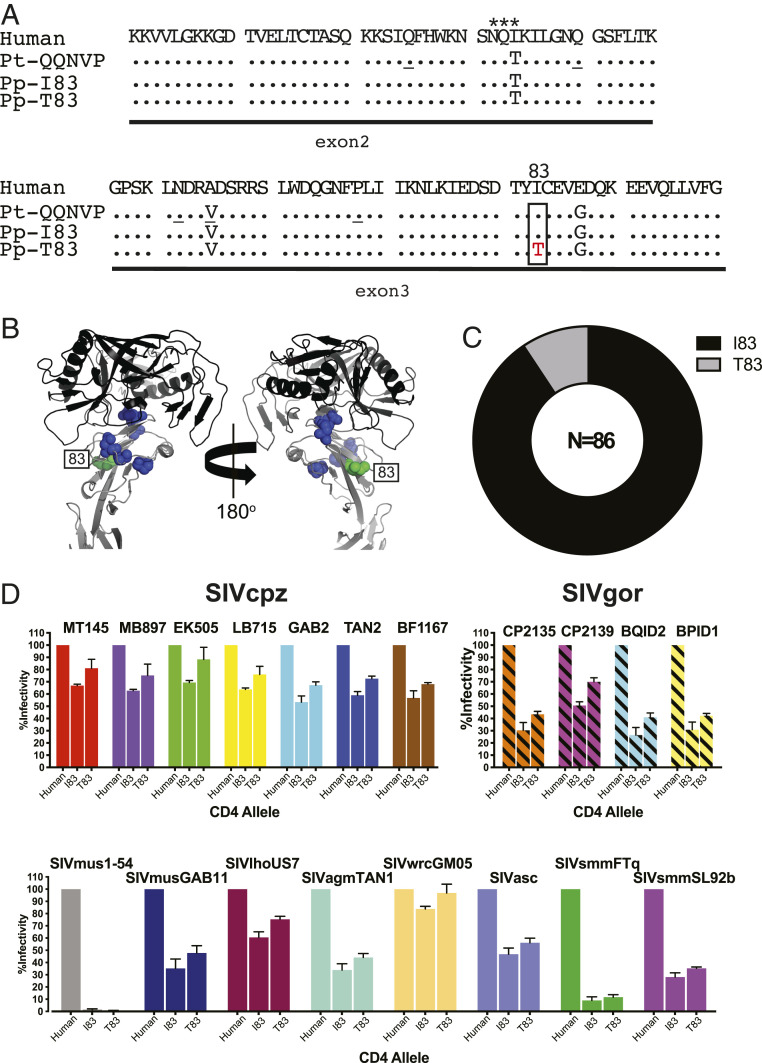
To determine the full extent of the bonobo CD4 diversity, we used fecal samples from wild populations collected previously for molecular epidemiological studies of SIVcpz and ape malaria (58, 59). Specimens were available from 86 genotyped individuals sampled at 8 field sites (SI Appendix, Table S1), including from a genetically isolated population (TL) east of the Lomami River (SI Appendix, Fig. S1). Since the CD4 gene is comprised of multiple exons and introns spanning nearly 20 Kb on chromosome 12, we limited our analysis to exons 2 (165 bp) and 3 (159 bp), which together encode the entire D1 domain. Each exon was amplified separately because of a large intervening intron (>13 Kb) and sequenced without fragmentation to maintain linkage between polymorphic sites. In addition, each exon was amplified up to eight times to exclude allelic dropout (SI Appendix, Table S1). While exon 2 sequences were identical in all animals, exon 3 sequences differed by a single nonsynonymous substitution, which led to an amino acid replacement at position 83 (I83T) (Fig. 1A). This polymorphism was located at the periphery of the Env binding region at a site not previously reported to be under positive selection (Fig. 1B). Among the animals studied, the T83 variant was relatively uncommon (found in 14 bonobos at 4 field sites), while the I83 variant represented the predominant form (found in 72 bonobos at all field sites) (Fig. 1C and SI Appendix, Fig. S1). Thus, in contrast to chimpanzees, bonobos exhibit limited D1 domain diversity, with only two coding variants circulating in wild populations (Table 1), the most common of which represents the inferred ancestral CD4 allele of both chimpanzees and bonobos.
Fig. 1.
Allelic diversity of CD4 in bonobos. (A) CD4 coding variants identified in wild bonobo populations. D1 domain variants of bonobos (P. paniscus, Pp; derived from exon 2 and 3 sequences) are compared to human and chimpanzee CD4, with dots indicating identity to the human reference. A single amino acid replacement (I83T) in the bonobo CD4 is boxed. A potential N-linked glycosylation site is indicated by asterisks. The ancestral chimpanzee (P. troglodytes; Pt) allele is shown for reference, with positions that are polymorphic in chimpanzees underlined. (B) Crystal structure of the HIV-1 gp120 envelope domain (black) bound to human CD4 (gray) (PDB ID code 4R2G), with polymorphic positions indicated for chimpanzees (blue) and bonobos (green). (C) CD4 allele frequency in wild bonobos. The number of tested individuals is indicated. (D) Effects of bonobo CD4 polymorphisms on SIV Env mediated cell entry. The infectivity of pseudoviruses carrying different SIV Envs is shown for transiently transfected cells expressing human and bonobo CD4 variants and the cognate CC5 receptors. Values are scaled relative to the human CD4 (set to 100%). Bars represent the average of three independent transfections, each performed in triplicate, with SDs shown (fold-changes of Env infectivity for different CD4 alleles are shown in SI Appendix, Table S2).
Table 1.
D1 domain genotyping of African primates
| Primate species | No. of individuals* | No. of D1 variants | SIV infection† | Non synonymous changes | Synonymous changes | |
| Apes | ||||||
| Chimpanzee (Pan troglodytes) | 544‡ | 9 | + | 6 | 1 | |
| Bonobo (Pan paniscus) | 100§ | 2 | − | 1 | 0 | |
| Western gorilla (Gorilla gorilla) | 97§ | 3 | + | 3 | 0 | |
| Eastern gorilla (Gorilla beringei) | 25§ | 2 | − | 3 | 0 | |
| Cercopithecine monkeys | ||||||
| L’Hoest’s monkey (Allochrocebus lhoesti) | 3 | 2 | + | 2 | 0 | |
| Sun-tailed monkey (Allochrocebus solatus) | 2 | 3 | + | 2 | 1 | |
| Tantalus monkey (Chlorocebus tantalus | 11§ | 3 | + | 3 | 0 | |
| Vervet monkey (Chlorocebus pygerythrus) | 63§ | 4 | + | 4 | 0 | |
| Grivet (Chlorocebus aethiops) | 26§ | 3 | + | 3 | 0 | |
| Green monkey (Chlorocebus sabaeus) | 53§ | 4 | + | 3 | 0 | |
| Malbrouck (Chlorocebus cynosuros) | 16§ | 4 | + | 3 | 0 | |
| Patas monkey (Erythrocebus patas) | 5§ | 1 | − | 0 | 0 | |
| Red-tailed monkey (Cercopithecus ascanius) | 4¶ | 2 | + | 2 | 1 | |
| Mustached monkey (Cercopithecus cephus) | 15 | 11 | + | 8 | 0 | |
| Greater spot-nosed monkey (Cercopithecus nictitans) | 17 | 9 | + | 8 | 1 | |
| Sykes’ monkey (Cercopithecus albogularis) | 22 | 3 | + | 3 | 0 | |
| Blue monkey (Cercopithecus mitis) | 7 | 3 | + | 3 | 0 | |
| De Brazza’s monkey (Cercopithecus neglectus) | 3 | 1 | + | 0 | 0 | |
| Diana monkey (Cercopithecus diana) | 5 | 3 | ? | 2 | 0 | |
| Lesser spot-nosed monkey (Cercopithecus petaurista) | 4 | 2 | ? | 2 | 0 | |
| Crested mona monkey (Cercopithecus pogonias) | 2 | 2 | − | 1 | 0 | |
| Red-capped mangabey (Cercocebus torquatus) | 3 | 2 | + | 4# | 0 | |
| Sooty mangabey (Cercocebus atys) | 5‡ | 2 | + | 2# | 0 | |
| Mandrill (Mandrillus sphinx) | 7 | 2 | + | 1 | 1 | |
| Chacma baboon (Papio ursinus) | 10 | 1 | − | 0 | 0 | |
| Olive baboon (Papio anubis) | 13 | 1 | − | 0 | 0 | |
| Yellow baboon (Papio cynocephalus) | 3 | 1 | − | 0 | 0 | |
| Colobine monkeys | ||||||
| Ugandan red colobus (Piliocolobus tephrosceles) | 29§ | 6 | + | 7 | 1 | |
| Mantled guereza (Colobus guereza) | 4 | 2 | + | 1 | 1 |
In addition to the within-species diversity shown here, D1 domain sequences were also obtained for single individuals of the following species: Preuss’s monkey (Allochrocebus preussi), Hamlyn’s monkey Cercopithecus hamlyni), Lowe’s mona monkey (Cercopithecus lowei), Allen’s swamp monkey (Allenopithecus nigroviridis), Angolan talapoin (Miopithecus talapoin), Drill (Mandrillus leucophaeus), and Angola colobus (Colobus angolensis) (see SI Appendix, Table S5 for GenBank accession numbers).
Plus sign (+), naturally SIV infected; minus sign (−) not naturally SIV infected; question mark (?), insufficient sampling to determine SIV infection status.
D1 domain sequences were obtained from GenBank.
All or some D1 domain sequences were extracted from whole genome or RNA-seq datasets (24, 48, 68). Note that the number of D1 domain variants represent minimum estimates, since alleles with low quality support were discarded.
Minimum number of individuals from 25 samples.
In addition to substitutions, a 3 bp deletion was observed in alleles of some red-capped and sooty mangabeys (Fig. 4A).
Although not naturally infected with SIV, bonobos are likely exposed to primate lentiviruses through their hunting behavior (57). Since the viruses infecting these prey species have not been characterized, we selected a panel of SIV Envs from diverse viral lineages and tested their ability to mediate entry into cells expressing the bonobo CD4 receptors (8, 11, 55). These included Envs from various SIVcpz and SIVgor strains, as well as SIVs infecting mustached monkeys, red-tailed monkeys (SIVasc; Cercopithecus ascanius), western red colobus (SIVwrc; Piliocolobus badius), tantalus monkeys (SIVtan; Chlorocebus tantalus), l’Hoest’s monkeys (SIVlho; Allochrocebus lhoesti), and sooty mangabeys. Briefly, 293T cells were transfected with plasmids expressing human and bonobo CD4 alleles as well as their cognate CCR5 coreceptors, and then infected with viruses pseudotyped with the various SIV Envs. Compared to human CD4, both bonobo alleles were less efficient in mediating cell entry (Fig. 1D and SI Appendix, Table S2), most likely because of a glycan at position 32 (N32), which sterically hinders SIV Env interaction in chimpanzees (55). However, all SIVcpz and SIVgor Envs, as well as most monkey SIV Envs, were able to utilize both bonobo CD4 variants. The I83 allele was as efficient in mediating cell entry as the chimpanzee QQNVP allele (SI Appendix, Fig. S3), which is not surprising given that the ectodomains of these two CD4 molecules are identical (SI Appendix, Fig. S2). The less frequent T83 allele appeared to enhance SIV Env-mediated cell entry (SI Appendix, Table S2), although this increase was only modest (on average 1.3-fold). Thus, bonobos lack the extensive CD4 diversity that appears to protect chimpanzees against primate lentiviruses. However, two Envs from SIVmus and SIVsmm utilized the bonobo CD4 variants only poorly (Fig. 1D). Although bonobos are not naturally exposed to these viruses, this finding suggests that bonobos may be resistant to some SIVs.
Western and Eastern Gorillas Encode Multiple D1 Domain Alleles.
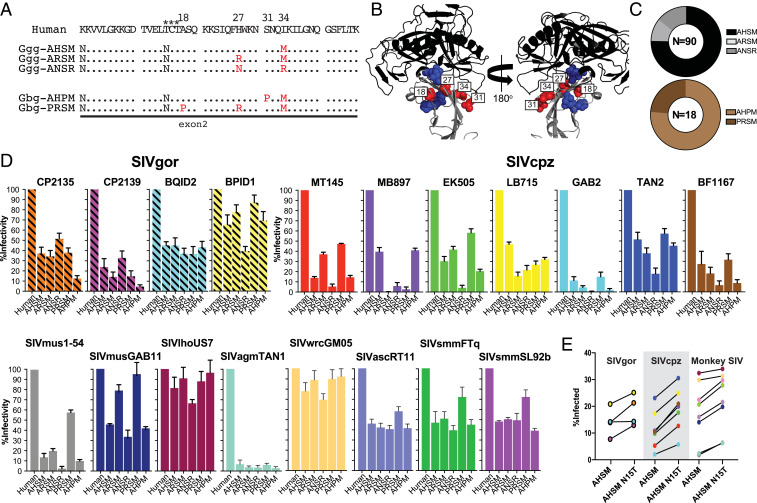
The paucity of D1 domain polymorphisms in bonobos prompted us to examine western and eastern (Gorilla beringei) gorillas, which occupy nonoverlapping ranges in sub-Saharan Africa (SI Appendix, Fig. S1). Of these, only western lowland gorillas in Cameroon are SIVgor infected, having acquired this virus only once from sympatric chimpanzees (8, 11). Mining publicly available genome databases (56, 60), we extracted complete CD4 protein sequences from 14 western and eastern lowland gorillas. An alignment of these sequences revealed five polymorphic sites, four of which were located in the D1 domain at positions 18 (A/P), 27 (H/N/R), 31 (S/P), and 34 (R/M), while the fifth was located at position 142 (K/R) in the D2 domain (SI Appendix, Fig. S4). All D1 domain polymorphisms were caused by SNPs in exon 2, which yielded five variants, three of which were only found in western gorillas (AHSM, ARSM, ANSR), while the other two were only found in eastern gorillas (AHPM, PRSM) (Table 1).
To determine whether these alleles represent the entirety of D1 domain diversity, we used fecal DNA to sequence this region in 90 additional western and 18 eastern lowland (G. beringei graueri) gorillas sampled at 14 and 6 field sites throughout Cameroon and the DRC, respectively (SI Appendix, Fig. S1). Specimens were selected based on their geographic origin and individual information, with both SIVgor+ SIVgor− western lowland gorillas included (SI Appendix, Table S3). Like for the bonobos, exons 2 and 3 were amplified separately because of the large intervening intron and sequenced without fragmentation. However, due to limited sample availability, some specimens could only be amplified once (SI Appendix, Table S3). Consistent with the genomic data, CD4 genotyping of wild gorillas confirmed the existence of AHSM, ARSM, and ANSR alleles in western and AHPM and PRSM alleles in eastern lowland gorillas, with no overlap between the two species (Fig. 2A). All D1 polymorphisms were located at the binding interface of Env and CD4 (Fig. 2B), with AHSM and AHPM representing the most frequent alleles in western and eastern gorillas, respectively (Fig. 2C). Although screening of wild gorillas did not uncover additional CD4 diversity, it is clear that additional intermediate alleles existed in these populations at some point in the past. For example, the ANSR allele of western gorillas is 2 amino acids different from its closest relatives, indicating three potential intermediates (AHSR, ANSM, ARSR), one of which must have existed in the past, if not now (SI Appendix, Fig. S5). Similarly, the AHPM and PRSM alleles in eastern gorillas differ by 3 amino acids, indicating six potential intermediates (AHSM, ARSM, ARPM, PHPM, PRPM, PHSM), two of which are present in western gorillas (SI Appendix, Fig. S5). Thus, there likely are additional CD4 alleles in both gorilla species that have not yet been identified.
Fig. 2.
Allelic diversity of CD4 in gorillas. (A) CD4 coding variants identified in western (G. gorilla gorilla; Ggg) and eastern (G. beringei graueri; Gbg) lowland gorillas. D1 domain variants in exon 2 are compared to the human CD4, with dots indicating sequence identity (exon 3 derived protein sequences were invariant). A potential N-linked glycosylation site is indicated by asterisks. Amino acid replacements are indicated in red, with allelic variants named based on the order of polymorphic amino acid residues. (B) Crystal structure of the HIV-1 gp120 envelope domain (black) bound to human CD4 (gray) (PDB ID code 4R2G) with polymorphic positions indicated for chimpanzees (blue) and gorillas (red). (C) CD4 allele frequencies in wild-living western (Upper) and eastern (Lower) gorillas. The number of tested individuals is indicated. (D) Effects of gorilla CD4 polymorphisms on SIV Env mediated cell entry. The infectivity of pseudoviruses carrying the SIV Envs indicated is shown for transiently transfected cells expressing human and gorilla CD4 variants and the cognate CCR5 receptors. Values are scaled relative to human CD4 (set to 100%). Bars represent the average of two or three independent transfections, each performed in triplicate, with SDs shown. MT145, MB897, EK505, LB715, and GAB2 represent SIVcpzPtt strains from central chimpanzees, while TAN2 and BF1167 represent SIVcpzPts strains from eastern chimpanzees. (E) Protective effect of the N15 glycan. The percentage of infected cells bearing the AHSM allele was compared to a mutant (N15T) lacking the N15 glycan (fold-changes of Env infectivity for different CD4 alleles are shown in SI Appendix, Table S4). Individual SIV Envs are color coded as in D.
To determine whether the D1 polymorphisms affected SIV Env-mediated cell entry, we synthesized one gorilla CD4 allele and generated the other D1 domain variants by site-directed mutagenesis. We then transfected 293T cells with plasmids expressing human and gorilla CD4, along with the corresponding CCR5 coreceptors, and infected these cells with pseudoviruses bearing the same SIV Envs used in the bonobo study. The results showed that all gorilla CD4 alleles were functional and facilitated pseudovirus entry, although infectivity levels were again lower compared to human CD4 (Fig. 2D). However, the five gorilla CD4 alleles differed in their ability to facilitate infection. For example, the ANSR allele mediated efficient entry of all SIVgor Envs, while the AHPM allele reduced infection of two of them (SI Appendix, Table S4). These allele-specific differences were even more pronounced when SIVcpz Envs were tested. For example, the MB897 Env utilized the AHSM and AHPM alleles, but was restricted by the ARSM, ANSR, and PRSM alleles, while the MT145 Env utilized the ARSM and PRSM alleles, but was restricted by the AHSM, ANSR, and AHPM alleles (Fig. 2D). Most, but not all, monkey SIV Envs utilized the five gorilla CD4 alleles with high efficiency (Fig. 2D and SI Appendix, Table S4). Thus, gorilla CD4 polymorphisms, like those in chimpanzees, inhibit SIV Env cell mediated entry in a strain and allele specific manner (55).
In addition to the five polymorphic sites, the gorilla CD4 encodes an invariant potential N-linked glycosylation site (PNGS) at position 15 (N15) that is absent in all other primate species (Fig. 2A). This PNGS, which has been experimentally confirmed to be glycosylated (61), appears to occupy a similar structural position as the chimpanzee glycan at position 66, which is known to interfere with SIV Env binding (55). To examine whether N15 has a protective effect, we changed the asparagine at position 15 in one representative gorilla CD4 allele (AHSM) to a threonine, which is present in all other African primates. Testing SIVgor, SIVcpz, and other SIV Envs, we found that removal of the N15 glycan increased Env mediated infectivity on average by 1.6-fold (SI Appendix, Table S4), with enhancement observed for nearly every Env (Fig. 2E). Thus, like the invariant N32 glycan in chimpanzees and bonobos, the invariant N15 glycan in gorillas provides some degree of protection against SIV infection.
Mustached Monkeys Exhibit an Extraordinary Degree of CD4 Diversity.
To examine whether CD4 diversity is unique to African apes, we next sequenced the D1 domain in mustached monkeys. We were particularly interested in this species because mustached monkeys harbor three different types of SIVmus, two of which represent recombinants with SIVgsn and other SIVs (14, 62, 63), but are infected at very low (0 to 6%) prevalence rates (2, 62, 64–66). Using remnant DNA from previous blood collections (2, 64), we CD4-genotyped 15 members of this species (Table 1). Sequence analysis of exons 2 and 3 identified eight nonsynonymous SNPs, which resulted in amino acid replacements at positions 30, 34, 39, 40, 41, 42, 50, and 90, the combination of which indicated a minimum of 11 different CD4 coding variants (Fig. 3A). Of these, only nine D1 domain alleles could be unambiguously inferred, because amplicons from the remaining samples contained polymorphic sites in both exons 2 and 3. The combination of the latter resulted in at least two more alleles as well as a possible third (Fig. 3A), although their exact sequences could not be determined due to different possibilities of exon 2 and 3 linkage. One polymorphic site (I34T) affected a PNGS at position 32. Thus, analysis of a very small number of mustached monkeys identified an extraordinary degree of D1 domain diversity, with 11 of the 15 animals exhibiting a heterozygous CD4 genotype (SI Appendix, Table S5).
Fig. 3.
Allelic diversity of CD4 in mustached monkeys. (A) CD4 coding variants identified in mustached monkeys. Mustached monkey D1 domain variants derived from both exons 2 and 3 (indicated on the bottom) are compared to one representative allele, with dots indicating identity to this reference (sequences are trimmed to the polymorphic region). Polymorphic positions are highlighted in red and their position is indicated on the top. Alleles 1 to 9 could be unambiguously inferred; the remaining alleles remain ambiguous because of polymorphisms in both exons 2 and 3, which could not be linked. For individual 10, permutations of exons 2 and 3 combinations resulted in one new allele (either 10a or 10b), which was paired with an allele already identified in other individuals. For individual 11, permutations resulted in either one new allele (11a) combined with an already known allele, or a combination of two new alleles (11b and 12?). An N-linked glycosylation site is indicated by asterisks. An arrow marks allele 2, which is the inferred ancestral allele. (B) Effects of mustached monkey CD4 polymorphisms on SIV Env mediated cell entry. A heatmap displays the percentage of cells expressing the indicated CD4 variant that were infected by the corresponding SIV Env, averaged across two or three experiments each performed in triplicate (fold-changes of Env infectivity for different CD4 alleles are shown in SI Appendix, Table S6).
To examine whether the D1 domain polymorphisms affected SIV Env-mediated cell entry, we generated a subset of mustached monkey CD4 alleles (alleles 1 to 7) and tested these, together with the cognate CCR5 coreceptor (67), in the single-round infection assay (Fig. 3B). The results showed that all mustached monkey CD4 variants mediated cell entry of the two available SIVmus Envs. However, infectivity of the other SIV Envs varied, with many utilizing only certain alleles (Fig. 3B and SI Appendix, Fig. S6). A comparison of alleles that differed only at position 34 (alleles 1 vs. 3 and 2 vs. 4) showed that a threonine resulted in less-efficient entry than an isoleucine (SI Appendix, Table S6), thus confirming the protective effect of the glycan at position 32. Of all mustached monkey CD4 variants, allele 2 (the inferred ancestral state) was the most permissive, mediating infection even of an SIVdeb Env from De Brazza’s monkeys (Cercopithecus neglectus), the only SIV Env that failed to use human CD4. In contrast, allele 7 was the most restrictive, with only three Envs being able to utilize this variant, two of which were from SIVmus strains (SI Appendix, Table S6). Interestingly, this allele encodes a proline at position 40, which is an Env contact residue on human CD4 and the site of a protective Q40R polymorphism in chimpanzees (55).
Within-Species CD4 Diversity Is Common among African Primates.
To determine how many other primates exhibit CD4 polymorphisms, we used remnant blood or fecal DNA from 124 members of 23 additional African monkey species to amplify and sequence the D1 domain (Table 1). These included samples from 22 Cercopithecine and one Colobine monkey species, 13 of which are known to be naturally SIV-infected (SI Appendix, Table S5). In addition, we extracted D1 domain sequences from published RNA-sequencing (RNA-seq) data of 29 Ugandan red colobus (48) and 4 patas monkeys (Erythrocebus patas) (47), and mined whole-genome databases from 163 African green monkeys belonging to 5 different species (24). Finally, we obtained CD4 sequences from an Angolan colobus (Colobus angolensis), a drill (Mandrillus leucophaeus), and five sooty mangabeys from GenBank. Together with the previously published chimpanzee data (55), we were able to compare D1 domain sequences from 1,101 African primates representing 36 different species.
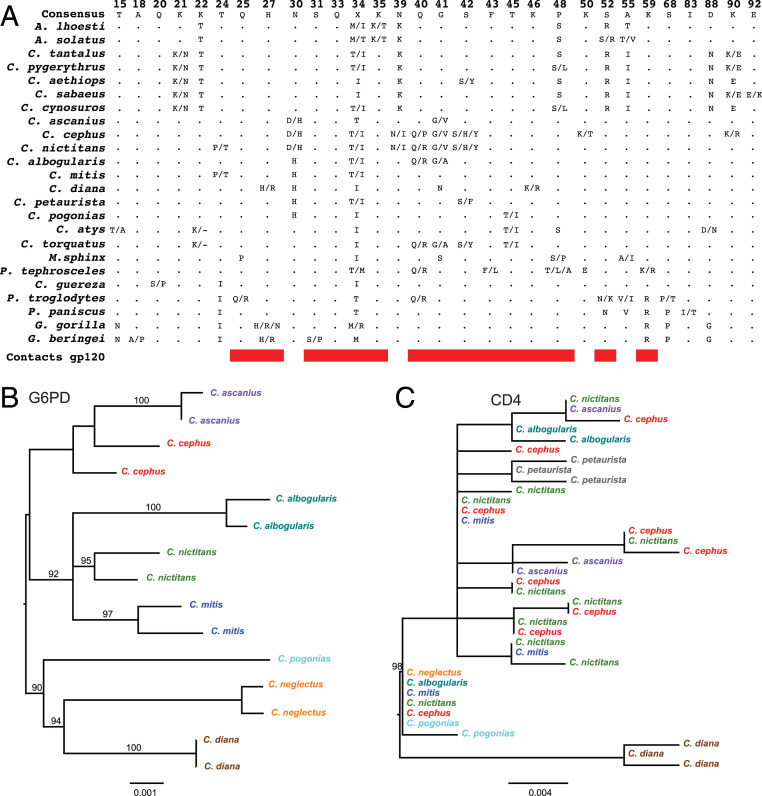
The various primate species exhibited a striking degree of CD4 diversity, both with respect to the number of species that had CD4 polymorphisms and the number of alleles that were detected. Of 29 species for which 2 or more individuals were genotyped (Table 1), 24 (83%) encoded more than 1 D1 domain variant, with many polymorphic sites located in putative Env–CD4 contact residues (Fig. 4A). In fact, the great majority of SNPs were nonsynonymous, with many species exhibiting no synonymous changes in the D1 domain (Table 1). Greater spot-nosed and mustached monkeys stood out as exhibiting the highest CD4 diversity, with the largest number of D1 variants identified (Table 1). Interestingly, identical polymorphisms were found in members of different species and genera. For example, the Q40R polymorphism, which was shown to affect Env–CD4 interaction in chimpanzees (55), was also found in two Cercopithecus, one Piliocolobus, and one Cercocebus species (Fig. 4A). In addition, two mustached monkey CD4 alleles exhibited a Q40P mutation (Fig. 3A), which was also highly restrictive in the infection assay (Fig. 3B). Although position 40 has not previously been reported to be under positive selection (52, 53), the frequent amino acid replacements at this site in distantly related primate species indicates parallel evolution, most likely driven by pathogenic SIV infections.
Fig. 4.
CD4 diversity in African primate species. (A) D1 domain positions exhibiting intraspecies polymorphisms. For each primate species, the positions of polymorphic residues are shown, with the amino acid residues indicated. Dots indicate identity to the CD4 consensus, while dashes indicate deletions. Contact residues between HIV-1 Env and human CD4 are indicated in red. (B and C) Maximum-likelihood trees of G6PD (B) and CD4 D1 domain (C) nucleotide sequences from different guenon species (color coded). Bootstrap values of >90% are shown; the scale bars indicate 0.001 and 0.004 nucleotide substitutions per site, respectively.
An alignment of D1 domain sequences also revealed that certain CD4 variants were present in multiple closely related primate species. L’Hoest’s and sun-tailed monkeys (genus Allochrocebus) shared certain D1 alleles, as did various African green monkey species, as well as different guenon species (SI Appendix, Fig. S7). To compare the genealogy of CD4 with that of a housekeeping gene, we used available genomic DNA to amplify a 1.6-kb fragment of the glucose-6-phosphate dehydrogenase (G6PD) gene from eight Cercopithecus species for which D1 domain sequences were also available (Fig. 4B). Phylogenetic analysis of these sequences revealed species-specific clustering and an overall topology that was very similar to phylogenies previously reported for these same primate species (68, 69). However, this was not the case for the corresponding CD4 D1 domain sequences (Fig. 4C). Unlike the G6PD sequences, most D1 domain sequences did not cluster according to their species, but were intermixed, with one D1 domain allele shared by six different species (Fig. 4C). These data suggest that the CD4 diversity found in these guenons predates their speciation, indicating trans-specific polymorphism. Since D1 domain sequences from different African green monkey species yielded similar results (SI Appendix, Fig. S8), it appears that CD4 variants have been maintained in various species by long-term balancing selection (70, 71).
Most Primate CD4 Receptors Are Glycan Shielded.
CD4 genotyping of the various primates revealed a total of six potential N-linked glycosylation sites within the D1 domain (SI Appendix, Fig. S7), all of which were predicted to project toward the Env trimer (Fig. 5A). A number of these seemed to be invariant within species, such as the N17 glycan that was present in the great majority of monkey species and the N15 glycan that was present in all eastern and western gorillas (Fig. 5B). Other glycans were polymorphic, such as the N21 glycan that was present in some, but not all, Chlorocebus species. In fact, African green monkeys exhibited three glycosylation sites in the D1 domain, but the maximal number in any one variant was only two (SI Appendix, Fig. S7). Certain glycans were unique to a particular species, including the N66 of chimpanzees and the N39 glycan of mandrills (Fig. 5B). The latter glycan is of interest since it is predicted to protrude directly into the Env binding site and may thus provide an explanation for the ability of SIVmnd1 to infect cells that do not express the CD4 receptor (72). Three guenon and two colobus species appeared to lack D1 domain glycans (Fig. 5B). However, these results are preliminary since in each case only very few individuals were genotyped. Thus, unlike humans, nearly all African primates encode at least one or more N-linked glycosylation sites in their D1 domain, which are positioned to impede SIV infection by creating steric clashes with glycans encoded by their Env glycoproteins.
Fig. 5.
D1 domain glycosylation in primates. (A) PNGS found in the D1 domain of different primate species. PNGS positions are modeled onto the structure of the HIV-1 envelope trimer, with different protomers highlighted in pink, green, and gray, respectively, and the bound human CD4 shown in black (PDB ID code 5U1F). (B) Phylogeny of African primate species adapted from Springer et al. (69). The tree highlights the presence of glycans within each species, which are color coded as in A. Solid and striped squares indicate invariant and polymorphic glycans, respectively. The scale bar indicates estimated primate divergence times as previously reported (69).
Discussion
Unlike human CD4, chimpanzee CD4 is highly polymorphic with nine D1 coding variants circulating in wild populations, all of which are the result of replacements in the outermost domain that binds the HIV/SIV Env trimer (55). Here, we show that CD4 diversity is not unique to chimpanzees, but common among African primates. Generating D1 domain sequences from over 500 monkeys and apes, we found polymorphic residues in 24 of 29 primate species, with as many as 11 different coding variants identified within a single species (Table 1). Moreover, D1 domain mutations altered SIV Env-mediated cell entry. Although the panel of Envs tested was not selected to examine specific SIV exposure risks, the infectivity data show that D1 domain polymorphisms alter CD4–Env interactions. We also found identical amino acid changes in species spanning multiple genera, such as the Q40R polymorphism, which was observed in chimpanzees, greater spot-nosed monkeys, Sykes’ monkeys, western red colobus, and red-capped mangabeys (Fig. 4A), suggesting similar selective pressures on CD4 in diverse African primates. Although not all D1 domain variants were functionally tested, those that reduce SIV infection in vitro likely do so by mechanisms similar to those described for chimpanzees (55). In contrast, mechanisms that confer protection in vivo, both at the individual and population level, are much less clear. It is possible that SIV Env trimers that have to bind two different CD4 variants mediate cell entry less efficiently than Env trimers that need to interact with only one, conferring a fitness advantage on individuals that are heterozygous for CD4 (55). Alternatively, a virus adapted to one set of CD4 alleles may be less able to infect an individual expressing a different set of CD4 alleles; then, as viruses adapt to a set of common CD4 variants, they may not be able to utilize rarer variants with similar efficiency. This could result in negative frequency-dependent selection (70), which could maintain CD4 diversity over a long timescale. Further studies are required to differentiate between these, and other, possibilities.
Unlike chimpanzees, bonobos are not naturally SIV-infected (58), which may explain their limited CD4 diversity (Fig. 1). However, bonobos are exposed to primate lentiviruses because they hunt SIV-infected monkeys. Thus, bonobos may have evolved mechanisms other than CD4 diversification to prevent cross-species infections, such as restriction factors that act downstream of the entry process. Alternatively, it is possible that our SIV Env panel is not sufficiently representative of the SIV strains that bonobos encounter. Although we identified two monkey SIV Envs that were restricted by both bonobo alleles, they do not represent relevant pathogens, because the ranges of their hosts do not overlap that of bonobos. To examine whether the two bonobo CD4 alleles have protective potential, it will be necessary to isolate and test SIV Envs from monkey species that are known to be hunted by bonobos, such as Thollon’s red colobus (Piliocolobus tholloni), Wolf’s monkey (Cercopithecus wolfii), and black crested mangabeys (Lophocebus aterrimus) (57, 73).
Gorilla species also exhibit CD4 diversity, with western and eastern gorillas encoding three and two CD4 variants, respectively (Fig. 2). In both species, the differences among the haplotypes indicate that additional variants have gone unsampled or must have existed at some point in the past (SI Appendix, Fig. S5). In particular, the AHPM and PRSM alleles in eastern gorillas necessitate at least two intermediate states, both of which (AHSM and ARSM) are currently found in western gorillas. The two species occupy disjunct ranges across central Africa, and it is thus likely that the latter two CD4 alleles predate the split of the two species (i.e., represent trans-specific polymorphisms). Since we sequenced the D1 domain from both SIVgor+ and SIVgor− western lowland gorillas, we asked whether there was an association between CD4 genotypes and SIVgor infection. Examining 60 gorillas from four sites (CP, BQ, DJ, BP), including 23 SIVgor-infected individuals, we failed to identify an excess of CD4 heterozygosity in these communities (SI Appendix, Table S3). It will thus be necessary to genotype larger numbers of both infected and uninfected gorillas to determine whether certain CD4 alleles, or allele combinations, are associated with lower SIVgor infection rates as was observed previously for SIVcpz infected chimpanzees (55). If protective polymorphisms are identified, it would then be important to determine how they confer protection at the individual level (e.g., by heterozygote advantage) or at the population level (e.g., by negative frequency dependent selection). Western gorillas are exposed to SIVgor, and possibly also to SIVcpzPtt from sympatric central chimpanzees through fights and other physical interactions (11). In contrast, eastern gorillas are not SIV-infected but may similarly be exposed to SIVcpzPts from sympatric eastern chimpanzees. Inhibition of the latter viruses by CD4 variants of eastern gorillas may explain why this species exhibits CD4 diversity. It will thus be important to test a larger number of SIVcpzPts strains for their ability to utilize the AHPM and PRSM variants.
In addition to apes, we characterized CD4 variants from mustached monkeys, which exhibited an extraordinary degree of CD4 diversity, with 11 D1 domain variants identified in just 15 individuals. A similarly high number of D1 domain variants was also found in greater spot-nosed monkeys, where 9 D1 domain variants were identified in 17 individuals (Table 1). Mustached monkeys forage in polyspecific troops with other SIV-infected guenons, including greater spot-nosed, and mona monkeys (74, 75). Consistent with this, mustached monkeys harbor three different types of SIVmus, at least two of which are recombinants with SIVgsn and other SIVs (14, 62). Despite their seemingly high SIV exposure risk, mustached, greater spot-nosed, and mona monkeys have very low infection rates, ranging from the complete absence of SIV in some communities to 6% prevalence in others (2, 62, 64–66). This is in contrast to other African primate species, such as western red colobus (P. badius), where prevalence rates of over 80% have been observed (76). It is thus tempting to speculate that the extraordinary CD4 diversity in mustached and greater spot-nosed monkeys reduces SIV transmission, both within and between these species, by forcing viruses to continuously adapt to different CD4 genotypes. However, since other restricting mechanisms have also been proposed (77), additional SIV prevalence and CD4 genotyping studies, including of rarely sampled mona monkeys, are necessary to determine to what extent CD4 diversity associates with SIV infection in these guenons.
All SIV Envs tested here and in our previous study (55) utilized human CD4 more efficiently than chimpanzee, bonobo, or gorilla CD4 alleles (Figs. 1 and 2), most likely because of the absence of D1 domain glycans in the human protein. However, one Env from a virus naturally infecting De Brazza’s monkeys failed to infect cells expressing human CD4, including TZM-bl cells, a HeLa cell line engineered to stably express human CD4, CCR5, and CXCR4 at high levels (78). Surprisingly, when this same SIVdeb Env was tested on cells transiently transfected with mustached monkey CD4, it was able to utilize two variants (alleles 2 and 5) efficiently, one of which (allele 2) is identical at the amino acid level to the De Brazza CD4 observed in this study (Fig. 4C). These data indicate that human CD4, despite lacking D1 domain polymorphisms and glycans, still serves as an entry barrier for some SIV Envs.
A comparison of CD4 sequences among the various primate species also provided evidence of trans-specific polymorphism. Within each of the three genera—Allochrocebus, Cercopithecus, and Chlorocebus—D1 domain sequences did not form species-specific clades (Fig. 4C and SI Appendix, Fig. S8). Instead, identical coding variants were identified in closely related species, suggesting that they predated the emergence of these species. In the case of very closely related species, such as African green monkeys, it could be argued that the sharing of D1 domain alleles is the result of incomplete lineage sorting. However, this explanation is highly unlikely for the Cercopithecus genus. Among 11 guenon species, all except Diana monkeys (Cercopithecus diana) shared identical D1 domain variants, including one that was found in nine different species (SI Appendix, Fig. S8). Interestingly, this variant (designated allele 2 in Fig. 3A) was the most permissive in mustached monkeys (Fig. 3B) and clustered ancestral to all other D1 domain alleles from these nine species (SI Appendix, Fig. S8). Its permissive phenotype may thus reflect the fact that SIVs have had the longest time to adapt to it. Among animals, only a limited number of trans-specific polymorphisms have been described, most of which involve immune genes where heterozygosity appears to confer resistance to pathogens, leading to long-term balancing selection (71, 79). The trans-specific polymorphisms at the CD4 locus in Cercopithecus involve species estimated to have shared a common ancestor 6 to 7 Mya (69, 80), which is consistent with selection maintaining CD4 diversity over a long timescale.
Although genotyping 26 individuals, we failed to detect CD4 polymorphisms in members of three baboon species (Table 1). Baboons are not naturally SIV-infected, but are susceptible to SIVagm, having acquired this virus from sympatric African green monkeys on rare occasions in the wild (81–83). Since all of these acquisitions appear to represent dead-end infections with no secondary spread, it is likely that baboons have acquired anti-SIV defenses downstream of the entry process. Humans, who also lack D1 domain diversity, have evolved restriction factors that target HIV/SIV at multiple steps in the viral life cycle. For example, tetherin, which limits HIV/SIV viral egress from infected cells, is believed to protect humans from zoonotic SIV infections because of a unique 5-amino acid deletion in the cytoplasmic domain that prevents the binding of SIV Nef proteins that counteract tetherin (84, 85). Hence, both humans and baboons may have evolved protective mechanisms that reduce the need for CD4 diversification, although in both species protection from SIV infection has not been absolute. It is also possible that in humans and baboons D1 domain polymorphisms are not compatible with some of the physiological functions of CD4, and are thus not tolerated.
Given that primate lentiviruses have zoonotic potential, it is important to identify and understand factors that limit their spread. The results of this and our previous study (55) suggest that CD4 receptor diversity reduces SIV infection at the population level and might even guard against cross-species infection. While several lines of evidence point to lentiviruses as the driving force of the CD4 diversification in primates, other CD4 tropic pathogens cannot be excluded. Indeed, human herpesvirus 7 (HHV-7) has been shown to require CD4 for entry into target cells (86) and related viruses infect wild apes (87). Although HHV-7 appears to be benign, there could be other pathogens, both extant and extinct, which require attachment to the D1 domain of CD4 to infect target cells. However, irrespective of the forces that have been driving CD4 diversification, it seems clear that existing polymorphisms alter SIV Env–CD4 interactions and in some instances afford protection from SIV infection. These data add to a growing body of evidence suggesting that primate lentiviruses have been exerting selective pressure on their hosts for millions of years. However, the retention of functional CD4 diversity also suggests that balancing selection is still ongoing and protects present day primates from pathogenic SIV infections.
Methods
Nonhuman Primate Samples.
CD4 and G6PD genotyping was performed using remnant DNA from blood or fecal samples collected previously from both wild-caught and captive primates for molecular epidemiological studies of SIV and Plasmodium infections. All samples were obtained with the approval of the respective Institutional Animal Care and Use Committees. International samples were shipped in compliance with Convention on International Trade in Endangered Species of Wild Fauna and Flora regulations and country-specific import and export permits. Relevant information for all samples is summarized in SI Appendix, Tables S1, S3, and S5.
D1 Domain Genotyping.
CD4 genotyping was carried out as previously described (55). Briefly, CD4 exon 2 (165 bp) and exon 3 (159 bp) were PCR-amplified using primers in adjacent introns resulting in amplicons that were 247 bp and 222 bp in length, respectively. Amplicons were MiSeq sequenced without fragmentation to ensure linkage of variable sites for each exon.
Assembly of CD4 Genes from Bonobo and Gorilla Genomic Databases.
Whole-genome Illumina sequencing reads from bonobos and gorillas were downloaded from the National Center for Biotechnology Information (NCBI) database (56) and from the European Nucleotide Archive (60).
Extraction of D1 Domain Sequences from Monkey RNA-Seq and Genomic Databases.
RNA-seq reads from red colobus monkeys (48) were downloaded from the NCBI Sequence Read Archive (SRA) from BioProject PRJNA413051 and aligned as described in SI Appendix. Reads from African green monkey genome datasets (24) were gathered from NCBI SRA BioProjects PRJNA168472, PRJNA168520, PRJNA168527, PRJNA168522, PRJNA168521, PRJNA362602, and PRJNA407948. RNA-seq reads from patas monkeys, aligned to the Macaca mulatta chromosome 11 (accession NC_027903.1), were provided by the authors (47). Consensus sequence extraction proceeded as for the African green monkeys.
Expression Plasmids and Pseudovirus Stocks.
The construction of an SIVcpz∆env reporter backbone and several SIV Envs were previously reported (55). Codon optimized SIVgor, SIVgsn, SIVmon, and SIVdeb env genes were synthesized and cloned into pcDNA3.1. SIV Env pseudotypes were produced in 293T cells by cotransfection of SIVcpz∆env-GFP and SIV env expression plasmids and titered on TZM-bl cells. Full-length chimpanzee CD4 was mutagenized to obtain the bonobo CD4 clone. Full-length gorilla CD4 was synthesized based on genomic sequences. Chimpanzee CCR5 was mutagenized to match bonobo (XM_034956280) and gorilla (XM_004034016) GenBank sequences. The generation of mustached guenon CD4 and CCR5 expression plasmids has previously been described (67). CD4 polymorphisms were introduced by site-directed mutagenesis into one parental plasmid to obtain multiple D1 domain variants. All CD4 and CCR5 alleles were cloned into pcDNA3.1.
Transient CD4 Transfection Assay.
Env-mediated entry was measured as previously described (55). Briefly, 293T cells were transfected with the various CD4 alleles, along with the species-matched CCR5, infected with SIV Env bearing pseudoviruses, and then analyzed for GFP expression by flow cytometry 2 d post infection. CD4 and CCR5 expression levels were determined at the time of infection using receptor specific antibodies (e.g., SI Appendix, Fig. S9).
Phylogenetic Analyses.
Sequence alignments were trimmed to the same start and end position (297 bp or 99 aa for CD4 D1 domain, 1,584 bp for G6PD). For G6PD sequences, regions with indels causing ambiguity in the alignment were removed. Maximum-likelihood trees were constructed using RaxML v8.2.12 (88) with 100 rapid bootstrap replicates, with model GTRGAMMA for nucleotide sequences and PROTGAMMAJTTF for amino acid sequences.
Statistical Analyses.
A hierarchical Bayesian model was used to assess the effects of CD4 polymorphisms on SIV Env-mediated cell entry (SI Appendix). The effects of the various alleles on the infectivity of each Env, as well as an overall allele effect, was estimated by Markov chain Monte Carlo sampling using Stan (89).
Supplementary Material
Acknowledgments
We thank Joseph Mudd and Jason Brenchley for providing curated RNA-sequencing data from patas monkeys; Estrelita Janse van Rensburg and Jason M. Mwenda for providing remnant DNA from various primate species; the Baltimore Zoo for providing samples from Diana monkeys; the staff of project PRESICA and Steve Ahuka Mundeke for fieldwork in Cameroon and the Democratic Republic of the Congo; field assistants from the Greater Mahale Ecosystem Research and Conservation Project for collecting fecal samples from red-tailed monkeys in Tanzania; and the Tanzania Commission for Science and Technology, the Tanzania Wildlife Research Institute, and the Tanzania National Parks Authority for their support and permission to conduct research in the Greater Mahale Ecosystem. This work was supported by grants from the NIH (R01 AI120810, R01 AI050529, R37 AI150590, P01 AI131251, P30 AI045008, R01 AI027698, P51 RR00164) and the Agence Nationale de Recherche pour le SIDA (ANRS 12325). R.M.R. was supported by a training grant (T32 AI 007632). The findings and conclusions of this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC). The use of trade names and commercial sources is for identification only and does not imply endorsement by CDC.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025914118/-/DCSupplemental.
Data Availability
Primate CD4 exons 2 and 3 sequences and G6PD sequences have been deposited in GenBank (accession nos. MW514379–MW514444, MW535772, and MW535773). Analysis code is archived on Zenodo (https://doi.org/10.5281/zenodo.4602351). All other study data are included in the article and SI Appendix.
References
- 1.Peeters M., Ma D., Liegeois F., Apetrei C., “Simian immunodeficiency virus infections in the wild” in Natural Hosts of SIV, Ansari A., Silvestri G., Eds. (Elsevier Inc., 2014), pp. 37–67. [Google Scholar]
- 2.Aghokeng A. F., et al., Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Infect. Genet. Evol. 10, 386–396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Courgnaud V., et al., Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76, 8298–8309 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Heuverswyn F., et al., Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444, 164 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Santiago M. L., et al., Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J. Virol. 77, 7545–7562 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharp P. M., Hahn B. H., Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1, a006841 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao F., et al., Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441 (1999). [DOI] [PubMed] [Google Scholar]
- 8.D’arc M., et al., Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc. Natl. Acad. Sci. U.S.A. 112, E1343–E1352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayouba A., et al., Evidence for continuing cross-species transmission of SIVsmm to humans: Characterization of a new HIV-2 lineage in rural Côte d’Ivoire. AIDS 27, 2488–2491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailes E., et al., Hybrid origin of SIV in chimpanzees. Science 300, 1713 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Takehisa J., et al., Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83, 1635–1648 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Souquière S., et al., Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75, 7086–7096 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin M. J., et al., Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 13, 2935–2947 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liégeois F., et al., Full-length genome analyses of two new simian immunodeficiency virus (SIV) strains from mustached monkeys (C. cephus) in Gabon illustrate a complex evolutionary history among the SIVmus/mon/gsn lineage. Viruses 6, 2880–2898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauck M., et al., Discovery and full genome characterization of two highly divergent simian immunodeficiency viruses infecting black-and-white colobus monkeys (Colobus guereza) in Kibale National Park, Uganda. Retrovirology 10, 107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gifford R. J., et al., A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 20362–20367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert C., Maxfield D. G., Goodman S. M., Feschotte C., Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 5, e1000425 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han G. Z., Worobey M., A primitive endogenous lentivirus in a colugo: Insights into the early evolution of lentiviruses. Mol. Biol. Evol. 32, 211–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katzourakis A., Tristem M., Pybus O. G., Gifford R. J., Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. U.S.A. 104, 6261–6265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keckesova Z., Ylinen L. M., Towers G. J., Gifford R. J., Katzourakis A., Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology 384, 7–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui J., Holmes E. C., Endogenous lentiviruses in the ferret genome. J. Virol. 86, 3383–3385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han G. Z., Worobey M., Endogenous lentiviral elements in the weasel family (Mustelidae). Mol. Biol. Evol. 29, 2905–2908 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allan J. S., et al., Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 65, 2816–2828 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svardal H., et al., Ancient hybridization and strong adaptation to viruses across African vervet monkey populations. Nat. Genet. 49, 1705–1713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Worobey M., et al., Island biogeography reveals the deep history of SIV. Science 329, 1487 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Beer B. E., et al., Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): Evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J. Virol. 73, 7734–7744 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Compton A. A., Emerman M., Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog. 9, e1003135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy K. R., Kirmaier A., Autissier P., Johnson W. E., Evolutionary and functional analysis of Old World primate TRIM5 reveals the ancient emergence of primate lentiviruses and convergent evolution targeting a conserved capsid interface. PLoS Pathog. 11, e1005085 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Compton A. A., Malik H. S., Emerman M., Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B., Molecular mechanism of HIV-1 entry. Trends Microbiol. 27, 878–891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janeway C. A. Jr., The T cell receptor as a multicomponent signalling machine: CD4/CD8 coreceptors and CD45 in T cell activation. Annu. Rev. Immunol. 10, 645–674 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Gay D., Buus S., Pasternak J., Kappler J., Marrack P., The T-cell accessory molecule CD4 recognizes a monomorphic determinant on isolated Ia. Proc. Natl. Acad. Sci. U.S.A. 85, 5629–5633 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clayton L. K., Sieh M., Pious D. A., Reinherz E. L., Identification of human CD4 residues affecting class II MHC versus HIV-1 gp120 binding. Nature 339, 548–551 (1989). [DOI] [PubMed] [Google Scholar]
- 34.Kwong P. D., et al., Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenchley J. M., et al., Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12, 1365–1371 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Bosinger S. E., Sodora D. L., Silvestri G., Generalized immune activation and innate immune responses in simian immunodeficiency virus infection. Curr. Opin. HIV AIDS 6, 411–418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keele B. F., et al., Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460, 515–519 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudicell R. S., et al., Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 6, e1001116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbian H. J., et al., Effective treatment of SIVcpz-induced immunodeficiency in a captive western chimpanzee. Retrovirology 14, 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandrea I., et al., Chronic SIV infection ultimately causes immunodeficiency in African non-human primates. AIDS 15, 2461–2462 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Apetrei C., et al., Direct inoculation of simian immunodeficiency virus from sooty mangabeys in black mangabeys (Lophocebus aterrimus): First evidence of AIDS in a heterologous African species and different pathologic outcomes of experimental infection. J. Virol. 78, 11506–11518 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling B., et al., Classic AIDS in a sooty mangabey after an 18-year natural infection. J. Virol. 78, 8902–8908 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joas S., et al., Species-specific host factors rather than virus-intrinsic virulence determine primate lentiviral pathogenicity. Nat. Commun. 9, 1371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandrea I., Silvestri G., Apetrei C., AIDS in African nonhuman primate hosts of SIVs: A new paradigm of SIV infection. Curr. HIV Res. 7, 57–72 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Beaumier C. M., et al., CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat. Med. 15, 879–885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palesch D., et al., Sooty mangabey genome sequence provides insight into AIDS resistance in a natural SIV host. Nature 553, 77–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mudd J. C., et al., Epigenetic silencing of CD4 expression in nonpathogenic SIV infection in African green monkeys. JCI Insight 5, 139043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simons N. D., et al., Genome-wide patterns of gene expression in a wild primate indicate species-specific mechanisms associated with tolerance to natural simian immunodeficiency virus infection. Genome Biol. Evol. 11, 1630–1643 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coffin J. M., Virions at the gates: Receptors and the host-virus arms race. PLoS Biol. 11, e1001574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demogines A., Abraham J., Choe H., Farzan M., Sawyer S. L., Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol. 11, e1001571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Demogines A., Farzan M., Sawyer S. L., Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J. Virol. 86, 6350–6353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z. D., Weinstock G., Gerstein M., Rapid evolution by positive Darwinian selection in T-cell antigen CD4 in primates. J. Mol. Evol. 66, 446–456 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Meyerson N. R., Sharma A., Wilkerson G. K., Overbaugh J., Sawyer S. L., Identification of owl monkey CD4 receptors broadly compatible with early-stage HIV-1 isolates. J. Virol. 89, 8611–8622 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyerson N. R., et al., Positive selection of primate genes that promote HIV-1 replication. Virology 454-455, 291–298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bibollet-Ruche F., et al., CD4 receptor diversity in chimpanzees protects against SIV infection. Proc. Natl. Acad. Sci. U.S.A. 116, 3229–3238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prado-Martinez J., et al., Great ape genetic diversity and population history. Nature 499, 471–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Surbeck M., Hohmann G., Primate hunting by bonobos at LuiKotale, Salonga National park. Curr. Biol. 18, R906–R907 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Li Y., et al., Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J. Virol. 86, 10776–10791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu W., et al., Wild bonobos host geographically restricted malaria parasites including a putative new Laverania species. Nat. Commun. 8, 1635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xue Y., et al., Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348, 242–245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warren C. J., et al., A glycan shield on chimpanzee CD4 protects against infection by primate lentiviruses (HIV/SIV). Proc. Natl. Acad. Sci. U.S.A. 116, 11460–11469 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aghokeng A. F., et al., Full-length sequence analysis of SIVmus in wild populations of mustached monkeys (Cercopithecus cephus) from Cameroon provides evidence for two co-circulating SIVmus lineages. Virology 360, 407–418 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liégeois F., et al., New STLV-3 strains and a divergent SIVmus strain identified in non-human primate bushmeat in Gabon. Retrovirology 9, 28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aghokeng A. F., et al., Widely varying SIV prevalence rates in naturally infected primate species from Cameroon. Virology 345, 174–189 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Schmidt F., et al., Phyloepidemiological analysis reveals that viral divergence led to the paucity of simian immunodeficiency virus SIVmus/gsn/mon infections in wild populations. J. Virol. 91, e01884-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steve A. M., et al., High prevalences and a wide genetic diversity of simian retroviruses in non-human primate bushmeat in rural areas of the Democratic Republic of Congo. EcoHealth 14, 100–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wetzel K. S., et al., Loss of CXCR6 coreceptor usage characterizes pathogenic lentiviruses. PLoS Pathog. 14, e1007003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hart J. A., et al., Lesula: A new species of Cercopithecus monkey endemic to the Democratic Republic of Congo and implications for conservation of Congo’s central basin. PLoS One 7, e44271 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Springer M. S., et al., Macroevolutionary dynamics and historical biogeography of primate diversification inferred from a species supermatrix. PLoS One 7, e49521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fijarczyk A., Babik W., Detecting balancing selection in genomes: Limits and prospects. Mol. Ecol. 24, 3529–3545 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Azevedo L., Serrano C., Amorim A., Cooper D. N., Trans-species polymorphism in humans and the great apes is generally maintained by balancing selection that modulates the host immune response. Hum. Genomics 9, 21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu H. Y., et al., CD4-Dependent and CD4-independent utilization of coreceptors by human immunodeficiency viruses type 2 and simian immunodeficiency viruses. Virology 278, 276–288 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Surbeck M., Fowler A., Deimel C., Hohmann G., Evidence for the consumption of arboreal, diurnal primates by bonobos (Pan paniscus). Am. J. Primatol. 71, 171–174 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Gautier-Hion A, Gautier J P, [Polyspecific associations of Cercopithecus species at M’passa Plateau (Gabon) (author’s transl)]. Folia Primatol (Basel) 22 (2–3), 134–77, 10.1159/000155623 (1974). [DOI] [PubMed] [Google Scholar]
- 75.Gautier-Hion A., Bourlière F., Gautier J.-P., Kingdon J., Eds., A Primate Radiation: Evolutionary Biology of the African Guenons (Cambridge University Press, Cambridge, UK, 1988). [Google Scholar]
- 76.Leendertz S. A., et al., High prevalence, coinfection rate, and genetic diversity of retroviruses in wild red colobus monkeys (Piliocolobus badius badius) in Tai National Park, Cote d’Ivoire. J. Virol. 84, 7427–7436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heigele A., et al., The potency of Nef-mediated SERINC5 antagonism correlates with the prevalence of primate lentiviruses in the wild. Cell Host Microbe 20, 381–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei X., et al., Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46, 1896–1905 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Těšický M., Vinkler M., Trans-species polymorphism in immune genes: General pattern or MHC-restricted phenomenon? J. Immunol. Res. 2015, 838035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perelman P., et al., A molecular phylogeny of living primates. PLoS Genet. 7, e1001342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jin M. J., et al., Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: Evidence for cross-species transmission in the wild. J. Virol. 68, 8454–8460 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kodama T., et al., Prevalence of antibodies to SIV in baboons in their native habitat. AIDS Res. Hum. Retroviruses 5, 337–343 (1989). [DOI] [PubMed] [Google Scholar]
- 83.van Rensburg E. J., et al., Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: Detection of a SIVagm variant from a chacma baboon. J. Gen. Virol. 79, 1809–1814 (1998). [DOI] [PubMed] [Google Scholar]
- 84.Jia B., et al., Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5, e1000429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sauter D., et al., Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lusso P., et al., CD4 is a critical component of the receptor for human herpesvirus 7: Interference with human immunodeficiency virus. Proc. Natl. Acad. Sci. U.S.A. 91, 3872–3876 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lavergne A., et al., African great apes are naturally infected with roseoloviruses closely related to human herpesvirus 7. J. Virol. 88, 13212–13220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carpenter B., et al., Stan: A probabilistic programming language. J. Stat. Softw. 76, 1–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primate CD4 exons 2 and 3 sequences and G6PD sequences have been deposited in GenBank (accession nos. MW514379–MW514444, MW535772, and MW535773). Analysis code is archived on Zenodo (https://doi.org/10.5281/zenodo.4602351). All other study data are included in the article and SI Appendix.