Significance
Social challenges are thought to be embedded in the genome, but little is known about how this adaptive modification of gene activity occurs in nature. We experimentally generated competition among free-living female tree swallows (Tachycineta bicolor) and measured both transcriptomic and epigenomic changes in the brain. Social competition rapidly altered gene expression and methylation, with effects that were enriched for hormone- and neurotransmitter-related processes also being found two full days later. These persistent effects on gene activity were correlated with epigenetic patterns as well as the degree of aggression experienced during the peak of competition. Thus, a single day of competition in the wild is encoded in the brain, both during competition and days later.
Keywords: gene network, MethylCap-seq, RNA-seq, dopamine, epigenetic
Abstract
Periods of social instability can elicit adaptive phenotypic plasticity to promote success in future competition. However, the underlying molecular mechanisms have primarily been studied in captive and laboratory-reared animals, leaving uncertainty as to how natural competition among free-living animals affects gene activity. Here, we experimentally generated social competition among wild, cavity-nesting female birds (tree swallows, Tachycineta bicolor). After territorial settlement, we reduced the availability of key breeding resources (i.e., nest boxes), generating heightened competition; within 24 h we reversed the manipulation, causing aggressive interactions to subside. We sampled females during the peak of competition and 48 h after it ended, along with date-matched controls. We measured transcriptomic and epigenomic responses to competition in two socially relevant brain regions (hypothalamus and ventromedial telencephalon). Gene network analyses suggest that processes related to energy mobilization and aggression (e.g., dopamine synthesis) were up-regulated during competition, the latter of which persisted 2 d after competition had ended. Cellular maintenance processes were also down-regulated after competition. Competition additionally altered methylation patterns, particularly in pathways related to hormonal signaling, suggesting those genes were transcriptionally poised to respond to future competition. Thus, experimental competition among free-living animals shifts gene expression in ways that may facilitate the demands of competition at the expense of self-maintenance. Further, some of these effects persisted after competition ended, demonstrating the potential for epigenetic biological embedding of the social environment in ways that may prime individuals for success in future social instability.
Competition is ubiquitous among social animals, and an individual’s behavioral and physiological response to competition can greatly impact fitness. For decades, research in this area was guided by hypotheses formed by studying males and was focused on steroid hormones (i.e., challenge hypothesis) (1); however, recent analyses indicate that androgen modulation is not necessarily a universal response to social competition (2–4). As more genome-wide studies are performed, it has become clear that responses to territorial aggression are mediated by changes in hundreds of genes in the brain (5–7), and yet, at least in vertebrates, this evidence largely comes from captive and laboratory-reared animals. Thus, whether the key aggression-related pathways discovered in this past work are relevant to animals responding to more naturalistic competition in the wild is unknown. This issue is especially important because competition in nature unfolds as a complex series of dynamic social interactions, often among multiple individuals in nondyadic, brief interactions. To date, we are aware of only one experiment examining genome-wide neural responses to territorial intrusions in a free-living animal; however, they used simulated intrusions involving an immobile intruder (8). There is evidence that resident–intruder paradigms may insufficiently mimic natural territorial encounters (9) and may not prompt a full neurogenomic response, particularly in contexts that would otherwise culminate in direct physical contact (10). Furthermore, control treatments in resident–intruder tests are often characterized by the absence of interactions, making it unclear if effects are due to agonistic interactions or are merely a by-product of any social exchange. Thus, despite the importance of competitive interactions in social systems, there is uncertainty regarding the genomic response to competition because prior experimental work may not sufficiently mimic natural social competition (11).
Competitive interactions may also have lasting effects if competitive experiences are encoded in the genome. Animals involved in recent territorial disputes tend to behave more aggressively and have a greater probability of winning future conflicts (i.e., winner effect) (12). However, little is known about how the accompanying neurogenomic responses also change over time, with only a handful of studies examining gene expression at multiple time points after a competitive experience, all sampled within a couple of hours (13, 14). This has created a gap in our understanding of how social experiences generate long-lasting effects on behavioral and physiological processes (i.e., biological embedding) (15). Epigenetic mechanisms, such as DNA methylation, are prime candidates to mediate the lasting effects of the social environment on gene activity (16, 17). To date, methylation has been shown to respond to territorial intrusions on the scale of hours (18); however, how competition-induced changes in methylation prime the genome over longer time periods has yet to be tested, much less in a free-living animal. This lends uncertainty as to whether and how naturalistic competition is biologically embedded to bring about lasting phenotypic changes.
Here, we experimentally generated a period of heightened territorial competition to explore the immediate and longer-lasting effects on gene activity in the female brain. We used free-living tree swallows (Tachycineta bicolor), which are ideal for studying ecologically relevant female–female competition. These songbirds are obligate secondary cavity nesters, meaning that a female’s reproductive success is dependent on her ability to acquire a cavity that she cannot excavate for herself (19). Nesting cavities are often limited, females must aggressively compete to obtain one, and female aggression is adaptive during territory establishment (20). Furthermore, nonterritory-holding female “floaters” intrude at territories throughout the early breeding season, and they can evict or even kill other females (21), demonstrating that competition after initial territorial establishment is a relevant threat for these birds. Using a population of free-living tree swallows breeding in artificial cavities (nest boxes), we experimentally increased competition by reducing nest box availability after initial territory settlement. We then returned boxes within 24 h to reverse the experiment. We collected females during the peak of competition or 48 h after the end of the competitive period, along with date-matched controls. We measured transcriptomic and epigenomic responses in brain regions containing the vertebrate social behavior network, the hypothalamus (HYPO) and ventromedial telencephalon (VmT) (22). Using gene coexpression network analyses, we identify pathways that were differentially regulated during and/or after competition, along with cooccurring changes in DNA methylation. These data shed light on the effects of female–female social competition in a context with clear consequences for reproductive success.
Results
Behavioral and Hormonal Responses to Competition.
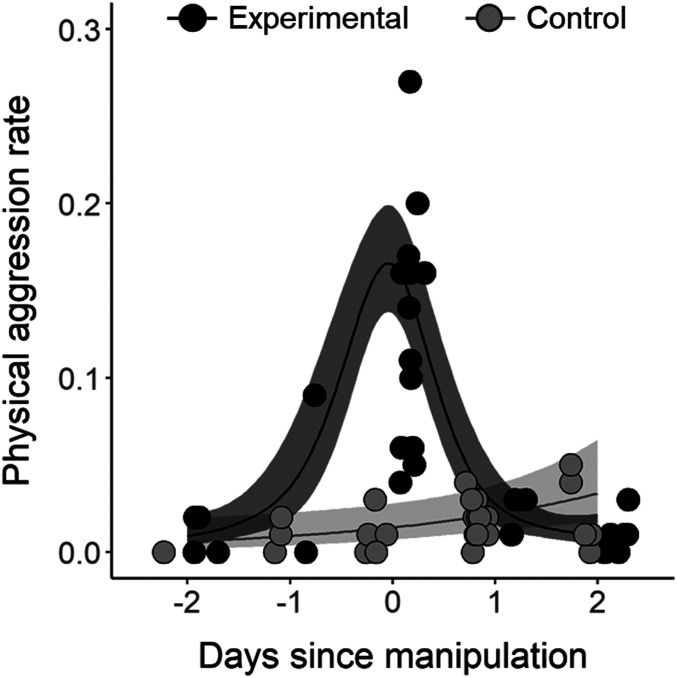
Fig. 1 summarizes the experimental design, which takes advantage of these birds’ tendency to defend the area ∼20 m around their nest box, including additional boxes (23). Our goal was to induce competition, concentrated near “focal” females, who were our primary study subjects. To achieve this, we removed half of the occupied nest boxes along with all unoccupied boxes at multiple experimental sites, thereby guaranteeing a minimum number of competitors (i.e., displaced females) in addition to any floater females also present. We erected new boxes to accommodate only half of the displaced females, placing them 20 m away from the untouched boxes of focal territory holders. This positioning was close enough to facilitate interactions between focal females and competitors vying for the nearby new boxes. Less than 24 h after the manipulation, we returned the removed boxes to their former positions, effectively reversing the experiment. Each experimental and unmanipulated control site was separated by >300 m, a distance that exceeds daily home ranges in this system (24). We performed daily behavioral observations during the experimental manipulation (day 0) and throughout the 48 h preceding (day −2) and following (day +2) the manipulation to quantify rates of physical aggression. A generalized additive model (GAM) was fit with rates of physical aggression as the dependent variable and the main effects of time of day, treatment (control vs. experimental sites), a smoothed effect of day, and a smoothed interaction term, with site as the random effect. The GAM explained 77.3% of the deviance in observed rates of physical aggression (R2 = 0.739). The smoothed term for the interaction between day and experimental sites was significant (effective degrees of freedom, EDF = 2.72, F = 33.09, P < 0.001), while the smoothed term for the interaction between day and control sites was not significant, suggestive of a more linear relationship (EDF = 1.00, F = 0.11, P = 0.75; Fig. 2). Rates of physical aggression also increased as time of day approached noon (β = 0.002 ± 0.001, t = 2.11, P = 0.040). Via radiofrequency identification (RFID) devices at nest boxes, we also showed that focal females did not lose ownership of their box, and in 40% of cases focal females were recorded at the nest entrance of the nearby (new) box, in addition to being detected at their original box (e.g., SI Appendix, Fig. S1). Our behavioral observations further confirmed that all focal females interacted with conspecifics at these new boxes; most aggressive interactions occur in flight and therefore cannot be detected by RFID devices at the nestbox. These data collectively demonstrate that focal females were involved in competition throughout the experimental period. Additionally, no displaced female RFID tags were detected at other sites, indicating the effect was localized.
Fig. 1.
Experimental design to generate heightened competition for focal females. The evening before day 0, 50% of the boxes were removed, leaving remaining “focal” boxes evenly spaced in dyads. A “new” box was placed equidistant to the focal boxes (∼20 m) to facilitate interactions between focal females and other conspecifics (displaced and/or floater females). Focal females were sampled on day 0 or day 2. Transparency denotes box removal/bird collection and letters indicate unique birds.
Fig. 2.
Smooth function for physical aggression rate (minutes in which physical aggression was observed per minutes observed per boxes occupied during daily observations). Circles represent individual observations and lines are the smooth function. Dark gray represents experimental sites and light gray represents control sites. Shading corresponds to 95% confidence intervals.
We collected focal females, those that experienced competition but did not have their box removed, at experimental sites during peak competition (day 0; n = 6) and 48 h after competition had ended (day 2; n = 4), along with date-matched control females (day 0, n = 5; day 2, n = 5). When a female was collected for molecular analyses, her box was claimed by a new, presumed floater female within 24 h (SI Appendix, Figs. S1 and S2). Plasma testosterone from all females was analyzed with a linear mixed model (random effect of site) and was not significantly affected by treatment (F1,4 = 1.64, P = 0.27), day (F1,12 = 0.002, P = 0.97), or their interaction (treatment × day: F1,12 = 1.48, P = 0.25). Average testosterone levels were 0.17 ng/mL ± 0.04 SE (n = 20), similar to previously reported baseline values in this species at this breeding stage (25).
Differentially Expressed Genes during and 2 d after Competition.
We performed RNA sequencing (RNA-seq) and differential expression analyses, comparing females collected at experimental and control sites within both time periods (day 0 and day 2) in two brain regions, the HYPO and VmT (SI Appendix, Figs. S3 and S4 and Dataset S1). The HYPO had n = 13 differentially expressed genes (DEGs) between control and experimental females on day 0 and the VmT had 43 DEGs. On day 2, the HYPO had 59 DEGs between control and experimental females and the VmT had 21 DEGs. The DEGs in the VmT on day 2 were enriched for the Gene Ontology (GO) process anatomical structure morphogenesis (GO:0009653; n = 11 genes, fold enrichment = 5.01, false discovery rate [FDR] = 0.035). No other significantly enriched functional annotations were identified for DEGs within each brain area or time point.
Overlap in DEGs between brain regions occurred within each time period (SI Appendix, Fig. S5). Two immediate early genes, ARC and EGR1 (also known as ZENK), were differentially expressed in both brain regions on day 0, during peak competition, suggesting these brain regions were stimulated by the manipulation (26). There were also four DEGs shared between brain regions on day 2.
Competition-Related Gene Modules.
Genes do not act alone, instead performing their functions in shared pathways and networks (27). For this reason, we performed a weighted gene coexpression network analysis (WGCNA) to identify modules of coexpressed genes across all samples (n = 20) within each brain region to determine how whole networks, rather than individual genes, responded to the manipulation (28). We tested for an association between modules and traits of interest (i.e., treatment, day, testosterone levels, and aggression), as well as potentially confounding variables (i.e., field sites and time of day collected).
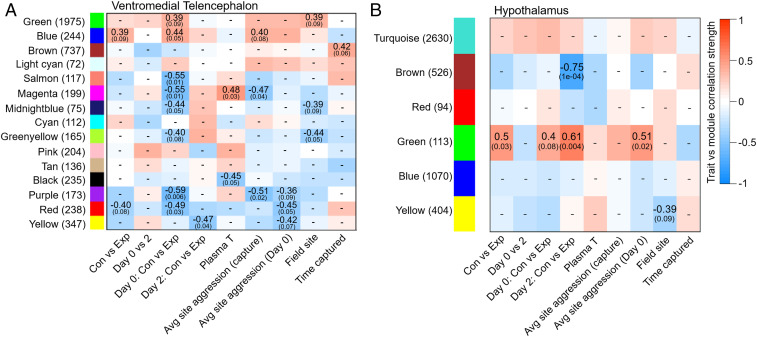
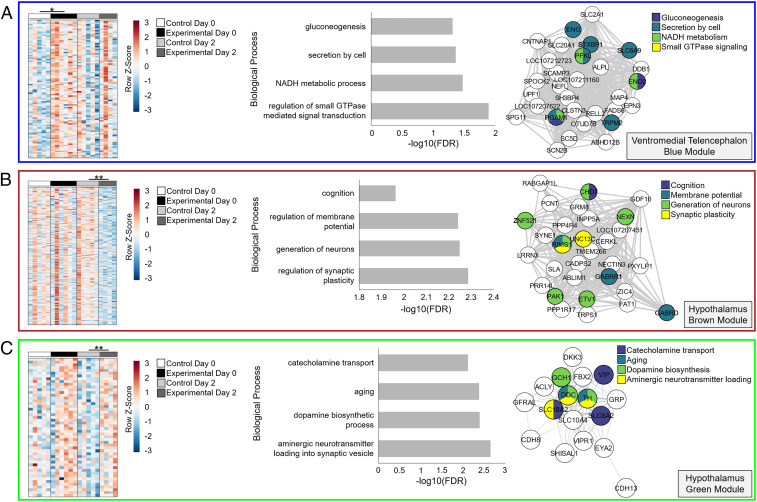
In the VmT, WGCNA constructed 15 modules, not including the gray module of “unassigned” genes (Dataset S2). Five modules (salmon, magenta, midnight blue, purple, and red) showed expression patterns that were negatively associated with the experimental treatment on day 0 (during peak competition), while one module (blue) was positively associated with treatment, indicating competition-induced up-regulation (Figs. 3A and 4A). Only the blue module had enriched functional annotations, including GO processes related to signal transduction, NADH (reduced nicotinamide-adenine dinucleotide) metabolic processes, and gluconeogenesis (Fig. 4A and SI Appendix, Table S1). The top intramodular hub genes were defined as those with a high trait-based gene significance (the absolute value of the correlation between the gene and trait; abs GS > 0.2) and high module membership (the correlation between the gene expression profile and module eigengene; MM > 0.6). Intramodular hub genes included key genes in gluconeogenesis, such as phosphofructokinase (PFKL; MM = 0.95; GS = 0.57), phosphoglycerate 1 (PGAM1; MM = 0.96; GS = 0.56), and enolase 2 (ENO2; MM = 0.88; GS = 0.47), along with a glucose transporter (SLC2A1; MM = 0.96; GS = 0.50) (Fig. 4A). Past work using simulated territorial intrusions identified several putative “toolkit” transcription factors that are thought to comprise an evolutionarily conserved response to social challenges (7). If these transcription factors are a conserved way in which animals regulate behavioral gene networks, we might expect these genes to be intramodular hubs in our competition-related modules. We identified 12 of these putative toolkit genes in the VmT, and 4 were hubs in modules associated with the experimental treatment (SI Appendix, Table S2). Toolkit genes were more likely to be hubs in competition-related modules than nontoolkit genes (33% vs. 6%; Fisher’s exact test, P = 0.004).
Fig. 3.
Module–trait relationships determined by biweight midcorrelation in a WGCNA. (A) Fifteen modules in the ventromedial telencephalon or (B) six modules in the hypothalamus were constructed from all expressed genes from 20 samples across two treatment groups and two time points. Statistical associations are presented for treatment (control [Con] and experimental [Exp]) at the peak of competition (day 0) and two days later (day 2), plasma testosterone (T), rates of physical aggression at the sites either the day females were collected or day 0, field site, and time of day collected. Presented are correlation coefficients and associated P values (only P < 0.10 is shown, within brackets).
Fig. 4.
Functional analysis of trait-associated modules. The (A) blue module in the ventromedial telencephalon and the (B) brown and (C) green modules in the hypothalamus were associated with the treatment. Heat maps depict genes with a trait-based gene significance (GS) > 0.2. Asterisks denote significant relationships (*P < 0.05; **P < 0.01). The top four GO terms are presented along with the top hub genes (GS > 0.2, module membership > 0.6, weight > 0.08). Node color indicates GO term inclusion.
In the HYPO, WGCNA constructed six modules (Dataset S3), two of which were trait-associated (Fig. 3B). The brown module was negatively associated with the experimental treatment on day 2, indicating these genes were down-regulated 2 d after competition (Fig. 4B). Genes in this module showed significant enrichment for GO terms related to synaptic plasticity, generation of neurons, and cognition (Fig. 4B and SI Appendix, Table S1). Hub genes included those related to the major excitatory and inhibitory neurotransmitters, including a gamma-aminobutyric acid receptor (GABRD; MM = 0.94; GS = −0.74) and a glutamate receptor (GRM8; MM = 0.98; GS = −0.74), along with several genes involved in the generation of neurons (Fig. 4B). The green module was positively associated with the experimental treatment regardless of day, but this relationship was stronger on day 2, indicating these genes were most strongly up-regulated 2 d after competition (Figs. 3B and 4C). Critically, the green module was also positively associated with the rate of physical aggression experienced on day 0 (Fig. 3B) and enriched for GO terms related to aminergic neurotransmitter loading and dopamine biosynthesis (Fig. 4C and SI Appendix, Table S1). The top intramodular hub genes included key genes in dopamine synthesis, like dopa decarboxylase (DDC; MM = 0.94; GS = 0.48) and tyrosine hydroxylase (TH; MM = 0.88; GS = 0.51), as well as genes important for catecholamine transport, like vasoactive intestinal peptide (VIP; MM = 0.83; GS = 0.52) and its receptor (VIPR1; MM = 0.70; GS = 0.53), monoamine transporter (SLC18A2; MM = 0.91; GS = 0.47), and norepinephrine transporter (SLC6A2; MM = 0.82; GS = 0.47) (Fig. 4C). Nine putative toolkit genes were identified in the HYPO, and one was a hub in a module associated with the experimental treatment (SI Appendix, Table S2). However, toolkit genes were not more likely to be hubs in competition-related modules than nontoolkit genes (11% vs. 5%; Fisher’s exact test, P = 0.353).
Differentially Methylated Genes during and 2 d after Competition.
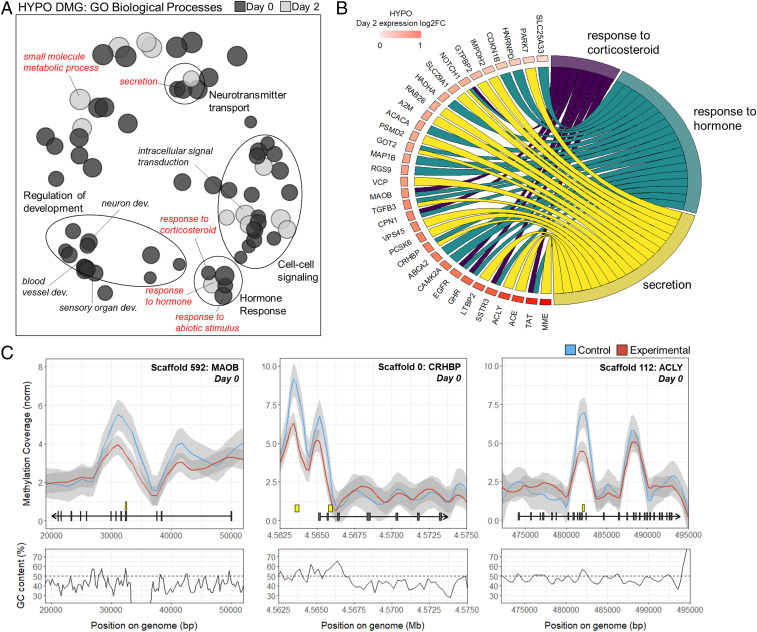
Due to the enduring transcriptomic effects of competition in the HYPO, we performed MethylCap sequencing (MethylCap-seq) in this tissue to identify differentially methylated regions (DMRs). We compared females collected at experimental and control sites within each time period (day 0 and day 2). At the peak of competition on day 0, there were n = 13,260 DMRs, and on day 2 after competition had subsided there were n = 7,382 DMRs. Further analysis was restricted to DMRs located 2 kb upstream of a gene (i.e., putative promoter) or within a gene body (including exons and introns). During competition (day 0), we observed 2,447 DMRs within gene bodies and 299 in the 2-kb upstream region; 1,963 unique genes had DMRs (SI Appendix, Fig. S6 and Dataset S4). Two days after competition (day 2), there were 1,249 DMRs within gene bodies and 191 in the 2-kb upstream region; 1,171 unique genes had DMRs at this time point (SI Appendix, Fig. S6 and Dataset S4). There was overlap in genes with DMRs between birds sampled on day 0 or day 2: Of the 1,963 unique genes with DMRs on day 0, 462 genes (23.5%) showed differential methylation on day 2 as well (SI Appendix, Fig. S7). These genes were enriched for processes including GTPase activity and neuron projection development (SI Appendix, Table S3). Genes with DMRs unique to day 0 were enriched for GO biological processes involving signaling, response to corticosteroid, development, and neurotransmitter transport (Fig. 5A and SI Appendix, Table S3). Notable genes with DMRs on day 0 included glucocorticoid receptors (NR3C1; also known as GR) and enzymes (HSD11B2) and monoamine oxidase B (MAOB) (Dataset S4). Genes with DMRs unique to day 2 were enriched for signal transduction, canonical Wnt signaling, and response to hormones (Fig. 5A and SI Appendix, Table S3). These hormone-related genes with DMRs on day 2 included gonadotropin-releasing (GNRHR) and corticotropin-releasing (CRHR1) hormone receptors, steroid enzymes (HSD3B1), somatostatin receptors (SSTR3), and opioid receptors (OPRL1) (Dataset S4).
Fig. 5.
Competition-induced changes in methylation. (A) Biological processes enriched in differentially methylated genes (DMGs) in response to competition in the hypothalamus on day 0 or 2 as revealed by GO analyses. Each circle represents a GO term; larger sizes indicate more significant (all FDR < 0.05). GO terms are clustered based on semantic similarity and labeled by AutoAnnotate in Cytoscape. (B) GOChord plot of leading-edge genes in biological processes found enriched in a gene set enrichment analysis of day-2 gene expression that also overlap with DMGs (genes also in red terms in A). Genes are linked to their assigned pathway via colored ribbons and ordered according to the observed log2 fold change (LogFC) on day 2 (red, higher expression in experimental females). (C) Key behavioral leading-edge genes in hormonal processes that also have differential methylation. (Upper) Normalized coverage and 95% confidence intervals (±2 kb of a gene) for control (blue) and experimental (red) females on day 0 or 2. Genes are indicated by an arrow and exons are gray rectangles. The yellow rectangle shows the DMR. (Lower) GC percentage with a dashed line indicating 50%.
Relationship between Gene Expression and Methylation over Time.
Globally, we found that competition-induced changes in methylation were correlated with gene expression (Spearman correlation of an individual’s treatment-induced log2 fold change in methylation vs. their treatment-induced log2 fold change in gene expression; only genes with DMRs were included). In particular, day-0 methylation in the gene body was weakly but significantly negatively correlated with gene expression at day 0 (rs = −0.14, padj < 0.001). Moreover, competition-induced methylation in the gene body on day 0 also inversely predicted gene expression two days later (rs = −0.09, padj = 0.013). There was also a trend for day-0 methylation patterns within the gene body to be positively correlated with day-2 gene body methylation patterns (rs = 0.12, padj = 0.087), indicative of persistent directional effects. Methylation in the 2-kb upstream region was unrelated to gene expression within or across time points (padj > 0.460; SI Appendix, Fig. S8).
We then asked how competition-induced changes in methylation may relate to subtle, but enduring, changes in gene expression at the functional level. First, we used a hypothesis-driven approach, asking whether biological processes enriched in genes with DMRs also contained an overrepresentation of genes with coordinately up- or down-regulated expression. We performed a gene set enrichment analysis (GSEA) within each time point, using all genes expressed in the HYPO ranked by their competition-induced change as gene lists. The gene sets tested (i.e., collection of genes associated with a specific biological process) were GO terms previously found to be enriched in genes with DMRs on day 0 and/or day 2 (found in SI Appendix, Table S3 and Fig. 5A). We found that five of the processes enriched with differentially methylated genes were also significantly enriched with up-regulated genes in the day-2 treatment comparison, including terms related to hormonal responses (response to corticosteroid and response to hormone) and neurotransmitter transport (secretion) (SI Appendix, Table S4). This suggests competition-induced epigenetic modifications may regulate the expression of genes in these processes, so we next looked for overlap between genes highlighted in the GSEA (i.e., leading-edge genes, those that contribute most to the enrichment) and those with competition-induced DMRs within these five processes (Fig. 5B). Genes with strong links to social behavior, like MAOB, corticotropin-releasing hormone binding protein (CRHBP), and ATP citrate lyase (ACLY) were among these overlapping genes; each of them showed lower methylation on day 0 (Fig. 5C), coupled with higher expression on day 2 (Fig. 5B). None of the biological processes enriched with differentially methylated genes were also enriched in genes whose expression was influenced by the treatment on day 0.
Next, we explored whether intramodular hub genes, which are potential drivers of the functional pathways highlighted in treatment-related modules in the HYPO, also contained DMRs (SI Appendix, Fig. S7). The brown module, which was characterized by processes involving synaptic plasticity and cognition, had several hub genes with established links to social behavior that also contained DMRs. For instance, reelin (RELN) and CRHR1 were among the brown module hub genes that also had DMRs on day 0 and day 2, respectively (Dataset S4). Another brown module gene, NR4A3 [a putative toolkit gene for social behavior (7)] also had a DMR located in the first exon on day 0 (SI Appendix, Fig. S9). The green module, which was characterized by processes related to aminergic neurotransmitter loading and dopamine biosynthesis, had hub genes ACLY and VIPR1 that also had DMRs on day 0 and day 2, respectively (Fig. 5C and Dataset S4). In sum, we found overlap in competition-induced methylation and potential drivers of functional patterns observed in modules of coexpressed genes in the HYPO.
Discussion
Experimental manipulation of competition among free-living female songbirds induced immediate and longer-lasting neurogenomic responses. Females at experimental sites experienced a temporary increase in physically aggressive interactions compared to controls, and this occurred without marked changes in plasma testosterone. Network analyses indicate that females quickly underwent a shift in gene expression that may have facilitated the energetic demands of competition at the expense of self-maintenance. Increased expression within aggression-related modules persisted for days after competition had ended. Furthermore, putative toolkit genes were more likely to be intramodular hubs in the VmT, suggesting these transcription factors are indeed a conserved way in which animals regulate behavioral gene networks in both simulated territorial encounters (7) and natural competition between free-living animals. Competition in the wild also altered methylation patterns, with changes occurring during competition having an apparent delayed effect on later expression patterns. Hormonal responses, neurotransmitter transport, and other behaviorally relevant signaling systems were enriched with epigenetically altered genes, suggesting these processes are transcriptionally poised to respond to future social instability. This study sheds light on the immediate and longer lasting genome-wide responses to competition among free-living animals.
The most prominent and immediate neurogenomic response to competition was a coordinated increase in processes related to signal transduction and metabolism. NADH metabolism plays a central role in integrating metabolic and signaling information to ensure the brain’s energy demands are met (29). Accordingly, hub genes associated with NADH metabolism overlapped with processes like cell secretion and gluconeogenesis. Notably, competition-induced increases in gluconeogenesis appear to be highly conserved (7). This pattern may reflect the brain’s dependence on glucose for energy to power synaptic activity (30). Neuronal metabolic needs likely increase during competition, particularly in the VmT because this brain region is linked with sensory inputs that are engaged during social interactions (31). However, this increase in neuronal metabolism was no longer present 2 d after competition had ended. Instead, it was replaced by a decrease in neuronal self-maintenance processes in the HYPO, including down-regulation of genes involved in cognition, neuron generation, and synaptic plasticity identified in the brown module. One of the putative transcription factor toolkit genes for social behavior (7), NR4A3, was identified as a brown module gene and showed competition-induced methylation during peak competition. RELN, a brown module hub gene, also displayed competition-induced changes in methylation, and epigenetic changes in this gene have been linked to psychiatric disorders (32). This suggests a female’s ability to meet the energetic demands of competition may trade off with long-term consequences for self-maintenance processes.
Females at experimental sites also experienced an increase in amine signaling in the HYPO, including catecholamine and dopamine, which persisted and became stronger days after competition had ended. An increase in catecholaminergic signaling is associated with aggression in birds (33) and amine signaling is also linked to territorial aggression in insects (5). In our analysis, competition-induced shifts in gene regulation appear to be driven by green module hub genes, including TH, DDC, and numerous catecholamine transporters. Many of these genes, like TH (the rate limiting enzyme in dopamine biosynthesis) and dopamine transporters, are activated via the winner effect, in which animals who have won territorial disputes behave more aggressively and are more likely to win in future conflicts (34, 35). Dopamine is also widely known for its role in motivation. Dopaminergic fibers are found throughout the social decision-making circuit, which contains the HYPO and VmT, and acts to interpret social stimuli and produce behavioral output, like aggression toward an intruder (36). Furthermore, as territory holders, the females in our study may have had a more robust response in this system, because residency status can influence motivation and fight outcomes (37, 38). Other HYPO green module hub genes included ACLY, a gene with myriad effects on signaling and energy metabolism, that also regulates acetylcholine production. Acetylcholine acts to coordinate neuron excitability in response to environmental stimuli (39) and influences social stress resilience (40). VIP and its receptor were also green module hubs and they can promote territorial aggression in songbirds (41). Both ACLY and VIPR1 displayed competition-induced changes in methylation. In short, numerous aggression-related genes showed up-regulation days after competition had ended, and their expression was related to the degree of aggression observed 2 d prior, consistent with the biological embedding of the competitive experience.
We found thousands of genes with differential methylation in the HYPO on the day of experimental competition (day 0) and 2 d later. Competition-induced methylation patterns have previously been detected on the timescale of hours (18), similar to what we found on day 0, but longer time scales (days) have yet to be explored. Interestingly, methylation patterns detected on day 0, as opposed to day 2, were more strongly negatively correlated with expression levels on day 2, such that genes with higher methylation on day 0 had lower expression on day 2, and vice versa. This suggests that competition-induced changes in methylation may have a delayed impact on future gene expression. DNA methylation can influence other facets of epigenetic regulation (e.g., histone modifications) (42) that could manifest as a delayed effect. Accordingly, subtle changes in gene expression occurring at the functional level and within module hub genes detected on day 2 were associated with DMRs on day 0. Hormonal responses and neurotransmitter transport were processes enriched in both differentially methylated genes on day 0 and concordantly up-regulated genes on day 2. Genes with lower levels of methylation on day 0 and higher expression on day 2 driving these patterns included CRHBP, related to corticosterone secretion, which can alter social approach vs. avoidance (43), and MAOB, which is strongly linked to stress responsiveness and aggression (44). Other glucocorticoid-related genes (NR3C1 and HSD11B2) also had lower levels of methylation during competition. This suggests that competition-induced changes in methylation may generate subtle, but enduring, changes in the expression of socially relevant genes and processes.
Although functional interpretations of DMRs are challenging in cases without subsequent changes in expression, these DMRs may nevertheless play a future role in social responsiveness. For example, canonical Wnt signaling, an evolutionarily conserved response to simulated territorial intrusions (7), was enriched with genes containing DMRs on day 2, suggesting this process may be poised for future transcriptional regulation (45, 46). Response to hormones was also enriched with key steroidogenic genes on day 2 that did not show changes in expression, like GNRHR and HSD3B1. We did not observe changes in circulating testosterone, agreeing with recent syntheses on social regulation of testosterone (2–4), but these epigenetic changes may nevertheless transcriptionally alter steroid responses for future challenges. Overall, our data show how natural competitive interactions influence the neurogenome, shedding light on the effects of competition when territorial aggression determines breeding opportunities for animals.
Materials and Methods
Study Population.
We monitored breeding tree swallows across eight sites near Bloomington, IN (39°9 N, 86°31 W) between March and April 2019. As part of continuing population monitoring, birds were captured in their nest boxes and given unique numbered US Geological Survey aluminum bands on one leg and a plastic color band on the other leg. Banding stopped 5 d prior to the experiment. Each plastic leg band contained a personal integrated transponder (PIT) tag (2.3-mm tag with an EM4102 transponder from Eccel Technologies) that transmits a unique identifying number when activated by an RFID reader/antenna located on nest boxes (SI Appendix, section A). This study was approved by the Bloomington Institutional Animal Care and Use Committee under protocol 18-004, as well as all relevant federal, state, and local permits.
Experimental Manipulation.
Our goal was to temporarily enhance competition by reducing nest box availability after territory establishment (i.e., once nest ownership could be confidently confirmed; SI Appendix, section A). In the evening, after birds had departed to roosting sites, all unoccupied boxes and 50% of occupied boxes were removed from experimental sites (Fig. 1). The remaining “focal” boxes were occupied by stable, known residents (i.e., consistent PIT tag readings, early signs of nest construction, and stable pair sightings). New boxes were erected to accommodate only half of the displaced females and were evenly dispersed ∼20 m between two focal boxes, but otherwise 100 m away from other focal or newly placed boxes. This placement was intended to facilitate interactions between focal females and conspecifics (displaced and/or floater females) competing for the nearby new boxes.
In the morning when females returned from roosting (day 0), we performed behavioral observations for 2 h to quantify rates of physical aggression, like grappling fights, in the vicinity of focal females (SI Appendix, section B and Movie S1). After this time, we collected roughly half of the focal females (those whose boxes we did not remove) at each experimental site. We also collected females from control sites, in which no box removal had occurred, on day 0. Control sites were >300 m from experimental sites, which is beyond the tree swallow home range (24). In the afternoon, after collections were complete, all boxes were returned to their initial (day −1) positions. Then, 48 h after the experimental manipulation (day 2), we collected the remaining focal females that had previously experienced competition, as well as a second set of date-matched controls at undisturbed control sites. Females collected on day 2 were all box owners on day 0, confirmed with RFID devices and observations (SI Appendix, Fig. S2). Collections occurred between 0800 and 1330 h (average 1052 h ± 22 min) from 16 to 18 April.
Sampled females were captured in their nestboxes (via nestbox trap) and immediately killed with an overdose of isoflurane, followed by decapitation. Trunk blood was collected and kept on ice before centrifugation to extract plasma and storage at −20 °C. Tissues, including whole brain, were immediately dissected and frozen on powdered dry ice in the field and transferred to −80 °C in the laboratory. No females had developed ovarian follicles, suggesting they were not yet reproductively active. Furthermore, females across all sites had completed ≤50% of nest construction at the time of the experiment and the earliest first egg date occurred ∼2 wk following the experiment, indicating no potential confound with reproduction per se (SI Appendix, Fig. S10). In total, we collected 26 females across day 0 (experimental, n = 11; control, n = 6) and day 2 (experimental, n = 4; control, n = 5). For downstream analyses, we excluded four young females in their first breeding year [determined by plumage color (47)] and five floater females captured in focal or new boxes that were not the owner, three of which were also young females (n = 6 total excluded). Our final sample size was 20 females collected across day 0 (experimental, n = 6; control, n = 5) and day 2 (experimental, n = 4; control, n = 5).
Plasma Testosterone.
Hormones were extracted from plasma using a triple ether extraction. We quantified testosterone levels using a competitive-binding commercial enzyme-linked immunosorbent assay (ADI 900-176; Enzo) as in ref. 25. Concentration was calculated by comparing a sample’s absorbance to the absorbance of the assay’s nine-point standard curve (Gen5 software, Biotek EPOCH plate reader). Intraplate variation was 3.75%.
Tissue Dissection.
Brains were macrodissected into functional regions, following Bentz et al. (48). After removing the optic tecta, optic chiasm, and hindbrain, we isolated the HYPO to the depth of the anterior commissure (including the preoptic area and ventromedial HYPO). We also collected the VmT by removing ∼1 mm of the ventromedial portion of the caudal telencephalon, based on the position of nucleus taeniae in songbirds (49). We focused on these tissues because they include nuclei that are involved in the regulation of social behavior and have been shown to respond to social challenges (6, 22).
RNA Isolation and RNA-Seq.
Total RNA was extracted using TRIzol (Invitrogen) and resuspended in water. Quality and quantity of RNA were analyzed with a TapeStation 2200 (Agilent Technologies). Total RNA was used for complementary DNA library construction using a TruSeq Stranded messenger RNA kit (Illumina). Sequencing was performed using an Illumina NextSEq. 500 Kit with a 75-bp sequencing module to generate paired-end reads. The resulting reads were cleaned using Trimmomatic v.0.36 (50) and mapped to the reference tree swallow transcriptome (51) using Bowtie2 v.2.3.4.3 (52). Results were filtered to only include reads mapped in proper pairs, sorted, and indexed using Samtools v.1.9 (53). Approximately 27.8 million read pairs per sample were mapped to the entire transcriptome, accounting for ∼90% (range 88 to 91%) of the total trimmed read pairs. When mapped against a high-quality protein-only subset of the transcriptome, approximately 8.8 million read pairs per sample were mapped with high confidence, which account for ∼29% of the total trimmed read pairs (SI Appendix, Table S5). By design, the high-quality subset maps directly to a specific gene. Differential gene expression and normalized values were determined using DESeq2 v.1.16.1 (54) in R/Bioconductor. Transcripts with fewer than 10 total reads across all samples were filtered out. P values were corrected using Benjamini–Hochberg corrections, and FDR ≤0.10 were considered differentially expressed (default value in DESeq2). Due to the nature of biological variation in the wild and our relatively small sample sizes, we provide SE for our log2 fold change calculations (Dataset S1), as well as gene-level plots for the top DEGs (SI Appendix, Fig. S4). RNA-seq data have been deposited into NCBI’s Gene Expression Omnibus (GSE155864).
Construction of Weighted Gene Coexpression Networks.
WGCNA was performed within a tissue (n = 20 samples per tissue) (28). Using the normalized counts from DESeq2, we filtered out genes with <10 norm counts in 90% of the samples and only used the 50% most variable genes (n = 5,353 genes in HYPO; n = 5,413 genes in VmT). We generated a signed hybrid network by selecting a soft threshold power (β) = 7 for the HYPO and β = 6 for the VmT in accordance with scale-free topology (SI Appendix, Fig. S11). We used a biweight midcorrelation (bicor) function and modules were calculated using a minimum module size of 30. A threshold of 0.25 was used to merge modules in Dynamic Tree Cut (SI Appendix, Fig. S12).
Finding Modules of Interest and Functional Annotation.
Expression levels for each module were summarized by the first principal component (module eigengene), which we used to test for an association between modules and traits of interest (i.e., treatment, day, testosterone level, and behavior) using the bicor function. We additionally included field site and time of capture to account for these confounding variables. For trait-associated modules, genes with an absolute GS value >0.2 and P value <0.05 were assessed for enrichment of biological process GO terms. We further define intramodular hub genes as those with a high traits-based GS (absolute value >0.2) and high MM (>0.6) with a threshold of P < 0.05 for both (55). The top hub genes were visualized in Cytoscape (56). We also used a Fisher’s exact test to examine whether putative toolkit genes involved in an evolutionarily conserved response to simulated territorial intrusions (7) were more likely to be hub genes in modules associated with the experimental treatment than nontoolkit genes.
DNA Isolation and MethylCap-Seq.
Genomic DNA was isolated from each HYPO using a Quick-DNA Microprep Plus Kit (Zymo Research). Integrity was checked with a TapeStation 2200 (Agilent Technologies) and 300 ng of DNA was sheared to an average size of 200 bp using a Covaris E220 sonicator (Covaris). DNA Methyl capture was performed using a MethylCap kit (Diagenode). A HYPO sample not included in downstream analyses was used as a negative control (the methyl binding domain protein was excluded during Methyl capture) and a fully methylated positive control (DNA treated with SssI CpG methyltransferase; New England Biolabs). Libraries were generated using the NextFlex MethylSeq Kit (Bioo Scientific) and paired-end sequencing was performed on an Illumina NextSEq. 500 platform with a 75-cycle sequencing module. The resulting reads were cleaned using Trimmomatic v.0.36 (50) and aligned to the reference tree swallow genome (57) using bwa v.0.7.17-r1188 (58). Potential PCR duplicates were marked using Picard v.2.20.3 (http://broadinstitute.github.io/picard/) and deduplicated, concordantly mapped read pair alignments were extracted using Samtools v.1.9 (53). On average, 13 million deduplicated inserts were obtained from the 17.6 million concordantly mapped read pairs per sample (∼68% of the total trimmed reads mapped) (SI Appendix, Table S6). For a given sample, we present normalized coverage as the average deduplicated insert coverage for each bin, calculated and normalized based on the total number of deduplicated inserts obtained for that sample.
Identification of DMRs.
We identified DMRs between control and experimental females on day 0 and day 2. Analyses were conducted for nonoverlapping local bins (width 250 bp) identified across the length of the genome. Differences in methylation for a given local bin were estimated as a difference in z-scores (the number of SDs by which the bin coverage varied from the larger 25-kb overlapping region’s mean coverage); the level of significance was computed as a difference in z-scores, defined by the z-ratio (SI Appendix, section C) (59). Bins with Benjamini–Hochberg-adjusted P ≤ 0.05 were identified as potential DMRs. We further filtered DMRs with low confidence (<3 CpG sites) and low coverage (<1 norm count in two-thirds of the samples) and additionally applied an absolute log2 fold change cut off of 0.5. Functional annotation for the filtered DMRs based on their positional overlap with gene bodies or 2-kb upstream regions was determined using FEATnotator v.1.1.3 (60). The CpG and GC content were calculated as the percent of CpG and GC respectively in each of 100 bp sliding windows. All MethylCap-seq data have been deposited into NCBI’s Gene Expression Omnibus (GSE155864).
Gene Ontology Enrichment Analysis.
We performed a GO enrichment analysis to identify overrepresented biological processes using PANTHER (61) with a Fisher’s exact test and a cutoff of FDR ≤0.05. The lists of GO terms were further summarized with REVIGO (http://revigo.irb.hr/), which clusters GO terms based on semantic similarity (similarity threshold = 0.7).
Gene Set Enrichment Analysis.
We performed separate GSEAs (62) to explore treatment effects on gene expression within each time point (day 0 or day 2) in the HYPO using the fgsea package in R (63). Gene lists included all genes identified in the expression dataset ranked by −log10P*sign(log2FC) for each comparison. Gene sets included all GO biological processes found enriched in genes with DMRs on day 0 and/or day 2 (SI Appendix, Table S3). Significance was assessed with 10,000 permutations, with a cutoff set as a Benjamini–Hochberg-adjusted P ≤ 0.05.
Supplementary Material
Acknowledgments
We thank S. E. Lipshutz, M. J. Woodruff, K. R. Content, T. A. Empson, K. R. Stansberry, S. Skrabalak, D. Galantsev, K. Brown, S. T. Myers, E. K. Dossey Curole, R. Walker, and K. Bear for facilitating fieldwork and three anonymous referees for helpful comments on earlier versions of this manuscript. We also acknowledge the Indiana University Research and Teaching Preserve, the Indiana Department of Natural Resources, and Cikana State Fish Hatchery for access to field sites and the Indiana University Center for Genomics and Bioinformatics for laboratory facilities. This work was funded by NSF Grant IOS-1656109. A.B.B., E.M.G., S.E.W., K.P.N., and K.A.R. were supported by NIH Grant T32HD049336. E.M.G. was also supported by an NSF Graduate Research Fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016154118/-/DCSupplemental.
Data Availability
RNA-sequencing and MethylCap-sequencing datasets can be obtained from the Gene Expression Omnibus database (GEO accession no. GSE155864). All study data are included in the article and/or supporting information.
References
- 1.Wingfield J. C., Hegner R. E., Dufty A. M. Jr, Ball G. F., The “challenge hypothesis”: Theoretical implications for patterns of testosterone secretion, mating systems, and breeding strategies. Am. Nat. 136, 829–846 (1990). [Google Scholar]
- 2.Goymann W., Moore I. T., Oliveira R. F., Challenge hypothesis 2.0: A fresh look at an established idea. Bioscience 69, 432–442 (2019). [Google Scholar]
- 3.Moore I. T., Hernandez J., Goymann W., Who rises to the challenge? Testing the challenge hypothesis in fish, amphibians, reptiles, and mammals. Horm. Behav. 123, 104537 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Rosvall K. A., Bentz A. B., George E. M., How research on female vertebrates contributes to an expanded challenge hypothesis. Horm. Behav. 123, 104565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alaux C., et al., Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl. Acad. Sci. U.S.A. 106, 15400–15405 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanogo Y. O., Band M., Blatti C., Sinha S., Bell A. M., Transcriptional regulation of brain gene expression in response to a territorial intrusion. Proc. Biol. Sci. 279, 4929–4938 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rittschof C. C., et al., Neuromolecular responses to social challenge: Common mechanisms across mouse, stickleback fish, and honey bee. Proc. Natl. Acad. Sci. U.S.A. 111, 17929–17934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukai M., et al., Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. PLoS One 4, e8182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goymann W., Social modulation of androgens in male birds. Gen. Comp. Endocrinol. 163, 149–157 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Oliveira R. F., et al., Assessment of fight outcome is needed to activate socially driven transcriptional changes in the zebrafish brain. Proc. Natl. Acad. Sci. U.S.A. 113, E654–E661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calisi R. M., Bentley G. E., Lab and field experiments: Are they the same animal? Horm. Behav. 56, 1–10 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Hsu Y., Earley R. L., Wolf L. L., Modulation of aggressive behaviour by fighting experience: Mechanisms and contest outcomes. Biol. Rev. Camb. Philos. Soc. 81, 33–74 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Bukhari S. A., et al., Temporal dynamics of neurogenomic plasticity in response to social interactions in male threespined sticklebacks. PLoS Genet. 13, e1006840 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shpigler H. Y., et al., Behavioral, transcriptomic and epigenetic responses to social challenge in honey bees. Genes Brain Behav. 16, 579–591 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Aristizabal M. J., et al., Biological embedding of experience: A primer on epigenetics. Proc. Natl. Acad. Sci. U.S.A., 1820838116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Day J. J., Sweatt J. D., DNA methylation and memory formation. Nat. Neurosci. 13, 1319–1323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott E., Ezra-Nevo G., Regev L., Neufeld-Cohen A., Chen A., Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat. Neurosci. 13, 1351–1353 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Herb B. R., Shook M. S., Fields C. J., Robinson G. E., Defense against territorial intrusion is associated with DNA methylation changes in the honey bee brain. BMC Genomics 19, 216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winkler D. W., et al., “Tree swallow (Tachycineta bicolor), version 2.0” in The Birds of North America, Poole A. F., Ed. (Cornell Lab of Ornithology, Ithaca, NY, 2011). [Google Scholar]
- 20.Rosvall K. A., Sexual selection on aggressiveness in females: Evidence from an experimental test with tree swallows. Anim. Behav. 75, 1603–1610 (2008). [Google Scholar]
- 21.Leffelaar D., Robertson R. J., Nest usurpation and female competition for breeding opportunities by tree swallows. Wilson Bull. 97, 221–224 (1985). [Google Scholar]
- 22.Goodson J. L., The vertebrate social behavior network: Evolutionary themes and variations. Horm. Behav. 48, 11–22 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muldal A., Gibbs H. L., Robertson R. J., Preferred nest spacing of an obligate cavity-nesting bird, the tree swallow. Condor 87, 356–363 (1985). [Google Scholar]
- 24.McCarty J. P., Winkler D. W., Foraging ecology and diet selectivity of tree swallows feeding nestlings. Condor 101, 246–254 (1999). [Google Scholar]
- 25.George E. M., Rosvall K. A., Testosterone production and social environment vary with breeding stage in a competitive female songbird. Horm. Behav. 103, 28–35 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Cullinan W. E., Herman J. P., Battaglia D. F., Akil H., Watson S. J., Pattern and time course of immediate early gene expression in rat brain following acute stress. Neuroscience 64, 477–505 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Sinha S., et al., Behavior-related gene regulatory networks: A new level of organization in the brain. Proc. Natl. Acad. Sci. U.S.A. 117, 23270–23279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler U., Hirrlinger J., Crosstalk of signaling and metabolism mediated by the NAD+/NADH redox state in brain cells. Neurochem. Res. 40, 2394–2401 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Mergenthaler P., Lindauer U., Dienel G. A., Meisel A., Sugar for the brain: The role of glucose in physiological and pathological brain function. Trends Neurosci. 36, 587–597 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardinal R. N., Parkinson J. A., Hall J., Everitt B. J., Emotion and motivation: The role of the amygdala, ventral striatum, and prefrontal cortex. Neurosci. Biobehav. Rev. 26, 321–352 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Tamura Y., Kunugi H., Ohashi J., Hohjoh H., Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol. Psychiatry 12, 519, 593–600 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Maney D. L., Goodson J. L., “Neurogenomic mechanisms of aggression in songbirds” in Advances in Genetics, Huber R., Bannasch D. L., Brennan P., Eds. (Academic Press, 2011), pp. 83–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filipenko M. L., Alekseyenko O. V., Beilina A. G., Kamynina T. P., Kudryavtseva N. N., Increase of tyrosine hydroxylase and dopamine transporter mRNA levels in ventral tegmental area of male mice under influence of repeated aggression experience. Brain Res. Mol. Brain Res. 96, 77–81 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Schwartzer J. J., Ricci L. A., Melloni R. H. Jr, Prior fighting experience increases aggression in Syrian hamsters: Implications for a role of dopamine in the winner effect. Aggress. Behav. 39, 290–300 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Kemp D. J., Wiklund C., Residency effects in animal contests. Proc. Biol. Sci. 271, 1707–1711 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuxjager M. J., Mast G., Becker E. A., Marler C. A., The ‘home advantage’ is necessary for a full winner effect and changes in post-encounter testosterone. Horm. Behav. 56, 214–219 (2009). [DOI] [PubMed] [Google Scholar]
- 38.O’Connell L. A., Hofmann H. A., The vertebrate mesolimbic reward system and social behavior network: A comparative synthesis. J. Comp. Neurol. 519, 3599–3639 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Picciotto M. R., Higley M. J., Mineur Y. S., Acetylcholine as a neuromodulator: Cholinergic signaling shapes nervous system function and behavior. Neuron 76, 116–129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mineur Y. S., et al., Cholinergic signaling in the hippocampus regulates social stress resilience and anxiety- and depression-like behavior. Proc. Natl. Acad. Sci. U.S.A. 110, 3573–3578 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodson J. L., Kelly A. M., Kingsbury M. A., Thompson R. R., An aggression-specific cell type in the anterior hypothalamus of finches. Proc. Natl. Acad. Sci. U.S.A. 109, 13847–13852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose N. R., Klose R. J., Understanding the relationship between DNA methylation and histone lysine methylation. Biochim. Biophys. Acta 1839, 1362–1372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly A. M., Vitousek M. N., Dynamic modulation of sociality and aggression: An examination of plasticity within endocrine and neuroendocrine systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shih J. C., Chen K., Ridd M. J., Monoamine oxidase: From genes to behavior. Annu. Rev. Neurosci. 22, 197–217 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam L. L., et al., Factors underlying variable DNA methylation in a human community cohort. Proc. Natl. Acad. Sci. U.S.A. 109 (suppl. 2), 17253–17260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Provençal N., et al., Glucocorticoid exposure during hippocampal neurogenesis primes future stress response by inducing changes in DNA methylation. Proc. Natl. Acad. Sci. U.S.A., 201820842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hussell D. J., Age and plumage color in female tree swallows. J. Field Ornithol. 54, 312–318 (1983). [Google Scholar]
- 48.Bentz A. B., Rusch D. B., Buechlein A., Rosvall K. A., The neurogenomic transition from territory establishment to parenting in a territorial female songbird. BMC Genomics 20, 819 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soma K. K., Bindra R. K., Gee J., Wingfield J. C., Schlinger B. A., Androgen-metabolizing enzymes show region-specific changes across the breeding season in the brain of a wild songbird. J. Neurobiol. 41, 176–188 (1999). [PubMed] [Google Scholar]
- 50.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bentz A. B., Thomas G. W. C., Rusch D. B., Rosvall K. A., Tissue-specific expression profiles and positive selection analysis in the tree swallow (Tachycineta bicolor) using a de novo transcriptome assembly. Sci. Rep. 9, 15849 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horvath S., Dong J., Geometric interpretation of gene coexpression network analysis. PLOS Comput. Biol. 4, e1000117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shannon P., et al., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taff C. C., Campagna L., Vitousek M. N., Genome-wide variation in DNA methylation is associated with stress resilience and plumage brightness in a wild bird. Mol. Ecol. 28, 3722–3737 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheadle C., Vawter M. P., Freed W. J., Becker K. G., Analysis of microarray data using Z score transformation. J. Mol. Diagn. 5, 73–81 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Podicheti R., Mockaitis K., FEATnotator: A tool for integrated annotation of sequence features and variation, facilitating interpretation in genomics experiments. Methods 79-80, 11–17 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sergushichev A. A., An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv [Preprint] (2016). https://www.biorxiv.org/content/10.1101/060012v1 (Accessed 15 August 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing and MethylCap-sequencing datasets can be obtained from the Gene Expression Omnibus database (GEO accession no. GSE155864). All study data are included in the article and/or supporting information.