Significance
Polyploidy can stimulate genetic and epigenetic changes that enhance the potential for plant adaptation and fitness under extreme environments, but the molecular basis for this is poorly understood. Here we show that tetraploid rice is more tolerant to salinity than diploid rice. Tetraploidy induces DNA hypomethylation which potentiates genomic loci coexistent with stress-responsive genes including those in jasmonate biosynthesis and signaling pathways for rapid and robust responses under stress. After salt stress, elevated expression of salt-responsive genes can induce hypermethylation and suppress adjacent TEs. This feedback regulation between polyploidy-induced DNA hypomethylation in rapid and strong stress response and stress-induced hypermethylation to repress TEs and/or TE-associated genes may provide evolutionary advantages for selection to enhance adaptation in polyploid plants and crops.
Keywords: polyploidy, gene expression, salt tolerance, DNA methylation, transposons
Abstract
Polyploidy is a prominent feature for genome evolution in many animals and all flowering plants. Plant polyploids often show enhanced fitness in diverse and extreme environments, but the molecular basis for this remains elusive. Soil salinity presents challenges for many plants including agricultural crops. Here we report that salt tolerance is enhanced in tetraploid rice through lower sodium uptake and correlates with epigenetic regulation of jasmonic acid (JA)–related genes. Polyploidy induces DNA hypomethylation and potentiates genomic loci coexistent with many stress-responsive genes, which are generally associated with proximal transposable elements (TEs). Under salt stress, the stress-responsive genes including those in the JA pathway are more rapidly induced and expressed at higher levels in tetraploid than in diploid rice, which is concurrent with increased jasmonoyl isoleucine (JA-Ile) content and JA signaling to confer stress tolerance. After stress, elevated expression of stress-responsive genes in tetraploid rice can induce hypermethylation and suppression of the TEs adjacent to stress-responsive genes. These induced responses are reproducible in a recurring round of salt stress and shared between two japonica tetraploid rice lines. The data collectively suggest a feedback relationship between polyploidy-induced hypomethylation in rapid and strong stress response and stress-induced hypermethylation to repress proximal TEs and/or TE-associated stress-responsive genes. This feedback regulation may provide a molecular basis for selection to enhance adaptation of polyploid plants and crops during evolution and domestication.
Polyploidy or whole-genome duplication (WGD) is a prominent process of evolution and speciation in some animals (1) and all flowering plants (2, 3). The common occurrence of polyploidy suggests an advantage for polyploid plants to enhance adaptation to diverse and extreme environments (4), consistent with the rapid expression divergence between duplicate stress-responsive genes after WGD in Arabidopsis (5, 6). In newly resynthesized Arabidopsis allotetraploids, expression divergence occurs in 5% or more genes, including those in photosynthesis, phytohormone, cell defense, and stress-responsive pathways (7); these gene expression changes lead to circadian-mediated growth vigor through increased energy and starch metabolic activities (8) and/or reduced energy costs for the defense response under nonstress conditions (9). From an ecological perspective, polyploid plants are more widely distributed under extreme environments including high latitudes and altitudes (2, 3, 10). In strawberries (Fragaria L.), allopolyploids exhibit higher fitness than diploid wild taxa by adapting to heterogenous environments (11). At the physiological level, both natural and resynthesized Arabidopsis autopolyploids have enhanced salinity tolerance by elevating potassium and reducing sodium uptake (12), while increased drought tolerance is associated with gene expression changes in abscisic acid (ABA) signaling and reactive oxygen species homeostasis (13).
Soil salinity is a severe abiotic stress, affecting more than 6% of the global total land area (14). Excessive levels of salt in soil reduce growth and yield of many crops including rice (15), one of the most important crops in the world (16). Jasmonic acid (JA) is a phytohormone and plays important roles in stress responses in plants (17). Many genes involved in JA biosynthesis and signaling pathways are activated under cold and salt conditions (18, 19). Exogenous application of JA can reduce the uptake of sodium by rice under salinity stress conditions (20).
Stress response in plants involves priming, which is an adaptative strategy to increase survival capacity in response to rapidly changing environments. Upon stress, plants can remember many changes at physiological, metabolic, transcriptional, and epigenetic levels. Subsequent stress conditions can allow plants to effectively augment responses to increase stress tolerance (21). Priming can occur at the chromatin level poised for transcription. For example, in Arabidopsis under dehydration stress, JA promotes accumulation of H3K4me3 histone marks and potentiates RD29B, an ABA-dependent dehydration gene, for transcription (22). DNA methylation is also involved in priming. In rdd (ros1/dml2/dml3) mutants, proximal transposable elements (TEs) of stress-related genes are hypermethylated to compromise their transcription against Fusarium oxysporum, resulting in susceptibility of plants to pathogen (23).
In plants, DNA methylation occurs in CG, CHG, and CHH (H = A, T, or C) contexts through distinct pathways. CG, CHG, and CHH methylation in Arabidopsis thaliana are catalyzed by METHYLTRANSFERASE 1 (MET1), CHROMOMETHYLASE 3 (CMT3), and CMT2, respectively, whereas de novo CHH methylation is mediated by RNA-directed DNA methylation (RdDM) pathway (24). DNA methylation can be reprogramed in response to biotic and abiotic stresses (25, 26). Polyploidization induces epigenetic changes (4), including DNA methylation that can be stably inherited as epialleles in allotetraploid cotton (27) and reversible during genome separation and merger in allohexaploid wheat (28); DNA methylation changes are also observed in autotetraploid rice (29). However, the relationship between DNA methylation changes and stress responses in polyploid plants is unknown. In this study, we analyzed ion contents, transcriptomes, and single-base resolution methylomes in diploid and autotetraploid rice under control and salinity conditions. We found a higher potassium/sodium ratio in tetraploid rice compared to diploid rice under salinity stress and identified extensive transcriptome changes in response to salt stress, including activation of JA-related genes. Tetraploidy reprogrammed CHH methylation of genomic loci coexistent with stress-responsive genes adjacent to TEs, and hypomethylation in tetraploid rice primed rapid and strong activation of these genes in JA biosynthesis and signaling pathways under salt stress. As a result, tetraploid rice has elevated JA-Ile content and enhanced JA signaling transmission. Thus, polyploidy-induced DNA methylation changes prime expression of JA-related genes, while stress-induced gene expression in turn promotes hypermethylation and suppresses proximal TEs and/or TE-associated stress-responsive genes. This feedback regulation may enhance stress tolerance and environmental adaptation in polyploid plants and crops.
Results
Tetraploid Rice Shows Enhanced Growth under Salt Stress.
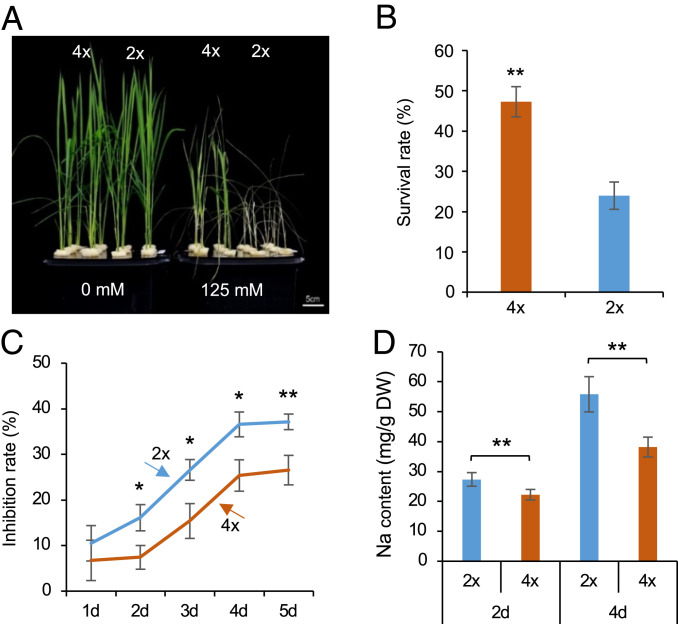
We investigated salt tolerance response in two sets of diploid (2×) and isogenic tetraploid (4×) rice (Oryza sativa L. ssp. japonica) (Materials and Methods). One set of O. sativa L. ssp. japonica cv. 02428 cytotype (30) was used for most studies, and another set of O. sativa L. ssp. japonica cv. Nipponbare cytotype (SI Appendix, Fig. S1) was used to validate survival rate, gene expression, and DNA methylation changes. Seedlings (02428) at the trefoil stage were cultured under a series of salt treatments for 6 d and then returned to nutrient solution for 5 d (Fig. 1A and SI Appendix, Fig. S2A). Based on survival rates, we selected 125 mM NaCl as an optimal treatment for further study, because the seedlings displayed severe necrosis at 150 mM or higher concentration and relatively mild effects in 100 mM NaCl (SI Appendix, Fig. S2A). Under 125 mM saline condition, tetraploid rice exhibited better growth (Fig. 1A) and higher survival rates (Fig. 1B) than diploid rice. Both fresh and dry weights of shoots or roots were reduced to a greater extent in diploid than in tetraploid rice (Fig. 1C and SI Appendix, Fig. S2 B–D). Physiologically, iron homeostasis and water uptake are maintained by low Na+ uptake and a high K+/Na+ ratio under saline conditions (14, 15). Consistent with the above notion, tetraploid rice had a lower Na+ uptake (Fig. 1D and SI Appendix, Fig. S2E) and a higher K+/Na+ ratio (SI Appendix, Fig. S2 H and I) than diploid rice in both shoots and roots, which persisted for 4 d tested. However, the total Na+ and K+ content was similar between tetraploid and diploid rice without salt stress (SI Appendix, Fig. S2 J–M), and the potassium content remained unchanged between diploid and tetraploid rice under the salt condition (SI Appendix, Fig. S2 F and G). This result was different from previous findings in Arabidopsis, in which tetraploids had a higher potassium uptake than diploids under salt stress (12), which may suggest different physiological responses to salt stress between monocots and eudicots.
Fig. 1.
Enhanced salinity tolerance in 02428 tetraploid rice. (A) Morphological changes in diploid (2×) and tetraploid (4×) rice in response to saline treatments (0 and 125 mM NaCl) for 6 d followed by recovery in water for 5 d. (Scale bar = 5 cm.) (B) Survival rates of diploid and tetraploid rice (n = 6 biological replicates, each replicate with 32 plants). (C) Inhibition rates of diploid and tetraploid rice after salt stress. Seedlings at trefoil stage were treated with 125 mM NaCl for 1 to 5 d. Dry weight of roots was used to calculate inhibition rates (n = 18 biological replicates, each replicate with 2 plants). (D) Sodium content in roots under the saline condition for 2 and 4 d (n = 12 biological replicates, each replicate with 2 plants). Single and double asterisks indicate statistical significance of P < 0.05 and P < 0.01, respectively (Student’s t test).
Tetraploid Rice Shows Specific Sets of Genes with Higher Expression Levels under Salt Stress.
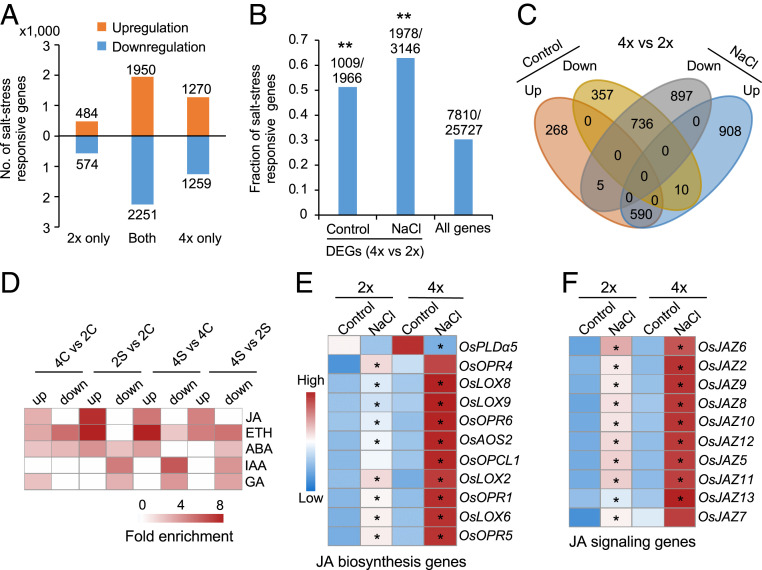
Roots serve as an interface for response to biotic and abiotic stresses (31), and the cytotype of roots determines salinity tolerance in Arabidopsis tetraploids (12). To determine the molecular basis for enhanced salt tolerance in tetraploid rice, we performed transcriptome analysis in roots of tetraploid and diploid rice under control and saline (125 mM for 24 h) conditions with three biological replicates (Dataset S1A). We identified 5,281 and 6,752 differentially expressed genes (DEGs) before and after salt stress in diploid and tetraploid rice, respectively, and these DEGs were called salt stress–responsive genes (Fig. 2A). Among them, 2,448 were up-regulated and 2,833 were down-regulated in diploid rice, while 3,228 were up-regulated and 3,524 were down-regulated in tetraploid rice (Fig. 2A); a large portion of the up-regulated (52%) and down-regulated (55%) genes were shared between the diploid and tetraploid rice. Interestingly, salt stress–responsive genes were significantly enriched in DEGs between tetraploid and diploid rice under control (51.3%) and salt (62.9%) conditions (Fig. 2B and Dataset S1B) (P < 0.01, hypergeometric test). Most (>66%) genes that were up-regulated (590/863) or down-regulated (736/1,103) between tetraploid and diploid rice in the control remained up-regulated or down-regulated after salt stress (Fig. 2C). These data suggest a conserved role for stress-induced gene expression in both diploid and tetraploid rice.
Fig. 2.
Polyploidy primes activation of JA-related genes under salt stress in 02428. (A) Number of DEGs in diploid and tetraploid rice under salt stress, which were divided into up- or down-regulated DEGs in diploid (2×) only, tetraploid (4×) only, or shared (both). (B) Ploidy-dependent genes are associated with salt stress–responsive genes. Y-axis shows fraction of salt responsive genes, with the numbers above columns indicating salt responsive genes over DEGs between tetraploid (4×) and diploid (2×) rice in the control or salt (NaCl) condition or all expressed genes. Double asterisks indicate the significance level of P < 0.01 (hypergeometric test). (C) Venn diagram showing overlap between ploidy-dependent genes without (control) or with (NaCl) salt stress, which are concerted up- or down-regulated in response to salt treatment. (D) Enrichment of phytohormone-related genes in ploidy-dependent and salt responsive genes in tetraploid or diploid rice without salt (control, 4C versus 2C) or with salt (4S versus 2S) stress. JA: jasmonic acid; ETH: ethylene; ABA: abscisic acid; IAA: indole-3-acetic acid; GA: gibberellic acid. (E and F) Expression levels of JA biosynthesis (E) and signaling (F) related genes in diploid (2×) and tetraploid (4×) rice without (control) or with salt (NaCl) treatment. A single asterisk indicates gene expression level difference before and after salt treatment in diploid and tetraploid rice, respectively (FDR < 0.05 and log2[fold change] ≥ 1).
Salt stress also induced a specific set of up-regulated (908, 60.2%) and down-regulated (897, 54.8%) genes between tetraploid and diploid rice (Fig. 2C), suggesting ploidy-dependent stress response. Moreover, tetraploid rice had 2.2- to 2.6-fold more up-regulated (1,270) and down-regulated (1,259) genes than diploid rice (484 up-regulated and 574 down-regulated) under salt stress (Fig. 2A). Among the shared genes between diploid and tetraploid rice, changes in average gene expression levels were higher in tetraploid than in diploid rice (P < 3e−16, Wilcoxon rank sum test) (SI Appendix, Fig. S3A). These stress-induced gene expression changes were not unique to 125 mM salt condition. In a lower salt condition (50 mM NaCl), most salt-responsive genes were also specific to diploid or tetraploid rice (SI Appendix, Fig. S3B). Moreover, ∼20% and ∼36% of these low salt-induced genes in tetraploid rice remained up-regulated (SI Appendix, Fig. S3C) and down-regulated (SI Appendix, Fig. S3D), respectively, under the higher salt (125 mM) condition. These data suggest that polyploidy has resulted in robust response of gene expression changes under both low and high salt stresses.
Stronger Activation of JA-Related Genes in Tetraploid under Salt Stress.
Gene Ontology analysis of DEGs using PageMan (32) showed overrepresentation of five hormone pathways (Fig. 2D). The JA-related genes under salt stress were overrepresented among the up-regulated genes in both diploid and tetraploid rice, whereas the genes related to indole-3-acetic acid or gibberellic acid pathway were enriched in the down-regulated genes (Fig. 2D). A similar number of up- or down-regulated genes after salt stress was enriched in ABA and ethylene pathways. The up-regulated genes in tetraploid rice were also enriched for JA pathways under both control and saline conditions (Fig. 2D), suggesting that both polyploidy and salt stress can act on JA-related genes.
Consistent with the above hypothesis, many genes in JA biosynthesis (Fig. 2E), signaling (Fig. 2F), and downstream (SI Appendix, Fig. S3E) pathways were induced in tetraploid rice at significantly higher levels under salt stress and slightly high levels under nonstress conditions. Expression levels for a subset of JA-related genes including OsLOX9, OsJAZ11, and OsJAMyb were validated by qRT-PCR (SI Appendix, Fig. S3F). OsLOX9, encoding a 13-lipoxygenase, is involved in JA biosynthesis induced by herbivore (33). Overexpression of OsJAMyb, a JA-inducible MYB transcription factor gene, enhances salt tolerance (34). Moreover, many of these JA-related genes are also activated under salt stress in various rice varieties, including Nipponbare, IR64, Pokkali, and N22 (35, 36) (SI Appendix, Fig. S3 G and H) and in other plants such as Arabidopsis and wheat (18, 37). The data suggest that activation of JA-related genes may enhance salt tolerance in tetraploid rice and other plants.
Polyploidy Induces CHH Hypomethylation, and Stress Induces CHH Hypermethylation.
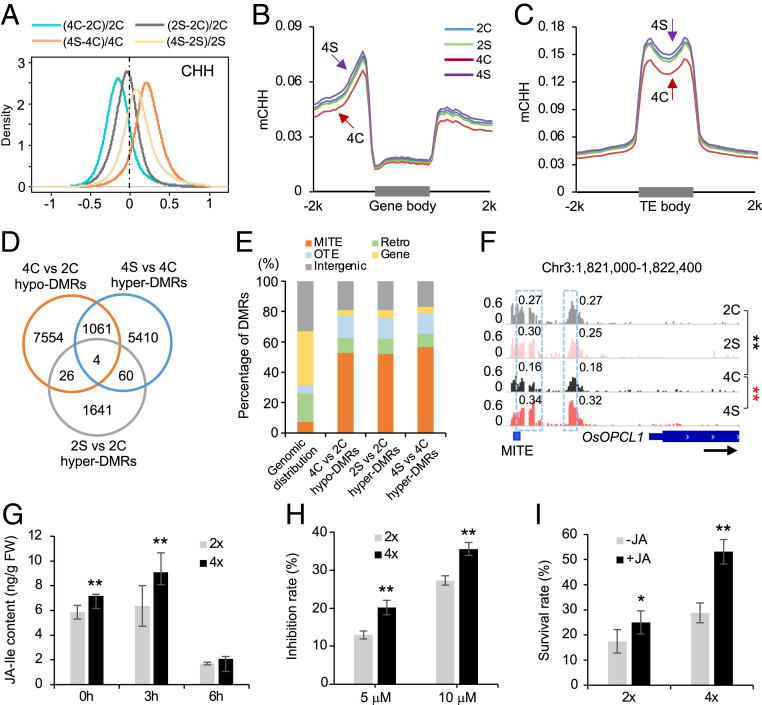
This polyploidy-dependent induction of stress-responsive genes may be associated with DNA methylation, which plays an important role in stress response (25, 26). To test this, we performed methylome sequencing (27) in roots of diploid and tetraploid rice under control and saline conditions. Pearson correlation coefficients were >0.95 between two biological replicates among all cytosine sites with an average coverage of ∼10-fold (∼85% cytosines), indicating a high reproducibility of the methylome data (Dataset S1A). To reduce technical variability, we identified 108,584,647 cytosines that are present in both cytotypes and two replicates for further analysis, accounting for ∼70% of cytosines of the rice genome. Under the control (nonstress) condition, CHH methylation levels were lower in tetraploid than in diploid (P < 2.2e−200, Wilcoxon rank sum test) (Fig. 3A and SI Appendix, Fig. S4A), which occurred in the flanking sequences of protein-coding genes (PCGs), TEs, and TE-encoded genes (TEGs) (Fig. 3 B and C and SI Appendix, Fig. S4B). However, CG and CHG methylation levels were not changed between tetraploid and diploid rice (SI Appendix, Fig. S4 A and C–J). A previous study reported that CHG and CHH methylation levels in some class II DNA transposons were slightly higher in tetraploid than in diploid rice (Oryza sativa L. ssp. indica cv. Aijiaonante) (29). This discrepancy may reflect different subspecies (japonica versus indica), tissues (root versus immature panicle), or sequencing-depths examined.
Fig. 3.
Dural roles of CHH methylation in activation of JA-related genes and repression of TEs in 02428. (A) Global CHH methylation changes after polyploidization and salt stress. Diploid rice is without (2C) or with salt (2S) treatment, and tetraploid rice is without (4C) or with salt (4S) stress. (B and C) Distributions of CHH methylation in protein-coding genes (B) and TEs (C). (D) Venn diagram showing overlap among ploidy-induced hypo-DMRs and salt-induced hyper-DMRs. (E) Distribution of stress-related hypo- or hyper-DMRs in miniature inverted transposable element (MITE), retrotransposon (Retro), other TE (OTE), gene, and intergenic regions, relative to their genomic distributions. (F) An example (genome browser view) of CHH methylation changes in the promoter of OsOPCL1. The gene and a MITE element are shown with a black arrow indicating transcriptional direction. Numbers in snapshot indicate CHH methylation levels of regions in dotted box. Double asterisks indicate the difference between 4C and 2C (black) and between 4S and 4C (red), respectively, at a statistical significance level of P < 0.01 (Fisher’s exact tests). (G) JA-Ile content in diploid and tetraploid rice under saline condition for 0, 3, and 6 h (n = 3 biological replicates). (H) Inhibition rates of diploid and tetraploid rice after treatments with 5 or 10 μM JA for 2 d (n = 30 biological replicates). (I) Survival rates of diploid and tetraploid rice after salt stress are increased by the treatment of 0 or 1 μM JA (n = 5 replicates, each replicate with 80 plants). Single and double asterisks indicate statistical significance of P < 0.05 and P < 0.01, respectively (Student’s t test).
Under salt stress, CHH methylation levels were increased from 3.3 to 3.9% in tetraploid rice (P < 2.2e−200, Wilcoxon rank sum test) but remained unchanged in diploid rice (Fig. 3A and SI Appendix, Fig. S4A), and the increased CHH methylation also occurred in the flanking regions of PCGs, TEs, and TEGs (Fig. 3 B and C and SI Appendix, Fig. S4B). Moreover, CHH methylation levels were higher in tetraploid than in diploid rice after salt treatment (P < 2.2e−86, Wilcoxon rank sum test), probably resulting from stress-induced hypermethylation (Fig. 3 A–C and SI Appendix, Fig. S4 A and B). These data suggest that salt stress promotes hypermethylation in tetraploid rice.
In rice, stress-induced gene expression can promote CHH hypermethylation in proximal TEs to silence TEs under phosphate starvation conditions (26). Consistent with this finding, we found increased TE expression (SI Appendix, Fig. S5A) and CHH methylation (SI Appendix, Fig. S5B) levels in tetraploid rice under salt stress. In diploid, TE expression (SI Appendix, Fig. S5C) and CHH methylation (SI Appendix, Fig. S5D) levels remained unchanged under salt stress. Under the control condition, TE expression (SI Appendix, Fig. S5E) and CHH methylation (SI Appendix, Fig. S5F) levels were lower in tetraploid than in diploid rice. These data suggest a feedback and feedforward relationship between TE expression and CHH methylation changes during polyploidization (diploid versus tetraploid) and salt stress (before and after). Reduced CHH methylation in tetraploid rice may prime a chromatin status poised for higher inducible levels of the stress-responsive genes with proximal TEs under salt stress, which in turn induce CHH methylation to repress stress-induced TE activities.
Stress-Related DMRs Occur in the TEs Proximal to the Genes.
To reveal how CHH methylation changes affect gene expression, we analyzed differentially methylated regions (DMRs) between tetraploid and diploid rice and between stress and control (nonstress) conditions. Among 8,645 hypo-DMRs (accounting for ∼0.23% of rice genome and ∼0.26% of conserved cytosines) identified in the control (Dataset S1C), 1,065 (12.3%, P < 2.2e−16, hypergeometric test) became hyper-DMRs in tetraploid rice under salt stress (Fig. 3D and Dataset S1C). In contrast, in diploid rice, only 30 (0.3%) of 8,645 hypo-DMRs became hyper-DMRs under salt stress. Indeed, the majority (75.6%, 6,535) of salt-induced hyper-DMRs (accounting for ∼0.18% of rice genome and ∼0.19% of conserved cytosines) in tetraploid rice was initially hypomethylated after polyploidization (SI Appendix, Fig. S4K). These data suggest that hypomethylation induced in tetraploid rice may potentiate a “priming” state for robust stress response (see below).
These hypo-DMRs in tetraploid rice and hyper-DMRs under salt stress were defined as stress-related DMRs, which were overrepresented in TEs, predominately in Class II DNA transposons such as MITEs (Fig. 3E), Harbinger, Helitron, and ToMar (SI Appendix, Fig. S6A) and a few Class I TEs such as SINE. Since MITEs were targeted by the RdDM pathway in rice (30, 38), we predicted that stress-related DMRs should overlap with RdDM targets. Using the published datasets (38), we found that most stress-related DMRs (∼72%) overlapped with DRM2-dependent targets (P < 2.2e−16, hypergeometric test) (SI Appendix, Fig. S6B), implying a role for the RdDM pathway in regulating stress-related CHH methylation. These stress-related DMRs (∼70%) occurred predominately in the flanking regions of TE-associated genes such as OsOPCL1 (Fig. 3F and SI Appendix, Fig. S6C), suggesting their roles in stress-induced gene expression (see below).
Polyploidy-Induced CHH Hypomethylation Primes JA-Related Genes.
The stress-related DMR overlapping genes frequently occurred in gene-rich regions and enriched in multiple gene families, such as PLD_C and lipoxygenase gene families, some of which were involved in JA biosynthesis (SI Appendix, Fig. S6 D and E). Using the Rice Expression Profile Database (ricexpro. dna.affrc.go.jp/), we identified 5,346 up-regulated genes after treatment by JA, of which 924 genes were associated with ploidy-induced hypo-DMRs. These hypo-DMRs overlapped with the genes that were overrepresented in JA-related pathways (SI Appendix, Fig. S7A). Among 816 salt-induced hyper-DMR overlapping JA-related genes in tetraploid rice, 347 (42.5%) were differentially expressed after salt treatment (Dataset S1D). This suggests that DNA methylation may affect expression of JA-related genes in tetraploid rice under salt stress.
We further analyzed expression of 54 JA-related genes, including 32 JA biosynthesis genes, 17 JA-signaling genes, and 5 JA downstream genes (Dataset S1E). Among them, 10 JA biosynthesis and 10 JA-signaling genes were activated in diploid, tetraploid rice or both after salt treatment for 24 h, displaying higher expression levels in tetraploid than in diploid rice after salt stress (Fig. 2 E and F). The majority including 80% (8/10) JA biosynthesis genes (SI Appendix, Fig. S7B) like OsLOX6 (SI Appendix, Fig. S7D) and 70% (7/10) JA-signaling genes (SI Appendix, Fig. S7C) like OsJAZ8 and OsJAZ12 (SI Appendix, Fig. S7 E and F) showed tetraploidy-induced hypomethylation, salt-induced hypermethylation, or both. Activation of JA-related genes is rapid after salt and cold stress in Arabidopsis (18, 19). Indeed, eight JA biosynthesis genes including OsPLDα4, OsPLDα5, OsAOS1, and OsAOC were induced at higher levels in tetraploid rice under saline condition for 3 and 6 h (SI Appendix, Fig. S8). Similarly, six JA-signaling genes including OsJAZ5, OsJAZ7, and OsJAZ8 and three JA pathway downstream genes including OsJAMyb, OsPR10 (aka RSOsPR10), and OsPR10a were rapidly induced by salt treatment at 3h and 6h (SI Appendix, Fig. S9). Interestingly, CHH methylation levels in the promoter regions of these genes (OsPLDα4, OsPLDα5, OsAOS1 and OsAOC) were decreased in tetraploid rice and increased after salt stress (SI Appendix, Figs. S7B and S8I). Thus, priming of CHH hypomethylation in tetraploid rice may lead to more rapid and robust response to salt stress than in diploid rice.
For example, AtOPCL1, encoding an OPC-8:0 CoA ligase, is responsible for wound-induced JA accumulation in Arabidopsis (39). Rice OsOPCL1 (Os03g04000) is a putative JA biosynthesis gene. Its promoter region was hypomethylated in tetraploid rice in control condition (Fig. 3F), while OsOPCL1 expression was dramatically induced under salt stress (Fig. 2E), suggesting that polyploidy-induced hypomethylation may prime a state ready for a robust response to stress stimuli. After salt stress, OsOPCL1 expression in tetraploid rice was highly induced and accompanied by the increased CHH methylation levels in its promoter region (Fig. 3F). The CHH methylation changes coincided with the location of a MITE in the protomer region, supporting the notion that stress-induced expression may in turn promote CHH hypermethylation to repress adjacent TEs.
To test if salt-induced gene expression preceded hypermethylation, we examined genome-wide changes in CHH methylation under 125 mM saline condition for 0 h, 3 h, and 24 h and expression (qRT-PCR) of JA pathway genes (SI Appendix, Fig. S10). In the genome-wide scale, CHH methylation levels were induced dramatically at 24 h but slightly or unchanged at 3 h (SI Appendix, Fig. S10A), including JA biosynthesis (SI Appendix, Fig. S10B) and signaling (SI Appendix, Fig. S10C) pathway genes, whereas expression of seven out of nine genes tested was induced at the highest levels in 3 h after salt treatment (SI Appendix, Fig. S10D). For example, expression of OsMYC2, a JA-signaling gene, was strongly induced at 3 h (SI Appendix, Fig. S10E), whereas CHH methylation levels remained unchanged but increased dramatically at 24 h (SI Appendix, Fig. S10F, when its expression level has decreased (SI Appendix, Fig. S10E). Similarly, expression of OsOPCL1, a JA biosynthesis gene, was dramatically increased at 3 h (SI Appendix, Fig. S10G), while its promoter methylation level has slightly increased and reached to a very high level at 24 h (SI Appendix, Fig. S10H), when its expression level tended to decrease (SI Appendix, Fig. S10G). These results support the notion that expression of salt-inducible genes precedes CHH methylation increase in promoter regions, consistent with the observation from phosphorus starvation in diploid rice (26).
Activation of JA Pathway Genes Enhanced Salt Tolerance in Tetraploid Rice.
To verify the role of jasmonate in salt tolerance in tetraploid rice, we measured JA and JA-Ile content under control and saline conditions. JA-Ile content in roots was higher in tetraploid than in diploid rice without salt stress (Fig. 3G). After salt stress for 3 h, JA-Ile content was increased in both but at higher levels in tetraploid than in diploid rice. After 6 h, the JA-Ile content remained higher in tetraploid rice but was reduced in both diploid and tetraploid rice relative to the control. JA content, however, remained at similar levels between diploid and tetraploid rice regardless of stress or nonstress conditions and was reduced after saline treatment for 3 and 6 h (SI Appendix, Fig. S11A). Consistent with increased JA-Ile content, OsJAR1, encoding a JA-Ile synthetase (40), was more highly induced in tetraploid than diploid rice under saline condition for 3 h (SI Appendix, Fig. S11B), which correlated with hypomethylation in its promoter region in tetraploid rice and hypermethylation after salt stress (SI Appendix, Fig. S11C). These data suggest that increased JA-Ile content may contribute to increased salt tolerance in tetraploid rice.
Root growth inhibition is an indicator for JA signaling response (18). The root length was more severely inhibited in tetraploid than in diploid rice after treatment with JA (5 or 10 μM) (Fig. 3H), suggesting a greater response of JA signaling in tetraploid rice. Consequently, the survival rate was increased in both diploid and tetraploid rice after JA treatment but more in tetraploid than in diploid (Fig. 3I). Consistent with the role of JA signaling in salt stress tolerance, the survival rate was decreased in the coi1-18 mutant of diploid rice (SI Appendix, Fig. S11D), in which JA signaling was disrupted through disrupting expression levels of OsCOI1a and OsCOI1b under salt stress (41). Collectively, these data suggest that JA pathways are strongly activated in tetraploid rice, contributing to increased salt stress tolerance.
JA-Related Genes Remain Reactivated in Response to Recurring Salt Stress.
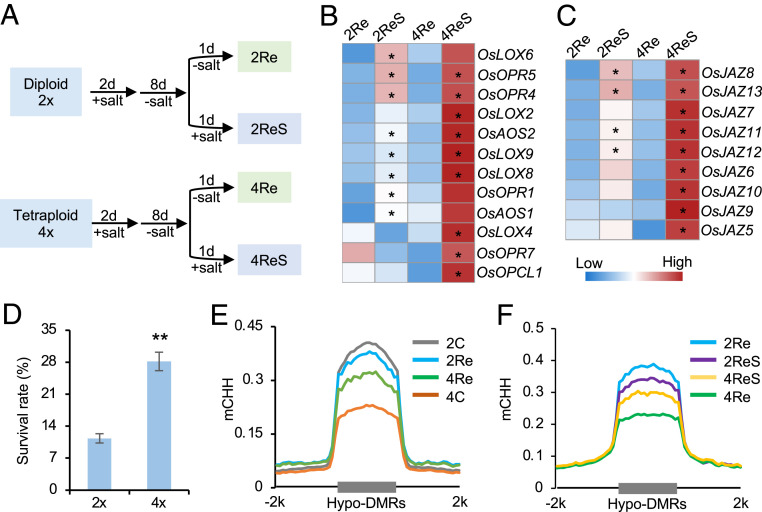
To test if JA-related genes remain activated after the initial stress treatment, we analyzed DNA methylation and gene expression changes in diploid and tetraploid rice after a second round of salt stress (Fig. 4A and Dataset S1F). These activated or repressed genes after the initial salt treatment remained up-regulated or down-regulated after the second salt treatment (SI Appendix, Fig. S12A). Moreover, expression level changes in the up-regulated or down-regulated genes were much higher in tetraploid than in diploid rice (P < 2.2e−6, Wilcoxon rank sum test) (SI Appendix, Fig. S12A). Consistent with global gene expression changes, many genes related to JA biosynthesis (Fig. 4B) and signaling (Fig. 4C) pathways were reactivated at much higher levels in tetraploid than in diploid rice, which correlated with increased survival rates in tetraploid rice (Fig. 4D). After their recovery in nutrient solution, CHH hypomethylation levels were partly retained in tetraploid rice (Fig. 4E and Dataset S1G), including genomic regions adjacent to JA-related genes (SI Appendix, Fig. S12B). After the second round of salt stress, these regions became hypermethylated in tetraploid rice (Fig. 4F), suggesting a positive reinforcement between salt-induced gene expression and consequent hypermethylation, as shown in rice under phosphate starvation (26) and in cotton during fiber cell development (42).
Fig. 4.
Stronger activation of JA-related genes upon recurring salt stress in 02428 tetraploid rice. (A) Experimental design for recurring (first and second rounds of) salt stress. After salt treatment for 2 d, seedlings were recovered in nutrient solution for 8 d (diploid, 2Re versus tetraploid, 4Re), and treated by a second round of salt stress for 1 d (diploid, 2ReS versus tetraploid, 4ReS). (B and C) Expression levels (RPM) of JA biosynthesis (B) and signaling (C)-related genes in diploid and tetraploid rice after the second round of salt stress. A single asterisk indicates gene expression level difference before and after salt treatment in diploid and tetraploid rice, respectively (P < 0.05 and log2[fold change] ≥ 1). (D) Survival rates of diploid and tetraploid rice after the second round of salt stress for 12 d and recovery in nutrient solution for 5 d (n = 4 biological replicates). Double asterisks indicate statistical significance of P < 0.01 (Student’s t test). (E) CHH methylation levels in all samples without salt treatment (4C versus 2C) and upon recovery (4Re versus 2Re) using the hypo-DMRs between diploid and tetraploid rice under the control condition (4C versus 2C). (F) CHH methylation levels in all samples after the second round of salt stress using the hypo-DMRs between tetraploid and diploid rice upon recovery (4Re versus 2Re).
Nipponbare and 02428 Tetraploids Share Responses to Ploidy and Salt Stress Changes.
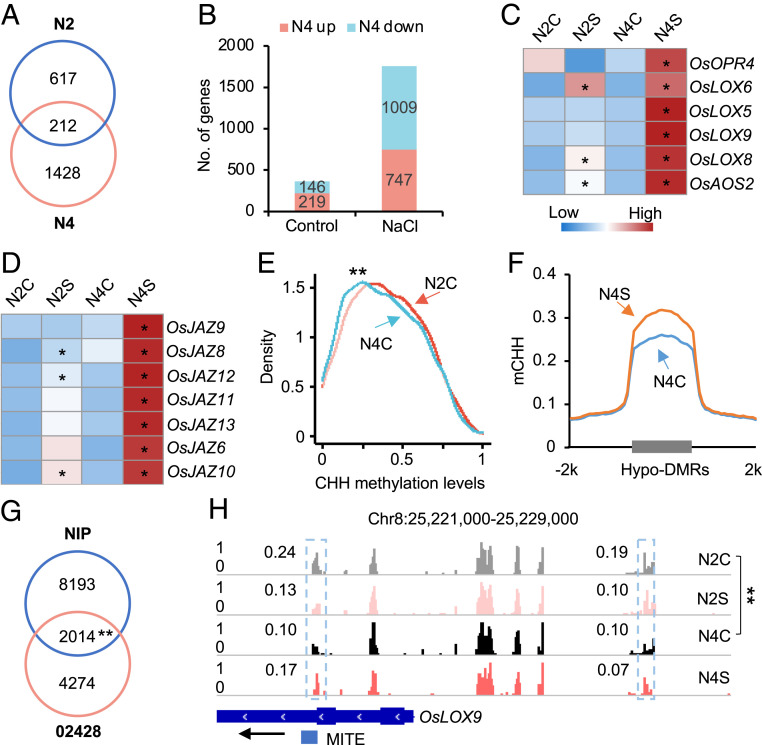
To explore if this “priming” mechanism is conserved among different tetraploid rice, we performed transcriptome and methylome analyses using another set of diploid and tetraploid (colchicine-induced) rice (Nipponbare) (SI Appendix, Fig. S1). Tetraploid (N4) rice showed higher survival rates than the diploid (N2) in saline condition (SI Appendix, Fig. S13A). After salt stress, more DEGs were identified in tetraploid (1,640) than in diploid (829) rice (Dataset S1H), and only 212 DEGs were shared between diploid and tetraploid rice (Fig. 5A). In addition, more genes were differentially expressed between tetraploid and diploid rice under the salt stress than control condition (Fig. 5B). In agreement with the findings in 02428, JA pathway-related genes were overrepresented in the salt-activated genes in tetraploid and diploid Nipponbare (SI Appendix, Fig. S13B), which were also enriched in tetraploidy-activated genes under salt stress. Indeed, many genes involved in JA biosynthesis (Fig. 5C), signaling (Fig. 5D), and downstream (SI Appendix, Fig. S13C) pathways were expressed at higher levels in tetraploid rice after salt stress, suggesting a shared pattern of transcriptional responses to polyploidy and salt stress in both sets of tetraploid rice.
Fig. 5.
Ploidy-induced CHH hypomethylation primes JA-related genes for stronger activation in Nipponbare tetraploid rice. (A) Venn diagram showing overlap of salt responsive genes between diploid (N2) and tetraploid (N4) rice. (B) Number of up- or down-regulated genes between diploid (N2) and tetraploid (N4) without (control) or with salt (NaCl) stress. (C and D) Expression levels of JA biosynthesis (C) and signaling (D)-related genes in diploid and tetraploid rice without (N2C versus N4C) or with salt (N2S versus N4S) treatment. Asterisks indicate expression level difference before and after salt treatment in diploid and tetraploid rice, respectively (P < 0.05 and log2[fold change] ≥ 1). (E) CHH methylation levels of ploidy-induced CHH hypo-DMRs (02428) in Nipponbare diploid (N2C) and tetraploid rice (N4C). Double asterisks indicate a statistical significance level of P < 7e−7 (Wilcoxon rank sum test). (F) CHH methylation changes in hypo-DMRs between N4C and N2C in tetraploid rice after salt stress (N4S versus N4C). (G) Venn diagram showing the overlap of ploidy-induced hypo-DMR genes between Nipponbare and 02428. Double asterisks indicate a statistical significance level of P < 3e−175 (Hypergeometric test). (H) A genome browser snapshot showing CHH methylation changes of OsLOX9. The gene and TE are shown with a black arrow indicating transcriptional direction. Numbers in snapshot indicate CHH methylation levels of regions in dotted box. Double asterisks indicate the difference between N4C and N2C at a statistical significance level of P < 0.01 (Fisher’s exact tests).
Between Nipponbare and 02428, among the hypo-DMRs in 02428 tetraploid rice, CHH methylation levels were also lower in tetraploid Nipponbare than in diploid (P < 3e−175, hypergeometric test) (Fig. 5E), which correlated with CHH methylation level increases in tetraploid Nipponbare after salt stress (Fig. 5F and Dataset S1I). Notably, 2,014 genes that overlapped with tetraploid-related hypo-DMRs were shared between the two genotypes (P < 3e−175, hypergeometric test) (Fig. 5G), implying a shared pattern of DNA methylation changes in the two sets of tetraploid rice. Moreover, phytohormone-related genes, like in JA pathways, were also overrepresented among those hypo-DMR genes in tetraploid Nipponbare (SI Appendix, Fig. S13D). For example, OsLOX9 (Fig. 5H), OsLOX5 (SI Appendix, Fig. S13E), and OsOPR4 (SI Appendix, Fig. S13F) were hypomethylated in tetraploid rice, and their expression levels were dramatically induced after salt stress. Together, these data indicate a shared mechanism for polyploidy-induced CHH hypomethylation to prime a more rapid and strong activation of JA-related genes in response to salt stress in two rice tetraploids than in diploids.
Discussion
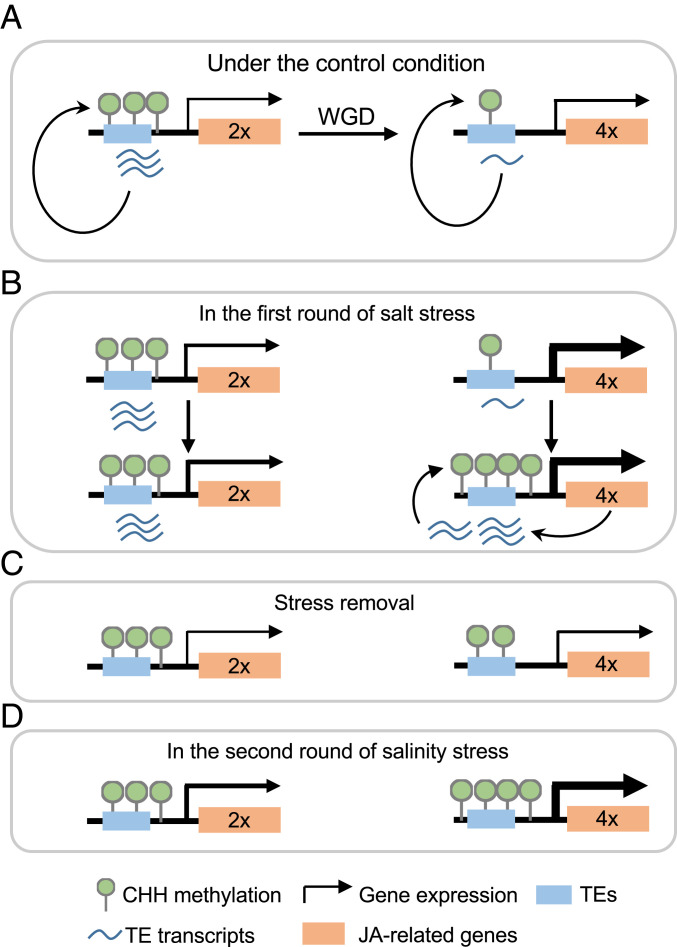
The molecular basis for enhanced adaptation and fitness under extreme environments in polyploids (10) is poorly understood. In this study, we provided experimental evidence that tetraploidy induces CHH hypomethylation and potentiates JA-related genes for higher induction levels in rapid and robust responses to salt stress, while the stress-induced gene expression in turn increases CHH methylation to repress neighboring TEs (Fig. 6).
Fig. 6.
Models for enhanced salt tolerance and fitness in tetraploid rice. (A) Polyploidy may repress proximal TEs through histone modifications, leading to CHH hypomethylation by RdDM pathway in tetraploid rice. (B) Under salt stress (the first salt treatment), CHH hypomethylation primes stress-related genes including those in JA biosynthesis and signaling pathways for more rapid and stronger activation of these genes to enhance salt tolerance in tetraploid (Upper) than in diploid rice. In turn, highly expressed stress-related genes may derepress neighboring TEs, triggering CHH hypermethylation to silence these TEs (Lower). (C) After stress removal, transcript abundance of JA-related genes in tetraploid returns to a similar level to diploid rice, while CHH hypomethylated status of tetraploid rice prior to the first salt treatment are also partly retained. (D) After recurring salt stress, stress-related genes of tetraploid rice are more strongly induced and subsequently hypermethylated in a process similar to that in B.
The hypomethylation status induced in tetraploid rice resembles the “priming state” in stress response, which has positioned plants for rapid and robust responses to environmental stimuli as an adaptive strategy to cope with abiotic and biotic stresses (43, 44). This priming state can be established with epigenetic modifications (45). For example, A. thaliana F1 hybrids show increased histone acetylation in promoters of salicylic acid pathway genes, priming more rapid and strong induction of defense-related genes than the parents upon pathogen attack (46). In this study, polyploidy induces CHH hypomethylation in TEs, which primes a chromatin status ready for activating JA-related genes with proximal TEs in rapid and robust responses to salt stress (Fig. 6). A higher JA-Ile content and enhanced JA-signaling cascade in tetraploid rice may result in a lower sodium uptake and a higher K+/Na+ ratio, as observed in diploid rice (20), to enhance salinity tolerance. Application of exogenous MeJA may keep stabilities of proteins like transporters to maintain homeostasis of potassium and sodium after salt stress.
Genome doubling (in both auto- and allopolyploids) and hybridization (in allopolyploids) can be viewed as potential causes of genomic stress (4) that in turn induces the internal “priming” status, which can potentiate activation of stress-related genes in rapid and robust responses to external stress. Alternatively, colocalization of TEs and stress-related genes may occur as part of genomic response of plants to ever changing environments. In Arabidopsis, stress-responsive genes tend to be preferentially retained after genome duplication (47), and the majority of these genes are associated with TEs. In rice, TEs tend to have a preference for insertion into 5′ flanking sequences of genes and adjacent to the stress-inducible genes (48). Under stress, TEs tend to be activated and jump into genomic regions with open chromatin including low DNA methylation. After stress, activated TE expression can induce DNA hypermethylation via RdDM to repress expression of TE-associated genes. This relationship between TEs and stress-responsive genes may confer an evolutionary advantage for adaptation to diverse environments. Mechanisms for priming and stress-induced gene expression in tetraploid rice are not well understood. One possibility may involve TEs, which comprise 35% or more of the rice genome (49), and TEs tend to be coregulated with expression of biotic stress-responsive genes in A. thaliana (25). Moreover, DNA demethylases (ROS1, DML2, and DML3) target TEs within or near promoter regions to positively regulate gene expression in response to biotic stress (23). Heat stress can induce expression and transposition activity of ONSEN, a copia-type retrotransposon, in A. thaliana, and heat-induced ONSEN transcript levels are higher in the mutants defective in RdDM pathways (50). These data suggest coregulation of stress responses and TE activities, which is probably because activation of TEs would be deleterious, and most activated TEs are silenced by DNA methylation in plants (51) after stress.
A positive correlation between CHH hypermethylation and expression of TE-associated genes, though counterintuitive, has been documented in maize (52), rice (26), and cotton (42). In rice, stress-responsive genes are activated after phosphate starvation, which in turn induces RdDM pathway and hypermethylation of these genes associated with TEs (26). In cotton, upregulation of ovule-related genes coincides with small interfering RNA (siRNA) abundance and RdDM-dependent CHH hypermethylation, and these genes are silenced by additional methylation via RdDM-independent (CMT2-dependent) pathway in late stages of fiber cell development (42). In tetraploid rice, polyploidy induces CHH hypomethylation in genomic loci coexistent with stress-responsive genes that are predominately associated with TEs, which may prime for rapid and robust responses to salt stress, while the stress-induced gene expression in turn promotes CHH hypermethylation to silence adjacent TEs (Fig. 6). Under nonstress condition, TEs are repressed in tetraploid rice, leading to reduced CHH methylation. It is also likely that other epigenetic modifications such as histone marks (H3K9me2 and H3K27me2) may repress TE expression, as reported in a recent study on polyploid wheat (53). Hypomethylation in tetraploid wheat after genome separation from hexaploid wheat is reversible after merger with another genome (28). Although the cause of hypomethylation in tetraploid rice remains to be elucidated, this positive feedback regulation of hypomethylation, induction of stress-responsive genes, and hypermethylation of adjacent TEs provides dynamic roles for TE-mediated DNA methylation in activation and repression of stress-responsive genes before and after biotic and abiotic stresses.
On one hand, polyploidization induces CHH hypomethylation in the genomeic loci coexistent with stress-responsive genes and adjacent TEs, which potentiates a “priming” state for transcriptional regulation. On the other hand, the primed stress-related genes are rapidly and strongly induced under salt stress, which in turn promotes the RdDM pathway to silence TEs and/or stress-responsive genes. These induced responses are reproducible in a recurring round of salt stress and shared among different tetraploid rice. This unique feedback regulation between polyploidy-induced epigenetic modifications and stress-responsive gene expression changes may provide a general mechanism for polyploid plants and crops to respond and adapt to environmental and climate challenges during evolution and domestication.
Materials and Methods
Plant Materials and Growth Conditions.
Isogenic diploid and tetraploid rice of 02428 (30) and Nipponbare were used for the study. Tetraploid of 02428 was from tissue culture and self-pollinated for 10 generations. Tetraploid of Nipponbare was artificially synthesized by colchicine treatment and self-pollinated for two generations. Chromosomes were counted as previously described (30). All plants were cultured in 0.5× Kimura B nutrient solution (54) in a growth chamber with 28 °C/25 °C (day/night) and 12 h/12 h (light/dark) cycles. Seedlings at the trefoil stage were treated by a series of 0, 50, 100, 150, and 200 mM NaCl for 6 d and recovered in the nutrient solution for 5 d to determine optimal concentrations. An optimal concentration of 125 mM saline was used for all other (first and second rounds) salt stress experiments (24 h) except for the low salt stress condition in which 50 mM salt was used. Roots of the plants treated with or without salt for 24 h were harvested from a random set of 12 individuals for each replicate and immediately frozen in liquid nitrogen. These samples were used for RNA-seq and MethylC-sequencing (MethylC-seq) analyses following the published methods (27).
Inhibition Rate Analysis.
Seedlings of diploid and tetraploid rice (02428) at the trefoil stage were treated by 125 mM salt solution for 1 to 5 d. Fresh weights of shoots or roots were measured immediately after harvesting in each day. Dry weights were measured after the samples were dried at 80 °C for 3 d. Inhibition rate was estimated using the ratio of the weight difference between the control and treated sample over the weight of the control sample. A total of 18 biological replicates were used for each time point per cytotype.
Tissue K+ and Na+ Quantification.
Seedlings at the trefoil stage of 02428 were cultured in a 0.5× Kimura B nutrient solution containing 125 mM NaCl for 2 and 4 d. Roots were washed three times with deionized water prior to harvest. Roots and shoots were separated and dried at 65 °C for 3 d. Dry samples were digested with a mixed solution of acids HNO3/HClO4 (85/15, vol/vol) (55). The contents of K+ and Na+ were measured by inductively coupled plasma mass spectrometry (PerkinElmer NexION-300×, Waltham, MA).
RNA Extraction, RNA-Seq, and Data Analysis.
Total RNA was extracted by mirVANA miRNA isolation kit (Ambion1561). Messenger RNA sequencing (mRNA-seq) libraries were constructed for each condition per cytotype with three biological replicates. In brief, mRNA was purified with Oligo-dT attached magnetic beads and randomly fragmented, which was reversely transcribed into complementary DNA (cDNA). After double-stranded cDNA synthesis, end repair, dA-tailing, and adapters ligation were performed following Illumina’s standard protocol. After PCR amplification, 400 to 500 bp fragments were recovered. Libraries were sequenced on the platform of Illumina HiSEq. 2500 with pair-end 125 bp.
The 3′ mRNA-seq libraries were performed for each condition per cytotype with two biological replicates. Briefly, after ∼1 μg RNA was fragmented, oligo-dT index primers were used to synthesize the first-strand cDNA. After synthesis of double-stranded DNA, end repair, dA-tailing, and adapter ligation were performed as previously described (56). Approximately 300 to 500 bp fragments were purified after PCR amplification. Libraries were sequenced using HiSeq X Ten platform for 150 bp pair-end reads.
For mRNA-seq, after removing low-quality reads, clean data were mapped onto O. sativa ssp. japonica cv. Nipponbare genome sequence (MSU v7.0) (57) using TopHat (v2.1.1) with at most two mismatches (58). Cufflinks (v2.2.1) was used to calculate transcripts levels (fragments per kilobase of transcript per million fragments, FPKM) (59). Differentially expressed genes were identified by R package edgeR (60) with P < 0.05, false discovery rate (FDR) < 0.05, and abs (log2-fold change) ≥ 1. For 3′ mRNA-seq, due to the sequential thymine in R2 reads, only R1 reads were used for further analysis. The genes with RPM (reads per million mapped) ≥ 5 were used to detect DEGs with P < 0.05 and abs (log2-fold change) ≥ 1 by edgeR. Significantly enriched pathways were identified by Pageman analysis (32).
Expression levels of TEs were estimated by TEtranscripts (61) with default normalized approach. All expressed TEs with normalized values ≥ 1 in one or more samples were used for boxplot analysis (also see mRNA-seq and 3′ mRNA-seq library information listed in Dataset S1A).
MethylC-Seq Library Construction and Data Analysis.
Genomic DNA was isolated using the cetyltrimethylammonium bromide method (62). Libraries were constructed using 1 μg of genomic DNA for each sample. After DNA was sheared, end repair, dA-tailing, and methylated adapter ligation were performed following the manufacturer’s instructions of NEBNext Ultra II DNA Library Prep Kit (NEB). Next, sodium bisulfite conversion was performed according the protocol of EZ DNA Methylation-Gold kit (ZYMO research, Irvine, CA). After amplification of 15 cycles, 350 to 500 bp fragments were purified. All libraries were sequenced on Illumina HiSeq X ten platform with 150 bp pair-end reads.
Clean reads were mapped onto the rice genome using Bismark (v0.15.1) software (63) with default parameters. Duplicates in unique mapped reads were removed to reduce PCR bias. Cytosines covered by at least three reads and conserved among all materials were used for further analysis. CHH DMRs were analyzed using 100 bp bins with at least eight CHH cytosines. Weighted methylation levels were calculated using the ratio of methylated cytosines (mC) over total cytosines (mC + nonmC) in each bin. For the samples with two replicates, CHH DMRs were identified by P < 0.05 (ANOVA test) and absolute methylation difference over 0.1. For the samples with one replicate, Fisher’s exact test was performed to detect CHH DMRs with FDR < 0.05 and absolute methylation difference over 0.1. Conversion frequency was estimated by Lambda genome mixed in the sample. The list of library information is shown in Dataset S1A. An example of hypergeometric tests is the following:
a = Stress-related DMRs overlap hypo-DMRs in drm2 mutants
b = stress-related DMRs
c = hypo-DMRs in drm2 mutants
d = all 100 bp windows in rice genome to identify DMRs
A hypergeometric test was performed by formula “hyper (a, b, d-b, c, lower.tail = FALSE, log.p = FALSE)” in R packages.
RNA Isolation and qRT-PCR Analysis.
Roots of diploid and tetraploid rice at trefoil stage were harvested after treatment with 125 mM NaCl for 0 h, 3 h, 6 h, and 24 h. Total RNA was isolated using TRIzol reagent (Invitrogen, San Diego, CA). Subsequently, RNA was converted to cDNA using HiScript II Q RT SuperMix for qPCR kit (Vazyme, R223-01, Nanjing, China), and qRT PCR was performed by AceQ qPCR SYBR Green Master Mix (Vazyme, Q111-02/03). All data were normalized to transcripts of Actin (LOC_Os03g50885), and relative expression levels were calculated using the 2-ΔΔCt method as previously described (64). ΔCt = Ct of genes − Ct of Actin; ΔΔCt = ΔCt of the test sample − ΔCt of the control (diploid without salt stress). Primer pairs are listed in Dataset S1J.
Quantification of JA and JA-Ile.
Roots of diploid and tetraploid rice at trefoil stage were collected after treatment with 125 mM NaCl for 0 h, 3 h, and 6 h and immediately frozen in liquid nitrogen. JA and JA-Ile contents were measured by liquid chromatography-tandem mass spectrometry (65). The amount of JA and JA-Ile was normalized to the fresh weight of roots with three biological replicates. Each sample (biological replicate) was measured three times as technical replicates.
JA Treatment and Root Growth Analysis.
Seven-day-old seedlings were treated with 5 μM or 10 μM JA (Sigma, J2500) or no JA (control). After JA treatment for 2 d, the length of roots was measured. Inhibition rates were calculated as noted above.
Supplementary Material
Acknowledgments
We thank Dr. Chuandeng Yi at Yangzhou University for providing diploid and tetraploid rice materials, Dr. Yufeng Wu in the Bioinformatics Center at Nanjing Agricultural University (NJAU) for providing the high-performance computing services, Dr. Donglei Yang at NJAU for providing the coi1-18 mutant line, and Dr. Wu Jiao at NJAU for assistance in data analysis. The work at NJAU was supported by the National Natural Science Foundation of China (91631302), Jiangsu Research and Education Innovation Consortium (2013 to 2015), and Jiangsu Collaborative Innovation Center for Modern Crop Production. The work at The University of Texas at Austin was supported by the NSF (IOS1739092) and the D. J. Sibley Centennial Professorship in Plant Molecular Genetics.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023981118/-/DCSupplemental.
Data Availability
The RNA-Seq and MethylC-seq data are deposited in the National Center for Biotechnology Information (NCBI) under the Bioproject accession no. PRJNA613869. All study data are included in the article and/or supporting information.
References
- 1.Muller H. J., Why polyploidy is rarer in animals than in plants. Am. Nat. 59, 346–353 (1925). [Google Scholar]
- 2.Soltis D. E., Visger C. J., Soltis P. S., The polyploidy revolution then…and now: Stebbins revisited. Am. J. Bot. 101, 1057–1078 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Van de Peer Y., Mizrachi E., Marchal K., The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Chen Z. J., Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biology 58, 377–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha M., Kim E. D., Chen Z. J., Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc. Natl. Acad. Sci. U.S.A. 106, 2295–2300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casneuf T., De Bodt S., Raes J., Maere S., Van de Peer Y., Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 7, R13 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., et al., Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172, 507–517 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni Z., et al., Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller M., Song Q., Shi X., Juenger T. E., Chen Z. J., Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat. Commun. 6, 7453 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Grant V., Plant Speciation (Columbia University Press, New York, ed. 2, 1981), p. 563. [Google Scholar]
- 11.Wei N., Cronn R., Liston A., Ashman T. L., Functional trait divergence and trait plasticity confer polyploid advantage in heterogeneous environments. New Phytol. 221, 2286–2297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao D. Y., et al., Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341, 658–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Pozo J. C., Ramirez-Parra E., Deciphering the molecular bases for drought tolerance in Arabidopsis autotetraploids. Plant Cell Environ. 37, 2722–2737 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Munns R., Tester M., Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Ismail A. M., Horie T., Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 68, 405–434 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Wing R. A., Purugganan M. D., Zhang Q., The rice genome revolution: From an ancient grain to green super rice. Nat. Rev. Genet. 19, 505–517 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Kazan K., Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Valenzuela C. E., et al., Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 67, 4209–4220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y., Jiang L., Wang F., Yu D., Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang D. J., et al., Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. J. Agron. Crop Sci. 191, 273–282 (2005). [Google Scholar]
- 21.Schwachtje J., et al., Induced, imprinted, and primed responses to changing environments: Does metabolism store and process information? Front Plant Sci 10, 106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu N., Avramova Z., Molecular mechanism of the priming by jasmonic acid of specific dehydration stress response genes in Arabidopsis. Epigenetics Chromatin 9, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le T. N., et al., DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 15, 458 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matzke M. A., Mosher R. A., RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Dowen R. H., et al., Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U.S.A. 109, E2183–E2191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Secco D., et al., Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4, e09343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Q., Zhang T., Stelly D. M., Chen Z. J., Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol. 18, 99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan J., et al., Dynamic and reversible DNA methylation changes induced by genome separation and merger of polyploid wheat. BMC Biol. 18, 171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., et al., Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. U.S.A. 112, E7022–E7029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L., et al., Rice interploidy crosses disrupt epigenetic regulation, gene expression, and seed development. Mol. Plant 11, 300–314 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Rellán-Álvarez R., Lobet G., Dinneny J. R., Environmental control of root system Biology. Annu. Rev. Plant Biol. 67, 619–642 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Usadel B., et al., PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7, 535 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou G., et al., Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 60, 638–648 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Yokotani N., et al., Role of the rice transcription factor JAmyb in abiotic stress response. J. Plant Res. 126, 131–139 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Mizuno H., et al., Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.). BMC Genomics 11, 683 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shankar R., Bhattacharjee A., Jain M., Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 6, 23719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y., et al., A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 164, 1068–1076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan F., et al., Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol. 171, 2041–2054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koo A. J., Chung H. S., Kobayashi Y., Howe G. A., Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281, 33511–33520 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Xiao Y., et al., OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol. Biol. 86, 19–33 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Yang D. L., et al., Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109, E1192–E1200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Q., Guan X., Chen Z. J., Dynamic roles for small RNAs and DNA methylation during ovule and fiber development in allotetraploid cotton. PLoS Genet. 11, e1005724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mauch-Mani B., Baccelli I., Luna E., Flors V., Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 68, 485–512 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Hilker M., Schmülling T., Stress priming, memory, and signalling in plants. Plant Cell Environ. 42, 753–761 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Friedrich T., Faivre L., Bäurle I., Schubert D., Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ. 42, 762–770 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Yang L., et al., Salicylic acid biosynthesis is enhanced and contributes to increased biotrophic pathogen resistance in Arabidopsis hybrids. Nat. Commun. 6, 7309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ha M., Li W. H., Chen Z. J., External factors accelerate expression divergence between duplicate genes. Trends Genet. 23, 162–166 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naito K., et al., Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461, 1130–1134 (2009). [DOI] [PubMed] [Google Scholar]
- 49.International Rice Genome Sequencing Project , The map-based sequence of the rice genome. Nature 436, 793–800 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Ito H., et al., An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Zhang H., Lang Z., Zhu J. K., Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Gent J. I., et al., CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23, 628–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y., et al., Histone H3K27 dimethylation landscapes contribute to genome stability and genetic recombination during wheat polyploidization. Plant J. 105, 678–690 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Chen J., Huang X. Y., Salt D. E., Zhao F. J., Mutation in OsCADT1 enhances cadmium tolerance and enriches selenium in rice grain. New Phytol. 226, 838–850 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Zhao F., Mcgrath S. P., Crosland A. R., Comparison of 3 wet digestion methods for the determination of plant sulfur by inductively-coupled plasma-atomic emission-spectroscopy (Icpaes). Commun. Soil Sci. Plant Anal. 25, 407–418 (1994). [Google Scholar]
- 56.Sanfilippo P., Miura P., Lai E. C., Genome-wide profiling of the 3′ ends of polyadenylated RNAs. Methods 126, 86–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawahara Y., et al., Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N. Y.) 6, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell C., et al., Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin Y., Tam O. H., Paniagua E., Hammell M., TEtranscripts: A package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31, 3593–3599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allen G. C., Flores-Vergara M. A., Krasynanski S., Kumar S., Thompson W. F., A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caldana C., Scheible W. R., Mueller-Roeber B., Ruzicic S., A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3, 7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balcke G. U., et al., An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 8, 47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq and MethylC-seq data are deposited in the National Center for Biotechnology Information (NCBI) under the Bioproject accession no. PRJNA613869. All study data are included in the article and/or supporting information.