Abstract
Estrogen receptor alpha gene (ESR1) mutations occur frequently in ER-positive metastatic breast cancer (MBC), and confer clinical resistance to aromatase inhibitors (AIs). Expression of the ESR1 Y537S mutation induced an epithelial-mesenchymal transition (EMT) with cells exhibiting enhanced migration and invasion potential in vitro. When small subpopulations of Y537S ESR1 mutant cells were injected along with WT parental cells, tumor growth was enhanced with mutant cells becoming the predominant population in distant metastases. Y537S mutant primary xenograft tumors were resistant to the antiestrogen tamoxifen (Tam) as well as to estrogen withdrawal. Y537S ESR1 mutant primary tumors metastasized efficiently in the absence of estrogen; however, Tam treatment significantly inhibited metastasis to distant sites. We identified a 9 gene expression signature which predicted clinical outcomes of ER-positive breast cancer patients, as well as breast cancer metastasis to the lung. Androgen receptor (AR) protein levels were increased in mutant models, and the AR agonist dihydrotestosterone (DHT) significantly inhibited estrogen-regulated gene expression, EMT, and distant metastasis in vivo, suggesting that AR may play a role in distant metastatic progression of ESR1 mutant tumors.
Introduction
Unfortunately the 5-year relative survival of women with metastatic breast cancer (MBC) is less than 25% [1], and new therapeutic strategies are needed. One of the prevailing models of metastasis holds that most primary tumor cells have low metastatic potential, but rare clones of cells within the primary tumor acquire metastatic competency through the acquisition of somatic mutations [2]. An aggressive clone whose mutations support a proliferative advantage and metastatic proclivity can dominate, and survive during metastatic dissemination. It is clear that estrogen can promote breast cancer metastasis. A major clinical question is how hormonally-responsive ER-positive breast cancers progress to a more aggressive and hormone-independent phenotype, and this development of acquired resistance to hormonal therapies is a major clinical problem.
Acquisition of somatic mutations in the ESR1 gene is a major contributor to resistance and poor clinical outcomes in MBC [3–6]. Mutations within the hormone binding domain (HBD) of ESR1 have been found at high frequencies in MBC patients [7], and can be subclonal [reviewed in [8]. We and others have reported that these mutations are less frequent, but present in primary breast tumors from patients before endocrine therapy, ostensibly in rare subclonal populations [9, 10]. Clinical data suggest that since the HBD ESR1 mutations are active in the absence of estrogen they could be “selected for” during the treatment of MBC with an AI [11]. In a retrospective study of patients treated with fulvestrant +/− the CDK4/6 inhibitor palbociclib, Y537S mutations continue to be acquired during treatment [12]. Recently it has also been shown that cells with HBD ESR1 mutations could metastasize in vivo, however, the biologic attributes associated with distant metastasis were not delineated [5, 13]. Using CRISPR-Cas9 knock-in of the ESR1 Y537S HBD mutation, we observed frequent spontaneous metastasis in vivo associated with the acquisition of an EMT phenotype. We have previously identified elevated AR expression as a mechanism of endocrine therapy resistance [14–16]. We herein demonstrate that AR protein was overexpressed in ESR1 mutant models and AR expression plays a role in EMT and distant metastasis.
Materials and methods
Cells and lentivirus infection
T47D (Parental, YS3 and YS53) and MCF-7 (BK parental, SF parental, YS1, YS30 and DG2) cells were cultured in RPMI 1640 and MEM medium respectively. Lentivirus infected cells (T47D WT, T47D YS and T47D DG) were generated as described [9] and are detailed in Supplemental Methods.
BrdU incorporation
Incorporation was performed as previously described [17] using 5% charcoal-stripped serum (CSS) as detailed in Supplemental Methods.
Xenograft studies
For metastasis studies, MCF-7 parental, CRISPR-Cas9 ESR1 mutant YS1, YS30, and DG2 clones, and T47D parental or T47D Y537S CRISPR clones were established as xenografts in the presence of estradiol (E2) as described [18] and detailed in Supplementary Methods. Mice were randomized to continue E2, E2 was withdrawn to stimulate treatment with an AI, or E2 withdrawal plus tamoxifen (Tam; 20mg/kg subcutaneously in peanut oil, 5 d/wk).
A Tam treatment study was performed with WHIM 20 PDX tumors as described [19] and detailed in Supplementary Methods. To compare hormonal agents, mice were first supplemented with E2, E2 was withdrawn when tumors reached 350 mm3 and randomized to receive Tam (20mg/kg), DHT (8mg in tubing), or enzalutamide (Enz, orally 50 mg/kg, 5 d/wk) treatment. An in vivo tumor cell mixing experiment was performed using MCF-7 parental and YS1 mutant cells grown in the absence of E2 as detailed in Supplementary Methods. Primary and metastatic tumors were analyzed using H&E, immunohistochemistry, immmunofluorescence, or in digital droplet PCR (ddPCR) assays as detailed in Supplemental Methods.
Cell invasion assays using IncuCyte live cell analysis
Cells were plated in 96 wells coated with Corning™ Collagen IV as recommended by the manufacturer’s protocol (Essen Bioscience, Ann Arbor MI), and grown in 5% CSS without E2. Images were taken every 2 hours and cells were examined for a total of 3 days after treatment. Fold change of wound healing area was calculated using IncuCyte software, and area changed was analyzed using GraphPad.
Generation of ESR1 point mutation knock-in (KI) breast carcinoma cell lines using CRISPR/Cas9
To introduce point mutations to generate Y537S and D538G ESR1 mutations, we first constructed ESR1 sgRNA vectors based on px458 [20]. Anti-sense oligonucleotide(s) (ssODNs) were synthesized with the intended mutations as well as silent mutations that served to prevent Cas9 re-cutting and facilitate screening. Details are provided in Supplemental Methods.
qRT-PCR
Cells were plated in regular serum-containing media for 24 hours, and for 48 hours in 5% CSS. Cells were treated for 72 hours with 4-OHT (100 nM) or DHT (10nM). RNA was extracted using RNeasy mini kits (Qiagen. Valencia, CA). qPCR primers from Bio-Rad and SYBR™ Green super mix were used according to company protocol. Assays were performed in triplicate and analyzed using CFX Maestro software.
ChIP-PCR
ChIP-Seq and ChIP-PCR experiments were conducted with cells grown in 5% CSS medium with or without hormonal supplementation as described in Supplemental Methods. [21, 22].
RNA expression analyses
For gene expression response to 4-OHT (100nM) treatment, cells were plated in serum-containing media and treated for 24 hours and compared to untreated control cells. For response to DHT (10nM), cells were incubated in 5% CSS for 48 hours then treated +/− DHT for 24 hours. RNA was extracted using the RNeasy kit (Qiagen. Valencia, CA). Labeled cRNA was hybridized onto Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays (Affymetrix Inc. Santa Clara, CA) in triplicate. Microarray data processing and analyses are described in Supplemental Methods.
Clinical biomarker analyses
We identified genes differentially overexpressed in MCF-7 YS1 cells compared with parental cells (> 4 fold), then filtered by removing a) genes that were estrogen-regulated (up-regulated with E2 treatment) in parental cells, b) genes overlapped with Estrogen Response Early and Late Hallmark gene sets in MSigDB, and c) genes reduced at least 20% by Tam treatment in MCF-7 YS1 mutant cells. The resultant 63 filtered genes were first evaluated using KM Plotter tool [23] (http://www.kmplot.com) for associations with recurrence free survival in ER+ breast cancer. 9 genes with HR values >1 and P values < 0.05 were identified with KM plotter (training set), and then validated as a gene signature using METABRIC dataset (1508 ER+ samples) [24]. We also evaluated site-specific breast cancer metastasis using EMC-MSK datasets (GSE2603, GSE5327, GSE2034 and GSE12276) [25]. Median cut-points were applied for all of the plots. Log-rank P values were calculated using “survdiff” command in the “survival” package of R. Kaplan-Meier curves were drawn with the “survfit” command in the same R package.
We compared the expression patterns of 68 mutant EMT related genes from YS1 ESR1 mutant cells, to express data from Affymetrix U133A microarrays of unmatched primary (n= 276) compared to metastases (n= 97) of ER/PR- positive, HER2 negative breast cancer samples from a MDACC cohort (GSE124647) [26]. Analyses are detailed in Supplemental Methods.
Statistical Analyses
There were three major types of statistical analysis used to analyze data for this study: those for the cell line experiments, for xenograft data and IncuCyte, and for survival analysis of breast tumor datasets; these are described in the Supplemental Methods.
Results
The Y537S ESR1 mutation enhances the frequency of distant metastasis in vivo
We observed differences in basal growth between ER-positive MCF-7 sublines; supplementary Fig. 1 shows a comparison of two MCF-7 sublines. The MCF-7 SF subline was more sensitive to E2 treatment compared with the MCF-7 BK subline in MTT growth assays (Supplementary Fig. 1A), and exhibited enhanced anchorage-independent growth (Supplementary Fig. 1B-D). Culturing cells as non-adherent mammospheres enriches for progenitor or tumorigenic cells [27]. We initiated tumor xenografts with both sublines, supplemented the animals with estrogen, and continued hormone treatments for one year. The MCF-7 SF subline exhibited more micrometastatic deposits to the lung (71%, 10/14 vs. 19%, 3/16 for MCF-7 BK; P= 0.0086, data not shown). We therefore used the parental MCF-7 SF subline (hereafter designated MCF-7) to develop CRISPR-Cas9 knock-in of two frequently-occurring ESR1 mutations, Y537S (CRISPR YS1 and YS30 clones), D538G (MCF-7 DG2) and T47D Y537S (YS53) (Supplementary Fig. 2A-D).
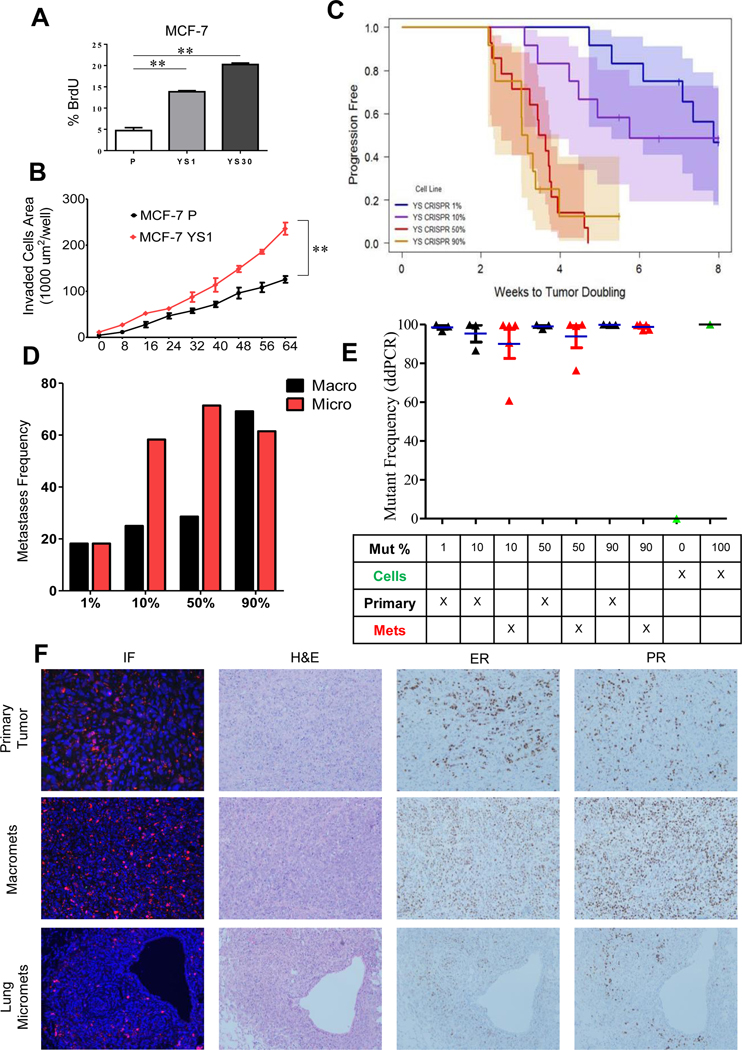
The MCF-7 Y537S CRISPR clones exhibited higher BrdU incorporation (Fig. 1A) consistent with a proliferative advantage in estrogen-free conditions. MCF-7 parental and YS1 mutant cells were tagged with GFP or mCherry, respectively. MCF-7 YS1 cells exhibited enhanced invasion in vitro compared with MCF-7 parental (Fig. 1B). We next mixed mutant cells at 1%, 10%, 50% or 90% with WT ESR1 cells before injection in mice. Tumors were allowed to take with E2 supplementation, and then switched to –E2 after reaching 250–350 mm3. The number of weeks to tumor doubling are shown in Fig. 1C. Multiple comparisons were performed to test significance among groups (Supplementary Table 1). Progression-free survival of the 1% and 10% mutant groups was similar to each other, but different from the 50% and 90% groups, which were shorter and similar to each other. All mice were harvested at 4 months, and macrometastasis to the adrenal glands and omentum, as well as lung micrometastases were observed. Metastatic frequencies were positively associated with the percentage of mutant cells injected suggesting that mutant cells enhanced distant metastasis (Fig. 1D, Chi-square test for trend P< 0.0001; metastatic frequencies are shown in Supplementary Table 2).
Fig. 1.
The Y537S ESR1 mutant becomes the predominant cell type in vivo. A. % BrdU incorporation was measured in MCF-7 parental (P), MCF-7 YS1 (YS1) and MCF-7 YS30 (YS-30) cells growing in the absence of E2. Data presented as mean ± SD in triplicate, with P values from the Student t test. **, P <0.01. B. IncuCyte invasion assay was performed in MCF-7 parental and YS1 mutant cells in triplicate in the absence of E2. Invaded cell surface area on the bottom membrane was calculated. Data presented as mean ± SD in triplicate with P values from two way ANOVA analysis. **, P <0.01. C. Primary tumor growth of MCF-7 parental and YS1 cells were analyzed (1%, 10%, 50%, and 90% mutant cells injected) using Kaplan-Meier progression-free survival curves. Mice were kept with E2 supplementation until tumor growth reached 250–350 mm3, and then switched to –E2 until the end of the study. Data are presented as tumor doubling estimated by the Kaplan-Meier method and compared using the Generalized Wilcoxon test for multiple comparisons (P values are shown in Supplementary Table. 1). D. Frequency of distant macrometastasis and lung micrometastasis per group is shown. Trend calculated using Chi-square test. *** P <0.001. E. ESR1 mutation frequency in primary and matched distant metastases (Met) between mixing groups was determined using ddPCR (n=3/group). Three mice per group were analyzed. Some mice had more than one macrometastasis (total number of tumor samples are labeled). There was insufficient tissue from the 1% metastasis group for analysis. MCF-7 parental and YS1 cells were used as negative and positive controls for ddPCR assays, respectively. F. Immunofluorescence staining was performed for GFP (parental) and mCherry-tagged (YS1) reporter plasmids to visualize ESR1 mutant or WT cells, respectively, within primary and metastatic tumors. Representative H&E, ER, and PR IHC staining of primary and metastatic tumors from Fig.1C are shown.
Mutant-specific droplet digital polymerase chain reaction (ddPCR) was used to determine the WT and mutant ESR1 allele frequencies in primary and metastatic tumors (Fig. 1E and F). Tumors exclusively contained ESR1 mutant cells when analyzed using ddPCR. Staining also confirmed the predominance of ER/PR-positive mutant cells (Fig. 1F; Allred scores for ER and PR staining are shown in Supplementary Table 2). These data, along with the enhanced cellular proliferation seen with BrdU staining (Fig. 1A), suggest that ESR1 mutant cells have a proliferative advantage in the absence of estrogen, and become the predominant cell type during tumor progression.
Induction of EMT in breast cancer cells with KI of the Y537S and D538G ESR1 mutations
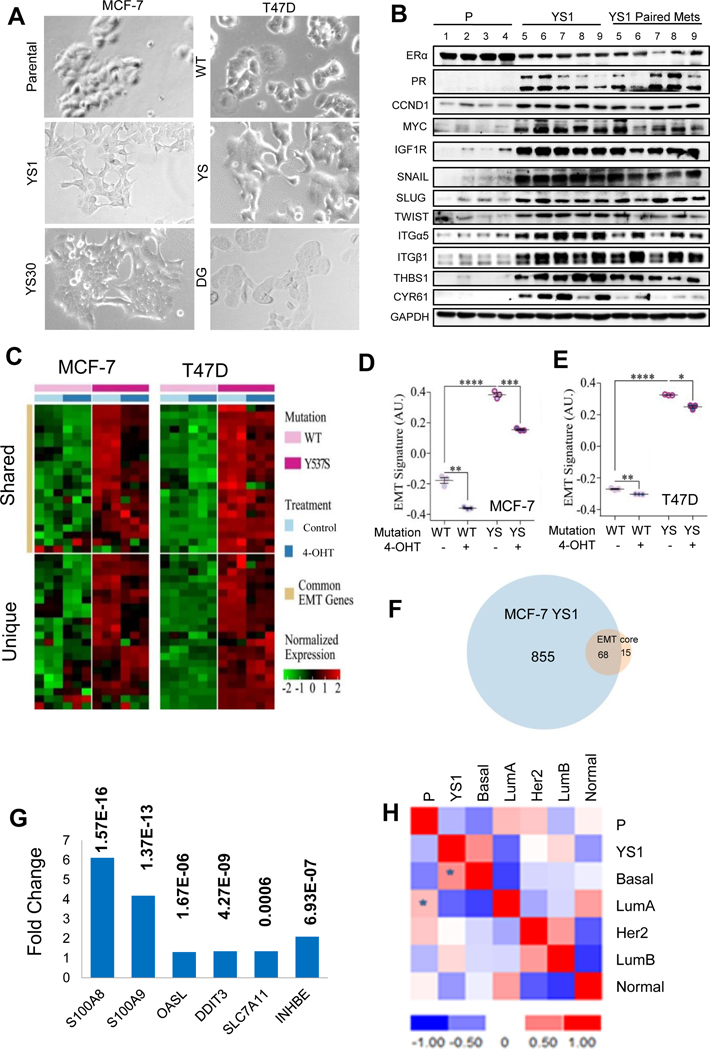
Both MCF-7 Y537S (CRISPR-Cas9) and T47D Y537S (Lentiviral) clones exhibited spindly fibroblast-like morphology characteristic of an EMT (Fig. 2A). As expected, we observed high constitutive ER transcription activity (Supplementary Figs 3A and B). Upregulation of SNAIL and other EMT proteins was observed in vitro in mutant models (Supplementary Fig. 3C). SNAIL expression is known to be estrogen-regulated in ovarian cancer [28]. MCF-7 parental and YS1 cells were grown as tumor xenografts in vivo; immunoblot analysis of showed enhanced levels of estrogen-regulated proteins such as PR, MYC, and CCND1 and a number of Hallmark EMT proteins including EMT transcription factors SNAIL, SLUG, TWIST, and other EMT Hallmark proteins Integrins α5 and β1 (ITGα5 and ITGß1), cysteine-rich angiogenic inducer 61 (CYR61) and thrombospondin (THBS1) in mutant tumors (Fig. 2B and Supplementary Fig. 4).
Fig. 2.
Y537S ESR1 mutants exhibit an EMT phenotype. A. Bright field images of ESR1 mutant MCF-7 and T47D cell lines growing in serum-containing medium. B. Immunoblot analysis of MCF-7 parental (n=4) and YS1 (n=5) primary /metastatic pairs growing after E2 withdrawal using indicated antibodies. C. Heat map representation of upregulated EMT genes expressed in common (upper panel) or unique (lower panel) in MCF-7 Y537S (YS1) and T47D Y537S cells compared to corresponding WT cells. D and E. EMT signature was shown from MCF-7 parental (WT) and YS1 (YS) or T47D Lenti WT and YS models. Cell were treated +/− 4-OHT for 24 hours in serum containing medium. The signature value is shown on the Y axis (AU arbitrary units): Data displayed as mean ± SD. Statistical significance calculated using Student t test, *, P <0.05 **, P <0.01 ***, P <0.001, ****, P <0.0001. F. Gene List Venn Diagram (http://genevenn.sourceforge.net/) was used to identify shared overexpressed genes in MCF-7 YS1 vs parental compared to the EMT core signature. G. Fold change average gene expression of six elevated genes in ER+/HER2- metastatic (n= 97) compared to primary (n= 276) breast cancers. P values are shown. Data analyses were performed in R version 3.5.1 and Bioconductor. Statistical significance was determined using the unpaired Student t-test. H, Gene expression of MCF-7 parental cells and MCF-7 YS1 cells grown in 5% CSS without E2 summarized in a correlation matrix were patterned with all five PAM50 centroids. Data calculated by Spearman’s Rank, with highest positive correlations denoted by *.
MCF-7 YS1 and T47D Y537S Lenti cells were treated with 4-OHT and RNA expression profiling was used to ER regulated expression. Gene set enrichment analysis (GSEA) was used to identify biological pathways associated with mutant expression, and the Molecular Signatures Database (MSigDB) repository was used to reveal Hallmark pathways (Supplementary Tables 3 and 4) and enriched KEGG terms (Supplementary Table 5) in mutant cells. The two models exhibited shared and unique EMT gene signatures that were decreased with 4-OHT treatment (Fig. 2C-E). Approximately half of the upregulated EMT genes were shared (Supplementary Table 6). Given 4-OHT’s effect in reducing EMT signature expression, and the shared upregulation of EMT genes in the Y537S models, mutant ESR1 may function as a master regulator of EMT transcription factors.
Genetic modulation of EMT-inducing transcription factors in ER-negative cell lines identified a core EMT signature that was significantly associated with the claudin-low/metaplastic subtypes of ER-negative tumors [29]. We compared genes differentially elevated in MCF-7 YS1 cells with the EMT core signature, and saw an overlap of 68 genes (Fig. 2F and Supplementary Table 7). We next explored the relevance of these 68 mutant EMT related genes in two MD Anderson Cancer Center breast cancer patient cohorts. Six genes (S100A8, S100A9, OASL, DDIT3, SLC7A11 and INHBE) were enriched in metastatic patients compared with primary breast cancers, suggesting that ESR1 mutant cells increase expression of selected metastatic genes (Fig. 2G).
These results demonstrate that Y537S ESR1 mutant cells exhibit a more “basal-like” phenotype. Indeed using centroid Spearman’s correlations with the PAM50-based subtype classifier [30], the highest positive correlation of the MCF-7 YS1 differential gene expression was with the basal subtype (Fig. 2H).
Tam treatment blocks distant metastasis of Y537S ESR1 mutant tumors
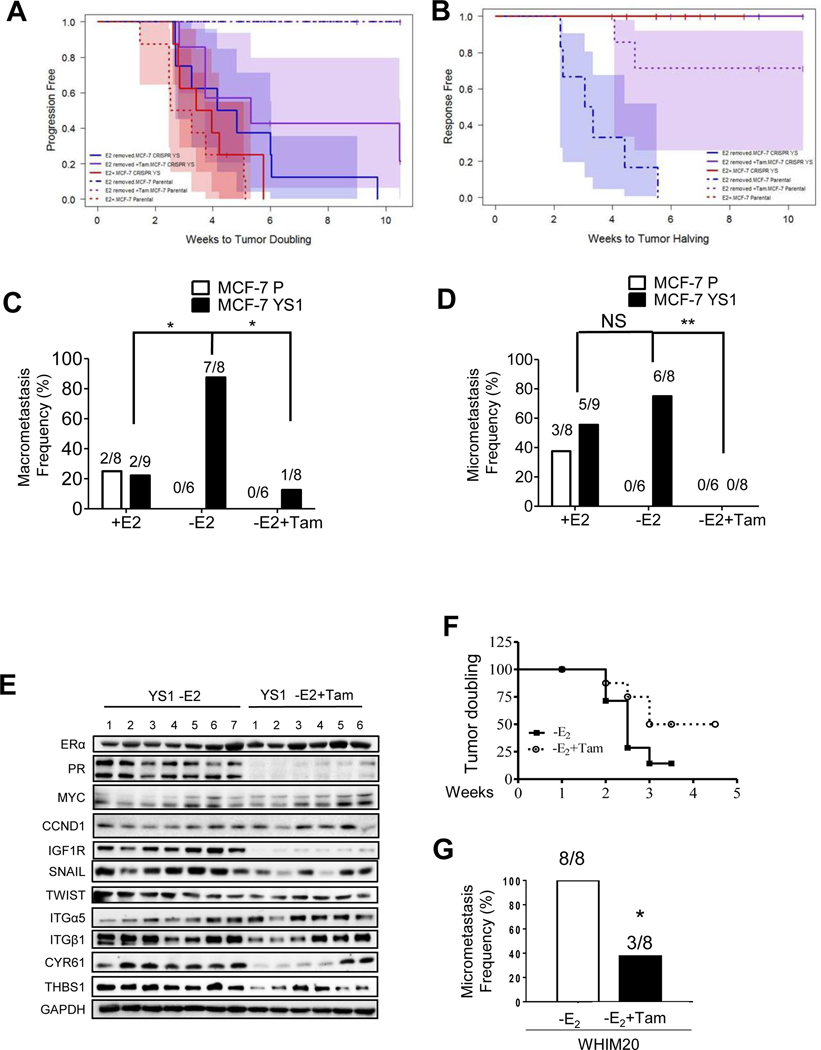
We next initiated an in vivo Tam treatment experiment; after tumor initiation using E2 supplementation, primary tumors were randomized to three treatment arms: E2, E2withdrawal, or withdrawal plus Tam. Growth curves are plotted by cell line for each animal and tumor size at baseline varied considerably. There was no difference in tumor size overall (P= 0.47 Kruskal-Wallis test). ER was expressed in all tumors (Supplementary Table 8). Estrogen-induced PR was observed in parental tumors, and lower PR levels were seen in the estrogen-withdrawal group. PR was constitutively higher, and Tam blocked PR expression in mutant tumors.
The primary outcomes were time to tumor doubling and time to response (tumor halving) by cell line and treatment. There was a difference in time to tumor doubling, with YS1 having a significantly faster time in both estrogen withdrawal (P=0.001) and Tam groups (P=0.008), but not the estrogen treated group (P=0.2) (Fig. 3A and Supplementary Table 9). In spite of estrogen withdrawal or Tam, the ESR1 mutant tumors grew. Time to treatment response (tumor halving) was different, with parental tumors exhibiting shorter time to response compared to mutant tumors (P<0.0001) (Fig. 3B and Supplementary Table 10). These results suggest that as expected from in vitro growth studies [9], the mutation rendered primary tumor xenografts resistant to the growth inhibitory effects of E2 withdrawal and Tam.
Fig. 3.
Tam treatment blocked metastasis in ESR1 mutant tumors. A and B. MCF-7 parental and YS1 mutant cells were injected in nude mice supplemented in E2 and randomized to three treatment armsto continue E2 (+E2), E2 removal (-E2) or E2 removal +Tam (-E2+Tam). Time to tumor doubling or halving are shown in A and B respectively using Kaplan-Meier method (P values showed in Supplementary Table 9 and 10). C and D. Number of mice that developed macrometastasis or lung micrometastasis in each treatment arm are shown. Data were analyzed using chi-square and Fisher’s exact test. * P <0.05, ** P <0.01, NS= no significant. E. Immunoblot analysis of primary tumors from MCF-7 YS1 mutant tumors treated with E2 withdrawal (-E2) (N=7) or -E2 +Tam (N=6). F. The frequency of lung micrometastasis were evaluated in WHIM20 PDX models grown in absence of E2 (-E2) or -E2+Tam. Time to tumor doubling was calculated using Kaplan-Meier method and analyzed using Log-rank (Mantel-Cox) Test. P=0.0978. G. After 5 months, mice were harvested and the frequency of lung micrometastasis was analyzed using chi-square and Fisher’s exact test. * P <0.05.
At 4 months mice developed aggressive distant macrometastases. There was a significant difference in the probability of macrometastasis between groups (P<0.001, Fisher’s exact test). E2 was required for macrometastasis of parental cells, and both exhibited similar metastatic frequencies with E2 (Fig. 3C). E2 withdrawal enhanced, and Tam treatment reduced metastatic frequency in mutant tumors. We observed E2-independent metastasis in vivo in other mutant clones (Supplementary Table 8); MCF-7 YS30 (50% metastasis frequency), MCF-7 DG2 (30%), and T47D Y537S (71.4%) These models validate the hypothesis that Y537S and D538G mutant cells have a metastatic advantage in the absence of E2. E2 was required for MCF-7 parental cells to metastasize to the lung; however, E2 withdrawal did not significantly increase lung micrometastases in ESR1 mutant mice (Fig. 3D). Although ER and PR expression was present in all MCF-7 macrometastases (Supplementary Fig. 5A), frequent ER-negative lung micrometastases were observed in ESR1 mutant tumors (Supplementary Table 8). Tam treatment reduced expression of estrogen-regulated proteins, and EMT expression in MCF-7 YS1 primary tumors (Fig. 3E and Supplementary Fig. 5B). Tam treatment significantly reduced distant metastatic frequency, but not growth in the mammary fat pad environment. We obtained similar results in the WHIM20 Y537S patient derived xenograft (PDX) model where Tam did not affect primary tumor growth (Fig. 3F, P value 0.0978), but blocked both lung macrometastases (Supplementary Fig 6A) and micrometastases (3G, * P value < 0.05; Supplementary Fig. 6B and C) [31]. These results demonstrate that Tam treatment is effective at reducing metastatic ESR1 mutant tumor burden in different genetic backgrounds.
9 gene signature predicts poor disease-free survival and metastatic outcomes in ER-positive patients
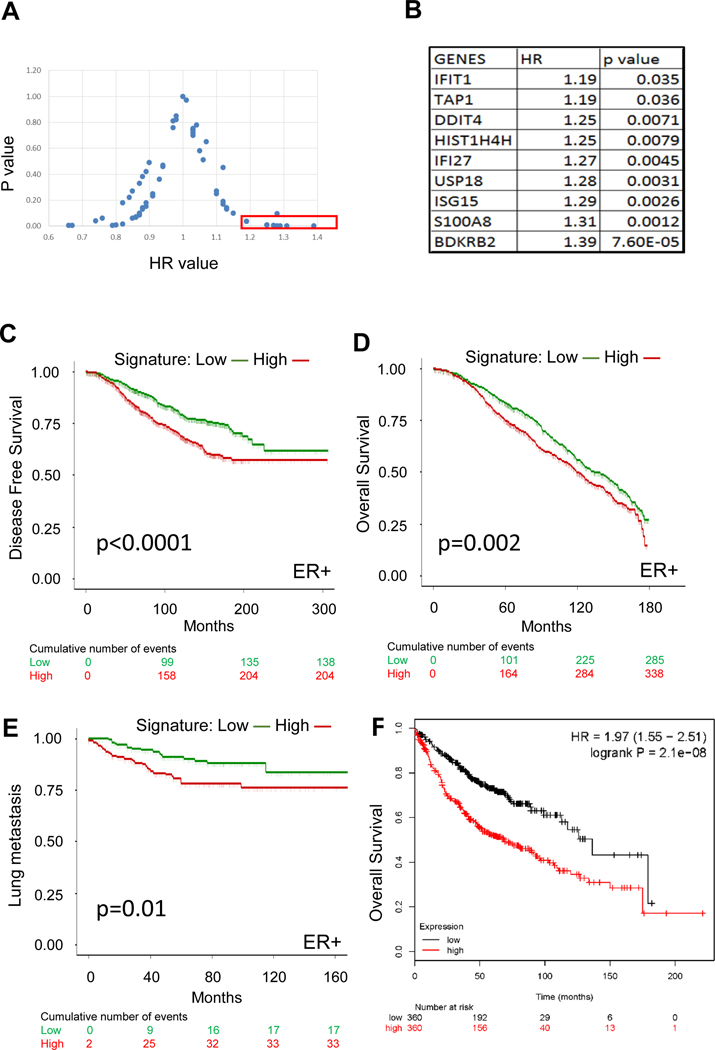
Gene expression signatures, such as the Oncotype Dx Breast Recurrence Score, are utilized for the clinical management of breast cancer to predict patient outcomes [32], and most of these signatures are related to cell proliferation. To determine whether genes differentially up-regulated in Y537S ESR1 mutant cells were associated with patient outcome, we identified a filtered 63 gene list and evaluated their associations with outcome using KM Plotter [23] (Fig. 4A). Nine genes associated with poor outcomes (HR>1 and P value< 0.05) are shown in Fig. 4B. This 9 gene signature contained 5 interferon α/γ response Hallmark pathway genes, suggesting an important role for cytokine signaling, as well as genes known to be involved in invasion and metastasis [33–37]. We then validated the association of this signature with ER-positive patient outcomes in METABRIC (Figs. 4C and D) [24]. We also evaluated the signature in a metastatic cohort from EMC-MSK; patients identified as signature “high” exhibited shorter lung-specific metastasis-free survival in ER-positive, but not ER-negative patients [25] (Fig. 4E and Supplementary Fig. 7, respectively); the signature did not predict bone or brain-specific breast cancer metastasis (data not shown). All of the Y537S ESR1 mutant models frequently metastasized to the lung, and the 9 gene signature was also associated with overall survival of lung adenocarcinoma using KM plotter (Fig. 4F). Thus, this poor outcome signature may consist of genes important for survival across tumor types.
Fig. 4.
The Y537S ESR1 mutant signature is prognostic of patient outcomes. A. The expression of 63 genes was evaluated for recurrence free survival using KM Plotter in ER-positive breast cancer patients. HR and P values for each gene are represented as a scatter plot. B. 9 genes from panel A with HR value >1 and p value <0.05 are shown. C and D. The predictive value of the 9 gene signature was evaluated for disease free (P value =1.97e−6, HR=1.691 (1.362–2.1)) and overall survival (P value =7.1e−4, HR=1.302 (1.117–1.617)) in ER-positive patients from the METABRIC dataset using Kaplan-Meier analysis [24]. E. Kaplan-Meier analysis were performed using the MCF-7 Y537S 9-gene signature in EMC-MSK breast cancer datasets [25]. Lung metastases as first site of metastasis were considered events. P value =0.0096, HR =2.128 (1.185–3.822) for ER-positive patient cohort. F. KM Plotter was used to correlate the 9 gene expression signature with patient overall survival using a lung adenocarcinoma cancer patient cohort [49]. Patients were separated into 2 groups (median cut off) based on their average expression of the 9 genes. HR was calculated to be 1.97 (1.55–2.51) with a significant P value of 2.1e-8. Low versus high signature stratification was calculated using sum of z-scores method. Statistical significance was calculated by log-rank test in Kaplan-Meier analyses. Hazard Ratios was calculated using Cox Proportional-Hazards model noted above with 95% confidence interval.
The AR agonist DHT inhibited metastasis in the WHIM20 PDX model
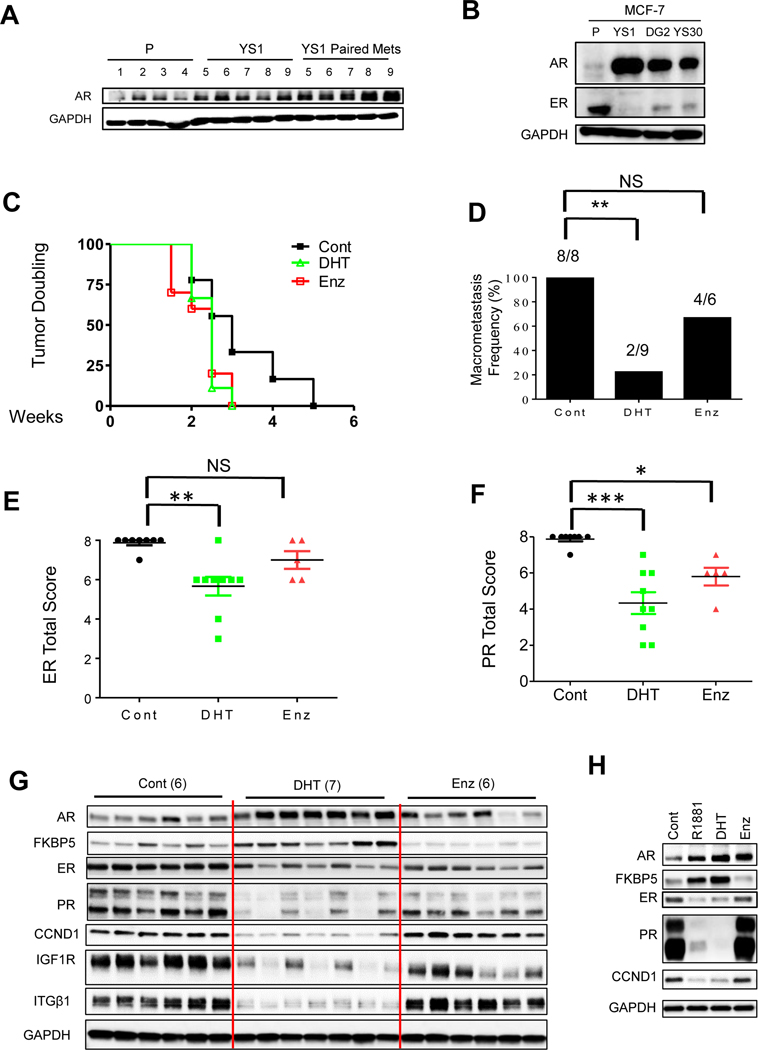
AR expression is elevated in Tam-resistant metastatic patients with WT ER, and AR overexpression confers resistance to both Tam and AI through cross-talk with the insulin like growth factor one receptor (IGF1R) [14, 16]. Recently we showed that IGF1R activation was a determining factor of response to Tam in Y537N, Y537S, and D538G ESR1 mutant models [9]. High levels of IGF1R and AR were observed in YS-1 primary xenograft and paired metastases (Fig. 2B and 5A), and ESR1 mutant CRISPR models expressed high levels of AR (Fig. 5B).
Fig. 5.
AR agonist treatment blocked Y537S mutant metastasis. A. Immunoblot analysis of MCF-7 parental (n=4) and YS1 (n=5) primary /metastatic pairs grown with E2 withdrawal. B. Western blot analysis of MCF-7 parental, YS1, DG, and YS30 clones cultured in 5% CSS media for 4 days GAPDH was used as loading control. C. Mice with WHIM20 PDX tumors were supplemented with E2, and randomized to –E2, -E2+DHT or –E2+Enz for total of 4 months. Time to tumor doubling was calculated using Kaplan-Meier method. Data were analyzed using Log-rank (Mantel-Cox) Test. P= 0.10 for DHT vs vehicle control and P= 0.07 for MDV vs vehicle control. D. Macrometastasis frequency was calculated and displayed chi-square and fisher exact analyses, * P =0.002. E and F. ER and PR total score IHC staining two-sided Student’s t-test was performed to test significance. *P <0.05, **P <0.01,* P <0.001, NS=Not significant. G. Western blot analyses of WHIM20 primary tumors; GAPDH was used as a loading control. H. Western blot analyses of MCF-7 YS1 cells; performed using in vitro cell line protein lysis and GAPDH was used as a loading control. Cells were starved for 48 hours in 5% CSS, and then treated with hormones for 24 hours.
We investigated the utility of AR targeted therapy using the WHIM20 Y537S ESR1 mutant PDX. AR selective modulators (SARMs) can inhibit WT or ESR1 mutant primary PDX tumor growth by modulating ER binding to its cistrome [38]. No significant differences were observed in primary tumor growth with SARM treatments, measured as time to tumor doubling in the WHIM20 ESR1 Y537S mutant PDX model (Fig.5C). All control WHIM20 primary tumors exhibited distant macrometastases (Fig. 5D). Treatment with the AR antagonist Enz numerically reduced, but the AR agonist DHT significantly reduced metastatic frequency (Fig. 5D), suggesting an unexpected hormonal therapeutic vulnerability of ESR1 mutant metastasis with DHT treatment. DHT significantly reduced ER and PR staining, while Enz treatment reduced PR staining in tumors (Fig. 5E, F). ER protein levels and estrogen–regulated protein expression (PR, CCND1, IGF1R) were more reduced with DHT treatment in WHIM20 primary tumors (Fig. 5G). AR levels and its downstream target FKBP5 were increased with agonist treatments in cells (Fig. 5H). These data suggested a novel molecular mechanism whereby AR agonist treatment leads to reduced mutant ER protein, in addition to its known effects on modulating the ER cistrome in the presence of estrogen [38].
DHT treatment blocks EMT gene expression via reduction in ER chromatin binding.
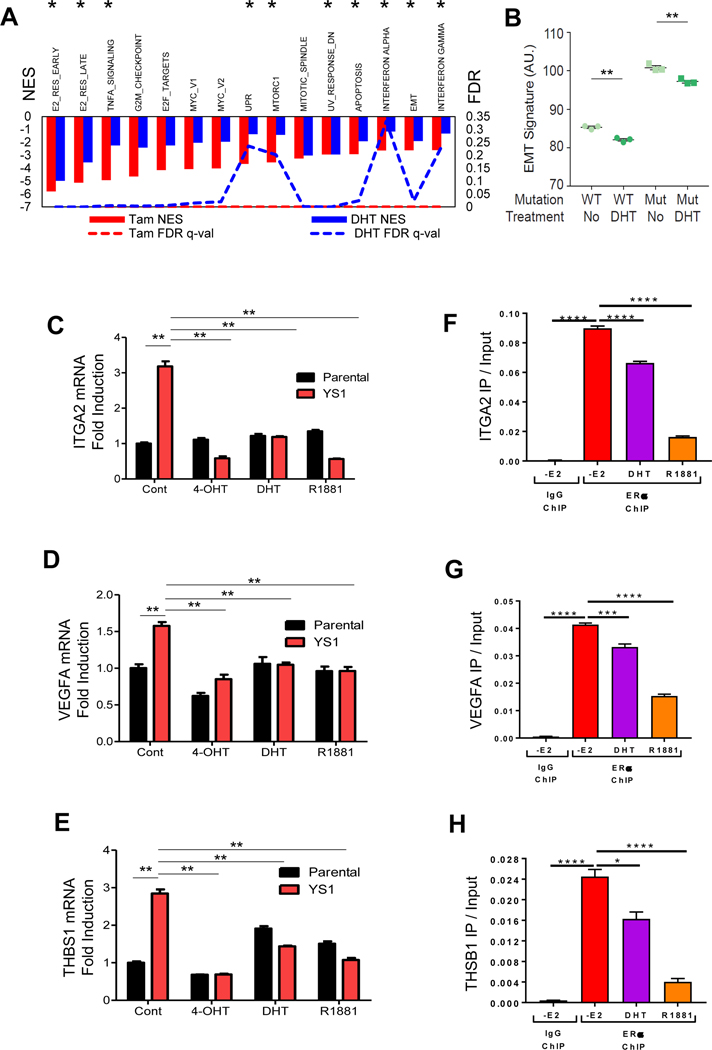
We observed that Tam and DHT treatment significantly reduced distant metastasis. We next evaluated their effects at the genomic level. Both 4-OHT and DHT down-regulated 10 of the top 15 Hallmark pathways increased in MCF-7 YS1 mutant cells (Fig. 6A). 4-OHT (as shown in Fig.2D and 2E), and DHT treatment reduced expression levels of the mutant EMT signature (Fig. 6B). Representative EMT Hallmark genes, ITGA2, VEGFA and THBS1, were used to validate gene expression by qRT-PCR and ChIP-PCR. Expression of these genes was upregulated in mutant cells with 4-OHT, DHT and R1881 reducing their gene expression (Figs. 6C-E). In addition, DHT and R1881 treatment reduced the binding of mutant ER to the transcription regulatory regions of these three EMT genes (Fig. 6F-H). These data suggest that AR activation may inhibit metastasis via a reduction in ER mutant protein binding to affect the EMT transcriptional program.
Fig. 6.
DHT treatment regulates mutant EMT gene expression through the ER cistrome. A. MCF-7 YS1 cells were grown in 5% CSS medium +/− DHT, or +/− 4-OHT in serum-containing media for 24 hours. Microarray analyses were performed and Hallmark pathways were identified. Hallmarks labeled with * were pathways constitutively upregulated in YS1 mutant cells (Supplementary Table 3). B. MCF-7 parental and YS1 cells were treated in 5% CSS medium +/− DHT and an EMT gene signature value was calculated (based on shared 21 EMT genes shown in Supplementary Table 6). DHT vs control P values were 0.0017 and 0.0052 for parental (WT) and YS1 (Mut) respectively. Data were analyzed using Student t test. C-E. MCF-7 YS1 cells were treated with either 4-OHT, DHT or R1881 in 5% CSS media for 72 hours. ITGA2, THBS1 and VEGFA mRNA expression was measured using qRT-PCR (Primers shown in Table 1) and normalized against 18S expression. Data were analyzed using Student t test. F-H. Binding of ER to EMT gene regulatory regions was determined using ChIP-PCR in YS1 cells treated with 100 nM 4-OHT, 10 nM DHT, or 10 nM R1881 for 4 hours (Primers shown in Table 2). Error bars represent the standard error of the mean (n = 3 technical replicates). Each set of technical replicates were independently validated with a second experiment (not shown). Data were analyzed by one-way ANOVA with Holm-Sidak Multiple comparisons test. ** P<0.01. * P<0.05, ** P<0.01, *** P<0.001, **** P< 0.0001.
DISCUSSION
One salient result of this study was the potential subclonal growth advantage of mutant cells in the presence of tumor cells expressing WT ER during estrogen withdrawal. ESR1 mutant tumor cells became the dominant cell type in the metastatic tumors, potentially out-competing WT cells in the absence of estrogen. These data also suggest that ESR1 mutant cells undergo rapid clonal expansion. Our results are consistent with the endogenous acquisition of mutations when WT ER-positive cells are grown in long-term estrogen deprivation conditions in vitro [39]. It has been hypothesized that the fittest tumor clone eventually dominates. Therefore, as we originally proposed [6], an estrogen-independent clone with aggressive potential would be an advantageous mutation. Reports of the emergence of polyclonal ESR1 mutations in clinical studies support our findings [12]. Estrogen removal (simulating treatment with an AI) imposes a selective pressure on cell subpopulations, thereby reducing the numbers of drug-sensitive (WT ER) tumor cell clones, but also selecting for endocrine-resistant (ESR1 mutant) subclones.
When Y537S ESR1 mutant cells were grown as primary tumor xenografts, estrogen withdrawal and Tam treatment did not reduce tumor growth; whereas parental primary tumors significantly responded to both treatments. In the presence of estrogen, there were no differences in metastatic frequency between the tumor groups, but a remarkably high rate of spontaneous distant metastasis was observed in mutant tumors when estrogen was withdrawn. We do not yet understand the mechanism behind the observation that estrogen treatment may negatively affect mutant’s ability to metastasize. One unanticipated result was the significant reduction in metastases observed in mutant xenograft and PDX models with Tam treatment. Since Tam did not affect primary ESR1 mutant tumor growth, reduction in distant metastases was not simply due to inhibition of proliferation in the primary tumor. Our data suggest that ESR1 mutations facilitate distant metastatic growth of ER-positive tumors, perhaps through remodeling of an EMT program that can unexpectedly be blocked by the use of Tam.
The role of EMT in ER-positive MBC progression has not been clearly defined. We observed an increase in Hallmark EMT programs in ESR1 mutant cells, and 4-OHT significantly inhibited EMT gene expression. We therefore hypothesize that EMT may play a role in the aggressive metastatic phenotype of ESR1 mutations. ER-positive metastatic clones may utilize an early EMT program to facilitate dissemination, as has been demonstrated in ER-negative basal breast cancer [40]. The role of EMT in endocrine therapy resistance is underexplored. Proteins involved in the transcriptional regulation of EMT include SNAIL, SLUG, TWIST, and ZEB1/2 [31], and significant elevated levels of SNAIL and TWIST were seen in mutant tumors. SNAIL or SLUG overexpression in ER-positive breast cancer cells can also lead to a loss of ER mRNA and protein [41, 42]. Thus, it will be important in future experiments to determine whether SNAIL upregulation could be involved in the evolution of some ER-negative lung metastases that were observed in our studies, and whether mutant ER could be a master regulator of EMT transcription factors.
The basal subtype of breast cancer displays a tropism for lung metastasis [43]. Y537S ESR1 mutant cells became basal-like using PAM50 classification, and all Y537S mutant models frequently metastasized to the lung. The 9 gene signature was predictive of breast cancer metastasis to the lung and lung adenocarcinoma overall survival, suggesting that these genes may underlie common oncogenic driver pathways. Several of the genes comprising the mutant signature, S100A8, ISG15, IFIT1, IFI27, and BDKRB2, are reported to be involved in invasion or metastasis [33–37]. Future investigations of the specific role of these genes in ESR1 mutant metastasis are warranted.
Our results suggest that one new therapeutic vulnerability of ESR1 mutant tumors maybe the use of AR agonists to selectively block metastasis. Moderate clinical benefit was recently reported for the combination of an AR antagonist Enz combined with the AI exemestane in a Phase II study of AR/ER-positive MBC patients, thus demonstrating the clinical potential of targeting AR in the appropriate biomarker-positive patient population [44]. AR protein was elevated in ESR1 mutant models compared with WT cells. Elevated AR levels are a known mechanism of endocrine therapy resistance in ER-positive metastatic breast cancer, along with activation of growth factor receptor signaling [15]. A long-standing conundrum in the AR field is the question of whether AR is a driver or a tumor suppressor in breast cancer (reviewed in [45]). ER-positive patients with an elevated AR/ER ratio experience a shorter recurrence-free interval [46], and exhibit aggressive biological features [47]. Our previous results suggest that AR collaborates with WT ER to mediate resistance to an AI [16], and here we demonstrate that AR plays a role in ESR1 mutant-mediated metastasis. A clinical question is how best to utilize AR targeting agents. The clinical utility of newer selective AR modulators [48] in combination with endocrine agents, or in combination with agents blocking mutant-mediated EMT expression in advanced breast cancer should be explored. The use of preclinical ER-positive models with efficient spontaneous metastasis in vivo should greatly improve our ability to predict the clinical utility of new targeted biotherapeutics in MBC.
Supplementary Material
Table 1.
Probe set for qPCR
| Gene name | BIO-RAD primers set ID |
|---|---|
| ITGA2 | ITGA2 SYBR Green assay qHsaCID0016134 |
| VEGFA | VEGFA SYBR Green assay qHsaCED0043454 |
| THBS1 | THBS1 SYBR Green assay qHsaCED0044787 |
Table 2.
Primer sequences for ChIP-PCR
| Gene and amplicon location | Primer sequences |
|---|---|
| ITGA2 (3’ Enhancer) hg19 chr5:52386157–52386278 | Forward 5’-TGCAGCTGAGCTCTCTGGATATAA-3’ |
| Reverse 3’-CTGCCCACGTCCTCTGATGC-5’ | |
| VEGFA (5’ Enhancer) hg19 chr6:43682221–43682320 | Forward 5’-TGCCCTCTTTCCACTACACC-3’ |
| Reverse 3’-TGTTTCTGCTGGCAAAGATCT-5’ | |
| THBS1 (5’ Enhancer) hg19 chr15:39826564–39826638 | Forward 5’-CCTTCCTGGTCTTCTTTGTCTG-3’ |
| Reverse 3’-GTTGAAAGACCAGGGGCACT-5’ |
Acknowledgments
The authors would like to thank Drs. Chandandeep Nagi for his pathology expertise, Sasha Pejerrey for initial edits of the manuscript. We acknowledge the joint participation with the Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with BCM and the Cancer Program. We acknowledge the joint collaboration with Dr. Symmans through the CPRIT MIRA grant mechanism.
Grant Support
Breast Cancer Research Foundation BCRF 19-055, NIH R01 CA207270, NIH R01CA072038, Cancer Prevention Institute of Texas CPRIT MIRA (Kelly Hunt, PI) to SAWF; Adrienne Helis Malvin Medical Research Foundation Cancer Program M-2017 to Charles E. Foulds; P30 CA125123-05 Dan L Duncan Cancer Center Cell-based assay screening core; Translational Breast Cancer Research Training Program T32-CA203690-02; Predoctoral Institutional Research Training Grant (T32) Training Grant 5T32GM008231-30; T32 2389301302, Cancer Prevention Institute of Texas (CPRIT) RP170005; NIEHS grants P30 ES030285, P42 ES027725; U01 CA217842, Susan G. Komen Breast Cancer Foundation SAC110052, Breast Cancer Research Foundation BCRF-19-110
Footnotes
Disclosure of Potential Conflicts of Interest
1. Dr. Charles E. Foulds discloses: an equity position in Coactigon, Inc.
2. Dr. Filip Janku discloses:
a) Grant/Research Funding (Institutional): Novartis, Genentech, BioMed Valley Discoveries, Plexxikon, Deciphera, Piqur, Symphogen, Bayer, FujiFilm Corporation and Upsher-Smith Laboratories, Astex, Asana, Astellas, Agios, Proximagen, Bristol-Myers Squibb;
b) Scientific Advisory Board: Deciphera, IFM Therapeutics, Synlogic, Guardant Health, Ideaya, PureTech Health;
c) Paid Consultant: Trovagene, Immunomet, Jazz Pharmaceuticals, Sotio;
d) Ownership Interests: Trovagene.
3. Dr. Gordon B. Mills discloses:
a) SAB/Consultant: AstraZeneca, Chrysallis Biotechnology, GSK, ImmunoMET, Ionis, Lilly, PDX Pharmaceuticals, Signalchem Lifesciences, Symphogen, Tarveda, Turbine, Zentalis Pharmaceuticals
b) Stock/Options/Financial: Catena Pharmaceuticals, ImmunoMet, SignalChem, Tarveda;
c) Licensed Technology: HRD assay to Myriad Genetics, DSP patents with Nanostring
4. Dr. Shunqiang Li discloses:
a) The Washington Unversity PDX development and trial center is supported by NIH 3U54CA224083-02S3.
b) Dr. Shunqiang Li has received license fee from Envigo. He received research funding from Pfizer, Takeda Oncology, and Zenopharm, outside of this project.
References
- 1.N.A. Howlader N, Krapcho M, Miller D, Bishop K, Altekruse SF, et al. , SEER Cancer Statistics Review, 1975–2013, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2013/, based on November 2015 SEER data submission, posted to the SEER web site, April 2016. [Google Scholar]
- 2.Klein CA, Selection and adaptation during metastatic cancer progression. Nature, 2013. 501(7467): p. 365–72. [DOI] [PubMed] [Google Scholar]
- 3.Fuqua SA, The role of estrogen receptors in breast cancer metastasis. J Mammary Gland Biol Neoplasia, 2001. 6(4): p. 407–17. [DOI] [PubMed] [Google Scholar]
- 4.Herynk MH and Fuqua SA, Estrogen receptor mutations in human disease. Endocr Rev, 2004. 25(6): p. 869–98. [DOI] [PubMed] [Google Scholar]
- 5.Jeselsohn R, Bergholz JS, Pun M, Cornwell M, Liu W, Nardone A, et al. , Allele-Specific Chromatin Recruitment and Therapeutic Vulnerabilities of ESR1 Activating Mutations. Cancer Cell, 2018. 33(2): p. 173–186 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang QX, Borg A, Wolf DM, Oesterreich S, and Fuqua SA, An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res, 1997. 57(7): p. 1244–9. [PubMed] [Google Scholar]
- 7.Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. , The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell, 2018. 34(3): p. 427–438 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuqua SA, Gu G, and Rechoum Y, Estrogen receptor (ER) alpha mutations in breast cancer: hidden in plain sight. Breast Cancer Res Treat, 2014. 144(1): p. 11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelsomino L, Gu G, Rechoum Y, Beyer AR, Pejerrey SM, Tsimelzon A, et al. , ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res Treat, 2016. 157(2): p. 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Inao T, Sueta A, Fujiwara S, et al. , Droplet digital polymerase chain reaction assay for screening of ESR1 mutations in 325 breast cancer specimens. Transl Res, 2015. 166(6): p. 540–553 e2. [DOI] [PubMed] [Google Scholar]
- 11.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, et al. , Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med, 2015. 7(313): p. 313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, et al. , The Genetic Landscape and Clonal Evolution of Breast Cancer Resistance to Palbociclib plus Fulvestrant in the PALOMA-3 Trial. Cancer Discov, 2018. 8(11): p. 1390–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, et al. , Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol, 2016. 2(10): p. 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Amicis F, Thirugnansampanthan J, Cui Y, Selever J, Beyer A, Parra I, et al. , Androgen receptor overexpression induces tamoxifen resistance in human breast cancer cells. Breast Cancer Res Treat, 2010. 121(1): p. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciupek A, Rechoum Y, Gu G, Gelsomino L, Beyer AR, Brusco L, et al. , Androgen receptor promotes tamoxifen agonist activity by activation of EGFR in ERalpha-positive breast cancer. Breast Cancer Res Treat, 2015. 154(2): p. 225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rechoum Y, Rovito D, Iacopetta D, Barone I, Ando S, Weigel NL, et al. , AR collaborates with ERalpha in aromatase inhibitor-resistant breast cancer. Breast Cancer Res Treat, 2014. 147(3): p. 473–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JA, Tan Y, Wang X, Cao X, Veeraraghavan J, Liang Y, et al. , Comprehensive functional analysis of the tousled-like kinase 2 frequently amplified in aggressive luminal breast cancers. Nat Commun, 2016. 7: p. 12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barone I, Brusco L, Gu G, Selever J, Beyer A, Covington KR, et al. , Loss of Rho GDIalpha and resistance to tamoxifen via effects on estrogen receptor alpha. J Natl Cancer Inst, 2011. 103(7): p. 538–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gates LA, Gu G, Chen Y, Rohira AD, Lei JT, Hamilton RA, et al. , Proteomic profiling identifies key coactivators utilized by mutant ERalpha proteins as potential new therapeutic targets. Oncogene, 2018. 37(33): p. 4581–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F, Genome engineering using the CRISPR-Cas9 system. Nat Protoc, 2013. 8(11): p. 2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kollipara RK and Kittler R, An integrated functional genomic analysis identifies the antitumorigenic mechanism of action for PPARgamma in lung cancer cells. Genom Data, 2015. 3: p. 80–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert JR and Nordeen SK, Analysis of Steroid Hormone-Induced Histone Acetylation by Chromatin Immunoprecipitation Assay, in Steroid Receptor Methods: Protocols and Assays, Lieberman BA, Editor. 2001, Humana Press: Totowa, NJ. p. 273–281. [DOI] [PubMed] [Google Scholar]
- 23.Mihaly Z, Kormos M, Lanczky A, Dank M, Budczies J, Szasz MA, et al. , A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res Treat, 2013. 140(2): p. 219–32. [DOI] [PubMed] [Google Scholar]
- 24.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. , The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature, 2012. 486(7403): p. 346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. , Genes that mediate breast cancer metastasis to the brain. Nature, 2009. 459(7249): p. 1005–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinn BV, Fu C, Lau R, Litton J, Tsai TH, Murthy R, et al. , SETER/PR: a robust 18-gene predictor for sensitivity to endocrine therapy for metastatic breast cancer. NPJ Breast Cancer, 2019. 5: p. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selever J, Gu G, Lewis MT, Beyer A, Herynk MH, Covington KR, et al. , Dicer-mediated upregulation of BCRP confers tamoxifen resistance in human breast cancer cells. Clinical cancer research : an official journal of the American Association for Cancer Research, 2011. 17(20): p. 6510–21 PMCID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SH, Cheung LW, Wong AS, and Leung PC, Estrogen regulates Snail and Slug in the down-regulation of E-cadherin and induces metastatic potential of ovarian cancer cells through estrogen receptor alpha. Mol Endocrinol, 2008. 22(9): p. 2085–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. , Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A, 2010. 107(35): p. 15449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, et al. , Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics, 2015. 8: p. 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, et al. , Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep, 2013. 4(6): p. 1116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, et al. , Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol, 2016. 34(10): p. 1134–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pidugu VK, Wu MM, Yen AH, Pidugu HB, Chang KW, Liu CJ, et al. , IFIT1 and IFIT3 promote oral squamous cell carcinoma metastasis and contribute to the anti-tumor effect of gefitinib via enhancing p-EGFR recycling. Oncogene, 2019. 38(17): p. 3232–3247. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Wang J, Zhang H, Zhu M, Chen F, Hu Y, et al. , Interferon-stimulated gene 15 (ISG15) is a trigger for tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget, 2014. 5(18): p. 8429–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiratsuka S, Watanabe A, Sakurai Y, Akashi-Takamura S, Ishibashi S, Miyake K, et al. , The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nat Cell Biol, 2008. 10(11): p. 1349–55. [DOI] [PubMed] [Google Scholar]
- 36.Chiang KC, Huang ST, Wu RC, Huang SC, Yeh TS, Chen MH, et al. , Interferon alpha-inducible protein 27 is an oncogene and highly expressed in cholangiocarcinoma patients with poor survival. Cancer Manag Res, 2019. 11: p. 1893–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang J, Liu W, Guo X, Zhang R, Zhi Q, Ji J, et al. , IRX1 influences peritoneal spreading and metastasis via inhibiting BDKRB2-dependent neovascularization on gastric cancer. Oncogene, 2011. 30(44): p. 4498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ponnusamy S, Asemota S, Schwartzberg LS, Guestini F, McNamara KM, Pierobon M, et al. , Androgen Receptor Is a Non-canonical Inhibitor of Wild-Type and Mutant Estrogen Receptors in Hormone Receptor-Positive Breast Cancers. iScience, 2019. 21: p. 341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin LA, Ribas R, Simigdala N, Schuster E, Pancholi S, Tenev T, et al. , Discovery of naturally occurring ESR1 mutations in breast cancer cell lines modelling endocrine resistance. Nat Commun, 2017. 8(1): p. 1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Felipe Lima J, Nofech-Mozes S, Bayani J, and Bartlett JM, EMT in Breast Carcinoma-A Review. J Clin Med, 2016. 5(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhasarathy A, Kajita M, and Wade PA, The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol Endocrinol, 2007. 21(12): p. 2907–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Wu Y, Abbatiello TC, Wu WL, Kim JR, Sarkissyan M, et al. , Slug contributes to cancer progression by direct regulation of ERalpha signaling pathway. Int J Oncol, 2015. 46(4): p. 1461–72. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Jin L, Han B, Siegel E, Cui Y, Giuliano A, and Cui X, Breast cancer lung metastasis: Molecular biology and therapeutic implications. Cancer Biol Ther, 2018. 19(10): p. 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A.V. Krop I, Colleoni M, Traina T, Holmes F, Estevez L, et al. ,, Results from a randomized placebo-controlled phase 2 trial evaluating exemestane ± enzalutamide in patients with hormone receptor–positive breast cancer [abstract]. , in In: Proceedings of the 2017 San Antonio Breast Cancer Symposium; 2017 Dec 5–9; San Antonio, TX. Philadelphia (PA): AACR; 2018. [Google Scholar]
- 45.Hickey TE, Robinson JL, Carroll JS, and Tilley WD, Minireview: The androgen receptor in breast tissues: growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol, 2012. 26(8): p. 1252–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cochrane DR, Bernales S, Jacobsen BM, Cittelly DM, Howe EN, D’Amato NC, et al. , Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Res, 2014. 16(1): p. R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rangel N, Rondon-Lagos M, Annaratone L, Osella-Abate S, Metovic J, Mano MP, et al. , The role of the AR/ER ratio in ER-positive breast cancer patients. Endocr Relat Cancer, 2018. 25(3): p. 163–172. [DOI] [PubMed] [Google Scholar]
- 48.Narayanan R, Coss CC, and Dalton JT, Development of selective androgen receptor modulators (SARMs). Mol Cell Endocrinol, 2018. 465: p. 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gyorffy B, Surowiak P, Budczies J, and Lanczky A, Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One, 2013. 8(12): p. e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.