Summary
In contrast to virtually all other tissues, the anatomy of differentiation in the bone marrow (BM) remains unknown. This is due to a lack of strategies to examine blood cell production in situ, which are required to understand differentiation, lineage commitment decisions, and define how spatial organizing cues inform tissue function. Here we developed approaches to image myelopoiesis and generated atlases of granulocyte and monocyte/dendritic cell differentiation. Granulopoiesis and dendritic/monopoiesis localize to different sinusoids and display lineage-specific spatial and clonal architectures. Acute systemic L. monocytogenes infection induces lineage-specific progenitor clusters through increased progenitor self-renewal, but the different lineages remain spatially separated. Monocyte dendritic cell progenitors (MDP) map with Ly6Clo monocytes and conventional dendritic cells; these localize to a subset of vessels expressing a major regulator of myelopoiesis1 colony-stimulating-factor 1 (CSF1/ M-CSF). Specific deletion of Csf1 in endothelium disrupted the architecture around MDP and their localization to sinusoids. Subsequently, there were reduced MDP numbers and differentiation ability, and loss of Ly6Clo monocytes and dendritic cells during homeostasis and infection. These data indicate that local cues produced by distinct blood vessels are responsible for specific spatial organization of definitive hematopoiesis.
In the BM, hematopoietic stem cells (HSC) and multipotent progenitors differentiate into lineage-specific progenitors that then generate all major blood cell lineages. scRNAseq2–4, and lineage tracing5–7 analyses have provided critical insight on the regulation and pathways of differentiation, but they require destruction of the organization of the tissue. With few exceptions most of our knowledge of the anatomy of blood differentiation still derives from classical studies using light or electron microscopy8–15. Because most myeloid cells cannot be uniquely identified based on morphology the anatomy of myelopoiesis and the structures that support myeloid cell production remain unknown.
Results
Strategies for imaging myelopoiesis
In myelopoiesis common myeloid progenitors (CMP) generate monocyte dendritic cell progenitors (MDP) or granulocyte-monocyte progenitors (GMP). MDP differentiate into Gfi1lo monocyte progenitors that generate Ly6Chi and Ly6Clo monocytes or dendritic progenitors (CDP and PreDC) that produce dendritic cells. GMP also generate monocytes via a Gfi1hi monocyte progenitor and neutrophils via a granulocyte progenitor (GP/proNeu)16–18 (Fig. 1a). Gfi1hi and Gfi1lo monocyte progenitors share cell surface markers and can only be distinguished transcriptionally or using Gfi1 reporter mice16. We refer here to the population containing both as MOP.
Figure 1. Stain development.
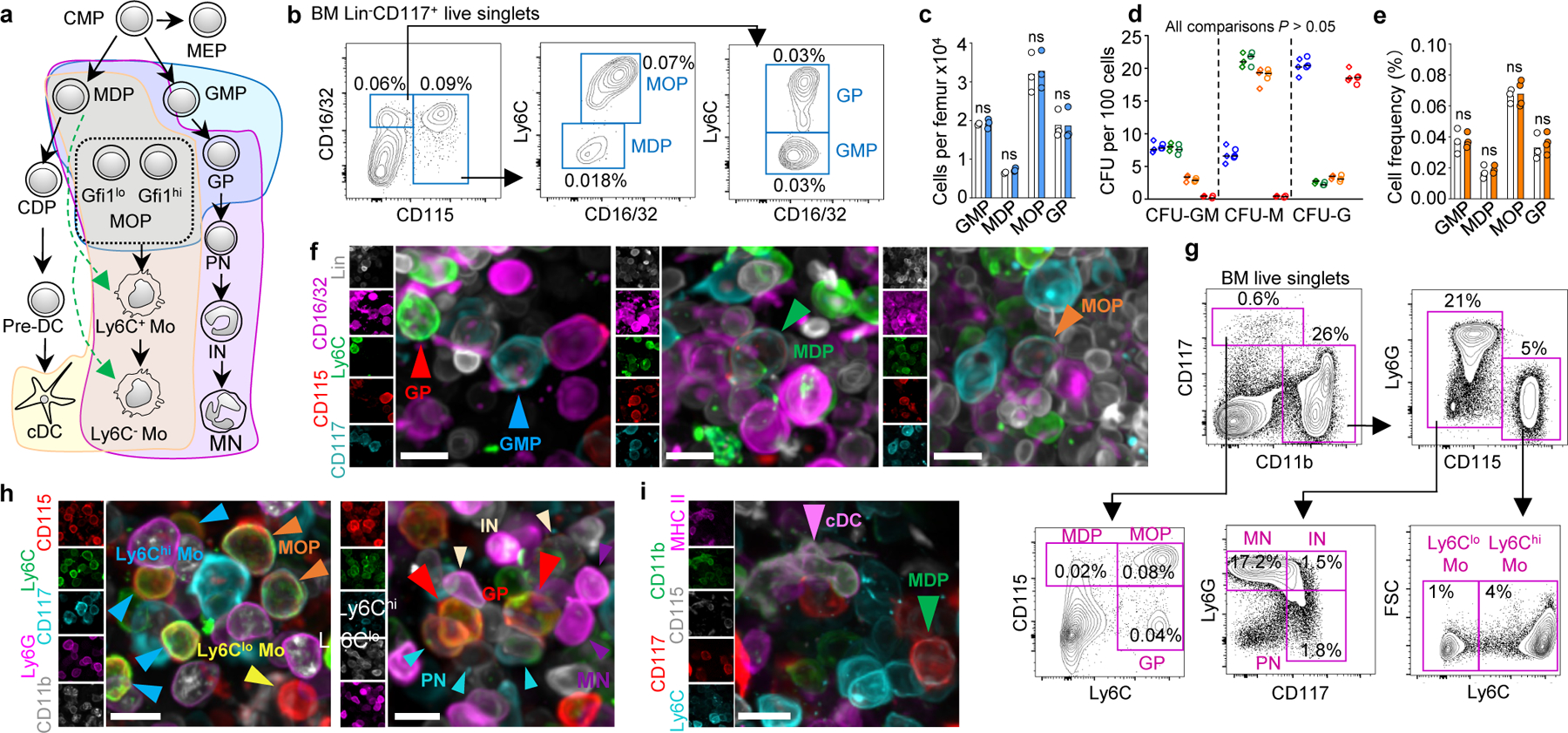
a, Schematic representation of myelopoiesis. b, FACS gates for isolation and imaging of the indicated progenitors. c, d, Cells per femur (c) and colonies produced (d) blue: GMP; green: MDP, orange: MOP, red: GP; n = total 3 mice in two experiments) for progenitors isolated as published16,19 (diamonds) or as shown in b (circles). e, Frequencies for the indicated populations in sternum by FACS (white) or imaging (orange). n = total 3 mice in three experiments. f, Images of the different progenitors. g, FACS gates for isolation and imaging of the indicated progenitors. h, i, Images showing the different myeloid cells. For all panels scale bar = 10 µm. Statistical differences were calculated using two-tailed Student’s T tests. ns = not significant.
To examine myelopoiesis in situ we developed three antibody stains (Fig. 1a shaded areas). The first stain combined two published approaches (16,19 and Extended Data Fig. 1a, b) to simultaneously detect MDP, GMP, MOP and GP (Fig. 1a-f). These progenitors showed same frequencies and colony-forming potential (Fig. 1c, d) as those in established FACS stains16,19 indicating that they label the same populations. We observed identical frequencies when using imaging or FACS (Fig. 1e, f), demonstrating that imaging detected that entire population of BM progenitors. Ly6C additionally allowed arteriole visualization (Extended Data Fig. 1c).
For the second stain we replaced the Lineage panel and CD16/32 with CD11b and Ly6G allowing simultaneous detection of MDP, MOP, GP, Ly6Chi and Ly6Clo monocytes, as well as pre-, immature, and mature neutrophils (PN, IN, and MN; Fig. 1a purple area, Fig. 1g, h and Extended Data Fig. 1d-h). Further validation came from selective GFP expression in MDP and MOP from Cx3cr1-gfp mice20; high Gfi1 or Irf8 expression in GP or MOP2,16; and cytospin analyses (Extended Data Fig. 1l,m). Conventional dendritic cells (cDC) can be detected as MHCII+ reticulated cells (Extended Data Fig. 1n-p). This allows replacing Ly6G with MHCII in Fig. 1g to simultaneously detect MDP, MOP, Ly6Chi and Ly6Clo monocytes, and cDC in the BM (Fig. 1a yellow area, Fig. 1i, and Extended Data Fig. 1q).
Atlas of steady state myelopoiesis
Whole-mounted sternum imaging (Fig. 2a, Extended Data Fig. 2a and Supplementary Video 1-5) showed that most GMP, MDP, MOP and GP are found as single cells throughout the marrow and minimally localize when compared to random simulations (Fig. 2a, Extended Data Fig. 2a,b and Supplementary Video 3). This agrees with previous data showing that Lin-CD117+CD16/32+ progenitors (GMP, GP, and MOP) were interspersed through the BM14. GP are closer to other GP than predicted from random (Fig. 2a, b and Extended Data Fig. 2b). One or two GP map with clusters of pre-neutrophils. Immature neutrophils are also present in the cluster but farther away from GP than pre-neutrophils (Fig. 2a-c, Extended Data Fig. 2c, d and Supplementary Video 5). Mature neutrophils are distributed throughout the BM and do not specifically associate with GP (Fig. 2a-c). To investigate the clonal organization of the clusters we used Ubc-creERT2:confetti mice21,22. In this model tamoxifen treatment leads to irreversible CFP, GFP, YFP or RFP expression in 7.3% of cells (Extended Data Fig. 2e). The experiments showed that preneutrophil clusters are oligoclonal. Confetti-labeled pre-neutrophils were closer than random to GP with the same color indicating a clonal relationship (Fig. 2d, e and Extended Data Fig. 2f, g). This suggests that GP are serially recruited to the cluster and quickly differentiate generating small bursts of pre-neutrophils. Half of pre-/immature neutrophils clusters lack GP and likely represent peripheral areas of larger clusters that locate above the sternum section (Fig. 2a blue squares and Extended Data Fig. 2c, d).
Figure 2. Atlas of myelopoiesis.
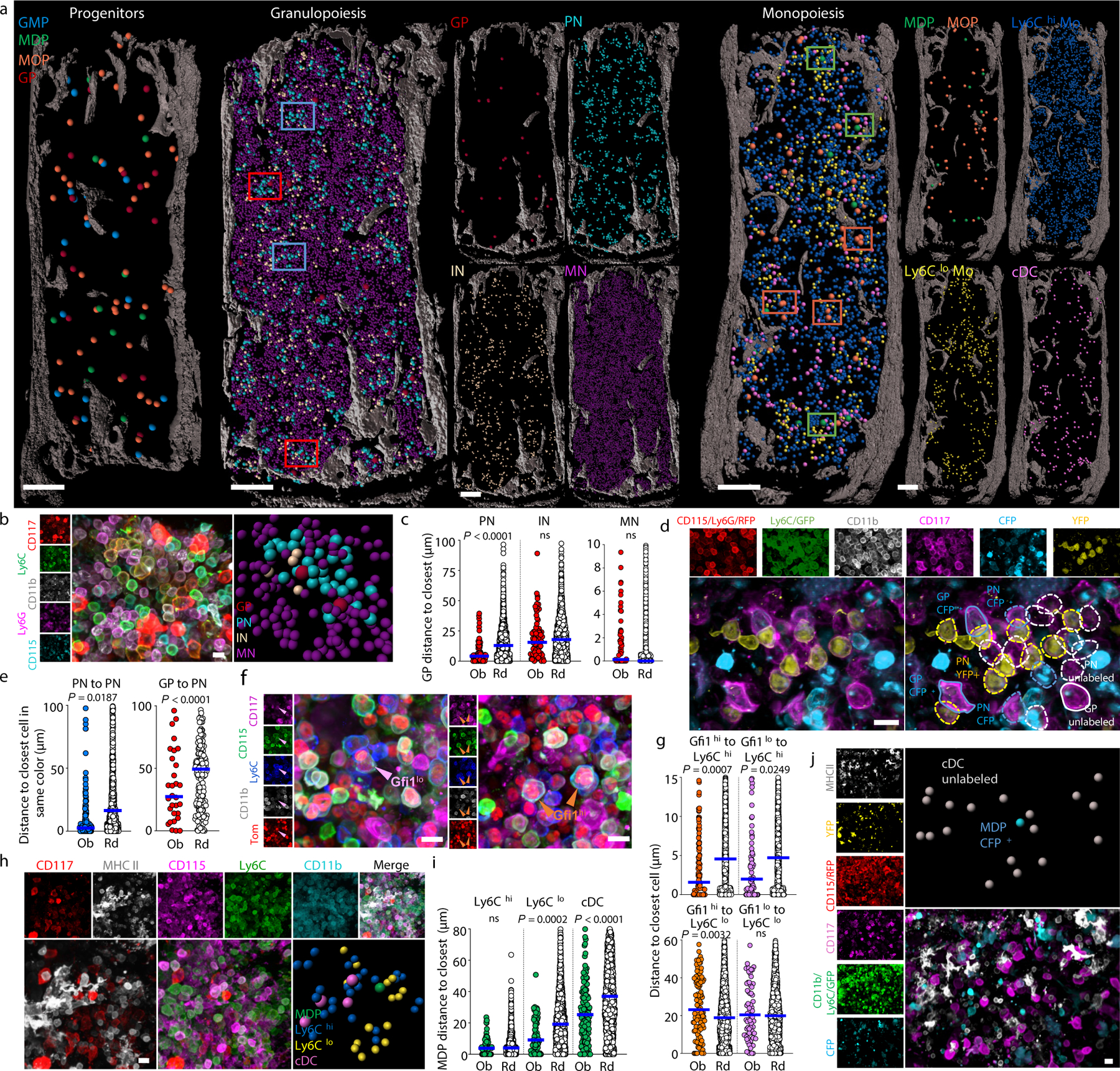
a, Maps of the indicated cells in a 35 µm optical slice of the sternum. Dots are 3x (progenitors), 1.5x (MN), or 2x (all other cells) the average size of the replaced cell. Color squares highlight representative PN clusters around GP (red) or without GP (blue) or areas around MDP (green) or MOP (orange). b, Neutrophil differentiation around a GP. c, Observed and random distances from each GP to the closest indicated cell in clusters associated with GP (n = 75 GP in total 3 sections of 3 mice). d, Oligoclonal preneutrophil cluster in confetti mice. Cells are YFP+, CFP+ or unlabeled CD11b-CD115-CD117+Ly6C+Ly6G- GP and CD11b+CD115-CD117+Ly6C+Ly6G- PN. e, Distances from each confetti-labeled GP or PN to the closest PN labeled in the same color (n = 30 GP in total 9 sections of 3 mice, n = 171 PN in total 3 sections of 3 mice). f, Images showing Gfi1hi and Gfi1lo MOP in Gfi1-tomato mice. g, Distances from each MOP to the closest indicated cell. (n = 113 Gfi1hi MOP and 72 Gfi1lo MOP in total 3 sections of 3 Gfi1-tomato mice). h, Ly6Clo monocyte and cDC localization near MDP. i, Distances from each MDP to the closest indicated cell (MDP-Ly6Chi or MDP-Ly6Clo monocyte, n = 67 MDP from total 6 sections of 4 mice; MDP-cDC, n = 139 MDP from total 11 sections of 6 mice). j, Image showing lack of contribution of a CFP+ MDP to surrounding cDC. In graphs one dot = one cell. Horizontal blue bars indicate the median distance. Unless indicated in all images each dot represents the size of the replaced cell. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant. For maps and images scale bars are 200 and 10 µm, respectively.
MOP contain both GMP-derived Gfi1hi progenitors and MDP-derived Gfi1lo progenitors16. The mixed MOP population showed close localization with Ly6Chi monocytes- that are spread through the BM (Fig. 2a and Supplementary Video 6)- but not with Ly6Clo monocytes or cDC (Extended Data Fig. 3a). We used Gfi1-tomato mice to map Gfi1hi and Gfi1lo MOP (Fig. 2f and Extended Data Fig. 3b, c). Both map near each other and MDP (Extended Data Fig. 3d) and are closer to Ly6Chi monocytes than random cells (Fig. 2g). Unexpectedly, the neighborhood of confetti-labeled MOP is devoid of monocytes labeled in the same color. These indicate that MOP do not contribute to surrounding monocytes and that monocytes and their progenitors quickly separate after differentiation (Extended Data Fig. 3e, f).
Ly6Clo monocytes are closer to each other than random cells (Fig. 2a and Extended Data Fig. 3g) generating Ly6Clo monocyte rich areas that are selectively enriched near MDP (Fig. 2a, h, i and Supplementary Video 6, 7). In agreement with previous studies23,24 cDC are also enriched in specific regions (Fig. 2a and Extended Data Fig. 3h). cDC also map with MDP (Fig. 2a, h, i and Supplementary Video 7). These suggested that MDP might cluster with their progeny. Fate mapping MDP was difficult as only 2 out of 125 MDP analyzed in 9 sternum segments were confetti-labeled. These showed no clonal relationship with surrounding cDC or Ly6Clo monocytes (Fig. 2j and Extended Data Fig. 3i). Together with the observation that the Ly6Clo monocytes and cDC are not clonally related between them (Extended Data Fig. 3j, k) these indicate that Ly6Clo monocytes and cDC are produced elsewhere and then recruited near MDP. These also demonstrate distinct spatial and clonal architectures for the granulocyte and mono/dendritic cell lineage. Note that these architectures are maintained between different samples and types of sections (Extended Data Fig. 4a-g).
Spatial segregation of myelopoiesis
GP identify areas of granulopoiesis and MDP localize with Ly6Clo monocytes and cDC respectively, but these progenitors do not colocalize. In agreement, preneutrophils, immature neutrophils, Ly6Clo monocytes and cDC are farther apart from each other than predicted from random distributions (Extended Data Fig. 5a-c) demonstrating segregation of myeloid lineages to different BM locations.
Sinusoids, arterioles, and the endosteum organize the BM via cytokine secretion25. Imaging analyses demonstrate that MDP, MOP, and GP are farther away from arterioles, show no specific localization with the endosteum, and are closer to sinusoids when compared to random progenitors (Fig. 3a, b, Extended Data Fig. 5d, e, and Supplementary Videos 8-10). Note that since MDP are rare cells they sometimes appear to concentrate towards one end of the sternum in some images (e.g. Fig. 3a) but they are equally distributed through the sternum (Extended Data Fig. 5f). Since sinusoids are a niche for HSC25 we mapped MDP - or a mixed population of GP and MOP- distances to HSC and found no differences when compared to random cells (Extended Data Fig. 5g-j). Together these indicate that myeloid progenitors do not reside in HSC niches and that sinusoids are the site of myelopoiesis. Since granulo- and mono/DC-poiesis do not overlap these raised the possibility that different sinusoids regulate specific myeloid lineages.
Figure 3. CSF1+ vessels organize myelopoiesis.
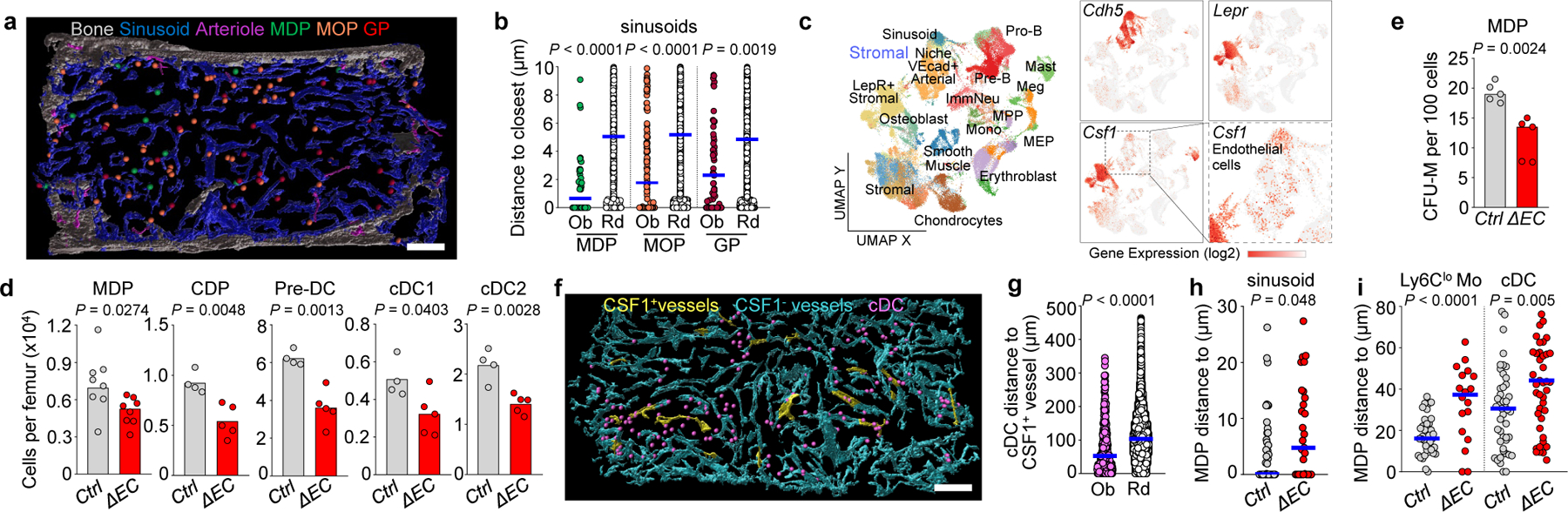
a, Map showing localization of MDP, MOP and GP to sinusoids. b, Progenitor distance to the closest sinusoid, n = 62 MDP, 218 MOP and 114 GP in total 6, 5 and 5 sections from 6 or 5 mice. c, UMAP of BM cells. Color coded clusters are reanalyses30 of published data27. Right panels: Cdh5, Lepr, and Csf1 expression. d, e, Cells per femur (d) or MDP-derived CFU-M (e) in control (pool of Cre:Csf1+/−, Csf1+/−, and Csf1fl/-) or Csf1ΔEC mice; 1 dot = 1 mouse. f, Map of CSF1+ and CSF1- vessels and cDC in Ctrl mice. g, cDC distance to the closest CSF1+ vessel (n = 442 cDC from total 4 sections of 3 WT mice). h, MDP distance to the closest vessel (n = 44 MDP from total 5 sections of 3 control mice; n = 29 MDP from total 5 sections of 3 Csf1ΔEC mice). i, MDP distance to closest Ly6Clo monocyte or cDC. For MDP-Ly6Clo monocyte, n = 37 MDP from total 4 sections of 3 control mice, n = 18 MDP from total 4 sections of 3 Csf1ΔEC mice. For MDP-cDC, n = 47 MDP from total 6 sections of 3 control mice, n = 47 MDP from total 9 sections of 3 Csf1ΔEC mice). Unless indicated 1 dot = 1 cell. Progenitor dots are thrice – and cDC dots twice- the size of the replaced cell. Scale bars = 200 μm. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant.
CSF1+ vessels organize myelopoiesis
We reclustered and analyzed published scRNA-Seq datasets26,27 (Extended Data Fig. 6a-l) and found that LepR+ perisinusoidal cells and small subsets of endothelial cells expressed Csf1 -a key regulator of myelopoiesis1- (Fig. 3c and Extended Data Fig. 6d, e). Conditional Csf1 deletion in LepR+ cells did not impact hematopoiesis (Extended Data Fig. 7a-e). In contrast, Csf1 deletion in endothelial cells (Extended Data Fig. 8a) caused reductions in MDP numbers that are further compounded by reductions in MDP-derived CFU-M, but not CFU-G or CFU-GM, colonies when compared to controls. Dendritic cells and progenitors downstream of MDP were also reduced. GMP and MOP numbers and function in Csf1ΔEC mice are minimally impaired (Fig. 3d, e and Extended Data Fig. 8b-d). Ly6Clo monocytes map near MDP (Fig. 2h, i) and showed a ~2-fold reduction in numbers in Csf1ΔEC mice in BM and blood whereas Ly6Chi monocytes - which do not associate with MDP - are unaffected (Extended Data Fig. 8e). Csf1ΔEC mice do not exhibit changes in BM cellularity, HSC, multipotent progenitors, CMP, GP, or mature cell numbers in BM or blood (Extended Data Fig. 8f-h). The vascular network is also indistinguishable from that of control mice (Extended Data Fig. 8i) indicating an intact vascular niche. CSF1 regulates HSC differentiation28 but competitive BM transplants revealed no differences in engraftment between recipients transplanted with Csf1ΔEC or control BM (Extended Data Fig. 8j). These indicate that endothelial-derived CSF1 does not regulate HSC function during homeostasis.
Imaging revealed widespread αCSF1 signal concentrated in endosteal and subendosteal regions. In contrast, only 8% of blood vessels express CSF1 (Fig. 3f and Extended Data Fig. 8k). The vascular signal is specific as it was almost completely abolished in the Csf1ΔEC mice (Extended Data Fig. 8l). The stain was compatible with αMHCII allowing detection of cDC but no other populations as the αCSF1 signal amplification protocol caused loss of most cell surface markers. cDC are specifically enriched around the CSF1+ vessels (Fig. 3f, g, Extended Data Fig. 8l-n, and Supplementary Video 11). Csf1 deletion in the vasculature causes MDP-but not cDC- to move away from sinusoids (Fig. 3h and Extended Data Fig. 8l, o, p); and disrupts the close localization between Ly6Clo monocytes and cDC and MDP (Fig. 3i and Extended Data Fig. 8q, r). Taken together, these indicate that CSF1+ vessels are a niche for MDP that regulates Ly6Clo monocytes and cDC production.
Effect of infection
We used Listeria monocytogenes to investigate stress-induced myelopoiesis. In agreement with previous studies29 we found loss of myeloid progenitors and mature cells 2 days after infection followed by massive cDC expansion (Fig. 4a and Extended Data Fig. 9a). Even during stress GP, MOP, and MDP remain spatially segregated (Fig. 4b and Extended Data Fig. 9b). While MDP are always found as single cells. GP and MOP recover via homogeneous clusters of 5–14 GP or 4–11 MOP at day 4 that mostly disaggregate by day 6 (Fig. 4b, c and Extended Data Fig. 9c). Almost all GP clusters are monoclonal indicating increased GP self-renewal. MOP clusters are bi- or oligo-clonal and contain both Gfi1hi and Gfi1lo MOP but are enriched in MDP-derived Gfi1lo MOPs (Fig. 4c and Extended Data Fig. 9d-g). Thus, both types of MOP are recruited to the same structure and then amplify via increased self-renewal. We were unable to image CD117dim pre-neutrophils and immature neutrophils as infection caused CD117 downregulation in these cells (Extended Data Fig. 9h). After infection, Gfi1hi and Gfi1lo MOP and MDP map closer to Ly6Chi –but farther from Ly6Clo- monocytes than in non-infected mice. Similarly, MDP also mapped closer to cDC (Fig. 4b and Extended Data Fig. 9j). This is expected as Ly6Chi monocytes and cDC recover quickly whereas Ly6Clo monocytes remain depleted through the course of the infection (Fig. 4a). Even in this setting of accelerated myelopoiesis neither confetti-labeled MOP nor MDP are clonally related to surrounding cells (Extended Data Fig. 9k-m) indicating that migration away from their progenitors is an intrinsic feature of monocytes and dendritic cells. Myeloid progenitors still mapped to sinusoids but farther away when compared with non-infected mice (Fig. 4d, e). In agreement, we found reduced Csf1–which causes MDP to localize to sinusoids Fig. 3h- in endothelial cells from infected mice (Extended Data Fig. 9n). This prevented CSF1+ vessels detection. Loss of endothelial derived CSF1 did not cause further MDP relocation away from sinusoids but disrupted the close localization of MDP with Ly6Clo monocytes and cDC and led to reduced recovery of MDP and downstream progenitors and cDC with other myeloid cells being less affected (Fig. 4f, g and Extended Data Fig. 9o-r). These indicate that endothelial-derived CSF1 is required for emergency DC production in response to infection. Extended Data Fig. 10 summarizes our findings.
Figure 4. Atlas of stress myelopoiesis.
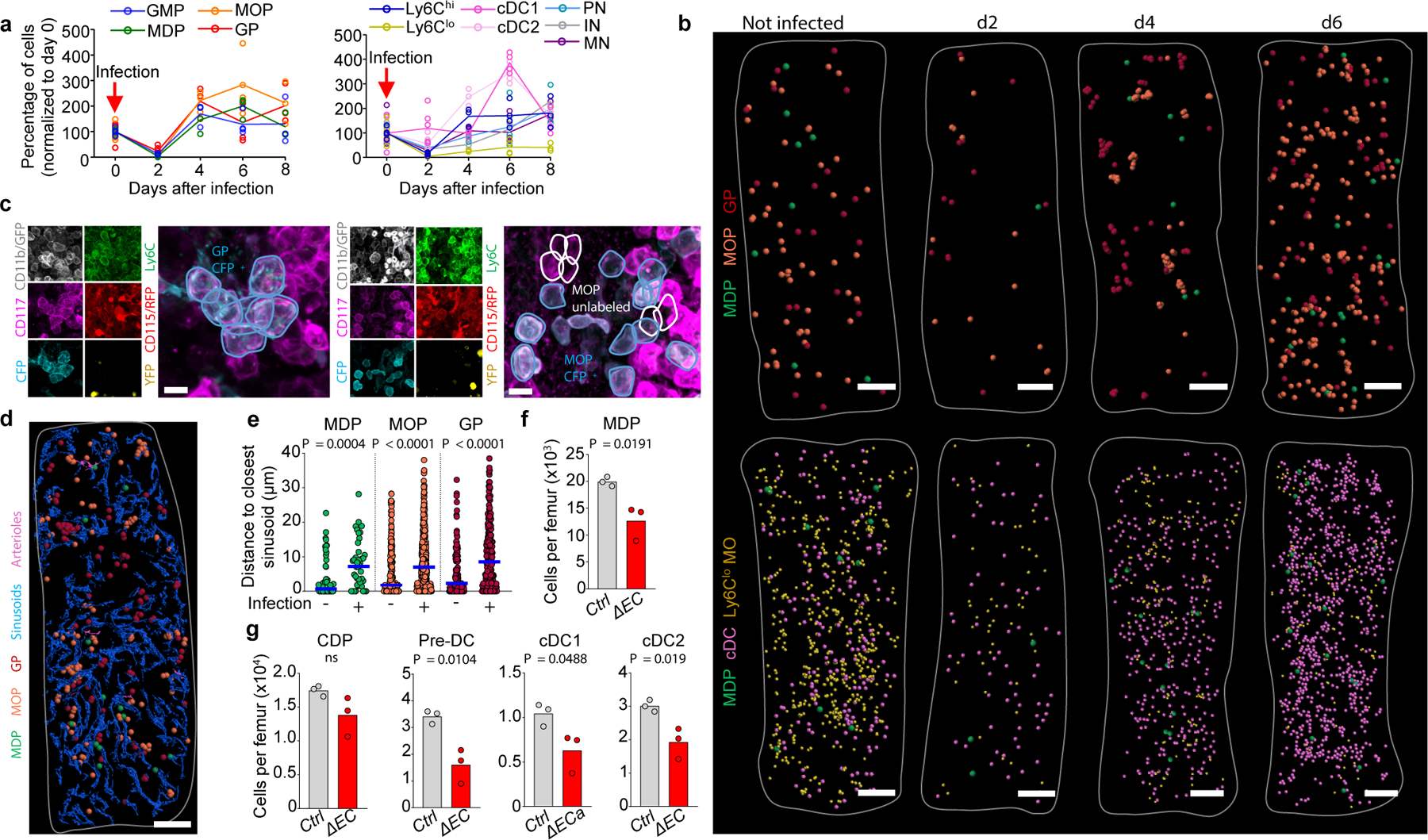
a, Average percentage of cells per femur (normalized to day 0) after L. monocytogenes infection (n = total 6, 6, 3, 4 and 3 mice for days 0, 2, 4, 6 and 8). b, Maps of myelopoiesis in infected mice. c, Image showing a monoclonal GP cluster (left) or oligoclonal MOP cluster (right) in confetti mice four days after L. monocytogenes infection. GP were tracked as YFP+, CFP+ or unlabeled CD11b-CD115-CD117+Ly6C+ cells. MOP were tracked as YFP+, CFP+, or unlabeled CD11b-CD115+CD117+Ly6C+ cells. d, e, Map (d) showing myeloid progenitors, sinusoids, and arterioles; and distances from each progenitor to the closest sinusoid (e) in WT mice uninfected (same as Fig. 3b) or four days after infection. n = 62 MDP from total 6 sternum sections of 6 uninfected wild-type mice and n = 41 MDP from total 3 sternum sections of 3 wild-type mice four days after infection; n = 218 MOP and n = 114 GP from total 5 sternum sections of 5 mice in wild-type mice; n = 463 MOP and n = 241 GP from total 3 sternum sections of 3 wild-type mice four days after infection (1 dot = 1 cell). Horizontal blue bars indicate the median. f, g, Number of the indicated cells per femur in control or Csf1ΔEC mice four days after infection (1 dot = 1 mouse in two independent experiments). For maps Scale bar = 200 μm, for images Scale bar = 10 μm. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant.
Discussion
The anatomy of differentiation in the BM remains largely unknown. Here we mapped myelopoiesis in situ and assessed clonal relationships between myeloid progenitors and surrounding cells. We demonstrate that there is spatial organization of myelopoiesis with granulo- and mono/dendritic cell -poiesis taking place in different sinusoids and displaying lineage-specific spatial and clonal architectures. CSF1+ vessels provide a unique niche that regulates MDP, monocytes, and cDC and is required for normal and stress-induced dendritic cell production. Thus, local cues produced by distinct blood vessels spatially organize and control myeloid differentiation. The study provides a new conceptual framework for understanding differentiation, dissecting hematopoiesis during disease, and designing organ systems capable of multilineage blood cell production ex vivo.
Methods
Mice
C57BL/6J-Ptprcb (CD45.2+), B6.SJL-PtprcaPepcb/BoyJ (CD45.1+), B6;129-Tg(Cdh5-cre)1Spe/J, B6.129(Cg)-Leprtm2(cre)Rck/J (LepR-cre), B6.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/1J (UBC-cre/ERT2), B6.129P2-Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle (confetti) and B6.129P2(Cg)-Cx3cr1tm1Litt/J (CX3CR1-GFP) mice were obtained from the Jackson Laboratory. Nestin-gfp mice51 were kindly provided by Paul. S. Frenette. Csf1fl/fl and Gfi1Tomato mice have been described previously31,32. To generate Csf1+/− mice we first bred Csf1fl/fl mice with Ubc-creERT2 mice22 to generate Ubc-creERT2:Csf1fl/+ mice. These were treated with tamoxifen leading to deletion of the Csf1fl allele in the germline. Tamoxifen treated Ubc-creERT2:Csf1fl/+ mice were then bred with wild-type mice to generate Csf1+/− mice. We mated Csf1+/− mice with Cdh5-cre:Csf1fl/+45 and LepR-cre:Csf1fl/+ mice44 to generate Cdh5-cre:Csf1fl/- and LepR-cre:Csf1fl/- mice. Mice received water and food ad libitum. All experiments -except confetti fate mapping- were performed in 8 to 14 weeks old male and female mice (Mus musculus). Mice were house at the vivarium at Cincinnati Children’s Hospital Medical Center under a 14h light: 10h darkness schedule, 30–70% humidity and at 22.2±1.1ºC. All animal experiments followed all relevant guidelines and regulations and were approved by the Animal Care Committee of Cincinnati Children’s Hospital Medical Center.
Tamoxifen treatment
Six week old B6.129P2-Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle (confetti)21 mice were treated with two pulses of tamoxifen in the diet (0.4g of Tamoxifen Citrate/Kg of rodent diet, Envigo). Each pulse was two weeks long and pulses were two weeks apart. Since GP, MDP and MOP do not persist in vivo for longer than two weeks16,33 we chased the mice for 12 weeks to ensure that all the structures originated from upstream progenitors.
Listeria monocytogenes infection and antibiotic treatment
For infection, Listeria monocytogenes WT strain 10403s was grown overnight, and then back-diluted to early log-phase growth (OD600 0.1) in Brain Heart Infusion media (Difco), washed and diluted in sterile saline, and injected intravenously into mice (1 × 104 CFUs/200 microliters per mouse). Csf1+/− heterozygous mice (Csf1ΔEC and controls) did not survive this dosage and were thus treated by supplementing the drinking water with ampicillin (2mg/ml) 48 hours after infection. This rescued death in 50% of the mice.
Bone marrow and peripheral-blood collection for FACS analyses.
Mice were euthanized by isoflurane inhalation followed by cervical dislocation. Bone marrow cells were harvested by flushing bones with 1 ml of ice-cold PEB buffer (2 mM EDTA and 0.5% bovine serum albumin in PBS). Blood was collected from the retro-orbital venous sinus in tubes containing EDTA. Red blood cells in peripheral blood were lysed by the addition of 1 ml of RBC lysis buffer (150 mM NH4Cl, 10 mM NaCO3 and 0.1 mM EDTA). Cells were immediately decanted by centrifugation, resuspended in ice-cold PEB and used in subsequent assays. We used CountBright™ Absolute Counting Beads (cat. no. C36950, Thermo Fisher Scientific) to count bone marrow and blood cell numbers in a flow cytometer according to the manufacturer’s protocol.
Isolation of bone marrow stromal cells.
Bone marrow digestion was performed as previously described34.
FACS analyses.
Cells were stained under dark for 30 min in PEB buffer containing antibodies, washed thrice with ice cold PBS and analyzed in a BD LSR II (BD Biosciences) instrument or FACS-purified with a BD FACS Aria II or a SH800S (Sony) cell sorter. Dead cells and doublets were excluded on the basis of FSC and SSC distribution and DAPI (Sigma) exclusion. Antibodies used were: B220 (clone RA3–6B2), CD3 (clone 145–2C11), CD8 (clone 53–6.7), CD11b (clone M1/70), CD11c (clone N418), CD16/32 (clone 93), CD24 (clone 30-F1), CD31 (clone A20), CD41 (clone MWReg30), CD45 (clone 30-F11), CD45.1 (clone A20), CD45.2 (clone 104), CD48 (clone HM48–1), CD105 (clone MJ7/18), CD115 (clone AFS98), CD135 (clone A2F10), CD144 (clone BV13), CD150 (clone TC15–12F12.2), CD169 (clone 3D6.112), CD172α (clone P84), F4/80 (clone BM8), Gr1 (clone RB6–8C5), Ly6C (clone HK1.4), Ly6-G (clone 1A8), Sca-1 (clone D7), Ter119 (clone TER-119), MHC II (clone M5/114.15.2), from BioLegend; CD34 (clone RAM34), CD117 (clone 2B8), and Siglec-H (clone eBio440c), from BioLegend or Thermo Fisher Scientific. Data was analyzed with FlowJo (Tree Star). Gating strategies for most analyses are shown in the main or Extended Data Figures. CDP33, pre-DC35, cDC1 and cDC235, CMP36 and macrophages37 are gated as described.
Cytospin preparation and analyses
FACS-purified cells were decanted in 30 slides using a Cytospin 4 Cytocentrifuge (Thermo Fisher Scientific) following manufacturer’s instructions. For histology analyses slides were stained with Camco Stain Pak (Cambridge Diagnostic Inc) according to the manufacturer’s instruction. This kit gives results similar to a Wright-Giemsa stain. Slides were analyzed on Zeiss AX10 inverted microscope (Carl Zeiss, Oberkochen, Germany).
Colony-forming unit (CFU) assay
The indicated number of FACS-purified cells were seeded into methylcellulose culture medium (M3534, StemCell Technologies), plated in 35 mm dishes and incubated at 37°C, in 5% CO2, with ≥ 95% humidity for 7–10 days. Colonies were identified and counted based on cluster size and cell morphology using Nikon Eclipse Ti inverted microscope (Nikon Instruments Inc., NY, USA).
RNA isolation and qPCR analyses.
RNA isolation was performed with a Dynabeads mRNA Direct Kit (Thermo Fisher Scientific, no. 61011) according to the manufacturer’s protocol. cDNA synthesis was performed with RNA to cDNA EcoDry Premix (Takara, 639546) according to the manufacturer’s instructions. qPCR was performed using SYBR Green Supermix (Quanta Biosciences, no. 95054) in an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Results were analyzed in SDS 2.4 software (Applied Biosystems). Primers were as follows: CSF1 forward, 5’ -ATGAGCAGGAGTATTGCCAAGG- 3’; CSF1 reverse, 5’ -TCCATTCCCAATCATGTGGCTA- 3’; IRF8 forward, 5’ -AGACCATGTTCCGTATCCCCT- 3’, IRF8 reverse, 5’ -CACAGCGTAACCTCGTCTTCC −3’; GFI1 forward, 5’ -AGAAGGCGCACAGCTATCAC- 3’, GFI1 reverse, 5’ -GGCTCCATTTTCGACTCGC- 3’; mGAPDH forward, 5’ -TGTGTCCGTCGTGGATCTGA- 3’; mGAPDH reverse, 5’-CCTGCTTCACCACCTTCTTGA-3’.
Competitive reconstitution assays in irradiated mice
Recipient mice were lethally conditioned with total 1175 rads irradiation dose (700 + 475 split dose 3 hours apart). 1 × 106 bone marrow cells of control or conditional knockout mice were mixed with 1 × 106 CD45.1+ competitor mouse bone marrow cells and transplanted by injection into the tail vein of CD45.1+ recipients.
Whole-mount immunostaining
In some cases we injected retro-orbitally with 10 μg of Alexa Fluor 647 anti-mouse CD31 (BioLegend, no. 110724) and 10 μg of Alexa Fluor 647 anti-mouse CD144 (BioLegend, no. 138006) ten minutes before euthanasia to visualize the BM vasculature as described34.
Whole-mount sternum stain has been described before38. Sterna were processed immediately after euthanasia. After dissection we removed all connective tissue by gentle scraping with a blade. Fragments with bone marrow cavity were dissected and sectioned along the sagittal or coronal plane to expose the BM as described. Each half of the sternum was fixed in 4% PFA (Electron Microscopy Sciences, no. 15710) in DPBS for 3 hours. Each fragment was washed thrice with DPBS, followed by 1 hour blocking in DPBS containing 10% (v/v) goat serum (Sigma-Aldrich, no. G9023). We stained each sample with 100 µl staining buffer (2% goat serum in DPBS and the indicated antibodies). All steps were performed at 4ºC.
CSF1 detection
After fixation and blocking as above, sternum was stained with 100 µl of staining buffer containing anti CSF1 (Thermo Fisher Scientific, no. PA5–95279, 1:200 dilution), anti MHCII (BioLegend, no. 107618) and 2% goat serum in DPBS at 4ºC for 12h. Each sternum was washed thrice with DPBS and then incubated with Superboost HRP conjugated anti-rabbit antibody (Invitrogen, no. B40943) in DPBS at room temperature for 1 hour. After washing three times each sternum was developed by incubation in 100 µl of tyramide solution (Invitrogen, no. B40943) for 8 minutes. Sterna were washed three additional times in DPBS and used for confocal analyses.
Confocal imaging
For upright confocal imaging we glued the periosteum to plastic plates positioning the exposed marrow facing up. Each sternum fragment is 300 µm deep (from the exposed BM surface to the external surface of the periosteum). All fluorophores can be confidently detected to a depth of 35 μm but we imaged to a depth of 250 µm to ensure full coverage of the sternum even if the BM surface was not completely parallel to the surface of the plate. We used a Nikon A1R GaAsP Multiphoton Upright Confocal Microscope. Specifications for the Upright Confocal: 405 nm, 488 nm, 561 nm, and 638 nm diode lasers, Coherent Chameleon II TiSapphire IR laser, tunable from 700–1000 nm, fully encoded scanning XY motorized stage, Piezo-Z nosepiece for high-speed Z-stack acquisition (100 μm/s), resonant and galvanometric scanners, four high-quantum efficiency, low-noise Hamamatsu photomultiplier tubes, a transmitted PMT for transmitted light for 400–820 nm detection, four high-quantum efficiency GaAsP non-descanned detectors for multiphoton imaging. We used a 16X Apo 0.8 NA LWD Water Immersion Objective with a 3.0 mm working distance. Images were taken using the resonant scanner with 8X line averaging, 1024 X 1024 pixel resolution, 2 μm Z-step, and pinhole at 21.7 μm. Bone signal was obtained from second harmonic generation (SHG) imaging with 840 nm excitation.
For inverted confocal imaging the sternum fragment was positioned with the exposed BM surface facing down on a glass plate and then covered with PBS. We imaged to a depth of 120µm using a Nikon A1R GaAsP Inverted Microscope. Specifications for the Nikon A1R GaAsP Inverted Confocal Microscope: 405 nm, 442 nm, 488 nm, 561 nm, 640 nm, and 730 nm diode lasers; Nikon A1R LUN-V Inverted Confocal Microscope: 405 nm, 445 nm, 488 nm, 514 nm, 561 nm, and 647 nm diode lasers with fully encoded scanning XY motorized stage, Piezo-Z stage insert for high-speed Z-stack acquisition (100 μm/s), resonant and galvanometric scanners, two high-quantum efficiency, low-noise Hamamatsu photomultiplier tubes, a transmitted PMT for transmitted light, and two very high-QE Gallium-Arsenide-Phosphide PMTs for overall 400–820 nm detection. We used a 40X Apo 1.15 NA LWD DIC-Water Immersion Objective with a 0.59 mm working distance for high power image. Images were taken using the resonant scanner with 8X line averaging, 1024 X 1024 pixel resolution, 0.5 μm Z-step, and pinhole at 50 μm. We also used a 20X Apo 0.95 NA LWD Water Immersion Objective with a 0.95 mm working distance. Images were taken using the resonant scanner with 8X line averaging, 1024 X 1024 pixel resolution, 2 μm Z-step.
Image and distance analyses
We used Nikon Elements software (5.20.02), Imaris x64 software (9.5) and Matlab software (2018a) installed in a HP Z4 64-bit workstation with Dual Intel Xeon Processor W-2145, 128GB ECC-RAM, 8 GB NVIDA Quadro RTX5000 Graphics card for all our analyses. We only analyzed images for which the whole sternum fragment (6–8 images) was successfully imaged. These images were stitched together using Nikon Elements. Stitched whole bone images were further processed by an artificial intelligence algorithm (Denoise.AI) for noise removal using Nikon elements. For confetti analyses we performed spectral unmixing of the GFP and YFP signal using the unmixing algorithm in Nikon elements. We manually identified each cell using relative intensities within a given focal plane. No cells are discarded from the analyses. Negative thresholds were defined using isotype controls and positive thresholds based on the positive signal detected using FACS analyses. Note that in all the images in the manuscript we adjusted the brightness and contrast of each channel to be able to detect negative, dim, and bright cells for each fluorescent signal. We used Imaris to replace each cell with a color-coded sphere and obtain its X, Y and Z coordinates. We also used Imaris to create surfaces for sinusoids, arterioles and the endosteal surface. For each cell type we measured the diameter of 50–150 cells of each type to obtain the mean diameter (HSC = 8.84 ± 1.56 μm, GMP = 12.68 ± 1.52 μm, MDP = 12.13 ± 1.19 μm, GP = 11.70 ± 0.99 μm, MOP = 11.49 ± 1.25 μm, PN = 10.21 ± 1.08 μm, IN = 8.72 ± 0.75 μm, MN = 8.10 ± 1.09 μm, Ly6Chi Mo = 9.25 ± 0.86 μm, Ly6Clo Mo = 9.30 ± 1.17 μm, cDC = 12.33 ± 2.69 μm). Since cDC have reticular shapes we measured the cell bodies to obtain the median diameter. We used Imaris to measure the distance from each cell to the closest vascular structure or the endosteum and then subtracted the mean radius for each cell type. To quantify cell to cell distance we exported the coordinates of the cells of interest to Matlab and then used an algorithm to quantify the distance from the center of each cell to the center of all other cells. We then subtracted the mean radius of each cell from these values.
Confetti fate mapping
In the confetti model GFP is detected in the nucleus whereas RFP is expressed in the cytoplasm35. By using antibodies conjugated to fluorochromes (Alexa Flour488 and Phycoerythrin, PE) that spectrally overlap with GFP or RFP but that stain only the membrane we could distinguish Ly6C+GFP- cells from GFP+ cells and CD115/Ly6G+RFP- cells from RFP+ cells. This allowed us to examine the relationships between YFP- or CFP-labeled cells. Since we could not distinguish the membrane signal from the nuclear/cytoplasmic signal in GFP+ or RFP+ cells these were discarded from the analyses.
Random simulations
We stained and imaged a sternum fragments with CD45 and Ter119 antibodies to detect hematopoietic cells; CD31, CD144 and Sca1 antibodies to detect sinusoids and arterioles; and used second harmonic generation to detect bone. These images were processed as above to obtain the coordinates of all hematopoietic cells (59,659 cells), vessels and bone in the sternum. We then used Research Randomizer39 to randomly select dots representing each type of myeloid cell at the same frequencies found in vivo through the bone marrow cavity and measured the distances between these random cells or with vessels and bone as above. Each random simulation was repeated 100–200 times.
For generating random distributions in fate mapping experiments using confetti mice we first obtained the coordinates and confetti color for each type of myeloid cell in each section analyzed. Then we used Research Randomizer39 to randomly place confetti labeled cells –at the same frequencies found in vivo- in the positions occupied for the myeloid cells in each section. We then measured the distances between these random cells. Each random simulation was repeated 100–200 times.
Calculation of cell frequencies in sternum sections
We compared the number for each type of myeloid cell detected in each sternum section with the average number of hematopoietic cells (59,659 cells) detected by CD45 and Ter119 in a whole mounted sternum section.
Stromal UMAP analysis
To identify diverse stromal, hematopoietic, and other cell populations we reanalyzed 19 independent 10x Genomics captures from two complementary stromal bone marrow scRNA-Seq datasets (GSE12842327) and (GSE10889126). For GSE108891, samples were re-reprocessed from the original FASTQ files, consistent with GSE128423 (Cell Ranger software). The merged counts files from these data were scaled, normalized and subjected to an unsupervised analysis with the in the software ICGS version 2 (AltAnalyze version 2.1.4). This analysis identified 26 transcriptionally distinct cell populations in 9,165 cells from the dataset from Tikhonova et al26 and 46 preliminary cell populations in 89,007 in the dataset from Baryawno et al.27 Preliminary annotations were produced using the ICGS2 BioMarker database, which includes marker genes from over 1,000 prior defined cell populations. To further annotate these populations and identify sub-clusters based on prior knowledge, we performed a secondary analysis using the supervised classification tool cellHarmony40, comparing all cells to reference hematopoietic (GSE12040926) and sorted stromal populations (GSE10889126), resulting in 61 final annotated cell populations for the dataset from Baryawno et al. Visualization of clusters and marker genes was performed using UMAP visualization in AltAnalyze. To allow others to explore side-by-side population specific markers in these two datasets, we provide an online viewer to query these and other genes in both datasets (http://www.altanalyze.org/Marrow-Stromal.html).
Statistics and reproducibility
For graphs quantifying cells in different mice we indicate the mean and each dot corresponds to one mouse. Statistical differences were calculated using two-tailed Student’s T tests and each mouse was considered biologically independent. For graphs showing distances between cells and structures each dot corresponds to one cell and the horizontal bar indicates the median. Statistical differences were calculated using two-tailed Student’s T tests and each cell was considered biologically independent. ns, not significant.
For cluster analyses we benchmarked K-means41 and spectral clustering approaches such as the DBSCAN42. We validated the clusters by sweeping the parameters on each algorithms and found an optimal clustering with K-means and by setting a minimum total count of 5 PN/IN cells in each cluster. We used the K-means++43 for centroid seeds with 3D Euclidean distance metric and obtained the optimal clusters with lower sum of within-PN/IN cluster, sum-of-squares point-to-cluster-centroid distances.
Numerous panels in the main and Extended Data figures show images that are representative of numerous independent experiments as follows. Figure 1f, n = total 4 mice in four experiments; Figure 1h, n = total 16 mice in twelve experiments; Figure 1i, n = total 12 mice in eleven experiments; Figure 2a, n = total 4 mice in three experiments for progenitors, n = total 4 mice in three experiments for granulopoiesis, n = total 6 mice in six experiments for monopoiesis; Figure 2b, n = total 16 mice in twelve experiments; Figure 2d, n = total 3 mice in three experiments; Figure 2f, n = total 3 mice in three experiments; Figure 2h, n = total 12 mice in eleven experiments; Figure 2j, n = total 3 mice in three experiments; Figure 3a, n = total 6 mice in six experiments; Figure 3f, n = total 3 mice in three experiments; Figure 4b, n = total 3 mice in three experiments for each timepoint; Figure 4c, n = total 2 mice in two experiments; Figure 4d, n = total 3 mice in three experiments; Extended data Figure 1c, n = total 3 mice in three experiments; Extended data Figure 1i, n = total 3 mice in three experiments; Extended data Figure 1n, n = total 3 mice in three experiments; Extended data Figure 1o, n = total 3 mice in three experiments; Extended data Figure 1p, n = total 3 mice in two experiments; Extended data Figure 2f, n = total 3 mice in three experiments; Extended data Figure 3b, n = total 3 mice in three experiments; Extended data Figure 3e, n = total 3 mice in three experiments; Extended data Figure 3i, n = total 3 mice in three experiments; Extended data Figure 3j, n = total 3 mice in three experiments; Extended data Figure 3k, n = total 3 mice in three experiments; Extended data Figure 4d, n = total 3 mice in three experiments for GP PN IN coronal sections, n = total 2 mice in two experiments for GP PN IN sagittal sections; Extended data Figure 4f, n = total 4 mice in four experiments for MDP Ly6Clo Mo cDC coronal sections, n = total 2 mice in two experiments for MDP Ly6Clo MO cDC sagittal sections; Extended data Figure 5a, n = total 3 mice in three experiments; Extended data Figure 5b, n = total 3 mice in three experiments; Extended data Figure 5d, n = total 6 mice in six experiments; Extended data Figure 5f, n = total 26 fragments in twenty-five experiments; Extended data Figure 5g, n = total 3 mice in three experiments; Extended data Figure 5i, n = total 3 mice in three experiments; Extended data Figure 5j, n = total 3 mice in three experiments for HSC MDP and n = total 3 mice in three experiments for HSC GP MOP; Extended data Figure 8k, n = total 3 mice in three experiments for αCSF1 and n = total 3 mice in three experiments for isotype control; Extended data Figure 8l, n = total 3 mice in three experiments; Extended data Figure 8m, n = total 3 mice in three experiments; Extended data Figure 8p, n = total 3 mice in three experiments for control and n = total 3 mice in three experiments for Csf1ΔEC; Extended data Figure 8q, n = total 3 mice in three experiments for control and n = total 3 mice in three experiments for Csf1ΔEC; Extended data Figure 9f, n = total 2 mice in two experiments; Extended data Figure 9i, n = total 2 mice in two experiments; Extended data Figure 9k, n = total 2 mice in two experiments; Extended data Figure 9m, n = total 2 mice in two experiments; Extended data Figure 9p, n = total 3 mice in three experiments for control and n = total 3 mice in three experiments for Csf1ΔEC.
Data reporting
Sample sizes were chosen based on previous studies. No statistical methods were used to predetermine sample size. All mice were included in the analyses. Mice were randomly allocated to the different groups on the basis of cage, genotype, and litter size. For all experiments, we aimed to have the same number of mice in the control and experimental groups. Most experiments were performed in unperturbed wild-type mice and thus these samples were not blinded. The phenotypes of mice with conditional deletion of Csf1 in endothelial cells or after L. monocytogenes infection were obvious in all imaging analyses making blinding moot.
Extended Data
Extended Data Figure 1. Validation of stains to detect myeloid cells.
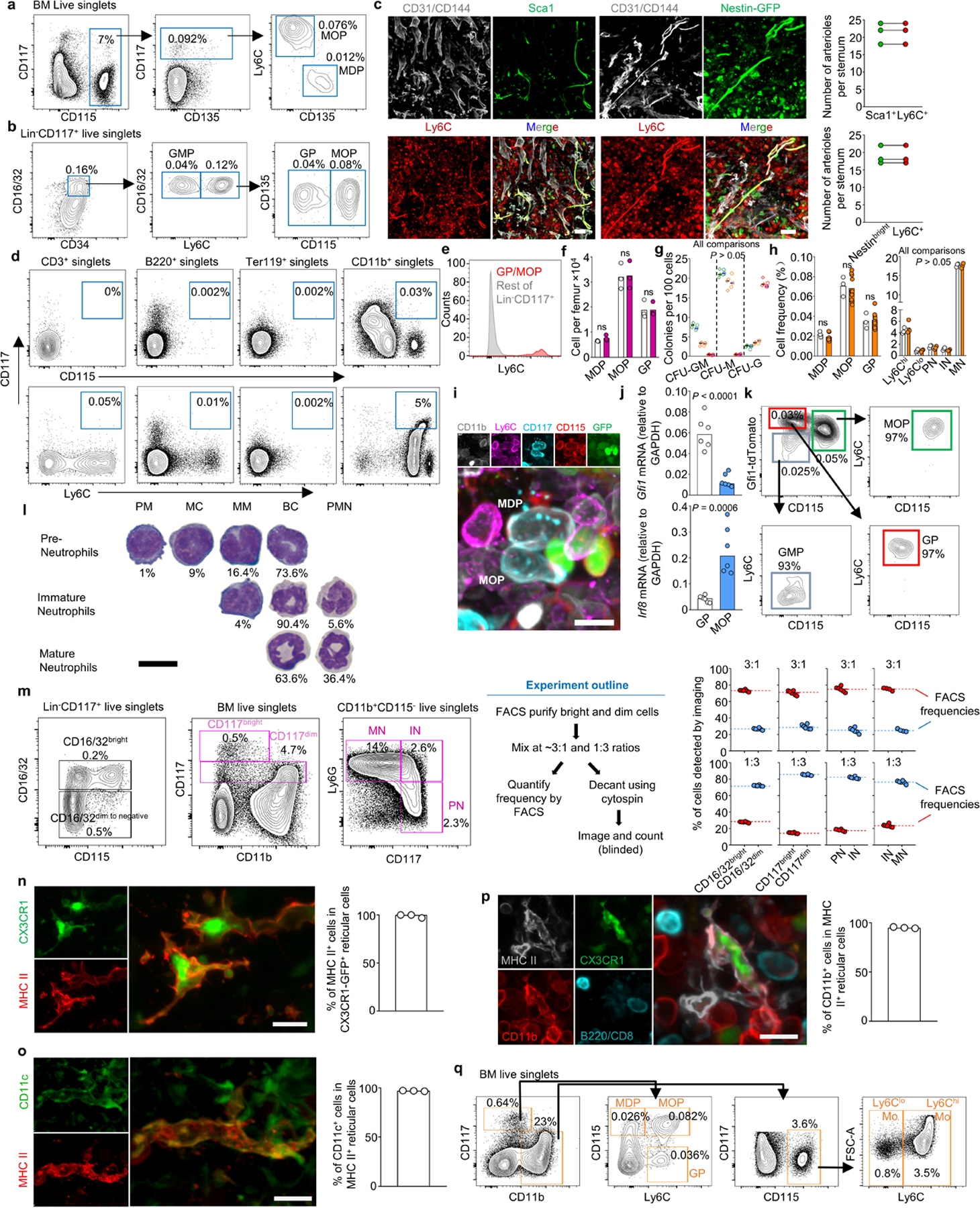
a, FACS plots showing the gating strategy to identify MDP and MOP (as described in reference19). b, Gating strategy to identify GMP, GP, and MOP (as described in reference16). The Lineage panel contains antibodies against Ly6G, CD11b, Ter119, B220 and CD3. c, Images showing that Ly6C labels arterioles detected as either CD31+CD144+Sca1bright in wild-type mice or CD31+CD144+Nestinbright structures in Nestin-GFP mice; the histograms show quantifications demonstrating that all Sca1+ arterioles or Nestin-GFPbright arterioles are also Ly6C+. Scale bars = 50 μm. d, e, FACS plots (d) showing that only the CD11b+ gate contains CD117+CD115+ or CD117+Ly6C+ cells and histogram (e) showing that GP and MOP are the only Ly6C+ cells in the Lin-CD117+ gate. Together these data indicate that CD11b alone can be used to replace the Lineage panel to exclude contamination of mature cells when detecting MDP, MOP and GP. f, g, Cell numbers per femur (f) and colony forming activity (g, green: MDP, orange: MOP, red: GP; n = total 3 mice in two experiments) of the indicated progenitors using previously described strategies16,19 (diamonds) or the one described in Fig1.g (circles). h, Frequency of total BM cells for each of the indicated populations in sternum when detected by FACS (white) or imaging (orange). i. Representative image showing that CD11b-CD117+CD115+Ly6C- MDP and CD11b-CD117+CD115+Ly6C+ MOP are GFP+ in Cx3cr1-gfp mice. Scale bar = 10 μm. j, qPCR showing Gfi1 and Irf8 expression (relative to Gapdh) in FACS-purified GP or MOP; n = total 6 mice in three experiments. k, FACS analyses in Gfi1-tdTomato mice showing differential tdTomato expression in GP and MOP in Gfi1-Tdtomato mice. l, Quantification of promyelocytes (PM), myelocytes (MC), metamyelocytes (MM), banded cells (BC) and polymorphonucleated neutrophils (PMN) in cytospin preparations of FACS-purified Pre-Neutrophils, Immature Neutrophils and Mature Neutrophils. n = total 2 mice. Scale bar = 10 μm. m, The stains require discrimination of CD16/32, CD117, and Ly6G bright and dim cells. The panels show the gating strategy, experimental design, and quantification of frequencies of decanted CD16/32 and CD117 bright and dim cells or IN, PN, and MN when compared to frequencies obtained by FACS prior cytospin. Each dot represents one image field from two experiments. n, o, Dendritic cells can be imaged as reticulated CX3CR1-GFP+ or CX3CR1-GFP+MHCII+ cells in Cx3cr1-gfp reporter mice23,24. The images and histograms show that all reticulated GFP+ cells were also MHCII+ and CD11c+ indicating that MHCII and cell shape are sufficient to unambiguously identify DC and distinguish them from macrophages that are CXCR1-GFP-CD11c- cells24; n = total 3 mice. Scale bar = 10 μm. p, Image and histogram showing that CX3CR1-GFP+MHCII+ dendritic cells are conventional dendritic cells as they are CD11b+ but do not express B220 or CD8. Scale bar = 10 μm. n = total 3 mice q, FACS gating strategy for isolation and imaging of the indicated cells. Dendritic cells are detected as MHCII+ reticulated cells in imaging analyses. In all bar graphs one dot corresponds to one mouse. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant.
Extended Data Figure 2. Strategies to map myelopoiesis in whole mounted sternum.
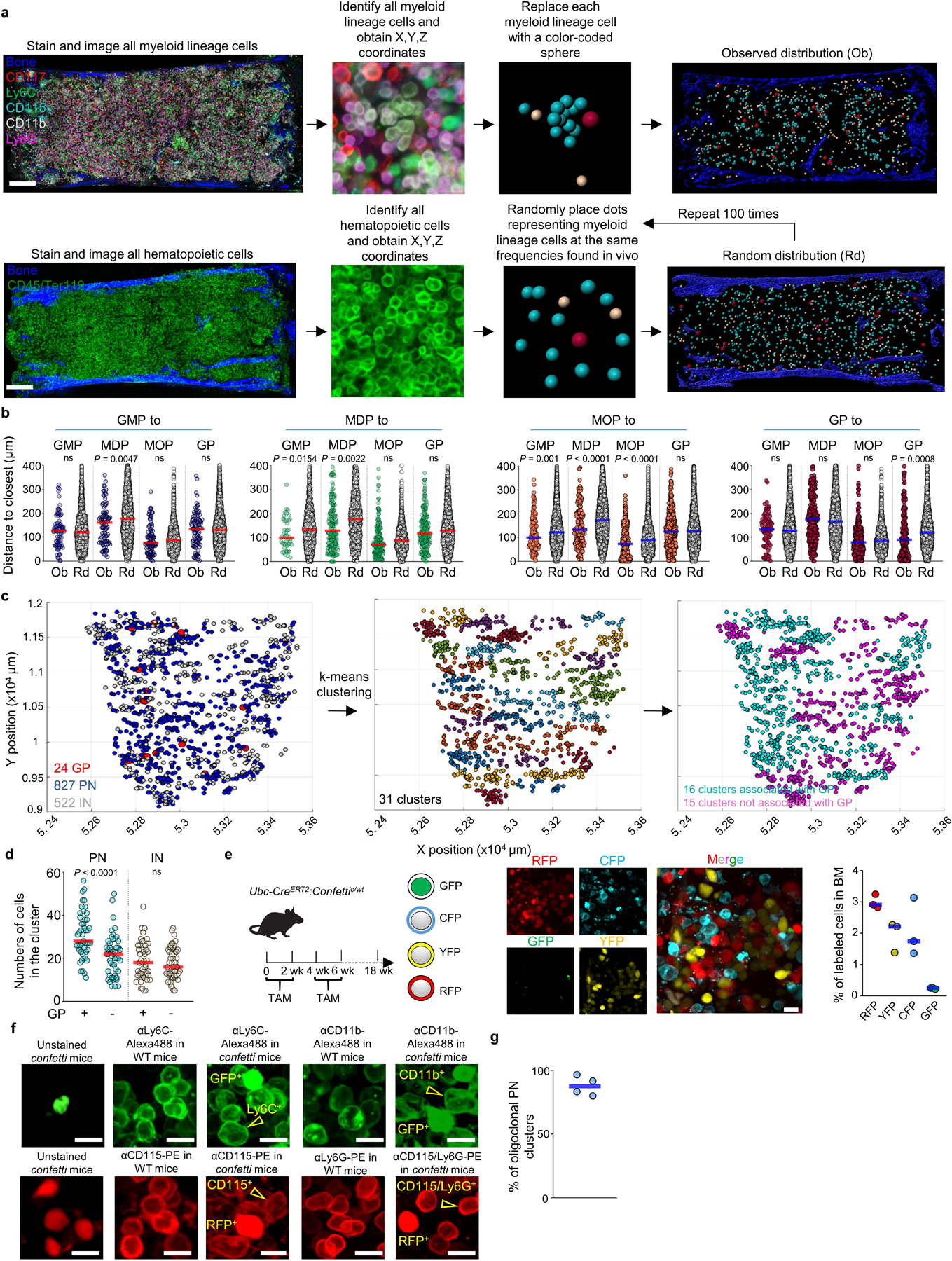
a, Scheme showing the experimental pipeline to identify, obtain the X, Y and Z coordinates, and replace each myeloid cell with a color-coded sphere centered in the cell to better visualize differentiation and to generate random distributions. Scale bar = 200 μm. b, Histograms showing the observed distribution of distances from each GMP (blue), MDP (green), MOP (orange) and GP (red) or random cells (white) to the closest indicated cell (n = 86 GMP from 4 sternum sections of 4 mice, n = 243 MDP from total 23 sternum sections of 15 mice, n = 458 MOP from total 11 sternum sections of 11 mice, n = 338 GP from total 15 sternum sections of 12 mice). c, xy graphs showing the location of GP, PN, and IN in mouse sternum sections (left); the different color coded PN/IN clusters identified using the K-means algorithm (center); and PN/IN clusters containing (pink) or not containing (blue) GP within the cluster. d, Number of PN and IN in each type of cluster. n = 1443 PN from total 3 sternum sections of 3 mice in cluster with GP, n = 1050 PN from total 3 sternum sections of 3 mice in cluster without GP. n = 880 IN from total 3 sternum sections of 3 mice in cluster with GP, n = 866 PN from total 3 sternum sections of 3 mice in cluster without GP. e, Experimental design, representative image, and histogram showing the percentage of CFP, GFP, RFP and YFP positive cells in fate mapping experiments using Ubc-creERT2:confetti mice. Scale bar = 10 μm. Each dot represents one sternum segment from total 3 confetti mice. f, In the confetti model GFP is detected in the nucleus whereas RFP is expressed in the cytoplasm21. The images show that by using antibodies conjugated to fluorochromes (Alexa Flour488 and Phycoerythrin, PE) that spectrally overlap with GFP or RFP but that stain only the membrane we could distinguish CD11b-Alexa488+GFP-, Ly6C-Alexa488+GFP- cells from GFP+ cells and CD115-PE+RFP-, and Ly6C-PE+RFP- from RFP+ cells. This allowed us to examine the relationships between YFP- or CFP-labeled cells. Since we could not distinguish the membrane signal from the nuclear/cytoplasmic signal in GFP+ or RFP+ cells these were discarded from the analyses. Scale bar = 10 μm. g, Percentage of PN clusters with at least 1 confetti-labeled PN that are oligoclonal (containing cells with at least two different origins: CFP+, YFP+, or no confetti label). Each dot represents one sternum segment from total 3 confetti mice. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant.
Extended Data Figure 3. Confetti analyses of mono/DCpoiesis.
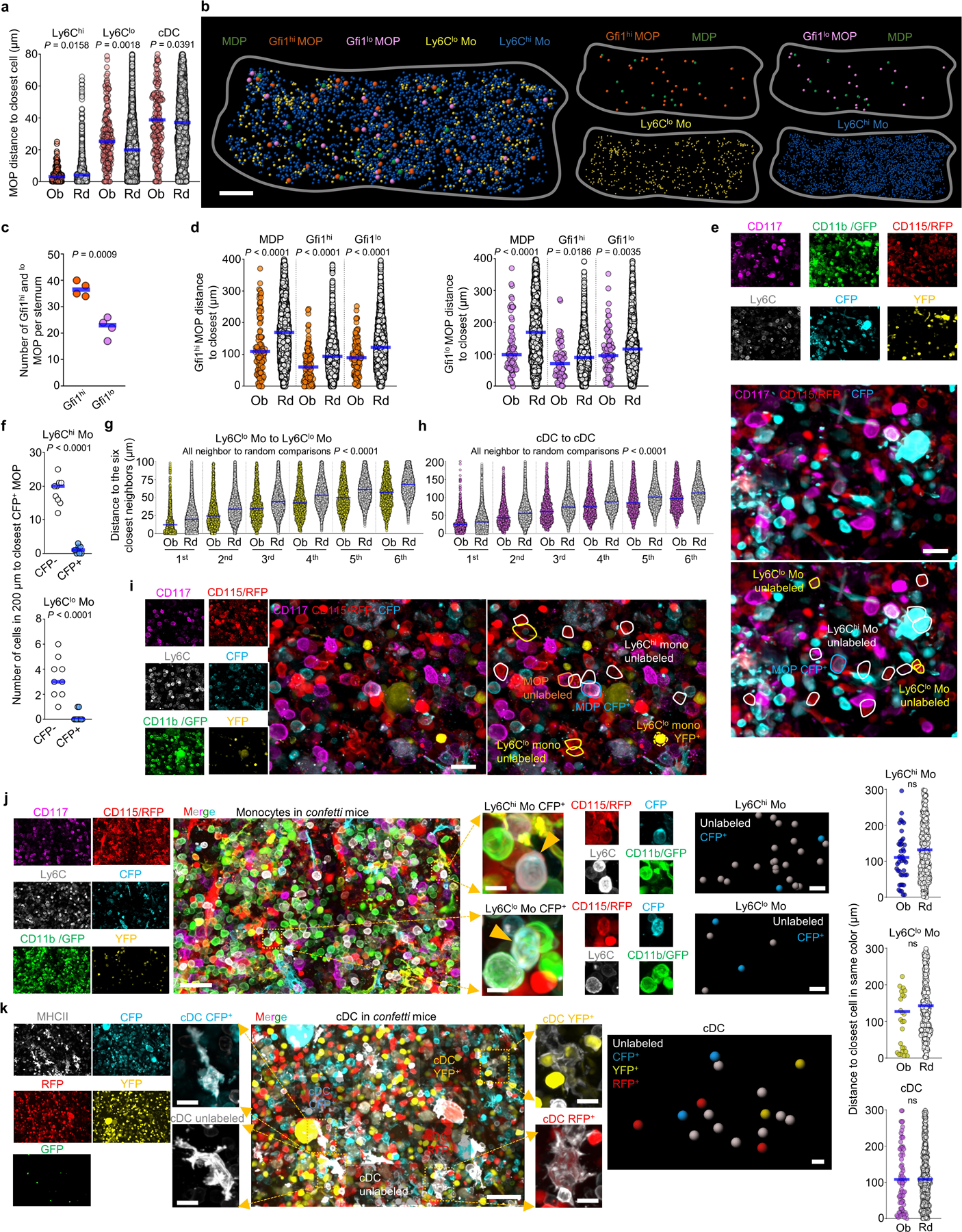
a, Histograms showing the observed (color) and random (white) distribution of distances from each MOP to the closest indicated cells (MOP-Ly6Chi monocyte, n = 137 MOP from 3 total sternum sections of 3 mice; MOP-Ly6Clo monocyte, n = 171 MOP from total 4 sternum sections of 4 mice; MOP-cDC, n = 200 MOP from total 5 sternum sections of 4 mice). b, Maps showing the location of Gfi1hi and Gfi1lo MOP, MDP, Ly6Chi, and Ly6Clo monocytes in a sternum segment from Gfi1-tomato mice. Scale bar = 200 μm. c, Number of Gfi1hi and Gfi1lo MOP per sternum segment. Each dot represents one sternum segment from 3 Gfi1-tomato mice in two experiments. d, Histograms showing the observed (color) and random (white) distribution of distances from each Gfi1hi MOP (orange) and Gfi1loMOP (purple) to the closest indicated cells. n = 113 Gfi1hi MOP and n = 72 Gfi1lo MOP from total 3 sternum segments from 3 mice. e, Representative image showing lack of contribution of a confetti-labeled MOP to surrounding monocytes. Tracked cells are YFP+, CFP+ or unlabeled CD117+CD115+CD11b-Ly6C+MOP, CD117-CD115+CD11b+Ly6Chi monocytes and CD117-CD115+CD11b+Ly6Clo monocytes. Scale bar = 20 μm. f, Quantification of cell numbers for CFP- (white) and CFP+ (blue) Ly6Chi monocytes (up) or Ly6Clo monocytes (down) found within the indicated distances to the closest CFP+ MOP cell. Each dot represents one CFP+ MOP (n = 9 MOP from total 6 sternum segments from 4 confetti mice). g, h, Histograms showing the observed (color) and random (white) distribution of distances from each Ly6Clo Mo (yellow) and cDC (pink) to the six closest indicated neighbor cells. n = 1603 Ly6Clo monocytes from total 3 sternum segments of 3 mice; n = 1228 cDC from total 6 sternum segments of 6 mice. i. Image showing lack of contribution of a CFP+ MDP to surrounding Ly6Clo monocytes. Tracked cells are YFP+, CFP+ or unlabeled CD117+CD115+CD11b-Ly6C-MDP, CD117+CD115+CD11b-Ly6C+ MOP, CD117-CD115+CD11b+Ly6Chi monocytes and CD117-CD115+CD11b+Ly6Clomonocytes. Scale bar = 20 μm. j, Image showing lack of association between confetti-labeled Ly6Chi and Ly6Clo monocytes. Tracked cells are YFP+, CFP+ or unlabeled CD11b+CD115+CD117-Ly6Chi monocytes and CD11b+CD115+CD117-Ly6Clo monocytes. Scale bar = 40 μm or 10 μm for zoomed in image. The histograms show the observed (color) and random (white) distribution of distances from each confetti-labeled Ly6Chi (blue) or Ly6Clo monocytes (yellow) to the closest Ly6Chi or Ly6Clo monocyte in the same color. n = 48 confetti-labeled Ly6Chi monocytes, and n=32 confetti-labeled Ly6Clo monocytes from total 3 sternum sections of 3 mice. k, Image showing lack of association between confetti labeled cDC. Tracked cells are RFP+, GFP+, YFP+, CFP+ or unlabeled MCHII+ reticulated cDC. Scale bar = 40 μm or 10 μm for zoomed in image. The histogram shows the observed (color) and random (white) distribution of distances from each confetti-labeled cDC (pink) to the closest cDC in the same color. n = 80 confetti-labeled cDC from total 3 sternum sections of 3 mice. Unless otherwise indicated for all graphs one dot corresponds to one cell. Horizontal blue bars indicate the median distance. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant.
Extended Data Figure 4. The architecture of myelopoiesis is similar in different sternum sections.
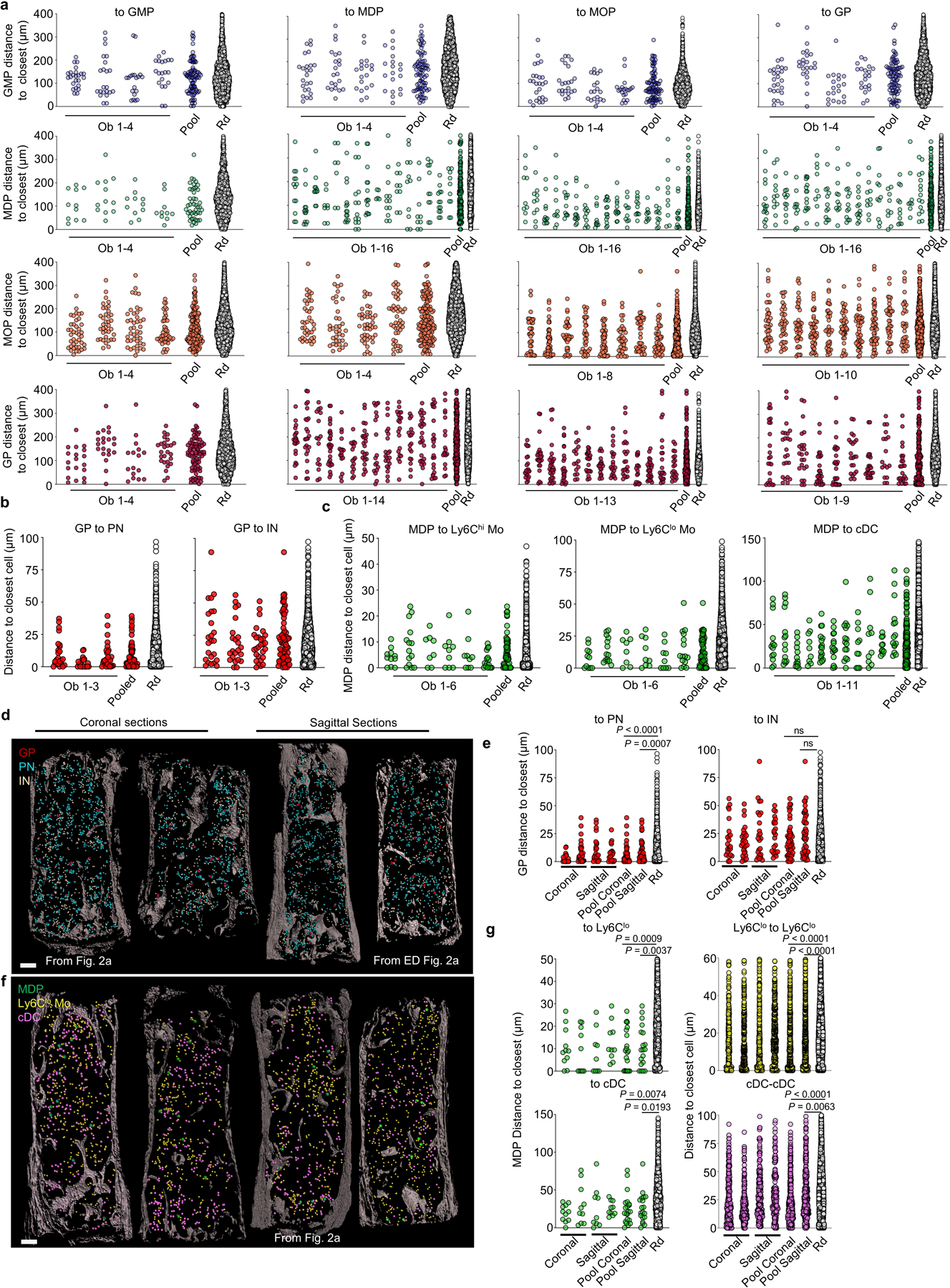
a, Observed distributions of distances for individual sternum sections (Ob), the pooled values, or random (Rd) simulations for the data shown in Extended Data Fig. 2b. For distance to GMP, n = 25, 22, 19, 20 GMP per sternum section from total 4 sternum sections in 4 mice; n = 11, 12, 11, 10 MDP per sternum section from total 4 sternum sections in 4 mice; n = 40, 37, 42, 46 MOP per sternum section from total 4 sternum sections in 4 mice; n = 19, 21, 17, 26 GP per sternum section from total 4 sternum sections in 4 mice. For distance to MDP, n = 25, 22, 19, 20 GMP per sternum section from total 4 sternum sections in 4 mice; n = 13, 14, 11, 9, 7, 14, 14, 14, 9, 11, 12, 10, 10, 11, 13, 11 MDP per sternum section from total 16 sternum sections in 16 mice; n = 40, 37, 42, 46 MOP per sternum section from total 4 sternum sections in 4 mice; n = 22, 24, 23, 28, 23, 16, 24, 18, 22, 21, 17, 26, 25, 20 GP per sternum section from total 14 sternum sections in 14 mice; For distance to MOP, n = 25, 22, 19, 20 GMP per sternum section from total 4 sternum sections in 4 mice; n = 9, 11, 12, 10, 11, 13, 11, 10, 14, 11, 13, 10, 9, 11, 10, 14 MDP per sternum section from total 16 sternum sections in 16 mice; n = 39, 49, 38, 33, 44, 36, 37, 44 MOP per sternum section from total 8 sternum sections in 8 mice; n = 22, 24, 28, 23, 16, 24, 18, 22, 21, 17, 26, 25, 20 GP per sternum section from total 13 sternum sections in 13 mice; For distance to GP, n = 25, 22, 19, 20 GMP per sternum section from total 4 sternum sections in 4 mice; n = 15, 12, 11, 10, 15, 9, 12, 11, 12, 11, 13, 10, 9, 11, 10, 15 MDP per sternum section from total 16 sternum sections in 16 mice; n = 40, 37, 42, 46, 39, 39, 49, 53, 38, 34 MOP per sternum section from total 10 sternum sections in 10 mice; n = 24, 23, 23, 28, 29, 22, 26, 17, 26 GP per sternum section from total 9 sternum sections in 9 mice. b, c, As “a” but corresponding to the data shown in Fig. 2c and Fig. 2i. n = 24, 23, 28 GP per sternum section from total 3 sternum sections in 3 mice; For MDP to Ly6Chi and lo Mo, n = 11, 14, 8, 9, 9, 16 MDP per sternum section from total 6 sternum sections in 4 mice. For MDP to cDC, n = 13, 14, 12, 9, 16, 15, 15, 10, 10, 10, 15 MDP per sternum section from total 11 sternum sections in 6 mice. d, e, Through the manuscript we have indistinctly used coronal and sagittal sternum sections. The maps show granulopoiesis in coronal and sagittal sternum sections shown or analyzed in Fig. 2a or Extended Data Fig. 2a (d) and observed and random distributions of distances for the indicated cells in each section or the pooled data for each type of section (e). n = 23 and 28 GP in coronal sections and 24 and 23 GP in sagittal sections. Scale bar = 200 μm. f, g, Representative maps of mono/DCpoiesis comparing coronal and sagittal sternum sections with those shown or analyzed in Fig. 2a (f) and observed and random distributions of distances for the indicated cells in coronal or sagittal sections (g). n = 11 or 10 MDP, 390 or 334 Ly6Clo Monocyte, and 218 or 258 cDC in coronal sections; n = 9 or 10 MDP, 419 or 380 Ly6Clo Monocyte and 183 or 159 cDC in sagittal sections. Scale bar = 200 μm. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant.
Extended Data Figure 5. Myeloid progenitor interaction with the microenvironment and HSC.
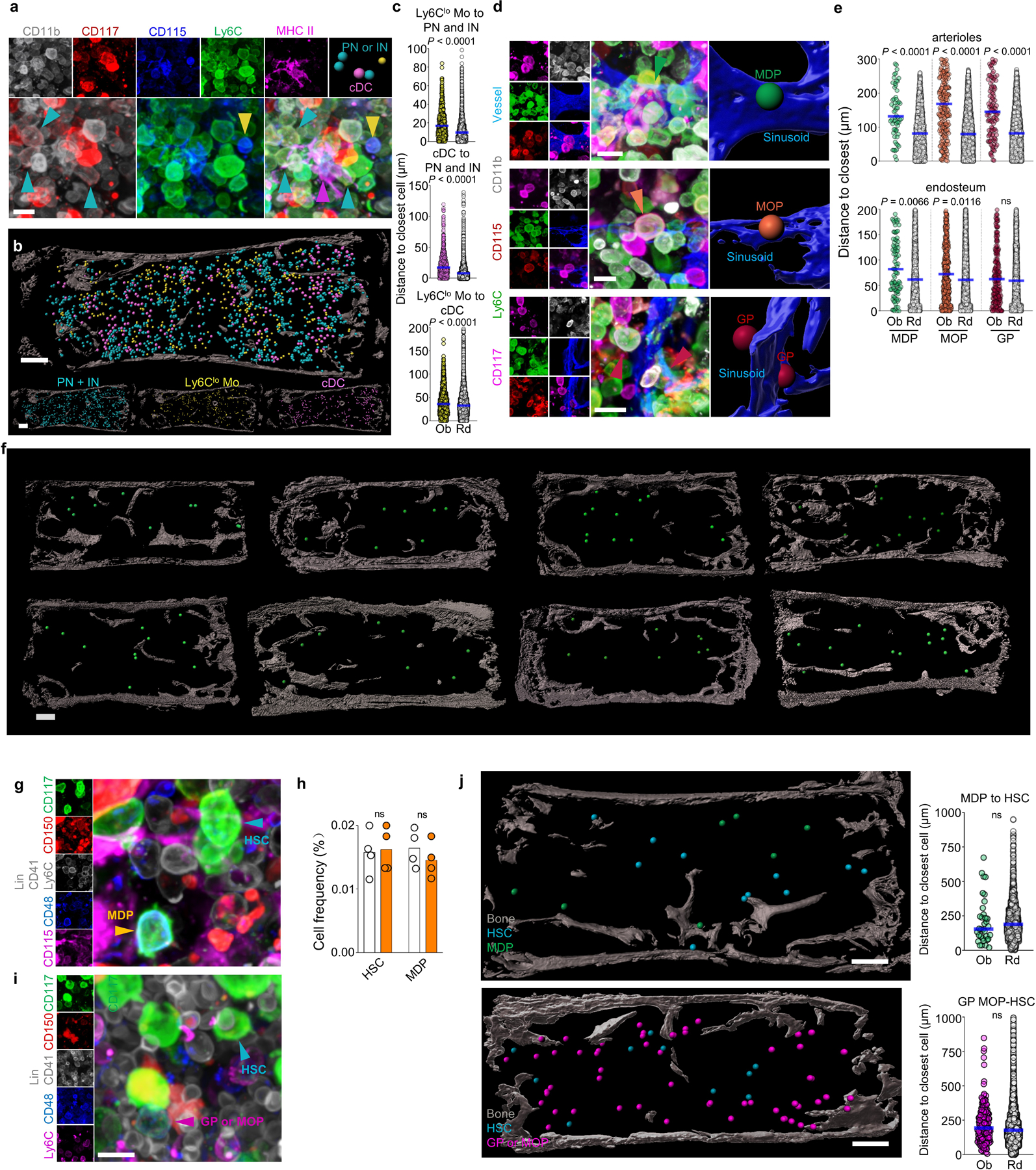
a, Representative image showing simultaneous detection of Pre- and Immature neutrophils, Ly6Clo monocytes and cDC. Scale bar = 10 μm. b, Map showing the location of the indicated cells in the bone marrow. Each dot corresponds to one cell. Note that the radius of each dot is 2x the average radius of the cell. Scale bar = 200 μm. c, Histograms showing the distance from each Ly6Clo Mo (yellow dots) or cDC (pink dots) and their random simulation (white dots) to the closest indicated cell (Ly6Clo to PN/IN, n = 500 Ly6Clo; cDC to PN/IN, n = 727 cDC; Ly6Clo to cDC, n = 1322 Ly6Clo Mo; from total 3 sternum sections of 3 mice). d, High-power images showing the relative positions of MDP, MOP, GP and sinusoids. Scale bar = 10 μm. e, Histograms showing the distance from each MDP (green dots), MOP (orange dots), GP (red dots) or random distribution (white dots) to the closest indicated structure (for distances to arterioles: n = 62 MDP from total 6 sternum sections of 6 mice; n = 218 MOP, and n = 114 GP from total 5 sternum sections of 5 mice; for distances to endosteal surface n = 98 MDP, n = 410 MOP, n = 217 GP, from total 9 sterna of 6 mice). f, Representative images of multiple sternum segments showing that MDP are evenly distributed through the bone marrow, consistent with their sinusoidal location. Scale bar = 200 μm. g, Representative image showing detection of HSC and MDP in a single stain. Scale bar = 10 μm. h, Quantification of MDP and Lin- CD117+CD48-CD41dimCD150+ HSC in femurs by FACS (white) or imaging (orange) analyses. Each dot corresponds to one mouse femur or sternum image. n = 4. i, Representative image showing detection of HSC and a population containing CD117+Ly6C+ GP and MOP in a single stain. Scale bar = 10 μm. j, Maps and histograms showing the relationships between HSC and MDP or GP/MOP in the bone marrow. In the map the dot radius is three times the average cell radius. Scale bar = 200 μm. (n = 35 MDP from total 4 sternum sections of 3 mice and n = 191 GP and MOP from total 3 sternum sections of 3 mice). Unless otherwise indicated for all graphs one dot corresponds to one cell. Horizontal blue bars indicate the median distance. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant.
Extended Data Figure 6. Broad comparison of stromal bone marrow compartments by single-cell RNA-Seq.

Comparative analyses of two previously described scRNA-Seq datasets profiling stromal and hematopoietic cell population in bone marrow (9,165 cells from Tikhonova et al. and 89,007 cells Baryawno et al.). a-c, Cell population predictions displayed on a UMAP plot from an unsupervised analysis of the two separate scRNA-Seq datasets (ICGS version 2). Distinct captures are denoted by the gating strategy (col2.3, niche col2.3, niche-LepR+, niche VEcad+). Populations are denoted as hematopoietic or stromal/mesenchymal based on prior-defined scRNA-Seq cell population marker gene signatures (ICGS – see cluster labels). d-l, Relative expression of ICGS2 cell-population marker genes (identified in both datasets) projected on to the two UMAP plots to verify cell identity, relative to capture strategy. Supplementary Table 2 shows the different ICGS2 marker genes and Cell barcode assignments for the difference cell clusters identified.
Extended Data Figure 7. CSF1 from LepR+ cells is dispensable for myelopoiesis.
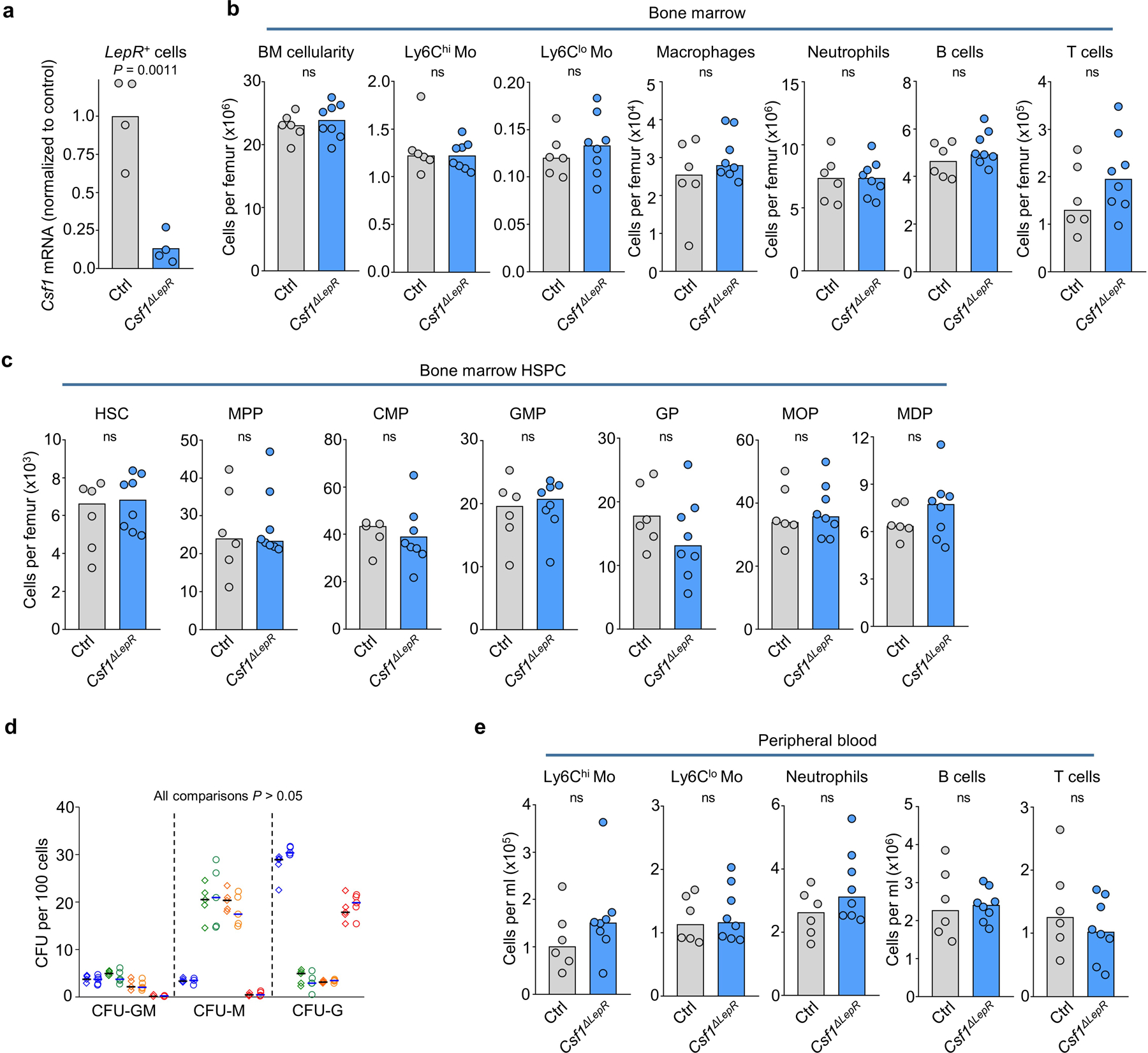
a. qPCR analyses showing Csf1 mRNA levels (relative to Gapdh) in Nestin-GFPdim perivascular cells (which largely overlap with LepR+ perivascular cells45). n = total 4 control mice and n = total 4 Csf1ΔLepR mice in four experiments. b, c, Number of BM cells or the indicated populations in the femur of control or Csf1ΔLepR mice. n = total 6 control and n = total 8 Csf1ΔLepR mice in six experiments. d, Colony forming activity (blue: GMP, green: MDP, orange: MOP, red: GP) of the indicated progenitors FACS-purified from control (diamonds, n = 5) or Csf1ΔLepR (circles, n = 5) total mice in four experiments. e, Number of the indicated populations in the blood of control or Csf1ΔLepR mice. For all panels one dot equals one mouse. n = total 6 control and n= total 8 Csf1ΔLepR mice in six experiments. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant.
Extended Data Figure 8. CSF1 from a subset of endothelial cells is necessary for DCpoiesis.
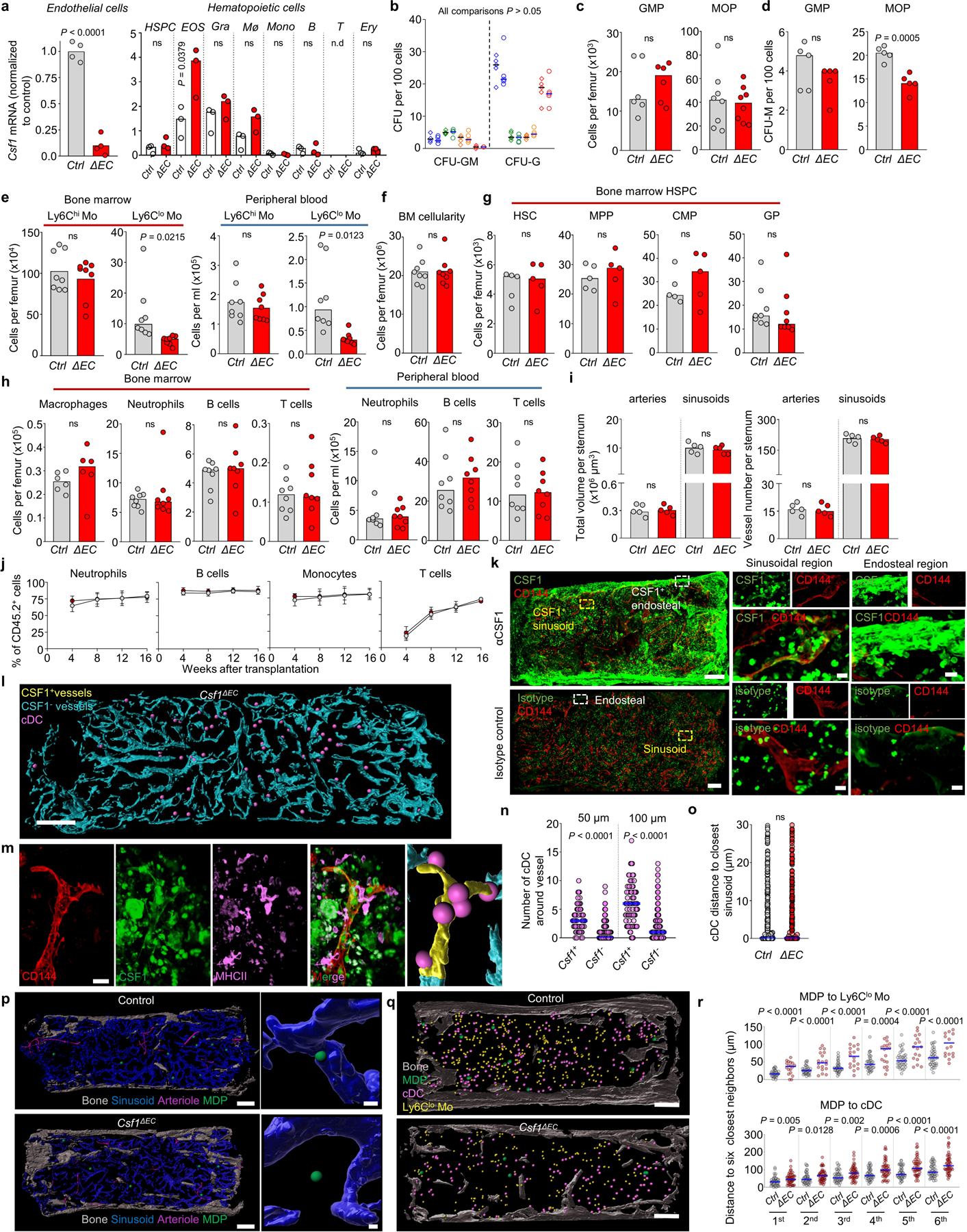
a, qPCR analyses showing Csf1 mRNA levels (normalized to endothelial Control) in FACS-purified endothelial cells and the indicated hematopoietic cells in control or Csf1ΔEC mice; n = total 4 control and n = total 4 Csf1ΔEC mice in four experiments. Note that although Cdh5-cre also recombines in subsets of hematopoietic cells it does not affect Csf1 expression in these cells. (n.d = none detected); n = total 3 control and n = total 3 Csf1ΔEC mice in three experiments. b, Colony forming activity (blue: GMP, green: MDP, orange: MOP, red: GP) of the indicated progenitors FACS-purified from control (diamonds, n = 5) or Csf1ΔEC (circles, n = 5) total mice in four experiments. c, d, Number (c) and CFU-M activity (d) of GMP or MOP from control or Csf1ΔEC mice. n = total 6 control and n = total 6 Csf1ΔEC mice in five experiments. e, Number of Ly6Chi and Ly6Clo monocytes in the bone marrow and peripheral blood of control or Csf1ΔEC mice. n = total 8 control and n = total 8 Csf1ΔEC mice in six experiments. f-h, Number of BM cells for the indicated populations in the femur or blood of control or Csf1ΔEC mice. n = total 8 control and n = total 8 Csf1ΔEC mice in six experiments. i, Volume and number of vessels in sternum sections of control or Csf1ΔEC mice. Each dot represents one sternum segment from n = 3 control and n = 3 Csf1ΔEC mice. j, The panels show the percentage of the indicated CD45.2+ cells in the blood of lethally irradiated CD45.1+ recipients after transplant of 106 BM cells purified from Ctrl ( white dots) or Csf1ΔEC mice (red dots) - both CD45.2+- together with 106 CD45.1+ competitor cells at the indicated time points after transplantation. The dots show the mean of 12 mice per group and the error bars show standard deviation. k, Representative images showing anti-CSF1 or isotype control stain in the BM of wild-type mice. Scale bars = 200 μm and 10 μm. l, Map of CSF1+ and CSF1- vessels and cDC in Csf1ΔEC mice. Scale bar = 200 μm. m, high-power image showing a CSF1+ vessel and cDC (pink dots) in Ctrl mice. The radius of the dot is 2x the average cDC radius. Scale bar = 20 μm. n, Number of cDC found within the indicated distances of CSF1+ and CSF1- vessels in wild-type (n = 76 CSF1+ vessels and n = 520 CSF1- vessels in total 4 sternum sections of 3 wild-type mice). o, Histograms showing the distance from each cDC to the closest sinusoid in control or Csf1ΔEC mice (n = 451 cDC in total 2 sternum sections of 2 control mice, n = 343 cDC in total 3 sternum sections of 3 Csf1ΔEC mice). p, Maps showing the relocation of MDP away from sinusoids in Csf1ΔEC mice. Scale bars = 200 μm and 10 μm. The radius of the dots is 3x (left map) or 1x (right images) the average radius of the MDP. q, Maps showing the distribution of MDP, Ly6Clo Mo, and cDC in the sternum of control or Csf1ΔEC mice. Scale bar = 200 μm. The radius of the dot is 3x (MDP) or 2x (all other cells) the average radius of the replaced cell. r, Histograms showing the distribution of distances from each MDP to the six closest Ly6Clo Mo or cDC in control or Csf1ΔEC mice (For MDP-Ly6Clo monocyte, n = 37 MDP from total 4 sternum sections of 3 Control mice, n = 18 MDP from total 4 sternum sections of 3 Csf1ΔEC mice. For MDP-cDC, n = 47 MDP from total 6 sternum sections of 3 Control mice, n = 47 MDP from total 9 sternum sections of 3 Csf1ΔEC mice). Unless otherwise indicated for panels a-i each dot corresponds to one mouse; for panels l-r each dot corresponds to one cell. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns= not significant.
Extended Data Figure 9. Changes in progenitor localization after infection.
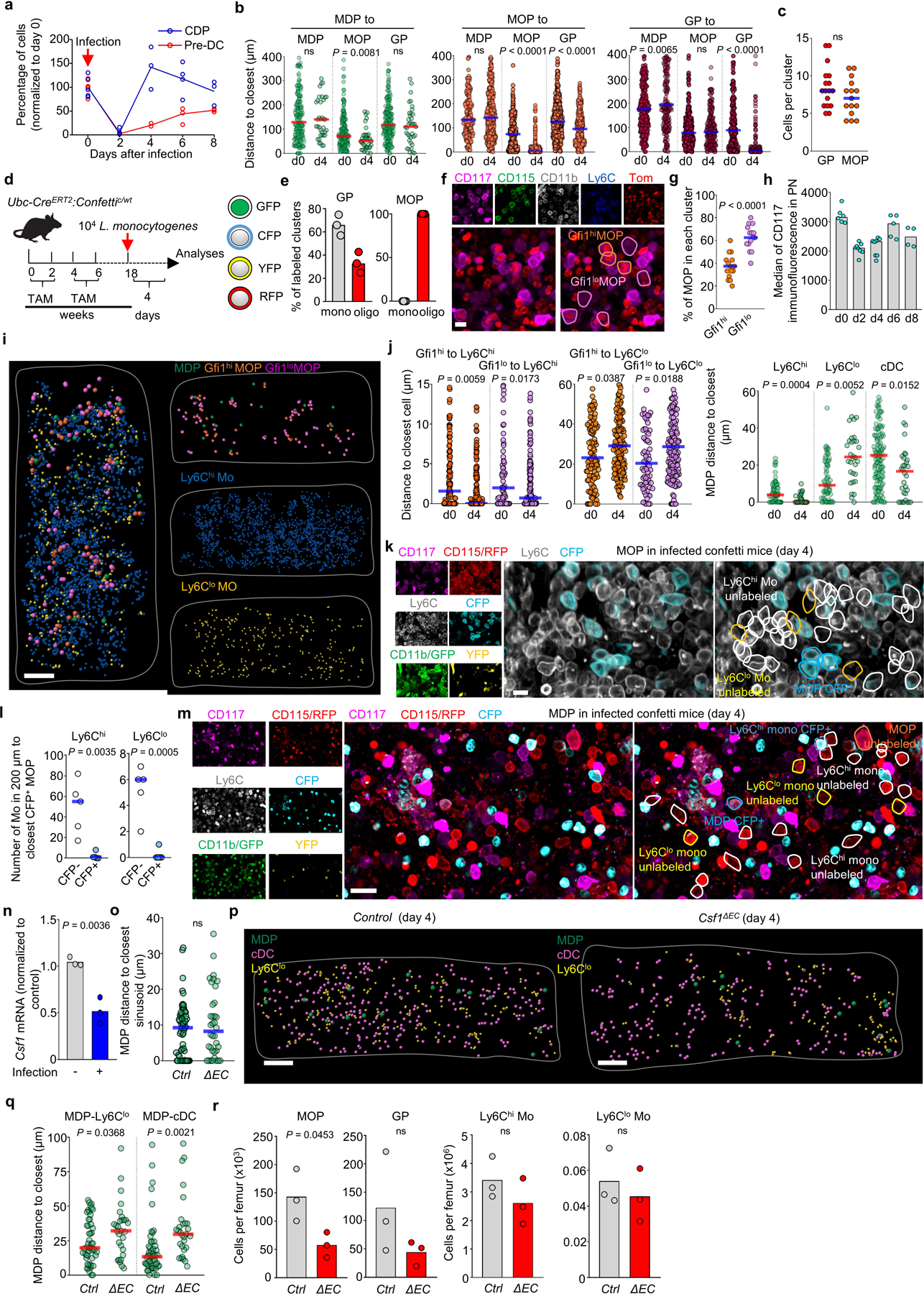
a, Average percentage of the indicated cells per femur (normalized to day 0) at the indicated time points after L. monocytogenes infection of wild-type mice. (n = 6 mice for day 0 and day 2, n = 3 mice for day 4, n = 4 mice for day 6 and n = 3 mice for day 8). b, Histograms showing the distribution of distances from each MDP (green, n = 188 MDP from total 16 sternum sections of 6 uninfected wild-type mice and n = 37 MDP from total 3 sternum sections of 3 wild-type mice four days after L. monocytogenes infection), MOP (orange, n = 165 MOP from total 4 sternum sections of 3 uninfected wild-type mice and n = 377 MOP from total 3 sternum sections of 3 wild-type mice four days after L. monocytogenes infection) or GP (red, n = 308 GP from total 14 sternum sections of 6 uninfected wild-type mice and n = 218 GP from total 3 sternum sections of 3 wild-type mice four days after L. monocytogenes infection) to the indicated progenitors four days after L. monocytogenes infection of wild-type mice. c, number of GP or MOP per cluster for the sternum sections analyzed in b. Each dot represents a cluster; n = 16 GP clusters and n = 15 MOP clusters from total 3 wild-type mice four days after L. monocytogenes infection. d, Experimental design for in vivo fate mapping using Ubc-creERT2:confetti mice. e, Percentage of GP or MOP clusters with at least 1 confetti labeled GP or MOP that are monoclonal (all cells in the cluster are labeled in the same confetti color) or oligoclonal (containing cells with at least two different origins: CFP+, YFP+, or no confetti label). Each dot represents a GP or MOP cluster from total 3 sternum segments from 2 confetti mice in two experiments four days after L. monocytogenes infection. f, Representative image showing a cluster composed of MDP-derived Gfi1lo and GMP-derived Gfi1hi MOP in Gfi1-tomato mice four days after L. monocytogenes infection. Scale bar = 10 μm. g, Percentage of GMP-derived Gfi1hi MOP (orange dots) or MDP-derived Gfi1lo MOP (purple dots) per cluster (each dot = 1 cluster, n = 15 clusters in total 3 sternum sections from 2 Gfi1-tomato mice four days after L. monocytogenes infection). h, Quantification of CD117 expression in PN of wild type mice at the indicated time points after infection. (n = 6 mice for day 0, n = 8 mice for day 2 and day 4, n = 5 mice for day 6 and n = 4 mice for day 8). i, Map showing the location of Gfi1hi and Gfi1lo MOP, MDP, Ly6Chi, and Ly6Clo monocytes in a sternum segment from Gfi1-tomato mice four days after infection. Scale bar = 200 μm. j, Histograms showing the distribution of distances from each Gfi1hi (orange) and Gfi1lo (purple) MOP and MDP (green) to the indicated cells in uninfected wild-type mice or four days after L. monocytogenes infection. Note that the d0 values are the same as shown in Fig. 2g,i. (n = 113 Gfi1hi MOP and n = 72 Gfi1lo MOP from total 3 sternum sections of 3 uninfected Gfi1-tomato mice; n = 155 Gfi1hi MOP and n = 144 Gfi1lo MOP from total 3 sternum sections of 2 Gfi1-tomato mice four days after L. monocytogenes infection; MDP-Ly6Chi monocyte, n = 67 MDP from total 6 sternum sections of 4 mice; MDP-Ly6Clo monocyte, n = 67 MDP from total 6 sternum sections of 4 wild-type uninfected mice; MDP-cDC, n = 139 MDP from total 11 sternum sections of 6 wild-type uninfected mice; and n = 32 MDP from total 3 sternum sections of 3 wild-type mice four days after L. monocytogenes infection). k, Representative image showing lack of contribution of a confetti-labeled MOP. Tracked cells are YFP+, CFP+ or unlabeled CD117+CD115+CD11b-Ly6C+ MOP, CD117-CD115+CD11b+Ly6Chi monocytes and CD117-CD115+CD11b+Ly6Clo monocytes. Scale bar = 10 μm. l, Quantification of cell numbers for CFP- (white) and CFP+ (blue) Ly6Chi monocytes (left) or Ly6Clo monocytes (right) found within the indicated distances to the closest CFP+ MOP cell in the confetti mice four days after L. monocytogenes infection. Each dot represents one CFP+ MOP from total 8 sternum segment of 2 confetti mice in two experiments four days after L. monocytogenes infection. m, Representative image showing lack of contribution of a confetti-labeled MDP to surrounding monocytes. Tracked cells are YFP+, CFP+ or unlabeled CD117+CD115+CD11b-Ly6C- MDP, CD117+CD115+CD11b-Ly6C+ MOP, CD117-CD115+CD11b+Ly6Chi monocytes and CD117-CD115+CD11b+Ly6Clo monocytes. Scale bar = 20 μm. n, qPCR analyses showing Csf1 mRNA levels (normalized to not infected) in BM endothelial cells FACS-purified from wild-type mice in the steady-state or 4 days after infection. n = total 3 uninfected mice and n = total 3 infected mice in two experiments. o, Histogram showing the distance from each MDP to the closest sinusoid in control (pool of Cre:Csf1+/−, Csf1+/−, and Csf1fl/-) or Csf1ΔEC mice 4 days after infection. n = 58 MDP from total 4 sternum sections of 3 control mice and n = 36 MDP from total 4 sternum sections of 3 Csf1ΔEC mice. p, q, Maps (p) showing the location of the indicated cells; and histogram (q) showing the distance from each MDP to the closest Ly6Clo monocyte and cDC in control or Csf1ΔEC mice 4 days after infection. n = 51 MDP from total 3 sternum sections of 3 control mice and n = 29 MDP from total 3 sternum sections of 3 Csf1ΔEC mice. r. Number of the indicated cells per femur in control or Csf1ΔEC mice 4 days after infection. Each dot indicates one mouse. n = 3 control and n = 3 Csf1ΔEC mice. Unless otherwise indicated for all panels one dot = one cell. Statistical differences were calculated using two-tailed Student’s T tests and p values are shown. ns = not significant.
Extended Data Figure 10. Architecture of myelopoiesis in the steady-state and after infection.
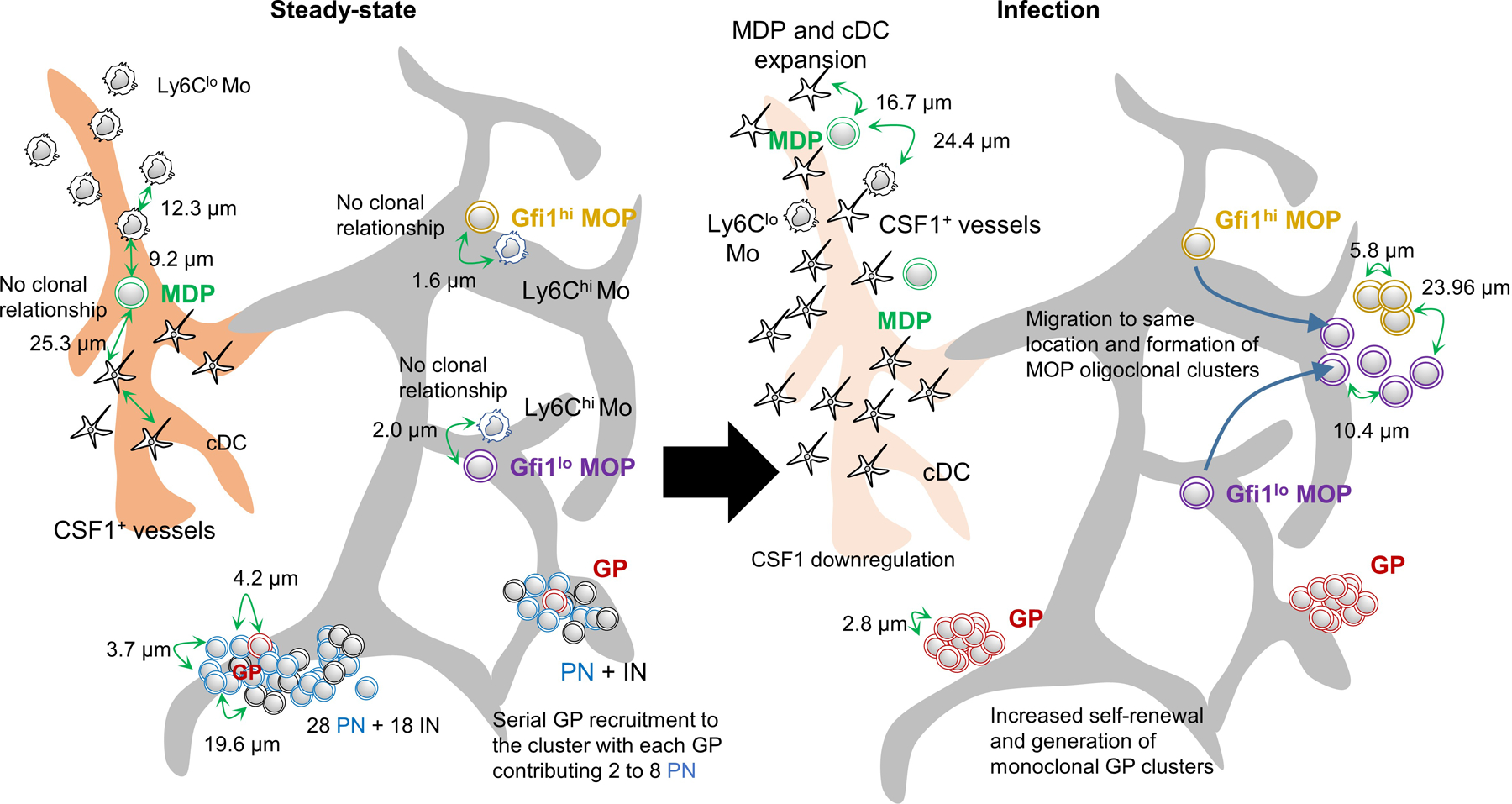
The models show the spatial distribution and average distances between the indicated cells in the steady-state and four days after infection with Listeria monocytogenes.
Supplementary Material
Acknowledgements
We would like to thank Drs. Jose Cancelas, Marie-Dominique Filippi, Damien Reynaud, Daniel Starczynowski, and Andres Hidalgo for valuable feedback on the manuscript. We are grateful to Lai Guan Ng, Immanuel Kwok, and Keith Leong for help in designing granulopoiesis experiments and reviewing the manuscript. We also thank the Confocal Imaging Core, the Research Flow Cytometry Core, and the Veterinary Services at the University of Michigan and Cincinnati Children’s Medical Center for experimental and technical assistance. This work was partially supported by the National Heart Lung and Blood Institute (R01HL122661 to H.L.G, and R01HL136529 to D.L.). VBSP is supported by NIH/NCATS U2CTR002818, NIH/NHLBI U24HL148865, and NIH/NIAID U01AI150748. N.S. is supported by the Cincinnati Pediatric Cell Atlas Center. LHF is supported by Department of Defense (DoD) through PRCRP Award W81XWH-20–1-0870(#CA191188). S.S.W. is supported by NIH through grants R01AI120202, R01AI124657 and DP1AI131080 and by the HHMI Faculty Scholar’s program, March of Dimes Ohio Collaborative for Prematurity Research, and Burroughs Wellcome Fund Investigator in the Pathogenesis Award. JXJ is supported by NIH/NIA AG045040 and Welch Foundation Grant AQ-1507. AS is supported by T32 AI118697/AI/NIAID NIH HHS/United States Data was generated using the SH800 cell sorter funded by NIH S10OD023410.
Footnotes
Declaration of Interest
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper. Supplementary Table 1 lists all the reagents and other resources used for the experiments described. Correspondence and requests for materials should be addressed to Daniel Lucas. Reprints and permissions information is available at www.nature.com/reprints. Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Source Data for quantifications described in the text or shown in graphs plotted in Figs. 1–4 and Extended Data Figs. 1-5 and 7-9 are available with the manuscript. scRNAseq data shown in Figure 3c and Extended Data Figure 6 are reanalyses of published datasets: (GSE12842327 URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128423) and (GSE10889126 URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108891). These were preliminary annotated based on ICGS2 BioMarker database40, followed by a secondary analysis using the supervised classification tool cellHarmony40, comparing all cells to reference hematopoietic (GSE120409 URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120409)44. Supplementary Table 2 shows the different ICGS2 marker genes and Cell barcode assignments for the difference cell clusters identified.
References
- 1.Stanley ER & Chitu V CSF-1 receptor signaling in myeloid cells. Cold Spring Harb Perspect Biol 6, doi: 10.1101/cshperspect.a021857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsson A et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702, doi: 10.1038/nature19348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nestorowa S et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–31, doi: 10.1182/blood-2016-05-716480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tusi BK et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60, doi: 10.1038/nature25741 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327, doi: 10.1038/nature13824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch K et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546, doi: 10.1038/nature14242 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Upadhaya S et al. Kinetics of adult hematopoietic stem cell differentiation in vivo. J Exp Med 215, 2815–2832, doi: 10.1084/jem.20180136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss L The structure of bone marrow. Functional interrelationships of vascular and hematopoietic compartments in experimental hemolytic anemia: an electron microscopic study. J Morphol 117, 467–537, doi: 10.1002/jmor.1051170308 (1965). [DOI] [PubMed] [Google Scholar]
- 9.Mohandas N & Prenant M Three-dimensional model of bone marrow. Blood 51, 633–643 (1978). [PubMed] [Google Scholar]
- 10.Westen H & Bainton DF Association of alkaline-phosphatase-positive reticulum cells in bone marrow with granulocytic precursors. J Exp Med 150, 919–937, doi: 10.1084/jem.150.4.919 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charbord P, Tavian M, Humeau L & Peault B Early ontogeny of the human marrow from long bones: an immunohistochemical study of hematopoiesis and its microenvironment. Blood 87, 4109–4119 (1996). [PubMed] [Google Scholar]
- 12.Ding L & Morrison SJ Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235, doi: 10.1038/nature11885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordeiro Gomes A et al. Hematopoietic Stem Cell Niches Produce Lineage-Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity 45, 1219–1231, doi: 10.1016/j.immuni.2016.11.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herault A et al. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature 544, 53–58, doi: 10.1038/nature21693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comazzetto S et al. Restricted Hematopoietic Progenitors and Erythropoiesis Require SCF from Leptin Receptor+ Niche Cells in the Bone Marrow. Cell Stem Cell 24, 477–486 e476, doi: 10.1016/j.stem.2018.11.022 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanez A et al. Granulocyte-Monocyte Progenitors and Monocyte-Dendritic Cell Progenitors Independently Produce Functionally Distinct Monocytes. Immunity 47, 890–902 e894, doi: 10.1016/j.immuni.2017.10.021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evrard M et al. Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity 48, 364–379 e368, doi: 10.1016/j.immuni.2018.02.002 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Kwok I et al. Combinatorial Single-Cell Analyses of Granulocyte-Monocyte Progenitor Heterogeneity Reveals an Early Uni-potent Neutrophil Progenitor. Immunity 53, 303–318 e305, doi: 10.1016/j.immuni.2020.06.005 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Hettinger J et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol 14, 821–830, doi: 10.1038/ni.2638 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Yona S et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91, doi: 10.1016/j.immuni.2012.12.001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snippert HJ et al. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. Cell 143, 134–144, doi: 10.1016/j.cell.2010.09.016 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Ruzankina Y et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126, doi: 10.1016/j.stem.2007.03.002 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sapoznikov A et al. Perivascular clusters of dendritic cells provide critical survival signals to B cells in bone marrow niches. Nat Immunol 9, 388–395, doi: 10.1038/ni1571 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Zhang B et al. Bone marrow niche trafficking of miR-126 controls the self-renewal of leukemia stem cells in chronic myelogenous leukemia. Nat Med 24, 450–462, doi: 10.1038/nm.4499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Q & Frenette PS Niches for Hematopoietic Stem Cells and Their Progeny. Immunity 48, 632–648, doi: 10.1016/j.immuni.2018.03.024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tikhonova AN et al. The bone marrow microenvironment at single-cell resolution. Nature 569, 222–228, doi: 10.1038/s41586-019-1104-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baryawno N et al. A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell 177, 1915–1932 e1916, doi: 10.1016/j.cell.2019.04.040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mossadegh-Keller N et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature 497, 239–243, doi: 10.1038/nature12026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serbina NV, Hohl TM, Cherny M & Pamer EG Selective expansion of the monocytic lineage directed by bacterial infection. J Immunol 183, 1900–1910, doi: 10.4049/jimmunol.0900612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkatasubramanian M, Chetal K, Schnell DJ, Atluri G & Salomonis N Resolving single-cell heterogeneity from hundreds of thousands of cells through sequential hybrid clustering and NMF. Bioinformatics 36, 3773–3780, doi: 10.1093/bioinformatics/btaa201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 31.Harris SE et al. Meox2Cre-mediated disruption of CSF-1 leads to osteopetrosis and osteocyte defects. Bone 50, 42–53, doi: 10.1016/j.bone.2011.09.038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thambyrajah R et al. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat Cell Biol 18, 21–32, doi: 10.1038/ncb3276 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Liu Z et al. Fate Mapping via Ms4a3-Expression History Traces Monocyte-Derived Cells. Cell 178, 1509–1525 e1519, doi: 10.1016/j.cell.2019.08.009 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Bowers E et al. Granulocyte-derived TNFalpha promotes vascular and hematopoietic regeneration in the bone marrow. Nat Med 24, 95–102, doi: 10.1038/nm.4448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlitzer A et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol 16, 718–728, doi: 10.1038/ni.3200 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Akashi K, Traver D, Miyamoto T & Weissman IL A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197, doi: 10.1038/35004599 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Chow A et al. CD169(+) macrophages provide a niche promoting erythropoiesis under homeostasis and stress. Nat Med 19, 429–436, doi: 10.1038/nm.3057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaku T et al. Hematopoiesis in 3 dimensions: human and murine bone marrow architecture visualized by confocal microscopy. Blood 116, e41–55, doi: 10.1182/blood-2010-02-268466 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urbaniak GC & Plous S Research Randomizer (Version 4.0). Computer Software Retrieved on June 22, 2013, from http://www.randomizer.org/ (2013). (2013). [Google Scholar]
- 40.DePasquale EAK et al. cellHarmony: cell-level matching and holistic comparison of single-cell transcriptomes. Nucleic Acids Res, doi: 10.1093/nar/gkz789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain AK, Murty MN & Flynn PJ Data clustering: A review. Acm Comput Surv 31, 264–323, doi:Doi 10.1145/331499.331504 (1999). [Google Scholar]
- 42.Ester M, Kriegel H, Sander J & Xiaowei X A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of 2nd International Conference on Knowledge Discovery and Data Mining (KDD) 96, 226–231 (1996). [Google Scholar]
- 43.Arthur D & Vassilvitskii S k-means plus plus : The Advantages of Careful Seeding. Proceedings of the Eighteenth Annual Acm-Siam Symposium on Discrete Algorithms, 1027–1035 (2007). [Google Scholar]
- 44.Muench DE. et al. Mouse models of neutropenia reveal progenitor-stage-specific defects. Nature 582(7810):109–114. doi: 10.1038/s41586-020-2227-7 (2020); [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinho S et al. PDGFRalpha and CD51 mark human nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J Exp Med 210, 1351–1367, doi: 10.1084/jem.20122252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


