Abstract
Chromosome ends are protected by guanosine-rich telomere DNA that forms stable G-quadruplex (G4) structures. The heterodimeric POT1-TPP1 complex interacts specifically with telomere DNA to shield it from illicit DNA damage repair and to resolve secondary structure that impedes telomere extension. The mechanism by which POT1-TPP1 accomplishes these tasks is poorly understood. Here, we establish the kinetic framework for POT1-TPP1 binding and unfolding of telomere G4 DNA. Our data identify two modes of POT1-TPP1 destabilization of G4 DNA that are governed by protein concentration. At low concentrations, POT1-TPP1 passively captures transiently unfolded G4s. At higher concentrations, POT1-TPP1 proteins bind to G4s to actively destabilize the DNA structures. Cancer-associated POT1-TPP1 mutations impair multiple reaction steps in this process, resulting in less efficient destabilization of G4 structures. The mechanistic insight highlights the importance of cell cycle dependent expression and localization of the POT1-TPP1 complex and distinguishes diverse functions of this complex in telomere maintenance.
Keywords: G4, DNA topology, kinetics, energy landscape
The structural topology and conformational dynamics of DNA play a critical role in regulating nearly all cellular processes.1 One regulatory topology found throughout the genome is a stable structure known as a G-quadruplex (G4).2–4 G4s are comprised of multiple sets of stacked G-tetrads that form Hoogsteen hydrogen bonds between planar guanines.5,6 The geometry and stability of G4 structures varies based on the nature and size of intercalating cations that support the G-tetrads. For instance, lithium ions have destabilizing effects, whereas potassium ions promote the formation of thermodynamically stable G4 structures.7,8 These G4 structures regulate a variety of cellular processes that include transcriptional regulation,9–11 DNA recombination,12 and telomere maintenance.13,14 Telomeres are specialized nucleoprotein complexes found at the ends of all eukaryotic chromosomes.15 Telomere DNA consists of tandem G-rich repeats (d(TTAGGG) in mammals) that end in a single-strand DNA (ssDNA) overhang, and play a vital role in successive rounds of cellular replication and DNA preservation.16,17 The G-rich telomere DNA is prone to forming G4 structures.18–22
The general DNA replication machinery is unable to copy the extreme ends of linear chromosomes, and therefore telomeres of most somatic cells get shorter over the course of cell divisions.23 Telomere attrition can be counterbalanced by telomerase, a ribonucleoprotein enzyme that adds telomeric repeat sequences to the ends of eukaryotic chromosomes.15 However, G4 structures in telomeres present a major hurdle for telomere elongation as they inhibit the binding of telomerase.14 This hurdle is overcome by POT1-TPP1, a specialized protein heterodimer, that specifically binds to telomere ssDNA, destabilizes G4 structures, and makes the ssDNA accessible to telomerase and potentially to other enzymes.13,24–30
Multiple POT1-TPP1 molecules coat long telomere ssDNA substrates at a periodicity of one heterodimeric protein complex for every two hexameric repeats of telomere ssDNA.31,32 Opening of G4s relies on the coordinated binding and conformational dynamics of individual domains within each POT1-TPP1 heterodimer. The X-ray crystal structure of the POT1 DNA-binding domain bound to 10 nucleotides of unstructured telomere ssDNA identifies tandem oligosaccharide-oligonu cleotide binding (OB) folds, each of which form molecular interactions with a hexameric repeat in the telomere DNA substrate.24 A single, intact POT1 OB-fold is sufficient for binding to telomere ssDNA, but the sequential and coordinated binding of both domains is necessary for unfolding G4 structures.27,33,34 While these findings identify physicochemical properties of POT1-TPP1 interactions with longer physiological lengths of telomere ssDNA, the individual contributions of POT1-TPP1 in telomere G4 binding versus destabilizing secondary structure have yet to be separated and characterized.
The POT1-TPP1 complex is extraordinarily sensitive to changes in gene sequence. Numerous mutations are associated with genomic instability. In fact, more than 300 single-nucleotide polymorphisms (SNPs) have been identified within the POT1 gene in patients diagnosed with varying types of cancer.35–39 The molecular consequence of each mutation is unlikely to be conserved as they reside in different regions throughout the gene and, therefore, code for diverse point mutations in distant domains at the protein level. These findings further indicate that binding and resolution of telomere ssDNA are related events that rely on precise and orchestrated interactions by the POT1-TPP1 heterodimer.
To understand the kinetic properties of POT1-TPP1 destabilization of G4s, we utilized a previously characterized telomere ssDNA sequence that adopts a defined homogenous G4 topology.40 Data from complementary biophysical techniques, including surface plasmon resonance (SPR), stopped flow techniques and binding studies revealed highly dynamic folding and unfolding of telomere ssDNA and G4s and allowed us to devise a detailed kinetic model of POT1-TPP1 interactions with G4 complexes. This model shows that two POT1-TPP1 complexes are required to fully destabilize G4s. The data reveal a passive and an active mode of POT1-TPP1 destabilization of G4 DNA, which uniquely depend on protein concentration. At low protein concentrations, POT1-TPP1 passively captures transiently unfolded G4s. At higher protein concentrations, two POT1-TPP1 proteins bind to G4s to actively destabilize the DNA structures. We also find that the cancer-related Q94R and H266L POT1 mutants exhibit disparate defects in telomere G4 binding and/or destabilization, which cumulatively lead to significant alterations in telomere ssDNA protection and telomere maintenance. Our results provide new insight into how POT1-TPP1 functions to maintain telomeres through binding and resolution of G4 structures in telomere ssDNA.
Formation of 6ThT22 G-quadruplex is a kinetically dynamic process
To systematically characterize the mechanism by which POT1-TPP1 destabilizes G4 structures, we set out to kinetically describe the differential modes of POT1-TPP1 binding versus unfolding of telomere G4 DNA. As the single-stranded overhang of telomeres is in the 50–200 nucleotide range,41–43 the G4s formed at this region are likely to represent diverse and heterogeneous topologies. Indeed, G4 morphology is highly dependent on DNA sequence, ssDNA length, and monovalent cation composition.20 Therefore, to avoid complications associated with structural heterogeneity, we used a well characterized telomere ssDNA construct (hT22; TAGGGTTAGGGTTAGGGTTAGG, guanines participating in G4 formation are underlined)40 with an added 5′ hexathymidine linker (6ThT22). The 5′ linker was introduced to prevent steric interactions between the telomere ssDNA and the immobilization surface used for subsequent protein-DNA binding kinetics. Circular dichroism (CD) confirmed the expected antiparallel topology of 6ThT22 DNA in buffer containing 90 mM K+ (Figure 1(A)).
Figure 1.
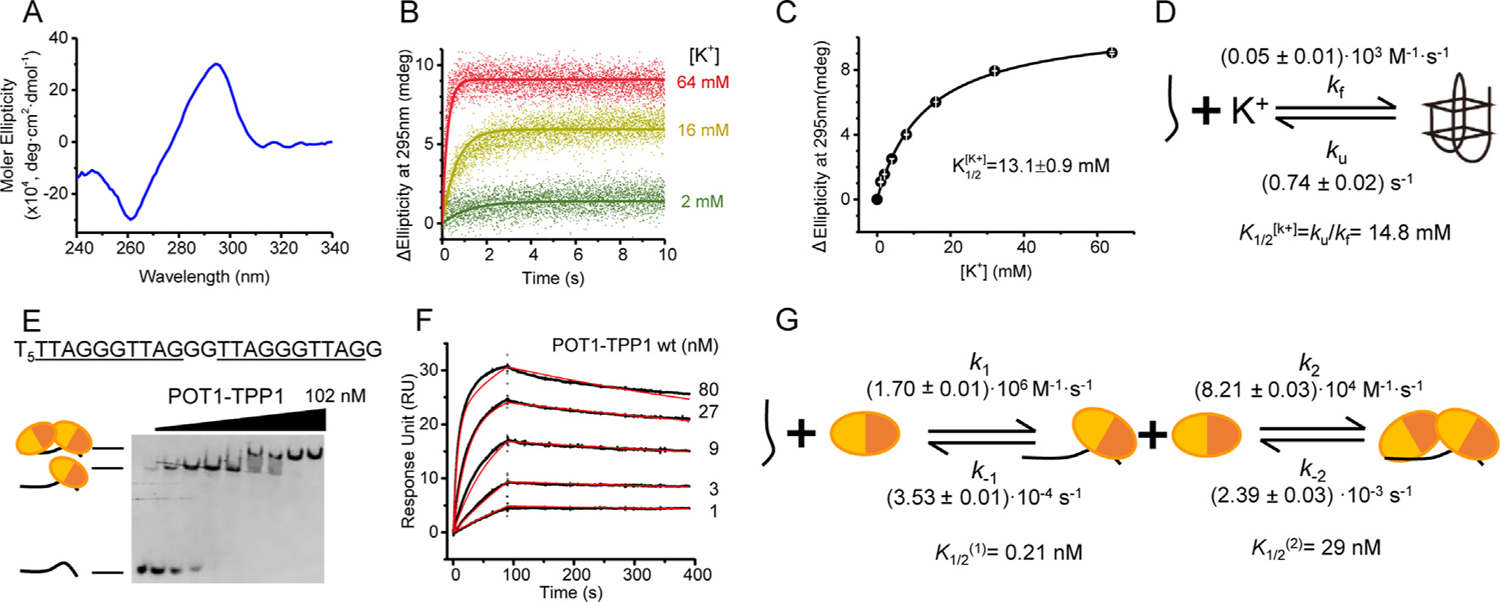
POT1-TPP1 binds unstructured 6ThT22 with two distinct binding affinities. (A) CD spectrum of 6ThT22 in K+ highlights signature peak (295 nm) and valley (260 nm) indicative of antiparallel G4 topology. (B) Time traces of ellipticity at 295 nm to monitor the folding of 6ThT22 into G4 structures with increasing concentrations of K+. (C) G4 formation of 6ThT22 structure, as monitored in changes at 295 nm ellipticity, as a function of [K+] with . (D) The model for 6ThT22 G4 formation upon mixing with K+ with . (E) EMSA analysis of POT1-TPP1 binding to unfolded 6ThT22 under equilibrium binding conditions in Li+ buffer. 1 nM 6ThT22 with 0–102 nM POT1-TPP1 protein. Substrate sequence of 6ThT22 (shown at top) with tandem POT1-TPP1 binding sites underlined. In the schematic, each circle represents an individual POT1-TPP1 heterodimer, with its separate DNA-binding domains colored in different shades of orange. (F) Sensorgrams displaying the interactions of immobilized 6ThT22 with POT1-TPP1 proteins in Li+ buffer (experimental data shown in black and fitted data shown in red). (G) Kinetic model of SPR data used for calculating individual rate constants of POT1-TPP1 binding to unfolded 6ThT22.
We first measured spontaneous folding and unfolding of 6ThT22 DNA using cation gradients coupled with CD analysis. These data indicate that 6ThT22 is maintained in an unfolded state in a solution devoid of monovalent cations, consistent with previous studies.7 The unfolded 6ThT22 DNA was then incubated with increasing concentrations of [K+) and G4 formation was monitored, in real-time, by CD at 295 nm44,45 (Figure 1 (B)). Plots of reaction amplitudes (A) were fit to a binding isotherm for 6ThT22 folding, yielding an apparent equilibrium dissociation constant for [K+] of in the presence of [K+] (Figure 1(C)). Time courses of the folding reaction (Figure 1(B)) were fit to a reversible binding reaction, yielding rate constants for 6ThT22 G4 folding (kf) and unfolding (ku) at each [K+] concentration (Figure 1(D), Figure S1). Consistent with prior reports,40 our data show that roughly 87% of 6ThT22 forms G4s in 90 mM KCl. The quadruplex spontaneously unfolds with considerable frequency (ku = 0.74 ± 0.02 s−1), indicating a lifetime of the folded G4 of τ = (ku)−1 = 1.35 s.
POT1-TPP1 binds unfolded 6ThT22 with two distinct binding affinities
We next examined binding of POT1-TPP1 proteins to unfolded 6ThT22 DNA. For binding and unwinding assessment, we used a well-studied, recombinant POT1-TPP1 complex that maintains both of its ssDNA-binding and telomerase recruitment capabilities.46,47 The proteins were co-expressed and purified using affinity and size-exclusion chromatography to achieve greater than 95% purity of the heterodimeric protein. An individual POT1 protein or POT1-TPP1 heterodimer binds to a ten nucleotide span of telomere ssDNA with nanomolar affinity.24,31,48,49 As such, the 6ThT22 substrate contains two consecutive POT1-TPP1 binding motifs. As a commonality to G4 formation,50 6ThT22 folds into a G4 structure in K+ but remains unfolded in Li+ buffer, which was confirmed by UV melting and size-exclusion chromatography analysis (Figure S2(A) and (B)). Electrophoretic mobility shift assays (EMSAs) of equilibrium binding reactions revealed association of two POT1-TPP1 complexes to the unfolded DNA, consistent with the presence of two POT1-TPP1 binding motifs in 6ThT22 (Figure 1(E)). Importantly, binding of POT1-TPP1 to 6ThT22 determined experimentally using EMSA in Li+ buffer correlated well with predicted binding data derived from the kinetic model (Figure S2(C)). Time-resolved surface plasmon resonance (SPR) data (Figure 1(F)) were also best fit to a two-site POT1-TPP1 binding model for unfolded 6ThT22 (Figure 1 (G), S3(A)–(D), Table S1). In addition, the SPR data revealed sequential binding of two POT1-TPP1 complexes (k1 > k2) and a significantly higher apparent affinity of the first POT1-TPP1 complex for unfolded 6ThT22 compared with the second complex (Table S2). These results demonstrate that the initial POT1-TPP1 binding has a high affinity for the unfolded 6ThT22, whereas the second binding event has a much lower affinity. These data are supported by previously published studies using different monovalent cations,28,46 suggesting that monovalent cation identity alone does not adversely affect POT1-TPP1 interactions with unstructured ssDNA substrates. Collectively, our data indicate that two POT1-TPP1 complexes bind unfolded 6ThT22 DNA with two distinct affinities.
Two POT1-TPP1 heterodimers are required for complete G4 destabilization
We next probed the interaction of POT1-TPP1 with 6ThT22 under conditions (90 mM K+), where the DNA folds into a G4 structure with an antiparallel topology.40 As seen for the unfolded DNA (Figure 1(E)), EMSA analysis revealed two POT1-TPP1 complexes bound to the G4 DNA (Figure 2(A)). SPR time courses were multi-phasic, and the data were best fit using two POT1-TPP1 binding events per G4 ssDNA substrate (Figure 2(B)). Interestingly, unlike the interactions with unfolded ssDNA (Figure 1(F)), the binding of two POT1-TPP1 proteins to preformed 6ThT22 G4 structures is characterized by a fast initial dissociation phase followed by a prolonged secondary dissociation phase. Next, stopped-flow CD was used to interrogate unfolding of the G4 structure upon addition of POT1-TPP1 protein by monitoring changes in ellipticity of the G4 spectral signature at 295 nm (Figure 2(C)). The loss of ellipticity at 295 nm indicates the opening of G4 upon POT1-TPP1 binding. The amplitude of the unfolding reaction scaled linearly with the POT1-TPP1 concentration until a protein:DNA ratio (r) of r = 2, indicating that two POT1-TPP1 proteins are required to fully unfold each quadruplex (Figure 2(D)).
Figure 2.
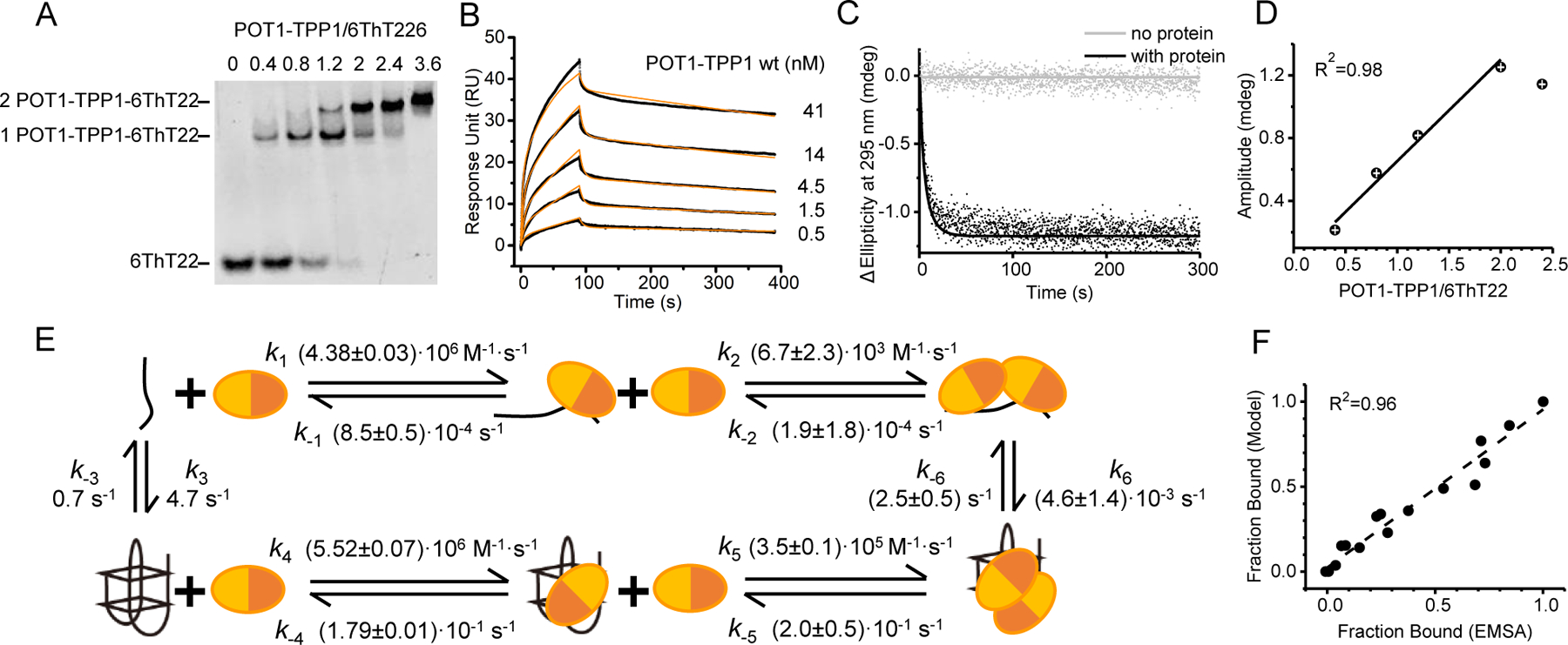
POT1-TPP1 binds and destabilizes 6ThT22 G4s. (A) Stoichiometric binding EMSAs of POT1-TPP1 with 6ThT22 in K+ buffer. (B) Experimental (black) and modeled (orange) sensorgrams to describe the interactions of immobilized 6ThT22 with POT1-TPP1 proteins in K+ buffer. (C) Time-dependent changes in ellipticity of 6ThT22 with or without POT1-TPP1. (D) Plot showing changes in CD amplitude (295 nm) of 6ThT22 DNA as a function of increasing molar ratio of POT1-TPP1 protein:DNA. (E) Kinetic model for protein binding and G4 unfolding of 6ThT22 in K+. (F) Correlation between EMSA experimental data and fitted data using kinetic model.
SPR and CD data were then globally fit to kinetic models of increasing complexity (Figure 2(E), S4, S5). The models incorporated data for spontaneous folding and unfolding of the G4 structure, as well as data for binding of POT1-TPP1 to the unfolded DNA (Figure 1). The minimal model fitting all datasets indicates binding of two POT1-TPP1 complexes to the G4 (Figure 2(E)). In order to verify the rate constants obtained from kinetic data, independent EMSAs were performed and compared the unbound, one POT1-TPP1 bound and two POT1-TPP1 bound fractions. These independently observed EMSA data are in excellent agreement with analogous values derived from the model (Figure 2(F)). The model shows that the first POT1-TPP1 complex binds to G4 and unfolded DNA with similar rate constants (k1 ~ k4). However, POT1-TPP1 dissociates significantly faster from G4 than from the unfolded DNA (k−1 < k−4). Accordingly, the first POT1-TPP1 binds to the unfolded DNA with markedly higher affinity than to the G4 . Binding of the second POT1-TPP1 protein exhibits higher affinity for the unfolded over G4 DNA, even though the affinity for both unfolded and G4 substrates is significantly lower than that of the first protein binding event. Binding of the POT1-TPP1 complexes results in a 3.6-times faster opening of the G4 complex when compared to the spontaneous opening rate constant of 6ThT22 G4 alone (k−6 > k−3). Refolding of G4 with two bound POT1-TPP1 occurs orders of magnitude less frequent than spontaneous refolding of naked G4s . These results suggest that there could be two distinct kinetic mechanisms by which POT1-TPP1 complexes regulate the binding and opening of these physiologically relevant, telomere ssDNA substrates.
POT1-TPP1 opens 6ThT22 formed G-quadruplex through both passive capture and active destabilization
Given the significantly divergent binding and dissociation kinetics of POT1-TPP1 for each DNA substrate (unwound versus G4), we sought to compile a kinetic model for the dynamic processes at play. Collectively, our kinetic analysis indicates that binding of two POT1-TPP1 complexes to 6ThT22 G4 DNA proceeds via two pathways (Figure 3). In one pathway, the first POT1-TPP1 captures spontaneously unfolding DNA, which allows a second POT1-TPP1 to bind and to stabilize the unfolded state. We propose to call this pathway the “passive” capture. In a second pathway, two POT1-TPP1 heterodimers bind consecutively to the G4 to facilitate its opening to an unfolded state that then remains bound by both POT1-TPP1 complexes. Based on the increase of the G4 opening rate observed, we propose to call this second pathway the “active” unfolding mechanism. The passive and active pathways described here are comparable to the terms ‘conformational selection’ and ‘induced fit’ mechanisms, respectively, that have been applied to other macromolecular interactions.51
Figure 3.
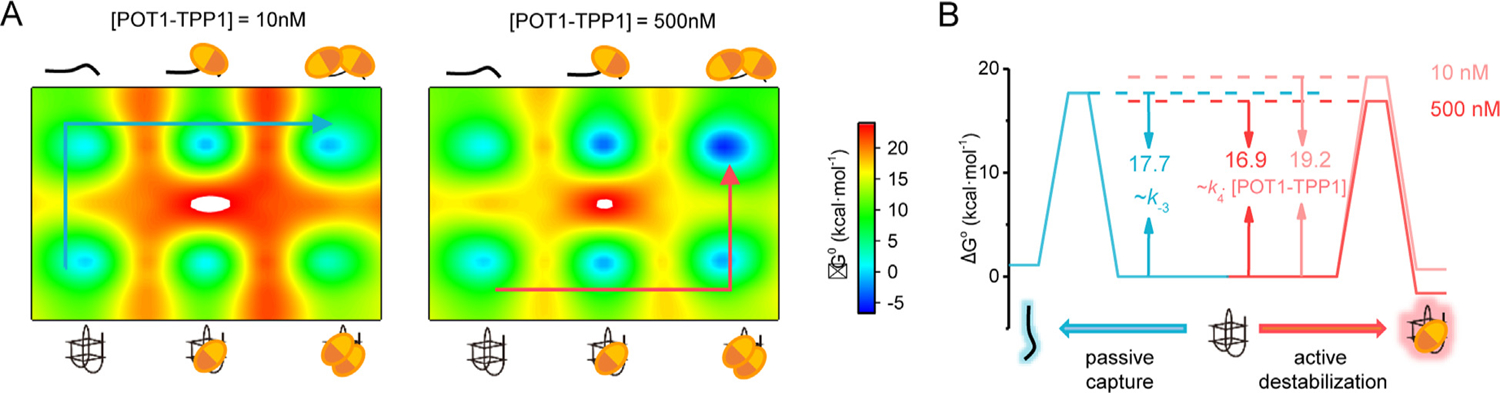
POT1-TPP1 promotes G4 destabilization through diverse mechanisms. (A) Free energy landscape of POT1-TPP1-DNA for different protein-bound states at low protein concentration (10 nM) on left and high protein concentration (500 nM) on right. The blue and red arrows indicate passive capture and active destabilization pathways, respectively. (B) Comparison of initial energy barriers of passive capture versus active destabilization pathways. The energy barrier between G4 and spontaneously unfolded DNA (blue) is depicted as a function of k−3. The energy barrier for POT1-TPP1 binding G4 (red) is depicted as a function of k4·[POT1-TPP1], which becomes favorable with increasing [POT1-TPP1]. The rate constants k−3 and k4 correspond to the respective steps labeled in Figure 2E.
The lower affinity of the first POT1-TPP1 complex for unfolded vs. G4 DNA indicates that the passive pathway is preferentially populated at low POT1-TPP1 concentrations ([POT1-TPP1] < 10 nM). Meanwhile, the active pathway becomes populated with increasing POT1-TPP1 concentrations ([POT1-TPP1] > 100 nM). Free energy landscapes of the reaction at low ([POT1-TPP1] = 10 nM) and high ([POT1-TPP1] = 500 nM) concentrations of POT1-TPP1 illuminate the change in pathway preference (Figure 3). At high POT1-TPP1 concentrations, the barriers between free DNA and POT1-TPP1 bound DNA, which are determined by the respective association rate constants (k1, k4, Figure 2(E)), are of roughly equal height. However, the G4 structure is favored over the unfolded DNA, and therefore the active pathway is more populated under these conditions (Figure 3). Use of the active pathway is thus determined by the height of the energy barrier between G4 and spontaneously unfolded DNA (k−3) relative to the height of the barrier between G4 and G4 bound to a single POT1-TPP1 heterodimer (k4·[POT1-TPP1], Figure 3(B)). These findings are of particular importance when translated to biologic systems, as the local telomeric concentrations of end-binding proteins, including POT1, are regulated throughout the cell cycle.52
Cancer related POT1 mutants impair different steps of 6ThT22 formed G4 binding and destabilization
Having established a kinetic framework to differentially assess the impact of POT1-TPP1 binding and destabilization of 6ThT22 G4, we next investigated how two pathogenic POT1 mutations, Q94R and H266L,35 affect the various steps within our model. We chose to investigate Q94 and H266 because they are located at distant and distinct regions of the POT1 DNA-binding domain. Moreover, both of these amino acids interact directly with guanosines of a bound 10-nucleotide, unstructured ssDNA telomere substrate24 (Figure S6(A)). The kinetic parameters of the POT1-TPP1 variants were determined using the same approach as described above for wild type (WT) POT1-TPP1. As compared to WT protein, both mutations significantly diminish overall binding and unfolding of 6ThT22. Interestingly, the two mutations differentially alter the kinetic landscape and the individual steps within the model (Figures 4, S3(E)–(L), S6–9, Tables S1–4).
Figure 4.
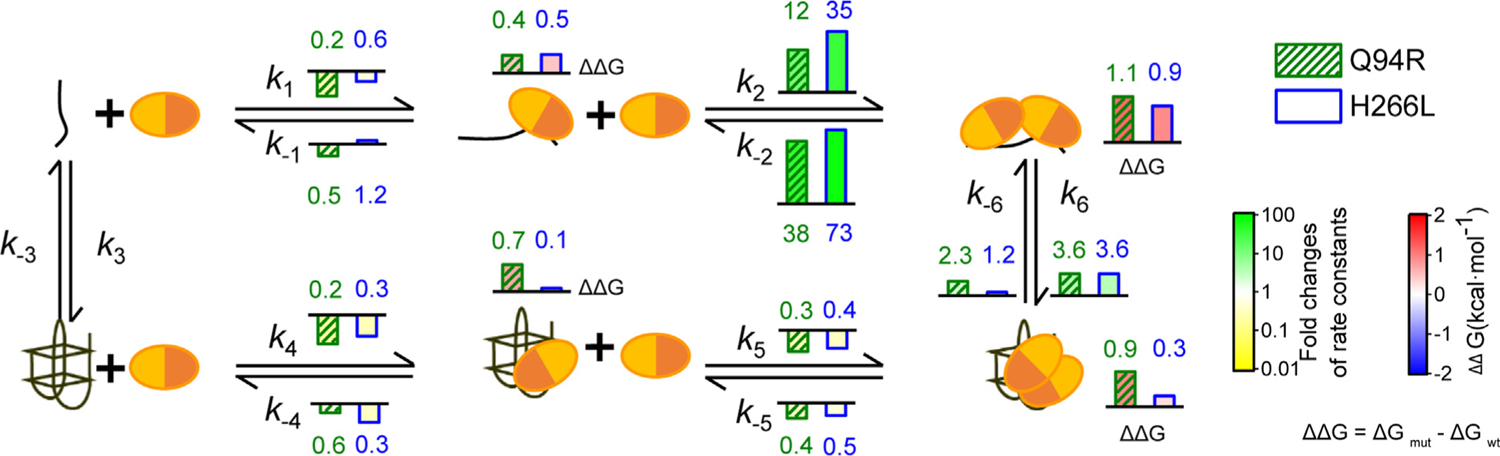
Pathogenic mutations differentially impair POT1-TPP1 binding and destabilization of telomere G4 DNA. Bar colors and heights represent fold changes in the values for POT1-TPP1 Q94R and H266L mutants as indicated in the top right corner. The relative change of individual rate constants and free energies (ΔΔG) for each mutation as compared to WT protein are indicated as individual values in the kinetic model.
In order to translate the kinetic data into a physiologically relevant context, we determined the impact of these mutations on the stability of the four POT1-TPP1 DNA complexes in our model (Figure 4). To accomplish this, we calculated ΔΔG values, which describe the difference in free binding energy of mutant vs. WT (ΔGmutant – ΔGwild type) for the four POT1-TPP1 DNA complexes in our model. A positive ΔΔG value indicates that the mutant requires more energy than the WT to bind the DNA; or in other words, the mutant has a weaker affinity than the WT protein for the DNA substrate.
We find that all protein-DNA bound states for both POT1 mutants are less energetically favorable than that of WT protein, as indicated by positive ΔΔGs. However, the degree by which each mutation destabilizes the DNA-bound complexes follows a more nuanced pattern. Both mutations similarly impact the stability of the protein complexes with the unfolded DNA, yet the stability of the DNA-complex with two units of POT1-TPP1 is affected significantly more than a complex with a single unit (Figure 4). This result reflects an additive impact of the mutations on DNA complex stability when two units are bound. Similarly, the stability of the POT1-TPP1 complexes with G4 DNA differ for each mutation. The Q94R mutation destabilizes the complexes with G4-DNA to a larger degree than the H266L mutation, which only slightly impacts the complex stability with G4 DNA, compared to WT POT1-TPP1 (Figure 4). The data collectively show that the Q94R mutation impacts DNA binding to both, unfolded and G4 DNA, while the H266L mutation mainly affects binding to unfolded DNA (Figure 4). The significant decrease in ΔΔG for two H266L mutant complexes bound to ssDNA compared to the G4 bound state further indicates that reduced biding to the unfolded ssDNA is a key factor in the inability of this mutation to efficiently destabilize G4 structures. Cumulatively, the differential defects identified for the Q94R and H266L mutations indicate that binding telomere DNA and destabilizing G4 structures are each important and distinct contributors of the cellular functions of POT1-TPP1.
Discussion
Our results indicate that unfolding of 6ThT22 G4s by POT1-TPP1 proceeds via two distinct pathways. At low protein concentrations of POT1-TPP1, the heterodimer exploits the spontaneous opening of G4s by “passively” capturing the unfolded DNA. Although this passive mode allows POT1-TPP1 to unfold the remaining G4 component and bind to the DNA in the unfolded state, we speculate that this pathway is insufficient to maintain unfolded G4 structures under physiological (K+ solvent) conditions29 to a degree necessary for unimpeded telomere function. Our data show that increased POT1-TPP1 concentrations accelerate G4 unfolding via an active pathway. This concentration-dependent mechanism bears direct physiologic relevance, since local telomere concentrations of POT1-TPP1 are thought to exceed 8 µM, given that there are approximately 50–100 copies of POT1-TPP1 per telomere53 localized within the cellular telomere volume of roughly 0.002–0.01 µm3.54,55 The accumulation of POT1-TPP1 at telomere DNA would function to actively destabilize telomere G4 to promote the binding and stabilization of telomerase.56
Thermodynamic measurements have fomented the view that G4 structures pose a significant energy barrier for their disassembly.57–59 Our kinetic measurements show that the tested G4, despite its high thermodynamic stability, transitions with previously unappreciated frequency between the folded and unfolded states. It is thus possible that other proteins which “unfold” G4s in a non-ATP dependent manner60,61 capture the unfolded state, as seen for the passive pathway of POT1-TPP1 and similarly described for other macromolecular interactions.51 G4 structures, despite their high thermodynamic stability, might thus be readily remodeled by an array of proteins. As seen for POT1-TPP1, even subtle aberrations in these proteins could have profound biological effects, given that G4 structures are often key elements for the regulation of gene expression.9,62 Furthermore, our data demonstrate that the cancer-related mutations, each occurring on different DNA binding domains (OB1, OB2), differentially impair POT1-TPP1 binding and/or opening of G4s. Whereas the Q94R mutant impairs binding of POT1-TPP1 to telomere DNA, the H266L mutation functions by reducing POT1-TPP1’s ability to open the G4 structure. Our model established for WT protein highlights the individual deficiencies associated with each mutant protein. However, the refined fitting deviates slightly for the mutant proteins investigated (e.g. Figure S6(E)), suggesting that other functional consequences, such as non-specific interactions or a loss of synergistic DNA-binding contributions arising from both OB-fold domains in the native protein, are associated with the mutant proteins. Taken together, these data implicate a coordinated binding of POT1-TPP1 that involves individual OB-folds. Specifically, our model is consistent with the tandem OB-fold domains of POT1-TPP1 binding to telomere ssDNA independently and functioning cooperatively to actively destabilize G4 structures. While both mutations affect multiple stages of the reaction, the largest defect of the Q94R mutation is for POT1-TPP1 binding to the G4 structure, while the largest defect of the H266L mutation is the opening step where two POT1-TPP1 proteins are bound to the G4 DNA.
While our investigations have focused exclusively on a single G4 topology and its relationship with telomere nucleoprotein complexes, our kinetic model can be expanded to define interactions with other G4 morphologies. For instance, G4s with different topologies exhibit different thermodynamic and kinetic properties,63 which may alter the rates of G4 destabilization. It is expected that the general trends described in our work will be applicable to other G4 structures, even though the inherent heterogeneity within specific populations (e.g. 64,65) will need to be considered in the case of hybrid G4 morphologies before absolute kinetic values can be derived for specific G4 structures. Additionally, the stability or instability of G4 structures located throughout the genome, in particular at promoter regions,11 are implicated in the transcriptional regulation of downstream genes. Thus, dysregulation of G4 binding partners/enzymes in either direction could conceivably result in aberrant expression of pathologic proteins or conversely, prevent the expression of cellular “guardians”, checkpoints, and tumor suppressors. Analogous to the disparate defects in G4 binding and unfolding we have identified for the Q94R and H266L POT1-TPP1 cancer mutations, it will be important to determine whether protein-DNA interactions are similarly affected by differences in G4 morphology and inherent stability.
Materials and Methods
Oligonucleotide preparation
Oligonucleotides used in this study were synthesized and HPLC purified by Integrated DNA Technologies (IDT). Unmodified oligos were used for circular dichroism spectroscopy, UV melting, size exclusion-HPLC, G4 formation and destabilization assays. IRD-700 labeled oligos were used for electrophoretic mobility shift assays (EMSAs), and biotinylated labeled oligos were used for surface plasmon resonance (SPR) assays. To pre-fold G4, oligos were prepared in in potassium phosphate-KCl buffer containing 90 mM K+ (pH 7.0) and heated to 95 °C for 5 min, then slowly cooled to 25 °C. G4s were allowed to stabilize for at least 4 hrs at 25 °C before proceeding to subsequent steps.
Circular dichroism spectroscopy
Circular dichroism spectra were obtained using a PiStar 180 spectrophotometer (Applied Photophysics) using 1 cm path length at 500 nM of G4 in 90 mM K+ potassium phosphate-KCl buffer (pH 7.0). Scans were performed at 25 °C spanning a range of 240–340 nm. Final spectra represent three independent scans that were smoothed and averaged. Each scan was recorded using a 4-nm bandwidth, 1-nm step size, and 1-s collection time per data point. A buffer only blank was subtracted from each spectrum with all data zero corrected at 340 nm.
Stopped flow measurement
Kinetics of G4 formation were measured with a PiStar 180 spectrophotometer (Applied Photophysics) upon rapid mixing with increasing concentration of K+. 6 µM unfolded 6ThT22 was prepared in 5 mM Tris buffer (pH 7.0) and rapidly mixed 1:1 with increasing concentrations of KCl in 5 mM Tris (pH7.0) to reach a final concentration of 3 µM oligo. G4 folding was monitored by ellipticity changes at 295 nm as a function of time (path length 1 cm, bandwidth 4 nm) for 10 s with 4000 data points. All data were zero corrected using the initial time point. A circulating water bath was used to maintain constant temperature at 25 °C. The averaged readings of eight mixing reactions were fitted by Kintek Explorer software to calculate rate constants of G4 folding and unfolding. To calculate the extent of G4 being folded upon K+ binding, the average readings were fitted to an exponential function (BoxLucas1 in Origin): , where A and kobs represent the amplitude and observed rate constants of G4 opening. The amplitude values were plotted against potassium concentration and fitted to a hyperbolic function:
| (1) |
where Amax and K1/2 represent the maximal amplitude and [K+] needed to fold half the oligonucleotide.
The pseudo first order G4 folding rate (kf90) at 90 mM K+ was determined with the following equation:
| (2) |
where kf is the folding rate constant obtained from global fitting to be 0.05 ± 0.01 mM−1 s−1 when the concentration of potassium, [K+], is at 90 mM.
The kinetics of G4 destabilization upon binding with POT1-TPP1 (WT and mutant) proteins were obtained by rapidly mixing 1 µM pre-folded G4 with varying concentrations of proteins as protein: oligo molar ratios at 0, 0.4, 0.8, 1.2, 2, 2.4. Both G4 and POT1-TPP1 proteins were prepared in 90 mM K+ potassium phosphate-KCl buffer (pH 7.0) and mixed 1:1 to reach a final G4 concentration of 500 nM. G4 destabilization was monitored by ellipticity changes at 295 nm as a function of time (path length 1 cm, bandwidth 4 nm) for 300 s with 900 data points at 25 °C, and data were zero corrected using the initial time point. Three to eight (typically more than five) mixing reactions were performed for each condition. The average readings were used for global fitting by Kintek Explorer software.
To calculate the extent of G4 being opened upon POT1-TPP1 binding, the average readings were fitted to an exponential function: , where A and kobs represent the amplitude and observed rate constants of G4 opening. The amplitude values were plotted against molar ratio of protein to oligo.
Protein expression and purification
Full-length POT1 (WT and mutant) with an N-terminal GST-tag and N-terminal 6 × His-tagged TPP1 (89–334) proteins were co-expressed using the recombinant baculovirus expression system to infect Spodoptera frugiperda 9 (Sf9) insect cells as described previously.24 Briefly, POT1-TPP1 protein complexes were first purified using affinity chromatography on GST-beads (Invitrogen). PreScission protease (GE Healthcare) was added to remove the N-terminal GST tag followed by size-exclusion chromatography (SEC) using a Superdex 200 10/300 chromatography column on an AKTA Purifier10 FPLC system (GE Healthcare) with either 20 mM Tris (pH 7.0), 90 mM LiCl or 90 mM K+ containing potassium phosphate-KCl buffer (pH 7.0) buffer. Proteins purified in Li+ buffer were used for assays with unfolded DNA, i.e., EMSAs and SPR under Li+ condition. Proteins purified in K+ buffer were used for assays with G4 formed DNA, i.e., EMSAs, G4 destabilization assays by stopped flow CD and for SPR under K+ buffer.
UV melting
The UV melting assays were carried out on a PiStar 180 spectrophotometer (Applied Photophysics). 16 µM of hT22 were prepared in either 20 mM Tris (pH 7.0), 90 mM LiCl or potassium phosphate-KCl buffer containing 90 mM K+ (pH 7.0) buffer. The absorbance was monitored using 2 mm path length while the temperature was increased from 10 °C to 90 °C in 1 °C/min increments. Absorbance changes at 295 nm were plotted against temperature to obtain UV melting profile under K+ or Li+ conditions.
Size exclusion HPLC
hT22 oligos were prepared at 10 µM in either Li+ or K+ buffer. For HPLC runs, 10 µL of DNA was injected into a Shimadzu HPLC and separated using a SEC-300 size-exclusion column (Thermo Scientific Acclaim). All SE-HPLC runs were conducted at 25 °C using a flow-rate of 0.2 ml/min and constant UV absorbance monitoring at a wavelength of 260 nm.
Electrophoretic mobility shift assays (EMSAs)
POT1-TPP1 binding to unfolded oligos was performed in buffer containing 20 mM Tris (pH 7.0), 90 mM LiCl, 5 mM DTT, 5 µg/mL BSA, 1.2 µg/mL tRNA and 6.25% glycerol. Reactions were performed using 1 nM IRD700-labeled 6ThT22 and variable concentrations of recombinant POT1-TPP1 protein. WT POT1-TPP1 were performed under the concentration range of 0 to 102 nM, while Q94R mutant concentrations ranged from 0 to 800 nM and H266L mutant POT1-TPP1 concentrations ranged from 0 to 512 nM. Binding reactions were incubated for 30 min at room temperature before 10 µL of the reaction was loaded onto a 5% Tris-borate non-denaturing gels (Bio-rad). Gels were run at 125 V for 35 min, then imaged with ODYSSEY imaging system (LI-COR). Image quantification was performed using Image Studio software. The data from EMSA experiments were fitted to a hyperbolic function: to obtain K1/2, which represent [POT1-TPP1] needed to obtain half bound shift. Averages and standard deviations from three measurements were used to define reported values.
POT1-TPP1 binding to pre-folded oligos was performed in the potassium phosphate-KCl buffer containing 90 mM K+ (pH 7.0), with 5 mM DTT, 5 µg/mL BSA, 1.2 µg/mL tRNA and 6.25% glycerol. Reactions were performed using 500 nM 6ThT22 with 4 nM IRD700-labeled 6ThT22 and variable concentrations of recombinant POT1-TPP1 protein to reach the protein:oligo ratios indicated. Binding reactions were incubated for 30 min at room temperature before 10 lL of the reaction was loaded onto a 5% Tris-borate non-denaturing gels (Bio-rad). Gels were run at 125 V for 35 min, then imaged with ODYSSEY imaging system (LI-COR). Image quantification was performed using Image Studio software. Unbound, one protein-bound and two protein-bound fractions were quantified and compared with the data simulated from global fitting performed using the same incubation times.
Surface plasmon resonance (SPR)
SPR analysis was performed using 5′ end labeled biotinylated 6ThT22 and WT or mutant POT1-TPP1 proteins on a BIAcore T200 instrument (GE Healthcare). Oligos were adsorbed onto an S series SA sensor chip (GE Healthcare) at a low density of approximately 5 response units (RUs). SPR experiments with unfolded oligo were performed in Li+ running buffer, 20 mM Tris (pH7.0), 90 mM LiCl, 0.05% Tween-20. POT1-TPP1 protein was similarly prepared in the Li+ running buffer and used at concentrations ranging from 0.33 to 80 nM for WT and H266L mutant proteins, and from 0.33 to 720 nM for Q94R mutant proteins. Series concentrations of proteins were injected and flowed over oligos at a rate of 30 µL/min. Surface was regenerated between injections using 2 M guanidinium chloride for 15 s and followed by Li+ running buffer for 60 s. SPR experiments using G4 folded oligo were performed in K+ running buffer, potassium phosphate-KCl buffer containing 90 mM K+ (pH 7.0), 5 mM MgCl2, 0.05% Tween-20. POT1-TPP1 proteins were prepared in the K+ running buffer and used at varying concentration that ranged from 0.5 to 40.5 nM for WT protein, 0.1 to 240 nM for H266L mutant protein, and 2.9 to 720 nM for Q94R mutant protein. Surface was regenerated between injections using water for 30 s, guanidinium chloride for 15 s, 2 M KCl for 60 s, and finally with K+ running buffer for 30 s.
Global data fitting and kinetic modeling
Kintek Global Kinetic Explorer program, Version 8.0, Kintek Corp, Snow Shoe, PA66,67 was used to fit: (i) Kinetic data for G4 folding and unfolding, (ii) POT1-TPP1 binding to unfolded telomere ssDNA (Li+ condition), and (iii) POT1-TPP1 binding and opening of G4 telomere ssDNA (K+ condition).
Kinetics of G4 folding/unfolding in absence of protein
G4 formation Data were fit to the model presented. Initial parameters were obtained using the Dynamic Simulation feature of Kintek which allows variation of rate constants with continuous simulation. The global fit was performed multiple times for alternating combinations of folding and unfolding rate constants (initial parameters) until an overall fit with the lowest possible χ2 value was reached. The folding and unfolding rate constants were used and fixed for the following POT1-TPP1 binding and opening G4 formed telomere ssDNA models.
POT1-TPP1 binding to unfolded telomere ssDNA (Li+ condition)
POT1-TPP1 binding to unfolded telomere ssDNA data were fit to the model presented. The and obtained from EMSAs under Li+ conditions, which corresponds to the ratio between dissociation rate constants and association rate constants for each step, were used as initial parameters. The global fit was performed multiple times for alternating combinations of fixed and floated variables until an overall fit with the lowest possible χ2 value was reached. The simulated sensorgrams calculated from obtained rate constants were plotted to compare with experimentally recorded sensorgram data.
POT1-TPP1 binding and opening of G4 telomere DNA (K+ condition).
Kinetic models were used for fitting POT1-TPP1 binding to G4 formed telomere ssDNA (SPR data under K+ conditions) and G4 destabilization upon POT1-TPP1 binding (stopped flow measurement). Values of the folding and unfolding rate constants obtained from G4 formation assays were used and fixed as k3 and k−3. Considering the possibility of difference due to the identity of monovalent ions, values of k1, k−1, k2, k−2 obtained from POT1-TPP1 binds to unfolded telomere ssDNA from Li+ conditions were used as initial parameters, then allowing those values to float to the best fit solution in the global fitting. Again, the global fitting was performed multiple times for alternating combinations of fixed and floating variables until an overall fit with the lowest possible χ2 value was reached. The simulated data points calculated from obtained rate constants were plotted to compare with experimental data points. FitSpace analysis was performed to determine the lower and upper boundaries for each kinetic parameter.
Free energy calculation
To obtain the free energy landscapes shown in Figures 3 and 4, the free energy for each state was determined according to the deviation of Eyring-Polanyi Equation: ΔG = RT ln (κB T/k h), where ΔG is the free energy barrier for any given step, R is the gas constant (1.98 cal/mol), T is the temperature in kelvin (298 K), κB and h are Boltzmann’s and Planck’s constants, respectively, and k is the rate constant determined for any given step. For any second order reaction, the association rate constant (k) was converted to a pseudo-first order rate constant (kpse) at a given protein concentration according to: kpse = k [protein], and kpse was used for calculating free energy. To obtain ΔΔG between POT1 WT and mutants, ΔΔG values were determined by: ΔΔG= ΔGmutant – ΔGWT, for each mutant and DNA-bound state.
Supplementary Material
Acknowledgements
This work was funded by grants from the NIH (R01 GM133841 and R01 CA240993 to DJT; R35 GM118088 to EJ).
Abbreviations:
- G4
G-quadruplex
- ssDNA
single-stranded DNA
- OB folds
oligosaccharide-oligonucleotide binding folds
- SNPs
single-nucleotide polymorphisms
- SPR
surface plasmon resonance
- CD
circular dichroism
- EMSA
electrophoretic mobility shift assay
Footnotes
Declaration of interests
The authors declare no conflicts of interest with the contents of this article.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2021.166846.
References
- 1.Travers A, Muskhelishvili G, (2015). DNA structure and function. FEBS J, 282, 2279–2295. 10.1111/febs.13307. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M, Lipsett MN, Davies DR, (1962). Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U. S. A, 48, 2013–2018. 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd AK, Johnston M, Neidle S, (2005). Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res, 33, 2901–2907. 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert JL, Balasubramanian S, (2005). Prevalence of quadruplexes in the human genome. Nucleic Acids Res, 33, 2908–2916. 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert DE, Feigon J, (1999). Multistranded DNA structures. Curr. Opin. Struct. Biol, 9, 305–314. 10.1016/S0959-440X(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 6.Simonsson T, (2001). G-quadruplex DNA structures–variations on a theme. Biol. Chem, 382, 621–628. 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya D, Mirihana Arachchilage G, Basu S, (2016). Metal cations in G-quadruplex folding and stability. Front Chem, 4, 38. 10.3389/fchem.2016.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardin CC, Watson T, Corregan M, Bailey C, (1992). Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG). Biochemistry, 31, 833–841. 10.1021/bi00118a028. [DOI] [PubMed] [Google Scholar]
- 9.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH, (2002). Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A, 99, 11593–11598. 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel DJ, Phan AT, Kuryavyi V, (2007). Human telomere, oncogenic promoter and 5’-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res, 35, 7429–7455. 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert JL, Balasubramanian S, (2007). G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res, 35, 406–413. 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen W, Gao L, Balakrishnan M, Bambara RA, (2009). A recombination hot spot in HIV-1 contains guanosine runs that can form a G-quartet structure and promote strand transfer in vitro. J. Biol. Chem, 284, 33883–33893. 10.1074/jbc.M109.055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaug AJ, Podell ER, Cech TR, (2005). Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. U. S. A, 102, 10864–10869. 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Liu JQ, Chen Z, Zheng KW, Chen CY, Hao YH, Tan Z, (2011). G-quadruplex formation at the 3’ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res, 39, 6229–6237. 10.1093/nar/gkr164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackburn EH, Greider CW, Szostak JW, (2006). Telomeres and telomerase: The path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med., 10.1038/nm1006-1133. [DOI] [PubMed]
- 16.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR, (1988). A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U. S. A, 85, 6622–6626 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=3413114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nandakumar J, Cech TR, (2013). Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell. Biol, 14, 69–82. 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D, (2006). Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res, 34, 2723–2735. 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT, (2009). Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc, 131, 4301–4309. 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ, (2006). Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc, 128, 9963–9970. 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phan AT, Luu KN, Patel DJ, (2006). Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res, 34, 5715–5719. 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan AT, Patel DJ, (2003). Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc, 125, 15021–15027. 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackburn EH, Epel ES, Lin J, (2015). Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science (80.-), 10.1126/science.aab3389. [DOI] [PubMed]
- 24.Lei M, Podell ER, Cech TR, (2004). Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol, 11, 1223–1229 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15558049. [DOI] [PubMed] [Google Scholar]
- 25.Baumann P, Cech TR, (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science (80-.), 292, 1171–1175 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11349150. [DOI] [PubMed] [Google Scholar]
- 26.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O’Connor MS, Songyang Z, (2007). TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature, 445, 559–562 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17237767. [DOI] [PubMed] [Google Scholar]
- 27.Hwang H, Buncher N, Opresko PL, Myong S, (2012). POT1-TPP1 regulates telomeric overhang structural dynamics. Structure, 20, 1872–1880. 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullins MR, Rajavel M, Hernandez-Sanchez W, de la Fuente M, Biendarra SM, Harris ME, Taylor DJ, (2016). POT1-TPP1 binding and unfolding of telomere DNA discriminates against structural polymorphism. J. Mol. Biol, 428, 2695–2708. 10.1016/j.jmb.2016.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H, (2014). G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc. Natl. Acad. Sci. U. S. A, 111, 2990–2995. 10.1073/pnas.1321436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Nora GJ, Ghodke H, Opresko PL, (2011). Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J. Biol. Chem, 286, 7479–7489. 10.1074/jbc.M110.205641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor DJ, Podell ER, Taatjes DJ, Cech TR, (2011). Multiple POT1-TPP1 proteins coat and compact long telomeric single-stranded DNA. J. Mol. Biol, 410, 10–17. 10.1016/j.jmb.2011.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corriveau M, Mullins MR, Baus D, Harris ME, Taylor DJ, (2013). Coordinated interactions of multiple POT1-TPP1 proteins with telomere DNA. J. Biol. Chem, 288, 16361–16370. 10.1074/jbc.M113.471896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M, Kiselar J, Whited TL, Hernandez-Sanchez W, Taylor DJ, (2019). POT1-TPP1 differentially regulates telomerase via POT1 His266 and as a function of single-stranded telomere DNA length. Proc. Natl. Acad. Sci. U. S. A, 116, 23527–23533. 10.1073/pnas.1905381116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi KH, Lakamp-Hawley AS, Kolar C, Yan Y, Borgstahl GEO, Ouellette MM, (2015). The OB-fold domain 1 of human POT1 recognizes both telomeric and non-telomeric DNA motifs. Biochimie, 10.1016/j.biochi.2015.04.015. [DOI] [PMC free article] [PubMed]
- 35.Ramsay AJ, Quesada V, Foronda M, Conde L, Martinez-Trillos A, Villamor N, Rodriguez D, Kwarciak A, et al. , (2013). POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet, 45, 526–530. 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 36.Shi J, Yang XR, Ballew B, Rotunno M, Calista D, Fargnoli MC, Ghiorzo P, Bressac-de Paillerets B, et al. , (2014). Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat. Genet, 46, 482–486. 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trigueros-Motos L, (2014). Mutations in POT1 predispose to familial cutaneous malignant melanoma. Clin. Genet, 86, 217–U109. 10.1111/cge.12416. [DOI] [PubMed] [Google Scholar]
- 38.Bainbridge MN, Armstrong GN, Gramatges MM, Bertuch AA, Jhangiani SN, Doddapaneni H, Lewis L, Tombrello J, et al. , (2015). Germline mutations in shelterin complex genes are associated with familial glioma. J. Natl. Cancer Inst, 107, 384. 10.1093/jnci/dju384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calvete O, Martinez P, Garcia-Pavia P, Benitez-Buelga C, Paumard-Hernandez B, Fernandez V, Dominguez F, et al. , (2015). A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nat. Commun, 6, 8383. 10.1038/ncomms9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galer P, Wang BF, Sket P, Plavec J, (2016). Reversible pH switch of two-quartet G-quadruplexes formed by human telomere. Angew. Chemie-Intl. Ed, 55, 1993–1997. 10.1002/anie.201507569. [DOI] [PubMed] [Google Scholar]
- 41.Makarov VL, Hirose Y, Langmore JP, (1997). Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9054505. [DOI] [PubMed] [Google Scholar]
- 42.McElligott R, Wellinger RJ, (1997). The terminal DNA structure of mammalian chromosomes. Embo J, 16, 3705–3714. 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW, (1997). Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev, 11, 2801–2809 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9353250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J, (2008). Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality, 20, 431–440. 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 45.Randazzo A, Spada GP, da Silva MW, (2013). Circular dichroism of quadruplex structures. Top. Curr. Chem, 330, 67–86. 10.1007/128_2012_331. [DOI] [PubMed] [Google Scholar]
- 46.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M, (2007). The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature, 445, 506–510 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17237768. [DOI] [PubMed] [Google Scholar]
- 47.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR, (2012). The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature, 492, 285–289. 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loayza D, Parsons H, Donigian J, Hoke K, de Lange T, (2004). DNA binding features of human POT1: a nonamer 5’-TAGGGTTAG-3’ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem, 279, 13241–13248. 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 49.Lei M, Zaug AJ, Podell ER, Cech TR, (2005). Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem, 280, 20449–20456. 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 50.Hardin CC, Perry AG, White K, (2000). Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers, 56, 147–194. . [DOI] [PubMed] [Google Scholar]
- 51.Hammes GG, Chang YC, Oas TG, (2009). Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. U. S. A., 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed]
- 52.Verdun RE, Crabbe L, Haggblom C, Karlseder J, (2005). Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell., 10.1016/j.molcel.2005.09.024. [DOI] [PubMed]
- 53.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T, (2010). In vivo stoichiometry of shelterin components. J. Biol. Chem, 285, 1457–1467. 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeynes JCG, Geraki K, Jeynes C, Zhaohong M, Bettiol AA, Latorre E, Harries LW, Soeller C, (2017). Nanoscale properties of human telomeres measured with a dual purpose X-ray fluorescence and super resolution microscopy gold nanoparticle probe. ACS Nano, 11, 12632–12640. 10.1021/acsnano.7b07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bandaria JN, Qin P, Berk V, Chu S, Yildiz A, (2016). Shelterin protects chromosome ends by compacting telomeric chromatin. Cell, 164, 735–746. 10.1016/j.cell.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jansson LI, Hentschel J, Parks JW, Chang TR, Lu C, Baral R, Bagshaw CR, Stone MD, (2019). Telomere DNA G-quadruplex folding within actively extending human telomerase. Proc. Natl. Acad. Sci. U. S. A, 116, 9350–9359. 10.1073/pnas.1814777116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaires JB, (2010). Human telomeric G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J, 277, 1098–1106. 10.1111/j.1742-4658.2009.07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang DH, Fujimoto T, Saxena S, Yu HQ, Miyoshi D, Sugimoto N, (2010). Monomorphic RNA G-quadruplex and polymorphic DNA G-quadruplex structures responding to cellular environmental factors. Biochemistry, 49, 4554–4563. 10.1021/bi1002822. [DOI] [PubMed] [Google Scholar]
- 59.Balagurumoorthy P, Brahmachari SK, (1994). Structure and stability of human telomeric sequence. J. Biol. Chem, 269, 21858–21869 http://www.ncbi.nlm.nih.gov/pubmed/8063830. [PubMed] [Google Scholar]
- 60.Byrd AK, Bell MR, Raney KD, (2018). Pif1 helicase unfolding of G-quadruplex DNA is highly dependent on sequence and reaction conditions. J. Biol. Chem, 293, 17792–17802. 10.1074/jbc.RA118.004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao J, Byrd AK, Zybailov BL, Marecki JC, Guderyon MJ, Edwards AD, Chib S, West KL, Waldrip ZJ, Mackintosh SG, Gao Z, Putnam AA, Jankowsky E, Raney KD, (2019). DEAD-box RNA helicases Dbp2, Ded1 and Mss116 bind to G-quadruplex nucleic acids and destabilize G-quadruplex RNA. Chem. Commun. (Camb.), 55, 4467–4470. 10.1039/c8cc10091h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernando H, Sewitz S, Darot J, Tavare S, Huppert JL, Balasubramanian S, (2009). Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res, 37, 6716–6722. 10.1093/nar/gkp740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang AY, Balasubramanian S, (2012). The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J. Am. Chem. Soc, 134, 19297–19308. 10.1021/ja309851t. [DOI] [PubMed] [Google Scholar]
- 64.Phan AT, Kuryavyi V, Luu KN, Patel DJ, (2007). Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res, 35, 6517–6525. 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai JX, Carver M, Punchihewa C, Jones RA, Yang DZ, (2007). Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res, 35, 4927–4940. 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson KA, Simpson ZB, Blom T, (2009). FitSpace explorer: an algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal. Biochem, 387, 30–41. 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 67.Johnson KA, Simpson ZB, Blom T, (2009). Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem, 387, 20–29. 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


