Abstract
Covid 19 caused by novel strain SARS- CoV-2 has become a pandemic due to its contagious nature of infection. It enters by binding with ACE2 receptor present on the outer surface of a cell by cleaving S1/S2 with proteolytic protein Furin. Further viral replication or transcription then takes place with the help of main protease 3CLpro and polymerase RdRp. This in silco study was carried out to block ACE2, Furin, 3CLpro and RdRP with various phytochemicals to prevent SARS- CoV-2 entry and replication or transcription. Twenty different phytochemicals were screened to understand the drug-likeliness obeying Lipinski’s rule 5 and further, molecular docking was performed using these phytochemicals to block their respective target proteins. All the phytochemicals follow Lipinski’s rule of five and molecular docking result shows best binding affinity of Podofilox − 7.54 kcal/mol with ACE2, Psoralidin − 8.04 kcal/mol with Furin, Ursolic acid − 8.88 kcal/mol with 3CLpro and Epiafzelechin − 8.26 kcal/mol with RdRp. Thus, blocking two human receptors ACE2 and Furin with Podofilox and Psoralidin respectively may prevent the viral entry into the cells. Also blocking viral proteins 3CLpro and RdRp with Ursolic acid and Epiafzelechin may prevent viral replication or transcription. Using this combination therapy of blocking the receptors responsible for viral entry and viral proteins responsible for replication or transcription may prevent SARS- CoV-2 infection.
Keywords: SARS- CoV- 2, ACE2, Furin, 3CLpro, RdRp, Phytochemicals, Drug-likeliness, Molecular docking
Introduction
The novel corona virus SARS- CoV-2 has become a pandemic since the time of its flare-up in China and is a contagious infection to humans. An effective drug for the treatment of individuals affected by this virus is still unavailable. To eradicate this virus different lines of therapy including drug designing and the use of antibodies are in progress [1]. Corona virus ailment 2019 (COVID-19) is an emerging infectious disease with no set up lab markers available to evaluate the severity of the illness. [2] Conducted a platelet count to see whether it could separate COVID-19 patients with severe diseases from those without severe disease. Studies were also carried out to assess whether thrombocytopenia is associated with extreme COVID-19 [2]. The coronavirus is a highly transmittable viral infection, which can be transferred by human to human contact [3]. On the basis of structural studies [4–6] and biochemical experiments [6]7, SARS CoV-2 is optimized for its binding to the ACE2 receptor in human. The most variable part of the genome of coronavirus is the receptor binding domain present in the spike protein. Six amino acids- L455, F486, Q493, S494, N501 and Y505 [7], have been shown to be important for optimized binding to ACE2 receptor and also for determining the host range. ACE2, a receptor acts by attaching to the outer surface of the cells situated in lungs, arteries, heart, kidney and intestines. It lowers the level of blood pressure by catalyzing angiotensin II cleavage into angiotensin 1–7. ACE2 acts as an entry point into cells of some Corona virus [8]. Another important feature of SARS CoV-2 is the presence of a polybasic cleavage site at the junction of the two subunits of the spike-S1 and S2. This promotes effective cleavage by furin and other proteases and also determines viral infectivity and host range [9]. In the media, it is regularly the “case fatality rate” that is mentioned when the danger of death from COVID-19 is discussed. This measure is in some cases called case fatality risk or case fatality ratio, or CFR. The CFR can be calculated easily. It is calculated by taking the number of individuals who have died due to the infection and dividing it by those who have been diagnosed as positive cases. For example if 10 individuals have died, and 100 individuals have been diagnosed as positive, the CFR is [10/100], or 10% [10]. Case Fatality Rate (CFR, in %) = (Number of Deaths from Disease X 100)/ Number of diagnosed cases of disease. Till now there are more than 8, 82,000 confirmed cases and more than 44,000 deaths due to SARS-CoV-2. COVID-19 infection can be described as a mellow upper respiratory tract disease or a lower respiratory tract infection including mild pneumonia, and full blown pneumonia with intense respiratory difficulty. It affects all age groups ranging from infants, to the aged people. Pregnant women and individuals above the age of 60 are at greater risk of succumbing to the infection. In pregnant women with COVID-19 infection there is no proof of vertical transmission of the infection, however an increase in the number of preterm deliveries has been noted [11]. No drugs or antiviral agents have been developed so far against SARS- CoV-2. Some in-vitro and limited clinical studies of few drugs like Hydroxychloroquine, Lopinavir, Ritonavir, Remdesivir, Erythromycin, Tocilizumab, have shown potential benefit. However there are several associated safety concerns like cardiac arrhythmias, retinal damage, risk of diabetes, hepatotoxicity, and GI perforation [12]. The aim of this study was to target the ACE2 receptor which is the entry point of the virus, furin a protease that helps in activation of SARS COV-2 by cleavage of S protein, main protease (M pro, also called 3CL pro) [13] -a dimer of two identical subunits that together form two active sites, which play an essential role in viral replication and RNA dependent RNA polymerase RdRp which synthesizes a full length negative strength RNA template to be used by RdRp to make More viral genomic RNA [14] using phytochemicals. By targeting these three sites the viral replication as well as viral entry can easily be prevented. An added advantage is that there are less safety concerns. In this study 5 different phytochemicals have been assessed for their ability to target the ACE2 receptor, 3CLpro, RdRp, and Furin using autodock 4.2. The result has been analysed based on the best binding energy followed by hydrogen bonds.
Materials and methodology
Protein database
UniProt i.e. Universal Protein Resource is a protein database which provides a stable, easily accessible, cyclopedic information about the protein sequence and functional annotation. UniProt database was formed by collaboration among Protein Information Resource, Swiss Institute of Bioinformatics and European Bioinformatics Information. This database was used to gather information about protein targets used in this in-silico study. UniProt Consortium (2008)—The universal protein resource (UniProt). [15]
Plants used in the study and the rationale behind it
The plants from which these phytochemicals were extracted are Leptospermum scoparium (red tea), Viscum album (mistletoe), Citrus sinensis (orange), Podophyllum hexandrum (Indian podphyllum, bankakri), Syringa vulgaris (common lilac), Polygonum cuspidatum (common knotweed), Psoralea corylifolia (babchi), Peganum harmala (harmal), Boswellia serrate (Indian frankincense), Morinda citrifolia (Indian mulberry), Oldenlandia diffusa (snake needle grass), Acacia mearnsii (black wattle), Plumbago zeylanica (Ceylon leadword), Prosopis juliflora (vlyati babool), Silybum marianum(milk thistle), Curcuma longa (turmeric), Marrubium peregrinum (horehound), Bauhinia pentandra (mountain ebony), Actinidia chinensis (Kiwi fruit), Vitex trifolia (nichinda).
The phytochemicals present in these plants were reported to have significant antimicrobial property against various microorganisms. Some of the phytochemicals that have anti-cancer, anti-oxidant, anti-hypertensive and anti-diabetic property have been used for binding with ACE-2 and Furin. The other phytochemicals with anti-microbial property were used for binding with 3CLpro and RdRP.[16–19].
Exploiting the antiviral mechanisms of bioactive constituents present in these plants could provide an insight in to their modes of action towards viral lifecycle, invasion, penetration, replication, assembly and release. One of the plants used in this study, Curcuma longa (turmeric) is a spice used in many Indian cuisines. It has known medicinal properties [20]
Structural database
RCSB PDB i.e. Research Collaboratory for Structural Bioinformatics. Protein Data Bank is a universal platform for storage of 3D structures of proteins, nucleic and their complexes. It uses the US data centre for global PDB storage and also allows users to enter and utilize data free of cost without any limitation with regard to usage. PDB helps in research, education in various fields with help tools, structure, and information. The proteins with PDB IDs 1R42, 6M71, 6NUR and 4OMC are used as target proteins in this in-silico approach. [21]
Homology modeling
Modeller 9.24 has been used for modeling four target proteins and molProbity was used to analyze best modelled structure. [22]
Compound database
PubChem is an open chemistry data base i.e. one can upload scientific information in Pubchem and others can access it with ease. It is available in the National Institute of Health (NIH). PubChem mainly consists of small molecules along with macromolecules and chemically modified structures. All the Phytochemicals and related information which have been used as ligand in this computational study were obtained from PubChem [23].
Swiss ADME
Swiss ADME is a freely accessible web tool which allows computation of physico chemical descriptors along with prediction of ADME parameters, druglike nature, pharmacokinetic properties and medicinal likeliness of molecules to support drug discovery. It uses canonicals SMILES format of chemical compounds as input. In this in-silico study was used to determine the drug likeliness (Lipinski’s Rule of five) and bioavailability of the phytochemical compounds [24].
Molecular docking
AutoDock version 4.2[25] was used for analysing the extent of protein–ligand interactions [26].Target proteins were first prepared by the addition of polar hydrogen, followed by the Kollman charge and Gasteiger charge. Ligands were prepared by adopting the necessary steps. After fixing the active sites with specific residues, a grid box of 90 × 90 × 90 Å was used for the search. Lamarckian genetic algorithm was used for auto docking the ligands with the proteins [27]. Analysis of hydrogen bonds and binding energies were carried out using AutoDock 4.2, PyMol 2.3.2, and Protein–Ligand interaction profiler.
Results and discussion
Structure of target protein and virtual screening of the phytochemicals
Structure of four target proteins ACE2 (PDB ID 1R42), 3CLpro (PDB ID 6M71), Nsp12 (PDB ID 6NUR) and Furin (PDB ID 4OMC) was obtained from Protein Data Bank (PDB). Twenty novel phytochemicals have been selected to target three different proteins namely ACE2, 3CLpro, RdRp and Furin. These phytochemicals have been subjected to virtual screening based on their drug-likeliness following Lipinski’s Rule of five. This rule describes molecular properties which are important for a drug's pharmacokinetics in the human body, including their absorption, distribution, metabolism, and excretion. The rule is important for drug development where a pharmacologically active lead structure is optimized step-wise for increased activity and selectivity, as well as drug-like properties as described by Lipinski's rule. Lipinski's rule says that, in general, an orally active drug should have no more than one violation of the following criteria:(a) Not more than 5 hydrogen bond donors (nitrogen or oxygen atoms with one or more hydrogen atoms) (b) Not more than 10 hydrogen bond acceptors (nitrogen or oxygen atoms) (c) A molecular weight under 500 daltons (d) An octanol–water partition coefficient log P of less than 5. (Tables 1, 2, 3, 4) This signifies that all twenty phytochemicals are drug-able according to Lipinski’s rule of five. [28].
Table 1.
Drug-likeliness analysis results of 10 phytochemicals targeting ACE2
| Compound/Pubchem CompoundCID | Source | Properties | ||||
|---|---|---|---|---|---|---|
| Molecular weight (< 500 Da) | Logp (< 5) | H-Bond donor (< 5) | H-bond acceptor (< 10) | Violations | ||
| 5,7-Dimethoxyflavanone/378,567 | Leptospermum scoparium (red tea) | 284.31 | 1.76 | 0 | 4 | 0 |
| Coniferin/5,280,372 | Viscum album (mistletoe) | 342.34 | − 1.32 | 5 | 8 | 0 |
| Flavanone/10,251 | Citrus sinensis(orange) | 224.25 | 2.47 | 0 | 2 | 0 |
| Podofilox/10,607 | Podophyllum hexandrum (Indian podphyllum, bankakri) | 414.41 | 1.43 | 1 | 8 | 0 |
| Syringin/5,316,860 | Syringa vulgaris (common lilac) | 372.4 | − 1.59 | 5 | 9 | 0 |
Table 2.
Drug-likeliness analysis results of 10 phytochemicals targeting Furin
| Compound/Pubchem CompoundCID | Source | Properties | ||||
|---|---|---|---|---|---|---|
| Molecular weight (< 500 Da) | Logp (< 5) | H-Bond donor (< 5) | H-bond acceptor (< 10) | Violations | ||
| Resveratrol/445,154 | Polygonum cuspidatum(common knotweed) | 228.24 | 2.26 | 3 | 3 | 0 |
| Psoralidin/5,281,806 | Psoralea corylifolia (babchi) | 336.34 | 2.88 | 2 | 5 | 0 |
| Harmine/5,280,953 | Peganum harmala (harmal) | 212.25 | 1.56 | 1 | 2 | 0 |
| Boswellic acid/168,928 | Boswellia serrata(Indian frankincense) | 456.70 | 5.82 | 3 | 2 | 1 |
| Damnacanthal/2948 | Morinda citrifolia (Indian mulberry) | 282.25 | 0.27 | 1 | 5 | 0 |
Table 3.
Drug-likeliness analysis results of 10 phytochemicals targeting 3CLpro
| Compound/Pubchem CompoundCID | Source | Properties | ||||
|---|---|---|---|---|---|---|
| Molecular weight (< 500 Da) | Logp (< 5) | H-Bond donor (< 5) | H-bond acceptor (< 10) | Violations | ||
| Ursolic acid/64,945 | Oldenlandia diffusa(snake needle grass) | 456.70 | 5.82 | 2 | 3 | 1 |
| Robinetinidol/12,314,983 | Acacia mearnsii(black wattle) | 290.27 | 0.24 | 5 | 6 | 0 |
| Plumbagin/10,205 | Plumbago zeylanica(Ceylon leadword) | 188.18 | 0.59 | 1 | 3 | 0 |
| Mesquitol/1,103,382 | Prosopis juliflora (vlyati babool) | 290.27 | 0.24 | 5 | 6 | 0 |
| Legalon/5213 | Silybum marianum(milk thistle) | 482.44 | − 0.40 | 5 | 10 | 0 |
Table 4.
Drug-likeliness analysis results of 10 phytochemicals targeting RdRp
| Compound/Pubchem CompoundCID | Source | Properties | ||||
|---|---|---|---|---|---|---|
| Molecular weight (< 500 Da) | Logp (< 5) | H-Bond donor (< 5) | H-bond acceptor (< 10) | Violations | ||
| Curcumin/969,516 | Curcuma longa (turmeric) | 368.38 | 1.47 | 2 | 6 | 0 |
| Ladanein/3,084,066 | Marrubium peregrinum(horehound) | 314.29 | 0.47 | 2 | 6 | 0 |
| Fisetinidol/442,397 | Bauhinia pentandra(mountain ebony) | 274.27 | 0.79 | 4 | 5 | 0 |
| Epiafzelechin/443,639 | Actinidia chinensis(Kiwi fruit) | 274.27 | 0.79 | 4 | 5 | 0 |
| Vitex norditerpenoid 1/ 11,208,535 | Vitex trifolia (nichinda) | 290.44 | 3.59 | 0 | 2 | 0 |
Homology modeling analysis
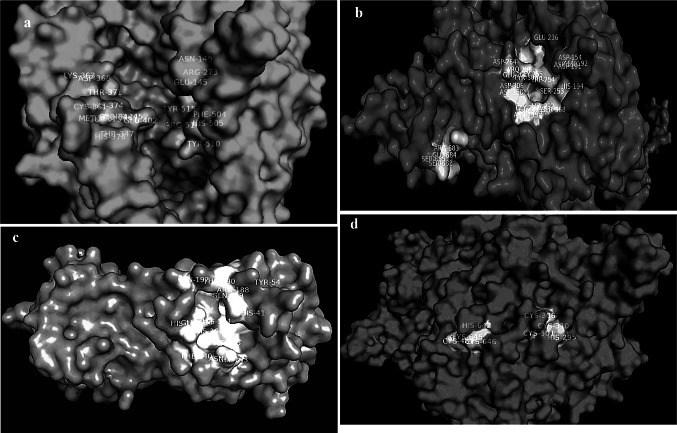
Homology modeling of two human proteins (ACE2 and Furin) and two viral proteins (3CLpro and RdRp) has been modeled to obtain the best structure for further study. The modeled structures are shown in Fig. 1a–d. The white area on the modeled structures indicates the active site of the target protein along with amino acid residue.
Fig. 1.
a Structure of ACE2 with its active sites in white colour, b Structure of Furin with its active sites in white colour, c Structure of 3CLpro with its active sites in white colour, d Structure of RdRp with its active sites in white colour
Molecular docking analysis
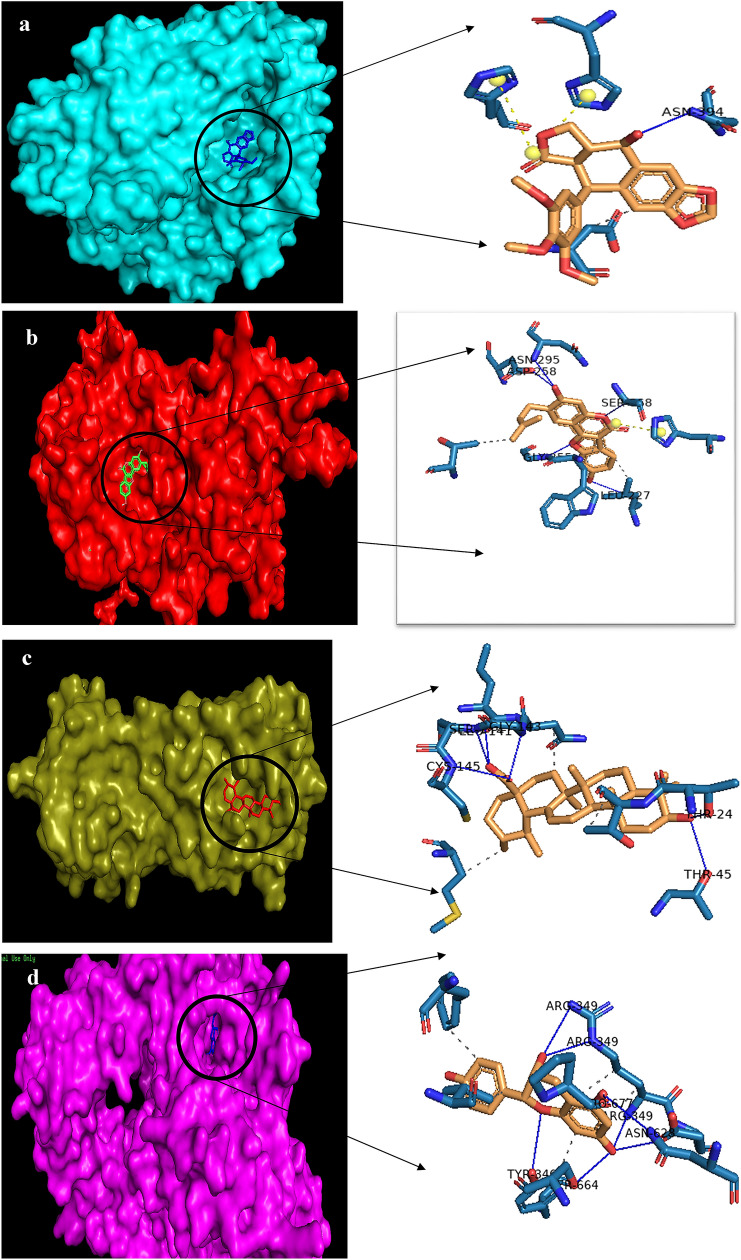
AutoDock 4.2 was used in this in-silico study to analyze the binding ability of twenty different phytochemicals with the four target proteins. Results of this study helped us to identify the phytochemicals which were able to block ACE2 and Furin (Table 5 and 6) and inhibit both 3CLpro and Nsp12 (Tables 7 and 8 respectively). Binding ability of phytochemicals is predicted based on the binding energy. The hydrogen bond is also considered when two or more compounds have similar binding energy. Docking results suggest that Podofilox with − 7.54 kcal/mol, Psoralidin with − 8.04 kcal/mol, Ursolic acid with -8.88 kcal/mol, and Epiafzelechin with −8.26 kcal/mol has best binding affinity against ACE2, Furin, 3CLpro and Nsp12 respectively. Four best protein–ligand complex structures were visualized using Pymol 2.3.2 (Fig. 2a–d). SARS-CoV-2 is optimized for its binding to the ACE2 receptor in human. ACE2, a carboxypeptidase, reduces the blood pressure level by removing a single amino acid residue of C-terminus of both angiotensin-II that converts angiotensin II into angiotensin 1–7 and Angiotensin-I into Angiotensin-(1–9). Angiotensin (1–7) acts as a vasodilator, thus reducing the blood pressure. ACE2 acts as an entry point for SARS-CoV-2 [29]. A functional polybasic (RRAR) cleavage site is present at the junction of two subunits S1 and S2 of the SARS-Cov-2 spike protein. This facilitates cleavage by furin [30] and other host proteases like TMPRSS2 effectively, which determines the viral host range and infectivity [31]. The main function of S1 subunit is to mediate attachment to the cell surface receptor whereas S2 subunit mediates the fusion of the viral membrane with the host cell membrane. [32]. It has been reported that loss of ACE2 in knockout mice showed no effect on blood pressure level. [33] High binding affinity of these phytochemicals towards ACE2 may result in blocking the attachment of receptor binding Domain of SARS-CoV-2 to ACE2, and blocking of Furin with phytochemicals may prevent cleavage of the spike protein and thus viral entry may be prevented without any effect on blood pressure level. The main protease of SARS-CoV-2 ie 3CLpro, also known as Nsp5, has 3 domains viz. domain 1(residue8-101),domain 2(residue 102–184) and domain 3(residue 201–303).Active site of 3CLpro is present between Domain 1 and domain 2 and has CYS HIS catalytic dyad. 3CLpro is naturally cleaved from poly-proteins to release mature enzymes and again cleaves downstream Nsps at 11 sites releasing Nsp4-Nsp16. It mainly deals with the maturation of Nsps which are essential for viral life cycle. Targeting 3CLpro with the high affinity binding phytochemicals may prevent the cleavage of the downstream Nsps eventually blocking the release of 11 Nsps, Nsp4-Nsp16 and their maturation which is essential for the virus. RdRp also known as Nsp12 is a RNA dependent RNA polymerase which plays an important role in viral replication/transcription. RdRp Domain has conserved region with SER-ASP-ASP motif present at C-terminus. The binding of Nsp12 to the RNA is brought about by the Nsp7-Nsp8 complex which enhances the enzyme activity of RdRp. By targeting RdRp, it’s enzyme activity may be blocked thereby inhibiting viral replication/transcription.
Table 5.
Molecular Docking of 5 molecules with ACE2
| Ligand name | Binding energy (Δg) | H- bond | Inhibition constant (µm) | Inter-molecular energy | VDW-H bond desolvation Energy | Ligand efficiency | Electrostatic energy |
|---|---|---|---|---|---|---|---|
| 5,7-Dimethoxyflavanone | − 7.02 | 1 | 7.13 | − 7.85 | − 7.88 | − 0.33 | 0.03 |
| Coniferin | − 4.93 | 7 | 244.86 | − 7.94 | − 7.9 | − 0.21 | − 0.04 |
| Flavanone | − 6.94 | 1 | 8.25 | − 7.21 | − 7.22 | − 0.41 | 0.01 |
| Podofilox | − 7.54 | 1 | 2.97 | − 8.91 | − 8.71 | − 0.25 | − 0.21 |
| Syringin | − 5.07 | 6 | 190.91 | − 8.37 | − 8.29 | − 0.2 | − 0.08 |
Table 6.
Molecular Docking of 5 molecules with Furin
| Ligand name | Binding energy (Δg) | H- bond | Inhibition constant (µm) | Inter-molecular energy | VDW-H bond desolvation Energy | Ligand efficiency | Electrostatic energy |
|---|---|---|---|---|---|---|---|
| Boswellic acid | − 7.49 | 4 | 3.22 | − 8.32 | − 7.65 | -0.23 | − 0.66 |
| Damnacanthal | − 5.95 | 5 | 43.88 | − 6.77 | − 6.43 | -0.28 | − 0.345 |
| Resveratrol | − 6.51 | 2 | 17.03 | − 6.78 | − 6.68 | -0.41 | − 0.1 |
| Psoralidin | − 8.04 | 5 | 1.28 | − 9.14 | − 8.95 | -0.32 | − 0.18 |
| Harmine | − 6.42 | 2 | 19.81 | − 6.69 | − 6.59 | -0.4 | − 0.1 |
Table 7.
Molecular Docking of 5 molecules with 3CLpro
| Ligand name | Binding energy (Δg) | H- bond | Inhibition constant (µm) | Inter- molecular energy | VDW-H bond desolvation Energy | Ligand efficiency | Electrostatic energy |
|---|---|---|---|---|---|---|---|
| Ursolic acid | − 8.88 | 6 | 310.63 | − 9.7 | − 9.54 | − 0.27 | − 0.16 |
| Robinetinidol | − 8.44 | 7 | 650.07 | − 10.09 | − 9.93 | − 0.4 | − 0.16 |
| Plumbagin | − 5.82 | 2 | 54.03 | − 6.1 | − 6.01 | − 0.42 | − 0.08 |
| Mesquitol | − 7.55 | 10 | 2.92 | − 9.2 | − 8.78 | − 0.36 | − 0.42 |
| Legalon | − 8.47 | 9 | 614.38 | − 10.94 | − 10.81 | − 0.24 | − 0.13 |
Table 8.
Molecular Docking of 5 molecules with RdRp
| Ligand name | Binding energy (Δg) | H- bond | Inhibition constant µm) | Inter-molecular Energy | VDW-H bond Desolvation Energy | Ligand efficiency | Electrostatic energy |
|---|---|---|---|---|---|---|---|
| Curcumin | − 6.05 | 4 | 36.81 | − 8.79 | − 8.84 | − 0.22 | 0.05 |
| Ladanein | − 6.29 | 7 | 24.35 | − 7.67 | − 7.06 | − 0.27 | − 0.61 |
| Fisetinidol | − 7.68 | 1 | 2.37 | − 9.05 | 9 | − 0.26 | − 0.04 |
| Epiafzelechin | − 8.26 | 8 | 886.38 | − 9.63 | − 9.38 | − 0,41 | − 0.24 |
| Vitex norditerpenoid 1 | − 4.02 | 1 | 1.13 | − 8.68 | − 8.29 | − 0.1 | − 0.4 |
Fig. 2.
a Molecular docking structure of Podofilox-ACE2, b Molecular docking structure of Psoralidin-Furin, c Molecular docking structure of Ursolic acid-3CLpro, d Molecular docking structure of Epiafzelechin -RdRp
Twenty novel phytochemicals on the basis of Lipinski’s rule of 5 which targeted two human proteins ACE2 and Furin and two viral proteins, 3CLpro and RdRp have been screened. Molecular Docking analysis reveals that Podofilox with ACE2, Psoralidin with Furin, Ursolic acid with 3CLpro and, Epiafzelechin with RdRp showed best binding affinity with respective target proteins. This is probably the first report of blocking Furin by the using the phytochemicals reported in the present study. By blocking two human proteins we may be able to prevent the entry of SARS- CoV-2 and by blocking two viral proteins we may be able to prevent viral replication/transcription. Combination therapy using phytochemicals mentioned in this study may finally be able to prevent SARS- CoV-2 infection.
Acknowledgement
The authors thank VIT, Vellore for providing facilities to carry out the work.
Compliance with ethical standard
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chavez S, Long B, Koyfman A, Liang SY. Coronavirus Disease (COVID-19): A primer for emergency physicians. Am J Emerg Med. 2020;S0735–6757(20):30178–30179. doi: 10.1016/j.ajem.2020.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin Chim Acta. 2020;506:145–148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol. 2020;94(7):e00127–e220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner AJ. ACE2 Cell Biology, Regulation, and Physiological Functions. Protect Arm Renin Angiotensin Syst RAS. 2015 [Google Scholar]
- 9.Sironi M, Hasnain SE, Rosenthal B, Phan T, Luciani F, Shaw MA, et al. SARS-CoV-2 and COVID-19: A genetic, epidemiological, and evolutionary perspective Infect. Genet Evol. 2020;84:104384. doi: 10.1016/j.meegid.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gondauri D, Mikautadze E, Batiashvili M. Research on COVID-19 virus spreading statistics based on the examples of the cases from different countries. Electron J Gen Med. 2020;174:209. [Google Scholar]
- 11.Liu H, Wang L, Zhao S, Kwak-kim J, Mor G, Liao A. Why are pregnant women susceptible to COVID-19 ? An immunological viewpoint J Reprod Immunol. 2020;139:103122. doi: 10.1016/j.jri.2020.103122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derwand R, Scholz M. Does zinc supplementation enhance the clinical efficacy of chloroquine/hydroxychloroquine to win today's battle against COVID-19? Med Hypotheses. 2020;142:109815. doi: 10.1016/j.mehy.2020.109815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nao N, Yamagishi J, Miyamoto H, Igarashi M, Manzoor R, Ohnuma A, et al. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus Hemagglutinin. mBio. 2017;8(1):e02298–e2316. doi: 10.1128/mBio.02298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UniProt Consortium. The universal protein resource (UniProt). Nucleic Acids Res2008;36(Database issue):D190–5. doi: 10.1093/nar/gkm895 [DOI] [PMC free article] [PubMed]
- 16.Nazaruk J, Orlikowski P. Phytochemical profile and therapeutic potential of Viscum album L. Nat Prod Res. 2016;30(4):373–385. doi: 10.1080/14786419.2015.1022776. [DOI] [PubMed] [Google Scholar]
- 17.Ashraf MA. Phytochemicals as Potential Anticancer Drugs: Time to Ponder Nature's Bounty. Biomed Res Int. 2020;31(2020):8602879. doi: 10.1155/2020/8602879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tohmé MJ, Giménez MC, Peralta A, Colombo MI, Delgui LR. Ursolic acid: A novel antiviral compound inhibiting rotavirus infection in vitro. Int J Antimicrob Agents. 2019;54(5):601–609. doi: 10.1016/j.ijantimicag.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Wang TY, Li Q, Bi KS. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J Pharm Sci. 2018;13(1):12–23. doi: 10.1016/j.ajps.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warrier, P.S., 1996. Indian medical plants- a compendium of 500 Species Volume 1 to 5 Orient Longman’
- 21.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 23.Bolton EE, Chen J, Kim S, Han L, He S, Shi W, et al. PubChem3D: a new resource for scientists. J Cheminform. 2011;3(1):32. doi: 10.1186/1758-2946-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11(5):905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43(W1):W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuhrmann J, Rurainski A, Lenhof HP, Neumann D. A new Lamarckian genetic algorithm for flexible ligand-receptor docking. J Comput Chem. 2010;31(9):1911–1918. doi: 10.1002/jcc.21478. [DOI] [PubMed] [Google Scholar]
- 28.Pollastri MP. Overview on the Rule of Five. Curr Protoc Pharmacol. 2010 Jun;Chapter 9:Unit 9.12. doi: 10.1002/0471141755.ph0912s49. PMID: 22294375. [DOI] [PubMed]
- 29.Ortega JT, Serrano ML, Pujol FH, Rangel HR. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z, et al. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Version 2. Signal Transduct Target Ther. 2020;5(1):92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Version 2. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Towler P, Staker B, Prasad SG, Menon S, Tang J, Parsons T, et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]