Abstract
When engineering microbes to overproduce a target molecule, engineers face multiple layers of trade-offs to allocate limited cellular resources between the target pathway and native cellular systems. These trade-offs arise from limited free ribosomes during translation, competition for metabolic precursors, as well as the negative relationship between production and growth rate. To achieve high production performance, microbes need to spontaneously make decisions in the dynamic and heterogeneous fermentation environment. In this review, we discuss recent advances in microbial control strategies that are used to manage these trade-offs and to improve microbial production. This review focuses on design principles and compares different implementations, with the hope to provide guidelines to future microbial engineering.
Introduction
Microbial fermentation has provided an environment-friendly and versatile platform for manufacturing various bio-based products. Typical fermentation products include alcohols, organic acids, amino acids, vitamins, commodity chemicals, antibiotics, antibodies, and industrial enzymes [1–4]. Recent advances in metabolic engineering and synthetic biology have added an increasing number of products to this list, including fragrances, pharmaceuticals, nutraceuticals, and advanced materials [5–10]. The ability to biologically produce diverse products could have profound societal impacts on multiple industries, only if cost-effective bioproduction can be achieved. This challenge demands the development of effective strategies to improve the production performance (i.e. titers, yields, productivities, and robustness) of engineered microbes.
When engineering microbes to overproduce a specific molecule at high productivity and yield, one needs to consider multiple layers of trade-offs rather than simply overproduce pathway molecules to the highest level. The first layer stems from ribosomal cost of translating target proteins (Figure 1a). Sequestered ribosomes by mRNAs of target proteins reduce a cell’s ribosomal budget to make native proteins for biomass generation and energy synthesis [11–14]. Furthermore, the allocation of limited translational power between multiple modules within a target pathway affects the overall catalytic efficiency of the pathway [15,16]. The second layer is metabolic trade-offs that involve both carbon cost and energy cost (Figure 1b). Conversion of precursor metabolites (e.g. acetyl-CoA) to target products can lead to insufficient material and/or energy supply for the synthesis of cellular structures [17]. Protein synthesis and enzymatic reactions from engineered pathways also consume energy molecules (i.e. ATP and NAD(P) H), which can be otherwise used to support cell growth. Besides, molecules in a target pathway can be toxic to cells, particularly when they are accumulated to high concentrations. Thus a balanced allocation of metabolites and energy molecules is required for optimizing microbial production [18]. The third layer of trade-offs comes between growth rate and product yield. High producers usually have a slower growth rate than low producers. The difference in single-cell growth rate caused by mutations or molecular noise allows low producers to accumulate, lowering overall yields (Figure 1c).
Figure 1.
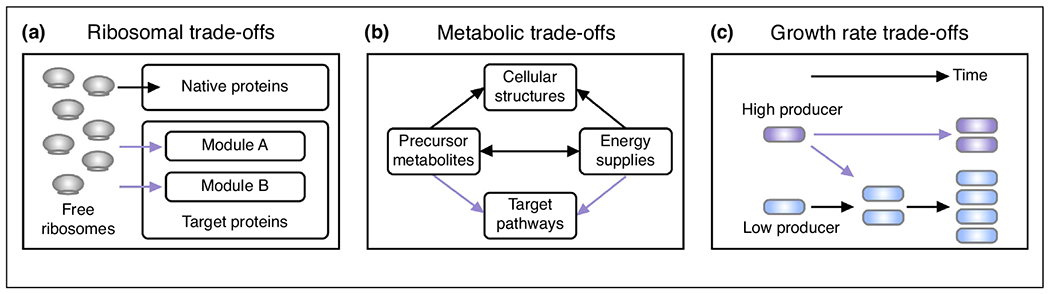
The presence of trade-offs during microbial production. (a) Ribosomal trade-offs. Overexpressing one protein decreases the number of free ribosomes available to synthesize native proteins and other target proteins within an engineered pathway. (b) Metabolic trade-offs. Precursor metabolites are used for the synthesis of cellular structures, energy supplies, as well as target pathways. Consumption of precursor metabolites by a target pathway can affect both cell growth and maintenance. (c) Growth rate trade-offs. Low producers usually grow faster than high producers. Without control, low producers gradually dominate the culture over time.
Over the past few years, many genetically encoded control strategies have been developed to improve microbial production by managing these trade-offs [19–23]. These control strategies help engineered cells to adjust their metabolism to combat dynamic and heterogeneous environments in large fermenters as well as stochastic cellular processes. In this review, we discuss recent research advances in control strategies with the focus on design principles. Here, we divide these control strategies into three categories based on their mode of operation: feedback control, two-stage metabolic switch, and population quality control.
Feedback control
Feedback control can alleviate the impact of ribosomal cost on the expression of specific genes of interest (GOIs). When the expression of GOI is uncontrolled, its protein level decreases as free ribosomes are depleted [14,24]. A common strategy in feedback control is to co-express a regulator with the GOI or to use an antithetic topology, where two components that sequester each other can be designed [25•,26,27]. Alternatively, orthogonal ribosomes can be engineered to feed back to the expression of orthogonal 16S rRNAs and to translate the GOI (Figure 2a) [28••], leading to robust expression of pathway enzymes and steady metabolic flux. Furthermore, in silico feedback control can be built using optogenetic regulation to achieve robust gene expression [29]. Additionally, stress-responsive promoters induced by burdensome genes can be used to control the expression of a single guide RNA (sgRNA), which in turn represses the expression of the burdensome gene via dCas9 (Figure 2b) [30•]. This CRISPR-based controller does not require genetic modification to the promoters of target genes, thus can be easily applied in many systems.
Figure 2.
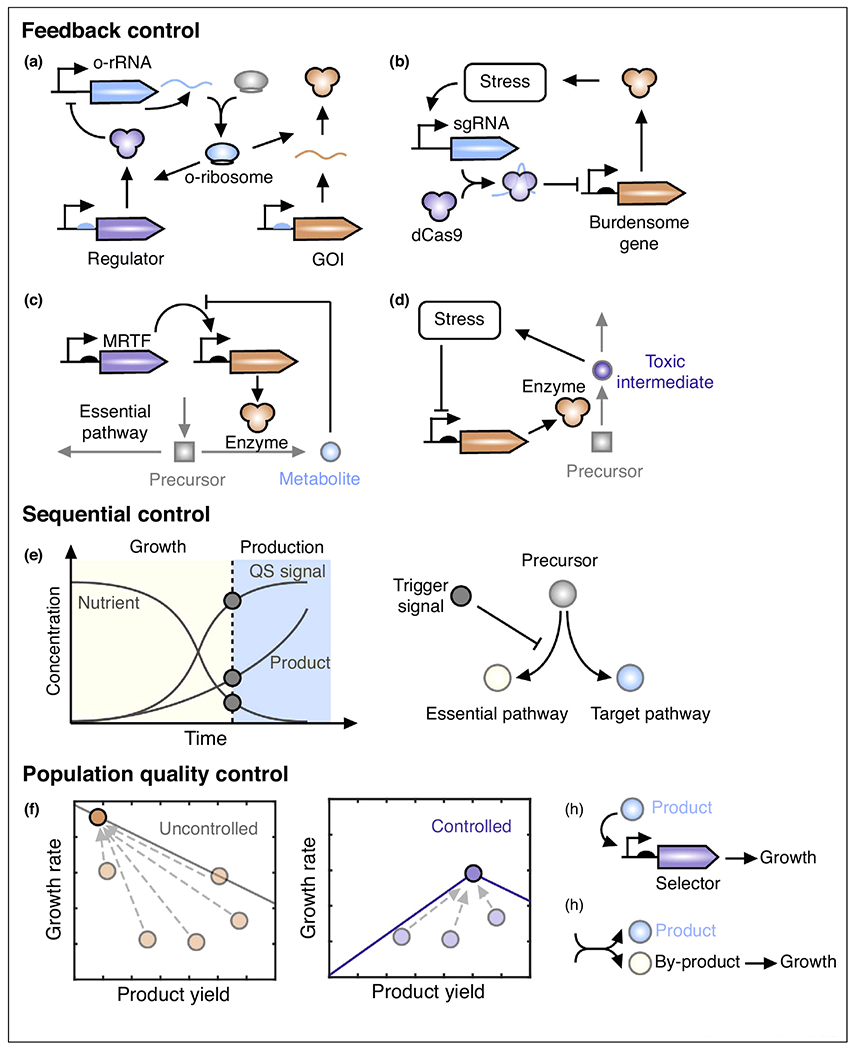
Design principles of control strategies that improve microbial production. (a) Feedback control of orthogonal rRNA provides robust gene expression from orthogonal ribosomes. (b) Stress-mediated feedback control to limit the burdensome protein synthesis. (c) Metabolic feedback control via MRTF to prevent metabolic overflow. (d) Stress-mediated feedback control to reduce the accumulation of the toxic intermediate. (e) Dynamics of trigger signals used to inhibit competing but essential pathways. (f) Setting an evolutionary stable point for high producers through population quality control. (g,h) Implement population quality control. Linking production and cell growth via sensor-selector (g) and by-product (h). GOI, gene of interests; o-rRNA, orthogonal rRNA; o-ribosome, orthogonal ribosome; MRTF, metabolite-responsive transcription factor; QS, quorum sensing.
Feedback control can also be built to mediate the metabolic costs raised by engineered metabolic pathways. In this case, a metabolite-responsive transcription factor (MRTF) can be used to detect metabolic overflow of an engineered pathway and then downregulate the expression of upstream enzymes, forming a metabolic feedback loop (Figure 2c). Such metabolic feedback controls have been developed for the malonyl-CoA biosynthetic pathway [31,32]. While malonyl-CoA is an essential metabolic precursor for many important pathways, excess malonyl-CoA produced from engineered pathways can inhibit cell growth. By using a malonyl-CoA-responsive transcription factor FapR, feedback controllers allowed excess malonyl-CoA to repress its own synthesis (via acetyl-CoA carboxylase), maintaining its cellular concentration at desirable levels. The feedback-controlled metabolic systems alleviated cellular toxicity otherwise caused by malonyl-CoA accumulation, leading to improved production of downstream metabolites, such as fatty acids [31,32]. Moreover, metabolic feedback loops can accelerate metabolite biosynthesis [33••] and recovery from metabolite depletion [34]. As demonstrated in a feedback-regulated fatty acid pathway, a layered negative metabolic loop can shorten the fatty acid rise-time (the time needed to reach half of the steady-state concentration) by as much as 12-fold [33••]. This negative feedback control topology has effectively accelerated metabolic response that would otherwise cause uncontrollable overproduction if a positive autoregulatory loop is used. In these metabolic feedback controls, the dose-response curve of MRTF can be rationally designed to coordinate the enzyme level and the metabolic flux [35]. If MRTFs are unavailable, stress-response promoters can be used to control the level of toxic intermediates. Stress-response promoters that are downregulated by the toxic intermediate can be used to drive the expression of enzymes leading to the synthesis of the intermediate to prevent its accumulation (Figure 2d) [36].
Several issues need to be considered to construct an effective feedback controller. First, the engineered feedback controller should be compatible with the natural regulatory network. Second, the feedback controller itself should not pose a significant burden on cell growth. Third, the controller should have suitable feedback strength. While too weak feedback can lead to a long delay, too strong feedback can cause overshoot and create noise [33••,37]. Fourth, the regulator should be easily scalable when multiple genes need control. Lastly, most existing feedback controls were developed and validated during exponential growth phase. Effective controls for use beyond exponential growth phase have not been extensively demonstrated and may require additional engineering efforts.
Metabolic switch in two-stage fermentation
During microbial production, metabolic flux can be spontaneously shifted from growth-related essential pathways to production pathways by a time-dependent trigger signal. Here we refer this strategy as metabolic switch that is often used in two-stage fermentation (Figure 2e). The trigger signal can be a quorum-sensing molecule that indicates the growth phase via cell density. Quorum-sensing metabolic switches have been developed in both Escherichia coli and Saccharomyces cerevisiae, showing improved production for multiple compounds [38–41]. Attention needs to be paid to avoid the accumulation of quorum-sensing molecules, which may cause problems in downstream purification processes. Beside quorum-sensing, the target product can also serve as the trigger signal. This has been demonstrated in muconic acid (MA) production with a metabolic switch embedding an MA sensor [42••]. As MA accumulated over time, cells gradually shift to the production phase by diverting flux from pyruvate and oxaloacetate to MA production. Other trigger signals include nutrient concentration [43,44], temperature change [45], and light [46]. For example, the native ergosterol pathway in S. cerevisiae was repressed by glucose limitation during the production phase, so that the common precursors, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) can be channeled to terpenoid production in engineered yeast [43,44]. While most metabolic switches control cell growth and pathway activity through transcriptional regulation by affecting promoter activities of target genes, other regulatory mechanisms, such as using RNAi [41,42••], mRNA attenuation [47], and protease-based regulation [48••] have also been developed. Among them, post-translational controls using protease-based regulation have the advantage of remaining functional in stationary phase, during which transcriptional and translation controls may not be effective. On the other hand, protease-based controls are associated with high ATP consumption during proteolysis and may compete with natural proteins for limited proteolysis machinery.
The kinetics of the trigger signal and the behavior of the controllers are essential to design a good metabolic switch. First, it is important to match the response range of the sensor with the concertation of the trigger molecule so that flux can be switched at the right time or growth stage [39]. Second, the promoter controlling the metabolic switch should have strong ‘on’ and ‘off’ modes (i.e. ultrasensitivity) [39]. This can be done by incorporating cooperative binding of the regulator at the promoter or using positive autoregulation [42••].
Population quality control
Another important aspect of microbial production is the relationship between growth and production, which is often negatively related. As a result, low producers, caused either by genetic mutations or molecular noise, grow faster than high producers and tend to dominate the culture over time, lowering overall titers and yields [49•,50–54]. This problem cannot be simply solved by the two types of controls discussed above but can be addressed by population quality control, which aims to alter the growth-production relationship (Figure 2f). Once high producers grow faster than low producers in a population, the overall titers and yields will be improved. Sensor-selector and growth-coupled production are two typical strategies to enable population quality control.
Sensor-selector
Sensor-selector leverages genetically encoded biosensors to sense intracellular metabolites and regulate genes that confer fitness advantages to cells enriched with the detected metabolites (Figure 2g). The selection can be achieved by controlling the expression of an antibiotic resistance gene, a toxin-antitoxin pair, or an auxotrophic marker [49•,50,55]. Sensor-selector has been traditionally used to select for the best genetic variant from large mutagenic libraries [56–58]. It has also been used to eliminate undesirable genetic mutants formed during long-term production, as shown for improved mevalonic acid production [49•]. Recent advances have exploited it to continuously select for high-producing variants from genetically identical populations. One of the first examples was demonstrated in fatty-acid producing E. coli, where enhanced fatty acid yield from the overall cell culture was achieved via enriching for non-genetic, high-producing cells using a fatty acid sensor-regulator [50]. The effect of selection on non-genetic variation suggests that some traits in the high producers could be epigenetically inherited to their offspring. With sensor-selector, low producers have a slow growth rate and are diminished during the selection, regardless of whether they are genetically different from high producers or not.
Sensor-selector has several limitations, some of which may be overcome by other methods. First, the method can be limited by the availability of a product-specific biosensor [59,60]. Instead of developing a new sensor, which can be challenging by itself, an alternative approach is to use a metabolite-dependent enzyme for auxotrophic selection. For example, a coenzyme B12-dependent methionine synthase, which links the end product B12 to cell growth, has been used to increase production [61]. Second, the cross-membrane transport of the product cannot be faster than the response time of sensing-selection. For example, if product transport is mediated by a membrane protein, strong expression of the transporter may allow low-producing cells to obtain product metabolites from high-producing cells. In this case, reducing the expression level of the transporter can be an effective strategy to eliminate crosstalk between single cells. Third, the sensor-selector strategy works effectively when sensing the end product. If the sensor-selector can only sense an intermediate metabolite of the target product, this strategy may select cells that accumulate the intermediate rather than those consuming the intermediate. This issue could be potentially solved by exploiting a co-culture system, with a second microbial strain converting the accumulated intermediate to a final product, as demonstrated in the production of phenol [62••] and tryptamine [63]. When using sensor-selectors in a co-culture system, additional attention needs to be paid to both the diffusion of the intermediate and competition between multiple populations [64].
Growth-coupled production
The design principle of growth-coupled production is to turn the product, an intermediate, or a by-product from the target pathway into a growth-essential molecule (Figure 2h). A common strategy to build growth-coupled production is to eliminate or over-accumulate an essential metabolite followed by rescuing this metabolite through the target pathway [65–68]. In the case of engineering E. coli for 1,4-butanediol production, model guided gene knockouts were performed to deliberately accumulate intracellular NADH to toxic levels, which reduced cell growth. The engineered 1,4-butanediol production pathway was then used to consume NADH and balance redox potential, thus providing a mechanism to positively relate production to cell growth [69]. In another example, an E. coli strain was engineered with pyruvate generated solely by biosynthesis of anthranilate [70•]. Introducing different anthranilate-derived biosynthetic pathways in this strain showed enhanced production of MA and tryptophan, with nearly no accumulation of anthranilate.
Cautions in the population quality control
When designing population quality control, stringent selection pressure can only be turned on after high producers have accumulated enough signal molecules to differentiate themselves from low producers. Furthermore, the controller should pose minimal additional burden on growth rate to prevent the sacrifice of productivity. The production threshold for high producers and the fraction of high producers in the population are important parameters determining the effect of the controller. In sensor-selector, these parameters can be tuned by engineering biosensors or applying external control to the selector [49•,50,71••]. In growth-coupled production, an iterative knockout approach may reach the same goal [69,72]. Measuring a distribution of production in single cells is beneficial in determining optimal parameters for the controller.
Conclusions
Considerable progress on various control strategies has been made to improve microbial production in recent years. Different control strategies were developed to solve different problems in bioproduction. Feedback control is a versatile tool to provide robust production of burdensome proteins and metabolites, avoid the accumulation of toxic intermediates, and shorten the rise-time of metabolites. Metabolic switch can replace the traditional inducible system used in two-stage batch fermentation to avoid resource competition between cell growth and engineered metabolic pathways. Population quality control can effectively prevent low-producing cells from dominating cell cultures, thus improving overall yields and titers, especially during large-scale production. Understanding the strengths and weaknesses of each control strategy can help obtain the most effective control for optimal performance enhancements. Different types of control strategies can also be potentially combined in one engineered strain to fulfill distinct functions for bioproduction. As demonstrated in a recent study on naringenin production, a metabolic feedback control was used to prevent overflow from malonyl-CoA to lipid biosynthesis, while a sensor-selector was used to couple naringenin production with cell growth [71••]. Combination of these two control strategies has increased both naringenin titer and strain stability. With the development of more sophisticated control technologies, the era of intelligent manufacturing in biology is coming.
Acknowledgements
This work is supported by the National Institute of General Medical Sciences of the National Institutes of Health [grant number R35GM133797]. YH is supported by the National Human Genome Research Institute training grant [grant number T32 HG000045].
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Peralta-Yahya PP, Zhang F, Del Cardayre SB, Keasling JD: Microbial engineering for the production of advanced biofuels. Nature 2012, 488:320–328. [DOI] [PubMed] [Google Scholar]
- 2.Bai W, Geng W, Wang S, Zhang F: Biosynthesis, regulation, and engineering of microbially produced branched biofuels. Biotechnol Biofuels 2019, 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdel-Mawgoud AM, Markham KA, Palmer CM, Liu N, Stephanopoulos G, Alper HS: Metabolic engineering in the host Yarrowia lipolytica. Metab Eng 2018, 50:192–208. [DOI] [PubMed] [Google Scholar]
- 4.Jiang W, Gu P, Zhang F: Steps towards ‘drop-in’ biofuels: focusing on metabolic pathways. Curr Opin Biotechnol 2018, 53:26–32. [DOI] [PubMed] [Google Scholar]
- 5.Galanie S, Thodey K, Trenchard IJ, Interrante MF, Smolke CD: Complete biosynthesis of opioids in yeast. Science 2015, 349:1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo X, Reiter MA, d‘Espaux L, Wong J, Denby CM, Lechner A, Zhang Y, Grzybowski AT, Harth S, Lin W et al. : Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 2019, 567:123–126. [DOI] [PubMed] [Google Scholar]
- 7.Bowen CH, Dai B, Sargent CJ, Bai W, Ladiwala P, Feng H, Huang W, Kaplan DL, Galazka JM, Zhang F: Recombinant spidroins fully replicate primary mechanical properties of natural spider silk. Biomacromolecules 2018, 19:3853–3860. [DOI] [PubMed] [Google Scholar]
- 8.Bowen CH, Reed TJ, Sargent CJ, Mpamo B, Galazka JM, Zhang F: Seeded chain-growth polymerization of proteins in living bacterial cells. ACS Synth Biol 2019, 8:2651–2658. [DOI] [PubMed] [Google Scholar]
- 9.Ekas H, Deaner M, Alper HS: Recent advancements in fungal-derived fuel and chemical production and commercialization. Curr Opin Biotechnol 2019, 57:1–9. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen PQ, Courchesne NMD, Duraj-Thatte A, Praveschotinunt P, Joshi NS: Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv Mater 2018, 30:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T: Interdependence of cell growth and gene expression: origins and consequences. Science 2010, 330:1099–1102. [DOI] [PubMed] [Google Scholar]
- 12.Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, Hwa T: Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 2015, 528:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceroni F, Algar R, Stan GB, Ellis T: Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods 2015, 12:415–418. [DOI] [PubMed] [Google Scholar]
- 14.Han Y, Zhang F: Heterogeneity coordinates bacterial multi-gene expression in single cells. PLoS Comput Biol 2020, 16:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee ME, Aswani A, Han AS, Tomlin CJ, Dueber JE: Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res 2013, 41:10668–10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biggs BW, Lim CG, Sagliani K, Shankar S, Stephanopoulos G, De Mey M, Ajikumar PK: Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli. Proc Natl Acad Sci U S A 2016, 113:3209–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu G, Yan Q, Jones JA, Tang YJ, Fong SS, Koffas MAG: Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol 2016, 34:652–664. [DOI] [PubMed] [Google Scholar]
- 18.Wu SG, He L, Wang Q, Tang YJ: An ancient Chinese wisdom for metabolic engineering: Yin-Yang. Microb Cell Fact 2015,14:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu P: Production of chemicals using dynamic control of metabolic fluxes. Curr Opin Biotechnol 2018, 53:12–19. [DOI] [PubMed] [Google Scholar]
- 20.Venayak N, Anesiadis N, Cluett WR, Mahadevan R: Engineering metabolism through dynamic control. Curr Opin Biotechnol 2015, 34:142–152. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Mannan AA, Han Y, Oyarzún DA, Zhang F: Dynamic metabolic control: towards precision engineering of metabolism. J Ind Microbiol Biotechnol 2018, 45:535–543. [DOI] [PubMed] [Google Scholar]
- 22.Tan SZ, Prather KL: Dynamic pathway regulation: recent advances and methods of construction. Curr Opin Chem Biol 2017, 41:28–35. [DOI] [PubMed] [Google Scholar]
- 23.Lv Y, Qian S, Du G, Chen J, Zhou J, Xu P: Coupling feedback genetic circuits with growth phenotype for dynamic population control and intelligent bioproduction. Metab Eng 2019, 54:109–116. [DOI] [PubMed] [Google Scholar]
- 24.Gyorgy A, Jiménez JI, Yazbek J, Huang HH, Chung H, Weiss R, Del Vecchio D: Isocost lines describe the cellular economy of genetic circuits. Biophys J 2015, 109:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.•.Aoki SK, Lillacci G, Gupta A, Baumschlager A, Schweingruber D, Khammash M: A universal biomolecular integral feedback controller for robust perfect adaptation. Nature 2019, 570:533–537 [DOI] [PubMed] [Google Scholar]; This study proved that an integral feedback can be implemented by embedding an antithetic feedback-control topology. Based on this theory, the authors engineered a synthetic controller and demonstrated its tunability and adaptative properties.
- 26.Dublanche Y, Michalodimitrakis K, Kümmerer N, Foglierini M, Serrano L: Noise in transcription negative feedback loops: Simulation and experimental analysis. Mol Syst Biol 2006, 2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briat C, Gupta A, Khammash M: Antithetic integral feedback ensures robust perfect adaptation in noisy bimolecular networks. Cell Syst 2016, 2:15–26. [DOI] [PubMed] [Google Scholar]
- 28.••.Darlington APS, Kim J, Jiménez JI, Bates DG: Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes. Nat Commun 2018, 9:695. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study utilized orthogonal ribosomes with negative feedback control to alleviate competition effects between genes translated by the orthogonal ribosomes. The authors also showed how to use the controller to optimize a multi-step metabolic pathway.
- 29.Milias-Argeitis A, Rullan M, Aoki SK, Buchmann P, Khammash M: Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nat Commun 2016, 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.•.Ceroni F, Boo A, Furini S, Gorochowski TE, Borkowski O, Ladak YN, Awan AR, Gilbert C, Stan GB, Ellis T: Burden-driven feedback control of gene expression. Nat Methods 2018, 15:387–393 [DOI] [PubMed] [Google Scholar]; The authors identified promoters that can be induced by the expression of burdensome genes from RNA-seq and employed one of these promoters to drive the expression of sgRNA, which together with dCas9 represses heterologous gene expression.
- 31.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M: Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci U S A 2014, 111:11299–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D, Xiao Y, Evans BS, Zhang F: Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator. ACS Synth Biol 2015, 4:132–140. [DOI] [PubMed] [Google Scholar]
- 33.••.Liu D, Zhang F: Metabolic feedback circuits provide rapid control of metabolite dynamics. ACS Synth Biol 2018, 7:347–356 [DOI] [PubMed] [Google Scholar]; The authors studied metabolic feedback circuits with three regulatory architectures. All metabolic feedback circuits accelerate metabolite dynamics. Different architectures provide distinct properties in dynamics, such as different degrees of acceleration and metabolic overshoot.
- 34.Hartline CJ, Mannan AA, Liu D, Zhang F, Oyarzún DA: Metabolite sequestration enables rapid recovery from fatty acid depletion in Escherichia coli. mBio 2020, 11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mannan AA, Liu D, Zhang F, Oyarzún DA: Fundamental design principles for transcription-factor-based metabolite biosensors. ACS Synth Biol 2017, 6:1851–1859. [DOI] [PubMed] [Google Scholar]
- 36.Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD et al. : Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol 2013, 31:1039–1046. [DOI] [PubMed] [Google Scholar]
- 37.Olsman N, Xiao F, Doyle JC: Architectural principles for characterizing the performance of antithetic integral feedback networks. iScience 2019, 14:277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soma Y, Hanai T: Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng 2015, 30:7–15. [DOI] [PubMed] [Google Scholar]
- 39.Gupta A, Reizman IMB, Reisch CR, Prather KLJ: Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 2017, 35:273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doong SJ, Gupta A, Prather KLJ: Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc Natl Acad Sci U S A 2018, 115:2964–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams TC, Averesch NJH, Winter G, Plan MR, Vickers CE, Nielsen LK, Krömer JO: Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab Eng 2015, 29:124–134. [DOI] [PubMed] [Google Scholar]
- 42.••.Yang Y, Lin Y, Wang J, Wu Y, Zhang R, Cheng M, Shen X, Wang J, Chen Z, Li C et al. : Sensor-regulator and RNAi based bifunctional dynamic control network for engineered microbial synthesis. Nat Commun 2018, 9:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors engineered a muconic acid sensor with antisense RNA to control the carbon flux from phosphoenolpyruvate. As muconic acid accumulated over time, cells gradually increased the flux toward muconic acid production and decreased the flux into the TCA cycle.
- 43.Scalcinati G, Knuf C, Partow S, Chen Y, Maury J, Schalk M, Daviet L, Nielsen J, Siewers V: Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene α-santalene in a fed-batch mode. Metab Eng 2012, 14:91–103. [DOI] [PubMed] [Google Scholar]
- 44.Xie W, Ye L, Lv X, Xu H, Yu H: Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab Eng 2015, 28:8–18. [DOI] [PubMed] [Google Scholar]
- 45.Harder BJ, Bettenbrock K, Klamt S: Temperature-dependent dynamic control of the TCA cycle increases volumetric productivity of itaconic acid production by Escherichia coli. Biotechnol Bioeng 2018, 115:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao EM, Zhang Y, Mehl J, Park H, Lalwani MA, Toettcher JE, Avalos JL: Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 2018, 555:683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C, Li YY, Ma J, Liu Y, He J, Li YY, Zhu F, Meng J, Zhan J, Li Z et al. : High production of 4-hydroxyisoleucine in Corynebacterium glutamicum by multistep metabolic engineering. Metab Eng 2018, 49:287–298. [DOI] [PubMed] [Google Scholar]
- 48.••.Gao C, Hou J, Xu P, Guo L, Chen X, Hu G, Ye C, Edwards H, Chen J, Chen W et al. : Programmable biomolecular switches for rewiring flux in Escherichia coli. Nat Commun 2019,10:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors developed three types of protease-based circuits, a dynamic regulatory circuit, an inverter, and an oscillator. These circuits have rapid kinetic response and were used for dynamic control in metabolic engineering.
- 49.•.Rugbjerg P, Sarup-Lytzen K, Nagy M, Sommer MOA: Synthetic addiction extends the productive life time of engineered Escherichia coli populations. Proc Natl Acad Sci U S A 2018, 115:2347–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]; This work linked mevalonic acid production to the expression of two growth regulators, glmM and folP. So cells with low mevalonic acid production will be eliminated due to their low growth rate.
- 50.Xiao Y, Bowen CH, Liu D, Zhang F: Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat Chem Biol 2016, 12:339–344. [DOI] [PubMed] [Google Scholar]
- 51.Rugbjerg P, Sommer MOA: Overcoming genetic heterogeneity in industrial fermentations. Nat Biotechnol 2019, 37:869–876. [DOI] [PubMed] [Google Scholar]
- 52.Schmitz AC, Hartline CJ, Zhang F: Engineering microbial metabolite dynamics and heterogeneity. Biotechnol J 2017, 12:1700422. [DOI] [PubMed] [Google Scholar]
- 53.Wang T, Dunlop MJ: Controlling and exploiting cell-to-cell variation in metabolic engineering. Curr Opin Biotechnol 2019, 57:10–16. [DOI] [PubMed] [Google Scholar]
- 54.Evans TD, Zhang F: Bacterial metabolic heterogeneity: origins and applications in engineering and infectious disease. Curr Opin Biotechnol 2020, 64:183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Cabales A, Li Z, Zhang H: Biosensor-assisted high performing cell selection using an E. coli toxin/antitoxin system. Biochem Eng J 2019, 144:110–118. [Google Scholar]
- 56.Raman S, Rogers JK, Taylor ND, Church GM: Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci U S A 2014, 111:17803–17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Snoek T, Romero-Suarez D, Zhang J, Ambri F, Skjoedt ML, Sudarsan S, Jensen MK, Keasling JD: An orthogonal and pH-tunable sensor-selector for muconic acid biosynthesis in yeast. ACS Synth Biol 2018, 7:995–1003. [DOI] [PubMed] [Google Scholar]
- 58.Flachbart LK, Sokolowsky S, Marienhagen J: Displaced by deceivers: prevention of biosensor cross-talk is pivotal for successful biosensor-based high-throughput screening campaigns. ACS Synth Biol 2019, 8:1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu D, Evans T, Zhang F: Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng 2015, 31:35–43. [DOI] [PubMed] [Google Scholar]
- 60.Zhang F, Keasling J: Biosensors and their applications in microbial metabolic engineering. Trends Microbiol 2011, 19:323–329. [DOI] [PubMed] [Google Scholar]
- 61.Noh MH, Lim HG, Moon D, Park S, Jung GY: Auxotrophic selection strategy for improved production of coenzyme B12 in Escherichia coli. iScience 2020, 23:100890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.••.Guo X, Li Z, Wang X, Wang J, Chala J, Lu Y, Zhang H: De novo phenol bioproduction from glucose using biosensor-assisted microbial coculture engineering. Biotechnol Bioeng 2019, 116:3349–3359 [DOI] [PubMed] [Google Scholar]; This study combined co-culture systems with sensor-selector for phenol production. An intermediate was first enriched by sensor-selector in upstream strains and then converted to phenol in downstream strains.
- 63.Wang X, Policarpio L, Prajapati D, Li Z, Zhang H: Developing E. coli-E. coli co-cultures to overcome barriers of heterologous tryptamine biosynthesis. Metab Eng Commun 2020,10:e00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsoi R, Wu F, Zhang C, Bewick S, Karig D, You L: Metabolic division of labor in microbial systems. Proc Natl Acad Sci U S A 2018, 115:2526–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burgard AP, Pharkya P, Maranas CD: OptKnock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 2003, 84:647–657. [DOI] [PubMed] [Google Scholar]
- 66.Von Kamp A, Klamt S: Growth-coupled overproduction is feasible for almost all metabolites in five major production organisms. Nat Commun 2017, 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feist AM, Zielinski DC, Orth JD, Schellenberger J, Herrgard MJ, Palsson BO: Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab Eng 2010, 12:173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klamt S, Mahadevan R: On the feasibility of growth-coupled product synthesis in microbial strains. Metab Eng 2015,30:166–178. [DOI] [PubMed] [Google Scholar]
- 69.Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R et al. : Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol 2011, 7:445–452. [DOI] [PubMed] [Google Scholar]
- 70.•.Wang J, Zhang R, Zhang Y, Yang Y, Lin Y, Yan Y: Developing a pyruvate-driven metabolic scenario for growth-coupled microbial production. Metab Eng 2019, 55:191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors engineered an E. coli strain with pyruvate generated solely by biosynthesis of anthranilate. This strain has a high flux towards anthranilate, which can be used to produce anthranilate-derived products.
- 71.••.Lv Y, Gu Y, Xu J, Zhou J, Xu P: Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield. Metab Eng 2020, 61:79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors integrate both a negative feedback loop and a product-based sensor-actuator to increase naringenin production and strain stability. The negative feedback loop prevented metabolic overflow from malonyl-CoA to lipid biosynthesis, and the naringenin-based sensor-actuator coupled cell growth to naringenin production.
- 72.Harder BJ, Bettenbrock K, Klamt S: Model-based metabolic engineering enables high yield itaconic acid production by Escherichia coli. Metab Eng 2016, 38:29–37. [DOI] [PubMed] [Google Scholar]


