Abstract
MicroRNAs are involved in various pathologies including cancer. The aim of the study was to assess the level of expression of miR-96-5p, -134-5p, -181b-5p, -200b-3p in FFPE samples of prostate cancer, adjacent cancer-free tissue, and benign prostatic hyperplasia. Samples of 23 FFPE prostate cancer and 22 benign prostatic hyperplasias were dissected and HE stained. Compartments of tumor tissue and adjacent healthy glandular tissue were isolated from each sample using Laser Capture Microdissection. Total RNA was isolated from dissected tissues. Expression of miR-96-5p, miR-134-5p, 181b-5p, and miR-200b-3p was determined by real-time RT-qPCR method. The expression of miR-200b-3p was significantly higher in cancerous prostate: both in adenocarcinomatous glands and in the adjacent, apparently unaffected glands compared to BPH samples. The expression of miR-181b-5p was lower in in both prostate cancer tissues and adjacent tissue compared to BPH samples. Expression of miR-96-5p and miR-134-5p was lower in prostate cancer tissues compared to BPH. Levels of miR-96-5p, miR-134-5p, and 181b-5p negatively correlated with the Gleason score. Given further studies, miR-96-5p, miR-134-5p and especially miR-200b-3p and miR-181b-5p may differentiate BPH and PC.
Electronic supplementary material
The online version of this article (10.1007/s00418-020-01941-2) contains supplementary material, which is available to authorized users.
Keywords: Prostate cancer, microRNA, Benign prostate hyperplasia, Laser capture microdissection
Introduction
Prostate cancer (PC) is classified as an adenocarcinoma in over 95% cases and preferably locates in the peripheral region of the prostate gland (Oh 2003). PC is the most common cancer in males and accounts for 20% of new cancer diagnoses. Next to lung cancer PC is responsible for the largest number of deaths. It is characterized by a relative high 5-year survival (98%), mainly due to frequent over diagnosis (Siegel et al. 2019). PC over diagnosis is one of the major problems of clinical medicine, that leads to the unnecessary therapy of indolent cancers (Lomas and Ahmed 2020; Costello 2020).
In 1966, Donald Gleason proposed histopathological grading scale for prostatic adenocarcinoma (Gleason 1966). It assesses dominant morphology and the second most common pattern (Oh 2003). It is broadly used as it correlates with prognosis and staging and as well as guides further therapy.
PC usually exhibits indolent growth, however, high-risk or metastatic PC is characterised by 50% recurrence rate. This group of patients requires intensive PSA (prostate-specific antigen) monitoring and/or adjuvant treatment with androgen deprivation therapy (ADT) (Sequeiros et al. 2013). PSA is constitutively produced by prostate gland cells and is used in PC screening tests and as a PC recurrence monitoring marker (Kanwal et al. 2017). Although PSA screening increased detection of PC, results may be biased by non-malignant pathologies, including prostatitis, benign prostatic hyperplasia (BPH) or preanalytical errors, all of which lead to false-positive results. Due to the limited specificity of PSA, novel biomarkers are in demand (Nogueira et al. 2009).
miRNAs are small non-coding molecules which consist of about 18–22 nucleotides. They regulate gene expression by suppressing mRNA translation or affecting mRNA stability in a sequence-specific manner (Aghdam et al. 2018; Kaminska et al. 2018). Therefore, miRNA regulate many aspects of cell biology. In cancer, miRNAs may either suppress tumor growth (tumor suppressor miRs) or promote oncogenesis and tumor-progression (oncomiRs) (Grzywa et al. 2019).
Many authors show an important impact of miRNAs on the pathogenesis of prostate cancer, as well as their role as a diagnostic marker (Kanwal et al. 2017; Sequeiros et al. 2013; Walter et al. 2013b). Recent studies showed miRNAs may serve as diagnostic and prognostic biomarkers in different cancers (Rapado-Gonzalez et al. 2019; Delangle et al. 2019; Butz and Patocs 2019; Bhat et al. 2019). Importantly, miRNAs can be detected in formalin-fixed tissues (FFPE), therefore, they may be potentially an extension of conventional histopathological diagnosis (Klopfleisch et al. 2011; Grzywa et al. 2020). Numerous studies evaluating miRNA expression in PC led to inconclusive results possibly due to the highly heterogeneous structure of the tumor (Yadav et al. 2018; Grzywa et al. 2017). Laser capture microdissection (LCM) overcomes this limitation since it enables to evaluate miRNA expression only within precisely dissected fragments of a sample. We chose four miRNAs that exhibited explicit down- or upregulation in PC in other studies, hsa-miR-96-5p, hsa-miR-134-5p, hsa-miR-181b-5p, hsa-miR -200b-3p (Sequeiros et al. 2013; Janiak et al. 2017; Walter et al. 2013b).
The study aimed to determine miRNA expression in prostates in which cancer has been diagnosed: both in cancerous and morphologically normal, adjacent tissue, as well as in benign prostatic hyperplasia cases from the archival formalin-fixed paraffin-embedded (FFPE) samples.
Materials and methods
Archival samples and preparation for LCM
Samples of 23 PC and 22 BPH have been obtained from the Department of Pathology, Medical University of Warsaw. PC patients included previously untreated primary prostate cancer. Each patient with PC underwent a radical prostatectomy in 2014–2020 in the Department of Urology, Medical University of Warsaw. Clinical patients’ data are presented in Table 1. Resected tumors were formalin-fixed and paraffin-embedded according to the standard protocol in the tissue processor. Thereafter the samples were cut on microtome and HE-stained for the pathologist examination (Gleason score assessment). Only fragments with confirmed presence of both neoplastic primary prostate cancer and unaffected prostate gland architecture were included in the study.
Table 1.
Clinical data of prostate cancer-bearing patients included in study
| Case number | Gleason score | Dominant tissue architecture in dissected neoplastic area | Age (years) | Lymphadenectomy | TNM |
|---|---|---|---|---|---|
| 101 | 4 + 3 | 4 | 60 | 0 | T2b |
| 102 | 4 + 3 | 4 | 65 | 0 | T1c |
| 103 | 4 + 3 | 4 | 60 | 0 | T2b |
| 104 | 4 + 3 | 4 | 65 | 0 | T2c |
| 105 | 4 + 3 | 3 | 63 | 0 | T2c |
| 106 | 4 + 3 | 3 | 76 | 1 | T3a |
| 107 | 4 + 3 | 3 | 73 | 0 | T3b |
| 108 | 4 + 3 | 4 | 73 | 1 | T3a |
| 110 | 3 + 4 | 3 | 61 | 0 | T1c |
| 112 | 3 + 4 | 3 | 59 | 0 | T2c |
| 113 | 5 + 5 | 5 | 69 | 1 | T3b |
| 114 | 3 + 4 | 3 | 66 | 1 | T2c |
| 115 | 3 + 4 | 3 | 63 | 1 | T1c |
| 116 | 4 + 3 | 3 | 68 | 0 | T1c |
| 117 | 3 + 4 | 4 | 50 | 0 | T1c |
| 118 | 3 + 3 | 3 | 68 | 0 | T2c |
| 119 | 3 + 3 | 3 | 57 | 0 | T1c |
| 120 | 4 + 3 | 4 | 67 | 0 | T2c |
| 121 | 3 + 4 | 3 | 71 | 1 | T3a |
| 122 | 3 + 2 | 2 | 74 | 1 | T2a |
| 123 | 4 + 4 | 4 | 61 | 0 | T2a |
| 124 | 4 + 5 | 4 | 70 | 0 | T2c |
| 126 | 4 + 3 | 4 | 59 | 0 | T2c |
| N | 23 | 23 | 7 | 23 | |
| Mean (± SD) | – | 65.31 ± 6.4 | – | – | |
| Median | – | 65.5 | – | T2c |
All samples were cut with a microtome to 10 µm slices (Leica, RM2055 model) and were mounted on glass slides (SuperFrost Ultra Plus, Menzel Gläser) with a drop of DNAse/RNAse-free water. Preceding optimization experiments indicated more efficient dissection on SuperFrost® glass slides comparing to dedicated membrane glass slides. Non-membrane slides provided better slices’ adherence and a possibility to dissect sufficient tissue area from surrounding compartments. Then, samples were incubated in a fume hood at 56 °C overnight to increase slices’ adherence. Mounted slices were HE stained according to the standard protocol in a set of stains, alcohol solutions, and xylene. Slides were immediately subjected to LCM.
Laser capture microdissection
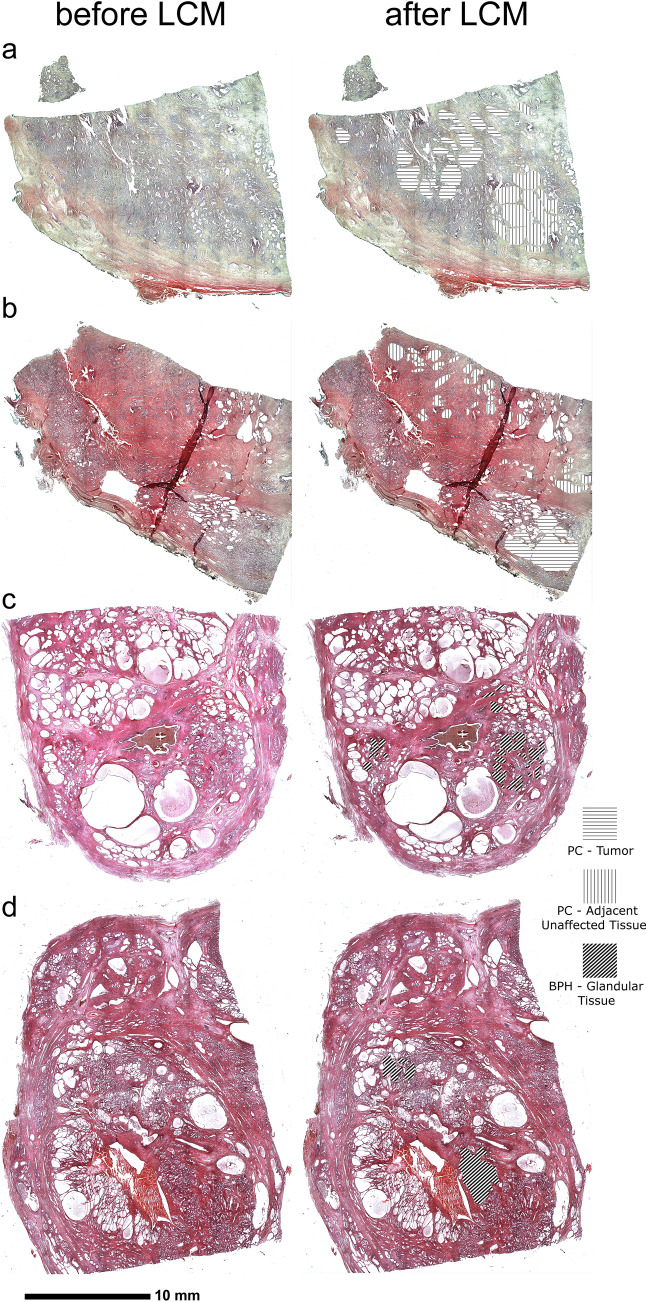
Stained and dehydrated sections of PC were subjected to LCM-aided dissection of two regions—engaged by neoplastic process and adjacent tissue that contained only glands of normal morphology, which was confirmed by IHC staining. These regions were selected, in each section, by a board-certified pathologist (Fig. 1a–d). In the case of BPH, only glandular tissue was highlighted (Fig. 1e–h). Subsequently, 10 mm2 of each region were marked to dissect with LCM system (Nonn et al. 2010; Hoefig and Heissmeyer 2010) (PALM Robo, Zeiss, Germany). Optimization assays indicated that Laser Pressure Capture mode (Auto-LPC) alone with non-membrane slides is sufficient for the dissection of tissues for further analysis. LCM was performed under following conditions: LCP energy—80–90, LCP spot distance—25 μm, magnification—5 ×, tissue collected in 20 μl of Digestion Buffer (RecoverAll, Ambion, Thermofisher) in 500 μl sterile PCR-tube cap. Each LCM was preceded by optimization of LCP energy and spot distance to provide a full dissection of marked areas. Caps were sealed back with tubes, centrifuged briefly and placed on wet ice until further steps.
Fig. 1.
Whole slide images of areas that underwent laser capture microdissection (LCM). Left side of the image represents sections before LCM, right side after LCM. a, b Samples of prostate cancer (PC). c, d Samples of benign prostate hyperplasia (BPH). Horizontal lines—tumor area of a PC sample; vertical lines—area of adjacent tissue of PC; diagonal lines—glandular tissue of a BPH sample
RNA isolation
Since FFPE treated nucleic acid are degraded and presence of protein crosslinks (Evers et al. 2011) hinder proper extraction, RecoverAll for FFPE kit (Ambion, Thermofisher, USA) was chosen for further analyses on the basis of prior optimization (Suppl. Figure 1). Total RNA extraction was conducted according to the manufacturer guidelines (100 µl Digestion Buffer volume and skipped deparaffinization). RNA was eluted with 60 µl ultrapure, molecular-grade water and stored in − 80 °C until further steps (Paskal et al. 2018).
RT-qPCR
Extracted RNA was quantified with NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) with an assessment of A260/A280 (min > 1.7). 100 ng of RNA was used for reverse transcription assay (TaqMan® MicroRNA Reverse Transcription Kit, Thermofisher) with primers for snU6, RNU43, miR-96-5p, miR-134-5p, miR-181b-5p and miR-200b-3p (TaqMan®, Thermofisher). cDNA from previous reactions was used for quantitative Real-Time PCR (qPCR). qPCR was set up as it follows: 2 µl cDNA, 7,5 µl qPCR master mix SensiFAST Probe Lo-Rox (BIO-84020, Bioline, UK), 0,5 µl microRNA-specific TaqMan assays snU6, RNU43, hsa-miR-96-5p, hsa-miR-134-5p, hsa-miR-181b-5p, hsa-miR-200b-3p (Assay no.: 001973, 001,095, 000,186, 001,186, 001,098, 002,251) (TaqMan®, Thermofisher, USA); molecular-grade water to final volume: 15 µl. U6 and RNU43 expression was used for miRNAs normalization between samples. Reactions were performed in triplicates. qPCR reaction was performed on Applied Biosystems® 7500 Real-Time PCR System (Thermofisher, USA) with the following setup: 1 × 95 °C 5 min, 45 × cycles 95 °C 10 s and 60 °C 50 s.
Data processing and analysis
Data were collected and processed with Excel 2016 (Microsoft, USA). Statistical analyses were conducted with GraphPad Prism 8.4.3 (GraphPad Software Inc.) using the following tests: Mann-U-Whitney, Chi-square, R-Spearman correlation coefficient, Wilcoxon signed-rank test. A p value of < 0.05 was considered statistically significant. The heatmap was generated using Graphpad using the − ΔCt values and Z-score. ROC curves analysis was calculated in GraphPad Prism.
Results
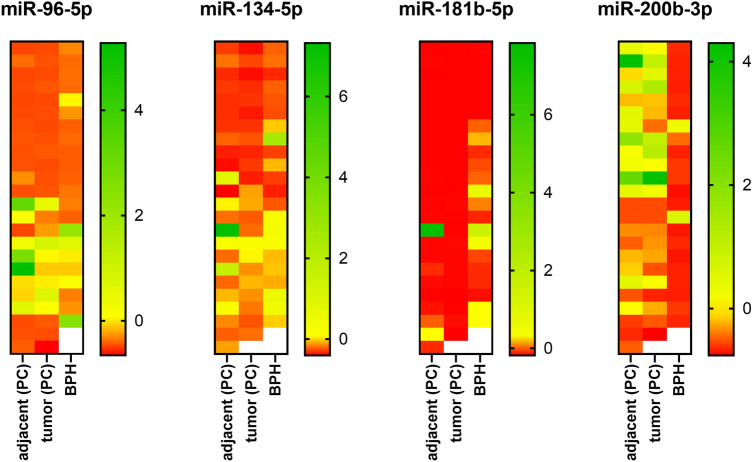
The expression of miR-96-5p, miR-134-5p, 181b-5p, and miR-200b-3p was analysed in 23 PC and 22 BPH samples. From each FFPE sample of PC, tumor tissue and adjacent morphologically healthy tissue were dissected using Laser Capture Microdissection (Fig. 1). In BPH, only glandular tissue was dissected from each FFPE sample. We found that the microRNA expression profile of BPH differs from PC (Fig. 2).
Fig. 2.
Relative expression of miR-96-5p, miR-134-5p, miR-181b-5p, miR-200b-3p in tumor tissue of prostate cancer (PC), adjacent unaffected tissue and in patients with benign prostatic hyperplasia (BPH). Relative expression is depicted as Z-score of relative expression (log10 of 2−ΔCt). Red represents the lowest expression, green represents the highest expression
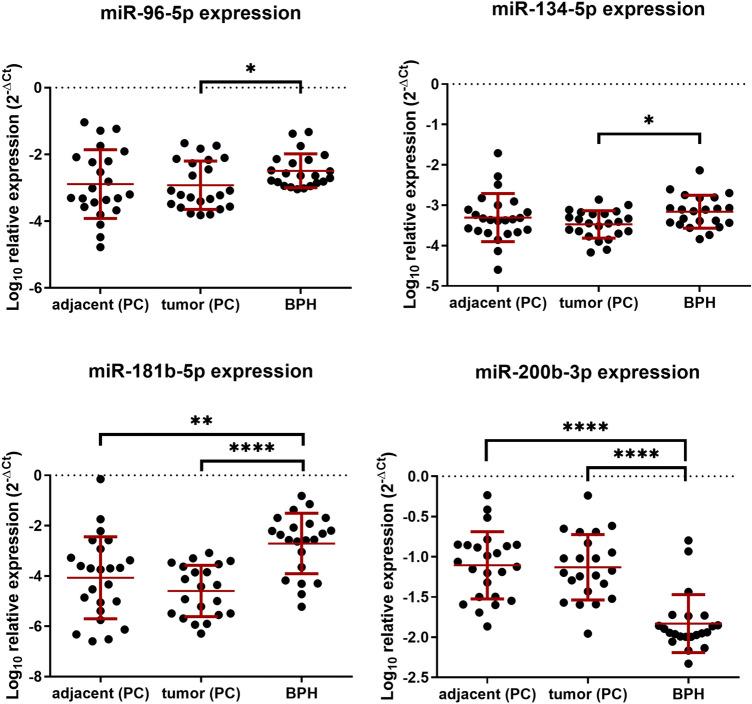
In PC the expression of miR-96-5p and miR-134-5p was downregulated compared to BPH (fold change 3.78, p = 0.0257; fold change 2.09, p = 0.0111, Fig. 3). miR-181b-5p was downregulated 93 times in PC and 19 times in adjacent tissue samples compared to BPH samples (p < 0.0001 and p = 0.0014, respectively). Moreover, there was a reverse correlation between miR-96-5p, miR-134-5p, 181b-5p expression and Gleason score (Table 2). No other parameter (age, lymphadenectomy, TNM) significantly correlated with any of examined microRNAs’ expression (p < 0.05, data not shown). Conversely, miR-200b-3p was upregulated nearly six times in PC samples compared to BPH and seven times compared to adjacent tissue (p < 0.0001 and p < 0.0001, respectively).
Fig. 3.
Relative expression of miR-96-5p, miR-134-5p, miR-181b-5p, miR-200b-3p, depicted as log10 of 2−ΔCt. Each of prostate cancer (PC) sample has had dissected tumor cells (tumor) and adjacent, unaffected tissue (adjacent). Benign prostatic hyperplasia (BPH) samples have had glandular tissue alone dissected. Tumor and adjacent samples’ expression was tested with the Wilcoxon signed-rank test, the difference between tumor/adjacent and BPH was tested with Mann-U-Whitney test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Table 2.
Results of R-spearman correlation coefficient of Gleason score and miR-96-5p, miR-134-5p, miR-181b-5p, miR-200b-3p expression
| R-Spearman correlation coefficient of Gleason score and mirs expression | ||||
|---|---|---|---|---|
| miR-96-5p | miR-134-5p | miR-181b-5p | miR-200b-3p | |
| Gleason score | − 0.599* | − 0.517* | − 0.448* | 0.259 |
Gleason score was summed up to obtain interval data
*p < 0.05
Despite precise dissection of tumor tissue and histologically healthy tissue, we did not observe any differences in miR-96-5p, miR-134-5p, miR-181b-5p and miR-200b-3p expression between tumor and adjacent tissue (Fig. 3).
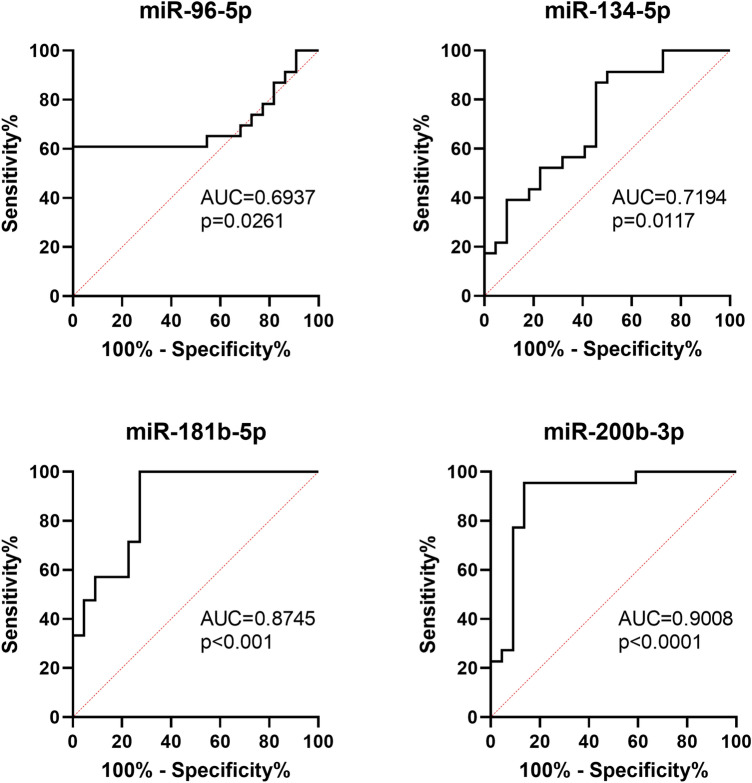
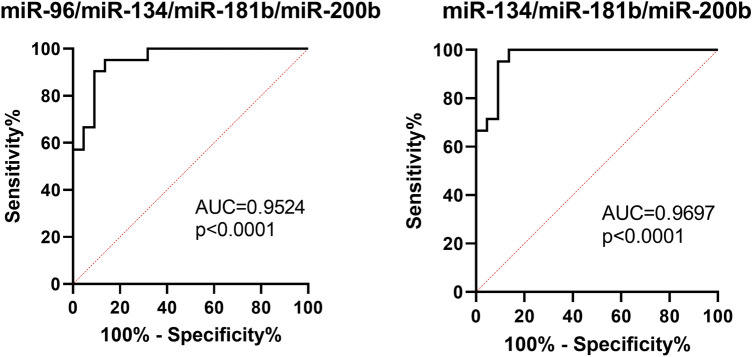
To analyse the possible utility of investigated miRNAs in differential diagnosis between PC and BPH, we performed ROC curves analysis (Fig. 4). From four analysed miRNAs, the expression of miR-200b-3p was the most specific and sensitive indicator of PC (AUC 0.9008 (95% CI 0.7987–1.000), p < 0.0001). Log10 relative expression of miR-200b-3p higher than − 1.658 favours diagnosis to PC than BPH with sensitivity 95.45% and specificity 86.38%. Analysis of combined expression of more than one miRNA revealed that all four miRNAs may be used to support differential diagnosis. For four-miRNA panel (4-miR), log10 relative expression was used according to the Eq. 4—miR = − [miR-96-5p + miR-134-5p + miR-181b-5p – (5 × miR-200b-3p)]. 4-miR was characterized by high AUC (0.9524, 95% CI 0.8946–1.00, p < 0.0001). For values lower than − 1.287, sensitivity was 90.48% and specificity 90.91% (Fig. 5). Panel of three miRNAs (3-miR) was calculated according to the Eq. 3—miR = − [miR-134-5p + miR-181b-5p – (5 × miR-200b-3p)] was characterized by AUC = 0.9697 (95% CI 0.9259–1.000, p < 0.0001) and sensitivity 95.24% and specificity 90.91% for values lower than 1.243 (Fig. 5).
Fig. 4.
ROC curves analysis for distinction of prostate cancer from benign prostate hyperplasia based on single microRNA expression
Fig. 5.
ROC curves analysis for distinction of prostate cancer from benign prostate hyperplasia based on combined microRNA expression including panel of four miRNAs (4-miR) and three miRNAs (3-miR)
Discussion
Routine core needle biopsies of prostate tumor consist of both cancer cells and adjacent non-cancerous prostate tissue. Such specimens are often used in RNA expression studies. Substantial contamination of RNA derived from non-cancerous cells may significantly alter the miRNA expression pattern. Thus, in this study, we dissected either PC tissue or adjacent, apparently healthy prostate gland. We compared the expression of given microRNA in PC tissue or adjacent healthy tissue in one prostate specimen. Our study was limited to four miRs of explicit expression in PC, basing on aforementioned studies.
We found that low expression of miR-200b-3p and the high expression of miR-181b-5p may favour the diagnosis towards BPH. Panel of combined expression of miR-134-5p, miR-181b-5p, and miR-200b is a promising tool to support differential diagnosis between PC and BPH. We found that expression of these four microRNAs remains comparable in cancer and non-affected tissue in PC samples. It suggests that changes of the microRNA expression profile are not limited to cancer cells, but also include adjacent, morphologically non-affected tissues. Multiple tumor-secreted microRNA were reported to modify tumor microenvironment (Pan et al. 2020), for instance to reprogram fibroblasts to become cancer-associated fibroblasts (Mitra et al. 2012). The diffusive feature of microRNA expression in tissues was firstly reported by Levine et al. (Levine et al. 2007). Further consideration and studies confirmed that microRNA may diffuse from tumor and act as tumor frontline invasion mediators (Vasilescu et al. 2020). All above explains the reported presence of similar microRNA expression pattern in both tumor and tissue within close distance to the tumor that was histologically unaffected. On the other hand, the design of our study did not include an additional external control of intrapatient microRNA expression pattern, e.g., different type of tissue or blood sample. Such analysis could reveal if microRNA expression pattern was patient-leaned and may have influenced the results. To minimize the effect of the origin of a samples, we employed proper statistical analysis for dependent (PC tumor vs adjacent) and for independent samples (PC vs BPH). Also, as it is depicted on heatmap, (Fig. 2) in case of miR-96-5p, miR-134-5p and miR-200b-3p, we observed that 1/3 to 1/2 of samples had highly similar expression in tumor vs adjacent tissue. Higher number of samples could reveal if the phenomenon is patient-dependent, disease-dependent or microRNA target-dependent. Conversely, miR-181b-5p was evenly expressed in PC-derived groups.
In recent years, several studies showed the clinical significance of miRNAs in PC pathogenesis. Most of the microRNAs act as tumor suppressors and are downregulated in PC cancer cells, e.g., Let-7 family, miR-221, miR-200a (Sequeiros et al. 2013). On the other side, some miRNAs are potently overexpressed in PC and promote tumor development (Sequeiros et al. 2013).
miR-96-5p promotes prostate cancer cells proliferation by targeting tumor suppressor gene FOXO1 (Yu et al. 2014b; Haflidadottir et al. 2013; Fendler et al. 2013). Xu et al. showed that miR-96-5p promoted colony formation, proliferation, and invasiveness of PC cells by targeting MTSS1 (Xu et al. 2016). Another study presented a mechanism in which epidermal growth factor receptor (EGFR) induced the expression of miR-96-5p. It targeted ETV6, the tumor suppressor, which leads to PC progression (Tsai et al. 2017). Moreover, miR-96-5p regulated autophagy under hypoxia in PC cells by targeting mTOR or ATG7 (Ma et al. 2014). It was shown that miR-96-5p modulates androgen signalling (Long et al. 2019) and takes part in prostate bone metastasis formation (Siu et al. 2015). However, its role in PC pathogenesis and interaction between PC cells and tumor microenvironment is complex and need to be further investigated. Several studies showed upregulation of miR-96-5p in prostate cancer tissues (Mihelich et al. 2011; Yu et al. 2014b; Haflidadottir et al. 2013; Navon et al. 2009; Larne et al. 2013). On the contrary, Kang et al. did not observe a correlation between the level of miR-96-5p expression and any clinicopathologic parameter (Kang et al. 2012) while in our cohort, levels of miR-96-5p negatively correlated with Gleason score. Walter et al. showed downregulation of miR-96-5p in high-grade PC tumors, what stays in line with our findings that, we found miR-96-5p showed lower expression in PC compared to BPH (Walter et al. 2013a).
So far, little is known about the role of miR-134 in PC biology. Presented data suggest the tumor suppressive role of miR-134-5p in human cancers. By targeting various genes, it influences oncogenic signalling pathways, e.g., MAPK/ERK pathway, Notch pathway, and EGFR. Upregulated-miR-134 inhibits the expression of cyclin D/cyclin D2/CDK4, KRAS, EGFR, POGLUT1, and STAT5B thus decreases cells’ proliferation. Since miR-134 targets and inactivates KRAS, Nanog mRNA, HNF4α, EGFR, ITGB1, and FOXM1, it also inhibits tumor invasion and metastasis (Pan et al. 2017). Ngalame et al. showed the negative correlation of miR-134 with RAS oncogenes. Downregulation of miR-134 led to the activation of RAS/ERK and PI3K/PTEN/AKT signalling pathways in human prostate epithelial and stem cells (Ngalame et al. 2014). Our study showed the downregulation of miR-134-5p in prostate cancer compared to BPH and negative correlation with Gleason score.
Tong et al. reported overexpression of miR-181 in prostate cancer. In this study, miR-181 promoted cells proliferation and tumor growth in mice via targeting DAX-1, a negative regulator of androgen receptor in PC (Tong et al. 2014). DAX-1 inhibits aromatase expression (Lanzino et al. 2013), but its role in PC cancer rather relies on modulation of carcinogenesis than sex-steroids mediated pathway (Nakamura et al. 2009). Analysis of RNA circularization in localized PC demonstrated that circCSNK-1 interacted with miR-181 and promoted cell growth (Chen et al. 2019).
In our study, expression of miR-181b-5p was significantly lower in PC and adjacent tissue compared with BPH. Moreover, the level of expression of miR-181b-5p correlated with Gleason score.
Several studies show the tumor-suppressive role of miR-200b in PC by targeting different genes, e.g., ZEB1, ZEB2 (Kong et al. 2009; Williams et al. 2013) and Bmi-1 (Yu et al. 2014a). ZEB1/2 proteins are hallmarks of epithelial-mesenchymal transition (EMT) and cofactors chemoresistance in PC (Orellana-Serradell et al. 2019) while BMI1 promotes cell proliferation, EMT and is critical for the development of castration-resistance in PC (Zhu et al. 2020). Katz et al. showed a link between low expression of miR-200b, the Gleason score as well as shorter survival (Katz et al. 2014) and pointed at its possible role as a potential prognostic marker. On the other hand, overexpression of circulating miR-200b in plasma was associated with bone metastasis, high PSA and bilateral tumor (Souza et al. 2017). MiR-200b was downregulated in PC tissue compared with healthy tissue and in PC cell lines compared to normal epithelial prostatic cells in the study of Yu et al. (Yu et al. 2014a). On the contrary, Hart et al. revealed upregulation of miR-200b in samples of PC (Hart et al. 2014). Moreover, in our previous study, we have shown that expression of miR-200b was higher in PC than in BPH samples (Janiak et al. 2017). In this study, we demonstrate that miR-200b-3p is downregulated in BPH samples compared to PC tissue but also to the adjacent, morphologically healthy tissue. These controversies between molecularly confirmed suppressive role of miR-200b and clinically opposite observations raise question what is the source of miR-200b in examined tissues—PC or invaded tissues? Explanation requires further research in spatial context of tissues.
miRs may be analyzed in tumor tissues as well as in body fluids. Exosomal PC-derived miRs are intensively researched as they seem to appear as more stable and promising non-invasive biomarkers of PC (Moustafa et al. 2018; Brase et al. 2011). Several studies showed the potential of microRNAs in urine or blood as diagnostic markers to discriminate PC from BPH (Haj-Ahmad et al. 2014; Al-Kafaji et al. 2018; Cochetti et al. 2016). Haj-Ahmad et al. showed different expression of miR-1825 and miR-484 in urine samples from healthy males and patients with BPH which may be valuable for PC and BPH differentiation (Haj-Ahmad et al. 2014). Although many studies analyzed the role of microRNAs in PC, the data vary and there is a need for further investigation (Sharma and Baruah 2019). In our opinion, assessment of miR-200b-3p and miR-181b-5p levels in blood and urine of PC and BPH patients may be a non-invasive diagnostic approach that is worth further studies.
Discordant results of various studies together with our findings indicates the need for further large-scale studies to answer whether indeed low level of miR-200b-3p or high level of miR-181b-5p indicates BPH. Moreover, our panel of combined expression of three or four miRNAs requires verification on larger cohort of PC and BPH patients along with correlation with clinical data.
Conclusions
miR-200b-3p expression was higher and miR 181b-5p was lower in PC tissues in comparison with BPH. miR-96-5b and miR-134b-5p are downregulated in PC compared with BPH. Thus, these microRNAs may differentiate BPH and PC. Further studies are needed to assess the clinical usefulness of these microRNA. There are no differences between levels of miR-96-5p, miR-134-5p, miR-181b-5p and miR-200b-3p in prostate cancer and adjacent tissue. miR-96-5p, miR-134-5p and miR-181b-5p correlate negatively with the Gleason score.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Funding
This work was supported by the First Faculty of Medicine, Medical University of Warsaw—Grant No. 1M15/NM2/14 and Statutory funds. The funder had no involvement in the study design, nor the collection, analysis, and interpretation of data.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest in relation to this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aghdam SG, Ebrazeh M, Hemmatzadeh M, Seyfizadeh N, Shabgah AG, Azizi G, Ebrahimi N, Babaie F, Mohammadi H. The role of microRNAs in prostate cancer migration, invasion, and metastasis. J Cell Physiol. 2018 doi: 10.1002/jcp.27948. [DOI] [PubMed] [Google Scholar]
- Al-Kafaji G, Said HM, Alam MA, Al Naieb ZT. Blood-based microRNAs as diagnostic biomarkers to discriminate localized prostate cancer from benign prostatic hyperplasia and allow cancer–risk stratification. Oncol Lett. 2018;16(1):1357–1365. doi: 10.3892/ol.2018.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat SA, Majid S, Rehman MU. Scenario and future prospects of microRNAs in gastric cancer: a review. Iran J Basic Med Sci. 2019;22(4):345–352. doi: 10.22038/ijbms.2019.32399.7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128(3):608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- Butz H, Patocs A. MicroRNAs in endocrine tumors. Ejifcc. 2019;30(2):146–164. [PMC free article] [PubMed] [Google Scholar]
- Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, Zeng Y, Hua JT, Petricca J, Guo H, Wang M, Yousif F, Zhang Y, Donmez N, Ahmed M, Volik S, Lapuk A, Chua MLK, Heisler LE, Foucal A, Fox NS, Fraser M, Bhandari V, Shiah YJ, Guan J, Li J, Orain M, Picard V, Hovington H, Bergeron A, Lacombe L, Fradet Y, Têtu B, Liu S, Feng F, Wu X, Shao YW, Komor MA, Sahinalp C, Collins C, Hoogstrate Y, de Jong M, Fijneman RJA, Fei T, Jenster G, van der Kwast T, Bristow RG, Boutros PC, He HH. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176(4):831–843.e822. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- Cochetti G, Poli G, Guelfi G, Boni A, Egidi MG, Mearini E. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: evaluation of potential diagnostic and prognostic role. Onco Targets Ther. 2016;9:7545–7553. doi: 10.2147/ott.S119027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello AJ. Considering the role of radical prostatectomy in 21st century prostate cancer care. Nat Rev Urol. 2020;17(3):177–188. doi: 10.1038/s41585-020-0287-y. [DOI] [PubMed] [Google Scholar]
- Delangle R, De Foucher T, Larsen AK, Sabbah M, Azaïs H, Bendifallah S, Daraï E, Ballester M, Mehats C, Uzan C, Canlorbe G. The use of microRNAs in the management of endometrial cancer: a meta-analysis. Cancers (Basel) 2019;11(6):832. doi: 10.3390/cancers11060832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers DL, Fowler CB, Cunningham BR, Mason JT, O'Leary TJ. The effect of formaldehyde fixation on RNA: optimization of formaldehyde adduct removal. J Mol Diagn. 2011;13(3):282–288. doi: 10.1016/j.jmoldx.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendler A, Jung M, Stephan C, Erbersdobler A, Jung K, Yousef GM. The antiapoptotic function of miR-96 in prostate cancer by inhibition of FOXO1. PLoS ONE. 2013;8(11):e80807. doi: 10.1371/journal.pone.0080807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason DF. Classification of prostatic carcinomas. Cancer Chemother Rep. 1966;50(3):125–128. [PubMed] [Google Scholar]
- Grzywa TM, Paskal W, Wlodarski PK. Intratumor and intertumor heterogeneity in melanoma. Transl Oncol. 2017;10(6):956–975. doi: 10.1016/j.tranon.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzywa TM, Klicka K, Rak B, Mehlich D, Garbicz F, Zielinski G, Maksymowicz M, Sajjad E, Wlodarski PK. Lineage-dependent role of miR-410-3p as oncomiR in gonadotroph and corticotroph pituitary adenomas or tumor suppressor miR in somatotroph adenomas via MAPK, PTEN/AKT, and STAT3 signaling pathways. Endocrine. 2019 doi: 10.1007/s12020-019-01960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzywa TM, Klicka K, Paskal W, Dudkiewicz J, Wejman J, Pyzlak M, Włodarski PK. miR-410-3p is induced by vemurafenib via ER stress and contributes to resistance to BRAF inhibitor in melanoma. PLoS ONE. 2020;15(6):e0234707. doi: 10.1371/journal.pone.0234707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haflidadottir BS, Larne O, Martin M, Persson M, Edsjo A, Bjartell A, Ceder Y. Upregulation of miR-96 enhances cellular proliferation of prostate cancer cells through FOXO1. PLoS ONE. 2013;8(8):e72400. doi: 10.1371/journal.pone.0072400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haj-Ahmad TA, Abdalla MA, Haj-Ahmad Y. Potential urinary miRNA biomarker candidates for the accurate detection of prostate cancer among benign prostatic hyperplasia patients. J Cancer. 2014;5(3):182–191. doi: 10.7150/jca.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart M, Nolte E, Wach S, Szczyrba J, Taubert H, Rau TT, Hartmann A, Grasser FA, Wullich B. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol Cancer Res. 2014;12(2):250–263. doi: 10.1158/1541-7786.Mcr-13-0230. [DOI] [PubMed] [Google Scholar]
- Hoefig KP, Heissmeyer V. Measuring microRNA expression in size-limited FACS-sorted and microdissected samples. Methods Mol Biol. 2010;667:47–63. doi: 10.1007/978-1-60761-811-9_4. [DOI] [PubMed] [Google Scholar]
- Janiak M, Paskal W, Rak B, Garbicz F, Jarema R, Sikora K, Wlodarski P. TIMP4 expression is regulated by miR-200b-3p in prostate cancer cells. Apmis. 2017;125(2):101–105. doi: 10.1111/apm.12638. [DOI] [PubMed] [Google Scholar]
- Kaminska K, Nalejska E, Kubiak M, Wojtysiak J, Zolna L, Kowalewski J, Lewandowska MA. Prognostic and predictive epigenetic biomarkers in oncology. Mol Diagn Ther. 2018 doi: 10.1007/s40291-018-0371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Ha YR, Kim SJ, Kang SH, Park HS, Lee JG, Cheon J, Kim CH. Do microRNA 96, 145 and 221 expressions really aid in the prognosis of prostate carcinoma? Asian J Androl. 2012;14(5):752–757. doi: 10.1038/aja.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwal R, Plaga AR, Liu X, Shukla GC, Gupta S. MicroRNAs in prostate cancer: functional role as biomarkers. Cancer Lett. 2017;407:9–20. doi: 10.1016/j.canlet.2017.08.011. [DOI] [PubMed] [Google Scholar]
- Katz B, Reis ST, Viana NI, Morais DR, Moura CM, Dip N, Silva IA, Iscaife A, Srougi M, Leite KR. Comprehensive study of gene and microRNA expression related to epithelial-mesenchymal transition in prostate cancer. PLoS ONE. 2014;9(11):e113700. doi: 10.1371/journal.pone.0113700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfleisch R, Weiss AT, Gruber AD. Excavation of a buried treasure—DNA, mRNA, miRNA and protein analysis in formalin fixed, paraffin embedded tissues. Histol Histopathol. 2011;26(6):797–810. doi: 10.14670/hh-26.797. [DOI] [PubMed] [Google Scholar]
- Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, Sarkar FH. miR-200 regulates PDGF-D-mediated epithelial–mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27(8):1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzino M, Maris P, Sirianni R, Barone I, Casaburi I, Chimento A, Giordano C, Morelli C, Sisci D, Rizza P, Bonofiglio D, Catalano S, Andò S. DAX-1, as an androgen-target gene, inhibits aromatase expression: a novel mechanism blocking estrogen-dependent breast cancer cell proliferation. Cell Death Dis. 2013;4(7):e724. doi: 10.1038/cddis.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larne O, Martens-Uzunova E, Hagman Z, Edsjo A, Lippolis G, den Berg MS, Bjartell A, Jenster G, Ceder Y. miQ—a novel microRNA based diagnostic and prognostic tool for prostate cancer. Int J Cancer. 2013;132(12):2867–2875. doi: 10.1002/ijc.27973. [DOI] [PubMed] [Google Scholar]
- Levine E, McHale P, Levine H. Small regulatory RNAs may sharpen spatial expression patterns. PLoS Comput Biol. 2007;3(11):e233. doi: 10.1371/journal.pcbi.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas DJ, Ahmed HU. All change in the prostate cancer diagnostic pathway. Nat Rev Clin Oncol. 2020;17(6):372–381. doi: 10.1038/s41571-020-0332-z. [DOI] [PubMed] [Google Scholar]
- Long MD, Singh PK, Russell JR, Llimos G, Rosario S, Rizvi A, van den Berg PR, Kirk J, Sucheston-Campbell LE, Smiraglia DJ, Campbell MJ. The miR-96 and RARγ signaling axis governs androgen signaling and prostate cancer progression. Oncogene. 2019;38(3):421–444. doi: 10.1038/s41388-018-0450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Yang HZ, Dong BJ, Zou HB, Zhou Y, Kong XM, Huang YR. Biphasic regulation of autophagy by miR-96 in prostate cancer cells under hypoxia. Oncotarget. 2014;5(19):9169–9182. doi: 10.18632/oncotarget.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihelich BL, Khramtsova EA, Arva N, Vaishnav A, Johnson DN, Giangreco AA, Martens-Uzunova E, Bagasra O, Kajdacsy-Balla A, Nonn L. miR-183-96-182 cluster is overexpressed in prostate tissue and regulates zinc homeostasis in prostate cells. J Biol Chem. 2011;286(52):44503–44511. doi: 10.1074/jbc.M111.262915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME, Lengyel E. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. J Cancer Discov. 2012;2(12):1100–1108. doi: 10.1158/2159-8290.CD-12-0206%JCancerDiscovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa AA, Kim H, Albeltagy RS, El-Habit OH, Abdel-Mageed AB. MicroRNAs in prostate cancer: from function to biomarker discovery. Exp Biol Med (Maywood) 2018;243(10):817–825. doi: 10.1177/1535370218775657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Suzuki T, Arai Y, Sasano H. Nuclear receptor DAX1 in human prostate cancer: a novel independent biological modulator. Endocr J. 2009;56(1):39–44. doi: 10.1507/endocrj.k08e-177. [DOI] [PubMed] [Google Scholar]
- Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS ONE. 2009;4(11):e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngalame NN, Tokar EJ, Person RJ, Xu Y, Waalkes MP. Aberrant microRNA expression likely controls RAS oncogene activation during malignant transformation of human prostate epithelial and stem cells by arsenic. Toxicol Sci. 2014;138(2):268–277. doi: 10.1093/toxsci/kfu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira L, Corradi R, Eastham JA. Prostatic specific antigen for prostate cancer detection. Int Braz J Urol. 2009;35(5):521–529. doi: 10.1590/S1677-55382009000500003. [DOI] [PubMed] [Google Scholar]
- Nonn L, Vaishnav A, Gallagher L, Gann PH. mRNA and micro-RNA expression analysis in laser-capture microdissected prostate biopsies: valuable tool for risk assessment and prevention trials. Exp Mol Pathol. 2010;88(1):45–51. doi: 10.1016/j.yexmp.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana-Serradell O, Herrera D, Castellón EA, Contreras HR. The transcription factor ZEB1 promotes chemoresistance in prostate cancer cell lines. Asian J Androl. 2019;21(5):460–467. doi: 10.4103/aja.aja_1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JY, Zhang F, Sun CC, Li SJ, Li G, Gong FY, Bo T, He J, Hua RX, Hu WD, Yuan ZP, Wang X, He QQ, Li DJ. miR-134: a human cancer suppressor? Mol Ther Nucleic Acids. 2017;6:140–149. doi: 10.1016/j.omtn.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Tian Y, Niu G, Cao C. Role of microRNAs in remodeling the tumor microenvironment (review) Int J Oncol. 2020;56(2):407–416. doi: 10.3892/ijo.2019.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paskal W, Paskal AM, Debski T, Gryziak M, Jaworowski J. Aspects of modern biobank activity—comprehensive review. Pathol Oncol Res. 2018;24(4):771–785. doi: 10.1007/s12253-018-0418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapado-Gonzalez O, Alvarez-Castro A, Lopez-Lopez R, Iglesias-Canle J, Suarez-Cunqueiro MM, Muinelo-Romay L. Circulating microRNAs as promising biomarkers in colorectal cancer. Cancers (Basel) 2019;11(7):898. doi: 10.3390/cancers11070898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeiros T, Garcia M, Montes M, Olivan M, Rigau M, Colas E, de Torres I, Morote J, Reventos J, Doll A. Molecular markers for prostate cancer in formalin-fixed paraffin-embedded tissues. Biomed Res Int. 2013;2013:283635. doi: 10.1155/2013/283635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Baruah MM. The microRNA signatures: aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol. 2019;21(2):126–144. doi: 10.1007/s12094-018-1910-8. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019 doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Siu MK, Tsai YC, Chang YS, Yin JJ, Suau F, Chen WY, Liu YN. Transforming growth factor-β promotes prostate bone metastasis through induction of microRNA-96 and activation of the mTOR pathway. Oncogene. 2015;34(36):4767–4776. doi: 10.1038/onc.2014.414. [DOI] [PubMed] [Google Scholar]
- Souza MF, Kuasne H, Barros-Filho MC, Ciliao HL, Marchi FA, Fuganti PE, Paschoal AR, Rogatto SR, Colus IMS. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS ONE. 2017;12(9):e0184094. doi: 10.1371/journal.pone.0184094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SJ, Liu J, Wang X, Qu LX. microRNA-181 promotes prostate cancer cell proliferation by regulating DAX-1 expression. Exp Ther Med. 2014;8(4):1296–1300. doi: 10.3892/etm.2014.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Chen WY, Siu MK, Tsai HY, Yin JJ, Huang J, Liu YN. Epidermal growth factor receptor signaling promotes metastatic prostate cancer through microRNA-96-mediated downregulation of the tumor suppressor ETV6. Cancer Lett. 2017;384:1–8. doi: 10.1016/j.canlet.2016.10.014. [DOI] [PubMed] [Google Scholar]
- Vasilescu C, Tanase M, Giza D, Procopiuc L, Dragomir MP, Calin AGA. How does a tumor get its shape? MicroRNAs act as morphogens at the cancer invasion front. Noncoding RNA. 2020;6(2):23. doi: 10.3390/ncrna6020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter BA, Valera VA, Pinto PA, Merino MJ. Comprehensive microRNA profiling of prostate cancer. J Cancer. 2013;4(5):350–357. doi: 10.7150/jca.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter BA, Valera VA, Pinto PA, Merino MJ. Comprehensive microRNA profiling of prostate cancer. J Cancer. 2013;4(5):350–357. doi: 10.7150/jca.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WK, Hurwitz M, D'Amico AV, et al. et al. Biology of prostate cancer, chapter 111. In: Kufe DW, Pollock RE, Weichselbaum RR, et al.et al., editors. Holland-frei cancer medicine. 6. Hamilton, ON: BC Decker; 2003. [Google Scholar]
- Williams LV, Veliceasa D, Vinokour E, Volpert OV. miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS ONE. 2013;8(12):e83991. doi: 10.1371/journal.pone.0083991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhong J, Guo B, Zhu Q, Liang H, Wen N, Yun W, Zhang L. miR-96 promotes the growth of prostate carcinoma cells by suppressing MTSS1. Tumour Biol. 2016;37(9):12023–12032. doi: 10.1007/s13277-016-5058-2. [DOI] [PubMed] [Google Scholar]
- Yadav SS, Stockert JA, Hackert V, Yadav KK, Tewari AK. Intratumor heterogeneity in prostate cancer. Urol Oncol. 2018;36(8):349–360. doi: 10.1016/j.urolonc.2018.05.008. [DOI] [PubMed] [Google Scholar]
- Yu J, Lu Y, Cui D, Li E, Zhu Y, Zhao Y, Zhao F, Xia S. miR-200b suppresses cell proliferation, migration and enhances chemosensitivity in prostate cancer by regulating Bmi-1. Oncol Rep. 2014;31(2):910–918. doi: 10.3892/or.2013.2897. [DOI] [PubMed] [Google Scholar]
- Yu JJ, Wu YX, Zhao FJ, Xia SJ. miR-96 promotes cell proliferation and clonogenicity by down-regulating of FOXO1 in prostate cancer cells. Med Oncol. 2014;31(4):910. doi: 10.1007/s12032-014-0910-y. [DOI] [PubMed] [Google Scholar]
- Zhu S, Zhao D, Li C, Li Q, Jiang W, Liu Q, Wang R, Fazli L, Li Y, Zhang L, Yi Y, Meng Q, Wang W, Wang G, Zhang M, Zu X, Zhao W, Deng T, Yu J, Dong X, Chen K, Cao Q. BMI1 is directly regulated by androgen receptor to promote castration-resistance in prostate cancer. Oncogene. 2020;39(1):17–29. doi: 10.1038/s41388-019-0966-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.