Abstract
Background
Strategies to increase nucleic acid vaccine immunogenicity are needed to move towards clinical applications in oncology. In this study, we designed a new generation of DNA vaccines, encoding an engineered vesicular stomatitis virus glycoprotein as a carrier of foreign T cell tumor epitopes (plasmid to deliver T cell epitopes, pTOP). We hypothesized that pTOP could activate a more potent response compared with the traditional DNA-based immunotherapies, due to both the innate immune properties of the viral protein and the specific induction of CD4 and CD8 T cells targeting tumor antigens. This could improve the outcome in different tumor models, especially when the DNA-based immunotherapy is combined with a rational therapeutic strategy.
Methods
The ability of pTOP DNA vaccine to activate a specific CD4 and CD8 response and the antitumor efficacy were tested in a B16F10-OVA melanoma (subcutaneous model) and GL261 glioblastoma (subcutaneous and orthotopic models).
Results
In B16F10-OVA melanoma, pTOP promoted immune recognition by adequate processing of both MHC-I and MHC-II epitopes and had a higher antigen-specific cytotoxic T cell (CTL) killing activity. In a GL261 orthotopic glioblastoma, pTOP immunization prior to tumor debulking resulted in 78% durable remission and long-term survival and induced a decrease of the number of immunosuppressive cells and an increase of immunologically active CTLs in the brain. The combination of pTOP with immune checkpoint blockade or with tumor resection improved the survival of mice bearing, a subcutaneous melanoma or an orthotopic glioblastoma, respectively.
Conclusions
In this work, we showed that pTOP plasmids encoding an engineered vesicular stomatitis virus glycoprotein, and containing various foreign T cell tumor epitopes, successfully triggered innate immunity and effectively promoted immune recognition by adequate processing of both MHC-I and MHC-II epitopes. These results highlight the potential of DNA-based immunotherapies coding for viral proteins to induce potent and specific antitumor responses.
Keywords: vaccination, adaptive immunity, antigens, immunity, cellular, immunotherapy, active
Introduction
Immunotherapy is a well-established treatment for many cancers. Until now, many efforts have been made to identify (neo)antigen targets1 and to develop checkpoint inhibitors2 that are able to circumvent tumor immunosuppressive mechanisms, opening the way to novel combined treatments.3 Immune checkpoint inhibitors that initially received approval for melanoma are now used and assessed to treat various types of malignancies. However, there are still challenges, as only a minority of cancer patients responds to current immunotherapy treatments. The identification of predictive biomarkers and the emergence of new treatment modalities could improve both the proportion of responding patients and the range of tumors that can be treated.
In the expanding field of cancer immunotherapy, the development of vaccines able to promote potent and specific antitumor T cell responses remains to be achieved. Among the technologies developed to enhance tumor antigen recognition, DNA vaccines are attractive due to their low cost, easy production, ability to induce a broad immune response and stability.4 In recent years, many efforts have been made to improve the immunogenicity and clinical potential of DNA vaccines5 by relying on the use of electroporation,6 codon optimization of plasmid constructs7 or co-administration of adjuvants.8 An ideal technology for cancer vaccines should allow the codelivery of multiple CD8 and CD4 T cell epitopes from several cancer antigens.9 Indeed, cancer vaccine strategies were initially focused on eliciting CD8 cytotoxic T cells (CTLs),10 but the central role of CD4 T helper cells in cancer immunity and immunotherapy has also been demonstrated over the years.11 12 In addition, targeting epitopes derived from different tumor antigens may overcome the risk of selecting tumor cells that have downregulated the vaccine target.13
The immune system has the ability to provide defense against both tumors and pathogens, and it has thus been postulated that elucidating and harnessing the shared mechanisms of immunity that underlie cancer and infectious disease could lead to new therapeutic developments.14 Indeed, several components derived from bacteria or viruses were shown to interact with the cancer vaccine response. For instance, unmethylated CpG motifs that are present on bacterial DNA can be used to modulate the immunogenicity of DNA vaccines.7 15 Coadministration of a plasmid encoding the HIV-1 Gag viral capsid protein improved the efficacy of a cancer DNA vaccine,16 and another plasmid encoding the vesicular stomatitis virus glycoprotein (VSV-G) has shown adjuvant properties.17 VSV-G is a viral fusion glycoprotein that is recognized by the immune system and activates the innate immune response.18 Some studies have previously suggested mechanisms by which VSV-G could modulate the immune response. VSV-G was shown to induce an immunogenic form of cellular fusion and necrosis, which could induce immune cell recruitment at the delivery site.17 19 20 VSV-G was also shown to enhance cross-presentation of antigens.17 21 22 Recently, VSV-G has been suggested to induce autophagy23 and thus may be involved in modulation of antigen processing pathways.24 Mutants have been constructed by inserting targeting ligands to alter the tropism of viral vectors,25 and it was described that this glycoprotein possesses permissive sites in which several amino acids can be inserted without altering protein function.26
For efficiently inducing T cell responses and ensuring therapeutic efficiency against tumor, an ideal vaccine should combine four key features: (1) it mimics native pathogen structure for an effective presentation to the immune system, (2) it has intrinsic adjuvant properties or incorporates adjuvants to stimulate immunity, (3) it has the ability to target antigen presenting cells and (4) it presents several cancer antigens containing both CD4 and CD8 epitopes or neoepitopes.9 27 28 Structural viral (glyco)proteins, such as VSV-G, combine by nature most of these key features and their flexibility can be harnessed to design systems that could entirely fill the gaps, resulting in potent cancer vaccine strategies. Hence, we developed a new DNA vaccine technology, called ‘pTOP’ (plasmid to deliver T cell epitopes), which encodes a modified VSV-G viral protein, engineered by inserting foreign T cell epitopes, to serve as a vaccine platform to deliver tumor epitopes. We hypothesized that such modified DNA vaccine would retain some intrinsic viral immunogenicity due to the presence of VSV-G, while promoting tumor specificity through the induction of antiepitope T cell responses, due to the antigen epitopes inserted in the viral protein.
The ability of pTOP DNA vaccine to activate a specific CD4 and CD8 response, was tested in a B16F10-OVA melanoma. B16 melanomas are very aggressive and highly metastatic tumors, which present different tumor antigens, such as gp100 and TRP2.29–31 In particular, the B16F10-OVA is a modified B16F10 cell line to express also the ovalbumin (OVA) antigen. Next, we assessed the ability of pTOP to cure mice bearing an orthotopic GL261 glioblastoma (GBM), considered as a poorly immunogenic tumor.32 Immunotherapy has recently been reconsidered as a promising option for GBM since the brain is no longer considered an ‘immune privileged’ organ. Indeed, the central nervous system possesses lymphatic vessels along the dural sinuses and meningeal arteries,33 34 and the blood–brain barrier is compromised in GBM patients, allowing immune cells to infiltrate from the peripheral circulation.35 Surgical debulking of the accessible tumor is the mainstay treatment in all eligible patients,36 who subsequently undergo radiotherapy and chemotherapy.37 However, the intrinsic properties of GBM (ie, high migratory and infiltrative patterns and the presence of tumor microtubes and glioma cancer stem cells) promote the formation of recurrences that inevitably lead to patient death.38 39 We hypothesized that the combination of the pTOP DNA vaccine and the GBM resection could improve the outcome, by helping the immune cells generated by the vaccine to reach the brain.
Material and methods
Plasmids
Codon-optimized gene sequences of plasmid OVA (pOVA) and VSV-G (pVSVG) were designed using GeneOptimizer and obtained by standard gene synthesis from GeneArt (Thermo Fisher Scientific, Waltham, Massachusetts, USA). These sequences were subcloned in the pVAX2 vector using cohesive-ends cloning. pTOP refers to the plasmids encoding VSV-G in which the foreign epitopes were inserted (figure 1B, C). Codon-optimized gene sequences were designed using GeneOptimizer and obtained by standard gene synthesis from GeneArt (Thermo Fisher Scientific, USA). The sequences were subcloned in the pVAX2 vector using cohesive-end cloning with BamHI and NotI restriction sites. To allow easy modifications of the epitopes, several restriction sites were added. Digestion by BamHI and HindIII or by SpeI and EcoRI allows insertion in position (18) or (191), respectively. The positions into which the epitopes are inserted are defined by the amino acid residue directly after the insertion site. In other words, insertion position (18) corresponds to the region between amino acid residues 17 and 18. For the epitopes inserted at the N terminus (ie, position (18), just after the signal peptide), an additional lysine residue was included.25 Overlapping phosphorylated oligonucleotides that encoded the restricted epitope (IDT-DNA, Belgium) were incorporated in the digested vector using cohesive-end cloning. pEmpty is an empty pVAX2 vector. The plasmids were prepared using the EndoFree Plasmid Mega or Giga Kit (Qiagen, Germany) and diluted in phosphate-buffered saline (PBS). The quality of the purified plasmid was assessed by the ratio of optical densities and by 1% agarose gel electrophoresis. Plasmids were sequenced by Sanger DNA sequencing (Genewiz, UK) and stored at −20°C. A list of the plasmids used in this study can be found in figure 1C. Three other plasmids (pTOP4, 5 and 6) have been developed and used against other cancer models, not shown in this study.
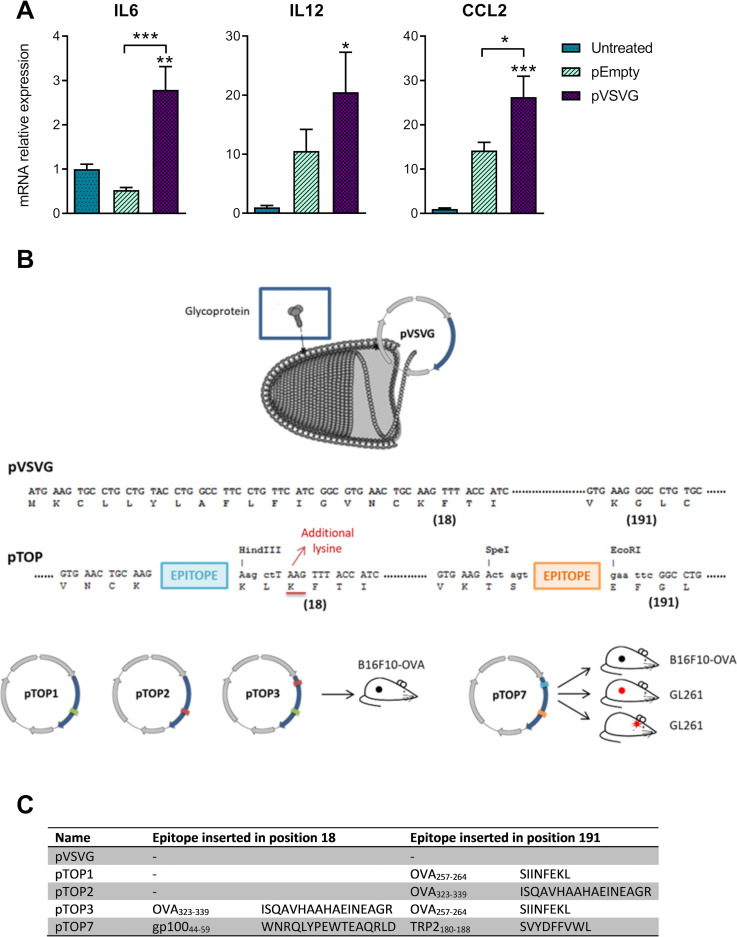
Figure 1.
Inflammatory cytokines expressed after intramuscular electroporation of pEmpty or pVSVG and a description of the pTOP technology and vaccine constructs. (A) mRNA expression of IL6, IL12 and CCL2 48 hours after the injection of 1 µg plasmid. Mice that did not receive any plasmid (untreated) were used as a control. The error bars represent the mean±SEM; n=2, n=3–4. Statistical analysis: One-way ANOVA with Tukey’s multiple comparisons test. *p<0.05, **p<0.01 and ***p<0.001 compared with untreated or to the specified group. (B) Graphical representation of the versatile pTOP technology (vesiculovirus picture adapted from ViralZone: www.expasy.org/viralzone, Sib Swiss Institute of bioinformatics). (C) pTOP vaccines that were used throughout the paper. The positions into which the epitopes were inserted are defined by the amino acid residue directly after the insertion site. ANOVA, analysis of variance; IL6, interleukin 6; pTOP, plasmid to deliver T cell epitopes; pVSVG, plasmid vesicular stomatitis virus glycoprotein.
Immunization
Intramuscular electroporation: After the mouse hair was removed using a rodent shaver (AgnTho’s, Lidingö, Sweden), 30 µL of a PBS solution containing 1 µg of plasmid was injected into the tibial cranial muscle. The leg was placed between 4 mm spaced plate electrodes, and 8 square-wave electric pulses (200 V/cm, 20 ms, 2 Hz) were delivered. For therapeutic immunization, the vaccine was administered 2, 9 and 16 days after subcutaneous tumor injection or 16, 23 and 29 days after GL261 orthotopic injection. Immune checkpoint blockade antibodies directed against CTLA4 (clone 9D9) and PD1 (clone 29 F.A12) were purchased from Bioconnect (Netherlands) and mice were injected intraperitoneally with 100 µg of each antibody in 100 µl of PBS 3, 6 and 9 days after tumor injection.
Ear pinna electroporation: A 30 µL DNA solution containing 1 µg of plasmid was injected into the pinna of both ears (two injections per mouse). The 2 mm spaced plate electrodes were applied around the ear to deliver 10 square-wave electric pulses (500 V/cm, 20 ms, 1 Hz).
For all electroporation protocols, electric pulses were generated by a Gemini System generator and delivered with BTX Caliper Electrodes (BTX; both from VWR International, Belgium). A conductive gel was used to ensure electrical contact with the skin (Aquasonic 100; Parker Laboratories, USA).
Quantification of inflammatory cytokines
To quantify inflammatory cytokines, mice were sacrificed 48 hours after intramuscular electroporation. The muscles were collected and kept in RNA later solution. Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific). The quality and quantity of RNA were evaluated using a NanoDrop 2000 (Thermo Fisher Scientific). RNA (1 µg) was reverse transcribed using a first standard synthesis system (SuperScript, Thermo Fisher Scientific) and oligo(dT) primers according to the supplier’s protocol. Primers were designed using Primer Blast software based on the consensus of sequences from GenBank (online supplemental table 1). SYBR green real-time qPCR (RT-qPCR) (GoTaq qPCR MasterMix kit, Promega, USA) was conducted on a StepOne Plus Real-Time RT-PCR System (Thermo Fisher Scientific). Analysis of the melting curves was performed to ensure the purity of PCR products. The results were analyzed with the StepOne Software V.2.1. The mRNA expression was calculated relative to the expression of corresponding β-actin, according to the delta-delta Ct method.
jitc-2020-001243supp001.pdf (511.9KB, pdf)
OT-I and OT-II proliferation
A single-cell suspension was prepared from the spleens and lymph nodes of transgenic OT-I and OT-II mice (Ghent University, Ghent, Belgium). T cells were isolated using a CD8a+ and CD4+ T cell isolation kit for mouse and an AutoMACS Pro Separator (Miltenyi Biotec, Germany). Subsequently, the isolated T cells were labeled with (carboxyfluorescein diacetate succinimidyl ester (CFSE); Thermo Fisher Scientific) by incubating 50×106 cells/mL PBS with 5 µM of CFSE for 7 min at 37°C. The reaction was blocked by adding ice-cold PBS +10% FBS (Life Technologies). A total of 2×106 OT-I or OT-II cells were injected into the tail vein of C57BL/6 recipient mice. The mice were treated 2 days later with plasmid injection and electroporation to the ear pinna. Mice were sacrificed 4 days later to collect the draining lymph nodes for single-cell suspension preparation. Flow cytometric measurement was performed after staining the cells with Aqua Live/Dead (Thermo Fisher), CD19 APC-Cy7, CD8 PerCP (for OT-I proliferation), CD4 eFluor 450 and Valpha2 TCR PE (for OT-II proliferation) and Fc block (all BD Biosciences, USA). Dextramer SIINFEKL H-2kb PE (Immudex, Denmark) was added for OT-I proliferation measurement. The read out was performed using a BD FACSVerse (BD Biosciences, USA).
In vivo killing assay
Mice were prophylactically immunized by intramuscular DNA electroporation (three plasmid injections one priming and two boosts, biweekly). Three weeks after the last vaccine administration, splenocytes (2.5×106 cells/mL PBS) from naive mice were pulsed with 1 µg/mL PBS of SIINFEKL or an irrelevant peptide for 1 hour at 37°C. The splenocytes pulsed with the OVA peptide were stained with 5 µM (high) CFSE, while the splenocytes pulsed with the irrelevant peptide were stained with 0.5 µM (low) CFSE. The two labeled cell populations were mixed at a 1:1 ratio, and a total of 1×107 cells were adoptively transferred into the immunized mice by intravenous injection. Two days after the transfer, the host mouse spleens were isolated and analyzed by flow cytometry after staining with Aqua Live/Dead (Thermo Fisher), Fc Block and α-F4/80 (BD Biosciences). The percentage of antigen-specific killing was determined using the formula: 100–100*((% CFSEhigh cells/% CFSElow cells)immunized mice/(% CFSEhigh cells/% CFSElow cells)non-immunized mice), where CSFElow cells are cells stimulated with the irrelevant peptide.
Orthotopic GL261 GBM syngeneic model and surgical resection of the GL261 tumor mass
For orthotopic GBM tumor grafting, C57BL/6 mice were anesthetized and fixed in a stereotactic frame. A surgical high-speed drill (Vellman, Belgium) was used to perform a hole in the right frontal lobe and 5×104 GL261 cells were slowly injected using a Hamilton syringe fitted with a 26S needle. To obtain cortical tumors, the injection coordinates were 0.5 mm posterior, 2.1 mm lateral from the bregma and 2.2 mm deep from the outer border of the cranium. The presence, volume and location of the tumors were determined by MRI (online supplemental methods), and the animals presenting GL261 tumors were randomly divided into four groups.
At day 17 post-tumor inoculation, the tumor mass was surgically removed by using the biopsy-punch resection technique (online supplemental methods).40 All animals were monitored daily and an MRI follow-up was performed 27 days after surgery. Eight to nine animals per group were sacrificed 29 days post-tumor inoculation for immunological analysis (FACS and PCR). The spleen and the brain of the animals were collected for further analysis. The remaining animals were sacrificed when they reached the previously mentioned end points.
Flow cytometry analysis of immune cells
Tumor-associated macrophages (TAM), myeloid-derived suppressor cell (MDSC), CD4 and CD8 T cell populations in brains and spleens removed 29 days after GL261 orthotopical cell injection were analyzed by FACS. Cells were passed through a 70 µm cell strainer (BD Falcon, New Jersey, USA), collected, counted using an automatic cell counter (Invitrogen, California) and washed with PBS, before adding the blocking solution with anti-CD16/CD32 antibody for 10 min on ice (clone 93, Biolegend, San Diego, California, USA). Cells were washed and incubated for 60 min at 4°C with the following antibodies: anti-CD3-APC-Cy7 (Biolegend, San Diego, California), anti-CD4-PE (BD bioscience, UK), anti-CD8-BV421 (Biolegend, San Diego, California, USA) for CD4 and CD8 T cell detection; with anti-CD11b-FITC (BD bioscience, UK), anti-F4/80-AF647 (BD bioscience, UK), anti-CD206-BV421 (Biolegend,) and anti-Gr1-PE (BD bioscience) for TAMs and MDSCs; with anti-CD3-APC-Cy7, anti-CD8-FITC (Proimmune, UK) and Pentamers-TRP2-PE (Proimmune, UK) for the detection of TRP-2-specific CD8 T cells. For staining with antiFoxP3-AF488 (BD bioscience) or anti-interferon gamma (IFNg)-APC (Biolegend), cells were previously incubated overnight at 4°C with a permeabilization/fixation solution (eBioscience Foxp3/Transcription Factor Staining Buffer Set, Thermo Fisher, Waltham, Massachusetts, USA). Cells were then incubated with anti-CD16/CD32 antibody for 10 min on ice (Biolegend), washed and incubated for 60 min at 4°C with anti-IFNg-APC or antiFoxP3-AF488 diluted in the permeabilization/fixation solution. Samples were washed with PBS fixed for 10 min with 4% formalin and, then, suspended in PBS. Sample data were acquired with FACSVerse (BD bioscience) and analyzed with FlowJo software (FlowJo, Ashland, Oregon, USA).
Enzyme-linked immunospot (ELISpot)
ELISpot was performed according to the manufacturer’s instruction (Immunospot, the ELISPOT source, Germany). Briefly, 3×105 fresh splenocytes diluted in 100 µL CTL-Test medium (Immunospot, the ELISPOT source) were cultured overnight at 37°C in anti-IFNg-coated 96 well plate. For stimulation, 10 ng/µl of TRP2180-188 peptide (SVYDFFVWL) was added to the splenocytes and incubated for 2 days. As positive control for splenocyte activation, Cell Stimulation Cocktail (Invitrogen, California) was used; PBS and a P815 irrelevant peptide (LPYLGWLVF) were used as negative control. The development of the ELISpot plate followed the manufacturer’s instruction and pots were counted by using an ELISPOT reader system (the ELISPOT source).
Statistical analysis
Statistical analyses were performed using GraphPad Prism V.7 for Windows. P values lower than 0.05 were considered statistically significant.
Results
The VSV-G promotes the innate immune response
To verify the viral protein-mediated activation of innate immunity, we evaluated whether innate cytokines were produced after intramuscular electroporation of an empty plasmid (pEmpty) or a plasmid coding for VSV-G (pVSVG). Mice that did not receive any plasmid were used as controls. Forty-eight hours after the electroporation of pVSVG, the expression levels of interleukin 6 (IL6), IL12 and CCL2 were increased in the injection site. In particular, the levels of CCL2 and IL6 were higher in pVSVG-treated mice compared with mice that received the pEmpty plasmid (figure 1A). To leverage the intrinsic immunogenicity of VSV-G, several pTOP plasmids were designed by inserting different tumor epitopes in two of the permissive insertion sites of the VSV-G protein (figure 1B, C).
pTOP induces a specific CD8 and CD4 response against OVA MHC class I and II epitopes, respectively
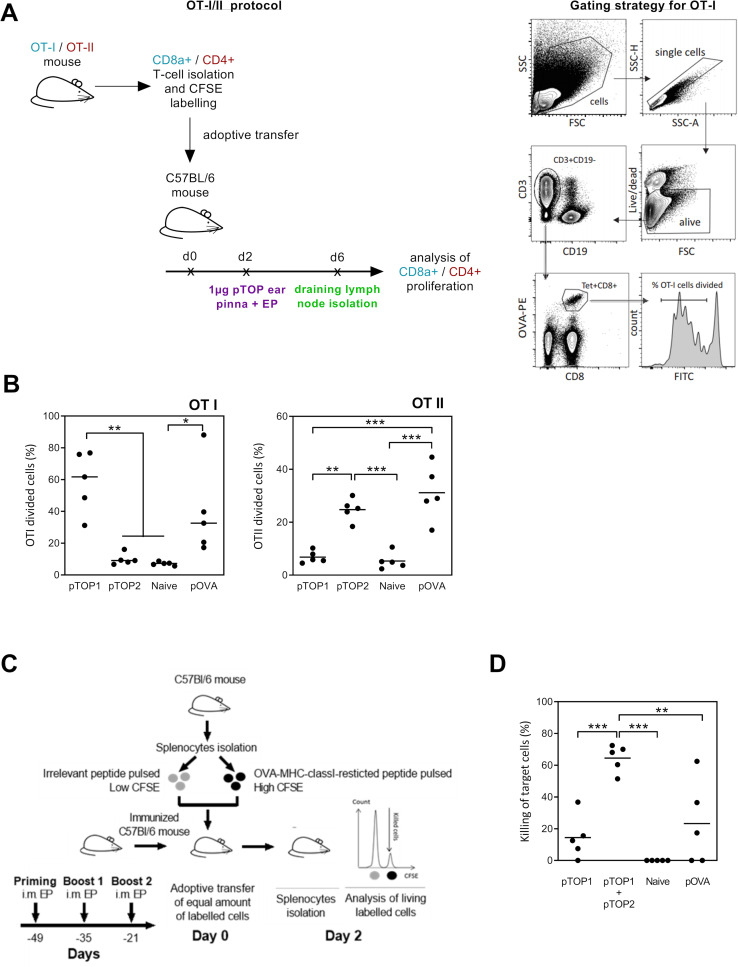
To characterize the immunogenic potential of pTOP, OVA was initially used as a model antigen. pTOP1 and pTOP2 were constructed by inserting an OVA-major histocompatibility complex (MHC) class I-restricted epitope (OVA257-264) and an OVA-MHC class II-restricted epitope (OVA323-339), respectively. pTOP1 and pTOP2 were tested for their ability to induce the proliferation of CFSE-labeled lymphocytes that were isolated from OT-I and OT-II transgenic mice and adoptively transferred to recipient mice. The recipient mice were immunized, and T lymphocytes were collected from the draining lymph nodes and analyzed by flow cytometry (figure 2A). As expected, the OVA-MHC class I-restricted epitope inserted in pTOP efficiently activated the specific CD8 T cell proliferation, while the OVA-MHC class II-restricted epitope induced division of the specific CD4 T cells (figure 2B). This response was similar to the proliferation induced by a classical pOVA vaccine, encoding the full-length OVA antigen in a pVAX2 vector, for both CD8 and CD4 T cells (figure 2B). pTOP plasmids induce a higher CTL killing activity compared with classical DNA vaccines.
Figure 2.
Evaluation of the OVA-specific cellular immune response and CTL killing activity. (A) Schematic representation of OT-I/II proliferation studies protocol. Purified CD8 or CD4 T cells from transgenic OTI and OTII mice were CFSE-labeled and adoptively transferred to C57BL/6 mice. The mice were treated 2 days later with 1 µg pTOP injection into the ear Pinna, followed by electroporation and sacrificed 4 days later to collect the draining lymph nodes for preparation of single-cell suspension and FACS analysis. (B) Quantification of the divided OTI and OTII cells is shown; n=5. (C) In vivo OVA-specific cytotoxic CD8 T cell killing assay protocol. C57BL/6 mice were first vaccinated three times by intramuscular electroporation of 1 µg pTOP every 2 weeks. Three weeks after the last vaccine administration, the mice received labeled splenocytes from naive mice that were pulsed with either SIINFEKL (the OVA peptide) or an irrelevant peptide (ie, a peptide that should not induce an immune response). Two days after transfer, splenocytes were collected and analyzed by FACS to determine the antigen-specific killing; n=5. (D) Percentage of the CTL killing activity for the different groups. Statistical analysis: one-way ANOVA with Tukey’s multiple comparisons test. *p<0.05, **p<0.01, ***p<0.001 compared with untreated or to the specified group (n=5). ANOVA, analysis of variance; CTL, cytotoxic T cell; CFSE, carboxyfluorescein diacetate succinimidyl ester; FSC, forward scatter (cell diameter); SSC, side scatter (cell granulometry); OVA, ovalbumin; pTOP, plasmid to deliver T cell epitopes; pOVA, plasmid OVA.
To assess the CTL response triggered by pTOP compared with the traditional DNA vaccination, an in vivo killing assay was performed. This experiment evaluates the ability of vaccinated mice (one priming and two boosts) to recognize and eliminate peptide-pulsed target cells, marked with a high amount of CFSE. A schematic representation of the CTL killing protocol is shown in figure 2C, where the irrelevant peptide is a peptide that should not induce an immune response, that is, no killing activity. The absence of the generation of an immune response following the stimulation with an irrelevant peptide, underlines the specificity of the pTOP vaccination. A higher CTL killing activity was observed in the presence of both pTOP1 and pTOP2, compared with pTOP1 alone or to the classical pOVA plasmid (figure 2D). This result indicates: (1) the necessity of a helper T cell response to improve the CTL activity, (2) an efficient helper T cell response induced by the codelivery of pTOP1 and pTOP2 and (3) the higher efficacy of the two pTOP codelivered plasmids, compared with the classical pOVA DNA vaccination.
Therapeutic vaccination with pTOP outperforms classical DNA vaccine coding for the full-length antigen
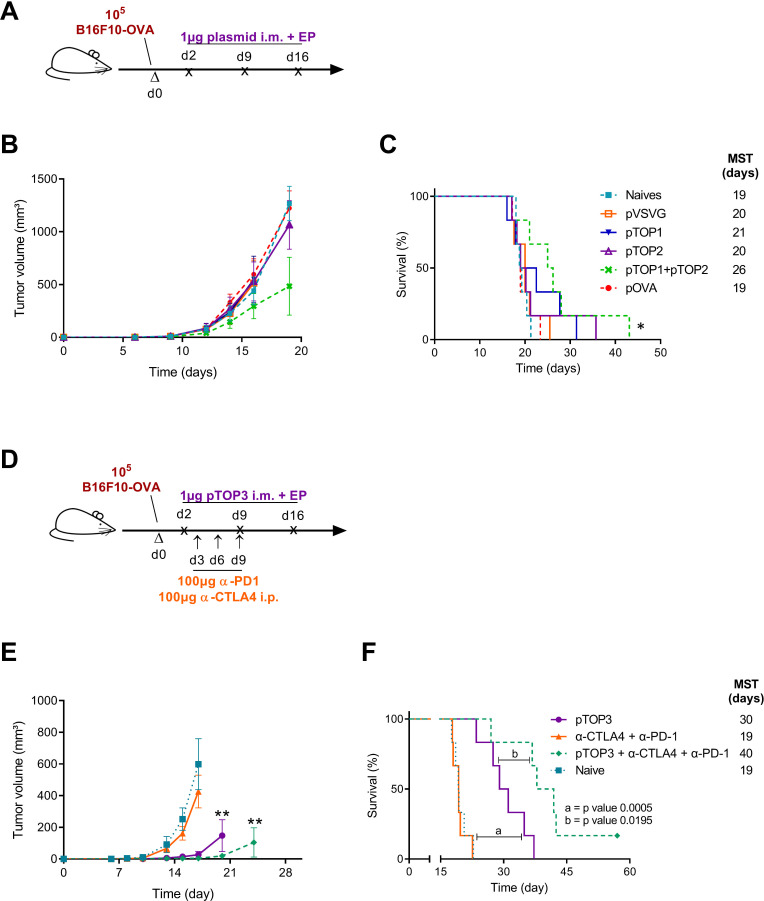
To further demonstrate the efficacy of the pTOP vaccination over the classical approach, C57Bl/6 mice have been therapeutically vaccinated 2, 9 and 16 days after the injection of 1×105 B16F10-OVA tumor cells (figure 3A and online supplemental methods). Only 1 µg of the vaccine has been used for each vaccination. A naïve group of mice and a group treated with the pVSVG plasmid encoding the VSV-G but without inserted epitopes have been used as a control. Compared with all the other groups, pTOP1 +pTOP2 was the only treatment that delayed the tumor growth, even if the difference with the other groups was not significant (figure 3B). Nevertheless, these two codelivered plasmids significantly increased the median survival time of mice (from 19 to 26 days), as showed in figure 3C. This result confirms that the delivery of pTOP vaccines confers a better immunity compared with the pOVA classical DNA vaccine and allows a more efficient antitumor response.
Figure 3.
B16F10-OVA therapeutic vaccination comparing pTOP vaccines and classical DNA vaccination, and combination with immune checkpoint blockade. (A) Schematic protocol of the B16F10-OVA injection and therapeutic DNA vaccination. C57BL/6 mice were first injected with B16F10-OVA. The DNA vaccines were intramuscularly electroporated 2, 9 and 16 days after injection of the tumor cells. (B) Tumor growth curves for the different groups. (C) percentage of survival as a function of time. The error bars represent the mean±SEM; n=6. Statistical analysis: one-way ANOVA with Tukey’s multiple comparisons test, two-way ANOVA with Bonferroni post-tests or Mantel-Cox test for comparison of survival curves. *p<0.05, compared with naive or to the specified group. (D) schematic protocol of the B16F10-OVA injection, therapeutic DNA vaccination and immune checkpoint blockade administration. C57BL/6 mice were first injected with B16F10-OVA. the DNA vaccines were intramuscularly electroporated 2, 9 and 16 days after injection of the tumor cells. Immune checkpoint blockade antibodies against CTLA4 (100 µg) and PD1 (100 µg) were injected intraperitoneally 3, 6 and 9 days after tumor injection. (E) tumor growth curves for the different groups. (F) percentage of survival as a function of time. The error bars represent the mean±SEM; n=6. Statistical analysis: One-way ANOVA with Tukey’s multiple comparisons test, two-way ANOVA with Bonferroni post-tests or Mantel-Cox test for comparison of survival curves. **p<0.01, compared with naive or to the specified group. ANOVA, analysis of variance; MST, median survival time; OVA, ovalbumin; pOVA, plasmid OVA; pTOP, plasmid to deliver T cell epitopes; pVSVG, plasmid vesicular stomatitis virus glycoprotein.
pTOP synergizes with immune checkpoint inhibitors
pTOP synergizes with immune checkpoint inhibitors pTOP3 was designed by inserting both OVA257-264 (CD8 epitope) and OVA323-339 (CD4 epitope) in the VSV-G sequence. To assess the ability of pTOP to promote therapeutic vaccination, pTOP3 was used to immunize mice that had been previously subcutaneously injected with 1×105 B16F10-OVA cells (figure 3D). Therapeutic immunization with 3 weekly administrations of 1 µg pTOP3, starting from 2 days after the injection of tumor cells, significantly delayed tumor growth and the median survival time reached 30 days, compared with 19 days for untreated (naives) mice. Three administrations at days 3, 6 and 9 of 100 µg anti-CTLA4 and 100 µg anti-PD1 antibodies alone (figure 3D) did not increase the survival of the mice but improved the efficacy of pTOP3. Indeed, the combination of pTOP3 and the two immune checkpoint blockers allowed a significant delay in the tumor growth, a dramatic increase of the median survival time and the eradication of the tumor for one out of six mice (figure 3E, F).
Insertion of gp100 and TRP2 epitopes in the pTOP plasmid allows therapeutic vaccination in multiple cancer models
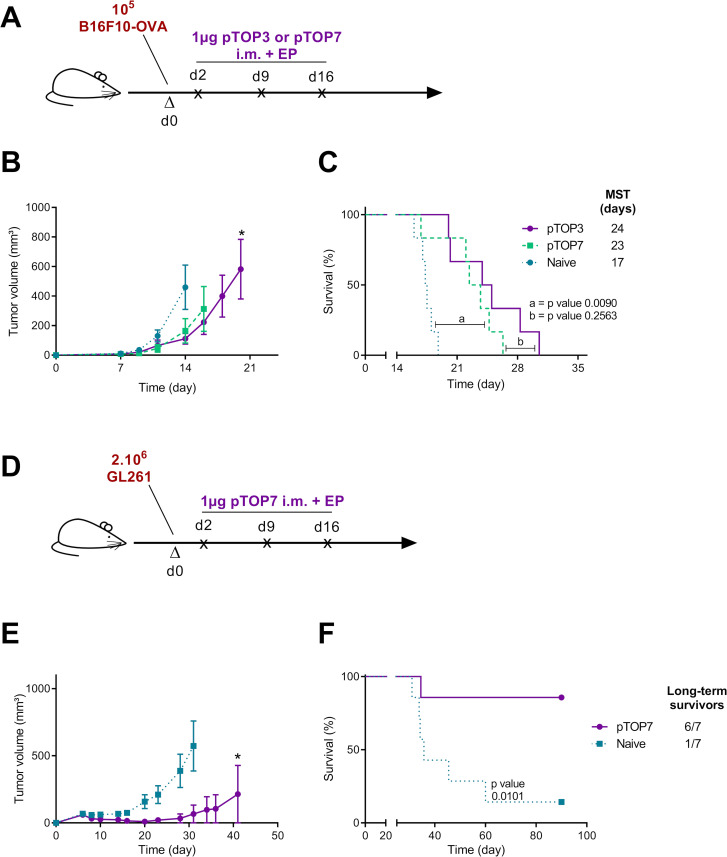
Next, we assessed whether pTOP could be used for the delivery of other tumor epitopes and then to evaluate its efficacy in a different cancer model. In particular, pTOP7 was obtained by inserting two tumor epitopes (TRP2180-188, a CD8 epitope, and gp10044-59, a CD4 epitope) in the VSV-G sequence and evaluated as a therapeutic vaccine delivered at days 2, 9 and 16 after tumor cell injection (figure 4A). First, we investigated its efficacy in a B16F10-OVA melanoma. We demonstrated that pTOP7 and pTOP3, which contained two OVA epitopes, were equally efficient against B16F10-OVA tumors (figure 4B, C). Then, we evaluated the efficacy of pTOP7, originally designed to treat B16F10 tumors, against GL261 GBM tumor model. Two million of GL261 cells have been injected subcutaneously at day 0 (a schematic representation of the vaccination protocol is shown in figure 4D). Indeed, TRP-2 and gp100 antigens are shared between melanoma and GBM due to the common prenatal origin of melanocytes and glial cells from the neural ectoderm.41 The expression of TRP2 and gp100 in GL261 cells was verified by RT-PCR (online supplemental figure 1). Tumor growth in vaccinated mice was significantly delayed compared with that of the untreated (naïve) group, and six out of seven mice were considered long-term survivors (figure 4E, F). Furthermore, long-term survivor vaccinated mice have been rechallenged with B16F10 (on the left flank) and with B16F1 (on the right flank). As expected, these tumors have been rejected or did not grow at all in the rechallenged mice (data not shown).
Figure 4.
Therapeutic immunization with pTOP vaccines in melanoma and glioblastoma models. (A) Schematic protocol, the development of tumor volume and the survival curves are shown for each tumor model. C57BL/6 mice were first injected with B16F10-OVA (A) or GL261 cells (D). The tumor growth and the survival curves for melanoma (C) and for GBM (E, F) are also shown. The pTOP vaccine was intramuscularly electroporated 2, 9 and 16 days after injection of the tumor cells. The error bars represent the mean±SEM; n=6–7. Statistical analysis: two-way ANOVA with Bonferroni post-tests or Mantel-Cox test for comparison of survival curves. *p<0.05 compared with naive. ANOVA, analysis of variance; GBM, glioblastoma; MST, median survival time; OVA, ovalbumin; pTOP, plasmid to deliver T cell epitopes.
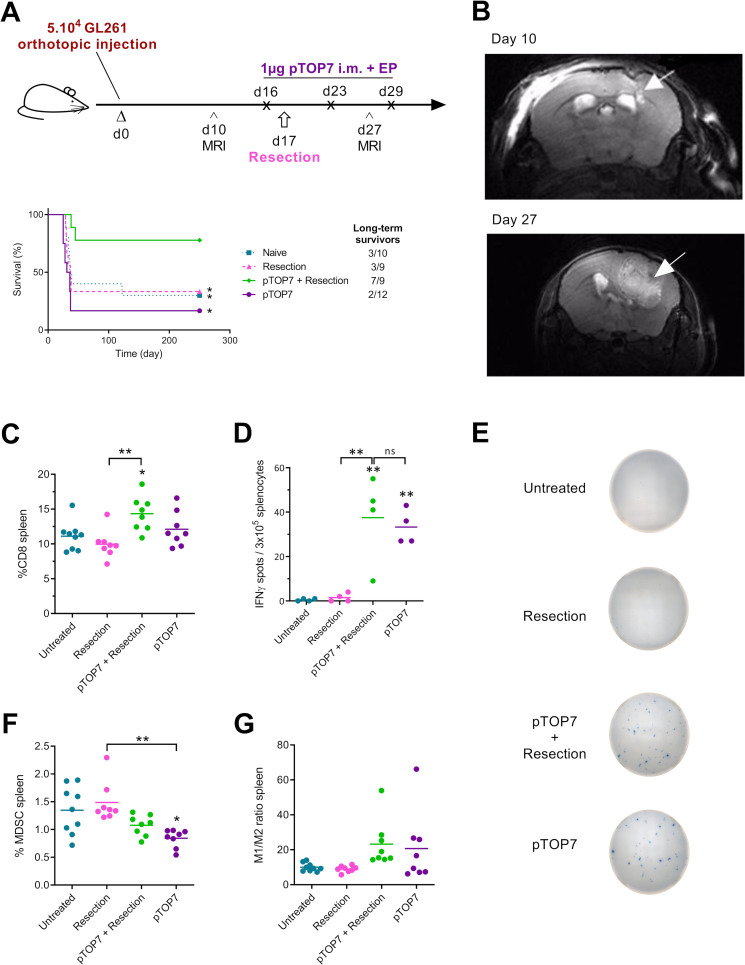
In a GL261 orthotopic model, tumor resection and pTOP vaccination significantly prolong mouse survival
As pTOP7 was highly efficient against the subcutaneously implanted GL261 tumors, we checked whether this DNA vaccine could prevent recurrences in a murine orthotopic GBM model when administered just before the surgical resection (figure 5A). Indeed, we hypothesized that the resection could help the vaccine-induced immune response to reach the brain, both by generating a local inflammatory microenvironment and by further disrupting the blood brain barrier. Tumorous lesions in GL261-bearing mice were observed between the cortex and the striatum in all implanted animals at day 10 postinoculation by MRI, as showed in figure 5B. Mice were vaccinated with 1 µg of pTOP7, at days 16, 23 and 29, and the tumor was resected 17 days after GL261 orthotopic inoculation. In the control groups (naive, resection or pTOP7), the general state of health started deteriorating from day 27–30 after the tumor injection, and the median survival time was less than 40 days. However, when resection was combined with therapeutic immunization with pTOP7, 78% of the mice survived for at least 250 days and thus were considered long-term survivors (figure 5A). MRI performed 27 days after tumor inoculation confirmed the presence of infiltrative and aggressive recurrences in the control groups (figure 5B). Due to the infiltrative patterns of the GL261 tumors, we were unable to provide adequate volume estimation of the tumor lesions at the designated time points, but the presence of the tumors and the infiltrative nature were confirmed postmortem by H&E staining (H/E) (online supplemental figure 2).
Figure 5.
Therapeutic immunization and resection in an orthotopic glioblastoma model and development of the systemic immune response. (A) Schematic protocol and survival curves for the therapeutic immunization. C57BL/6 mice first received an intracranial injection of GL261 cells at day 0. MRI was used to monitor brain tumors on days 10 and 27. The pTOP vaccine was intramuscularly electroporated 16, 23 and 19 days after the injection of tumor cells, and resection of the tumor was performed on day 17; n=9–12. (B) representative axial T2-weighted MRI image of an untreated (naïve) mouse brain before (day 10) and after tumor resection (day 27). The white arrows indicate the GL261 primary and recurrent tumors. (C–H) analysis of immune cells in the spleen 29 days after GL261 inoculation. The percentage of CD8 T cells is shown for all the groups (C), and the production of IFNγ by splenocytes stimulated with TRP2 peptide was assessed by ELISPOT (D, E). The percentage of MDSCs (F) and the ratio of M1/M2 macrophages (G) are displayed. The error bars represent the mean±SEM; n=7–9. Statistical analysis: one-way ANOVA with Tukey’s multiple comparisons test or Mantel-Cox test for comparison of survival curves. *p<0.05, **p<0.01 compared with naive or to the specified group. ANOVA, analysis of variance; IFNγ, interferon-γ; ns, not significant; MDSCs, myeloid-derived suppressor cell; pTOP, plasmid to deliver T cell epitopes.
pTOP induces a systemic antigen-specific immune response and modulates the number of immune cells in the spleen
We then evaluated systemic immune activity after resection and/or immunization with pTOP7. Splenocytes were collected 29 days after tumor challenge and analyzed by flow cytometry and ELISpot. When resection and the pTOP7 treatments were combined, the CD8 infiltration was significantly higher compared with that of the untreated (naïve) mice and those that underwent resection alone (figure 5C). The activation of TRP2-specific T cells was assessed by IFNγ ELISpot. In the absence of vaccination, only a few spots were detected, whereas the splenocytes from pTOP7-treated mice showed a significantly higher number of spots (figure 5D, E). Furthermore, the cell stimulation by using an irrelevant peptide (in the current experiment, the P815 peptide) were comparable to the stimulation with PBS (negative control) and showed no IFNγ production (data not shown). This aspect demonstrates the specificity of the pTOP7 vaccination against TRP2 peptide. In addition, the number of MDSCs was reduced when mice were treated with pTOP7 (figure 5F), and a modulation (although not statistically significant) of the M1/M2 macrophage ratio was observed in mice treated with pTOP7 with or without tumor resection (figure 5G).
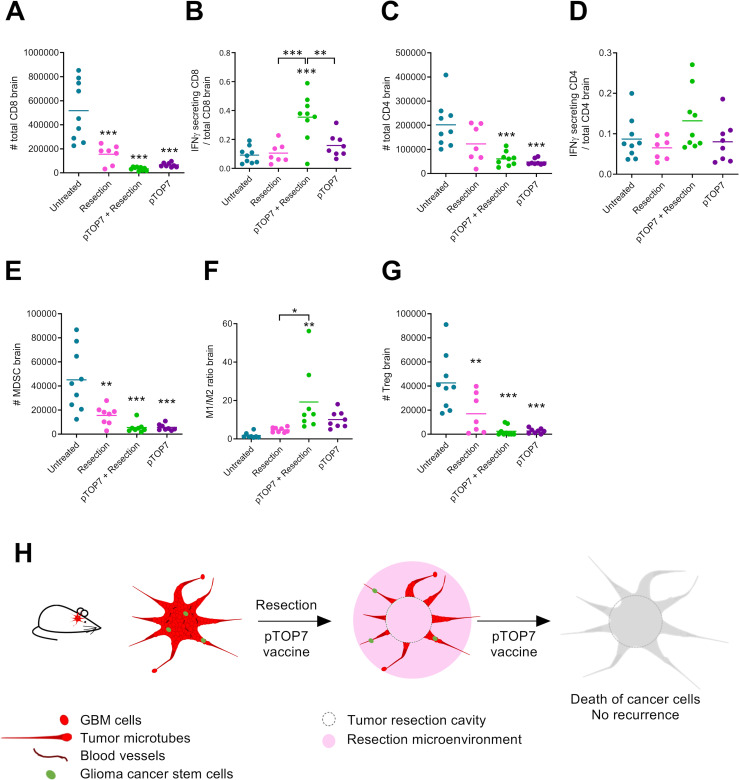
pTOP and tumor resection enhance the activity of immune cells and reduced the number of infiltrated immunosuppressive cells in the brain
To study the mechanism underlying the synergy between pTOP7 and the resection of GL261 tumors and their contribution to prolonging mouse survival, the infiltration of different immune cells was assessed in the mouse brains 29 days after tumor inoculation. Decreased infiltration of CD8 T cells was observed in the treated groups, especially in the combination group (figure 6A). However, when analyzing the activity of those cells, we observed that 35% of infiltrated CD8 T cells in the combination group produced IFNγ compared with only 9%–16% for the other groups (figure 6B). The same trend was observed for the number of CD4 T cells (figure 6C) and IFNγ-secreting CD4 T cells, but the slight increase observed in the combination group was not significantly different from that of the other groups (figure 6D). The flow cytometry analysis also revealed a significant decrease in the infiltrated immunosuppressive cells for all the groups compared with that of the untreated (naïve) group. This effect was seen for MDSCs (figure 6E), the M1/M2 macrophage ratio (figure 6F) and regulatory T cells (Tregs, figure 6G). The greatest effect on infiltrated immunosuppressive cells was observed when pTOP7 was used alone or in combination with resection, and the M1/M2 macrophage ratio was significantly higher for mice treated with the combination compared with that of untreated (naïve) mice or mice with resected tumors.
Figure 6.
Development of immune cells and immunosuppressive cells in the brain 29 days after GL261 inoculation. (A, B) Total number of CD8 T cells and the ratio of IFNγ-secreting CD8/total CD8 T cells. (C, D) Total number of CD4 T cells and the ratio of IFNγ-secreting CD4/total CD4 T cells. (E, F) percentage of MDSCs and the ratio of M1/M2 macrophages in the brain. (G) Number of Tregs in the brain. The error bars represent the mean±SEM; n=7–9. Statistical analysis: one-way ANOVA with Tukey’s multiple comparisons test. *p<0.05, **p<0.01, ***p<0.001 compared with naive or to the specified group. (H) Principle of combining resection and pTOP7 vaccination for preventing GL261 recurrences. ANOVA, analysis of variance; GBM, glioblastoma; IFNγ, interferon-γ; MDSCs, myeloid-derived suppressor cells; pTOP, plasmid to deliver T cell epitopes.
Discussion
Immunotherapy is emerging as a new therapeutic option for different tumors, and cancer vaccines represent a good approach for the induction of a specific and long-lasting immune response. However, new strategies are needed to improve the immunogenicity of the DNA vaccines and to overcome the immunosuppressive tumor microenvironment, thus activating a broad and potent immune response against cancer.
In the current study, we tested the efficacy of pTOP, a plasmid encoding a modified VSV-G protein with defined T cell epitopes, in different cancer models. We hypothesized that pTOP could: (1) activate a potent innate immune response, due to the presence of the viral protein; (2) induce a specific T cell immune response against the encoded antigens, thus potentiating the global DNA vaccine activity and (3) improve the outcome in different tumor models, when the vaccine is combined with a rational therapeutic strategy.
We demonstrated that the electroporation of a plasmid encoding the VSV-G viral protein induced a higher expression of IL12, IL6 and CCL2 cytokines and chemokines, compared with the classical pVAX2 empty plasmid (pEmpty). This suggests a strong activation of the innate immune system in the presence of VSV-G and, in particular, of the response linked to the viral recognition. Indeed, these cytokines and chemokines are involved in the viral clearance, but play also a key role in the switch from the innate to the adaptive immunity,42–46 a crucial aspect for an effective cancer vaccination. Hence, this innate response must be followed by an adaptive T cell response specifically directed against cancer antigens. We showed that the insertion of foreign epitopes in specific positions of the VSV-G sequence triggered correct antigen processing and presentation by both types of MHC molecules as well as the CTL response. Furthermore, when compared with a classical pOVA vaccine, the pTOP vaccine increased the CTL antigen-specific killing activity (figure 2D). All these aspects could explain the delay in the B16F10-OVA tumor growth and the higher median survival time observed with the pTOP vaccine, compared with the classical DNA vaccines. Interestingly, pTOP vaccine was administered after the tumor injection (therapeutic vaccination) and was active at a low dose. Indeed, only 1 µg of pTOP plasmid was delivered, while previous DNA vaccination protocols in mice have usually reported doses between 25 and 100 µg.47 48 Our experiments also demonstrated the specificity of the pTOP vaccination has by a killing assay (figure 2) and an ELISpot experiments (figure 5D).
In the same melanoma model, we demonstrated that the therapeutic vaccination with pTOP enhanced the efficiency of anti-CTLA4 and anti-PD1 antibodies. These immune checkpoint blockers were totally ineffective when used alone, but drastically increased the survival of mice when combined with the vaccine. We have already reported that the combination between pTOP DNA-based immunotherapy and an oncolytic adenovirus improved the outcome of B16F1 melanoma-bearing mice.49 Indeed, the oncolytic virus enhanced the immune response generated by the vaccine and drove the immune cells into the tumor site.49 Following these results, we hypothesized that the administration of the pTOP vaccine in combination with tumor resection could be effective against an orthotopic model of GBM. Indeed, the inflammation generated by the tumor removal could drive in the tumor microenvironment the immune response generated by the previous vaccination. pTOP was able to generate a specific and long-lasting immune response against GBM and to target residual cells in mice that underwent surgical resection (figure 6H). However, in contrast to melanoma B16F10 and GBM GL261 implanted subcutaneously, when the pTOP treatment was used as a single therapy for orthotopically implanted GL261, the survival was not improved. Even if the GBM disrupts the blood brain barrier, the presence of this blood brain barrier and the low amounts of lymphatic vessels regulate the entry of the immune cells. Furthermore, its high heterogeneity, the presence of glioma stem cells, immunologically ‘cold’ tumor environment with low tumor mutation burden and few tumor infiltrating lymphocytes as well as the suppressive TME facilitate immune escape. This absence of effect of pTOP monotherapy on survival seems consistent with clinical trials, as several studies reported that cancer vaccines are able to induce an antigen-specific immune response but do not have a good impact on the overall survival of patients.50 51 Interestingly, a recent clinical study evaluated the efficacy of an anti-PD1 therapy delivered before surgical GBM tumor resection. The tumor-infiltrating lymphocytes produced an IFNγ response within the TME and increased the median survival time.52 Here, we observed that the ratio of IFNγ-producing CD8/total CD8 T cells was significantly higher for the group combining the pTOP treatment and resection, indicating the presence of active infiltrated CD8 T cells that, although low in number, seemed not to be exhausted.53 In addition, the immunosuppressive activity was reduced, as fewer MDSCs, M2 macrophages and Tregs were observed. To our knowledge, we are the first to report the combination of GBM surgical resection and vaccine immunotherapy being performed before tumor debulking. These results confirm our hypothesis that a vaccine administration prior to surgery might take advantage of the acute inflammatory response induced by GBM resection (ie, induction of an excessive healing response, production of inflammatory cytokines and recruitment of both M1 and M2 macrophages,54–57 leading to a dramatic improvement in the therapeutic efficiency.
In the field of cancer immunotherapy, we have entered into an era of combined treatments,3 and the development of potent therapeutic anticancer vaccines may be the missing element for being able to efficiently treat more patients and a wider range of tumors. There is a strong rationale for combining cancer vaccines with other immunotherapy drugs, such as immune checkpoint inhibitors or oncolytic viruses. Perhaps more surprisingly, our results indicated that combining cancer vaccine and tumor resection allowed the infiltration of activated T cells to the resection site with a strong impact on mouse survival in an aggressive GBM preclinical model.
Taken together, this study highlights the potential of pTOP DNA-based immunotherapy as an innovative and versatile technology to induce a specific and more potent antitumor response compared with classical DNA vaccination. Furthermore, for the first time, we demonstrated that the administration of a DNA vaccine followed by the tumor resection could improve the outcome in a GBM model. All these strategies open new perspectives in the treatment of cancer.
Acknowledgments
We are grateful to Johan Grooten and Ans De Beuckelaer for providing us with the B16F10-OVA cells and the transgenic OT-I and OT-II mice. Sophie Lucas is acknowledged for the kind gift of the GL261 cells. The authors would also like to thank Olivier Dedobbeler and Nicolas Dauguet (De Duve Institute, Brussels, Belgium) for their help with the AutoMACS Pro Separator and the FACSVerse, as well as Rebecca Gould for her experimental help.
Footnotes
AL and CB contributed equally.
Contributors: AL, CB and GV designed the work, performed experiments and analysed data. SL, MB, LL and BG performed or contributed to specific experiments. AL, CB and GV prepared the manuscript. KV, SL and BU provided technical supports. VP and GV supervised the project.
Funding: GV is supported by a FIRST spin-off grant (1610437) from the Walloon Region. This work is also funded by a Proof of Concept grant (1780012) from the Walloon Region. LL was a research fellow of the FSR-FNRS (Belgium). The work was also partly supported by Fondation contre le cancer (Belgium) and FSR-FNRS (Belgium).
Competing interests: GV, LL and VP are coapplicants of a patent: 'Modified VSV-G and vaccines thereof', PCT/EP2017/073119. GV, AL, CB and VP are coapplicants of a patent: 'Modified vesicular stomatitis virus glycoprotein and uses thereof for the treatment of brain tumors', EP19199334.4. Both patents are pending.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
All animal experiments were performed following the Belgian national regulations guidelines in accordance with EU Directive 2010/63/EU, and were approved by the ethical committee for animal care of the faculty of medicine of the Université Catholique de Louvain (2014/UCL/MD/004 and 2016/UCL/MD/001).
References
- 1. Yarchoan M, Johnson BA, Lutz ER, et al. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer 2017;17:569. 10.1038/nrc.2017.74 [DOI] [PubMed] [Google Scholar]
- 2. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252–64. 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fares CM, Van Allen EM, Drake CG, et al. Mechanisms of resistance to immune checkpoint blockade: why does checkpoint inhibitor immunotherapy not work for all patients? Am Soc Clin Oncol Educ Book 2019;39:147–64. 10.1200/EDBK_240837 [DOI] [PubMed] [Google Scholar]
- 4. Yang B, Jeang J, Yang A, et al. Dna vaccine for cancer immunotherapy. Hum Vaccin Immunother 2014;10:3153–64. 10.4161/21645515.2014.980686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saade F, Petrovsky N. Technologies for enhanced efficacy of DNA vaccines. Expert Rev Vaccines 2012;11:189–209. 10.1586/erv.11.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lambricht L, Lopes A, Kos S, et al. Clinical potential of electroporation for gene therapy and DNA vaccine delivery. Expert Opin Drug Deliv 2016;13:295–310. 10.1517/17425247.2016.1121990 [DOI] [PubMed] [Google Scholar]
- 7. Lopes A, Vanvarenberg K, Préat V, et al. Codon-Optimized P1A-Encoding DNA vaccine: toward a therapeutic vaccination against P815 mastocytoma. Mol Ther Nucleic Acids 2017;8:404–15. 10.1016/j.omtn.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li L, Petrovsky N. Molecular adjuvants for DNA vaccines. Curr Issues Mol Biol 2017;22:17–40. 10.21775/cimb.022.017 [DOI] [PubMed] [Google Scholar]
- 9. Lopes A, Vandermeulen G, Préat V. Cancer DNA vaccines: current preclinical and clinical developments and future perspectives. J Exp Clin Cancer Res 2019;38:146. 10.1186/s13046-019-1154-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coulie PG, Van den Eynde BJ, van der Bruggen P, et al. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 2014;14:135–46. 10.1038/nrc3670 [DOI] [PubMed] [Google Scholar]
- 11. Knutson KL, Disis ML. Tumor antigen-specific T helper cells in cancer immunity and immunotherapy. Cancer Immunol Immunother 2005;54:721–8. 10.1007/s00262-004-0653-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ivanova EA, Orekhov AN. T helper lymphocyte subsets and plasticity in autoimmunity and cancer: an overview. Biomed Res Int 2015;2015:1–9. 10.1155/2015/327470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Slingluff CL, Petroni GR, Yamshchikov GV, et al. Immunologic and clinical outcomes of vaccination with a multiepitope melanoma peptide vaccine plus low-dose interleukin-2 administered either concurrently or on a delayed schedule. J Clin Oncol 2004;22:4474–85. 10.1200/JCO.2004.10.212 [DOI] [PubMed] [Google Scholar]
- 14. Vance RE, Eichberg MJ, Portnoy DA, et al. Listening to each other: infectious disease and cancer immunology. Sci Immunol 2017;2. 10.1126/sciimmunol.aai9339. [Epub ahead of print: 13 Jan 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bode C, Zhao G, Steinhagen F, et al. Cpg DNA as a vaccine adjuvant. Expert Rev Vaccines 2011;10:499–511. 10.1586/erv.10.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambricht L, Vanvarenberg K, De Beuckelaer A, et al. Coadministration of a plasmid encoding HIV-1 Gag enhances the efficacy of cancer DNA vaccines. Mol Ther 2016;24:1686–96. 10.1038/mt.2016.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mao C-P, Hung C-F, Kang TH, et al. Combined administration with DNA encoding vesicular stomatitis virus G protein enhances DNA vaccine potency. J Virol 2010;84:2331–9. 10.1128/JVI.01954-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Georgel P, Jiang Z, Kunz S, et al. Vesicular stomatitis virus glycoprotein G activates a specific antiviral Toll-like receptor 4-dependent pathway. Virology 2007;362:304–13. 10.1016/j.virol.2006.12.032 [DOI] [PubMed] [Google Scholar]
- 19. Errington F, Jones J, Merrick A, et al. Fusogenic membrane glycoprotein-mediated tumour cell fusion activates human dendritic cells for enhanced IL-12 production and T-cell priming. Gene Ther 2006;13:138–49. 10.1038/sj.gt.3302609 [DOI] [PubMed] [Google Scholar]
- 20. Bateman A, Bullough F, Murphy S, et al. Fusogenic membrane glycoproteins as a novel class of genes for the local and immune-mediated control of tumor growth. Cancer Res 2000;60:1492–7. [PubMed] [Google Scholar]
- 21. Bateman AR, Harrington KJ, Kottke T, et al. Viral fusogenic membrane glycoproteins kill solid tumor cells by nonapoptotic mechanisms that promote cross presentation of tumor antigens by dendritic cells. Cancer Res 2002;62:6566–78. [PubMed] [Google Scholar]
- 22. Linardakis E, Bateman A, Phan V, et al. Enhancing the efficacy of a weak allogeneic melanoma vaccine by viral fusogenic membrane glycoprotein-mediated tumor cell-tumor cell fusion. Cancer Res 2002;62:5495–504. [PubMed] [Google Scholar]
- 23. García-Valtanen P, Ortega-Villaizán MDM, Martínez-López A, et al. Autophagy-Inducing peptides from mammalian VSV and fish VHSV rhabdoviral G glycoproteins (G) as models for the development of new therapeutic molecules. Autophagy 2014;10:1666–80. 10.4161/auto.29557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gannage M, Münz C. Mhc presentation via autophagy and how viruses escape from it. Semin Immunopathol 2010;32:373–81. 10.1007/s00281-010-0227-7 [DOI] [PubMed] [Google Scholar]
- 25. Ammayappan A, Peng K-W, Russell SJ. Characteristics of oncolytic vesicular stomatitis virus displaying tumor-targeting ligands. J Virol 2013;87:13543–55. 10.1128/JVI.02240-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schlehuber LD, Rose JK. Prediction and identification of a permissive epitope insertion site in the vesicular stomatitis virus glycoprotein. J Virol 2004;78:5079–87. 10.1128/JVI.78.10.5079-5087.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu H, Zhuang Q, Xu J, et al. Cell-Penetrating peptide enhanced antigen presentation for cancer immunotherapy. Bioconjug Chem 2019;30:2115–26. 10.1021/acs.bioconjchem.9b00245 [DOI] [PubMed] [Google Scholar]
- 28. Belnoue E, Mayol J-F, Carboni S. Targeting self and neo-epitopes with a modular self-adjuvanting cancer vaccine. JCI Insight 2019;5. [Epub ahead of print: 23 Apr 2019]. 10.1172/jci.insight.127305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Celik C, Lewis DA, Goldrosen MH. Demonstration of immunogenicity with the poorly immunogenic B16 melanoma. Cancer Res 1983;43:3507–10. [PubMed] [Google Scholar]
- 30. Tüting T. T cell immunotherapy for melanoma from bedside to bench to barn and back: how conceptual advances in experimental mouse models can be translated into clinical benefit for patients. Pigment Cell Melanoma Res 2013;26:441–56. 10.1111/pcmr.12111 [DOI] [PubMed] [Google Scholar]
- 31. Overwijk WW, Restifo NP. B16 as a mouse model for human melanoma. Curr Protoc Immunol 2001;Chapter 20:21. 10.1002/0471142735.im2001s39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tivnan A, Heilinger T, Lavelle EC, et al. Advances in immunotherapy for the treatment of glioblastoma. J Neurooncol 2017;131:1–9. 10.1007/s11060-016-2299-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Absinta M, Ha S-K, Nair G, et al. Human and nonhuman primate meninges harbor lymphatic vessels that can be visualized noninvasively by MRI. Elife 2017;6. 10.7554/eLife.29738. [Epub ahead of print: 03 Oct 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Louveau A, Harris TH, Kipnis J. Revisiting the mechanisms of CNS immune privilege. Trends Immunol 2015;36:569–77. 10.1016/j.it.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell 2017;31:326–41. 10.1016/j.ccell.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yabroff KR, Harlan L, Zeruto C, et al. Patterns of care and survival for patients with glioblastoma multiforme diagnosed during 2006. Neuro Oncol 2012;14:351–9. 10.1093/neuonc/nor218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weller M, van den Bent M, Tonn JC, et al. European association for neuro-oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol 2017;18:e315–29. 10.1016/S1470-2045(17)30194-8 [DOI] [PubMed] [Google Scholar]
- 38. Wick W, Platten M. Understanding and treating glioblastoma. Neurol Clin 2018;36:485–99. 10.1016/j.ncl.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 39. Osswald M, Jung E, Sahm F, et al. Brain tumour cells interconnect to a functional and resistant network. Nature 2015;528:93–8. 10.1038/nature16071 [DOI] [PubMed] [Google Scholar]
- 40. Bianco J, Bastiancich C, Joudiou N, et al. Novel model of orthotopic U-87 Mg glioblastoma resection in athymic nude mice. J Neurosci Methods 2017;284:96–102. 10.1016/j.jneumeth.2017.04.019 [DOI] [PubMed] [Google Scholar]
- 41. Chi DD, Merchant RE, Rand R, et al. Molecular detection of tumor-associated antigens shared by human cutaneous melanomas and gliomas. Am J Pathol 1997;150:2143–52. [PMC free article] [PubMed] [Google Scholar]
- 42. Dienz O, Rud JG, Eaton SM, et al. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol 2012;5:258–66. 10.1038/mi.2012.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hurst SM, Wilkinson TS, McLoughlin RM, et al. Il-6 and its soluble receptor orchestrate a temporal switch in the pattern of leukocyte recruitment seen during acute inflammation. Immunity 2001;14:705–14. 10.1016/S1074-7613(01)00151-0 [DOI] [PubMed] [Google Scholar]
- 44. Monteiro JM, Harvey C, Trinchieri G. Role of interleukin-12 in primary influenza virus infection. J Virol 1998;72:4825–31. 10.1128/JVI.72.6.4825-4831.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stubblefield Park SR, Widness M, Levine AD, et al. T cell-, interleukin-12-, and gamma interferon-driven viral clearance in measles virus-infected brain tissue. J Virol 2011;85:3664–76. 10.1128/JVI.01496-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perussia B, Chan SH, D'Andrea A, et al. Natural killer (NK) cell stimulatory factor or IL-12 has differential effects on the proliferation of TCR-alpha beta+, TCR-gamma delta+ T lymphocytes, and NK cells. J Immunol 1992;149:3495. [PubMed] [Google Scholar]
- 47. Vandermeulen G, Vanvarenberg K, De Beuckelaer A, et al. The site of administration influences both the type and the magnitude of the immune response induced by DNA vaccine electroporation. Vaccine 2015;33:3179–85. 10.1016/j.vaccine.2015.05.005 [DOI] [PubMed] [Google Scholar]
- 48. Thalmensi J, Pliquet E, Liard C, et al. Anticancer DNA vaccine based on human telomerase reverse transcriptase generates a strong and specific T cell immune response. Oncoimmunology 2016;5:e1083670. 10.1080/2162402X.2015.1083670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lopes A, Feola S, Ligot S, et al. Oncolytic adenovirus drives specific immune response generated by a poly-epitope pDNA vaccine encoding melanoma neoantigens into the tumor site. J Immunother Cancer 2019;7:174 10.1186/s40425-019-0644-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Swartz AM, Shen SH, Salgado MA, et al. Promising vaccines for treating glioblastoma. Expert Opin Biol Ther 2018;18:1159–70. 10.1080/14712598.2018.1531846 [DOI] [PubMed] [Google Scholar]
- 51. Young JS, Dayani F, Morshed RA. Immunotherapy for high grade gliomas: a clinical update and practical considerations for neurosurgeons. World Neurosurg 2019. 10.1016/j.wneu.2018.12.222. [Epub ahead of print: 21 Jan 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cloughesy TF, Mochizuki AY, Orpilla JR, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med 2019;25:477–86. 10.1038/s41591-018-0337-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486–99. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Okolie O, Bago JR, Schmid RS, et al. Reactive astrocytes potentiate tumor aggressiveness in a murine glioma resection and recurrence model. Neuro Oncol 2016;18:1622–33. 10.1093/neuonc/now117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weil S, Osswald M, Solecki G, et al. Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro Oncol 2017;19:1316–26. 10.1093/neuonc/nox070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu H, Leiss L, Yang N, et al. Surgical debulking promotes recruitment of macrophages and triggers glioblastoma phagocytosis in combination with CD47 blocking immunotherapy. Oncotarget 2017;8:12145–57. 10.18632/oncotarget.14553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hamard L, Ratel D, Selek L, et al. The brain tissue response to surgical injury and its possible contribution to glioma recurrence. J Neurooncol 2016;128:1–8. 10.1007/s11060-016-2096-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001243supp001.pdf (511.9KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information.