Abstract
Within the field of mycology, macrofungi have been relatively well-studied when compared to microfungi. However, the diversity and distribution of microfungi inhabiting woody material have not received the same degree of research attention, especially in relatively unexplored regions, such as Yunnan Province, China. To help address this knowledge gap, we collected and examined fungal specimens from different plants at various locations across Yunnan Province. Our investigation led to the discovery of four species that are clearly distinct from extant ones. These taxonomic novelties were recognized based on morphological comparisons coupled with phylogenetic analyses of multiple gene sequences (non-translated loci and protein-coding regions). The monotypic genus Neoheleiosa gen. nov. (type: N. lincangensis) is introduced in Monoblastiaceae (Monoblastiales) for a woody-based saprobic ascomycete that possesses globose to subglobose or obpyriform ascomata with centric or eccentric, papillate ostioles, an ascomatal wall with thin-walled cells of textura globulosa, cylindric, pedicellate asci with an ocular chamber, and 1-septate, brown, guttulate, longitudinally striated, bicellular ascospores. Neoheleiosa has a close phylogenetic affinity to Heleiosa, nevertheless, it is morphologically dissimilar by its peridium cells and ornamented ascospores. Acrocalymma hongheense and A. yuxiense are described and illustrated as new species in Acrocalymmaceae. Acrocalymma hongheense is introduced with sexual and asexual (coelomycetous) features. The sexual morph is characterized by globose to subglobose, ostiolate ascomata, a peridium with textura angularis cells, cylindric-clavate asci with a furcate to truncate pedicel and an ocular chamber, hyaline, fusiform, 1-septate ascospores which are surrounded by a thick, distinct sheath, and the asexual morph is featured by pycnidial conidiomata, subcylindrical, hyaline, smooth, annelledic, conidiogenous cells, hyaline, guttulate, subcylindrical, aseptate conidia with mucoid ooze at the apex and with a rounded hilum at the base. Acrocalymma yuxiense is phylogenetically distinct from other extant species of Acrocalymma and differs from other taxa in Acrocalymma in having conidia with three vertical eusepta. Magnibotryascoma kunmingense sp. nov. is accommodated in Teichosporaceae based on its coelomycetous asexual morph which is characterized by pycnidial, globose to subglobose, papillate conidiomata, enteroblastic, annelledic, discrete, cylindrical to oblong, hyaline conidiogenous cells arising from the inner layer of pycnidium wall, subglobose, oval, guttulate, pale brown and unicelled conidia.
Keywords: Ascomycota, coelomycetes, Honghe, Kunming, Pleosporales, Yuxi
Introduction
Dothideomycetes is the largest and most ecologically diverse class of Ascomycota (Hongsanan et al., 2020a), consisting of 28,729 species (Kirk, 2019). This class comprises saprobes, human and plant pathogens, endophytes, epiphytes, lichens, lichenicolous, nematode-trapping and rock-inhabiting members (Jeewon et al., 2017, 2018; Zhang J.F. et al., 2019; Hongsanan et al., 2020a). Hyde et al. (2013) provided the first comprehensive monograph of the families in Dothideomycetes. Since then, the taxonomies of Dothideomycetes have been updated with new taxa in several journal series, e.g., Fungal Diversity notes, Fungal planet description sheets, Mycosphere notes, Fungal Biodiversity Profiles, Fungal Systematics and Evolution, New and Interesting Fungi. Wijayawardene et al. (2014) provided an outline for the proposals of protection or suppression of generic names of Dothideomycetes. Consistent with the “one fungus-one name” concept, Rossman et al. (2015) provided recommendations for the nomenclature of pleomorphic genera in the class. Attributable to the continual changes of the taxa in this class, the taxonomy of Dothideomycetes is in a perpetual transitional state (Pem et al., 2019a), and as such, the outline of this class has been frequently revised (Wijayawardene et al., 2017, 2020). Recent publications by Hongsanan et al. (2020a, b) expanded the taxonomic concepts of families in the Dothideomycetidae, Pleosporomycetidae, and orders and families incertae sedis in Dothideomycetes. Hongsanan et al. (2020a, b) have accepted 38 orders and 210 families in Dothideomycetes. In order to provide a suitable platform to bring these data together, the website https://www.dothideomycetes.org was established by Pem et al. (2019a). Liu et al. (2017) proposed divergence time estimates as additional evidence for rearranging the internal classification of this class and this has been helpful to establish new families and species (Zhang S.N. et al., 2019, Bhunjun et al., 2021). The most recent order-level multi-gene phylogeny for Dothideomycetes is provided by Maharachchikumbura et al. (2021), which also introduced another two orders viz. Homortomycetales and Holmiellales, bringing the total number of orders to 40 in the class.
Monoblastiaceae is the only family in Monoblastiales comprising both lichenized and non-lichenized ascomycetes. Wijayawardene et al. (2020) accepted six genera in this family. Initiated by Hyde et al. (2020); Hongsanan et al. (2020b) synonymized Eriomycetaceae under Monoblastiaceae. Accordingly, this family currently includes 11 genera (Hongsanan et al., 2020b). The majority of these fungi grow on bark in tropical forests, but the family is also commonly found in leaf-inhabiting lichen communities (Aptroot and Sipman, 1993; Lücking, 2008). These foliicolous lichens can be useful in monitoring the environmental health of tropical forest ecosystems (Hongsanan et al., 2020b).
Alcorn and Irwin (1987) introduced Acrocalymma to accommodate A. medicaginis, which was previously identified as Stagonospora meliloti, known for causing root and foliar rot of Medicago sativa in Australia. In the phylogenetic analyses of Trakunyingcharoen et al. (2014), Acrocalymma species (A. aquatica, A. cycadis, A. fici, A. medicaginis, and A. vagum) represented an undefined lineage in Dothideomycetes for which the family name Acrocalymmaceae was introduced. They also showed that Massarina walkeri and A. medicaginis are congeneric and thus introduced a new combination, A. walkeri. Recently, Jayasiri et al. (2019) introduced another species, Acrocalymma pterocarpi, to this family. Dong et al. (2020) introduced the most recent species, Acrocalymma bipolare, a freshwater species recovered from submerged wood in the Nile River, Egypt.
Teichosporaceae was established by Barr (2002) based on morphological similarities of Bertiella, Byssothecium, Chaetomastia, Immotthia, Loculohypoxylon, Moristroma, Sinodidymella, and the type genus Teichospora. However, Moristroma, Byssothecium and Bertiella were excluded from the family by Lumbsch and Huhndorf (2010). Jaklitsch et al. (2016) revised Teichosporaceae and accepted only Teichospora in the family. The family Floricolaceae was also synonymized under Teichosporaceae by Jaklitsch et al. (2016), and every genus within this family became a synonym of Teichospora. however this current taxonomic rearrangement needs to be verified with wider sampling. In addition subsequent outlines did not follow the monotypic nature of Teichosporaceae (Wijayawardene et al., 2018). Currently, Teichosporaceae contains thirteen accepted genera (Hongsanan et al., 2020a).
The present research paper introduces two new species in Acrocalymmaceae, one new genus and a new species in Monoblastiaceae and one new species in Teichosporaceae from fifteen specimens collected from Honghe, Kunming, Lincang, Qujing, and Yuxi in Yunnan Province, China.
Materials and Methods
Isolates and Specimens
Fungal materials were collected from various deciduous trees and dried woody litter in Yunnan Province, China during the dry season. Collected samples were brought to the laboratory in Zip lock plastic bags. Samples were examined with an Olympus SZ61 Series microscope. Single ascospore isolation was carried out following the method described in Senanayake et al. (2020). Germinated spores were individually transferred to potato dextrose agar (PDA) plates and grown at 20°C in daylight. The living cultures were deposited at the Kunming Institute of Botany Culture Collection (KUMCC), Kunming, China, and duplicated at the China General Microbiological Culture Collection Center (CGMCC). Dry herbarium materials have been stored in the herbarium of Cryptogams Kunming Institute of Botany, Academia Sinica (KUN-HKAS). MycoBank numbers have been registered as outlined in MycoBank1.
Morphological Observations
In hand sections of the ascomata/conidiomata, which were mounted in distilled water, the following characteristics were evaluated: ascomata/conidiomata diameter, height, color, and shape; width of peridium; and height and diameter of ostioles. Length and width (at the widest point) of asci, ascospores, conidiophores and conidia were also measured. Images were captured with a Canon EOS 600D digital camera fitted to a Nikon ECLIPSE Ni compound microscope. Measurements were made with the Tarosoft (R) Image Frame Work program, and images used for figures were processed with Adobe Photoshop CS5 Extended version 10.0 software (Adobe Systems, United States).
DNA Extraction, PCR Amplifications, and Sequencing
Genomic DNA was extracted from the axenic mycelium as described by Phookamsak et al. (2017). When the spores failed to germinate in culture, DNA was extracted directly from the fruiting bodies of the fungus as outlined by Wanasinghe et al. (2018b). DNA to be used as templates for Polymerase chain reaction (PCR) were stored at 4°C for use in regular work and duplicated at -20°C for long-term storage.
The primers and PCR protocols for each gene were conducted by following Thiyagaraja et al. (2020a) and Wanasinghe et al. (2020). PCR was carried out at a volume of 25 μl, which contained 12.5 μl of 2× Power Taq PCR MasterMix (Bioteke Co., China), 1 μl of each primer (10 μM), 1 μl genomic DNA and 9.5 μl deionized water. The amplified PCR fragments were sent to a commercial sequencing provider (BGI, Ltd., Shenzhen, China). The nucleotide sequence data acquired were deposited in GenBank (Table 1).
TABLE 1.
Taxa used in the phylogenetic analysis of Monoblastiaceae and their corresponding GenBank numbers.
| Species | Strain no. | GenBank accession no. |
||||
| ITS | LSU | SSU | tef1 | mtSSU | ||
| Acrocordia subglobosa | HTL940 | NA | JN887392 | JN887373 | JN887417 | GU327681 |
| Anisomeridium cf. willeyanum | MPN549 | NA | JN887393 | NA | NA | JN887407 |
| Anisomeridium phaeospermum | MPN539 | NA | JN887394 | JN887374 | NA | NA |
| Anisomeridium sp. | MPN533 | NA | JN887395 | JN887375 | JN887419 | NA |
| Anisomeridium sp. | MPN540 | NA | JN887397 | JN887377 | JN887420 | JN887409 |
| Anisomeridium sp. | MPN534 | NA | JN887396 | JN887376 | NA | JN887408 |
| Anisomeridium sp. | MPN542 | NA | JN887398 | JN887378 | NA | JN887410 |
| Anisomeridium ubianum | MPN94 | NA | GU327709 | JN887379 | NA | GU327682 |
| Elsinoe centrolobii | CBS 222.50 | NR_148132 | KX886969 | NG_062717 | DQ677934 | NA |
| Elsinoe phaseoli | CBS 165.31 | NR_148161 | DQ678095 | NG_062718 | DQ677935 | NA |
| Eriomyces heveae | MFLUCC 17-2232 | NR_169673 | MH109524 | NA | NA | NA |
| Funbolia dimorpha | CPC 14170 | JF951136 | JF951156 | JF951136 | NA | NA |
| Funbolia dimorpha | CBS 126491 | NA | NG_064276 | NA | NA | NA |
| Heleiosa barbatula | JK 5548I | NA | GU479787 | GU479753 | NA | NA |
| Italiofungus phillyreae | CPC 35566 | MT223804 | MT223899 | NA | NA | NA |
| Megalotremis verrucosa | Lucking 26316 | NA | GU327718 | JN887383 | NA | GU327694 |
| Myriangium hispanicum | CBS 247.33 | KX887304 | GU301854 | GU296180 | GU349055 | NA |
| Neoheleiosa lincangensis | HKAS 111911 | MW424766 | MW424781 | MW424796 | MW430102 | MW422272 |
| Neoheleiosa lincangensis | HKAS 111912 | MW424765 | MW424780 | MW424795 | MW430101 | MW422273 |
| Neoheleiosa lincangensis | HKAS 111913 | MW424767 | MW424782 | MW424797 | MW430103 | MW422274 |
| Neoheleiosa lincangensis | HKAS 111914 | MW424764 | MW424779 | MW424794 | MW430100 | MW422275 |
| Phellinocrescentia guianensis | CBS 138913 | NR_137935 | NG_058119 | NA | NA | NA |
| Pseudopassalora gouriqua | CBS 101954 | NR_160207 | NG_067272 | NA | NA | NA |
| Pseudopassalora gouriqua | CPC 1811 | NA | JN712565 | NA | NA | NA |
| Trypetheliopsis kalbii | MPN243 | NA | JN887406 | JN887391 | NA | GU327703 |
The newly generated sequences are indicated in bold. NA: Sequence data not available in GenBank.
Molecular Phylogenetic Analyses
Sequencing and Sequence Alignment
Sequences generated from different primers were analyzed along with sequences retrieved from GenBank (Tables 1–3). Sequences with high similarity indices were determined from a BLAST search to find the closest matches with taxa in Pleosporales, and from recently published data (Thambugala et al., 2015; Jaklitsch et al., 2016; Jayasiri et al., 2019; Hongsanan et al., 2020b). The multiple alignments of all consensus sequences, as well as the reference sequences were automatically generated with MAFFT v. 7 (Kuraku et al., 2013; Katoh et al., 2019)2, and improved manually when necessary using BioEdit v. 7.0.9.0 (Hall, 1999).
TABLE 3.
Taxa used in the phylogenetic analysis of Teichosporaceae and their corresponding GenBank numbers.
| Species | Strain no. | GenBank accession no. |
||||
| ITS | LSU | SSU | rpb2 | tef1 | ||
| Asymmetrispora mariae | C134m | KU601580 | KU601580 | NA | NA | KU601614 |
| Asymmetrispora mariae | C139 | KU601582 | KU601582 | NA | NA | KU601615 |
| Asymmetrispora mariae | C144 | KU601583 | KU601583 | NA | NA | KU601613 |
| Asymmetrispora mariae | C159 | KU601584 | KU601584 | NA | NA | KU601612 |
| Asymmetrispora mariae | CBS 124079 | NA | JN851819 | NA | NA | KR075166 |
| Asymmetrispora tennesseensis | ANM 911 | NA | GU385207 | NA | NA | GU327769 |
| Aurantiascoma minimum | ANM 60 | NA | GU385182 | NA | NA | NA |
| Aurantiascoma minimum | ANM 933 | NA | GU385195 | NA | NA | NA |
| Aurantiascoma minimum | GKM 169N | NA | GU385165 | NA | NA | GU327768 |
| Aurantiascoma minimum | SMH 2448 | NA | GU385166 | NA | NA | NA |
| Floricola clematidis | MFLUCC 17–2182 | MT310638 | MT214594 | MT226706 | NA | MT394651 |
| Floricola striata | JK 5603K | NA | GU479785 | GU479751 | NA | NA |
| Floricola striata | JK 5678I | NA | GU301813 | GU296149 | GU371758 | GU479852 |
| Floricola viticola | IT-2178 | KT305997 | KT305993 | KT305995 | NA | NA |
| Magnibotryascoma acaciae | CPC 24801 | KR611877 | KR611898 | NA | NA | NA |
| Magnibotryascoma kunmingense | KUMCC 20-0254 | MW424769 | MW424784 | MW424799 | MW430112 | MW430105 |
| Magnibotryascoma kunmingense | KUMCC 20-0255 | MW424770 | MW424785 | MW424800 | MW430113 | MW430106 |
| Magnibotryascoma kunmingense | KUMCC 20-0256 | MW424768 | MW424783 | MW424798 | MW430111 | MW430104 |
| Magnibotryascoma kunmingense | KUMCC 20-0257 | MW424771 | MW424786 | MW424801 | MW430114 | MW430107 |
| Magnibotryascoma kunmingense | KUMCC 20-0259 | MW424772 | MW424787 | MW424802 | MW430115 | MW430108 |
| Magnibotryascoma kunmingense | KUMCC 20-0260 | MW424773 | MW424788 | MW424803 | MW430116 | MW430109 |
| Magnibotryascoma kunmingense | KUMCC 20-0261 | MW424774 | MW424789 | MW424804 | MW430117 | MW430110 |
| Magnibotryascoma mali | MFLUCC 17-0933 | MF173433 | MF173429 | MF173431 | MF173437 | MF173435 |
| Magnibotryascoma melanommoides | MP5 | KU601585 | KU601585 | NA | NA | KU601610 |
| Magnibotryascoma rubriostiolatum | C158 | KU601587 | KU601587 | NA | KU601596 | KU601607 |
| Magnibotryascoma rubriostiolatum | C158x | KU601588 | KU601588 | NA | KU601597 | KU601608 |
| Magnibotryascoma rubriostiolatum | TR5 | KU601589 | KU601589 | NA | KU601598 | KU601606 |
| Magnibotryascoma rubriostiolatum | TR7 | KU601590 | KU601590 | NA | KU601599 | KU601609 |
| Magnibotryascoma sp. | MFLUCC 12-0088 | NA | KF531927 | KF531928 | NA | NA |
| Magnibotryascoma uniseriatum | ANM 909 | NA | GU385206 | NA | NA | NA |
| Misturatosphaeria aurantonotata | ATCC 42522 | NA | U43479 | U43461 | AY485625 | NA |
| Misturatosphaeria aurantonotata | GKM 1238 | NA | GU385173 | NA | NA | GU327761 |
| Misturatosphaeria aurantonotata | GKM 1280 | NA | GU385174 | NA | NA | GU327762 |
| Misturatosphaeria aurantonotata | SMH 4330 | NA | GU385167 | NA | NA | GU327770 |
| Misturatosphaeria sp. | SMH 3747 | NA | GU385196 | NA | NA | NA |
| Paulkirkia arundinis | MFLUCC 12-0328 | NA | KU848206 | NA | NA | NA |
| Pseudoaurantiascoma kenyense | GKM 1195 | NA | GU385194 | NA | NA | GU327767 |
| Pseudoaurantiascoma kenyense | GKM 234N | NA | GU385188 | NA | NA | GU327765 |
| Pseudoaurantiascoma kenyense | GKM L100Na | NA | GU385189 | NA | NA | GU327766 |
| Ramusculicola clematidis | MFLUCC 17-2146 | MT310640 | MT214596 | MT226707 | MT394707 | MT394652 |
| Ramusculicola thailandica | MFLUCC 10-0126 | KP899138 | KP888644 | KP899130 | NA | KR075170 |
| Ramusculicola thailandica | MFLUCC 13-0284 | KP899141 | KP888647 | KP899131 | NA | KR075167 |
| Teichospora auroafricana | CBS 119330 | EU552115 | EU552115 | NA | NA | NA |
| Teichospora auroafricana | CBS 122674 | MH863228 | MH874755 | NA | NA | NA |
| Teichospora claviformis | GKM 1210 | NA | GU385212 | NA | NA | GU327763 |
| Teichospora grandicipis | CPC 1852 | JN712456 | JN712520 | NA | NA | NA |
| Teichospora grandicipis | CPC 1853 | JN712457 | JN712521 | NA | NA | NA |
| Teichospora kingiae | CPC 29104 | KY173468 | KY173557 | NA | NA | NA |
| Teichospora mariae | C136 | KU601581 | KU601581 | NA | KU601595 | KU601611 |
| Teichospora nephelii | CPC 27539 | KY173469 | KY173558 | NA | NA | NA |
| Teichospora proteae | CBS 122675 | NA | EU552117 | NA | NA | NA |
| Teichospora pusilla | C140 | KU601586 | KU601586 | NA | NA | KU601605 |
| Teichospora quercus | CBS 143396 | MH107920 | MH107966 | NA | MH108010 | MH108030 |
| Teichospora trabicola | C134 | KU601591 | KU601591 | NA | KU601600 | KU601601 |
| Teichospora trabicola | C141 | KU601592 | KU601592 | NA | NA | KU601603 |
| Teichospora trabicola | C160 | KU601594 | KU601594 | NA | NA | KU601602 |
| Torula chromolaenae | MFLUCC 17-1514 | MT214383 | MT214477 | MT214428 | MT235831 | MT235792 |
| Torula chromolaenae | MFLUCC 17-1504 | MT214384 | MT214478 | MT214429 | MT235832 | MT235793 |
The newly generated sequences are indicated in bold. NA: Sequence data not available in GenBank.
TABLE 2.
Taxa used in the phylogenetic analysis of Acrocalymmaceae and their corresponding GenBank numbers.
| Species | Strain no. | GenBank accession no. |
||
| SSU | LSU | ITS | ||
| Acrocalymma ampeli | MFLU 19-2734 | MW079341 | MW063211 | MW063150 |
| Acrocalymma ampeli | NCYU19-0008 | MW079342 | MW063212 | MW063151 |
| Acrocalymma aquaticum | MFLUCC 11-0208 | JX276953 | NG_042698 | NR_121544 |
| Acrocalymma fici | CBS 317.76 | NA | NG_057056 | NR_137953 |
| Acrocalymma hongheense | HKAS 111907 | MW424792 | MW424777 | MW424763 |
| Acrocalymma hongheense | HKAS 111908 | MW424791 | MW424776 | MW424762 |
| Acrocalymma hongheense | HKAS 111909 | MW424790 | MW424775 | MW424761 |
| Acrocalymma medicaginis | MFLUCC 17-1439 | MT214388 | MT214433 | MT214339 |
| Acrocalymma medicaginis | MFLUCC 17-1423 | MT214387 | MT214432 | MT214338 |
| Acrocalymma medicaginis | CPC 24340 | NA | KP170713 | KP170620 |
| Acrocalymma pterocarpi | MFLUCC 17-0926 | MK347840 | NG_066306 | NA |
| Acrocalymma pterocarpi | NC13-171 | NA | LC517881 | LC517880 |
| Acrocalymma vagum | CPC 24226 | NA | NA | KP170636 |
| Acrocalymma vagum | CPC 24225 | NA | NA | KP170635 |
| Acrocalymma walkeri | UTHSC DI16-195 | NA | LN907338 | NA |
| Acrocalymma yuxiense | HKAS 111910 | MW424793 | MW424778 | NA |
| Ascocylindrica marina | MD6011 | KT252905 | KT252907 | NA |
| Ascocylindrica marina | MF416 | MK007124 | MK007123 | NA |
| Boeremia exigua | CBS 431.74 | EU754084 | EU754183 | FJ427001 |
The newly generated sequences are indicated in bold. NA: Sequence data not available in GenBank.
Phylogenetic Analyses
The single-locus datasets were examined for topological incongruences among loci for members of the analyses. The alignments were concatenated into a multi-locus alignment that was subjected to maximum-likelihood (ML) and Bayesian (BI) phylogenetic analyses.
The CIPRES Science Gateway platform (Miller et al., 2010) was used to perform RAxML and Bayesian analyses. ML analyses were made with RAxML-HPC2 on XSEDE v. 8.2.10 (Stamatakis, 2014) using GTR + GAMMA swap model with 1,000 bootstrap repetitions.
Evolutionary models for Bayesian analysis were selected independently for each locus using MrModeltest v. 2.3 (Nylander et al., 2008) under the Akaike Information Criterion (AIC) implemented in both PAUP v. 4.0b10 and GTR + I + G was selected as the best fit model for all three analyses. MrBayes analyses were performed setting GTR + I + G, 5 M generations, sampling every 1,000 generations, ending the run automatically when standard deviation of split frequencies dropped below 0.01 with a burn-in fraction of 0.25. ML bootstrap values equal or greater than 60% and BYPP greater than 0.95 are given above each node of every trees.
Phylograms were visualized with FigTree v1.4.0 program (Rambaut, 2014) and reorganized in Microsoft power point (2007) and Adobe Illustrator® CS5 (Version 15.0.0, Adobe®, San Jose, CA). The finalized alignments and trees were deposited in TreeBASE, submission ID: 275993.
Results
Phylogenetic Analyses
Three analyses were performed in this study; the first is an updated phylogeny of the genera in Monoblastiaceae (Figure 1), whereas the remaining two datasets represent taxa in Acrocalymmaceae (Figure 2) and genera treated in Teichosporaceae (Figure 3), respectively.
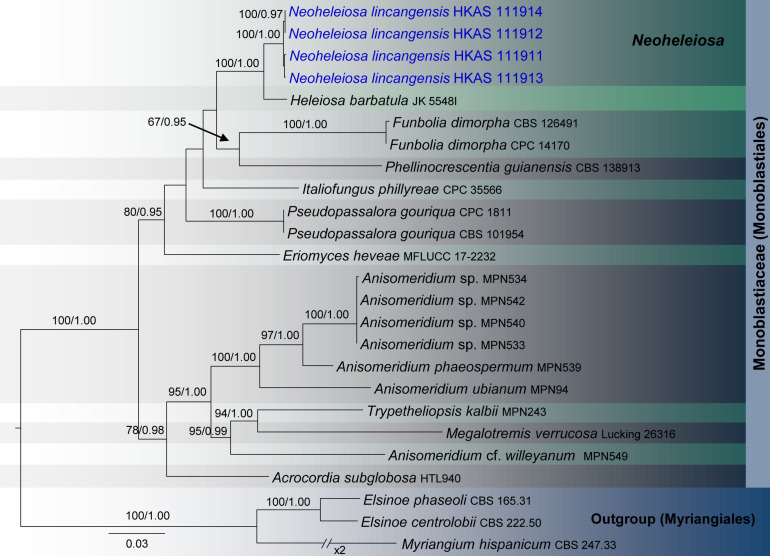
FIGURE 1.
RAxML tree based on a combined dataset of partial SSU, LSU, ITS, tef1, and mtSSU sequence analyses in Monoblastiaceae. Bootstrap support values for ML equal to or greater than 60%, Bayesian posterior probabilities (BYPP) equal to or greater than 0.95 are shown as ML/BI above the nodes. The new isolates are in blue. The scale bar represents the expected number of nucleotide substitutions per site.
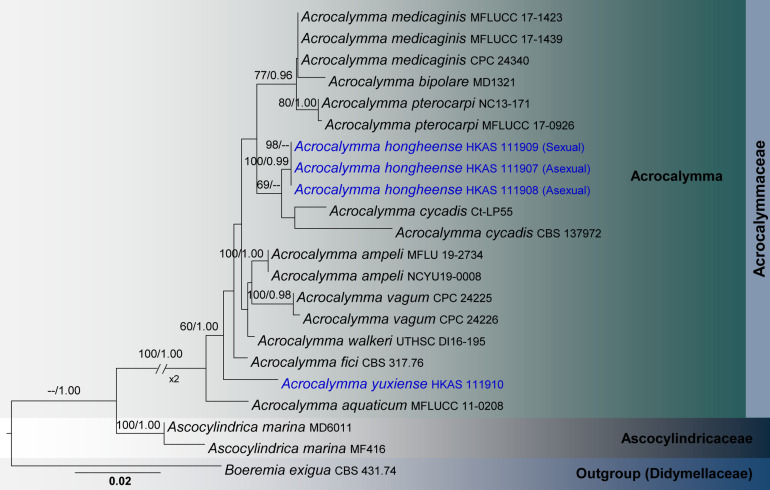
FIGURE 2.
RAxML tree based on a combined dataset of partial SSU, LSU and ITS sequence analyses in Acrocalymmaceae. Bootstrap support values for ML equal to or greater than 60%, Bayesian posterior probabilities (BYPP) equal to or greater than 0.95 are shown as ML/BI above the nodes. The new isolates are in blue. The scale bar represents the expected number of nucleotide substitutions per site.
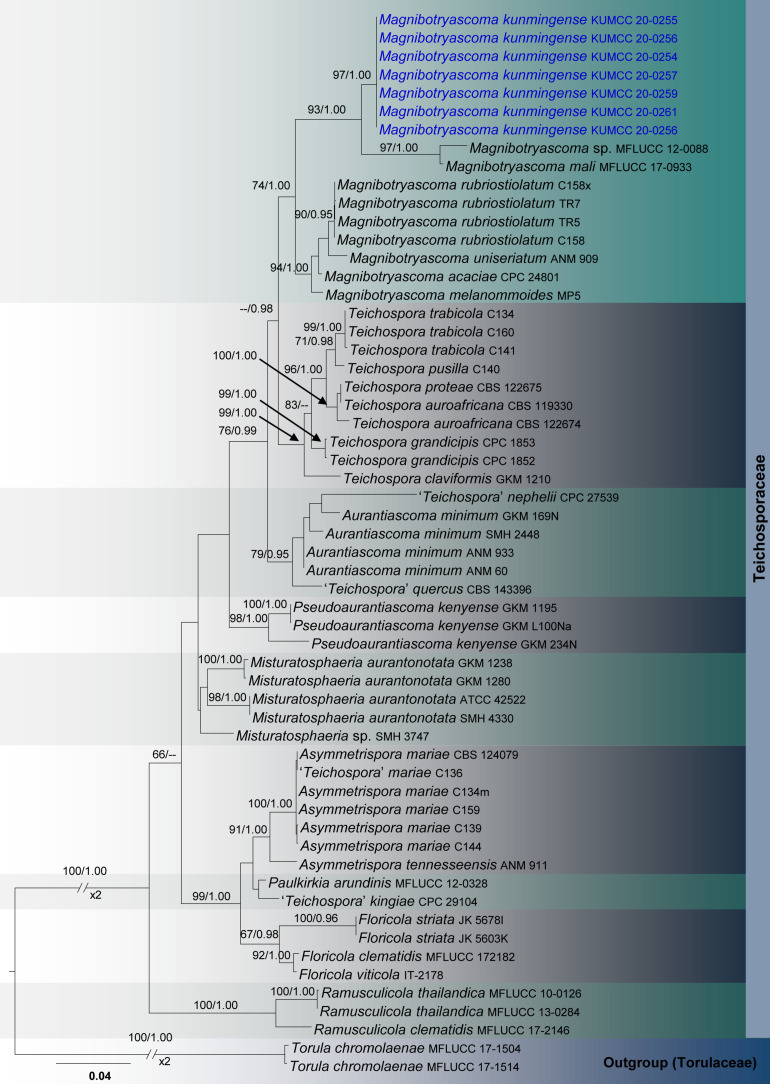
FIGURE 3.
RAxML tree based on a combined dataset of partial SSU, LSU, ITS, tef1, and rpb2 sequence analyses in Teichosporaceae. Bootstrap support values for ML equal to or greater than 60%, Bayesian posterior probabilities (BYPP) equal to or greater than 0.95 are shown as ML/BI above the nodes. The new isolates are in blue. The scale bar represents the expected number of nucleotide substitutions per site.
Monoblastiaceae SSU, LSU, ITS, tef 1, and mtSSU phylogeny (Figure 1): The alignment contained 25 isolates and the tree was rooted to Elsinoe centrolobii (CBS 222.50) and E. phaseoli (CBS 165.31). The final alignment contained a total of 4,062 characters used for the phylogenetic analyses, including alignment gaps. The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of −14276.596302. The matrix had 1,076 distinct alignment patterns, with 49.73% of undetermined characters or gaps. Parameters for the GTR + I + G model of the combined amplicons were as follows: Estimated base frequencies; A = 0.249541, C = 0.238483, G = 0.280978, T = 0.230998; substitution rates AC = 0.975103, AG = 2.399794, AT = 1.850660, CG = 1.423793, CT = 6.985604, GT = 1.000; proportion of invariable sites I = 0.411774; gamma distribution shape parameter α = 0.563137. The Bayesian analysis ran 125,000 generations before the average standard deviation for split frequencies reached below 0.01 (0.008726). The analysis generated 1,251 trees (saved every 100 generations) from which 939 were sampled after 25% of the trees were discarded as burn-in. The alignment contained a total of 1,077 unique site patterns.
Acrocalymmaceae SSU, LSU, and ITS phylogeny (Figure 2): The alignment contained 22 isolates and the tree was rooted to Boeremia exigua (CBS 431.74). The final alignment contained a total of 2,317 characters used for the phylogenetic analyses, including alignment gaps. The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of −5083.5038. The matrix had 265 distinct alignment patterns, with 31.12% of undetermined characters or gaps. Parameters for the GTR + I + G model of the combined amplicons were as follows: Estimated base frequencies; A = 0.250179, C = 0.212903, G = 0.27372, T = 0.263198; substitution rates AC = 2.467522, AG = 3.254238, AT = 3.774672, CG = 0.816185, CT = 11.170637, GT = 1.000; proportion of invariable sites I = 0.737361; gamma distribution shape parameter α = 0.95506. The Bayesian analysis ran 420,000 generations before the average standard deviation for split frequencies reached below 0.01 (0.009821). The analysis generated 4,201 trees (saved every 100 generations) from which 3,151 were sampled after 25% of the trees were discarded as burn-in. The alignment contained a total of 266 unique site patterns.
Teichosporaceae SSU, LSU, ITS, tef1, and rpb2 phylogeny (Figure 3): The alignment contained 58 isolates and the tree was rooted to Torula chromolaenae (MFLUCC 17-1504 and MFLUCC 17-1514). The final alignment contained a total of 4,474 characters used for the phylogenetic analyses, including alignment gaps. The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of −16749.008095. The matrix had 1,159 distinct alignment patterns, with 47.5% of undetermined characters or gaps. Parameters for the GTR + I + G model of the combined amplicons were as follows: Estimated base frequencies; A = 0.242588, C = 0.25624, G = 0.278572, T = 0.222599; substitution rates AC = 1.47144, AG = 3.993923, AT = 1.82804, CG = 1.343882, CT = 11.37556, GT = 1.000; proportion of invariable sites I = 0.495477; gamma distribution shape parameter α = 0.4815. The Bayesian analysis ran 390,000 generations before the average standard deviation for split frequencies reached below 0.01 (0.009894). The analysis generated 3,901 trees (saved every 100 generations) from which 2,926 were sampled after 25% of the trees were discarded as burn-in. The alignment contained a total of 1,162 unique site patterns.
The phylogenetic results obtained for each dataset are discussed where applicable in the descriptive notes below.
Taxonomy
In the present study, one new genus and four new species were found. These taxa are subsequently described in the orders Monoblastiales and Pleosporales below.
Monoblastiaceae Walt. Watson, New Phytologist 28: 106 (1929)
Notes
Hongsanan et al. (2020b) accepted 11 genera viz. Acrocordia, Anisomeridium, Caprettia, Eriomyces, Funbolia, Heleiosa, Megalotremis, Monoblastia, Phellinocrescentia, Pseudopassalora, and Trypetheliopsis in Monoblastiaceae. Meanwhile, Crous et al. (2020) added Italiofungus to accommodate Italiofungus phillyreae, which was collected on leaves of Phillyrea latifolia from Italy. In this study, we add another genus, Neoheleiosa gen. nov. to Monoblastiaceae based on multi-gene phylogenetic evidences.
Neoheleiosa Mortimer gen. nov.
MycoBank: MB838517
Etymology: The generic epithet reflects the similarity to Heleiosa.
Saprobic on dead wood. Sexual morph: Ascomata, solitary, scattered, immersed to erumpent, globose to subglobose or obpyriform, dark brown to black, coriaceous, ostiolate. Ostiole centric or eccentric, papillate, black, smooth, filled with hyaline cells when mature. Peridium comprising blackish to dark brown, thin-walled cells of textura globulosa. Hamathecium comprising numerous, branched, septate, pseudoparaphyses. Asci 8-spored, bitunicate, fissitunicate, cylindric, pedicellate, rounded, and thick-walled at the apex, with an ocular chamber. Ascospores overlapping uniseriate, narrowly ovoid to clavate, 1-septate, initially hyaline, becoming dark brown at maturity, with conically rounded ends, guttulate, thick walled, faintly longitudinally striated, lacking a mucilaginous sheath. Asexual morph: Undetermined.
Type species: Neoheleiosa lincangensis
Neoheleiosa lincangensis Mortimer sp. nov. Figure 4.
MycoBank: MB838518
Etymology: The specific epithet reflects Lincang, from where the holotype was collected.
Holotype: HKAS 111914
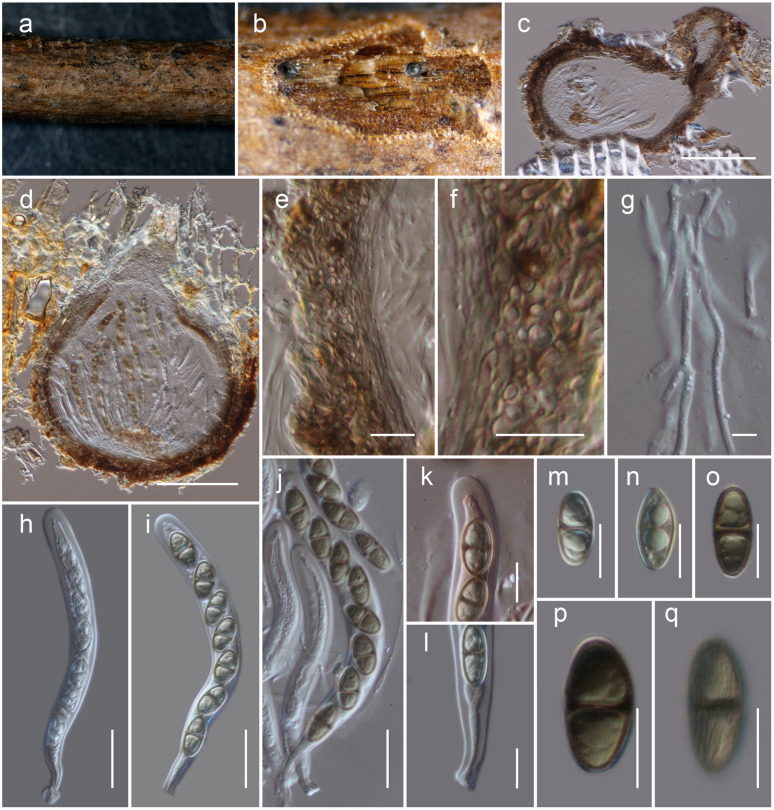
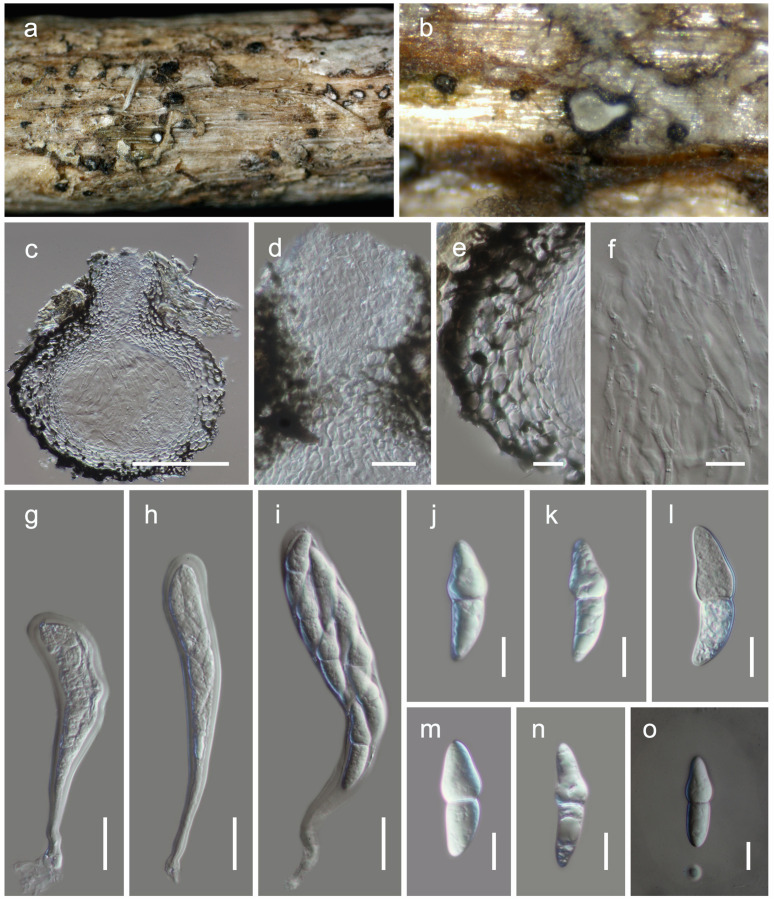
FIGURE 4.
Neoheleiosa lincangensis (HKAS 111914). (a) Ascomata observed on host substrate. (b,d) Horizontal sections of ascomata. (c) Vertical sections through an ascoma. (e,f) Cells of peridium. (g) Pseudoparaphyses. (h–j) Asci. (k) Ocular chamber. (l) Pedicel. (m–q) Ascospores (note: q longitudinally striated). Scale bars: (c,d) = 100 μm, (e, f, k–q) = 10 μm, (g) = 5 μm, (h–j) = 20 μm.
Habitat terrestrial, saprobic on dead twigs of Pittosporum sp. Sexual morph: Ascomata, 130–160 μm high, 200–250 μm diam. ( = 146 × 217 μm, n = 5), solitary, scattered, immersed to erumpent, globose to subglobose or obpyriform, dark brown to black, coriaceous, ostiolate. Ostiole 70–110 μm long, 30–50 μm diam. ( = 90 × 40 μm, n = 5), eccentric, papillate, black, smooth, filled with hyaline cells when mature. Peridium 8–12 μm thick at the base, 15–25 μm wide at sides, comprising blackish to dark brown, thin-walled cells of textura globulosa. Hamathecium 2–2.5 μm wide, comprising numerous, filamentous, branched, septate, pseudoparaphyses. Asci 115–135 × 10–12 μm ( = 124 × 10.6 μm, n = 30), 8-spored, bitunicate, fissitunicate, cylindrical, pedicel furcate, rounded and thick-walled at the apex, with an ocular chamber. Ascospores 16.5–17.5 × 7–9 μm ( = 17.1 × 8.2 μm, n = 30), overlapping uniseriate, narrowly ovoid to clavate, 1-septate, initially hyaline, becoming dark brown at maturity, with conically rounded ends, guttulate, thick walled, faintly longitudinally striated, lacking a mucilaginous sheath. Asexual morph: Undetermined.
Material Examined
China, Yunnan Province, Lincang, Yongde County, Bankaxiang, (N: 23.997479, E: 99.480670), on dead twigs of Pittosporum sp., 8 April 2019, D.N. Wanasinghe, DW0738-07 (HKAS 111914, holotype); ibid. DW0738-09 (HKAS 111911); DW0738-11 (HKAS 111912); DW0738-12 (HKAS 111913).
Notes
Four specimens with 1-septate, brown ascospores species were collected on dead twigs of Pittosporum from Bankaxiang in Yunnan Province. We made several attempts to obtain a culture via single spore isolations and direct isolation from fungal tissues (Senanayake et al., 2020). However, we were unable to get a pure culture, and DNA was extracted directly from the fruiting bodies. Sequence data of these four collections grouped in a strongly supported monophyletic clade in the concatenated SSU, LSU, ITS, tef 1 and mtSSU sequence analyses (Figure 1). Morphologically all these collections were identical and there were no differences in their DNA sequence data. Thus, we recognize that all of these specimens belong to a single species, Neoheleiosa lincangensis sp. nov.
Acrocalymmaceae Crous and Trakun., IMA Fungus 5 (2): 404 (2014)
MycoBank: MB838519
Etymology: The specific epithet reflects Honghe, from where the holotype was collected.
Holotype: HKAS 111909
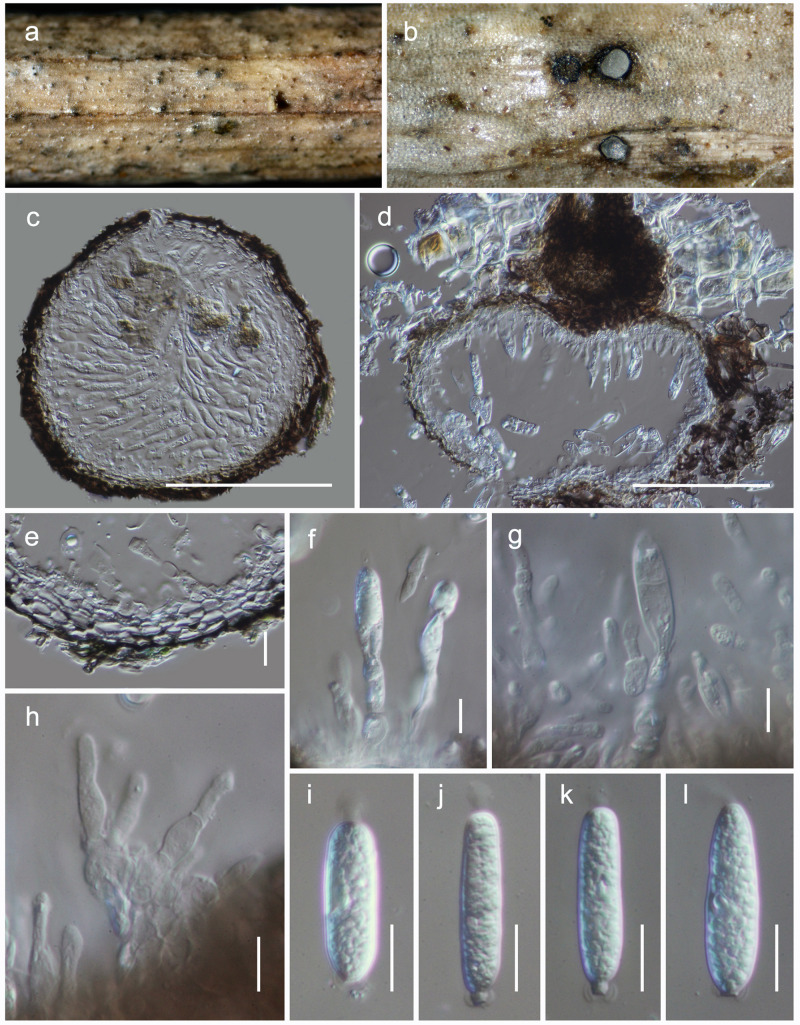
FIGURE 5.
Sexual morph of Acrocalymma hongheense (HKAS 111909). (a) Ascomata observed on host substrate. (b) Horizontal section of an ascoma. (c) Vertical sections through an ascoma. (d) Close up of ostiole. (e) Cells of peridium. (f) Pseudoparaphyses. (g–i) Asci. (i–o) Ascospores (note: o stained with Indian Ink). Scale bars: (c) = 50 μm, (d,g–i) = 20 μm, (e,f,j–o) = 10 μm.
FIGURE 6.
Asexual morph of Acrocalymma hongheense (HKAS 111908). (a) Conidiomata observed on host substrate. (b) Horizontal section of a conidioma. (c,d) Vertical sections through conidiomata. (e) Cells of pycnidia wall. (f–h) Conidiogenous cells. (i–l) Conidia (note the mucoid mass oozing out from apex of all conidia). Scale bars: (c,d) = 100 μm, (e–l) = 10 μm.
Habitat terrestrial, saprobic on dead woody litter. Sexual morph: Ascomata 180–220 μm high, 160–200 μm diam. ( = 196 × 184 μm, n = 5), dark brown, gregarious, immersed beneath host epidermis, visible as numerous, raised, dome-shaped areas on host surface, globose to subglobose, uni-loculate, glabrous with rough walls, coriaceous, ostiolate. Ostioles 50–80 μm long, 30–50 μm diam. ( = 70 × 40 μm, n = 5), centrally located, filled with hyaline cells. Peridium 25–40 μm wide, of unequal thickness, thick on sides toward the apex, composed of dark brown to black cells, arranged in textura angularis. Hamathecium composed of 1–2.5 μm wide, numerous, filamentous, branched, septate, pseudoparaphyses. Asci 100–140 × 15–22 μm ( = 118.9 × 18.5 μm, n = 30), 8-spored, bitunicate, cylindric-clavate, with a furcate to truncate pedicel, apically rounded with an ocular chamber. Ascospores 25–35 × 9.5–11 μm ( = 31.8 × 9.8 μm, n = 40), overlapping bi-seriate, hyaline, fusiform with acute ends, 1-septate, constricted at the septum, upper cell wider than lower cell, smooth-walled, surrounded by a thick, distinct sheath. Asexual morph: coelomycetous. Conidiomata 150–200 μm high, 180–220 μm diam. ( = 166 × 214 μm, n = 5), pycnidial, dark brown, immersed to semi-erumpent, globose, with a central ostiole. Pycnidia wall 10–20 μm wide, of unequal thickness, composed of dark brown to black cells, arranged in textura angularis. Conidiophores reduced to conidiogenous cells or with a single supporting cell. Conidiogenous cells 8–17 × 3.5–7 μm ( = 11.5 × 5.3 μm, n = 40), lining the inner cavity, subcylindrical, hyaline, smooth, annelledic, proliferating percurrently at apex, with prominent periclinal thickening at the apex. Conidia 20–35 × 7–9 μm ( = 27.3 × 7.5 μm, n = 40), hyaline, smooth, guttulate, solitary, subcylindrical, straight, obtusely rounded and with mucoid ooze at the apex, protuberant and with a rounded hilum at base, aseptate, guttulate, sometimes conidia becoming 1-septate.
Material Examined
China, Yunnan Province, Honghe Hani and Yi Autonomous Prefecture, Mengzi, (N: 23.185677, E: 103.413816), on woody litter, 16 March 2019, D.N. Wanasinghe, DW0375-004 (HKAS 111909, holotype); ibid. DW0375-005 (HKAS 111908); ibid. Yuxi, Yi and Dai Autonomous County, Yuanjiang Hani, (N: 23.740730, E: 103.413816), 24 May 2019, DW0636-014 (HKAS 111907).
Notes
During our investigation on the diversity of woody-based Dothideomycetes in Yunnan Province, three isolates (HKAS 111907, HKAS 111908, HKAS 111909) were recovered from decaying woody litter in Honghe and Yuxi counties. Two of them had asexual morphs while the third specimen had a sexual morph. Conidiomata, conidiophores and conidia of these asexual fungi morphologically resemble the remaining taxa in Acrocalymma (Alcorn and Irwin, 1987; Zhang et al., 2012; Crous et al., 2014; Trakunyingcharoen et al., 2014). The sexual morph was similar to Acrocalymma pterocarpi, which was the only known sexual morph in the genus, by its ascomata and asci features (Jayasiri et al., 2019). Phylogenetically, HKAS 111907, HKAS 111908, and HKAS 111909 are monophyletic with strong bootstrap supports (100% ML/0.99 BYPP, Figure 2). This clade has a sister relationship to Acrocalymma cycadis (CBS 137972 and Ct-LP55), but was not statistically supported. Within our new isolates, there were no nucleotide differences in ITS, LSU and SSU gene regions. Therefore, we recognize these three isolates belong to one species (Jeewon and Hyde, 2016), which we introduce as a new species herein.
Acrocalymma yuxiense Mortimer sp. nov. Figure 7.
MycoBank: MB838520
Etymology: The specific epithet reflects Yuxi, from where the holotype was collected.
Holotype: HKAS 111910
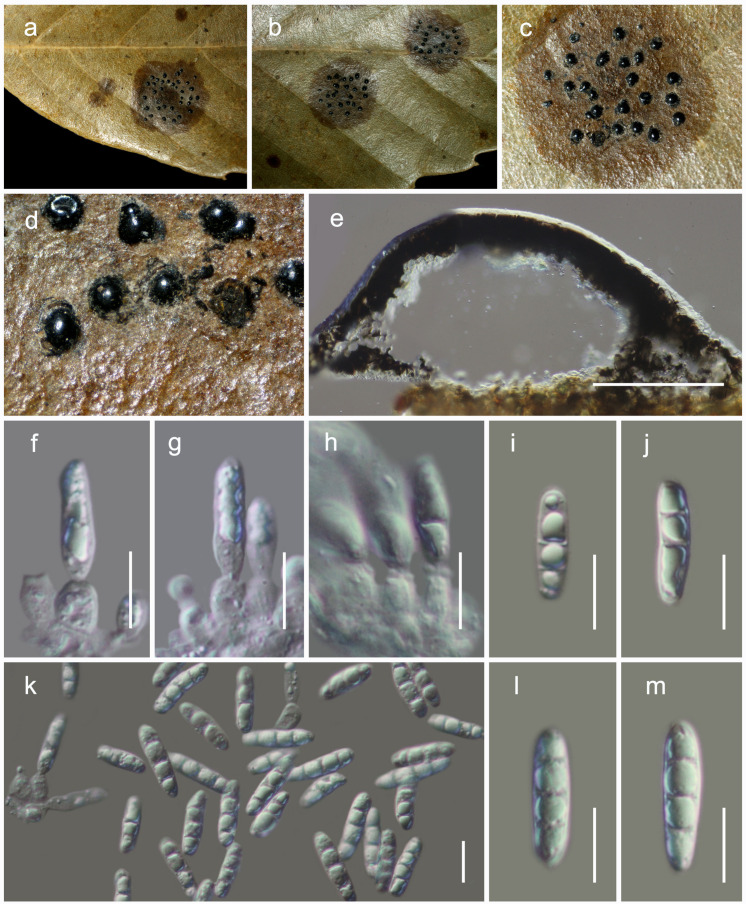
FIGURE 7.
Acrocalymma yuxiense (HKAS 111910). (a) Conidiomata observed on dried leaves of Quercus sp. (e) Vertical sections through a conidioma. (f–h) Conidiogenous cells. (i–m) Conidia. Scale bars: (e) = 100 μm, (f–m) = 10 μm.
Habitat terrestrial, saprobic or weakly pathogenic on dried leaves of Quercus sp. Sexual morph: Undetermined. Asexual morph: coelomycetous, Conidiomata 130–170 μm high, 220–260 μm diam. ( = 155 × 242 μm, n = 5), pycnidial, dark brown, immersed to semi-erumpent, globose, with central ostiole. Pycnidia wall 10–30 μm wide, of unequal thickness, composed of dark brown to black cells, arranged in textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 4–8 × 2.5–4.5 μm ( = 6.3 × 3.4 μm, n = 20), lining the inner cavity, cylindrical to subcylindrical, hyaline, smooth, percurrently proliferating 1–2 times at apex, with prominent periclinal thickening at apex. Conidia 15–21 × 4–5 μm ( = 18.4 × 4.6 μm, n = 30), hyaline, smooth, guttulate, solitary, subcylindrical, straight, obtusely rounded at apex and base, 3-euseptate, guttulate, conidia sometimes 1-septate.
Material Examined
China, Yunnan Province, Yuxi (N: 24.210342, E: 102.540022), on dried leaves of Quercus glauca, 14 March 2019, D.N. Wanasinghe, DW0886-01 (HKAS 111910, holotype).
Notes
Acrocalymma yuxiense, collected from dried leaves of Quercus glauca in Yunnan, is in an independent lineage with weak support and is phylogenetically distinct from other extant species of Acrocalymma (Figure 2). This new species differs from other taxa in Acrocalymma in having conidia with 3 vertical eusepta while other species produce aseptate conidia.
Teichosporaceae M.E. Barr, Mycotaxon 82: 374 (2002)
Magnibotryascoma kunmingense Mortimer sp. nov. Figure 8
MycoBank: MB838522
Etymology: The specific epithet reflects Kunming, from where the holotype was collected.
Holotype: HKAS 111919
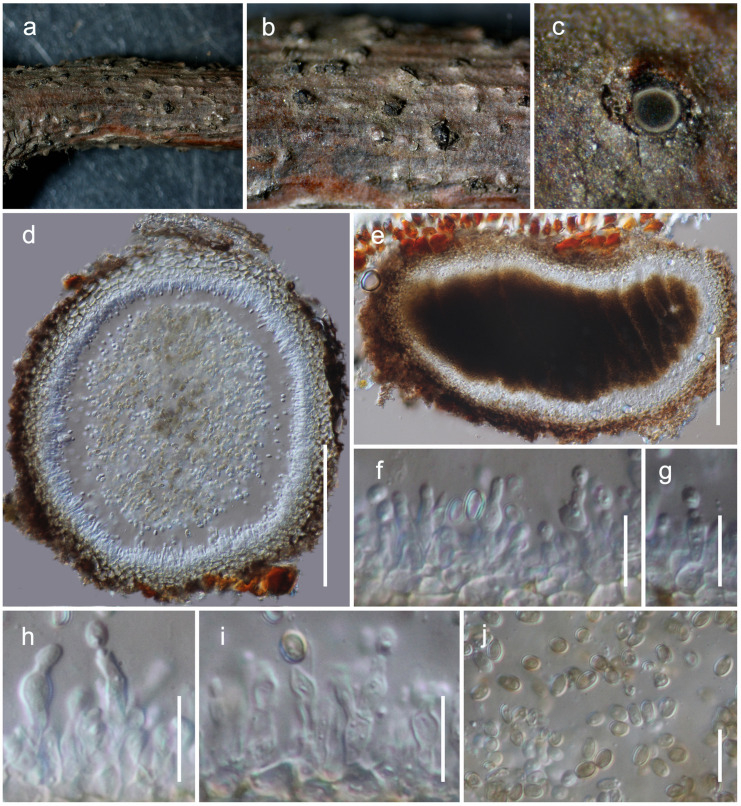
FIGURE 8.
Magnibotryascoma kunmingense (HKAS 111919). (a,b) Conidiomata observed on host (c) Horizontal section of a conidioma. (e) Vertical sections through a conidioma. (f–i) Conidiogenous cells. (j) Conidia. Scale bars: (d,e) = 100 μm, (f–j) = 10 μm.
Habitat terrestrial, saprobic on dead twigs. Sexual morph: Undetermined. Asexual morph: coelomycetous, Conidiomata 150–180 μm high, 250–380 μm diam. ( = 161 × 323 μm, n = 5) pycnidial, solitary, aggregated, uniloculate, immersed, globose to subglobose, coriaceous, dark brown to brown, papillate, with a central ostiole. Pycnidia wall 20–30 μm wide, thick, 2-layered, with outer layer composed of light brown to brown cells of textura angularis, lined with a hyaline innermost layer bearing conidiogenous cells. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–7 × 2–4.5 μm ( = 4.9 × 3.1 μm, n = 20), enteroblastic, annelledic, discrete, cylindrical to oblong, hyaline, arising from the inner layer of pycnidium wall. Conidia 3.8–5.1 × 2.5–3.4 μm ( = 4.4 × 3 μm, n = 30), subglobose, oval, guttulate, hyaline when immature, pale brown at maturity, aseptate, smooth-walled.
Culture Characteristics
Colonies grew on PDA at 20°C in the dark reached 2 cm diam., within 14 days, dense, circular, slightly raised, surface smooth, entire margin, white in surface view and off-white to gray in reverse.
Material Examined
China, Yunnan Province, Kunming, Panlong District, (N: 25.139854, E: 102.737896), on dead twigs of Machilus yunnanensis Lecomte, 26 June 2020, D.N. Wanasinghe, DWKIB20-013B (HKAS 111919, holotype), ex-type culture (KUMCC 20-0254); ibid. Acer cappadocicum var. sinicum Rehd., DWKIB20-039 (HKAS 111916), culture (KUMCC 20-0255); ibid. DWKIB20-027 (HKAS 111921), culture (KUMCC 20-0256); ibid. DWKIB20-041 (HKAS 111915), culture (KUMCC 20-0257); ibid. Qujing, Luoping County, Changdixiang (N: 25.018503, E: 104.406763), DW1303-1 (HKAS 111917), culture (KUMCC 20-0261); ibid. Kunming, Xishan, (N: 25.043763, E: 102.482118), 18 July 2019, DW1287-9 (HKAS 111920), culture (KUMCC 20-0259); ibid. Kunming, Xishan, (N: 25.119848, E: 102.546979), 19 July 2019, DW1344-5 (HKAS 111918), culture (KUMCC 20-0260).
Notes
Seven isolates of Magnibotryascoma kunmingense were obtained from Acer cappadocicum var. sinicum, Machilus yunnanensis and decaying woody litter in Kunming (Panlong and Xishan) and Qujing (Changdixiang). All these specimens were morphologically similar to Magnibotryascoma mali in terms of their conidial and conidiomatal characteristics. Phylogenetically, all strains of Magnibotryascoma kunmingense are monophyletic with 97% ML and 1.00 BI statistical support values (Figure 3). This clade constitutes a sister clade to Magnibotryascoma sp. (MFLUCC 12-0088) and M. mali (MFLUCC 17-0933).
Discussion
The mountainous region of Yunnan Province, China is an important global biodiversity hotspot for studying the evolution of plants, animals, and fungi (Feng and Yang, 2018). The mountains of Southwest China; Eastern Himalaya-Nepal-India and Indo-Burma-India-Myanmar are included in the world’s 35 biodiversity hotspots, and these three hotspot regions intersect in Yunnan Province (Myers et al., 2000; Mittermeier et al., 2005). As a result of the diverse landscape and climatic conditions within Yunnan Province, fungi in Yunnan Province have higher rates of endemism; however, they also share evolutionary connections with species from other regions of the world (Feng and Yang, 2018). Among them, wood-decaying Basidiomycota in tropical China are well studied, and many new species have been documented (Dai et al., 2003, 2004, 2011; Cui et al., 2009; Wang et al., 2011; Yuan and Dai, 2012). This has facilitated better understanding of species diversity and the systematics of woody-based basidomycetous groups, such as Polyporales. However, woody-based microfungi such as Dothideomycetes are relatively neglected compared to the level of research conducted on Basidiomycetes (Wanasinghe et al., 2020). In the last few years, there has been increasing attention on woody-based microfungal occurrences, and more studies are reporting on the microfungal diversity, especially in Dothideomycetes, of Yunnan Province (Bao et al., 2018; Huang et al., 2018; Luo et al., 2018; Wanasinghe et al., 2018a, 2020, 2021; Rathnayaka et al., 2019; Qiao et al., 2020; Thiyagaraja et al., 2020b; Yasanthika et al., 2020).
In this study, we added taxonomic novelties in Acrocalymmaceae, Monoblastiaceae, and Teichosporaceae from Yunnan Province. To the best of our knowledge, these are the first accounts of the species in these three families from Yunnan Province. Within the broader region of China, there is one report of Acrocalymma medicaginis (Acrocalymmaceae) on Trachycarpus fortunei (Taylor and Hyde, 2003) and seven species from Teichosporaceae viz. Magnibotryascoma mali (on Malus halliana), Sinodidymella verrucose (on Salsola gemmascens), Teichospora borealis (on Salix tianschanica), T. lonicerae (on Lonicera stenanthera), T. populi (on Populus sp.), T. solitaria (on Nitraria sibirica), and T. winteriana (on Hippophae rhamnoides) currently documented (Yuan and Barr, 1995; Teng, 1996; Zhuang, 2005; Zhang et al., 2012; Hyde et al., 2017).
Neoheleiosa has a close phylogenetic affinity to Heleiosa with 100% ML and 1.00 BI support values (Figure 1). Kohlmeyer et al. (1996) introduced Heleiosa as a monotypic genus to accommodate Heleiosa barbatula, which is characterized by cylindrical asci with a short pedicel, 1-septate, ellipsoid ascospores with appendages. This fungus was found on senescent leaves of Juncus in salt marshes on the Atlantic coast of the United States (North Carolina, Virginia). Ascomata, asci and ascospore shapes of both genera were shown to be similar. However, the ascospores of Neoheleiosa do not have any appendages (Figures 4m–q), whereas Heleiosa have 10 or more short and curved hair-like subapical appendages at each end. Ascospores with a gelatinous sheath or appendages may help fungi to attach to plant substrates in aquatic or marine habitats (Shearer, 1993; Hyde and Goh, 2003; Jones, 2006; Devadatha et al., 2018; Hashimoto et al., 2018). The subapical appendages of Heleiosa ascospores could be an adaptation for its marine-based habitat and loss of appendaged ascospores in Neoheleiosa is potentially advantageous to adaptation to a non-aquatic habitat.
The surface ornamentation of spores, such as the presence of or absence of appendages, is often used in ascomycetous taxonomy to delineate species or genera (Jeewon et al., 2003; Liu et al., 2014; Phookamsak et al., 2014; Wijayawardene et al., 2016; Paz et al., 2017; Réblová et al., 2015, 2018; Pem et al., 2019b). Abbott and Currah (1997) and Villegas et al. (2005) reported spore ornamentation as a useful character in differentiating various genera within Helvellaceae and Gomphales. Jeewon et al. (2002) clearly demonstrated that species bearing appendages can form distinct phylogenetic lineages and discussed how this character can be a significant phylogenetic marker at the intergeneric level. Considering the similarities shown between our newly proposed genus and Heleiosa, we based our placement of these specimens in a new genus due to the lack of any appendages on the ascospores, a significant variation in morphology compared to the spores of Heleiosa. The ascospores of Neoheleiosa are ornamented with longitudinal streaks (Figure 4q), whereas Heleiosa have smooth-walled ascospores. Furthermore, the arrangement of peridium cells has been reported to be useful to demarcate different genera (Hyde et al., 2013; Tian et al., 2015; Ekanayaka et al., 2017; Paz et al., 2017; Senanayake et al., 2018). In our study, we observed that the peridium cells of Heleiosa are textura angularis, while Neoheleiosa have cells of textura globulosa. It could be possible that with wider collections in the future, species from these two genera will constitute distinct lineages that will further substantiate their generic placement, a scenario which has been reported in other studies (Huang et al., 2017; Wanasinghe et al., 2018b; Jaklitsch and Voglmayr, 2020).
Acrocalymmaceae includes a single genus with eight species viz. Acrocalymma aquaticum, A. bipolare, A. cycadis, A. fici, A. medicaginis, A. pterocarpi, A. vagum and A. walker. These are reported from terrestrial habitats, excluding Acrocalymma aquatica and A. bipolare (Zhang et al., 2012; Dong et al., 2020). The majority of these species are saprobic on various host substrates (Hongsanan et al., 2020a,b; Tennakoon et al., 2021). Only Acrocalymma medicaginis and A. vagum are reported as pathogens (Crous et al., 2014; Trakunyingcharoen et al., 2014). Moreover, Acrocalymma medicaginis is known as the causal agent of root and crown rot of Medicago sativa (Alcorn and Irwin, 1987). Even though there are only eight described species, more than 200 ITS sequences have been deposited from these species in GenBank. Endophytic strains of Acrocalymma vagum have the highest number of reported sequences, followed by A. medicaginis.
Thambugala et al. (2015) introduced Magnabotrioscoma to accommodate M. uniseriata (≡Misturatosphaeria uniseriate), which is morphologically distinct from the remaining genera in Floricolaceae. Magnabotrioscoma species have been reported as woody-based saprobes on Clematis vitalba, Malus halliana, Ribes sanguineum, Robinia pseudoacacia, Salix sp., and Vaccinium myrtillus from Belgium, China, Germany, Norway and the United Kingdom (Jaklitsch et al., 2016; Hyde et al., 2017; Phukhamsakda et al., 2020). In this study, we classified another species in this genus, Magnibotryascoma kunmingensis, growing on the decaying woody litter of Acer cappadocicum var. sinicum and Machilus yunnanensis. The sexual morph of the genus is characterized by erumpent to superficial ascomata lacking a subiculum and fusiform to elliptical and guttulate ascospores (Mugambi and Huhndorf, 2009; Thambugala et al., 2015; Phukhamsakda et al., 2020), and the asexual morph has pycnidial conidiomata featuring aseptate and brown conidia (Crous et al., 2015; Jaklitsch et al., 2016). One interesting finding in this genus is the close connection between uncultured fungal strains (i.e., JF945459, KC978028, KX515906, LS994072) and endophytic strains (i.e., EF419935, EF419972, EF419996) with our new strains in BLAST search results. These strains are not linked to morphological details, and it is therefore difficult to provide further insights into their morpho-phylo relationships. This study reveals that there are potentially many new species awaiting discovery in this region (Hyde et al., 2018).
Data Availability Statement
The datasets generated for this study can be found in the NCBI GenBank, MycoBank and TreeBASE.
Author Contributions
PM, DW, and RJ designed the study. DW did the sample collection. PM and DW were involved in the phylogenetic analyses. SL and J-CX contributed for the research funds. All authors contributed to the writing, preparation, and submission of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Austin G. Smith at World Agroforestry (ICRAF), Kunming Institute of Botany, China, is thanked for English editing. Shaun Pennycook is thanked for nomenclatural advices. Lu Wen Hua and Li Qin Xian are thanked for their invaluable assistance. We express our sincere gratitude to D. J. Baht for his advices on fungal descriptions. PM thank Chiang Mai University for partial supported of this work. We acknowledge Kunming Institute of Botany, Chinese Academy of Sciences for providing the laboratories and instruments for molecular work. RJ thanks the University of Mauritius for support.
Funding. The research was funded by the Key Research Project, Agroforestry Systems for Restoration and Bio-industry Technology Development (Grant No. 2017YFC0505101), Ministry of Sciences and Technology of China (Grant No. 2017YFC0505100). DW would like to thank the CAS President’s International Fellowship Initiative (No. 2021FYB0005), the 64th batch of China Postdoctoral Science Foundation (Grant No. 2018M643549), and Postdoctoral Fund from the Human Resources and Social Security Bureau of Yunnan Province.
References
- Abbott S. P., Currah R. S. (1997). The helvellaceae: systematic revision and occurrence in northern and northwestern North America. Mycotaxon 61 1–125. [Google Scholar]
- Alcorn J. L., Irwin J. A. G. (1987). Acrocalymma medicaginis gen. et sp. nov. causing root and crown rot of Medicago sativa in Australia. Trans. Brit. Mycol. Soc. 88 163–167. 10.1016/s0007-1536(87)80211-5 [DOI] [Google Scholar]
- Aptroot A., Sipman H. J. M. (1993). Musaespora, a genus of pyrenocarpous lichens with campylidia, and other additions to the Foliicolous lichen flora of New Guinea. Lichenologist 25 121–135. 10.1017/s0024282993000192 [DOI] [Google Scholar]
- Bao D. F., Luo Z. L., Jiu J. K., Bhat D. J., Sarunya N., Li W. L., et al. (2018). Lignicolous freshwater fungi in China III: three new species and a new record of Kirschsteiniothelia from northwestern yunnan province. Mycosphere 9 755–768. 10.5943/mycosphere/9/4/4 [DOI] [Google Scholar]
- Barr M. E. (2002). Teichosporaceae, another family in the Pleosporales. Mycologia 82 373–389. [Google Scholar]
- Bhunjun C. S., Phukhamsakda C., Jeewon R., Promputtha I., Hyde K. D. (2021). Integrating different lines of evidence to establish a novel ascomycete genus and family (Anastomitrabeculia, Anastomitrabeculiaceae) in Pleosporales. J. Fungi 7:94. 10.3390/jof7020094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P. W., Schumacher R. K., Wingfield M. J., Lombard L., Giraldo A., Christensen M., et al. (2015). Fungal systematics and evolution: FUSE 1. Sydowia 67 81–118. [Google Scholar]
- Crous P. W., Shivas R. G., Quaedvlieg W., van der Bank M., Zhang Y., Summerell B. A., et al. (2014). Fungal planet description sheets: 214–280. Persoonia 32 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P. W., Wingfield M. J., Schumacher R. K., Akulov A., Bulgakov T. S., Carnegie A. J., et al. (2020). New and interesting fungi. 3. FUSE 6 157–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B. K., Dai Y., Bao H. Y. (2009). Wood-inhabiting fungi in southern China 3. a new species of Phellinus (Hymenochaetales) from tropical China. Mycotaxon 110 125–130. 10.5248/110.125 [DOI] [Google Scholar]
- Dai Y. C., Cui B. K., Yuan H. S., He S. H., Wei Y. L., Qin W. M., et al. (2011). Wood-inhabiting fungi in southern China 4. polypores from hainan province. Ann. Bot. Fenn. 48 219–231. 10.5735/085.048.0302 [DOI] [Google Scholar]
- Dai Y., Härkönen M., Niemelä T. (2003). Wood-inhabiting fungi in southern China 1. polypores from hunan province. Ann. Bot. Fenn. 40 381–393. [Google Scholar]
- Dai Y., Wei Y., Wang Z. (2004). Wood-inhabiting fungi in southern China 2. polypores from sichuan province. Ann. Bot. Fenn. 41 319–329. [Google Scholar]
- Devadatha B., Sarma V. V., Jeewon R., Wanasinghe D. N., Hyde K. D., Jones E. B. G. (2018). Thyridariella, a novel marine fungal genus from India: morphological characterization and phylogeny inferred from multigene DNA sequence analyses. Mycol. Prog. 17 791–804. 10.1007/s11557-018-1387-4 [DOI] [Google Scholar]
- Dong W., Wang B., Hyde K. D., McKenzie E. H. C., Raja H. A., Tanaka K., et al. (2020). Freshwater Dothideomycetes. Fungal Divers. 105 319–575. [Google Scholar]
- Ekanayaka A. H., Ariyawansa H., Hyde K., Jones E., Daranagama D., Phillips A. J. L., et al. (2017). DISCOMYCETES: the apothecial representatives of the phylum ascomycota. Fungal Divers. 87 237–298. 10.1007/s13225-017-0389-x [DOI] [Google Scholar]
- Feng B., Yang Z. (2018). Studies on diversity of higher fungi in Yunnan, southwestern China: a review. Plant Divers. 40 165–171. 10.1016/j.pld.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acid. Symp. Ser. 41 95–98. [Google Scholar]
- Hashimoto A., Hirayama K., Takahashi H., Matsumura M., Okada G., Chen C. Y., et al. (2018). Resolving the Lophiostoma bipolare complex: generic delimitations within Lophiostomataceae. Stud. Mycol. 90 161–189. 10.1016/j.simyco.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongsanan S., Hyde K. D., Phookamsak R., Wanasinghe D. N., McKenzie E. H. C., Sarma V. V., et al. (2020a). Refined families of Dothideomycetes: dothideomycetidae and pleosporomycetidae. Mycosphere 11 1553–2107. 10.5943/mycosphere/11/1/13 [DOI] [Google Scholar]
- Hongsanan S., Hyde K. D., Phookamsak R., Wanasinghe D. N., McKenzie E. H. C., Sarma V. V., et al. (2020b). Refined families of Dothideomycetes: orders and families incertae sedis. Fungal Divers. 105 17–318. [Google Scholar]
- Huang S., Jeewon R., Wanasinghe D. N., Manawasinghe I., Bulgakov T. S., Hyde K. D., et al. (2017). Phylogenetic taxonomy of Dematiopleospora fusiformis sp. nov. (Phaeosphaeriaceae) from Russia. Phytotaxa 316 239–249. 10.11646/phytotaxa.316.3.3 [DOI] [Google Scholar]
- Huang S., Maharachchikumbura S. S. N., Jeewon R., Bhat D. J., Chomnunti P., Hyde K. D., et al. (2018). Morphological and molecular taxonomy of Jahnula dianchia sp. nov. (Jahnulales) from submerged wood in Dianchi Lake, Yunnan China. Mycol. Prog. 17 547–555. 10.1007/s11557-018-1390-9 [DOI] [Google Scholar]
- Hyde K. D., Goh T. K. (2003). “Adaptations for dispersal in filamentous freshwater fungi,” in Freshwater Mycology, eds Tsui K. M., Hyde K. D. (Hong Kong: Fungal Diversity Press; ), 231–258. [Google Scholar]
- Hyde K. D., Dong Y., Phookamsak R., Jeewon R., Bhat D. J., Jones E. B. G., et al. (2020). Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 100 5–277. [Google Scholar]
- Hyde K. D., Jones E. B. G., Liu J. K., Ariyawansa H. A., Boehm E., Boonmee S., et al. (2013). Families of Dothideomycetes. Fungal Divers. 63 1–313. 10.1007/978-3-319-23534-9_1 [DOI] [Google Scholar]
- Hyde K. D., Norphanphoun C., Abreu V. P., Bazzicalupo A., Chethana T. K. W., Clericuzio M., et al. (2017). Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 87 1–235. [Google Scholar]
- Hyde K. D., Norphanphoun C., Chen J., Dissanayake A. J., Doilom M., Hongsanan S., et al. (2018). Thailand’s amazing diversity: up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 93 215–239. 10.1007/s13225-018-0415-7 [DOI] [Google Scholar]
- Jaklitsch W. M., Voglmayr H. (2020). Fenestelloid clades of the cucurbitariaceae. Persoonia 44 1–40. 10.3767/persoonia.2020.44.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch W. M., Olariaga I., Voglmayr H. (2016). Teichospora and the teichosporaceae. Mycol. Prog. 15 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasiri S. C., Hyde K. D., Jones E. B. G., McKenzie E. H. C., Jeewon R., Phillips A. J. L., et al. (2019). Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10 1–186. 10.5943/mycosphere/10/1/1 [DOI] [Google Scholar]
- Jeewon R., Hyde K. D. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Jeewon R., Cai L., Zhang K., Hyde K. D. (2003). Dyrithiopsis lakefuxianensis gen et sp. nov. from Fuxian Lake, Yunnan, China and notes on the taxonomic confusion surrounding Dyrithium. Mycologia 95 911–920. 10.2307/3762019 [DOI] [PubMed] [Google Scholar]
- Jeewon R., Liew E. C., Hyde K. D. (2002). Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Mol. Phylogenet. Evol. 25 378–392. 10.1016/s1055-7903(02)00422-0 [DOI] [PubMed] [Google Scholar]
- Jeewon R., Wanasinghe D. N., Rampadaruth S., Puchooa D., Zhou L., Liu A., et al. (2017). Nomenclatural and identification pitfalls of endophytic mycota based on DNA sequence analyses of ribosomal and protein genes phylogenetic markers: a taxonomic dead end? Mycosphere 8 1802–1817. 10.5943/mycosphere/8/10/7 [DOI] [Google Scholar]
- Jeewon R., Yeung Q. S. Y., Wanasinghe D. N., Rampadarath S., Puchooa D., Wang H. (2018). Hidden mycota of pine needles: molecular signatures from PCR-DGGE and Ribosomal DNA phylogenetic characterization of novel phylotypes. Sci. Rep. 8:18053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. B. G. (2006). Form and function of fungal spore appendages. Mycoscience 47 167–183. 10.1007/s10267-006-0295-7 [DOI] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformatics. 20 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk P. M. (2019). Catalogue of Life. Available online at: http://www.catalogueoflife.org (accessed January 16, 2021). [Google Scholar]
- Kohlmeyer J., Volkmann-Kohlmeyer B., Eriksson O. E. (1996). Fungi on Juncus roemerianus. 8. New bitunicate ascomycetes. Can. J. Bot. 74 1830–1840. 10.1139/b96-220 [DOI] [Google Scholar]
- Kuraku S., Zmasek C. M., Nishimura O., Katoh K. (2013). aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41 W22–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. K., Hyde K. D., Jeewon R., Phillips A. J., Maharachchikumbura S. S. N., Ryberg M., et al. (2017). Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Divers. 84 75–99. 10.1007/s13225-017-0385-1 [DOI] [Google Scholar]
- Liu J. K., Phookamsak R., Dai D. Q., Tanaka K., Jones E. B. G., Xu J. C., et al. (2014). Roussoellaceae, a new pleosporalean family to accommodate the genera Neoroussoella gen. nov., Roussoella and Roussoellopsis. Phytotaxa 181 001–033. [Google Scholar]
- Lücking R. (2008). Foliicolous Lichenized Fungi. Flora Neotropica. New York: New York Botanical Garden Press; 103 1–867. [Google Scholar]
- Lumbsch H. T., Huhndorf S. M. (2010). Outline of Ascomycota 2009. Myconet 14 1–64. [Google Scholar]
- Luo Z. L., Hyde K. D., Liu J. K., Bhat D. J., Bao D. F., Li W. L., et al. (2018). Lignicolous freshwater fungi from China II: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 9 444–461. 10.5943/mycosphere/9/3/2 [DOI] [Google Scholar]
- Maharachchikumbura S. S. N., Wanasinghe D. N., Cheewangkoon R., Al-Sadi A. M. (2021). Uncovering the hidden taxonomic diversity of fungi in Oman. Fungal Divers. In press. 10.1007/s13225-020-00467-1 [DOI] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE) 14 Nov 2010, (New Orleans, LA: Institute of Electrical and Electronics Engineers; ), 1–8. [Google Scholar]
- Mittermeier R. A., Gil P. R., Hoffman M., Brookscristina P., Goettsch Mittermeier G. (2005). Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions. Mexico: Cemex. [Google Scholar]
- Mugambi G. K., Huhndorf S. M. (2009). Molecular phylogenetics of Pleosporales: Melanommataceae and Lophiostomataceae re-circumscribed (Pleosporomycetidae, Dothideomycetes, Ascomycota). Stud. Mycol. 64 103–121. 10.3114/sim.2009.64.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N., Mittermeier R. A., Mittermeier C. G., da Fonseca G. A., Kent J. (2000). Biodiversity hotspots for conservation priorities. Nature 403 853–858. 10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- Nylander J. A. A., Wilgenbusch J. C., Warren D. L., Swofford D. L. (2008). AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24 581–583. 10.1093/bioinformatics/btm388 [DOI] [PubMed] [Google Scholar]
- Paz A., Bellanger J. M., Lavoise C., Molia A., ławrynowicz M., Larsson E., et al. (2017). The genus Elaphomyces (Ascomycota, Eurotiales): a ribosomal DNA-based phylogeny and revised systematics of European ‘deer truffles’. Persoonia 38 197–239. 10.3767/003158517x697309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pem D., Hongsanan S., Doilom M., Tibpromma S., Wanasinghe D. N., Dong W., et al. (2019a). https://www.dothideomycetes.org: an online taxonomic resource for the classification, identification, and nomenclature of Dothideomycetes. Asian J. Mycol. 2 287–297. 10.5943/ajom/2/1/19 [DOI] [Google Scholar]
- Pem D., Hyde K. D., Doilom M., Camporesi E., Hongsanan S., Rampadarath S., et al. (2019b). Multigene phylogenetic analyses to establish new Valsaria species and taxonomic significance of spore ornamentation. PLoS One 14:e0217982. 10.1371/journal.pone.0217982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phookamsak R., Liu J. K., McKenzie E., Manamgoda D., Ariyawansa H. A., Thambugala K. M., et al. (2014). Revision of Phaeosphaeriaceae. Fungal Divers. 68 159–238. [Google Scholar]
- Phookamsak R., Wanasinghe D. N., Hongsanan S., Phukhamsakda C., Huang S. K., Tennakoon D. S., et al. (2017). Towards a natural classification of Ophiobolus and ophiobolus-like taxa; introducing three novel genera Ophiobolopsis, Paraophiobolus and Pseudoophiobolus in Phaeosphaeriaceae (Pleosporales). Fungal Divers. 87 299–339. 10.1007/s13225-017-0393-1 [DOI] [Google Scholar]
- Phukhamsakda C., McKenzie E. H. C., Phillips A. J. L., Jones E. B. G., Bhat D. J., Stadler M., et al. (2020). Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 101 1–204. 10.1007/s13225-020-00448-4 [DOI] [Google Scholar]
- Qiao M., Hua Z., Lv R., Yu Z. (2020). Neodactylariales, Neodactylariaceae (Dothideomycetes, Ascomycota): new order and family, with a new species from China. MycoKeys 73 69–85. 10.3897/mycokeys.73.54054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2014). FigTree v1.4: Tree Figure Drawing Tool. Available online at: http://treebio.ed.ac.uk/software/figtree/ (accessed January 9, 2021). [Google Scholar]
- Rathnayaka A. R., Dayarathne M. C., Maharachchikumbura S. S. N., Liu J. K., Tennakoon D. S., Hyde K. D. (2019). Introducing Seriascoma yunnanense sp. nov. (Occultibambusaceae, Pleosporales) based on evidence from morphology and phylogeny. Asian J. Mycol. 2 245–253. 10.5943/ajom/2/1/15 [DOI] [Google Scholar]
- Réblová M., Fournier J., Štěpánek V. (2015). Pisorisporiales, a new order of aquatic and terrestrial fungi for Achroceratosphaeria and Pisorisporium gen. nov. in the Sordariomycetes. Persoonia 34 40–49. 10.3767/003158515x685544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M., Miller A. N., Réblová K., Štěpánek V. (2018). Phylogenetic classification and generic delineation of Calyptosphaeria gen. nov., Lentomitella, Spadicoides and Torrentispora (Sordariomycetes). Stud. Mycol. 89 1–62. 10.1016/j.simyco.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman A., Crous P. W., Hyde K. D., Hawksworth D. L., Aptroot A., Bezerra J. L., et al. (2015). Recommended names for pleomorphic genera in Dothideomycetes. IMA Fungus 6 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake I. C., Jeewon R., Chomnunti P., Wanasinghe D. N., Norphanphoun C., Karunarathna A., et al. (2018). Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Divers. 93 241–443. 10.1007/s13225-018-0410-z [DOI] [Google Scholar]
- Senanayake I. C., Rathnayaka A. R., Marasinghe D. S., Calabon M. S., Gentekaki E., Lee H. B., et al. (2020). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11 2678–2754. 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Shearer C. A. (1993). The freshwater ascomycetes. Nova Hedwigia 56 1–33. [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. E., Hyde K. D. (2003). Microfungi of Tropical and Temperate Palms. Hong Kong: Fungal Divers Press, 1–459. [Google Scholar]
- Teng S. C. (1996). Fungi of China. Ithaca, NY: Mycotaxon, Ltd, 1–586. [Google Scholar]
- Tennakoon D. S., Kuo C. H., Maharachchikumbura S. S. N., Thambugala K. M., Gentekaki E., Phillips A. J. L., et al. (2021). Taxonomic and Phylogenetic Contributions to Leaf Litter Inhabiting Microfungi Associated with Celtis Formosana, Ficus Ampelas, f. septica, Macaranga tanarius and Morus australis. Hong Kong: Fungal Divers. [Google Scholar]
- Thambugala K. M., Hyde K. D., Tanaka K., Tian Q., Wanasinghe D. N., Ariyawansa H. A., et al. (2015). Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers. 74 199–266. 10.1007/s13225-015-0348-3 [DOI] [Google Scholar]
- Thiyagaraja V., Lücking R., Ertz D., Wanasinghe D. N., Karunarathna S. C., Camporesi E., et al. (2020a). Evolution of non-lichenized, saprotrophic species of Arthonia (Ascomycota, Arthoniales) and resurrection of Naevia, with notes on Mycoporum. Fungal Divers. 102 205–224. 10.1007/s13225-020-00451-9 [DOI] [Google Scholar]
- Thiyagaraja V., Wanasinghe D. N., Worthy F., Karunarathna S. C. (2020b). Addition to Melanommataceae: a new geographical record of Alpinaria rhododendri from Shangri La, China. Asian J. Mycol. 3 335–344. 10.5943/ajom/3/1/8 [DOI] [Google Scholar]
- Tian Q., Liu J. K., Hyde K. D., Wanasinghe D. N., Boonmee S., Jayasiri S. C., et al. (2015). Phylogenetic relationships and morphological reappraisal of Melanommataceae (Pleosporales). Fungal Divers. 74 267–324. 10.1007/s13225-015-0350-9 [DOI] [Google Scholar]
- Trakunyingcharoen T., Lombard L., Groenewald J. Z., Cheewangkoon R., To-anun C., Alfenas A. C., et al. (2014). Mycoparasitic species of Sphaerellopsis, and allied lichenicolous and other genera. IMA Fungus 5 391–414. 10.5598/imafungus.2014.05.02.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas M., Cifuentes J., Estrada-Torres A. (2005). Sporal characters in Gomphales and their significance for phylogenetics. Fungal Divers. 18 157–175. [Google Scholar]
- Wanasinghe D. N., Jeewon R., Gareth Jones E. B., Boonmee S., Kaewchai S., Manawasinghe I. S., et al. (2018a). Novel palmicolous taxa within Pleosporales: multigene phylogeny and taxonomic circumscription. Mycol. Prog. 17 571–590. 10.1007/s11557-018-1379-4 [DOI] [Google Scholar]
- Wanasinghe D. N., Mortimer P. E., Xu J. (2021). Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaea viscosa in Honghe (China). J. Fungi 7:180. 10.3390/jof7030180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe D. N., Phukhamsakda C., Hyde K. D., Jeewon R., Lee H. B., Jones E. B. G., et al. (2018b). Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 89 1–236. 10.1007/s13225-018-0395-7 [DOI] [Google Scholar]
- Wanasinghe D. N., Wijayawardene N. N., Xu J., Cheewangkoon R., Mortimer P. E. (2020). Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China). PLoS One 15:e0235855. 10.1371/journal.pone.0235855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Cui B. K., Li H. J., Du P., Jia B. S. (2011). Wood-rotting fungi in eastern China 5. polypore diversity in Jiangxi Province. Ann. Bot. Fenn. 48 237–246. 10.5735/085.048.0304 [DOI] [Google Scholar]
- Wijayawardene N. N., Crous P. W., Kirk P. M., Hawksworth D. L., Boonmee S., Braun U., et al. (2014). Naming and outline of Dothideomycetes-2014 including proposals for the protection or suppression of generic names. Fungal Divers. 69 1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Al-Ani L. K. T., Tedersoo L., Haelewaters D., Rajeshkumar K. C., et al. (2020). Outline of fungi and fungus-like taxa. Mycosphere 11 1060–1456. 10.5943/mycosphere/11/1/8 10549654 [DOI] [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Lumbsch H. T., Liu J. K., Maharachchikumbura S. S. N., Ekanayaka A. H., et al. (2018). Outline of ascomycota: 2017. Fungal Divers. 88 167–263. 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Rajeshkumar K. C., Hawksworth D. L., Madrid H., Kirk P. M., et al. (2017). Notes for genera: ascomycota. Fungal Divers. 86 1–594. [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Wanasinghe D. N., Papizadeh M., Goonasekara I. D., Camporesi E., et al. (2016). Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers. 77 1–316. 10.1007/s13225-016-0360-2 [DOI] [Google Scholar]
- Yasanthika E., Dissanayake L. S., Wanasinghe D. N., Karunarathna S. C., Mortimer P. E., Samarakoon B. C., et al. (2020). Lonicericola fuyuanensis (Parabambusicolaceae) a new terrestrial pleosporalean ascomycete from Yunnan province, China. Phytotaxa 446 103–113. 10.11646/phytotaxa.446.2.3 [DOI] [Google Scholar]
- Yuan H., Dai Y. (2012). Wood-inhabiting fungi in southern China. 6. polypores from guangxi autonomous region. Ann. Bot. Fenn. 49 341–351. 10.5735/085.049.0605 [DOI] [Google Scholar]
- Yuan Z. Q., Barr M. E. (1995). Some new taxa and new records of the genera Pleospora and Teichospora from Xinjiang. China. Mycotaxon 54 111–116. [Google Scholar]
- Zhang J. F., Liu J. K., Jeewon R., Wanasinghe D. N., Liu Z. Y. (2019). Fungi from Asian Karst formations III. molecular and morphological characterization reveal new taxa in Phaeosphaeriaceae. Mycosphere 10 202–220. 10.5943/mycosphere/10/1/3 [DOI] [Google Scholar]
- Zhang S.-N., Hyde K. D., Jones E. B. G., Jeewon R., Cheewangkoon R., Liu J.-K. (2019). Striatiguttulaceae, a new pleosporalean family to accommodate the genera Longicorpus gen. nov. and Striatiguttula gen. nov., from palms in mangrove ecosystem. MycoKeys 49 99–129. 10.3897/mycokeys.49.30886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Crous P. W., Schoch C. L., Hyde K. D. (2012). Pleosporales. Fungal Divers. 53 1–221. 10.1007/978-3-319-23534-9_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang W. Y. (2005). Fungi of Northwestern China. Ithaca, NY: Mycotaxon, Ltd, 1–430. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in the NCBI GenBank, MycoBank and TreeBASE.