Abstract
Twin studies indicate that there is a significant genetic contribution to the risk of developing alcohol use disorder (AUD). With the exception of coding variants in ADH1B and ALDH2, little is known about the molecular effects of AUD-associated loci. We previously reported that the AUD-associated synonymous polymorphism rs279858 within the GABAA α2 receptor subunit gene, GABRA2, was associated with gene expression of the chr4p12 GABAA subunit gene cluster in induced pluripotent stem cell (iPSC)-derived neural cultures. Based on this and other studies that showed changes in GABRA2 DNA methylation associated with schizophrenia and aging, we examined methylation in GABRA2. Specifically, using 69 iPSC lines and neural cultures derived from 47 of them, we examined whether GABRA2 rs279858 genotype predicted methylation levels and whether methylation was related to GABAA receptor subunit gene expression. We found that the GABRA2 CpG island undergoes random stochastic methylation during reprogramming and that methylation is associated with decreased GABRA2 gene expression, an effect that extends to the GABRB1 gene over 600 kb distal to GABRA2. Further, we identified additive effects of GABRA2 CpG methylation and GABRA2 rs279858 genotype on expression of the GABRB1 subunit gene in iPSC-derived neural cultures.
Keywords: alcohol use disorder, epigenetics, GABAA receptor subunit genes, promoter methylation, stem cell-derived neural cultures
1 |. INTRODUCTION
Approximately 8.5% of American adults meet criteria for moderate or severe alcohol use disorder (AUD) (Grant et al., 2017). AUD is a complex disorder involving genetic and environmental components, with an estimated heritability of 50–60% (Verhulst, Neale, & Kendler, 2015). Prior genetic investigations including genome-wide association studies have shown that AUD is polygenic, with many genes having small individual effects (Sullivan et al., 2018) that may synergize with each other and interact with social and environmental factors (Tawa, Hall, & Lohoff, 2016).
Linkage analysis performed by the Collaborative Study on the Genetics of Alcoholism identified the association of GABRA2 with increased risk of AUD in European Americans (EAs) (Edenberg et al., 2004). The association was confirmed in other case–control studies (Covault, Gelernter, Hesselbrock, Nellissery, & Kranzler, 2004; Covault, Gelernter, Jensen, Anton, & Kranzler, 2008; Enoch, 2008; Fehr et al., 2006; Lappalainen et al., 2005), and more recently in a meta-analysis that included over 3,000 subjects (Li et al., 2014). A commonly studied tag-single nucleotide polymorphism (SNP) in the AUD-associated yin-yang haplotype block is the synonymous GABRA2 Exon 5 T-to-C rs279858 variant, with the C (minor) allele associated with risk for AUD in EAs. There are 129 SNPs in strong linkage disequilibrium (LD) (r2 ≥ .8) with rs279858 spanning a 101 kb haplotype block (hg19 chr4:46,260,587–46,361,545). It is unknown which of these variants are causal for the increased genetic risk for AUD. This AUD-associated haplotype block in GABRA2 has also been reported to be associated with a number of behavioral and physiologic endophenotypes, including childhood conduct disorder (Dick et al., 2006), greater activation of the insular cortex during anticipation of a monetary reward (Villafuerte et al., 2012), and greater frequency of electroencephalographic beta-wave oscillations (Edenberg et al., 2004)—which has been associated with an increased risk of relapse to problematic drinking (Bauer, 2001). These endophenotypes are not dependent on direct effects of alcohol and suggest that variants in this region could alter connectivity or patterning during neural development that may subsequently place individuals at increased genetic risk for developing AUD.
GABA is the major inhibitory neurotransmitter in the adult human brain, and binds to ligand-gated GABAA and GABAC receptors to mediate chloride influx into mature neurons. GABAA receptors are typically composed of two α, two β, and either a γ or δ subunit, with α1 and α2 being the most highly expressed GABAA subunits throughout the brain (Barnard et al., 1998; Pirker, Schwarzer, Wieselthaler, Sieghart, & Sperk, 2000). The majority of genes encoding GABAA receptor subunits are found within four clusters on chromosomes 4p12 (β1, α4, α2, γ1), 5q34 (β2, α6, α1, γ2), 15q11 (β3, α5, γ3), and Xq28 (θ, α3, ε) (Steiger & Russek, 2004), allowing for their coordinated developmental regulation via common regulatory elements (Barnard et al., 1998; Fillman, Duncan, Webster, Elashoff, & Weickert, 2010). In B6J mice, it was recently reported that a single base pair change in Gabra2 adjacent to a splice acceptor site was significantly associated with lower expression of Gabra2 mRNA and protein in many brain regions compared with other inbred strains (Mulligan et al., 2019). Correction of this single base pair deletion restored expression of Gabra2 mRNA and protein, suggesting that this Gabra2 sequence variant in B6J mice may explain strain differences in anxiety-like endophenotypes and behavioral sensitization to alcohol (Mulligan et al., 2019).
Our laboratory has used an induced pluripotent stem cell (iPSC)-derived neural culture model to investigate the relationship between rs279858 genotype and RNA levels for GABRA2 and other three other GABAA receptor subunit genes at chr4p12. The transcription of GABRA2 in neural cultures was significantly lower in rs279858 C-allele carrier lines than in T-allele homozygotes (Lieberman, Kranzler, Joshi, Shin, & Covault, 2015). Further, expression of the other three GABAA genes in the chr4p12 cluster was significantly correlated with GABRA2 expression despite being outside the rs279858-tagged haplotype block (Lieberman et al., 2015). These results suggest that there are shared regulatory elements that modify transcription of chr4p12 GABAA receptor subunit genes.
Regulatory mechanisms that could modulate transcription within this region include promoter-proximal CpG island methylation. Promoter-proximal CpG island methylation is typically negatively correlated with gene expression (Baylin et al., 2001; Hake, Xiao, & Allis, 2004; Leung et al., 2015). GABRA2 promoter-proximal CpG island methylation was reported to be increased in samples from patients with schizophrenia (Rukova et al., 2014), and the GABRA2 CpG island was identified as one of eight sites out of 50 loci examined using human brain tissue that underwent a progressive increase in DNA methylation over the lifespan (Siegmund, Schumm-Draeger, Antoni, & Bibra, 2007). Further, there have been recent reports that the GABRA2 CpG island may be subject to aberrant methylation during reprogramming somatic cells to pluripotency (Panopoulos et al., 2017). In light of these reports, we examined the contribution of GABRA2 promoter-proximal CpG island methylation to the transcriptional endophenotype observed in iPSC-derived neural cultures.
Specifically we examined: (a) whether the process of reprogramming fibroblasts to iPSCs has an effect on the methylation level at this promoter-proximal CpG site and its relation to genotype at the AUD-associated tag-SNP rs279858, (b) whether the level of methylation is maintained following the differentiation of iPSCs to neural cultures, (c) whether the level of methylation is associated with the level of transcription in neural cultures for chr4p12 GABAA receptor subunit genes, and (d) whether effects of CpG-island methylation and rs279858 on RNA levels are additive or interactive.
2 |. METHODS
2.1 |. Dermal fibroblast cell culture
Fibroblast cell lines were generated from skin punch biopsies donated by participants enrolled in a study of the effects of alcohol combined with dutasteride in male social drinkers (Milivojevic, Feinn, Kranzler, & Covault, 2014) or in a clinical trial of pharmacotherapy for heavy drinkers (Kranzler et al., 2014). Fibroblasts were cultured in DMEM (ThermoFisher Scientific; Waltham, MA) with 10% vol/vol FBS. Fibroblast DNA was genotyped at rs279858 using a commercial TaqMan genotyping assay (C__2073557_1_, ThermoFisher Scientific).
2.2 |. Culture of human embryonic stem cell lines
Five human embryonic stem cell (hESC) lines (H1, H9, CT2, CT3, and CT4) obtained from the UConn Stem Cell Core were grown on irradiated mouse embryonic fibroblasts (MEFs) using hESC media containing DMEM with F12 (DMEM/F12, 1:1 ratio, ThermoFisher Scientific) supplemented with 20% Knockout Serum Replacer (ThermoFisher Scientific), 1X nonessential amino acids, 1 mM l-glutamine (ThermoFisher Scientific), 0.1 mM β-mercaptoethanol (MP Biomedicals; Santa Ana, CA), and 4 ng/ml of basic fibroblast growth factor (bFGF, Millipore-Sigma; Burlington, MA). Cells were passaged weekly. DNA was harvested from cultures that were 60–80% confluent.
2.3 |. Generation of iPSC lines
Fibroblasts were reprogrammed into iPSCs by the Stem Cell core at UConn Health using retroviral vectors to express five factors (OCT4, SOX2, KLF4, c-MYC, and LIN28) or Sendai virus to express four factors (OCT4, SOX2, KLF4, and c-MYC). Seventeen fibroblast lines were reprogrammed using retroviral vectors and 12 lines using Sendai virus once that protocol became available at the UConn Stem Cell core. iPSCs were grown on irradiated MEFs using hESC media containing DMEM with F12 (DMEM/F12, 1:1 ratio, ThermoFisher Scientific) supplemented with 20% Knockout Serum Replacer (ThermoFisher Scientific), 1X nonessential amino acids, 1 mM l-glutamine (ThermoFisher Scientific), 0.1 mM β-mercaptoethanol (MP Biomedicals), and 4 ng/ml of bFGF (Millipore-Sigma). All clones used in this study were derived from Caucasian fibroblast donors. We examined 69 iPSC clones (19 T/T, 27 T/C, and 23 C/C, genotyped at rs279858) generated from 29 donors (mean age = 42.9 (SD = 10.6)); including eight clones from three female donors. iPSC cells were passaged weekly and DNA was harvested from cultures once they had reached 60–80% confluency.
2.4 |. iPSC differentiation into forebrain neurons
iPSCs were differentiated into forebrain-lineage neural cultures utilizing methods that we have previously described in detail (Lieberman, Levine, Kranzler, Abreu, & Covault, 2012) which were adapted from a protocol developed by the WiCell Institute (SOP-CH-207; www.wicell.org, Madison, WI). In brief, the protocol utilizes an embryoid body-based protocol wherein iPSC colonies are cultured in suspension for 4 days prior to neural induction. Neuroepithelial cells were then generated by culturing cells for 3 weeks in neural induction media containing 1X N2 supplement (Life Technologies) and 2 μg/ml heparin (Sigma Aldrich), following which cells were dissociated and plated in 24-well plates on poly-l-ornithine and Matrigel (BD Biosciences, Bedford, MA) coated glass coverslips in neural differentiation media containing 1X B27 supplement (Life Technologies), 1 μg/ml laminin (Sigma-Aldrich), and 10 ng/ml each of brain-derived neurotrophic factor (Peprotech, Rocky Hill, NJ), glial-derived neurotrophic factor (Peprotech), and insulin-like growth factor 1 (Peprotech). Mature cultures, operationally defined as 12 weeks post neural cell plating, contain a mixture of neurons and glial cells. With this protocol, approximately 50% of cells show immunostaining for the glutamatergic neuron transcription factor TBR1 and 25% for GABA-ergic markers (Fink et al., 2017; Lieberman et al., 2020; Lieberman, Kranzler, Levine, & Covault, 2018). Neurons in these cultures demonstrate spontaneous synaptic activity, functional ionotropic GABAA and glutamatergic receptors, and an ability to generate trains of action potentials in response to current injection (Lieberman et al., 2012) Fink et al., 2017; Lieberman et al., 2020). A comparison of transcriptome profiles for human iPSC-derived forebrain lineage neural cultures and human postmortem brain tissue indicated that 12-week iPSC-derived neural cultures are most similar to that of first trimester forebrain tissue (Brennand et al., 2015; Mariani et al., 2012). Normalized RNA-seq expression levels (fragments per kilobase of transcript per million mapped reads, FPKM) for iPSC-derived neural cultures for the chr4p12 GABAA receptor subunit genes (Lieberman et al., 2015) are similar to those reported for 12-week post-conception human cerebral cortex (Lindsay et al., 2016) (GABRG1 = 0.7 vs. 0.8; GABRA2 = 3.5 vs. 3.0; GABRA4 = 0.4 vs. 0.1, and GABRB1 = 0.3 vs. 0.3). We have also reported that our iPSC neural differentiation methods yield cultures with expression quantitative trait loci (eQTLs) correlating more similarly with those identified in postmortem human cerebral cortex than eQTLs identified for subcortical brain regions or whole blood (Jensen et al., 2019).
Neural cells generated from 10 iPSC clones (derived from 8 donors) were examined for GABRA2 CpG island methylation and neural cultures from 47 clones (26 donors) were available for measures of RNA levels.
2.5 |. DNA extraction
DNA from fibroblasts, hES cultures, iPSCs, and 12-week old neural cultures was extracted using the Gentra Puregene Cell Kit (Qiagen; Germantown, MD) following the manufacturer’s instructions. DNA was rehydrated in 100 μl of DNA Hydration Solution and stored at −20°C until use.
2.6 |. Methylation-sensitive restriction endonuclease-coupled quantitative polymerase chain reaction assay for GABRA2 CpG island methylation
This protocol was adapted from the EpiTect Methyl II DNA Methylation Assay (Qiagen), which is a refinement of a quantitative DNA methylation assay using methylation-sensitive and dependent restriction enzymes (REs) described by Oakes, La Salle, Robaire, and Trasler (2006). Genomic DNA is digested with a mCpG-sensitive (HhaI) or mCpG-dependent (McrBC) RE and then polymerase chain reaction (PCR) amplified using primers flanking the region of interest to allow SYBR Green real-time quantitative PCR (qPCR) to measure levels of methylated (i.e., HhaI digest-resistant) or unmethylated (i.e., McrBC digest-resistant) DNA relative to sham digested DNA. A 252 bp amplicon (hg19 chr4:46392251–46392502) encompassing multiple CpG sites in the GABRA2 promoter-proximal CpG island was designed using the Primer3 software program (Rozen & Skaletsky, 2000) and included five HhaI and multiple McrBC sites. Genomic DNA (60 ng) was digested overnight at 37°C in 30 μl reactions containing 1X NEBuffer2, 1 mM GTP, 200 μg/ml BSA, and 2 U HhaI or McrBC RE (New England Biolabs, Ipswich, MA). Restriction enzymes were inactivated by incubation at 65°C for 20 min. Mock digests omitting REs, and a double digest with both REs were included to provide for each sample a PCR reference Ct for amount of total target DNA and amount of total digestable DNA, respectively. Then, 4 μl of DNA restriction digests were PCR amplified in 20 μl reactions containing 600 nM each of the forward (CTCGATCCCTCGATCCCTGGTAACT) and reverse (CGCCACACTACATTCCCGGCTTTA) primers and 1X Qiagen RT2 SYBR Green qPCR MasterMix. Real-time PCR was conducted using an ABI 7500 thermal cycler with SYBR as the detector and ROX as the passive dye using this thermal profile: 10 min at 95°C; 3 cycles of 30 s at 99°C followed by 1 min at 72°C; and then 40 cycles of 15 s at 97°C followed by 1 min at 72°C. PCR threshold Ct values for each reaction condition were used together with the Qiagen EpiTect Methyl II PCR assay excel spreadsheet to calculate the percentage of PCR amplicons for the surveyed region that are unmethylated versus methylated at the respective RE sites. The entire assay was repeated in duplicate or triplicate for 75% of samples with a mean SD for methylation level in a given sample of 3.8%, consistent with reproducibility represented in Oakes et al. (2006).
2.7 |. Reverse transcription and qPCR
Gene expression data for 32 neural cultures were available as part of our prior report (Lieberman et al., 2015), and qPCR data from an additional 15 neural cultures were generated using the same methods as described in Lieberman et al. (2015) and briefly described here. RNA was extracted from mature neural cultures using TRIzol reagent (Life Technologies) per the manufacturer’s instructions. RNA concentrations were determined using a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific) and cDNA synthesized using a High Capacity cDNA Reverse Transcription Kit (Life Technologies). Relative transcript levels represented in the resulting cDNA were measured by real-time qPCR using FAM-labeled TaqMan probe and primer sets (Life Technologies) for GABRG1 (Hs00381554_m1), GABRA2 (Hs00168069_m1), GABRA4 (Hs00608034_m1), and GABRB1 (Hs00181306_m1). A VIC-labeled GUSB probe and primer set (β-glucoronidase, 4326320E) was used as a within-well reference gene for normalization. CT values for target genes and the internal reference gene were converted to relative expression levels using the standard curve method, which allows for comparison across assay plates by using a reference RNA sample whose RNA level for each probe is defined as 1 (Wong, Bottiglieri, & Snead III, 2003; Wong & Medrano, 2005).
2.8 |. Allelic expression imbalance qPCR
TaqMan (Life Technologies) FAM- and VIC-labeled allelic discrimination probe and primer sets targeting exonic SNPs in GABRG1 (Exon 3 synonymous rs976156, C__8723970_1), GABRA2 (Exon 5 synonymous rs279858, C_2073557_1), GABRA4 (3′ UTR rs7660336, C__11278069_10), and GABRB1 (3′ UTR rs10028945, C__2119811_30) were used to examine allelic expression bias for chr4p12 GABAA cluster subunit genes for samples heterozygous at one or more of these SNPs, as described previously (Lieberman et al., 2015). The threshold used for allelic imbalance was a 50% greater abundance of RNA from 1 of the 2 alleles, consistent with the threshold used for other studies (Lin et al., 2012; Serre et al., 2008).
2.9 |. Statistical analysis
Statistical analysis was performed using SPSS (v25, IBM, Armonk, NY). One-way analysis of variance (ANOVA) was used to compare the mean GABRA2 promoter-proximal CpG island methylation level by genotype at rs279858, sex, and reprogramming method. We used Pearson correlation (r) to examine the degree of association of donor age and iPSC GABRA2 CpG island methylation. We used chi-square analysis and pairwise comparisons to examine differences in the concordance of GABRA2 methylation levels for multiple clones generated from the same donor. We used chi-square analysis to compare the GABRA2 promoter-proximal CpG island methylation level (<25, 25–75, and >75%) with neural cell monoallelic expression (MAE) (yes vs. no) at one or more chr4p12 GABAA subunit genes. We also used simple linear regression to examine the correlations between GABRA2 methylation in iPSCs and expression of GABRA2 and GABRB1 in neural cultures and between GABRA2 methylation in iPSCs and neural cultures. We used two-way ANOVA to examine whether genotype at rs279858 and methylation level at the GABRA2 promoter-proximal CpG island were independent or interactive in predicting RNA expression of chr4p12 GABAA receptor subunit genes. Statistical significance was defined as p < .05.
3 |. RESULTS
We examined DNA methylation at the GABRA2 gene promoter region CpG island in the context of the rs279858 tag-SNP associated with AUD, (Figure 1). We assayed methylation levels in five hES cell lines, 28 fibroblast lines, 69 iPSC lines generated from 29 donor subjects, and 10 iPSC-derived neural cultures. Four of five hES cell lines had no methylation while one line, CT2, had a methylation level of 50%, consistent with one of the two chromosomes being highly methylated and the other unmethylated. All 28 fibroblast lines examined had less than 3% methylation (mean 1.3%). In contrast, the mean degree of methylation in the 69 human iPSC lines was much higher (58.5%), with iPSC methylation having a tri-phasic distribution <25, 25–75, and >75%, as illustrated in Figure 2, Panel (a). The distribution is consistent with low levels of methylation (<25%) for both chromosomes in 19 samples, high levels (>75%) for both chromosomes in 31 samples and one chromosome each with low versus high methylation for 19 samples, resulting in an intermediate level of total methylation. There was no association of mean percentage methylation with rs279858 genotype (Figure 2, Panel (b)) (F = 0.2, df = 2, p = .81), sex (F = 1.8, df = 1, p = .18) or with the reprogramming method used (retrovirus vs. Sendai virus, 60.5 and 53.7%, respectively; F = 0.47, df = 1, p = .50). There was no correlation between methylation at this site and donor age (r = −.06; p = .60).
FIGURE 1.
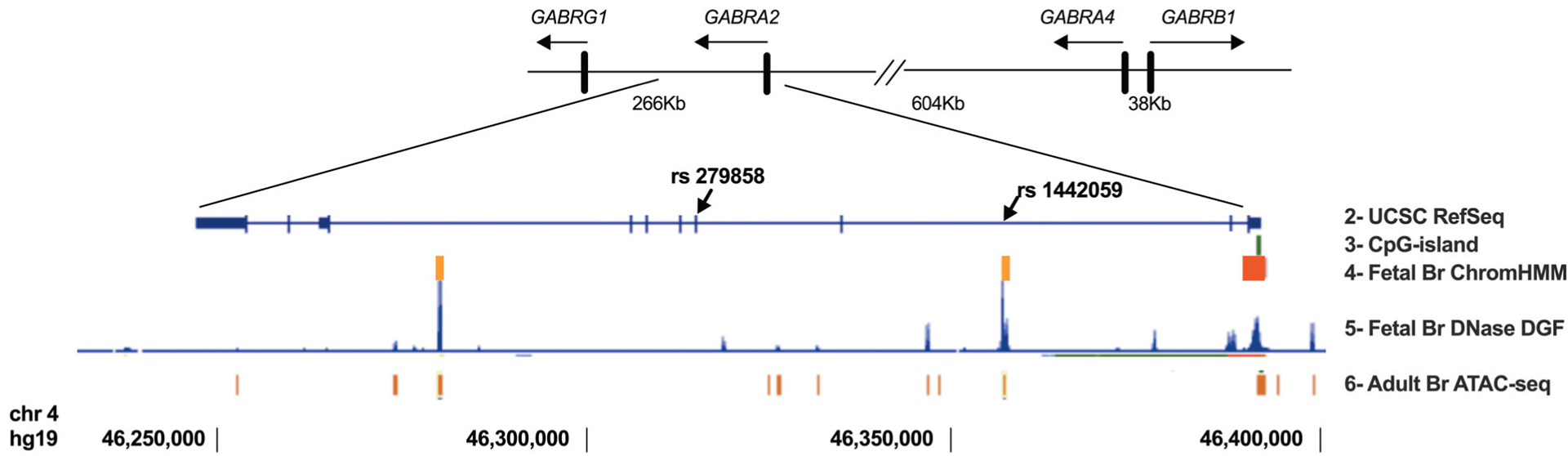
Schematic of the chromosome 4p12 cluster GABRG1, GABRA2, GABRA4, and GABRB1 genes including distances between promoters of each of the four GABAA receptor subunit genes. The second line displays the UCSC RefSeq ideogram of GABRA2 and the location of rs279858 within Exon 5 of GABRA2. The third line depicts the CpG island assayed in the methylation analysis, proximal to the GABRA2 promoter. Lines 4–6 contain chromatin landscape features that serve as guides to candidate functional single nucleotide polymorphisms (SNPs) in LD with rs279858 (e.g., rs1442059 in IVS3). The fourth line depicts fetal brain 15-state ChromHMM predictions for the locations of the GABRA2 promoter (in red) and putative enhancers (in orange), Line 5, fetal brain DNase digital genomic footprints (ENCODE Univ. Washington) highlighting potential regulatory factor binding sites and on Line 6 adult hippocampus ATAC-seq peaks (BOCA: Atlas of chromatin accessibility)
FIGURE 2.
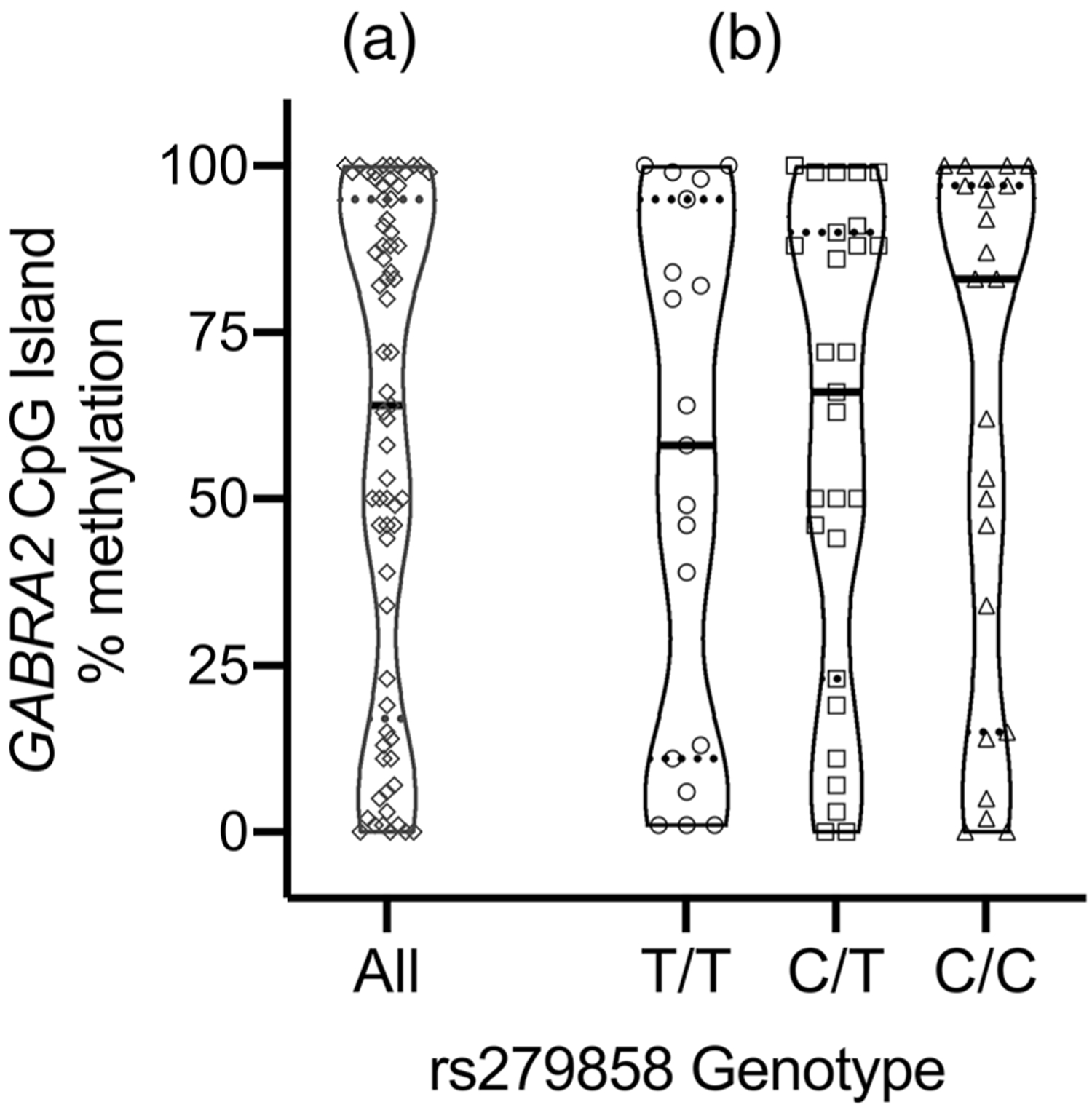
Violin plot of GABRA2 promoter region mCpG levels for 69 iPSC clones. Panel (a) Triphasic distribution of GABRA2 methylation values with samples clustered below 25% (n = 19), between 25 and 75% (n = 19), or above 75% (n = 31). Panel (b) Violin plots of methylation stratified by rs279858 genotype. There was a nonsignificant step-wise trend for increasing levels of methylation associated with the number of alcohol use disorder (AUD)-associated C alleles. Median methylation level, represented by the black horizontal bar in each violin plot, was lowest for rs279858 T/T iPSCs (57.7%) and highest for rs279858 C/C iPSCs (82.7%). Quartiles are demarcated by dashed horizontal lines. ◊ = All sample (n = 69); ○ = T/T (n = 19); □ = C/T (n = 27); Δ = C/C (n = 23)
We examined multiple independent iPSC clones from several fibroblast lines to determine whether mCpG variation in iPSCs was associated with the fibroblast line methylation differences or was a random clonal event. In analyzing GABRA2 promoter-proximal CpG island methylation for multiple iPSC clones generated from an individual donor, we found that independent clones often had discordant levels of methylation (Figure 3). In pair-wise comparisons between clones derived from the same donor, we observed a 43% concordance rate for sharing the same relative methylation level (low, intermediate, or high), which did not differ significantly from chance (χ2 = 0.8, df = 1, p = .37). This suggests the presence of strong, random, clone-specific effects of reprograming on methylation of the GABRA2 promoter-proximal CpG island.
FIGURE 3.
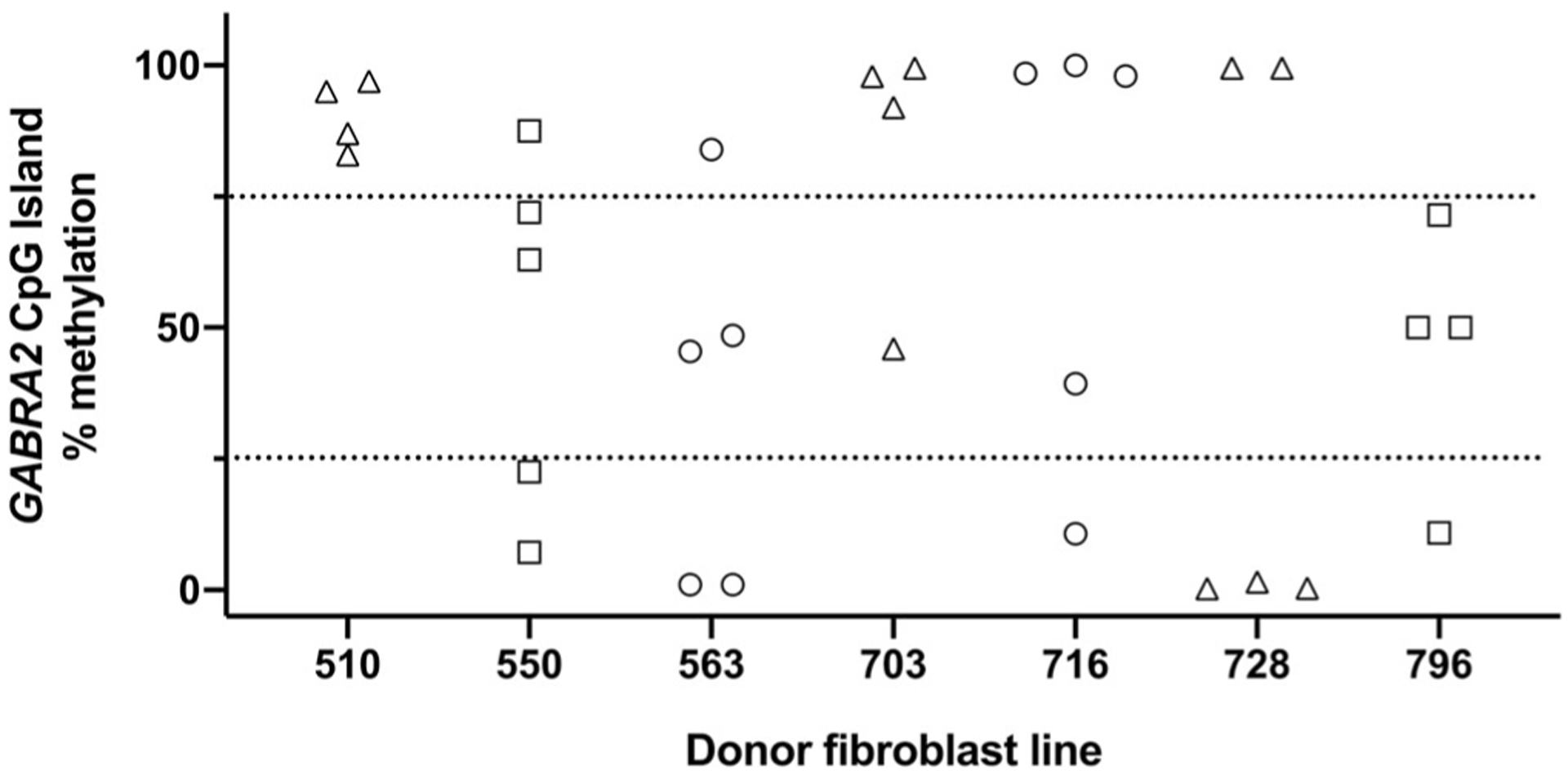
GABRA2 methylation levels are illustrated for multiple iPSC clones (4–5) generated from seven different fibroblast lines. Clones derived from an individual donor fibroblast line were frequently discordant for level of methylation (i.e., low, intermediate, or high) in the GABRA2 promoter-proximal CpG island, suggesting a strong clone-specific methylation effect at the GABRA2 locus as a result of reprogramming. Donor genotype at rs279858: ○ = T/T; □ = C/T; Δ = C/C
We examined whether GABRA2 methylation levels persisted over the course of neural differentiation by comparing GABRA2 methylation levels in 10 independent iPSC clones and their 12-week old neural progeny. As illustrated in Figure 4, there was a strong positive correlation between the methylation level observed in individual iPSC lines and neural cells derived from each line (r = .97; p < .0001). The strong correlation between GABRA2 promoter-proximal CpG island methylation in iPSCs and clone-matched neural cultures allowed for comparison of GABRA2 mCpG level in iPSCs to mRNA expression of the chr4p12 GABAA subunits in neural cultures. There was a highly significant negative correlation between GABRA2 percent methylation in iPSCs and GABAA subunit mRNA in neural cultures for GABRG1 (r = −.65), GABRA2 (r = −.82), GABRA4 (r = −.57), and GABRB1 (r = −.67). Figure 5a illustrates this relationship for GABRA2 subunit RNA and for expression of the GABRB1 gene (Figure 5, Panel (b)) which is located over 600 kb distal from the GABRA2 promoter region CpG island. In contrast, there was no correlation between GABRA2 methylation and expression of COMMD8, 25 kb distal to GABRB1 (r = .05).
FIGURE 4.
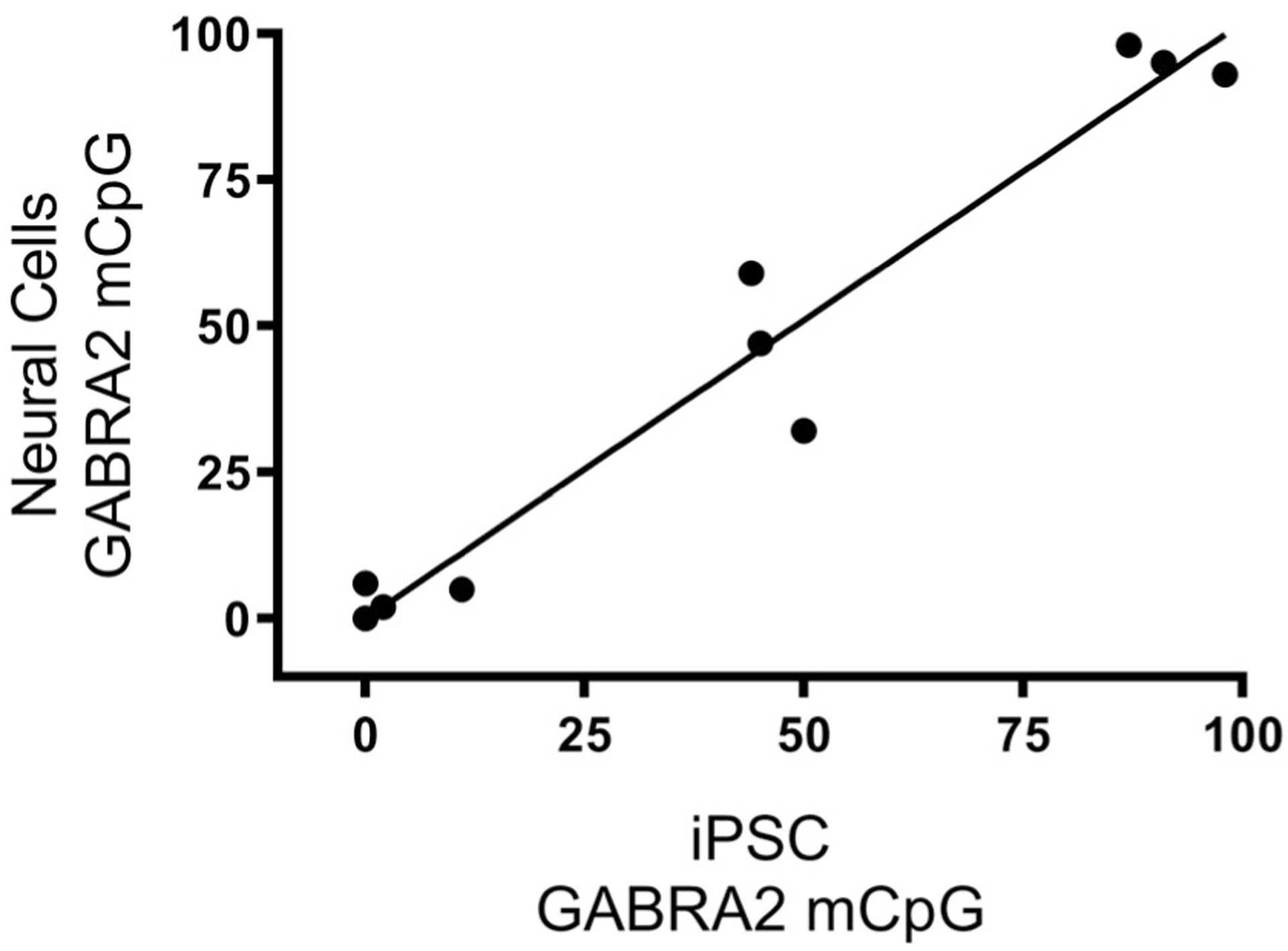
GABRA2 mCpG levels were highly correlated between individual iPSC lines and neural cultures derived from each line (r = .97; p < .0001)
FIGURE 5.
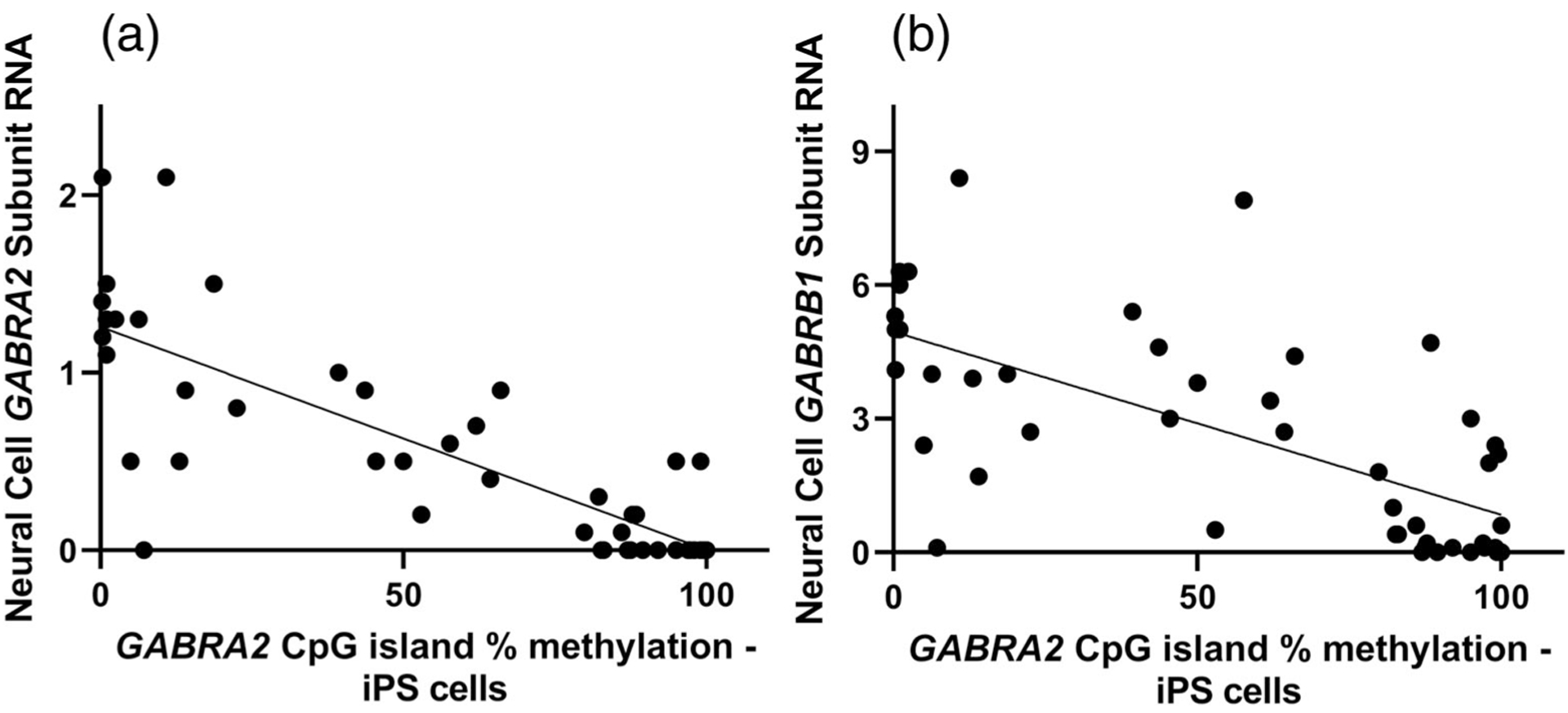
GABRA2 (a) and GABRB1 (b) expression in forebrain-lineage neural cells is negatively correlated with GABRA2 promoter-proximal CpG island methylation. GABRA2 r = −.82; p < .0001. GABRB1 r = −.67; p < .0001
As noted earlier, 19 of 69 (28%) iPSC lines had an intermediate level of GABRA2 methylation (mean = 53% for this group, which represents the average for two chromosomes) as does the hES line CT2 (with 50% methylation). These results are consistent with the hypothesis that intermediate methylation is due to one chromosome having an unmethylated GABRA2 CpG island and the partner chromosome having a high level of GABRA2 methylation. Due to the negative correlation between iPSC GABRA2 methylation and neural cell chr4p12 GABAA subunit gene expression (Figure 5), we would predict that neural cells generated from iPSCs with intermediate methylation would be more likely to show allelic expression bias of the four chr4p12 GABAA subunit genes. Examination of allelic bias in RNA expression for neural cultures heterozygous at an exonic SNP in one or more of the chr4p12 GABAA subunit genes revealed allele-biased expression in 7 of 16 lines. Consistent with our hypothesis, 83% of lines with intermediate methylation had allelic expression bias versus 14 and 25%, respectively, for lines with low or high methylation levels (χ2 = 6.4, df = 2, p = .040) (Table 1).
TABLE 1.
GABRA2 iPSC CpG island methylation levels and neural cell monoallelic RNA expression at one or more chr4p12 GABAA subunit genes
| GABRA2 iPSC methylation level | |||
|---|---|---|---|
| Count (frequency within methylation level)a | |||
| MAE | Low (<25%) | Intermediate (25% –75%) | High (>75%) |
| Yes | 1 (0.14) | 5 (0.83) | 1 (0.33) |
| No | 6 (0.86) | 1 (0.17) | 2 (0.67) |
Abbreviations: iPSC, induced pluripotent stem cell; MAE, monoallelic expression.
MAE × CpG methylation level χ2 = 6.4, df = 2, p = .040.
Finally, we used two-way ANOVA models to examine whether there were independent or interactive effects of the AUD-associated haplotype block tag-SNP rs279858 and GABRA2 promoter CpG island methylation on mRNA levels of the chr4p12 GABAA subunit genes. In models that included rs279858 genotype and GABRA2 methylation and their interaction both rs279858 genotype and methylation level were significant (F = 4.3, df = 2, p = .02 and F = 19.7, df = 2, p < .001, respectively) for GABRB1 mRNA expression but not for other subunit genes. There was no interaction of mCpG level × genotype (F = 0.44, df = 4, p = .78). Higher levels of GABRA2 methylation and the number of rs279858 AUD-associated C-alleles were additive in nature; the lowest GABRB1 neural cell mRNA levels were in C/C genotype lines with high GABRA2 methylation and the highest expression of GABRB1 was found in T/T genotype lines with low or intermediate levels of methylation (Figure 6).
FIGURE 6.
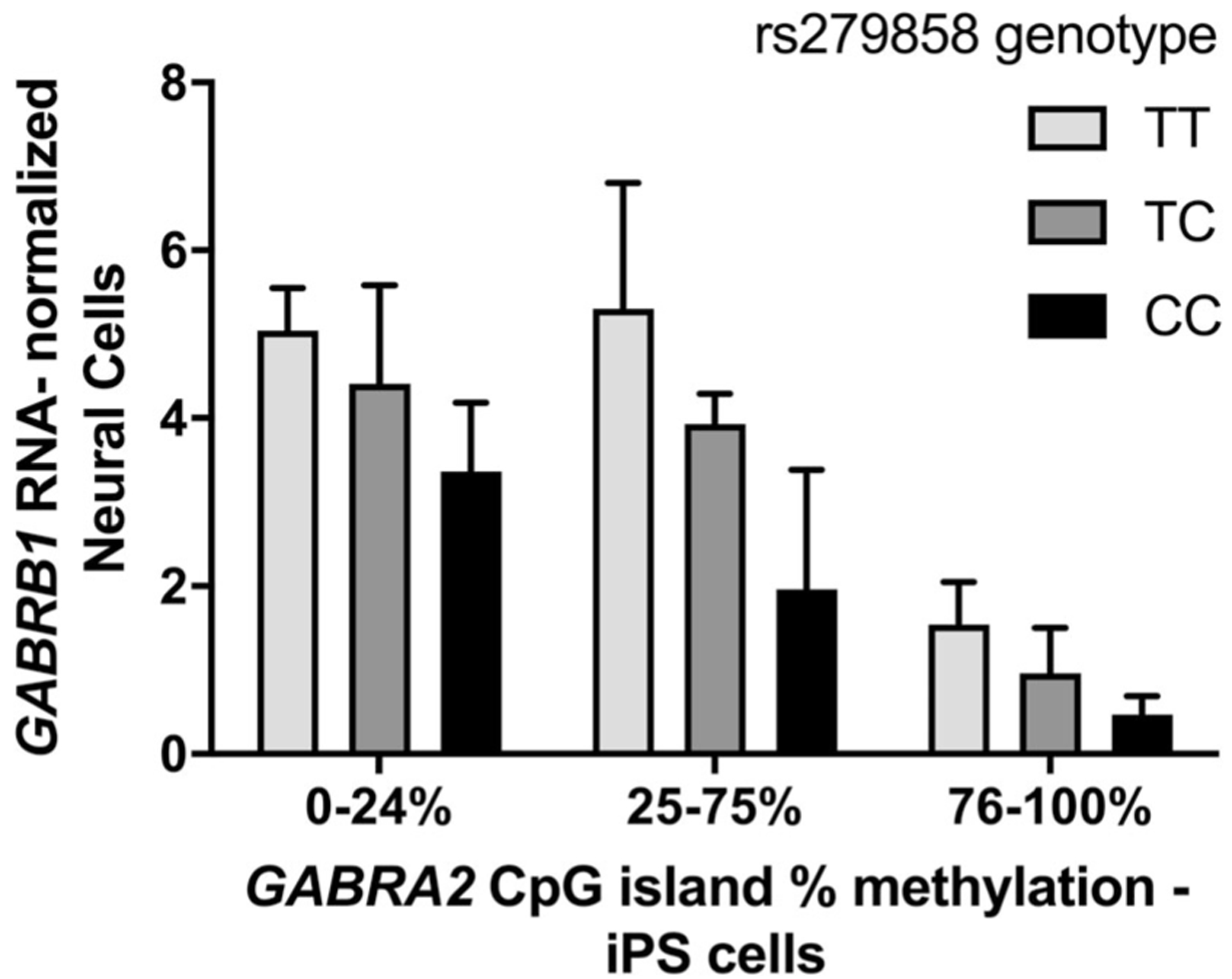
Grouped histogram of neural cell GABRB1 mRNA expression normalized to GUSB for 13 T/T, 19 C/T, and 15 C/C iPSC clones, stratified by genotype and methylation level at the GABRA2 promoter-proximal CpG island. GABRB1 expression is significantly associated with both GABRA2 promoter CpG island methylation and rs279858 genotype despite being over 600 kb distal from the GABRA2 promoter region and alcohol use disorder (AUD)-associated haplotype block
4 |. DISCUSSION
Our laboratory has used an iPSC-derived neural culture model to investigate molecular correlates of the AUD-associated GABRA2 synonymous SNP rs279858, and previously reported an endophenotype in which the transcription of chr4p12 GABAA subunit genes was significantly lower in rs279858 C-allele carriers than T-allele homozygotes (Lieberman et al., 2015). In the current study, we examined whether promoter-proximal CpG methylation at GABRA2 is a factor linking rs279858 genotype to the previously observed transcriptional endophenotype in this human neural culture model system. The regulation of gene expression by promoter-proximal CpG methylation has been proposed as one molecular mechanism mediating stable adaptations (or in the case of neuropsychiatric disorders, maladaptations) in the brain (Rukova et al., 2014; Tsankova, Renthal, Kumar, & Nestler, 2007). Other mechanisms to consider but that were not directly examined here include SNP effects on enhancer functions or intrachromosomal interaction sites frequently associated with open chromatin states (see Forrest et al., 2017 for an example of the use of iPSC derived neurons guided by open chromatin regions identified by ATAC-seq in evaluating disease associated SNPs) (Forrest et al., 2017).
We found that methylation at the GABRA2 promoter-proximal CpG island was randomly established during reprogramming of fibroblasts (which uniformly lack methylation at this site) into iPSCs such that 58% of chromosomes experienced de novo methylation of the GABRA2 CpG island during reprogramming. There was no significant association of rs279858 genotype with methylation at the GABRA2 CpG island suggesting that the effects of functional loci linked to the rs279858 tag-SNP on GABAA gene expression in this model are independent of promoter methylation. Of note, rs279858 was not identified as a methylation QTL (meQTL) in postmortem brain, (meQTL database (Gaunt et al., 2016), BIOS QTL browser (Bonder et al., 2017)). Consistent with a random clone specific methylation event at this locus during iPSC reprogramming, we observed that sister clones generated from the same donor were frequently discordant in methylation levels. A question raised in the stem cell biology literature is whether the clonal variation in CpG island methylation observed in iPSCs is the result of reprogramming a mosaic fibroblast culture or if resetting of this methylation occurs during reprogramming itself; our results support the latter for the GABRA2 locus. Within an iPSC clone, methylation levels were stable over time in culture and clonal variation in GABRA2 CpG methylation was maintained during neural differentiation of iPSCs. This stability in GABRA2 methylation levels allowed us to compare GABRA2 CpG island methylation in iPSCs with mRNA expression of chr4p12 GABAA receptor subunit genes in clonally matched differentiated neural cell cultures. Consistent with a broad literature that extends from cancer biology to studies of neuronal gene expression (Baylin et al., 2001; Leung et al., 2015) and shows that promoter region CpG island methylation is associated with reduced gene expression, we observed a significant negative correlation between GABRA2 CpG island methylation in iPSCs and GABRA2 mRNA levels in neural cultures. We also observed a significant negative correlation between GABRA2 CpG island methylation in iPSCs and expression of the three other chr4p12 GABAA receptor subunit genes in neural cultures. This correlation did not extend to the COMMD8 gene, 25 kb distal to GABRB1, suggesting the existence of a GABAA subunit gene cluster specific cis-regulatory effect of GABRA2 methylation in human cells. Consistent with this observation, mRNA for the chr4 GABAA subunit genes γ1, α2, and β1 have been reported to decrease in parallel in the prefrontal cortex over the course of development from the neonatal period to young adulthood (Fillman et al., 2010; Hashimoto et al., 2009).
In the samples where we observed intermediate methylation (i.e., 25–75%), there was a significant association of MAE bias for one or more chr4p12 GABAA receptor subunit genes in iPSC-derived neural cultures with intermediate methylation level at the GABRA2 promoter. In ongoing research, preliminary findings show a paucity of ATAC-seq reads in the chr4p12 GABAA receptor subunit gene region in a neural culture derived from a highly methylated iPSC line, which contrasts with neural cultures from iPSCs with low levels of GABRA2 promoter CpG-island methylation. This finding suggests that methylation at the GABRA2 locus during iPSC reprogramming leads to a recruitment of factors that convert this chromosomal region into a relatively closed heterochromatin state. We hypothesize that GABRA2 promoter methylation and subsequent inactivation of the region encompassing the GABAA receptor subunit gene cluster for one but not both chromosomes during reprogramming in clones with intermediate levels of methylation results in the MAE bias observed in our transcriptomic analysis of iPSC-derived neural cultures for lines (Table 1). While MAE was originally defined in immune cells (T and B) or in relation to X chromosome inactivation, random MAE has subsequently been described in autosomes in various cell types (Gimelbrant, Hutchinson, Thompson, & Chess, 2007; Savol et al., 2017). Further, MAE of neural-specific genes may be associated with genetic risk for developing neuropsychiatric disorders (Jeffries et al., 2013; Lin et al., 2012).
Results from a study of aberrantly methylated CpG islands using human iPSCs generated from three pairs of female Caucasian monozygotic twins (Panopoulos et al., 2017) are of interest in considering our results. In this study, aberrant methylation was defined as CpG island methylation that differed from what was expected in comparing human iPSCs to a panel of 15 hESC lines. Clone-specific aberrantly methylated CpG sites were identified in 463 genes, including the chr4 GABRA2, GABRA4, and GABRB1, but not GABAA receptor subunit genes on other chromosomes (Panopoulos et al., 2017). Aberrantly methylated CpG sites were enriched in regulatory regions marked by MYC-family transcription factor binding sites (TFBS; Panopoulos et al., 2017). Consistent with Panopoulos et al. (2017), the TFBS identified by ChIP-seq data for H1 hES cells from ENCODE datasets (UT Austin, Release August 2, 2011) show prominent c-Myc binding flanking the GABRA2 promoter region CpG island (Consortium et al., 2007; Euskirchen et al., 2007).
Myc-transcription factors are typically included as part of Yamanaka-based iPSC reprogramming factor protocols as cMyc-activated genes enhance cell proliferation and immortality, while also modulating cellular metabolism to support these processes (Nakagawa et al., 2008; Nakagawa, Takizawa, Narita, Ichisaka, & Yamanaka, 2010). The inclusion of cMyc as one of the reprogramming transcription factors could have contributed to the random clone-specific GABRA2 CpG island methylation that we observed in our iPSC samples. Although Myc family factors are widely used in protocols for reprogramming somatic cells, OCT4, SOX2, NANOG, and LIN28A were reported by Yu et al. (2007) to be sufficient for reprogramming human adult somatic cells into pluripotent stem cells. A study that directly compared reprogramming using Yamanaka with the Yu et al. (2007) factors found that fibroblasts reprogrammed using the more common Yamanaka factors resulted in greater failure to demethylate fibroblast specific CpG’s in contrast to the Yu et al. (2007) factors, which showed a relative failure to methylate stem cell-relevant CpGs (Planello et al., 2014). No examination of the differences between methods for aberrant methylation in Myc-binding regions or GABRA2 promoter-associated CpG-island methylation was reported.
While our results identifying GABRA2 promoter region CpG methylation to be markedly greater on average in iPSC compared with hES cell lines confirm those of Panopoulos et al. (2017), two observations warrant noting relative to the question of whether methylation we observed at this site is simply an artifact of the exogenous cMyc used in reprogramming. First, one of the five hESC lines that we examined, CT2, demonstrated an intermediate (50%) level of methylation at the GABRA2 promoter-proximal CpG island. The observation of an intermediate level of methylation in an hESC line that had not undergone reprogramming indicates that exposure to exogenous cMyc is not required for establishment of GABRA2 promoter-proximal CpG island methylation. Second, Siegmund et al. (2007) reported that GABRA2 promoter CpG-island methylation increases with age in neurosurgical samples of human cerebral cortex, lending support to the hypothesis that reprogramming is not the only trigger for methylation of the GABRA2 promoter CpG-island. It is notable that the amplicon (hg19 chr4:46,392,368-chr4:46,392,443) used to assess GABRA2 CpG-island methylation in the Siegmund et al. (2007) study is within the amplicon that we used in our assay. Together with our results, these findings suggest that GABRA2 promoter methylation state should be considered when researchers use stem cells and their downstream derivatives to explore molecular correlates of the chr4 GABAA subunit genomic region and that preference be considered for use of iPSC and neural culture lines with low levels of GABRA2 methylation in genetic experiments in which the interpretation involves measuring mRNAs for GABAA receptor subunit genes.
Independent of the strong negative correlation between methylation at GABRA2 and chr4p12 GABAA subunit gene expression, in models that include both genotype and methylation levels, we observed a significant relationship between rs279858 genotype and mRNA levels for GABRB1 but not GABRG1, GABRA2, or GABRA4, such that neural cultures from rs279858 C-allele carrier donors had significantly lower GABRB1 mRNA expression than T-allele homozygotes. The effects on GABRB1 transcription in neural cell cultures of methylation at the GABRA2 CpG island and the rs279858 genotype appeared to be additive. These results are consistent with the presence of long-range intrachromosomal interactions between the GABRA2 AUD-associated yin-yang haplotype region and the GABRB1 gene. Virtual circular chromatin conformation capture (4C-seq) data from human dorsolateral prefrontal cortex suggest the presence of such an interaction (Schmitt et al., 2016). Functional AUD-associated variants in GABRA2 may in part act by regulating the GABRB1 gene. Similar phenomena in which SNPs alter the function of regulatory elements that affect distal sites rather than the nearest gene have been identified in normal development, such as of hair, skin, and eye pigmentation (Visser, Kayser, & Palstra, 2012), and in the predisposition to, or severity of, specific health conditions including cardiovascular disease (Montefiori et al., 2018), prostate cancer (Qian et al., 2019), obesity (Smemo et al., 2014), and asthma (Tang et al., 2015). The tag-SNP rs279858 is unlikely to be the functional genetic element underlying the association of the region with AUD. The identification of candidate functional SNPs will benefit from considering regions of intrachromosomal contact and the chromatin landscape in the AUD-associated haplotype region including open chromatin and enhancer histone marks. Although rs279858 is not in a region with open chromatin or regulatory element binding sites, other SNPs in LD with rs279858 are (e.g., rs1442059, which is located in an open chromatin/enhancer-marked region in GABRA2 IVS3, Figure 1) and provide candidates to examine by combining gene editing with this iPSC-derived neural cell culture model.
There are several limitations of this study that should be noted. First, while we have analyzed 69 clones from 29 donors, only 3 of those donors (contributing 8 clones) were female. While there was no significant correlation between GABRA2 promoter-proximal CpG methylation and sex, the limited number of female donors limits the strength of this finding. Second, the two reprogramming methods used in this study both included c-Myc which may promote methylation near MYC-factor binding sites (Panopoulos et al., 2017) such as the GABRA2 promoter-proximal CpG island. Examination of GABRA2 promoter methylation for iPSCs generated without c-Myc would be informative. Third, as the transcriptome profile of iPSC-derived neural cells is similar to first-trimester fetal brain (Brennand et al., 2015; Mariani et al., 2012), our results with this system may reflect developmentally constrained regulatory effects rather than adult brain regulatory effects. Indeed, we found no association of rs279858 genotype with chromosome 4p12 GABAA gene expression in postmortem adult cortex in either the BrainCloud or Braineac postmortem adult brain datasets (Lieberman et al., 2015) and rs279858 is not annotated as an eQTL in the adult brain in the Genotype-Tissue Expression Portal. Though rs279858 is not associated with the expression of chromosome 4p12 GABAA genes in adult postmortem brain, functional genetic loci in this region could increase risk for AUD via developmental mechanisms. Fourth, our results provide only a limited advance in understanding of the nature of a GABRA2 functional genetic locus contributing to the risk of AUD. Finally, our results documenting that methylation at the GABRA2 promoter is randomly reset as part of widely used iPSC reprograming methods indicate that methylation at this site should be considered a possible confounder in studies assessing the chromatin and regulatory landscape in this region in iPSC derived cells. The use of iPSC and iPSC-derived neural cultures with low levels of GABRA2 methylation should be considered in studies seeking to identify putative functional SNPs in LD with the AUD-associated synonymous tag-SNP rs279858 in this model system.
In sum, we show that random GABRA2 CpG methylation set during the reprogramming of fibroblasts to iPSCs is stable over differentiation and associated with mRNA expression of GABRA2 but also GABRB1, a gene ~600 kb away from GABRA2. Independently of this de novo reprogramming associated DNA methylation, AUD-associated genetic variation in GABRA2 is correlated with GABRB1 transcript expression in models that control for separate effects of reprogramming associated methylation (Figure 6). Our findings complement previous studies in alcohol research that examined the chr4p12 GABA receptor subunit gene locus, but are also of broad interest to other groups utilizing iPSC reprogramming for the study of neuropsychiatric disorders in general. Future studies aimed at identifying and characterizing functional variants that mediate regulatory effects of chromatin loop formation between GABRA2 and GABRB1 could help to explain the transcriptional endophenotype that we observed in iPSC-derived neural cultures associated with the AUD-associated tag-SNP rs279858.
ACKNOWLEDGMENTS
The authors would like to thank Leann Crandall for her valued assistance in generating iPSC lines and Nicholas Wasko and Austen Katz for help developing methylation assays. This work was supported by NIAAA grant P60 AA03510 to the UConn Health Alcohol Research Center; the Connecticut Department of Health Regenerative Medicine Fund grant 15-RMB-UCHC-04 to J. C.; NIAAA grant F30 AA027153 to A. G.; and the VISN 4 Mental Illness Research, Education and Clinical Center of the Department of Veteran’s Affairs.
CONFLICTS OF INTEREST
Dr H.R. K. is a member of an advisory board for Dicerna and the Alcohol Clinical Trials Group of the American Society of Clinical Pharmacology, which was sponsored in the past 3 years by AbbVie, Alkermes, Amygdala Neurosciences, Arbor Pharmaceuticals, Dicerna, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, and Pfizer. He is also named as an inventor on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. The other authors declare no conflicts of interest.
Footnotes
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Barnard EA, Skolnick P, Olsen RW, Mohler H, Sieghart W, Biggio G, … Langer SZ (1998). International Union of Pharmacology. XV. Subtypes of gamma-aminobutyric acid A receptors: Classification on the basis of subunit structure and receptor function. Pharmacological Reviews, 50(2), 291–313. [PubMed] [Google Scholar]
- Bauer LO (2001). Predicting relapse to alcohol and drug abuse via quantitative electroencephalography. Neuropsychopharmacology, 25(3), 332–340. 10.1016/S0893-133X(01)00236-6 [DOI] [PubMed] [Google Scholar]
- Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, & Herman JG (2001). Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Human Molecular Genetics, 10(7), 687–692. 10.1093/hmg/10.7.687 [DOI] [PubMed] [Google Scholar]
- Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, … Heijmans BT (2017). Disease variants alter transcription factor levels and methylation of their binding sites. Nature Genetics, 49(1), 131–138. 10.1038/ng.3721 [DOI] [PubMed] [Google Scholar]
- Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, … Gage FH (2015). Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Molecular Psychiatry, 20(3), 361–368. 10.1038/mp.2014.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, … de Jong PJ (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature, 447(7146), 799–816. 10.1038/nature05874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, & Kranzler HR (2004). Allelic and haplotypic association of GABRA2 with alcohol dependence. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 129B(1), 104–109. 10.1002/ajmg.b.30091 [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, & Kranzler HR (2008). Markers in the 5′-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology, 33(4), 837–848. 10.1038/sj.npp.1301456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Bierut L, Hinrichs A, Fox L, Bucholz KK, Kramer J, … Foroud T (2006). The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behavior Genetics, 36(4), 577–590. 10.1007/s10519-005-9041-8 [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, … Begleiter H (2004). Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. American Journal of Human Genetics, 74(4), 705–714. 10.1086/383283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA (2008). The role of GABA(A) receptors in the development of alcoholism. Pharmacology, Biochemistry, and Behavior, 90(1), 95–104. 10.1016/j.pbb.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, … Snyder M (2007). Mapping of transcription factor binding regions in mammalian cells by ChIP: Comparison of array- and sequencing-based technologies. Genome Research, 17(6), 898–909. 10.1101/gr.5583007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, … Szegedi A (2006). Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatric Genetics, 16(1), 9–17. 10.1097/01.ypg.0000185027.89816.d9 [DOI] [PubMed] [Google Scholar]
- Fillman SG, Duncan CE, Webster MJ, Elashoff M, & Weickert CS (2010). Developmental co-regulation of the beta and gamma GABAA receptor subunits with distinct alpha subunits in the human dorsolateral prefrontal cortex. International Journal of Developmental Neuroscience, 28(6), 513–519. 10.1016/j.ijdevneu.2010.05.004 [DOI] [PubMed] [Google Scholar]
- Fink JJ, Robinson TM, Germain ND, Sirois CL, Bolduc KA, Ward AJ, … Levine ES (2017). Disrupted neuronal maturation in Angelman syndrome-derived induced pluripotent stem cells. Nature Communications, 8, 15038. 10.1038/ncomms15038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest MP, Zhang H, Moy W, McGowan H, Leites C, Dionisio LE, … Duan J (2017). Open chromatin profiling in hiPSC-derived neurons prioritizes functional noncoding psychiatric risk variants and highlights neurodevelopmental loci. Cell Stem Cell, 21(3), 305–318 e308. 10.1016/j.stem.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, … Relton CL (2016). Systematic identification of genetic influences on methylation across the human life course. Genome Biology, 17, 61. 10.1186/s13059-016-0926-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, & Chess A (2007). Widespread monoallelic expression on human autosomes. Science, 318(5853), 1136–1140. 10.1126/science.1148910 [DOI] [PubMed] [Google Scholar]
- Grant BF, Chou SP, Saha TD, Pickering RP, Kerridge BT, Ruan WJ, … Hasin DS (2017). Prevalence of 12-month alcohol use, high-risk drinking, and DSM-IV alcohol use disorder in the United States, 2001–2002 to 2012–2013: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. JAMA Psychiatry, 74(9), 911–923. 10.1001/jamapsychiatry.2017.2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Xiao A, & Allis CD (2004). Linking the epigenetic ‘language’ of covalent histone modifications to cancer. British Journal of Cancer, 90(4), 761–769. 10.1038/sj.bjc.6601575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Nguyen QL, Rotaru D, Keenan T, Arion D, Beneyto M, … Lewis DA (2009). Protracted developmental trajectories of GABAA receptor alpha1 and alpha2 subunit expression in primate prefrontal cortex. Biological Psychiatry, 65(12), 1015–1023. 10.1016/j.biopsych.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries AR, Collier DA, Vassos E, Curran S, Ogilvie CM, & Price J (2013). Random or stochastic monoallelic expressed genes are enriched for neurodevelopmental disorder candidate genes. PLoS One, 8(12), e85093. 10.1371/journal.pone.0085093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KP, Lieberman R, Kranzler HR, Gelernter J, Clinton K, & Covault J (2019). Alcohol-responsive genes identified in human iPSC-derived neural cultures. Translational Psychiatry, 9(1), 96. 10.1038/s41398-019-0426-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Wetherill R, Feinn R, Pond T, Gelernter J, & Covault J (2014). Posttreatment effects of topiramate treatment for heavy drinking. Alcoholism, Clinical and Experimental Research, 38(12), 3017–3023. 10.1111/acer.12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, … Gelernter J (2005). Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcoholism, Clinical and Experimental Research, 29(4), 493–498. 10.1097/01.alc.0000158938.97464.90 [DOI] [PubMed] [Google Scholar]
- Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, … Ren B (2015). Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature, 518(7539), 350–354. 10.1038/nature14217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Sulovari A, Cheng C, Zhao H, Kranzler HR, & Gelernter J (2014). Association of gamma-aminobutyric acid A receptor alpha2 gene (GABRA2) with alcohol use disorder. Neuropsychopharmacology, 39(4), 907–918. 10.1038/npp.2013.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman R, Jensen KP, Clinton K, Levine ES, Kranzler HR, & Covault J (2020). Molecular correlates of topiramate and GRIK1 rs2832407 genotype in pluripotent stem cell-derived neural cultures. Alcoholism, Clinical and Experimental Research, 44, 1561–1570. 10.1111/acer.14399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman R, Kranzler HR, Joshi P, Shin DG, & Covault J (2015). GABRA2 alcohol dependence risk allele is associated with reduced expression of chromosome 4p12 GABAA subunit genes in human neural cultures. Alcoholism, Clinical and Experimental Research, 39(9), 1654–1664. 10.1111/acer.12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman R, Kranzler HR, Levine ES, & Covault J (2018). Examining the effects of alcohol on GABAA receptor mRNA expression and function in neural cultures generated from control and alcohol dependent donor induced pluripotent stem cells. Alcohol, 66, 45–53. 10.1016/j.alcohol.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman R, Levine ES, Kranzler HR, Abreu C, & Covault J (2012). Pilot study of iPS-derived neural cells to examine biologic effects of alcohol on human neurons in vitro. Alcoholism, Clinical and Experimental Research, 36(10), 1678–1687. 10.1111/j.1530-0277.2012.01792.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Hrabovsky A, Pedrosa E, Wang T, Zheng D, & Lachman HM (2012). Allele-biased expression in differentiating human neurons: Implications for neuropsychiatric disorders. PLoS One, 7(8), e44017. 10.1371/journal.pone.0044017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay SJ, Xu Y, Lisgo SN, Harkin LF, Copp AJ, Gerrelli D, … Chinnery PF (2016). HDBR expression: A unique resource for global and individual gene expression studies during early human brain development. Frontiers in Neuroanatomy, 10, 86. 10.3389/fnana.2016.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, … Vaccarino FM (2012). Modeling human cortical development in vitro using induced pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America, 109(31), 12770–12775. 10.1073/pnas.1202944109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milivojevic V, Feinn R, Kranzler HR, & Covault J (2014). Variation in AKR1C3, which encodes the neuroactive steroid synthetic enzyme 3alpha-HSD type 2 (17beta-HSD type 5), moderates the subjective effects of alcohol. Psychopharmacology (Berl), 231(17), 3597–3608. 10.1007/s00213-014-3614-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori LE, Sobreira DR, Sakabe NJ, Aneas I, Joslin AC, Hansen GT, … Nobrega MA (2018). A promoter interaction map for cardiovascular disease genetics. eLife, 7, e35788. 10.7554/eLife.35788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Abreo T, Neuner SM, Parks C, Watkins CE, Houseal MT, … Williams RW (2019). Identification of a functional non-coding variant in the GABA a receptor alpha2 subunit of the C57BL/6J mouse reference genome: Major implications for neuroscience research. Frontiers in Genetics, 10, 188. 10.3389/fgene.2019.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, … Yamanaka S (2008). Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature Biotechnology, 26(1), 101–106. 10.1038/nbt1374 [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Takizawa N, Narita M, Ichisaka T, & Yamanaka S (2010). Promotion of direct reprogramming by transformation-deficient Myc. Proceedings of the National Academy of Sciences of the United States of America, 107(32), 14152–14157. 10.1073/pnas.1009374107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, la Salle S, Robaire B, & Trasler JM (2006). Evaluation of a quantitative DNA methylation analysis technique using methylation-sensitive/dependent restriction enzymes and real-time PCR. Epigenetics, 1(3), 146–152. 10.4161/epi.1.3.3392 [DOI] [PubMed] [Google Scholar]
- Panopoulos AD, Smith EN, Arias AD, Shepard PJ, Hishida Y, Modesto V, … Frazer KA (2017). Aberrant DNA methylation in human iPSCs associates with MYC-binding motifs in a clone-specific manner independent of genetics. Cell Stem Cell, 20(4), 505–517 e506. 10.1016/j.stem.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirker S, Schwarzer C, Wieselthaler A, Sieghart W, & Sperk G (2000). GABA(A) receptors: Immunocytochemical distribution of 13 subunits in the adult rat brain. Neuroscience, 101(4), 815–850. 10.1016/s0306-4522(00)00442-5 [DOI] [PubMed] [Google Scholar]
- Planello AC, Ji J, Sharma V, Singhania R, Mbabaali F, Muller F, … Batada NN (2014). Aberrant DNA methylation reprogramming during induced pluripotent stem cell generation is dependent on the choice of reprogramming factors. Cell Regeneration, 3(1), 4. 10.1186/2045-9769-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Zhang L, Cai M, Li H, Xu H, Yang H, … Lu W (2019). The prostate cancer risk variant rs55958994 regulates multiple gene expression through extreme long-range chromatin interaction to control tumor progression. Science Advances, 5(7), eaaw6710. 10.1126/sciadv.aaw6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, & Skaletsky H (2000). Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology, 132, 365–386. 10.1385/1-59259-192-2:365 [DOI] [PubMed] [Google Scholar]
- Rukova B, Staneva R, Hadjidekova S, Stamenov G, Milanova t., & Toncheva D (2014). Genome-wide methylation profiling of schizophrenia. Balkan Journal of Medical Genetics: BJMG, 17(2), 15–23. 10.2478/bjmg-2014-0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savol AJ, Wang PI, Jeon Y, Colognori D, Yildirim E, Pinter SF, … Sadreyev RI (2017). Genome-wide identification of autosomal genes with allelic imbalance of chromatin state. PLoS One, 12(8), e0182568. 10.1371/journal.pone.0182568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, … Ren B (2016). A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Reports, 17(8), 2042–2059. 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, … Hudson TJ (2008). Differential allelic expression in the human genome: A robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genetics, 4(2), e1000006. 10.1371/journal.pgen.1000006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund T, Schumm-Draeger PM, Antoni D, & Bibra HV (2007). Beneficial effects of ramipril on myocardial diastolic function in patients with type 2 diabetes mellitus, normal LV systolic function and without coronary artery disease: A prospective study using tissue Doppler. Diabetes & Vascular Disease Research, 4(4), 358–364. 10.3132/dvdr.2007.065 [DOI] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, … Nobrega MA (2014). Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature, 507(7492), 371–375. 10.1038/nature13138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger JL, & Russek SJ (2004). GABAA receptors: Building the bridge between subunit mRNAs, their promoters, and cognate transcription factors. Pharmacology & Therapeutics, 101(3), 259–281. 10.1016/j.pharmthera.2003.12.002 [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum AD, Breen G, … Psychiatric Genomics Consortium. (2018). Psychiatric genomics: An update and an agenda. The American Journal of Psychiatry, 175(1), 15–27. 10.1176/appi.ajp.2017.17030283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, … Ruan Y (2015). CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell, 163(7), 1611–1627. 10.1016/j.cell.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawa EA, Hall SD, & Lohoff FW (2016). Overview of the genetics of alcohol use disorder. Alcohol and Alcoholism, 51(5), 507–514. 10.1093/alcalc/agw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, & Nestler EJ (2007). Epigenetic regulation in psychiatric disorders. Nature Reviews. Neuroscience, 8(5), 355–367. 10.1038/nrn2132 [DOI] [PubMed] [Google Scholar]
- Verhulst B, Neale MC, & Kendler KS (2015). The heritability of alcohol use disorders: A meta-analysis of twin and adoption studies. Psychological Medicine, 45(5), 1061–1072. 10.1017/S0033291714002165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villafuerte S, Heitzeg MM, Foley S, Yau WY, Majczenko K, Zubieta JK, … Burmeister M (2012). Impulsiveness and insula activation during reward anticipation are associated with genetic variants in GABRA2 in a family sample enriched for alcoholism. Molecular Psychiatry, 17(5), 511–519. 10.1038/mp.2011.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M, Kayser M, & Palstra RJ (2012). HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Research, 22(3), 446–455. 10.1101/gr.128652.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CG, Bottiglieri T, & Snead OC III. (2003). GABA, gamma-hydroxybutyric acid, and neurological disease. Annals of Neurology, 54(Suppl 6), S3–S12. 10.1002/ana.10696 [DOI] [PubMed] [Google Scholar]
- Wong ML, & Medrano JF (2005). Real-time PCR for mRNA quantitation. BioTechniques, 39(1), 75–85. 10.2144/05391RV01 [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, & Thomson JA (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318(5858), 1917–1920. 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]


