Summary
With the advent of deep generative models in computational chemistry, in-silico drug design is undergoing an unprecedented transformation. Although deep learning approaches have shown potential in generating compounds with desired chemical properties, they disregard the cellular environment of target diseases. Bridging systems biology and drug design, we present a reinforcement learning method for de novo molecular design from gene expression profiles. We construct a hybrid Variational Autoencoder that tailors molecules to target-specific transcriptomic profiles, using an anticancer drug sensitivity prediction model (PaccMann) as reward function. Without incorporating information about anticancer drugs, the molecule generation is biased toward compounds with high predicted efficacy against cell lines or cancer types. The generation can be further refined by subsidiary constraints such as toxicity. Our cancer-type-specific candidate drugs are similar to cancer drugs in drug-likeness, synthesizability, and solubility and frequently exhibit the highest structural similarity to compounds with known efficacy against these cancer types.
Subject areas: Complex System Biology, Systems Biology, Transcriptomics, Computer Science
Graphical abstract

Highlights
-
•
A conditional generative model for de novo design of anticancer hit molecules is devised
-
•
Drug sensitivity and toxicity models steer the molecule design via reinforcement learning
-
•
Molecules are designed to target individual transcriptomic profiles of cell lines
-
•
Targeted, hit-like molecules are generated more frequently, even for unseen cell lines
-
•
In silico, the molecules exhibit similar physicochemical properties to real cancer drugs
Complex System Biology; Systems Biology; Transcriptomics; Computer Science
Introduction
Drug discovery
Eroom's law describes the observation that the productivity of the drug discovery pipeline, as measured by the number of FDA-approved drugs per billion US dollar invested, has been halved every 9 years since the 1950s (Scannell et al., 2012). Indeed, only a minimal portion of all synthesized drug candidates obtain market approval (less than 0.01%), with an estimated 10–15 years until market release and costs that range between one (Scannell et al., 2012) and three billion dollars per drug (DiMasi et al., 2016). This low efficiency has been attributed to the high dropout rate of candidate molecules in the early stages of the pipeline, highlighting the need for more accurate in silico and in vitro models that produce more potent candidate drugs. In addition to the initial wet-lab validations, the discovery pipeline involves a sequential process that builds upon high-throughput screenings, ADMET-assessments, and a lengthy phase of clinical trials. The costs of the experimental and clinical phase can be prohibitive and any solution that helps to reduce the number of required experimental assays can provide a competitive advantage and reduce time to market. The problem's linchpin is on how to improve the exploration and navigation through the chemical space that has been estimated to contain ∼1030–1060 drug-like molecules with bioactive properties (Polishchuk et al., 2013).
Related work
Deep learning methods have recently gained popularity to aid drug discovery (Chen et al., 2018) and many have demonstrated the feasibility of in silico design of novel candidate compounds with desired chemical properties (Popova et al., 2018; Gomez-Bombarelli et al., 2018; You et al., 2018). In all of these models, the generative process is controlled by a structurally driven evaluator (or critic) that biases the generation of a chemical to satisfy the required chemical structural properties. Although very effective in generating compounds with desired chemical properties, these methods disregard system-level information, e.g. about the cellular environment in which the drug is intended to act. However, the two main causes of the increasing attrition rate in drug design are a lack in efficacy against the specific disease of interest and off-target cytotoxicity (Wehling, 2009), calling to bridge systems biology closer with drug discovery. Related methodology has been used for protein-targeting de novo generation (Zhavoronkov et al., 2019; Aumentado-Armstrong, 2018; Grechishnikova, 2021; Chenthamarakshan et al., 2020; Skalic et al., 2019; Krishnan et al., 2021). These contributions attempt to utilize deep learning methods for de novo design of compounds to specifically target a protein that has been implicated in tumor proliferation or treatment response (e.g. gene-knockout study). For example, the study by Zhavoronkov et al. (2019) curated and utilized, among others, patent data and several datasets about molecules (unspecific bioactive compounds, kinase inhibitors, DDR1 kinase inhibitors, molecules targeting non-kinase targets) specifically to develop DDR1 inhibitors. They synthesized and tested six drug candidates in cell assays. Two of them were found to be active, and one was even successfully validated in animal models. Envisioning a precision or even personalized medicine perspective, identifying protein targets is challenging, whereas sequencing and omics data are straightforward to gather.
Very recently, Méndez-Lucio et al. (2020) proposed a method for the de novo design of molecules against desired targets, represented by the gene expression signatures of knocked-out (suspected) targets. Notably, 97% of all anticancer candidate drugs fail in clinical trials and never receive FDA approval, questioning the current approaches of target identification for the discovery of pharmaceuticals (Wong et al., 2019). Taking ten drug-indication pairs from ongoing clinical trials, Lin et al. (2019) found that the proposed mechanism of action (MOA) for all of them were incorrect; knocking out the target genes did not ever hamper cancer fitness. Although the wrong target genes were identified through RNA interference with siRNA, seemingly silencing essential off-target genes, all drugs retained their anticancer effect through target-independent mechanisms. The fact that off-target cytotoxicity is a common MOA of anticancer drugs in clinical trials corroborates the need to to scrutinize current lead compound discovery strategies and calls to develop novel methodology with unconventional approaches. It is for this reason that we herein propose a novel framework to generate lead compound candidates solely based on a tumor's metabolic signature, as opposed to attempting to target a specific protein or incorporating information about potential targets directly into the design process. Here, we guide the learning process solely by transcriptomic profiles of cancer cells, which thus act as metabolic signatures. Transcriptomic data have been successfully used for de novo drug identification (Verbist et al., 2015; De Wolf et al., 2018) and has been advocated for a pivotal role in the future of drug discovery (Dopazo, 2014). Related work has addressed the generation of anticancer candidate drugs by conditionally sampling from an IC50 vector (Joo et al., 2020).
Our contribution
We present a novel framework for molecule generation based on deep generative models and reinforcement learning that, for the first time, enables the generation of molecules while taking into account the disease context encoded in the form of gene expression profile (GEP) data (for a graphical illustration see Figure 1A). Our framework is depicted in Figure 1B and consists of a conditional molecule generator (embodied by two separate Variational Autoencoders) and a critic module that evaluates the efficacy of proposed compounds on the target profile (see Figure 1D). The training procedure is split into two stages. In the first stage, the models are trained independently; one VAE is trained on gene expression data (in the following called profile VAE or just PVAE) from TCGA (Weinstein et al., 2013), and another VAE (in the following called SMILES VAE or just SVAE) is trained on bioactive small molecules from ChEMBL (Bento et al., 2013) (see Figure 1C). As a critic, we use PaccMann, a multimodal drug sensitivity prediction model developed and validated in our previous work (Manica et al., 2019; Cadow et al., 2020). In the second stage, the encoder of the profile VAE is combined with the decoder of the molecule VAE and exposed to a joint retraining that is optimized using a policy gradient regime with a reward coming from the critic module. The goal of the optimization is to tune the generative model such that it generates (novel) compounds that have maximal efficacy against a given biomolecular profile that is characteristic for a cancer site, a patient subgroup, or even an individual. By efficacy, we refer to predict cellular IC50 (i.e. the micromolar concentration necessary to inhibit 50% of the cells) as opposed to e.g. enzymatic IC50. This efficacy is a joint property of a drug-cell-pair, as treatment response to a compound heavily varies depending on the tumor's genomic and transcriptomic makeup (Geeleher et al., 2016). In this work, we focus on profile-specific compound generation and optimize the generator with IC50 as critic, but we would like to note that our framework can be extended to more complex reward functions and include a case study on the concurrent optimization of (predicted) drug efficacy and toxicity.
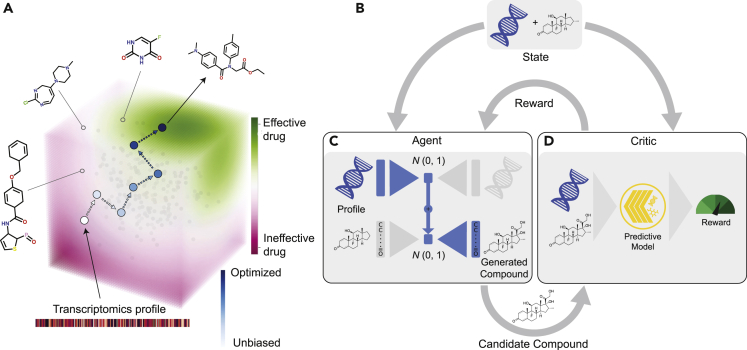
Figure 1.
The proposed framework for anticancer compound design against specific cancer profiles
(A) Conceptually, the model performs a guided walk through the chemical space in order to find effective compounds. Starting from an unbiased molecule generator (trained only on a dataset of bioactive compounds without any information about cancer), compounds are sampled and screened in silico against the transcriptomic profiles of interest. The outcome of the screening guides the generator toward sampling from manifolds with more effective compounds.
(B–D) (B) The training process depicted in more detail. The conditional compound generator (called “agent”) is embodied through two initially separate VAEs. The compound generation starts with a biomolecular profile of interest e.g. a transcriptomic profile. Through a pretrained omics VAE, the profile is encoded into the latent space of gene expression profiles. The latent representation of the profile is decoded through the molecular decoder of a separately pretrained molecule VAE to produce a candidate compound (see C). This generative process can optionally be “primed” through encoding a known, effective compound or a functional group with the molecular encoder. The proposed compound is then evaluated by a critic. Our critic is represented by a multimodal drug sensitivity prediction model that ingests the compound and the target profile of interest (see D). The IC50 efficacy, as predicted by the critic, is interpreted as reward and is subject to maximization during the RL-based optimization. Over the course of training, the generator will thus learn to produce candidate compounds with higher and higher efficacy. <START> is the start and <END> is the end token.
Results
Pretraining profile VAE and SMILES VAE
In the first phase of training, the two components depicted in Figure 1C were trained independently. The profile VAE consisted of a set of stacked dense layers and was trained as a denoizing VAE to enhance generalization abilities. The purpose of the PVAE was to find a lower dimensional representation of the cell profiles that maintains structural similarity and later allows a fusion with the latent representation of molecules. The encoder of the PVAE learned to meaningfully embed gene expression profiles (bulk RNA-Seq from TCGA (Weinstein et al., 2013)) into a latent space, such that the decoder could reconstruct the profiles, but also generate novel, seemingly realistic gene expression profiles (GEP). In alignment with the reported consensus between transcriptomic data in TCGA and cancer cell line databases (Ghandi et al., 2019), we found the distributions of GEPs in GDSC (Yang et al., 2012) and TCGA to be sufficiently similar to justify our decision to perform the PVAE pretraining on ∼10k TCGA samples, whereas the reinforcement learning (RL) optimization was performed on GDSC cell lines (compare Figure S2 in supplementary material S2).
The SMILES VAE was pretrained for 10 epochs on ∼1.4 million structures from ChEMBL (Bento et al., 2013) (for details see transparent methods). Both encoder and decoder consisted of stack-augmented gated-recurrent units as used in Popova et al. (2018). The purpose of the SVAE was to learn the syntax of SMILES and general semantics about bioactive compounds. The novelty and diversity of the generated molecules were validated by sampling 10,000 molecules through decoding random points from the latent space. About 96.2% of the 10,000 generated molecules were valid molecular structures (surpassing the results of Popova et al. (2018) who used the same stack-augmented GRUs and reported 95% SMILES validity). Moreover, 99.72% of the valid molecules were unique across the 10,000 generations, and the novelty was 1, i.e. none of the generated compounds was present in the training dataset. Comparing the Tanimoto similarity (Tanimoto, 1958) of the ECFP molecular fingerprints (Rogers and Hahn, 2010) of 1,000 generated molecules with the training and test data from ChEMBL, we found that the vast majority had a Tanimoto similarity (τ), between 0.2 and 0.6 (on average 0.41 ± 0.1 for training and 0.38 ± 0.08 for testing molecules), suggesting that our model learned to propose novel molecular structures from the chemical space. In addition, an interactive visualization of the chemical space with Faerun (Probst and Reymond, 2018) shows the TMAP (Probst and Reymond, 2020) (i.e., an algorithm to represent high-dimensional data as minimum spanning trees) of ChEMBL and generated compounds through the TMAP algorithm. The generated molecules mix well with the training molecules into the chemical space. For detailed results of both the PVAE and the SVAE, see the supplementary material S3.
Omic-specific compound generation (RL training)
Here, we present the results of our molecule generator conditioned on gene expression profiles of cancer subtypes.
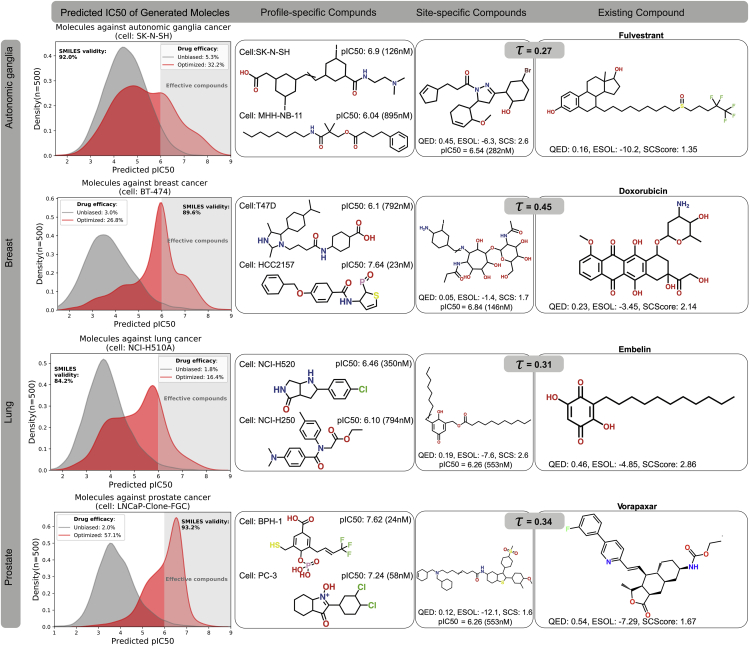
As a proof of concept, we show results for cancer in four different sites: breast (carcinoma), lung (carcinoma), prostate (carcinoma), and autonomic ganglia (neuroblastoma). The conditional generator was initialized as the SVAE, i.e. sampling from the unbiased generator yielded random molecules from the chemical space as learned from the ChEMBL data. For the evaluation, all generated compounds with a predicted IC50 value below 1 μM (i.e. pIC50 > 6) were considered to be effective. Moreover, within each cancer type (or site), 80% of the cell lines (breast: 50, lung: 169, prostate: 7, autonomic ganglia: 56) were considered as training cell lines and used to optimize the parameters of the conditional generator. We observed that over time the generator learned to produce more molecules with high predicted efficacy according to the critic. To test both the generalization abilities and whether the generator actually utilized the omics-profile for the generation, we used the remaining 20% of cell lines to verify whether conditioning the generator on unseen cell lines of the same site also leads to compounds with low IC50. As presented in Figure 2 (left column), our model learned to produce compounds with lower IC50 values, for unseen cell lines from the given cancer site. In other words, the IC50 distribution of candidate compounds proposed by the generative model were successfully shifted toward higher efficacy (lower IC50). The baseline model corresponds to the pretrained SVAE from which n = 500 molecules were randomly sampled. In all four cases, a significant portion (between 16% and 57%) of molecules generated from the optimized model were assigned an IC50 value below 1μM, whereas only 2%–5% of the candidates generated by the baseline model (i.e. the SVAE) were classified as effective. Moreover, in all cases the generator maintained an almost equal SMILES validity (84%–93%) compared with the baseline, much higher than what Méndez-Lucio et al. (2020) reported based on gene expression (8%–9%). The second column of Figure 2 shows generated molecules that are predicted to be effective against unseen cell lines from the respective cancer site. As opposed to the personalized regime in the second column, the third column of Figure 2 showcases a precision medicine regime. Here, novel molecules were designed specifically for each cancer site, i.e. a single, characteristic GEP. In all cases, the model generated compounds that exhibited high efficacy against the average cellular profile of the target site while maintaining efficacy against the majority of individual cell lines for that site. We have thus formulated a novel problem, namely how to drive a molecular generative model to produce molecules with low predicted efficacy against an omics profile of interest. To the best of our knowledge, this problem has not been tackled before, exacerbating a comparison to other works. In the most similar work, Méndez-Lucio et al. (2020) proposed a model that produces hit-like molecules to induce a desired gene expression profile.
Figure 2.
Sample results for profile-driven model optimization and anticancer compound generation
Each row illustrates the results of training the RL pipeline on cell lines from a specific cancer type: autonomic ganglia, breast, lung, and prostate cancer. The first column compares the distributions of pIC50 predictions given by the critic model for a set of n = 500 drug candidates generated with RL optimization and a set of 9,000 molecules without RL optimization. As demonstrated by the density plots, the RL optimization process leads to candidate compounds with a higher mean pIC50 for the target cancer, highlighting the successful optimization of the generative model toward the design of more effective compounds. The second column presents candidate compounds with a high predicted efficacy against a particular cell line that was not seen during training. The third column showcases generated compounds that were optimized to be effective against all cell line profiles of the given cancer type in each row. In the fourth column, we present an existing anticancer compound (approved against at least one type of cancer) that was in the top-3 neighborhood of the generated compound in the third column. The existing and generated compounds are compared in terms of Tanimoto structural similarity between RDKit fingerprints as well as three chemical scores crucial in drug design, namely druglikeness (QED, 0 worst, 1 best), synthetic complexity (SCS or SCScore, 1 best, 5 worst), and solubility (ESOL, given in M/L). The crossed carbon double bond in some molecule means that the E-Z configuration is undefined.
Investigation of nearest neighbors
For a more quantitative assessment, the last column of Figure 2 compares the four cancer-type-specific candidate compounds with one of their top-3 neighbors using the Tanimoto similarity score, τ, from several hundreds of existing anticancer compounds. It is well known that the Tanimoto similarity across compounds is highly correlated with their induced sensitivity patterns on cancer cell lines (Shivakumar and Krauthammer, 2009). The candidate compound proposed against breast cancer (Figure 2 second row, third column) resembles a collection of fused sugarlike moieties and has doxorubicin, a commonly used chemotherapeutical against breast cancer (Lao et al., 2013), as one of the top-3 nearest neighbors. The generated compound against lung cancer (Figure 2, third row, third column) presents similarities to embelin, an existing anticancer compound from the GDSC database. Comparing the two structures, it is evident that the generated compound and embelin share a long carbon chain and a single six-membered fully carbonic ring. Embelin was tested against 965 cell lines from GDSC/CCLE from which the highest reported efficacy is against a lung cell line (NT2-D1). Embelin is also known to be the only known non-peptide inhibitor of XIAP (Poojari, 2014), a protein that plays an important role in lung cancer development (Cheng et al., 2010). The closest neighbor of the prostate-specific generated compound (Figure 2 fourth row, third column) is vorapaxar. Its efficacy is highest against a prostate cancer cell line (DU_145) according to GDSC/CCLE. Vorapaxar is an antagonist of a protease-activated receptor (PAR-1) that is known to be overexpressed in various types of cancer, including prostate (Zhang et al., 2009). Lastly, the third closest neighbor of the generated compound against neuroblastoma (Figure 2 first row, third column) is fulvestrant, an antagonist/modulator of ERα that has recently been proposed as a novel anticancer agent for neuroblastoma (Gorska et al., 2016). The predicted pIC50 activity profile of fulvestrant and our candidate drug are highly correlated across all cell lines (ρ = 0.88), indicating that they may exhibit similar pharmacological properties. Similar observations are made for the lung and prostate cancer candidates and their neighbors embelin and vorapaxar (ρ = 0.55 and ρ = 0.69).
To summarize, for all four investigated cancer types, the proposed compounds showed the highest structural similarity to anticancer drugs that are, for each specific cancer type, either (1) already FDA approved (breast), (2) known inhibitors of relevant targets (lung, prostate), or (3) have been advocated for (neuroblastoma). This result is remarkable, especially as the generator was never exposed to any anticancer compounds. Indeed, only the critic had seen two out of the four compounds during training, highlighting that the generator has learned some structural characteristics that make a compound efficacious against a particular cancer type, according to our critique.
In the above comparisons, the search space was restricted to compounds with known anticancer properties. To investigate whether the proposed compounds generally had a higher similarity to drugs associated with cancer, we carried out a comparison with compounds from a broader pool of chemicals, namely ChEMBL (Bento et al., 2013), a database of >1.5 million bioactive molecules with drug-like properties. The nearest neighbor (τ = 0.54) of our breast cancer compound in the ChEMBL database is CHEMBL1093122, a conjugate of plumbagin and phenyl-2-amino-1-thioglucoside that inhibits the synthesis of mycothiol (Gammon et al., 2010). Plumbagin itself and many of its derivatives are widely studied anti-breast cancer compounds (Zhang et al., 2016; Kawiak et al., 2017; Dandawate et al., 2014). For our lung cancer compound, the nearest neighbor (τ = 0.48) is polyoxyethylene dioleate, a surfactant that has been patented for the treatment of eight types of cancer, including three types of lung cancer (small lung cell cancer, lung adenocarcinoma, and metastatic lung cancer, Girsh (2007)). It is also utilized in targeted drug delivery systems for drug-resistant lung cancer (Kaur et al., 2016). The nearest neighbor (τ = 0.31) of the prostate cancer compound is Clinolamide (or Linolexamide), which is included in a patent of diagnostic and/or therapeutically active compounds for several types of cancer, including prostate cancer (Klaveness et al., 2004). The nearest neighbor (τ = 0.35) of the proposed neuroblastoma compound is NSC-715466. NSC-715466 has been evaluated for anticancer effects in a recent release of the NCI-60 database (Shoemaker, 2006) and inhibits cancer cell growth by 65% ± 15% across all tested cell lines, with a below-average inhibition for cancer in the central nervous system (57% ± 9%). Regarding its efficacy, it only falls in the 51st percentile of all 53,217 compounds tested in NCI-60, which presumably prevented further investigations. The four discussed ChEMBL compounds as well as the analysis of NSC-715466 can be found in Figure S4 in supplementary material S4.
It is promising to observe that the molecules with the highest Tanimoto similarity to our compounds are associated with cancer (some even to the specific types of cancer our compounds were optimized for) even when using a larger database of bioactive compounds. However, it is worth keeping in mind that a high Tanimoto score to a known cancer drug is not necessary for anticancer drug efficacy. Oftentimes, cancer drugs used for the same cancer type or even drugs sharing the same mechanism of action exhibit low Tanimoto similarity, e.g. TKIs used for NSCLC such as crizotinib and erlotinib, τ = 0.11. Across all anticancer compounds in GDSC and CCLE databases, we note that the average Tanimoto similarity (τ = 0.149 ± 0.05) is not much lower than the average similarity of two compounds of a given site (τ = 0.154 ± 0.06). For the following results and discussion, the anticancer compounds from GDSC/CCLE were associated with the site where they had the highest average IC50 efficacy.
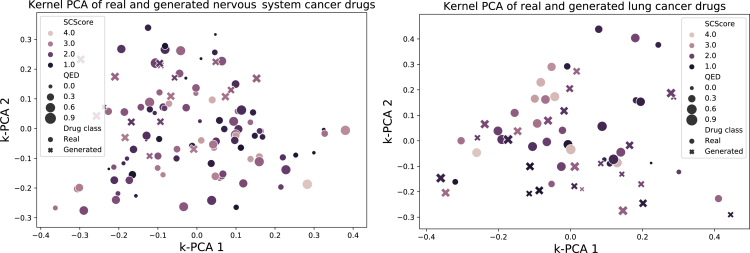
To better understand whether the generated drugs mimic the space of cancer-specific anticancer drugs, Figure 3 shows visualizations of real and generated cancer drugs for one specific cancer type, using kernel PCA based on Tanimoto similarity (Schölkopf et al., 1998). In addition to the class belongings, the plots also depict the QED score (Bickerton et al., 2012), a quantitative estimate of drug-likeness (0 worst, 1 best), and SCScore (Coley et al., 2018), an estimated score of synthetic complexity (1 best, 5 worst). The fact that the generated molecules are well intermingled with the real drugs suggests that the generator proposes diversified structures that mimic some properties of anticancer compounds. It is also curious to see that several real drugs have low QED and/or high synthetic complexity scores (the same holds true for the generated molecules). Moreover, we provide interactive TMAP visualizations of the site-specific generated compounds (links in the availability section).
Figure 3.
Visualization of generated molecules and real anticancer drugs
A kernel PCA of real and generated molecules based on Tanimoto similarity. The size of the points denotes the QED score, whereas the coloring represents the synthetic complexity score (SCScore). Overall, both generated and existing molecules are heterogeneously distributed in the 2D projection and do not form clear clusters.
Chemical properties of generated molecules
In this work, the conditional generator is trained using PaccMann as sole critic. However, besides inhibitory efficacy, there is a myriad of properties of a candidate drug that crucially influence its potential for becoming an anticancer compound.
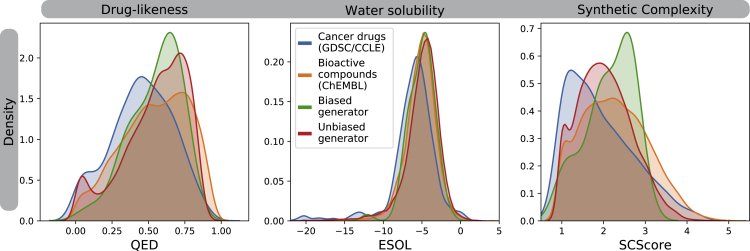
Some of these can be approximated in silico, e.g. water solubility (ESOL, Delaney (2004)), drug-likeness (QED, Bickerton et al. (2012)), and synthesizability (SCScore, Coley et al. (2018)). Figure 4 gives an overview of the distribution of QED, ESOL, and SCScore for sets of (1) known anticancer compounds (blue), (2) molecules from ChEMBL (orange), (3) compounds generated by the SVAE (red), and (4) compounds proposed by the conditional generator (green). Despite the fact that none of these properties were explicitly optimized, comparing the distributions reveals a good agreement overall. Interestingly, anticancer drugs exhibit, compared with the ChEMBL compounds, much less drug-like properties (lower QED) on average and seem to be more synthesizable (lower SCScore). This tendency of anticancer drugs toward synthetically less complex structures is likely to result from the high attrition rate in clinical trials and the corresponding cost reduction policies. It is also encouraging to see, on average, that the unbiased generator (SVAE) generates molecules with more desired properties compared with the data used for training (ChEMBL compounds have lower QED and higher SCScore). Moreover, the cancer drugs have, on average, a significantly lower QED than the other three sets in Figure 4 (0.45 ± 0.2 with >10% of GDSC/CCLE drugs even having a QED <0.2, whereas it is 0.55 for the other three sets). Indeed the QED scores of the other three sets were so similar that we failed to reject the null hypothesis that the QED scores of these three sets are from different distributions (Kruskal-Wallis test, α = 0.05). Regarding synthetic complexity, both the unbiased and the biased generator fail to produce molecules with SCScores as low as the anticancer drugs (MWU, p < 0.01), but they produce structures that are estimated to be less complex than the ChEMBL molecules (MWU, p < 0.01). Overall, we observe that the biased generator produces molecules with less desired properties than the unbiased generator (SVAE). This is to be expected because the unbiased generator was pretrained to mimic the data from ChEMBL, whereas no explicit optimization of chemical scores was performed during the RL optimization. For one generated compound, we exemplarily show a possible synthesis route that was assigned a high confidence score by the retrosynthesis model (Schwaller et al., 2020) and consists of four reactions and ten commercially available reactants (see Figure S6 in supplementary material S6 appendix (S8)).
Figure 4.
Comparison of chemical scores for real drugs in the GDSC and CCLE databases versus our generated compounds
We compared three chemical scores for druglikeness as assessed by QED score (0 worst, 1 best), for solubility as assessed via ESOL, given in log(M/L) (most drugs have a solubility between −8 and −2), and for synthetic accessibility as assessed by SAS (1 best, 10 worst). These three scores are computed for the panel of known anticancer drugs, bioactive molecules from ChEMBL, and molecules generated before (red) and after (green) RL optimization.
Savjani et al. (2012) reported that 40% of novel chemicals cause practical problems due to insolubility. Water solubility remains challenging to approximate in silico (Sorkun et al., 2019) and thus we treat the good agreement in the ESOL scores (see Figure 4 middle panel) with caution.
Finally, we would like to point out that utilizing an IC50 drug sensitivity prediction model as sole critic limits the performance of the framework, as the expressive power of the conditional generator is inherently “upper bounded” by the predictive power of the critic. Crucially, due to a lack of available data, the critic was only trained on the cancer candidate compounds in GDSC (Yang et al., 2012) but not on compounds without any inhibitory efficacy against cell lines. Notably, however, GDSC contains many samples with IC50 in millimolar scale, leading the critic to cover vast ranges of irrelevant molecules with 3 < pIC50 < 6. We additionally verified the generalization capabilities of the critic by comparing the predicted efficacy of cancer drugs (most of them were seen during training) and a “negative” set of molecules from ChEMBL across all 965 cell lines from GDSC. The results can be found in Figure S5 in supplementary material S5 and show that although 15.2% of the virtual drug screenings with anticancer compounds were positive (IC50<1μmol), only 7.7% of all the ChEMBL molecules showed potential anticancer effects. Moreover, the generated anticancer compounds were found to have a significantly higher Tanimoto similarity to anticancer drugs than to either ChEMBL molecules (p < 0.01, one-sided MWU) or molecules generated without the RL optimization, i.e. from the SVAE (p < 0.01, one-sided MWU). These two results are encouraging and suggest that PaccMann can seemingly drive the molecule generation away from ordinary bioactive compounds such as in ChEMBL, more toward mimicking the properties of anticancer drugs.
Toxicity: Case study on multi-objective optimization
In the previous sections, the RL optimization of the molecular generation was guided using the predicted IC50 on a given cell profile as sole reward. To investigate whether PaccMannRL is able to generate desired molecules in a multi-objective optimization setting, we performed RL optimization using an adjusted reward function that incorporates (1) low IC50 against a given omic profile (as in all other experiments), (2) low environmental toxicity, and (3) low adverse drug reactions, a measure of high-level phenotypic side-effect observations (for details see supplementary material S1.4). Similarly to IC50, toxicity and adverse drug reactions were also evaluated with predictive neural networks. The toxicity model was pretrained on Tox21 (Huang et al., 2016) (∼8k molecules measured across 12 toxicity endpoints) and the adverse reaction model was pretrained on SIDER (∼1.5k marketed drugs with 27 classes of reported adverse drug reactions). We note that the toxicity predictor achieved an ROC-AUC of 0.877 on Tox21 (surpassing the previous state-of-the-art) and an ROC-AUC of 0.835 on SIDER (Markert et al., 2020). For details on these predictive models see Markert et al. (2020).
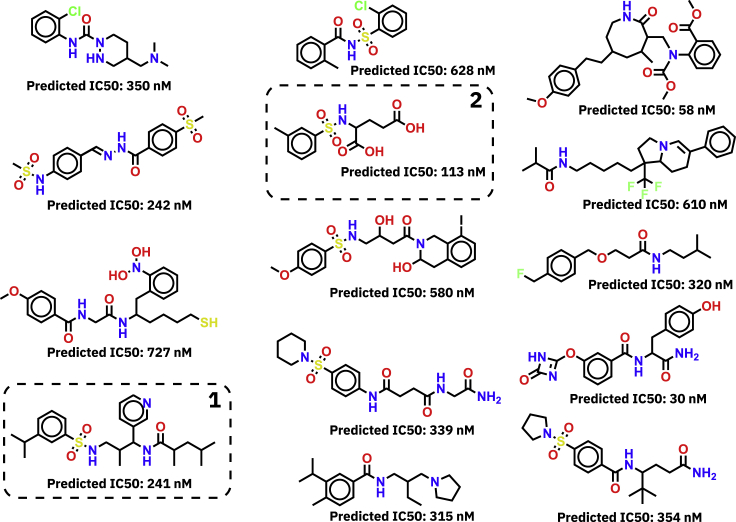
Figure 5 displays a collection of molecules that were generated against the NCI-H520 cell line (a human lung squamous cancer cell line) during an RL optimization that was restricted to lung cancer cell lines (similar to Figure 2, third row). Notably, the NCI-H520 cell line was in the test set for the RL optimization. The adjusted reward function (Equation 5) used low IC50 as primary and low toxicity scores as secondary objective. All displayed molecules fulfilled the multi-objective, i.e., they were predicted to be active on NCI-H520 (IC50<1μM) while not being predicted toxic in any of the 12 Tox21 toxicity assays. Furthermore, we computed Tanimoto similarities of the molecules shown in Figure 5 with approved cancer drugs, according to the National Cancer Institute. For molecules 1 and 2 we obtained Tanimoto coefficients >0.45 for several drugs that are FDA approved for the use against lung cancer, such as vinorelbine, vincristine, vinblastine, irinotecan, topotecan, and alectinib (Wishart et al., 2018). Notably, across all investigated FDA-approved cancer drugs, molecule 1 (which was generated against NCI-H520, an NSCLC cell line) exhibited the highest Tanimoto similarity with vinorelbine, a targeted drug for NSCLC.
Figure 5.
Case study on multi-objective optimization including toxicity
Molecules generated against the NCI-H520 cell-line with a multi-objective reward function based on a combination of low IC50 and low toxicity in Tox21 and SIDER tasks. For all fourteen molecules depicted, all Tox21 assay predictions are negative. Molecules 1 and 2 are further discussed in the text.
Discussion
Summary
In this work we presented PaccMannRL, a novel framework for molecular generation that enables us to condition on the transcriptomic profile of the target cell or cancer site. By using RL optimization we demonstrated that our proposed generative model is able to produce candidate compounds with high predicted efficacy (low IC50) against a given target profile, even if this profile was never seen during training. Notably, this was achieved despite the fact that the generator was never exposed to anticancer drugs explicitly but only pretrained on bioactive compounds from ChEMBL (the only component that was trained on drugs with known anticancer effects is the critic). Moreover, an analysis of the generated compounds for four different cancer types revealed that the predicted compounds share many structural similarities with known anticancer compounds for the same cancer types that the generation was optimized for. In a case study, we briefly investigated the ability of PaccMannRL to generate molecules that fulfill multiple objectives (specifically high activity against a lung cancer cell line and low toxicity) and found many candidates with desired predicted properties.
Interpretation and future work
Although our results are a modest step toward disease-specific molecular generation, a lot of further optimization is needed. For instance, subsidiary properties of a candidate drug that determine its potential for becoming a successful anticancer compound (e.g., water solubility, drug-likeness, synthesizability or off-target cytotoxicity) are not directly optimized. However, despite not explicitly incorporating them into the reward function, we find that the produced molecules exhibit desired properties in terms of drug-likeness, water solubility, and ease of synthesis. One possible future endeavor is to incorporate information in the reward function not only about drug efficacy but also about other drug-relevant chemical properties; one such example was shown in the concurrent optimization of molecules with a low IC50 and low toxicity. An additional investigation toward that end might be a substructure search for toxicophores within the generated molecules. This could help to winnow the promising molecules from those that exhibit low IC50 due to cytotoxicity caused by a toxicophore. Another observation we made is that the stability of the conditional generation can diminish over RL training time and result in overly simplistic molecules, especially if the learning rate is set too high. However, after pretraining of the SVAE (and before starting the RL training), the generated molecules were found to be reasonably similar to the training molecules (bioactive compounds from ChEMBL). We, therefore, conceive that for more targeted applications, an improved chemical space could be obtained by fine-tuning the SVAE on, e.g., anticancer drugs or even bioactive molecules with a shared mechanism of action, like JAK inhibitors. If the set of possible target molecules is small, SMILES augmentation can further be used to improve the quality of molecular generators (Arús-Pous et al., 2019). We also emphasize that the chemical space covered by the SVAE encompasses 86 distinct SMILES tokens, significantly more than in most related work and more than in landmark publications that produced de novo molecules with high efficacy in vitro and in vivo (Zhavoronkov et al., 2019).
The resulting multimodal objectives may be difficult to optimize due to possibly counteracting/interfering gradients. A possible approach to circumvent this challenge is potentially to use explicit compensation techniques (Yu et al., 2020) or defining gradient-free global objectives (Häse et al., 2018). Another challenge that needs to be overcome to improve the reliability and accuracy of the critic is the expected distributional differences between the data used for training (cancer cell lines) and the targeted data (human data from clinical trials). A possible approach that can be explored is the exploitation of transfer learning techniques, as suggested in Sharifi-Noghabi et al. (2020). Oftentimes, medical chemists do not start the drug design from scratch but from the scaffold of an approved drug. The goal of the scaffold hopping is to find a drug with similar effects (e.g. increased efficacy or reduced side effects). Although our framework enables users to incorporate prior knowledge into the design process by priming the latent code, we have not yet explored the full potential of this idea here. As the decoded molecule is not guaranteed to maintain similarity to the primer, the recently proposed “deep generative scaffold decorator” could be integrated into our framework to facilitate a more systematic exploration and the possibility of adding fragments to established drug scaffolds (Arús-Pous et al., 2020). An alternative future endeavor is to explore graph-based, instead of sequential, representations of molecules to directly generate a molecular graph from the context set using a conditional structure generation framework (Yang et al., 2019). For future work, one may conceive a generic framework where the molecule generation can be conditioned on possibly multimodal context information such as a (multi)omics profiles, a target protein, a primed drug, a drug scaffold, or a combination thereof. The resulting latent spaces, consisting of semantically distinct modalities, could then be jointly explored by machine learning techniques designed to operate on sets instead of fixed-length vectors, such as permutation-invariant operations (Zaheer et al., 2017).
Conclusion
Due to growing evidence of our insufficient understanding of the mechanisms of action of drug candidates even in clinical trials (Lin et al., 2019), the design of generative approaches that can, as shown here, bypass the need for a detailed characterization of drug targets and cytotoxicity mechanisms may be a novel, valuable approach within generative chemistry.
Limitations of the study
Without IC50 assays on the investigated cell lines, the in vitro efficacy of the proposed molecules remains unclear. Even in case of positive cell line drug sensitivity assays, these results may not translate to animal models due to numerous reasons (e.g., adverse reactions or drug delivery). Therefore, the potential for the presented molecules to become anticancer drugs cannot be conclusively assessed in this study. Furthermore, the quality of the molecular generator is inherently limited by the predictive power of the in silico IC50 evaluator and critically depends on how well this predictor generalizes across the chemical space. In addition, the proposed molecules are represented by SMILES sequences that are generated auto-regressively by using a stochastic sampling process, the common practice in SMILES generation. When the seed is not fixed, a point in the latent space does not necessarily map to a unique molecule.
Resource availability
Lead contact
Jannis Born, jab@zurich.ibm.com.
Materials availability
This study did not generate any new materials.
Data and code availability
Code: the source code is publicly available on: https://github.com/PaccMann/. For a detailed example see: https://github.com/PaccMann/paccmann_rl. To assess the critic, please see Manica et al. (2019).
Data: this study did not generate any new data and exclusively used publicly available databases. The transcriptomic data used to pretrain the Profile VAE and the molecular data used to train the SMILES VAE are processed variants of TCGA (Weinstein et al., 2013) and CheMBL (Bento et al., 2013) and available via: https://ibm.box.com/v/paccmann-pytoda-data. The GDSC (Yang et al., 2012) cell profiles as well as the pretrained models can be found under the same link.
The interactive TMAP visualizations of the molecules generated by the (unbiased) SVAE can be found on https://paccmann.github.io/rl/unbiased.html. The TMAPs of the cancer-site-specific candidate compounds are accessible on https://paccmann.github.io/.
Methods
All methods can be found in the accompanying transparent methods supplemental file.
Acknowledgments
The authors thank Prof. Dr. Karsten Borgwardt and Dr. Maria Gabrani for continuous support and useful discussions on the manuscript. The project leading to this publication has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement No 826121. J.B. acknowledges financial support from the German Academic Exchange Service (DAAD).
Author contributions
J.B, M.M., and A.O. contributed equally to this work. Conceptualization: M.M, J.B., and A.O.; Methodology: J.B., M.M., and A.O.; Software: J.B., M.M., A.O., J.C., and G.M.; Investigation: J.B., A.O., M.M., and G.M.; Data curation: M.M.; Writing—Original Draft: A.O., J.B., and M.M; Writing—Review & Editing: J.B.; Visualization: J.B., M.M., and A.O.; Project Administration: M.R.M.; Funding Acquisition: M.R.M.;
Declaration of interests
The authors declare that a patent associated to the methodology presented in this manuscript was granted (US20200365238A1).
Inclusion and diversity
While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: April 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102269.
Contributor Information
Jannis Born, Email: jab@zurich.ibm.com.
Matteo Manica, Email: tte@zurich.ibm.com.
María Rodríguez Martínez, Email: mrm@zurich.ibm.com.
Supplemental information
References
- Arús-Pous J., Johansson S.V., Prykhodko O., Bjerrum E.J., Tyrchan C., Reymond J.-L., Chen H., Engkvist O. Randomized smiles strings improve the quality of molecular generative models. J. Cheminform. 2019;11:1–13. doi: 10.1186/s13321-019-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arús-Pous J., Patronov A., Bjerrum E.J., Tyrchan C., Reymond J.-L., Chen H., Engkvist O. Smiles-based deep generative scaffold decorator for de-novo drug design. J. Cheminform. 2020;12:1–18. doi: 10.1186/s13321-020-00441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumentado-Armstrong T. arXiv; 2018. Latent Molecular Optimization for Targeted Therapeutic Design; p. 1809.02032. [Google Scholar]
- Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M., Krüger F.A., Light Y., Mak L., McGlinchey S. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2013;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickerton G.R., Paolini G.V., Besnard J., Muresan S., Hopkins A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012;4:90. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadow J., Born J., Manica M., Oskooei A., Rodríguez Martínez M. Paccmann: a web service for interpretable anticancer compound sensitivity prediction. Nucleic Acids Res. 2020;48:W502–W508. doi: 10.1093/nar/gkaa327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Engkvist O., Wang Y., Olivecrona M., Blaschke T. The rise of deep learning in drug discovery. Drug Discov. Today. 2018;23:1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- Cheng Y.-J., Jiang H.-S., Hsu S.-L., Lin L.-C., Wu C.-L., Ghanta V.K., Hsueh C.-M. Xiap-mediated protection of h460 lung cancer cells against cisplatin. Eur. J. Pharmacol. 2010;627:75–84. doi: 10.1016/j.ejphar.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Chenthamarakshan V., Das P., Hoffman S.C., Strobelt H., Padhi I., Lim K.W., Hoover B., Manica M., Born J., Laino T., Mojsilovic A. Cogmol: target-specific and selective drug design for COVID-19 using deep generative models. Adv. Neural Inf. Process. Syst. 2020;33 [Google Scholar]
- Coley C.W., Rogers L., Green W.H., Jensen K.F. Scscore: synthetic complexity learned from a reaction corpus. J. Chem. Inf. Model. 2018;58:252–261. doi: 10.1021/acs.jcim.7b00622. [DOI] [PubMed] [Google Scholar]
- Dandawate P., Ahmad A., Deshpande J., Swamy K.V., Khan E.M., Khetmalas M., Padhye S., Sarkar F. Anticancer phytochemical analogs 37: synthesis, characterization, molecular docking and cytotoxicity of novel plumbagin hydrazones against breast cancer cells. Bioorg. Med. Chem. Lett. 2014;24:2900–2904. doi: 10.1016/j.bmcl.2014.04.100. [DOI] [PubMed] [Google Scholar]
- De Wolf H., Cougnaud L., Van Hoorde K., De Bondt A., Wegner J.K., Ceulemans H., Göhlmann H. High-throughput gene expression profiles to define drug similarity and predict compound activity. Assay Drug Dev. Tech. 2018;16:162–176. doi: 10.1089/adt.2018.845. [DOI] [PubMed] [Google Scholar]
- Delaney J.S. Esol: estimating aqueous solubility directly from molecular structure. J. Chem. Inf. Comput. Sci. 2004;44:1000–1005. doi: 10.1021/ci034243x. [DOI] [PubMed] [Google Scholar]
- DiMasi J.A., Grabowski H.G., Hansen R.W. Innovation in the pharmaceutical industry: new estimates of r&d costs. J. Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Dopazo J. Genomics and transcriptomics in drug discovery. Drug Discov. Today. 2014;19:126–132. doi: 10.1016/j.drudis.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Gammon D.W., Steenkamp D.J., Mavumengwana V., Marakalala M.J., Mudzunga T.T., Hunter R., Munyololo M. Conjugates of plumbagin and phenyl-2-amino-1-thioglucoside inhibit mshb, a deacetylase involved in the biosynthesis of mycothiol. Bioorg. Med. Chem. 2010;18:2501–2514. doi: 10.1016/j.bmc.2010.02.049. [DOI] [PubMed] [Google Scholar]
- Geeleher P., Cox N.J., Huang R.S. Cancer biomarker discovery is improved by accounting for variability in general levels of drug sensitivity in pre-clinical models. Genome Biol. 2016;17:190. doi: 10.1186/s13059-016-1050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., Barretina J., Gelfand E.T., Bielski C.M., Li H. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569:503. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girsh, L. (2007). Lipid-containing compositions and methods of using them. US Patent App. 11/501,380.
- Gomez-Bombarelli R., Wei J.N., Duvenaud D., Hernandez-Lobato J.M., Sánchez-Lengeling B., Sheberla D., Aguilera-Iparraguirre J., Hirzel T.D., Adams R.P., Aspuru-Guzik A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 2018;4:268–276. doi: 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorska M., Kuban-Jankowska A., Milczarek R., Wozniak M. Nitro-oxidative stress is involved in anticancer activity of 17β-estradiol derivative in neuroblastoma cells. Anticancer Res. 2016;36:1693–1698. [PubMed] [Google Scholar]
- Grechishnikova D. Transformer neural network for protein-specific de novo drug generation as a machine translation problem. Sci. Rep. 2021;11:1–13. doi: 10.1038/s41598-020-79682-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häse F., Roch L.M., Aspuru-Guzik A. Chimera: enabling hierarchy based multi-objective optimization for self-driving laboratories. Chem. Sci. 2018;9:7642–7655. doi: 10.1039/c8sc02239a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Xia M., Nguyen D.-T., Zhao T., Sakamuru S., Zhao J., Shahane S.A., Rossoshek A., Simeonov A. Tox21challenge to build predictive models of nuclear receptor and stress response pathways as mediated by exposure to environmental chemicals and drugs. Front. Environ. Sci. 2016;3:85. [Google Scholar]
- Joo S., Kim M.S., Yang J., Park J. Generative model for proposing drug candidates satisfying anticancer properties using a conditional variational autoencoder. ACS Omega. 2020;5:18642–18650. doi: 10.1021/acsomega.0c01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P., Garg T., Rath G., Murthy R., Goyal A.K. Surfactant-based drug delivery systems for treating drug-resistant lung cancer. Drug Deliv. 2016;23:717–728. doi: 10.3109/10717544.2014.935530. [DOI] [PubMed] [Google Scholar]
- Kawiak A., Domachowska A., Jaworska A., Lojkowska E. Plumbagin sensitizes breast cancer cells to tamoxifen-induced cell death through grp78 inhibition and bik upregulation. Sci. Rep. 2017;7:43781. doi: 10.1038/srep43781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaveness J., Rongved P., Høgset A., Tolleshaug H., Cuthbertson A., Godal A., Hoff L., Gogstad G., Bryn K., Naevestad A. US Patent; 2004. Diagnostic/therapeutic Agents; p. 6,680,047. [Google Scholar]
- Krishnan S.R., Bung N., Bulusu G., Roy A. Accelerating de novo drug design against novel proteins using deep learning. J. Chem. Inf. Model. 2021;61:621–630. doi: 10.1021/acs.jcim.0c01060. [DOI] [PubMed] [Google Scholar]
- Lao J., Madani J., Puértolas T., Álvarez M., Hernández A., Pazo-Cid R., Artal Á., Antón Torres A. Liposomal doxorubicin in the treatment of breast cancer patients: a review. J. Drug Deliv. 2013;2013:456409. doi: 10.1155/2013/456409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Giuliano C.J., Palladino A., John K.M., Abramowicz C., Yuan M.L., Sausville E.L., Lukow D.A., Liu L., Chait A.R. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 2019;11:eaaw8412. doi: 10.1126/scitranslmed.aaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica M., Oskooei A., Born J., Subramanian V., Saez-Rodriguez J., Rodriguez Martinez M. Toward explainable anticancer compound sensitivity prediction via multimodal attention-based convolutional encoders. Mol. Pharm. 2019;16:4797–4806. doi: 10.1021/acs.molpharmaceut.9b00520. [DOI] [PubMed] [Google Scholar]
- Markert, G., Born, J., Manica, M., Schneider, G., and Rodriguez Martinez, M. (2020). Chemical representation learning for toxicity prediction. PharML Workshop at ECML-PKDD (European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases).
- Méndez-Lucio O., Baillif B., Clevert D.-A., Rouquié D., Wichard J. De novo generation of hit-like molecules from gene expression signatures using artificial intelligence. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-019-13807-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polishchuk P.G., Madzhidov T.I., Varnek A. Estimation of the size of drug-like chemical space based on gdb-17 data. J. Comput. Aided Mol. Des. 2013;27:675–679. doi: 10.1007/s10822-013-9672-4. [DOI] [PubMed] [Google Scholar]
- Poojari R. Embelin–a drug of antiquity: shifting the paradigm towards modern medicine. Expert Opin. Investig. Drugs. 2014;23:427–444. doi: 10.1517/13543784.2014.867016. [DOI] [PubMed] [Google Scholar]
- Popova M., Isayev O., Tropsha A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018;4:eaap7885. doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst D., Reymond J.-L. Fun: a framework for interactive visualizations of large, high-dimensional datasets on the web. Bioinformatics. 2018;34:1433–1435. doi: 10.1093/bioinformatics/btx760. [DOI] [PubMed] [Google Scholar]
- Probst D., Reymond J.-L. Visualization of very large high-dimensional data sets as minimum spanning trees. J. Cheminform. 2020;12:1–13. doi: 10.1186/s13321-020-0416-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D., Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Savjani K.T., Gajjar A.K., Savjani J.K. Drug solubility: importance and enhancement techniques. ISRN Pharm. 2012;2012:195727. doi: 10.5402/2012/195727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell J.W., Blanckley A., Boldon H., Warrington B. Diagnosing the decline in pharmaceutical r&d efficiency. Nat. Rev. Drug Discov. 2012;11:191. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- Schölkopf B., Smola A., Müller K.-R. Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput. 1998;10:1299–1319. [Google Scholar]
- Schwaller P., Petraglia R., Zullo V., Nair V.H., Haeuselmann R.A., Pisoni R., Bekas C., Iuliano A., Laino T. Predicting retrosynthetic pathways using transformer-based models and a hyper-graph exploration strategy. Chem. Sci. 2020;11:3316–3325. doi: 10.1039/c9sc05704h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi-Noghabi H., Peng S., Zolotareva O., Collins C.C., Ester M. Aitl: adversarial inductive transfer learning with input and output space adaptation for pharmacogenomics. Bioinformatics. 2020;36:i380–i388. doi: 10.1093/bioinformatics/btaa442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar P., Krauthammer M. volume 10. Springer; 2009. Structural similarity assessment for drug sensitivity prediction in cancer; p. S17. (BMC Bioinformatics). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R.H. The nci60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Skalic M., Sabbadin D., Sattarov B., Sciabola S., De Fabritiis G. From target to drug: generative modeling for the multimodal structure-based ligand design. Mol. Pharm. 2019;16:4282–4291. doi: 10.1021/acs.molpharmaceut.9b00634. [DOI] [PubMed] [Google Scholar]
- Sorkun M.C., Khetan A., Er S. Aqsoldb, a curated reference set of aqueous solubility and 2d descriptors for a diverse set of compounds. Sci. Data. 2019;6:1–8. doi: 10.1038/s41597-019-0151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto T.T. IBM Internal Report; 1958. Elementary Mathematical Theory of Classification and Prediction. [Google Scholar]
- Verbist B., Klambauer G., Vervoort L., Talloen W., Shkedy Z., Thas O., Bender A., Göhlmann H.W., Hochreiter S., Consortium Q. Using transcriptomics to guide lead optimization in drug discovery projects: lessons learned from the qstar project. Drug Discov. Today. 2015;20:505–513. doi: 10.1016/j.drudis.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Wehling M. Assessing the translatability of drug projects: what needs to be scored to predict success? Nature reviews. Drug Discov. 2009;8:541–546. doi: 10.1038/nrd2898. [DOI] [PubMed] [Google Scholar]
- Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M., Network C.G.A.R. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. Drugbank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.H., Siah K.W., Lo A.W. Estimation of clinical trial success rates and related parameters. Biostatistics. 2019;20:273–286. doi: 10.1093/biostatistics/kxx069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson IR Genomics of drug sensitivity in cancer (gdsc): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2012;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Zhuang P., Shi W., Luu A., Li P. Conditional structure generation through graph variational generative adversarial nets. Adv. Neural. Inf. Process. Syst. 2019;32:1338–1349. [Google Scholar]
- You J., Liu B., Ying Z., Pande V., Leskovec J. Graph convolutional policy network for goal-directed molecular graph generation. Adv. Neural Inf. Process. Syst. 2018;31:6410–6421. [Google Scholar]
- Yu T., Kumar S., Gupta A., Levine S., Hausman K., Finn C. Gradient surgery for multi-task learning. Adv. Neural. Inf. Process. Syst. 2020;33 [Google Scholar]
- Zaheer M., Kottur S., Ravanbakhsh S., Poczos B., Salakhutdinov R.R., Smola A.J. Deep sets. Adv. Neural. Inf. Process. Syst. 2017;30:3391–3401. [Google Scholar]
- Zhang X., Wang W., True L.D., Vessella R.L., Takayama T.K. Protease-activated receptor-1 is upregulated in reactive stroma of primary prostate cancer and bone metastasis. Prostate. 2009;69:727–736. doi: 10.1002/pros.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yang C., Rao X., Xiong J. Plumbagin shows anti-cancer activity in human breast cancer cells by the upregulation of p53 and p21 and suppression of g1 cell cycle regulators. Eur. J. Gynaecol. Oncol. 2016;37:30–35. [PubMed] [Google Scholar]
- Zhavoronkov A., Ivanenkov Y.A., Aliper A., Veselov M.S., Aladinskiy V.A., Aladinskaya A.V., Terentiev V.A., Polykovskiy D.A., Kuznetsov M.D., Asadulaev A. Deep learning enables rapid identification of potent ddr1 kinase inhibitors. Nat. Biotechnol. 2019;37:1038–1040. doi: 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code: the source code is publicly available on: https://github.com/PaccMann/. For a detailed example see: https://github.com/PaccMann/paccmann_rl. To assess the critic, please see Manica et al. (2019).
Data: this study did not generate any new data and exclusively used publicly available databases. The transcriptomic data used to pretrain the Profile VAE and the molecular data used to train the SMILES VAE are processed variants of TCGA (Weinstein et al., 2013) and CheMBL (Bento et al., 2013) and available via: https://ibm.box.com/v/paccmann-pytoda-data. The GDSC (Yang et al., 2012) cell profiles as well as the pretrained models can be found under the same link.
The interactive TMAP visualizations of the molecules generated by the (unbiased) SVAE can be found on https://paccmann.github.io/rl/unbiased.html. The TMAPs of the cancer-site-specific candidate compounds are accessible on https://paccmann.github.io/.