Highlights
-
•
Subsolid nodules have higher microbiome diversity compared with solid nodules
-
•
Microbiome composition of Subsolid nodules is distinct from that of solid nodules
-
•
Microbial signatures show robust performance to predict lung adenocarcinoma or subsolid nodules
-
•
Some lung microbial species are associated with clinical characteristics
Keywords: Lung adenocarcinoma, Microbiome, Subsolid nodules, Biomarker
Abstract
Objectives
Increasing evidence indicates that microbiota dysbiosis in the human body may play vital roles in carcinogenesis. However, the relationship between microbiome and lung cancer remains unclear. In this study, we aimed to characterize the microbiome in early stage of lung adenocarcinoma (LUAD), which presented as subsolid nodules (SSN) or solid nodules (SN).
Materials and Methods
We performed 16S rRNA sequencing of 35 pairs (10 SSN and 25 SN) of LUAD tumor tissues and paired adjacent normal tissues. Machine learning was used to identify microbial signatures and construct predictive models.
Results
SSN has higher microbiome richness and diversity compared with SN (richness p = 0.017, Shannon index p = 0.17), and the microbiome composition of SSN is distinct from that of SN (Bray-Curtis p = 0.013, unweighted unifrac p = 0.001). Phylum Chloroflexi (p = 0.009), Gemmatimonadetes (p = 0.018) and genus including Cloacibacterium (p = 0.003), Subdoligranulum (p = 0.002), and Mycobacterium (p = 0.034) were significantly increased in SSN. Tumor and normal tissues had similar richness and diversity, as well as overall microbiome composition. Probiotics with anti-cancer potential, like Lactobacillus, showed elevated levels in normal tissues (p = 0.018). A random forest model with 20 genera-based biomarkers achieved high accuracy for LUAD prediction (area under curve, AUC = 0.879). Meanwhile, a five genera-based signature can accurately discriminate SSN between SN (AUC = 0.950). Cross-validation of these two models also showed high predictive performance (LUAD AUC = 0.813, SSN AUC = 0.933).
Conclusions
This study demonstrates, for the first time, the tumor bacterial microbiome composition of LUAD manifested as SSN is distinct from that presented as SN, which adds new knowledge to SSN in the perspective of microbiome. Furthermore, microbiome signatures showed good performance to predict LUAD or SSN.
1. Introduction
Lung cancer accounts for the highest cancer-related mortality worldwide [1]. Despite encouraging results achieved with targeted therapy and immunotherapy in recent years, the 5-year overall survival remains poor, mainly as a result of late diagnosis and a high frequency of recurrence and metastasis [2]. Non-small cell lung cancer (NSCLC) accounts for over 80% of lung cancer while lung adenocarcinoma (LUAD) is the major subtype [3]. With the wide application of high-resolution CT in lung cancer screening, ground-glass opacity nodules (GGNs), which also called subsolid nodules (SSNs), have emerged as a new type of lung cancer [4]. Over 80% of early-stage lung cancers manifest as radiological SSN in clinic [4,5]. SSNs have been considered as a special clinical subtype and are less aggressive than pure solid LUAD. Unlike solid lung cancer, many GGNs did not harbor the driver mutations that commonly occurred in LUAD [6]. Meanwhile, clinical and epidemiologic studies have suggested a strong association among chronic infection, inflammation, and lung cancer [7,8]. Therefore, we hypothesis that microbiome may play vital roles in the initiation and progression of LUAD.
Human commensal microbiota plays essential roles in varieties of biological processes, which have an extensive impact on human health. Dysbiosis of the human microbiota was commonly found in metabolic diseases, such as obesity and diabetes [9,10]. Recently, more and more studies revealed that microbiota may contribute to carcinogenesis and affect response to cancer therapies through various biological mechanism, including inflammation, metabolism, and immune responses [11–13]. However, most studies focus on digestive system-related cancer [12,13], rare studies have been conducted to clarify the interplay between lung cancer and microbiota. A special bond between gut microbiome and lung was constructed as “Gut-Lung Axis” [14]. Dysbiosis of gut microbiota was found to correlate with lung cancer. As lungs are no longer considered sterile, emerging evidence suggests that microbiota in lungs may play a more important role in the lung carcinogenic process [15,16]. Therefore, a better understanding of the relationship between the lung microbiome and lung cancer will promote the development of novel diagnosis and treatment strategies.
Here, we compared the microbiota profiles between LUAD tumor and normal lung tissues, as well as solid LUAD and subsolid LUAD, through the 16 s ribosomal RNA (rRNA) gene sequencing analysis. We also identified the specific microbiome signatures potentially useful for the diagnosis of lung cancer.
2. Materials and methods
2.1. Patients and tissue samples
35 lung tumor tissues and paired normal lung tissues for 16 s rRNA sequencing were obtained from patients who underwent surgical resection at Peking University People's Hospital between April 2017 and 2018. Tumor and normal lung specimens were frozen in a liquid nitrogen tank immediately after resection in a sterile environment, and then transferred to −80 °C until processing for DNA extraction. Pathological diagnosis was confirmed by pathologists while staging was performed according to the TNM staging system of the IASLC, version 8 [17]. All patients were diagnosed as lung adenocarcinoma, and they were further divided into 2 groups based on thin-section CT findings including subsolid nodules (SSN group, n =10) and solid nodules (SN group, n = 25). Age, gender and body mass index (BMI) were matched between these two groups, and comprehensive clinical characteristics for the enrolled patients are summarized in Table 1. This study was approved by the ethics committee of Peking University People's Hospital (2018PHB012-01). Written informed consent was obtained from all patients.
Table 1.
Baseline clinical characteristics of objects in this study.
| Clinical characteristics | SSN (n =10) | SN (n =25) | P-value |
|---|---|---|---|
| Age (years; mean ± SD) | 57.80 ± 9.27 | 57.48 ± 11.46 | 0.938 |
| Gender (M/F) | 3/7 | 11/14 | 0.264 |
| BMI (kg/m2, mean ± SD) | 25.13 ± 3.21 | 23.24 ±2.41 | 0.065 |
| Smoking history (Y/N) | 2/8 | 4/21 | 1.000 |
| Location (upper/middle or lower lobe) | 3/7 | 13/12 | 0.285 |
| TNM stage (I/II/III) | 10/0/0 | 19/2/4 | 0.454 |
| Lymph node metastasis (N0/N1) | 10/0 | 19/6 | 0.152 |
| Pleural invasion (Y/N) | 10/0 | 17/8 | 0.073 |
| NLR (mean ± SD) | 2.16 ± 1.27 | 1.92 ± 0.89 | 0.540 |
| PLR (mean ± SD) | 114.93 ± 31.90 | 123.38 ± 42.47 | 0.575 |
| LMR (mean ± SD) | 4.15 ± 1.54 | 4.25 ±1.44 | 0.866 |
| AMC (109/L, mean ± SD) | 0.42 ± 0.09 | 0.44 ± 0.15 | 0.654 |
| CHOL (mmol/L, mean ± SD) | 5.00 ± 1.18 | 4.79 ± 1.06 | 0.604 |
| TG (mmol/L, mean ± SD) | 1.34 ±0.58 | 2.08 ±2.83 | 0.419 |
| HDL (mmol/L, mean ± SD) | 1.19 ± 0.37 | 1.10 ± 0.19 | 0.345 |
| LDL (mmol/L, mean ± SD) | 3.24 ± 0.84 | 3.04 ± 0.65 | 0.462 |
Abbreviations: SSN, subsolid nodules; SN, solid nodules; M/F, male/female; BMI, body mass index; Y/N: Yes/No; NLR: neutrophil-lymphocyte ratio; PLR, platelet-lymphocyte ratio; LMR, lymphocyte-monocyte ratio; AMC, absolute monocyte count; CHOL, total cholesterol; TG, triglyceride; HDL, high-density lipoprotein; LDL, low-density lipoprotein; SD, standard deviation.
2.2. DNA extraction and 16s gene sequencing
Microbial DNA was extracted from the samples using the E.Z.N.A.® soil DNA Kit(Omega Bio-tek, Norcross, GA, U.S.) according to manufacturer's protocols. The final DNA concentration and purification were determined by NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3–V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 338 F (5’- ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) by thermocycler PCR system (GeneAmp 9700, 110 ABI, USA). The amplified products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor ™ -ST (Promega, USA) according to the manufacturer’s protocol. Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina HiSeq 2500 platform (Illumina, San Diego, USA). To address potential environmental contaminations, we introduced 3 negative controls that were processed with the samples. qPCR was used to evaluated the bacterial load in negative controls with 16 s primers (Forward: CCTACGGGNGGCWGCAG; Reverse: GACTACHVGGGTATCTAATCC).
2.3. Microbiome sequences analysis
Raw fastq files were demultiplexed, quality-filtered by Trimmomatic software (version 0.33) [18] and merged by FLASH software (version 1.2.7) [19]. Chimeric sequences were detected and removed using UCHIME software (version 4.2) [20]. Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UCLUST in QIIME software (version 1.8.0) [21]. The OTUs were filtered with a threshold of 0.005% of all sequence numbers. The taxonomy of each 16 s rRNA gene sequence was analyzed by Ribosomal Database Project Classifier (version 2.2, http://sourceforge.net/projects/rdpclassifier/) algorithm [22] against the Silva 16 s rRNA database (Release128, http://www.arb-silva.de) [23] using a confidence threshold of 80%.
Alpha diversity is implemented to display the diversity of bacteria among the different samples. Richness (number of OTUs) and the Shannon diversity index in our samples were calculated with QIIME and displayed by R software (version 3.6.1). Beta diversity analysis was used to evaluate global differences between microbial community composition. Bray-Curtis and unweighted unifrac distance for principal co-ordinates analysis (PCoA) were carried out to visualize the Beta diversity between different groups. Permutational multivariate analysis of variance (PERMANOVA) test was performed to analyze microbiome similarity between different groups. We also performed non-parametric factorial Wilcoxon sum-rank test to detect species-related significant abundance differences in two groups. The linear discriminant analysis (LDA) effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/lefse/) [24] was used to detect microbial biomarkers that differentiate different groups. Significant correlations (P<0.05, and |correlation r| >0.3) between differential genera and clinical characteristics were calculated by Spearman's correlation analyses. The random forest (RF) algorithm was applied to construct predictive models based on microbiota signatures.
Finally, the microbiota functional compositions were predicted using PICRUSt [25]. The OTU abundance table was normalized and then the metagenome functional predictions were performed and mapped on Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologues pathways [26]. STAMP software (version 2.0.8) was used to differential analysis and visualize the results [27].
2.4. Statistical analyses
Statistical analyses were implemented using QIIME, R and STAMP software. Fisher's exact test was used to evaluate categorical variables. Unpaired sample Student's t-test or Wilcoxon rank-sum test was used to compare the differences of continuous variables between two groups. Statistical tests were all two-sided, and differences with P < 0.05 were considered statistically significant.
3. Results
3.1. Participant characteristics
The demographic and clinical characteristics of SSN patients and SNs are shown in Table 1. All patients are northern Chinese and clinical characteristics, including age (p = 0.938), gender (p = 0.264), BMI (p = 0.065), smoking history (p =1), tumor location (p = 0.285), and TNM stage (p = 0.454) were paired between these two groups. Although lymph node metastasis and pleural invasion were only found in SN group, the differences were not significant. Meanwhile, inflammation and lipid-related parameters also showed no significant difference.
Microbial profiling of lung adenocarcinoma and paired normal tissue
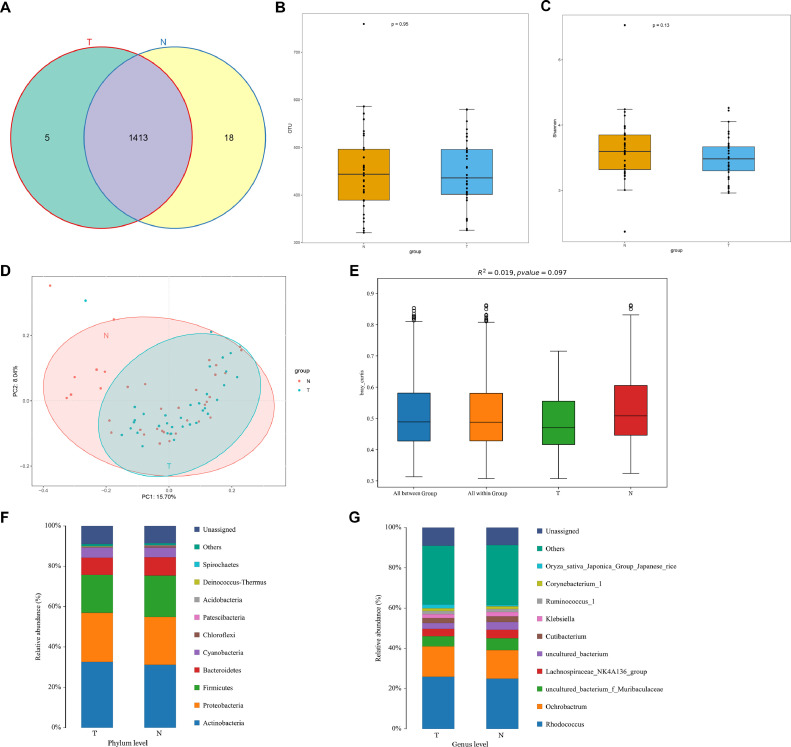
A total of 56,16,903 effective taxon tags were analyzed after sequence de-noising, trimming and chimera filtration. The reads were clustered into 1436 OTUs at 97% similarity cutoff, with 1418 OTUs in lung cancer tissues and 1431 in normal tissues (Fig. 1A). The sequencing depths were evaluated by plotting the rarefaction curve for each sample (Fig. S1A). Most of the samples reached plateau, indicating that the sequencing depth was adequate. Cq values of 16s were 39.6, 39.4 and not detectable in three negative controls, which indicated the influence of contamination could be neglected. Furthermore, we compared the alpha diversity between LUAD tumor and normal groups. LUAD tumor tissue samples tend to have a lower microbial richness (number of OTUs, p =0.95, Fig. 1B) and diversity (Shannon index, p =0.13, Fig. 1C, Fig. S1B) than paired normal tissues, although the differences were nonsignificant. We then assessed the overall microbiome composition between the two groups. PCoA and PERMANOVA analysis did not find significant differences between tumor and normal tissues when considering Bray Curtis (R2=0.019, p = 0.097, Fig. 1D–E) and unweighted unifrac distance (R2=0.013, p = 0.616, Fig. S2). However, normal tissue seemed to have a left shift in Fig. 1D.
Fig. 1.
Microbial profiles of lung adenocarcinoma and paired normal tissue. (A) Different operational taxonomic units (OTUs) between tumor and normal tissues. (B) Richness was evaluated by the number of OTUs. (C) Shannon index of tumor and normal tissues. (D) Principal coordinate analysis (PCoA) using Bray Curtis method. (E) PERMANOVA analysis using Bray Curtis method. (F) Species composition at phylum level. (G) Species composition at genus level.
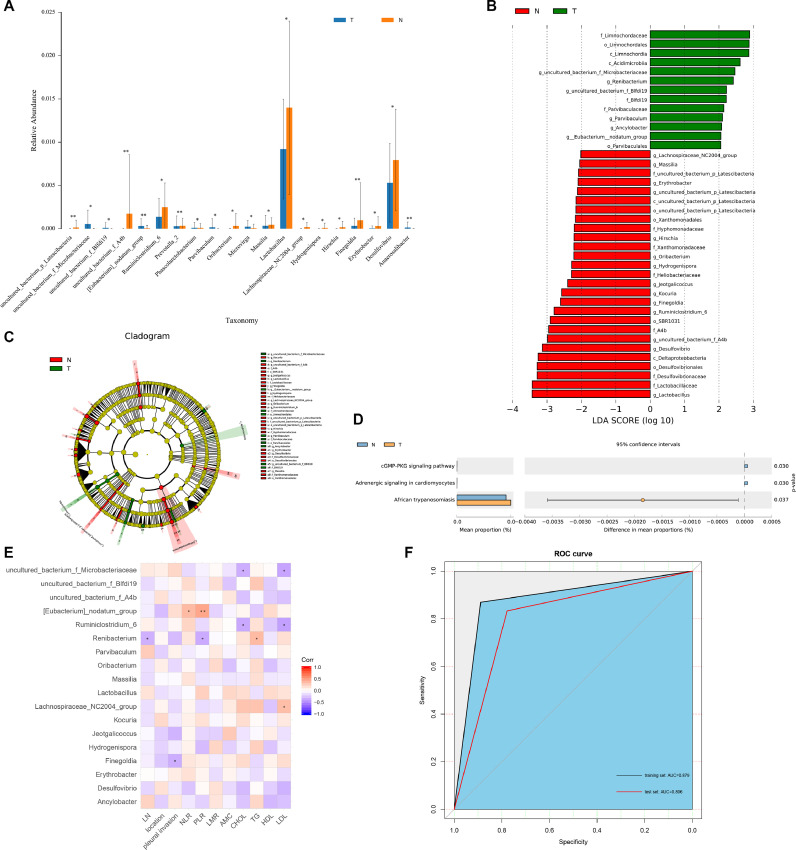
We next compared the relative abundance of microbiota composition of LUAD tumor and normal lung tissues. At phylum level, the proportions of Actinobacteria and Proteobacteria increased, but the proportions of Firmicutes (p = 0.036) and Bacteroidetes decreased in LUAD tumor tissues compared to that in normal tissues (Fig. 1F). At genus level, Rhodococcus and Ochrobactrum were main compositions in both tumor and normal tissues (Fig. 1G). Furthermore, we found 54 statistically significant different genera between these two groups (Fig. 2A). The relative abundance of probiotics, such as Lactobacillus (p = 0.018) showed a notable decline in tumor tissues. Moreover, we performed LEfSe analysis to identify biomarkers that may discriminate between LUAD tumor and normal tissues, which revealed 40 discriminative features (LDA > 2, p < 0.05, Fig. 2B–C) at the class (n = 4), order (n = 6), family (n = 10) and genus (n = 20). Interestingly, we found that tumor group was associated with Parvibaculales, Parvibaculaceae, and Parvibaculum at order, family and genus level, respectively. Renibacterium and Ancylobacter were prominent genus level biomarkers. For normal tissues, Lactobacillus was the strongest marker regardless of species levels.
Fig. 2.
Identification of predictive biomarkers for lung adenocarcinoma and functional prediction. (A) Top 20 differential genus between tumor and normal tissues. (B) LEfSe analysis identified 40 markers at different levels between tumor and normal tissues. The first letter indicates taxonomy level, p: phylum, c: class, o: order, f: family, g: genus. (C) Cladogram of bacterial lineages with significantly different representations in tumor and normal tissues. (D) Differential pathways at KEGG pathway level 3. (E) The relationships among clinical indicators and genera features. (F) ROC curve for the training set and test set. *p < 0.05, **p < 0.01. ROC, receiver operating characteristic; KEGG, Kyoto Encyclopedia of Genes and Genomes.
KEGG pathways enrichment analyses were performed to explore functional differences of the microbiome in tumor and normal tissues. Although the functional composition of these two groups was highly similar, cGMP-PKG signaling pathway was significantly less abundant in tumor than normal tissues (p = 0.03, Fig. 2D).
Associations between lung microbial species and clinical characteristics
To further explore the potential roles microbiota played in LUAD, we investigated the relationships between the relative abundance of genus features (n = 20) and several clinical indices using Spearman's correlation analyses (Fig. 2E). The abundance of LUAD-enriched genus, [Eubacterium]_nodatum_group, was positively correlated with neutrophil-lymphocyte ratio (NLR, r = 0.361, p = 0.033) and platelet-lymphocyte ratio (PLR, r = 0.458, r = 0.006). The abundance of Finegoldia was negatively correlated with pleural invasion (r = -0.353, p = 0.038). And we also found a negative correlation between Renibacterium and lymph node metastasis (r = -0.343, p = 0.043). In addition, the lipid-related parameters total cholesterol (CHOL), triglyceride (TG), and low-density lipoprotein (LDL) were positively or negatively correlated with different genera.
3.4. Detection of LUAD based on the lung microbiota
Based on features identified by LEfSe analysis, we constructed a random forest model to differentiate tumor and normal tissues. All 71 samples were randomly split into the training set (70%) and test set (30%) for model calibration and validation. All the 20 genera were selected based on the feature evaluation step. In the training set, this model had an AUC of 0.879 (95% CI, 0.786–0.972, Fig. 2F). We used the test set to further evaluate the model and reached acceptable predictive power (AUC = 0.806, 95% CI: 0.624–0.987, Fig. 2F), albeit lower than that in the training set. Moreover, 10-fold cross-validation was performed five times for model assessment, resulting in an average AUC of 0.813. These results suggest that our model has stable performance and may provide a possibility for clinical transformation.
3.5. Distinct tumor bacterial microbiome between subsolid and solid nodules
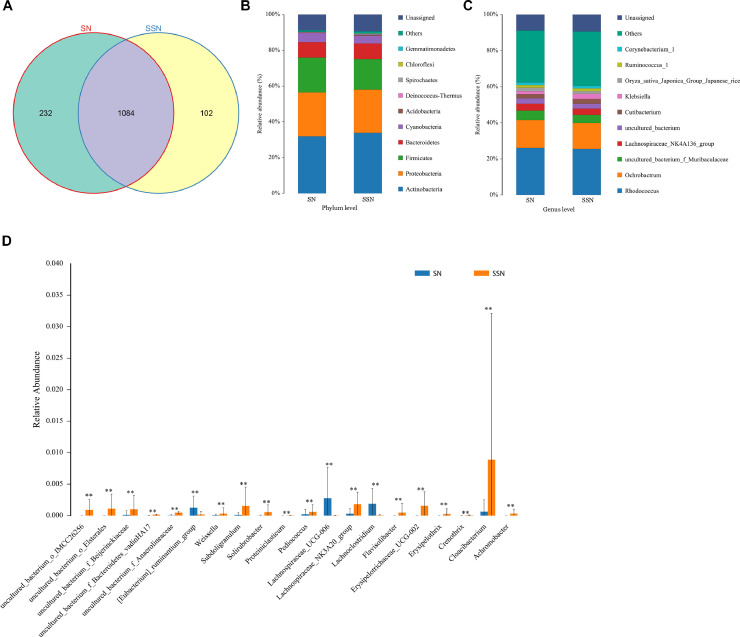
Microbiome profiles are different between SSN and SN. SSN group owns 102 distinctive OTUs while SN group has 232 unique OTUs (Fig. 3A, Fig. S3). At phylum level, both SSN and SN were mainly composed of Actinobacteria, Proteobacteria, Firmicutes, and Bacteroidetes (Fig. 3B). SSN had significantly higher abundances of Chloroflexi (p = 0.009) and Gemmatimonadetes (p = 0.018). At genus level, Rhodococcus and Ochrobactrum are main compositions in these two groups (Fig. 3C). Furthermore, 53 diverse abundance genera were found using Wilcoxon rank-sum test (Fig. 3D). Remarkably, SSN group has higher abundances of Cloacibacterium (p = 0.003), Subdoligranulum (p = 0.002), and Achromobacter (p = 0.001).
Fig. 3.
Microbiome profiles of LUAD manifested as SSN and SN. (A) Different OTUs between SSN and SN. (B) Species composition at phylum level. (C) Species composition at genus level. (D) Top 20 differential genus between SSN and SN. **p < 0.01. LUAD, lung adenocarcinoma; SSN, subsolid nodules; SN, solid nodules; OTUs: operational taxonomic units.
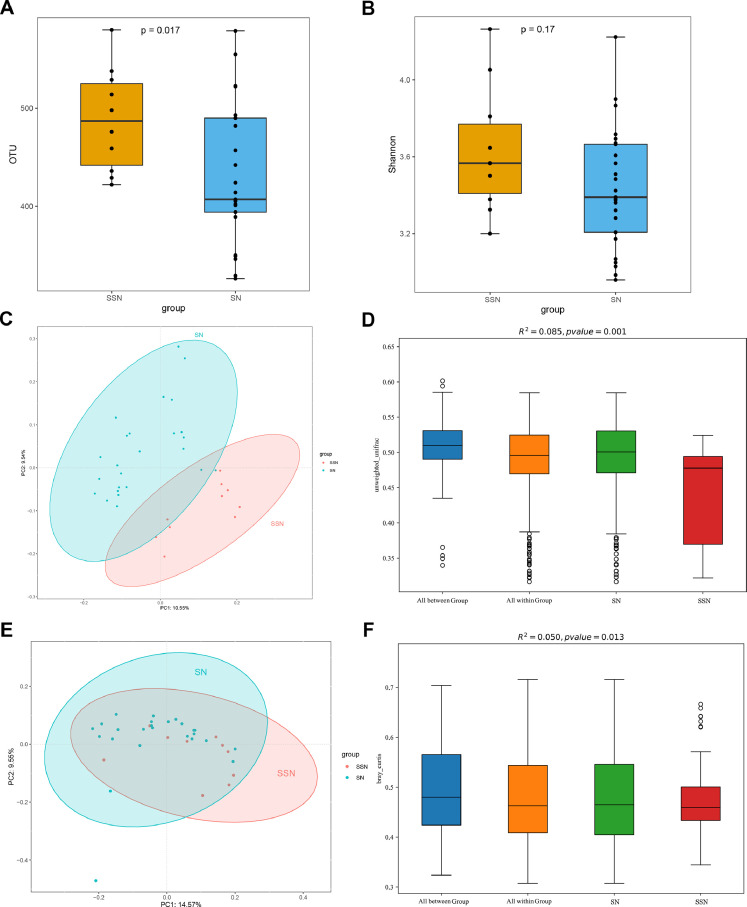
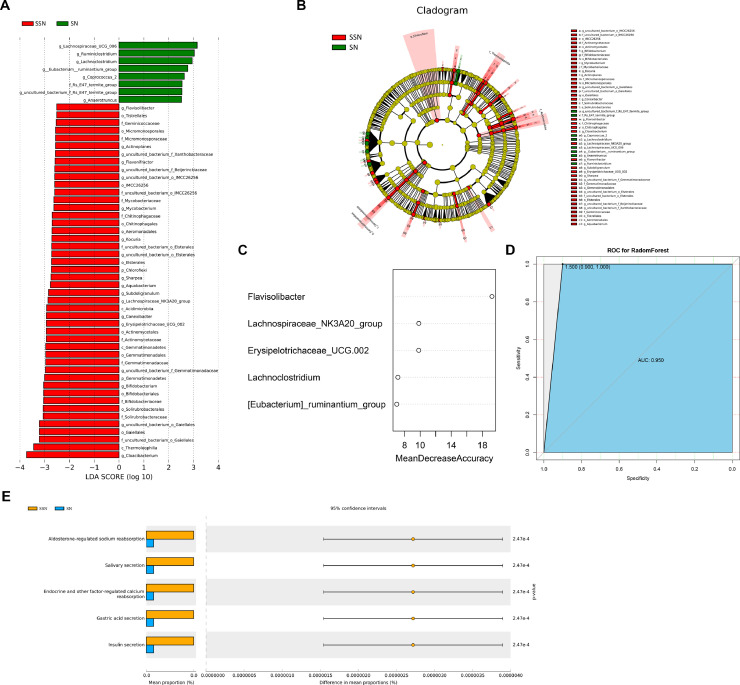
For alpha-diversity, SSN group has greater bacterial richness (p = 0.017, Fig. 4A) and diversity (p =0.17, Fig. 4B, Fig. S1C). We then assessed the microbiome composition of all tumor tissues with beta-diversity values. Both PERMANOVA and PCoA analyses confirmed SSN and SN have statistically different microbiome composition when considering unweighted unifrac distance (R2=0.085 p = 0.001, Fig. 4C–D) and Bray Curtis (R2=0.05, p = 0.013, Fig. 4E–F). The microbiome composition and species abundance similarity of all tumor tissues was shown in Fig. S4. Moreover, LEfSe analysis revealed 54 features that may discriminate SSN and SN (LDA > 2.5, p < 0.05, Fig. 5A–B). Consistent with composition differences, Chloroflexi and Gemmatimonadetes were detected as potential markers for SSN at phylum level. At genus level, increased bacteria such as Cloacibacterium, Subdoligranulum, and Mycobacterium and decreased bacteria like Lachnoclostridium were strong discriminative features for SSN.
Fig. 4.
Microbiome diversity between LUAD manifested as SSN and SN. (A) Richness was evaluated by the number of operational taxonomic units. (B) Shannon index of SSN and SN. (C) PCoA using unweighted unifrac method. (D) PERMANOVA analysis using unweighted unifrac method. (E) PCoA using Bray Curtis method. (F) PERMANOVA analysis using Bray Curtis method. LUAD, lung adenocarcinoma; SSN, subsolid nodules; SN, solid nodules; PCoA, Principal coordinate analysis.
Fig. 5.
Identification of biomarkers and functional prediction. (A) LEfSe analysis identified 54 markers at different levels between SSN and SN. The first letter indicates taxonomy level, p: phylum, c: class, o: order, f: family, g: genus. (B) Cladogram of bacterial lineages with significantly different representations in SSN and SN groups. (C) 5 genus signatures to construct the predictive model. (D) ROC curve for the random forest model. (E) Differential pathways at KEGG pathway level 3. SSN, subsolid nodules; SN, solid nodules; ROC, receiver operating characteristic; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Furthermore, we constructed a predictive model to discriminate SSN from SN. According to LEfSe result, 26 genus features were selected as candidate signatures. After feature elimination, the best performing model was selected as the final model, with five genera signatures (Fig. 5C). SSN could be accurately discriminated from SN using random forest algorithm with a sensitivity of 1, specificity of 0.8, and AUC of 0.950 (95% CI, 0.852–1.000, Fig. 5D). To further verify the accuracy of this model, we performed 10-fold cross-validation for five times, resulting in a mean AUC of 0.933.
16s functional predictive analysis also showed differences between SSN and SN groups. KEGG pathway comparisons found oral microbiome related pathways, salivary secretion (p<0.001) and gastric acid secretion (p<0.001) were significantly enriched in SSN but not in SN (Fig. 5E). In addition, endocrine and other factor-regulated calcium reabsorption and two other pathways were also different between these groups.
Discussion
This study demonstrates, for the first time, the distinct tumor bacterial microbiome between subsolid and solid nodules. Increasing evidence suggests that reduced microbiome diversity is associated with poor survival of cancer patients [28,29]. Our findings that SSN tissues have higher microbiome diversity than SN is consistent with the fact that SSN is less invasive compared with SN and patients with SSN have longer recurrence-free survival [4]. Therefore, this study provides new clues to the indolent nature of SSN in the perspective of microbiome. In addition, consistent with previous findings [16,30,31], we also found similar microbiome composition between LUAD tumor and normal tissues. Furthermore, microbiome signatures showed good performance to predict LUAD or SSN, which may provide a possibility and reliability for clinical transformation.
Most previous studies on the lung microbiota were based on bronchoalveolar lavage, bronchoscopic brushing, or sputum samples, and few directly characterized the microbiome in lung tumor tissues [16,32]. Increasing evidence revealed that alpha diversity is higher in non-malignant lung tissues than in tumor tissues [16,31]. Beta diversity, or overall microbiome composition is not significantly different between normal and tumor tissues. A study by Yu and colleagues demonstrated that Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria were core microbiota in lung tissues at phylum level [33]. In this study, we also found a declining alpha diversity in LUAD tumor tissues, and core microbiota was similar to Yu's finding. Although overall microbiome composition and functional pathways were similar between tumor and normal tissues, we still found some differences. Lactobacillus was reported to show anti-cancer effects in various cancers [34,35]. Lactobacillus could exert direct cytotoxic effects on cancer cells and has various immunoregulatory functions [34]. Our results showed an obviously decreased abundance of Lactobacillus in LUAD tumor tissues. cGMP-PKG signaling pathway was known to regulate cell apoptosis and proliferation [36]. Several studies demonstrated that cGMP-PKG signaling pathway could suppress Wnt/β-catenin signaling to inhibit tumor cell growth [36], [37], [38]. In this study, we found that cGMP-PKG signaling pathway was significantly less abundant in LUAD tumor than normal tissues.
Lung adenocarcinomas manifested as radiological subsolid nodules were defined as a special clinical subtype [4]. SSN are considered to be more indolent and less aggressive than LUAD manifested as SN. The prevalence of lymphatic metastasis was lower in patients with SSN than in those with SN. In this study, we also found the lymph node metastasis or pleural invasion were more frequency in SN. However, the microbiome properties of SSN remain unclear. Only one study tried to characterize the microbiota in SSN [39]. They found that Mycobacterium was the core microbiota found in SSN, and two functional pathways, Secondary Metabolism and the Serine Threonine protein kinase were significantly increased in SSN over normal tissue samples. Nevertheless, they did not compare the microbiome between SSN and SN. In the present study, we, for the first time, characterized the different microbiome between SSN and SN. Higher diversity of SSN than SN suggests SSN is less malignant, which also supports the observation that SSN is more indolent than SN. PCoA and PERMANOVA showed significantly different overall microbiome composition between SSN and SN, which also provided evidence that SSN is a special subtype. Moreover, we found elevated Chloroflexi and Gemmatimonadetes in SSN group. Interestingly, increased genera, including Cloacibacterium, Subdoligranulum, and Mycobacterium were detected as potential biomarkers for SSN. Mycobacterium tuberculosis and Mycobacterium avium complex are considered to relate to lung cancer [40,41]. Another study found that 6.5% of patients with pulmonary nontuberculous mycobacterial disease, also had lung cancer [42]. These epidemiologic links are mechanically interpreted by chronic inflammation-associated carcinogenesis [43]. The persistent infection of mycobacterial organisms in the lung can induce a pro-inflammatory response to local tissues and then trigger the release of factors that may support the outgrowth of premalignant cells. Besides, Cloacibacterium, Subdoligranulum were also reported to relate to chronic inflammation [44,45]. And Achromobacter infection may mimic lung cancer [46]. These results indicated that microbiota in the local microenvironment may also contribute to the initiation and progression of SSN. We also found function differences between SSN and SN. Salivary secretion and gastric acid secretion were significantly enriched in SSN but not in SN, which means that SSN may contain more oral-derived microbiota. Furthermore, endocrine and other factor-regulated calcium reabsorption was different between SSN and SN. Calcium ion is an important second messenger within cells, which can participate in the development of tumors in various ways [47]. Thus, the abnormal absorption of calcium ions in cells may associate with tumorigenesis.
Recently, Poore et al. proposed a novel cancer diagnostic approach with preferable accuracy through microbiome analyses of blood and tissues [48]. They found unique microbial signatures in tissue and blood within and between most major types of cancer. Likewise, in current study, we identified microbial signatures to differentiate LUAD tumor and normal tissues, as well as SSN and SN. LEfSe analysis was used to identify potential markers and machine learning was applied to identify microbial signatures. Although the microbiome diversity between LUAD tumor and normal tissues were similar, some species had different abundances between these two groups. A 20 genera-based signature showed a good diagnostic accuracy to discriminate LUAD from normal tissues in both training and test sets. Meanwhile, a five genera-based signature can accurately discriminate SSN and SN. In addition, cross-validation of these two models results in reasonably high predictive powers as well. Our results indicated that microbiota may become a novel tool for cancer diagnostic.
We also recognize several limitations of our study. Although 16s rRNA gene sequencing is widely used for microbiota characterization, it has great limitations in species level analysis and functional prediction. Further study using metagenomic sequencing may provide a deeper understanding of the characteristics and function of microbiota in lung cancer. In addition, the number of patients and samples included are insufficient. Bacterial derived metabolites play important roles in tumor development and may strengthen the accuracy to diagnosis lung adenocarcinoma. However, we didnot make metabolome analysis in this study. Future studies with large sample sizes and more rigorous designs are expected to further comprehend microbiota in lung cancer.
5. Conclusions
To our knowledge, this is the first study that investigated tumor microbiome diversity between subsolid and solid lung adenocarcinoma. Early stage LUAD manifested as radiological SSN has elevated microbiome richness and diversity compared with SN, and the microbiome composition of SSN is distinct from that of SN. Our finding is consistent with the fact that SSN is less invasive compared with SN. Therefore, this study provides new clues to the indolent nature of SSN in the perspective of microbiome. In addition, we found similar diversity and microbiome composition between LUAD tumor and normal tissues. We also uncover the microbial signatures associated with LUAD or SSN and develop accurate genera-based predictors for clinical diagnosis.
Funding
This study was supported by the National Natural Science Foundation of China (81871879, 81772469, and 61877001).
Authors' contributions
MQ, FY and GJ conceived and designed the study; YM, SW and SM conducted the experiments, collected and analyzed the data; YM, FY and MQ interpreted the data. YM, SW and MQ wrote and revised the manuscript.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101050.
Contributor Information
Mantang Qiu, Email: qiumantang@163.com.
Fan Yang, Email: yangfan@pkuph.edu.cn.
Guanchao Jiang, Email: jiangguanchao@263.net.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Polanski J., Jankowska-Polanska B., Rosinczuk J., Chabowski M., Szymanska-Chabowska A. Quality of life of patients with lung cancer. Onco Targets Ther. 2016;9:1023–1028. doi: 10.2147/OTT.S100685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molina J.R., Yang P., Cassivi S.D., Schild S.E., Adjei A.A. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye T., Deng L., Wang S., Xiang J., Zhang Y., Hu H., Sun Y., Li Y., Shen L., Xie L., Gu W., Zhao Y., Fu F., Peng W., Chen H., Shen Y. Lung adenocarcinomas manifesting as radiological part-solid nodules define a special clinical subtype. J. Thorac. Oncol. 2019;14(10):S546–S547. doi: 10.1016/j.jtho.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 5.Fan L., Wang Y., Zhou Y., Li Q., Yang W., Wang S., Shan F., Zhang X., Shi J., Chen W., Liu S.Y. Lung cancer screening with low-dose CT: baseline screening results in Shanghai. Acad. Radiol. 2019;26(10):1283–1291. doi: 10.1016/j.acra.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Ren Y.J., Huang S.J., Dai C.Y., Xie D., Zheng L., Xie H.K., Zheng H., She Y.L., Zhou F.Y., Wang Y., Li P.P., Fei K., Jiang G.N., Zhang Y., Su B., Sweet-Cordero E.A., Tran N.L., Yang Y.N., Patei J.N., Rolfo C., Rocco G., Cardona A.F., Tuzi A., Suter M.B., Yang P., Xu W.N., Chen C. Germline predisposition and copy number alteration in pre-stage lung adenocarcinomas presenting as ground-glass nodules. Front Oncol. 2019;9:288. doi: 10.3389/fonc.2019.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomes M., Teixeira A.L., Coelho A., Araujo A., Medeiros R. The role of inflammation in lung cancer. Inflamm. Cancer. 2014;816:1–23. doi: 10.1007/978-3-0348-0837-8_1. [DOI] [PubMed] [Google Scholar]
- 8.Engels E.A. Inflammation in the development of lung cancer: epidemiological evidence. Expert Rev. Anticancer. 2008;8(4):605–615. doi: 10.1586/14737140.8.4.605. [DOI] [PubMed] [Google Scholar]
- 9.Vangipurapu J., Silva L.F., Kuulasmaa T., Smith U., Laakso M. Microbiota-related metabolites and the risk of type 2 diabetes. Diabetes Care. 2020;43(6):1319–1325. doi: 10.2337/dc19-2533. [DOI] [PubMed] [Google Scholar]
- 10.Liu R.X., Hong J., Xu X.Q., Feng Q., Zhang D.Y., Gu Y.Y., Shi J., Zhao S.Q., Liu W., Wang X.K., Xia H.H., Liu Z.P., Cui B., Liang P.W., Xi L.Q., Jin J.B., Ying X.Y., Wang X.L., Zhao X.J., Li W.Y., Jia H.J., Lan Z., Li F.Y., Wang R., Sun Y.K., Yang M.L., Shen Y.X., Jie Z.Y., Li J.H., Chen X.M., Zhong H.Z., Xie H.L., Zhang Y.F., Gu W.Q., Deng X.X., Shen B.Y., Xu X., Yang H.M., Xu G.W., Bi Y.F., Lai S.H., Wang J., Qi L., Madsen L., Wang J.Q., Ning G., Kristiansen K., Wang W.Q. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 2017;23(7):859. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 11.Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillere R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., Fidelle M., Flament C., Poirier-Colame V., Opolon P., Klein C., Iribarren K., Mondragon L., Jacquelot N., Qu B., Ferrere G., Clemenson C., Mezquita L., Masip J.R., Naltet C., Brosseau S., Kaderbhai C., Richard C., Rizvi H., Levenez F., Galleron N., Quinquis B., Pons N., Ryffel B., Minard-Colin V., Gonin P., Soria J.C., Deutsch E., Loriot Y., Ghiringhelli F., Zalcman G., Goldwasser F., Escudier B., Hellmann M.D., Eggermont A., Raoult D., Albiges L., Kroemer G., Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 12.Mima K., Nakagawa S., Sawayama H., Ishimoto T., Imai K., Iwatsuki M., Hashimoto D., Baba Y., Yamashita Y., Yoshida N., Chikamoto A., Baba H. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. 2017;402:9–15. doi: 10.1016/j.canlet.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Nugent J.L., McCoy A.N., Addamo C.J., Jia W., Sandler R.S., Keku T.O. Altered tissue metabolites correlate with microbial dysbiosis in colorectal adenomas. J. Proteome Res. 2014;13(4):1921–1929. doi: 10.1021/pr4009783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bingula R., Filaire M., Radosevic-Robin N., Bey M., Berthon J.Y., Bernalier-Donadille A., Vasson M.P., Filaire E. Desired turbulence? gut-lung axis, immunity, and Lung cancer. J. Oncol. 2017;2017 doi: 10.1155/2017/5035371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramirez-Labrada A.G., Isla D., Artal A., Arias M., Rezusta A., Pardo J., Galvez E.M. The influence of lung microbiota on lung carcinogenesis, immunity, and immunotherapy. Trends Cancer. 2020;6(2):86–97. doi: 10.1016/j.trecan.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Mao Q.X., Jiang F., Yin R., Wang J., Xia W.J., Dong G.C., Ma W.D., Yang Y., Xu L., Hu J.Z. Interplay between the lung microbiome and lung cancer. Cancer Lett. 2018;415:40–48. doi: 10.1016/j.canlet.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 17.Detterbeck F.C., Bolejack V., Arenberg D.A., Crowley J., Donington J.S., Franklin W.A., Girard N., Marom E.M., Mazzone P.J., Nicholson A.G., Rusch V.W., Tanoue L.T., Travis W.D., Asamura H., Rami-Porta R., C I.S.P.F., Boards A., Workgrp M.P.S. The IASLC lung cancer staging project: background data and proposals for the classification of lung cancer with separate tumor nodules in the forthcoming eighth edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2016;11(5):681–692. doi: 10.1016/j.jtho.2015.12.114. [DOI] [PubMed] [Google Scholar]
- 18.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Tumbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glockner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6) doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas G.M., Beiko R.G., Langille M.G.I. Predicting the functional potential of the microbiome from marker genes using PICRUSt. Methods Mol. Biol. 2018;1849:169–177. doi: 10.1007/978-1-4939-8728-3_11. [DOI] [PubMed] [Google Scholar]
- 26.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks D.H., Tyson G.W., Hugenholtz P., Beiko R.G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin Y.P., Dong H., Xia L.L., Yang Y., Zhu Y.Q., Shen Y., Zheng H.J., Yao C.C., Wang Y., Lu S. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in Chinese patients with NSCLC. J. Thorac. Oncol. 2019;14(8):1378–1389. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Riquelme E., Zhang Y., Zhang L.L., Montiel M., Zoltan M., Dong W.L., Quesada P., Sahin I., Chandra V., San Lucas A., Scheet P., Xu H.W., Hanash S.M., Feng L., Burks J.K., Do K.A., Peterson C.B., Nejman D., Tzeng C.W.D., Kim M.P., Sears C.L., Ajami N., Petrosino J., Wood L.D., Maitra A., Straussman R., Katz M., White J.R., Jenq R., Wargo J., McAllister F. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bingula R., Filaire E., Molnar I., Delmas E., Berthon J.Y., Vasson M.P., Bernalier-Donadille A., Filaire M. Characterization of microbiota in saliva, bronchoalveolar lavage fluid, non-malignant, peritumoural and tumor tissue in non-small cell lung cancer patients: a cross-sectional clinical trial. Respir. Res. 2020;21(1):129. doi: 10.1186/s12931-020-01392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters B.A., Hayes R.B., Goparaju C., Reid C., Pass H.I., Ahn J. The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol. Biomark. 2019;28(4):731–740. doi: 10.1158/1055-9965.EPI-18-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patnaik S.K., Cortes E.G., Kannisto E.D., Punnanitinont A., Dhillon S.S., Liu S., Yendamuri S. Lower airway bacterial microbiome may influence recurrence after resection of early-stage non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2021;161(2):419–429. doi: 10.1016/j.jtcvs.2020.01.104. e16. [DOI] [PubMed] [Google Scholar]
- 33.Yu G., Gail M.H., Consonni D., Carugno M., Humphrys M., Pesatori A.C., Caporaso N.E., Goedert J.J., Ravel J., Landi M.T. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17(1):163. doi: 10.1186/s13059-016-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abedin-Do A., Taherian-Esfahani Z., Ghafouri-Fard S., Ghafouri-Fard S., Motevaseli E. Immunomodulatory effects of Lactobacillus strains: emphasis on their effects on cancer cells. Immunotherapy. 2015;7(12):1307–1329. doi: 10.2217/imt.15.92. [DOI] [PubMed] [Google Scholar]
- 35.Banna G.L., Torino F., Marletta F., Santagati M., Salemi R., Cannarozzo E., Falzone L., Ferrau F., Libra M. Lactobacillus rhamnosus GG: an overview to explore the rationale of its use in cancer. Front Pharmacol. 2017;8:603. doi: 10.3389/fphar.2017.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K., A.P. G The interaction between the Wnt/beta-catenin signaling cascade and PKG activation in cancer. J. Biomed. Res. 2017;31(3):189–196. doi: 10.7555/JBR.31.20160133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Browning D.D., Kwon I.K., Wang R. cGMP-dependent protein kinases as potential targets for colon cancer prevention and treatment. Future Med. Chem. 2010;2(1):65–80. doi: 10.4155/fmc.09.142. [DOI] [PubMed] [Google Scholar]
- 38.Li N., Xi Y., Tinsley H.N., Gurpinar E., Gary B.D., Zhu B., Li Y., Chen X., Keeton A.B., Abadi A.H., Moyer M.P., Grizzle W.E., Chang W.C., Clapper M.L., Piazza G.A. Sulindac selectively inhibits colon tumor cell growth by activating the cGMP/PKG pathway to suppress Wnt/β-catenin signaling. Mol. Cancer Ther. 2013;12(9):1848–1859. doi: 10.1158/1535-7163.MCT-13-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren Y.J., Su H., She Y.L., Dai C.Y., Xie D., Narrandes S., Huang S.J., Chen C., Xu W.N. Whole genome sequencing revealed microbiome in lung adenocarcinomas presented as ground-glass nodules. Transl. Lung Cancer R. 2019;8(3):235. doi: 10.21037/tlcr.2019.06.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilaniya V., Gera K., Kunal S., Shah A. Pulmonary tuberculosis masquerading as metastatic lung disease. Eur. Respir. Rev. 2016;25(139):97–98. doi: 10.1183/16000617.00002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lande L., Peterson D.D., Gogoi R., Daum G., Stampler K., Kwait R., Yankowski C., Hauler K., Danley J., Sawicki K., Sawicki J. Association between pulmonary mycobacterium avium complex infection and lung cancer. J. Thorac. Oncol. 2012;7(9):1345–1351. doi: 10.1097/JTO.0b013e31825abd49. [DOI] [PubMed] [Google Scholar]
- 42.Marras T.K., Daley C.L. Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin. Chest Med. 2002;23(3):553–567. doi: 10.1016/s0272-5231(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 43.Liang H.Y., Li X.L., Yu X.S., Guan P., Yin Z.H., He Q.C., Zhou B.S. Facts and fiction of the relationship between preexisting tuberculosis and lung cancer risk: a systematic review. Int. J. Cancer. 2009;125(12):2936–2944. doi: 10.1002/ijc.24636. [DOI] [PubMed] [Google Scholar]
- 44.Hirano A., Umeno J., Okamoto Y., Shibata H., Ogura Y., Moriyama T., Torisu T., Fujioka S., Fuyuno Y., Kawarabayasi Y., Matsumoto T., Kitazono T., Esaki M. Comparison of the microbial community structure between inflamed and non-inflamed sites in patients with ulcerative colitis. J. Gastroenterol. Hepatol. 2018 doi: 10.1111/jgh.14129. [DOI] [PubMed] [Google Scholar]
- 45.Sundin J., Rangel I., Repsilber D., Brummer R.J. Cytokine response after stimulation with key commensal bacteria differ in post-infectious irritable bowel syndrome (PI-IBS) patients compared to healthy controls. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0134836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Claassen S.L., Reese J.M., Mysliwiec V., Mahlen S.D. Achromobacter xylosoxidans infection presenting as a pulmonary nodule mimicking cancer. J. Clin. Microbiol. 2011;49(7):2751–2754. doi: 10.1128/JCM.02571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monteith G.R., Prevarskaya N., Roberts-Thomson S.J. The calcium-cancer signalling nexus. Nat. Rev. Cancer. 2017;17(6):367–380. doi: 10.1038/nrc.2017.18. [DOI] [PubMed] [Google Scholar]
- 48.Poore G.D., Kopylova E., Zhu Q.Y., Carpenter C., Fraraccio S., Wandro S., Kosciolek T., Janssen S., Metcalf J., Song S.J., Kanbar J., Miller-Montgomery S., Heaton R., Mckay R., Patel S.P., Swafford A.D., Knight R. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579(7800):567. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.