Abstract
Vimentin is an intermediate filament (IF) protein that is expressed in leukocytes, fibroblasts and endothelial cells of blood vessels. Vimentin filaments contribute to structural stability of the cell membrane, organelle positioning and protein transport. Vimentin self-assembles into a dimer that subsequently forms high-order structures, including tetramers and octamers. The details of IF assembly at crystallographic resolutions are limited to the tetrameric form. We describe a crystal structure of a fragment of a vimentin rod domain (coil 1B) with a dimer of tetramers in the asymmetric unit. Coil 1B in the crystal is in an infinitely high-order filamentous assembly state, in which the tetramers are packed against each other laterally in an antiparallel fashion across the crystal lattice. In one of the directions of lateral packing, the tetramers pack against each other strictly head-to-tail, and in the orthogonal direction the tetramers pack in a staggered manner. This organization of the tetramers of coil 1B in the crystal lattice, together with previously reported biochemical and structural data, yield a model of high-order vimentin filament assembly.
Keywords: Intermediate filament, oligomerization, crystal structure, coiled-coil, helical domain
Graphical abstract
A crystal structure of a filamentous state of coil 1B of human vimentin yields a model of assembly of a vimentin filament.
Introduction
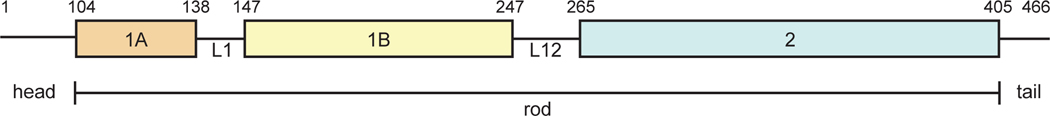
Mechanical stability of cells of vertebrate organisms is maintained by a filament system composed of microfilaments (6 nm diameter), microtubules (25 nm diameter) and intermediate filaments (IFs), which derive their name from their intermediate thickness (10 nm diameter) [1]. IFs are much more diverse in their sequences than microfilaments and microtubules. Prior studies categorized IFs into six different types [2, 3] based on their sequence: keratins (types I and II); vimentin (type III); neurofilaments (type IV); lamins (type V); and nestin (type VI). Cytoplasmic IFs (types I to IV) and nuclear lamins (type V) are genetically related, whereas type VI IFs are quite distinct [4]. Probing the structure of IFs, such as vimentin, provides insights into the roles and interrelationships among this remarkable family of structural proteins. Despite the sequence diversity of IF, IFs share a similar domain organization. An IF protein consists of a helical rod domain flanked by non-helical head and tail domains (Fig. 1). A fundamental building block of IFs is a dimer, in which the central, helical rod domains of two monomers interact to form a parallel coiled coil. Vimentin dimers are stable even in 6 M urea, due to stabilizing interactions between hydrophobic residues that are repeated in a nearly continuous periodic heptad pattern (Fig. 2) over the entire rod domain [5]. Crosslinking and biophysical studies of keratins and vimentins indicate that IF dimers associate with each other laterally in an anti-parallel manner [6–8]. Higher order assembly of IFs proceeds by association of these tetramers into a loose and dynamic lateral assembly of octamers [9]. Four octamers assemble into so-called unit-length filaments (ULFs). The assembly of a mature IF occurs spontaneously in vitro merely by changing buffer conditions [10, 11]. A kinetic study of the assembly of vimentin in vitro using electron and scanning force microscopy suggests that formation of vimentin filaments occurs in a two-step mechanism, where ULFs rapidly form and then anneal to each other longitudinally [12]. The final step in the formation of a mature IF is a compaction phase, in which a molecular rearrangement results in a significant decrease in the IF diameter from 17 to 10 nm [9, 13–17].
Figure 1.
The domain organization of vimentin. The region boundaries are indicated by residue numbers.
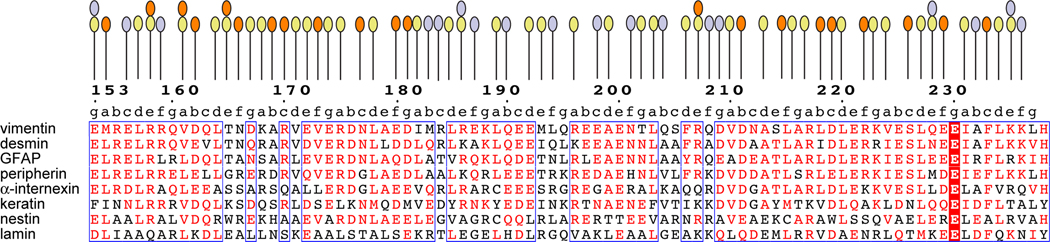
Figure 2.
A multiple sequence alignment of the ordered region of coil 1B sequences of human IF proteins. The heptad pattern is denoted by letters a-g above the alignment. Residues involved in the dimerization, tetramerization and higher-order interfaces are marked by yellow, orange and light-blue ovals, respectively. The boxes indicate the regions of the highest sequence similarity.
At the heart of studying structural features of IFs lies our interest in correlating structure with biological function and dysfunction. Although the functions of different IFs are somewhat redundant at normal conditions, their roles become individually important under specific stresses. For example, even though vimentin knockout mice did not display any apparent phenotype, wound healing in such mice was shown to be delayed [18]. Methylation of the vimentin gene and differential regulation of vimentin expression have been associated with cancer and its invasiveness, and with gastrointestinal diseases [19–24]. In addition, in both cancer cells and normal cells, vimentin sequesters a naturally occurring tumor suppressor, Prostate apoptosis response-4 (Par-4), capable of triggering apoptosis. Targeting vimentin to liberate Par-4 for its antitumor activity using secretagogues such as withaferin A or synthetically generated arylquins is a potential approach to developing new antineoplastic agents [25–27].
Elucidating IF structure and assembly represents a major challenge. Filamentous proteins are not readily amenable to crystallization. Obtaining a high-resolution structure of a full-length IF has not been possible due to the intrinsically flexible nature of long filaments. To circumvent this obstacle, crystals of relatively short and rigid helical segments of an IF could be obtained. Vimentin is the most extensively crystallographically characterized IF to date. Several crystal structures of overlapping segments of the vimentin rod domain (Table 1) [28–33] have shown that the rod domain of vimentin consists of two helical regions, commonly referred to as coils 1 and 2, with the first one subdivided into coils 1A and 1B (Fig. 1) [34]. These helical regions are separated by linker regions L1 (between 1A and 1B) and L12 (between 1B and 2), whose structures have not been rigorously established, likely due to the presence of dynamic elements that lack secondary structure. This “divide and conquer” approach has provided useful structural data on low-order assembly states of vimentin rod subdomains as dimers and, for one crystal structure of coil 1B, as its tetramer (PDB ID: 3UF1; [30]). Superimposition of structures of several overlapping fragments of vimentin yielded a model of the nearly complete rod domain in its dimeric form with the coil 1B region modeled as a tetramer [29]. Despite this progress, difficulties with obtaining crystals have precluded one from gaining experimental insight into high-order filamentous assembly at crystallographic resolutions. Therefore, the details of the subsequent vimentin associations to form octamers and the final ULFs as well as the details of the associations of IFs with other proteins (e.g., vimetin and Par-4) are a subject of continued interest. In this study, we report the first crystal structure of a high-order filamentous state of vimentin coil 1B.
Table 1.
Crystal structures of vimentin regions.
| PDB ID | Residues | Region | Number of monomers in asymmetric unit | Assembly state in the crystal |
|---|---|---|---|---|
| 5WHFa | 153 – 238 | 1B | 8 | Infinite filament |
| 3S4R [29] | 99 – 189 | 1A, L1, 1B (ΔC) | 2 | 2 (1B), 4 (1A), Sparse/swapped-coil filament |
| 3G1E [33] | 101 – 139 | 1A | 2 | 2, staggered filament |
| 1GK7 [36] | 102 – 138 | 1A | 1 | 1 |
| 3SSU [29] | 144 – 189 | 1B (ΔC) | 2 | 2 |
| 3UF1 [30] | 144 – 251 | 1B | 4 | 4 |
| 3SWK [29] | 153 – 238 | 1B | 2 | 2 |
| 4YV3 [28] | 161 – 238 | 1B (ΔN) | 3 | 3 (2- swapped coil) |
| 4YPC [28] | 161 – 243 | 1B (ΔN) | 1 | 3 (N-terminal coiled-coil), staggered filament |
| 1GK4 [36] | 328 – 411 | 2 (ΔN) | 6 | 4 |
| 3KLT [32] | 253 – 324 | part of L12, part of 2 | 4 | 4 (non-biological), 2- part of coil 2, filamentous |
| 3TRT [31] | 261 – 335 | part of L12, part of 2 | 2 | 2 |
| 1GK6 [36] | 385 – 412 | 2 (ΔN) | 2 | 2 |
This study.
Results
Crystal structure of vimentin coil 1B in a high-order filamentous assembly state
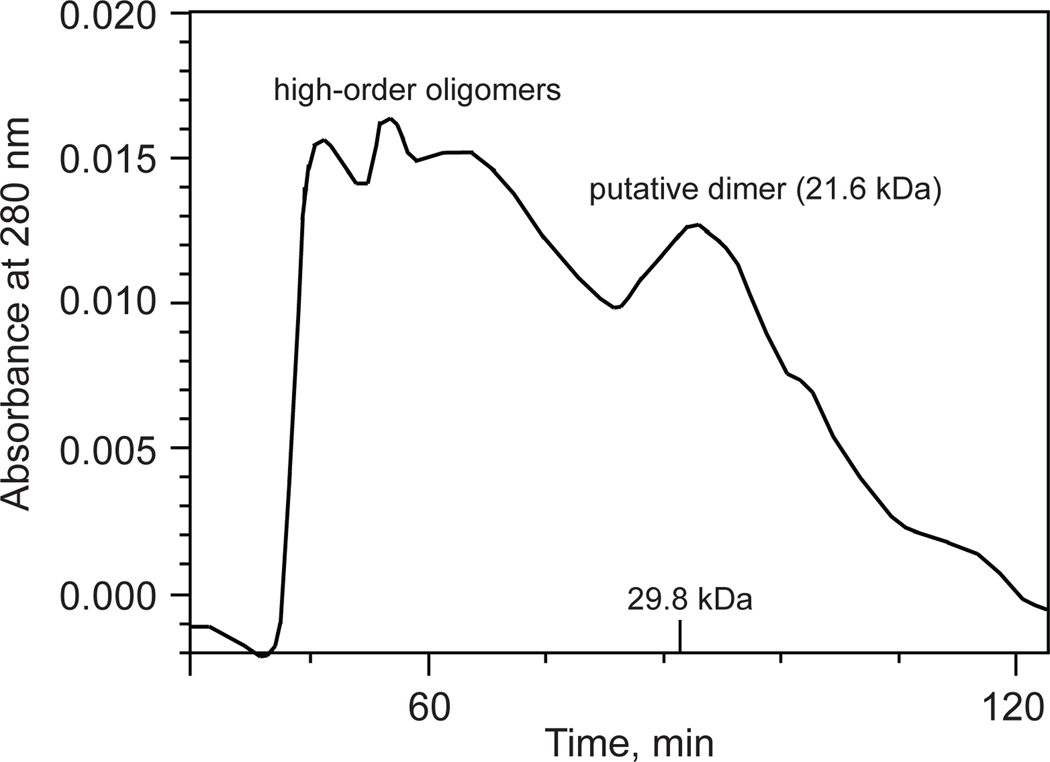
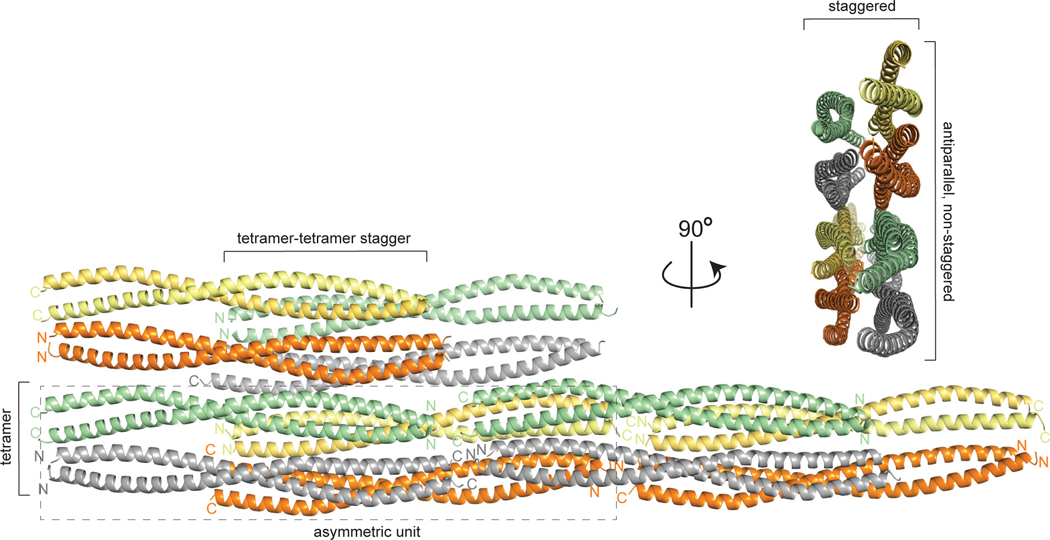
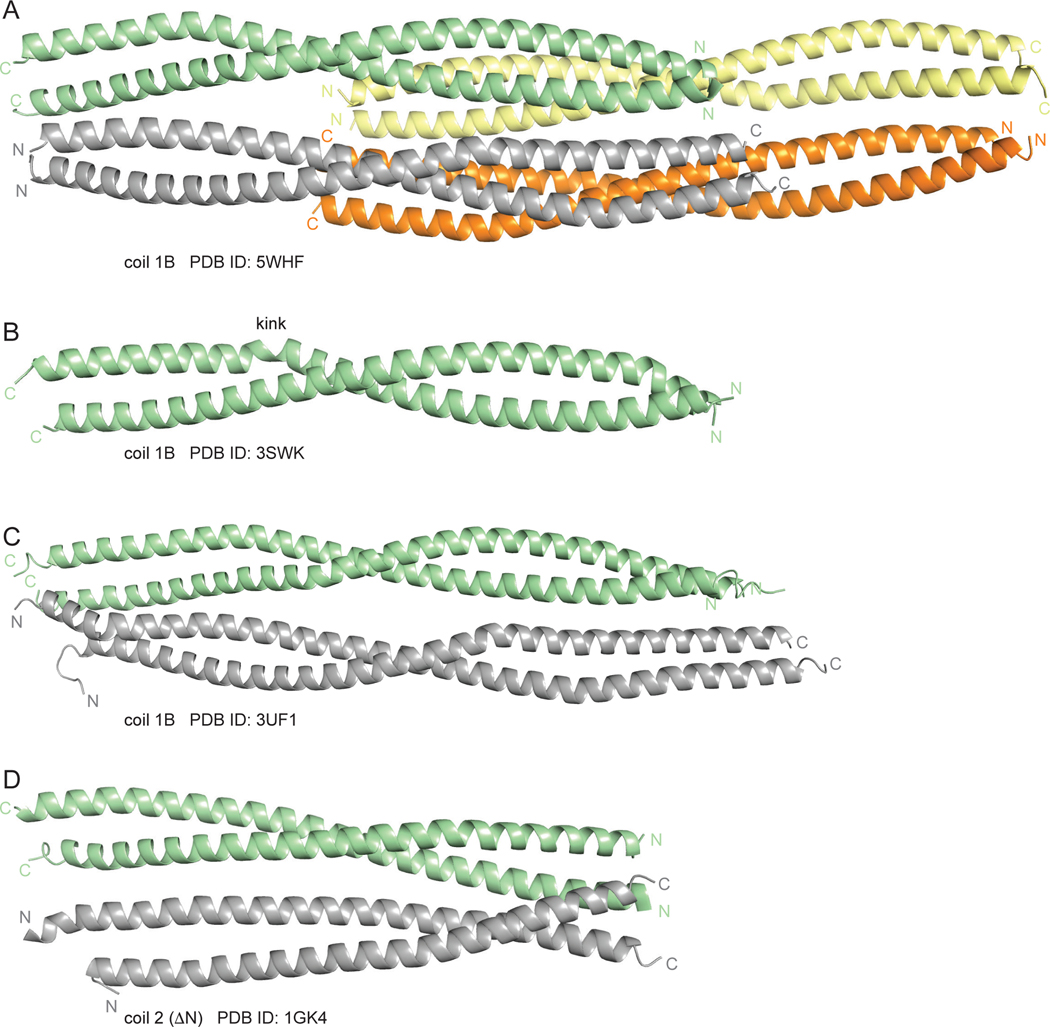
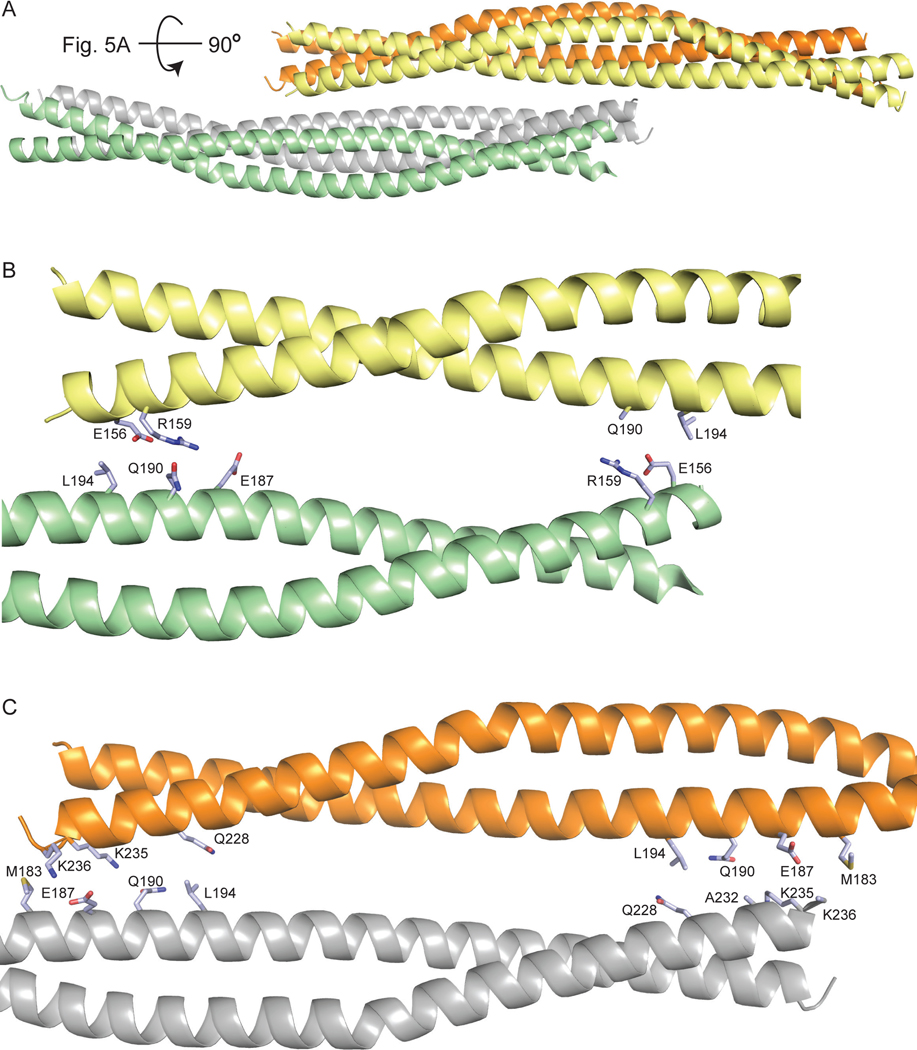
Recombinant vimentin coil 1B (residues 153–238) was overexpressed in E. coli and purified by a combination of Ni2+-chelating and size-exclusion chromatography. The purified protein eluted from the size-exclusion column distributed relatively evenly over multiple peaks, one likely corresponding to a dimer and the others to higher-order oligomeric forms (Fig. 3). In agreement with this observation, the same construct (with residue 153 at the N-terminus) was crystallized as a dimer [29], and a larger construct (residues 144–251) containing linker L1 and coil 1B formed somewhat unstable tetramers as well as higher-order oligomers at a higher salt concentration (100 mM NaCl; favoring oligomerization) than that in our experiment (40 mM NaCl) [30]. A search for crystallization conditions yielded a novel crystal form of coil 1B (Table 2). Molecular replacement by using a crystal structure of the dimer of coil 1B (PDB ID: 3SWK; [29]) as a search model did not yield a solution, indicating structural differences. However, molecular replacement with a tetramer of the larger construct (residues 144–251, PDB ID: 3UF1; [30]), where only residues 153–238 were used in the search model, yielded a solution with two tetramers of coil 1B in the asymmetric unit (Fig. 4). The crystal structure is characterized by strong translational noncrystallographic symmetry, with the Patterson peak height of 47 and the vector (0.0, 0.5, 0.0) in fractional coordinates, as indicated by the Xtriage analysis in the PHENIX package [35]. The refined 2.25 Å-resolution crystal structure reveals that the two tetramers of coil 1B in the asymmetric unit are laterally bound to each other in a staggered, antiparallel fashion (Figs. 4 and 5A). In the orthogonal direction perpendicular to the coiled coil axis, the crystal symmetry generates tetramers packed against each other in an antiparallel fashion without a stagger, i.e. in a periodic lattice of oppositely oriented dimers (Fig. 4). Along the coiled coil axis, the dimers are all packed head to tail. Overall, the lateral and longitudinal packing creates a filamentous lattice.
Figure 3.
A size-exclusion (HiPrep Sephacryl 26/60 S-200 HR column) chromatogram of purified vimentin coil 1B. The column was run in the gel filtration buffer, as specified in the Materials and Methods, at 2 mL/min. The 29.8 kDa mark on the x-axis serving as a calibration standard corresponds to the dimer of the DNA binding domain of FLI1 protein (with the dimer molecular weight of 29.8 kDa) [48].
Table 2.
X-ray diffraction data collection and refinement statistics for vimentin coil 1B.
| Data collection | |
| Space group | P21 |
| Unit cell dimensions | |
| a, b, c (Å) | 39.99, 77.27, 123.39 |
| α, β, γ (°) | 90, 95.03, 90 |
| Solvent content fraction | 0.47 |
| Resolution (Å) | 65.42–2.25 (2.32–2.25)a |
| <I/σ I> | 7.20 (3.50) |
| Completeness (%) | 89.8 (82.2) |
| Multiplicity | 3.0 (3.0) |
| Rmerge | 0.089 (0.305) |
| Rmeas | 0.124 (0.428) |
| CC1/2 | 0.994 (0.888) |
| Number of unique reflections | 32,022 |
| Structure refinement statistics | |
| Number of monomers per asymmetric unit | 8 |
| Resolution (Å) | 40.97–2.25 |
| Rwork (%) | 23.5 |
| Rfree (%) | 28.2 |
| R.m.s.d. from ideal | |
| bond lengths deviation (Å) | 0.007 |
| bond angles deviation (°) | 0.83 |
| Number of protein atoms | 5340 |
| Average B-factors (Å2) | |
| protein | 38 |
| water | 44 |
| glycerol | 51 |
| % of residues in | |
| favored regions | 99.25 |
| allowed regions | 0.75 |
| outlier regions | 0.00 (0 residues) |
Numbers in parentheses indicate the values in the highest-resolution shell.
Indicates MolProbity [47] statistics.
Figure 4.
High-order packing of vimentin coil 1B in filamentous crystals. The rectangle with the dashed line border denotes an octameric assembly of two staggered tetramers in an asymmetric unit.
Figure 5.
Assemblies of coils 1B and 2 in different crystal structures. A. The octameric assembly of coil 1B in the crystal structure reported in this study. B. A previously reported coiled-coil dimer of coil 1B (PDB ID: 3SWK). C. A previously reported tetramer of coil 1B (PDB ID: 3UF1). D. A tetramer of antiparallel coiled-coils of coil 2 ΔN (previously known as coil 2B; PDB ID: 1GK4). The PDB IDs of the respective structures are indicated under the images.
The dimerization and the tetramerization interfaces
In addition to the two prior crystal structures of the entire coil 1B mentioned above, there are two dimeric structures (PDB ID: 3S4R and 3SSU; [29]) that contain the N-terminal half of coil 1B (residues 153–189). This region of the parallel in-register coiled coil dimers in our structure is highly similar to the respective regions of these two structures (the Cα r.m.s.d. of 0.88 Å and 0.87 Å for 3S4R and 3SSU, respectively). The dimeric structure of the entire coil 1B (PDB ID: 3SWK) is somewhat conformationally different from our structure (the Cα r.m.s.d. of 3.21 Å). A salient difference is a helix-perturbing kink in the C-terminal portion of coil 1B in the former structure (residues 207–209; Fig. 5B). This kink is absent in our structure, in the structure of tetrameric coil 1B (PDB ID: 3UF1 [30]; Fig. 5C) and in the two structures of coil 1B lacking the N-terminal region needed for proper dimer formation (PDB ID: 4YV3 and 4YPC [28]). Therefore, this kink was likely a crystallization artifact. As expected, the conformation of the tetramers in our structure, where the in-register coiled coil dimers are bound to each other laterally and symmetrically in the antiparallel non-staggered fashion, closely resembles that of the prior tetramer structure (PDB ID: 3UF1; the Cα r.m.s.d. of 1.65 Å; Figs. 5A and 5C). To our surprise, in examining the previously reported crystal structures of vimentin regions (Table 1), we encountered a similar antiparallel non-staggered dimer-dimer assembly in the crystals of coil 2 lacking the N-terminal region, formerly known as coil 2B (PDB ID: 1GK4 [36]; Fig. 5D).
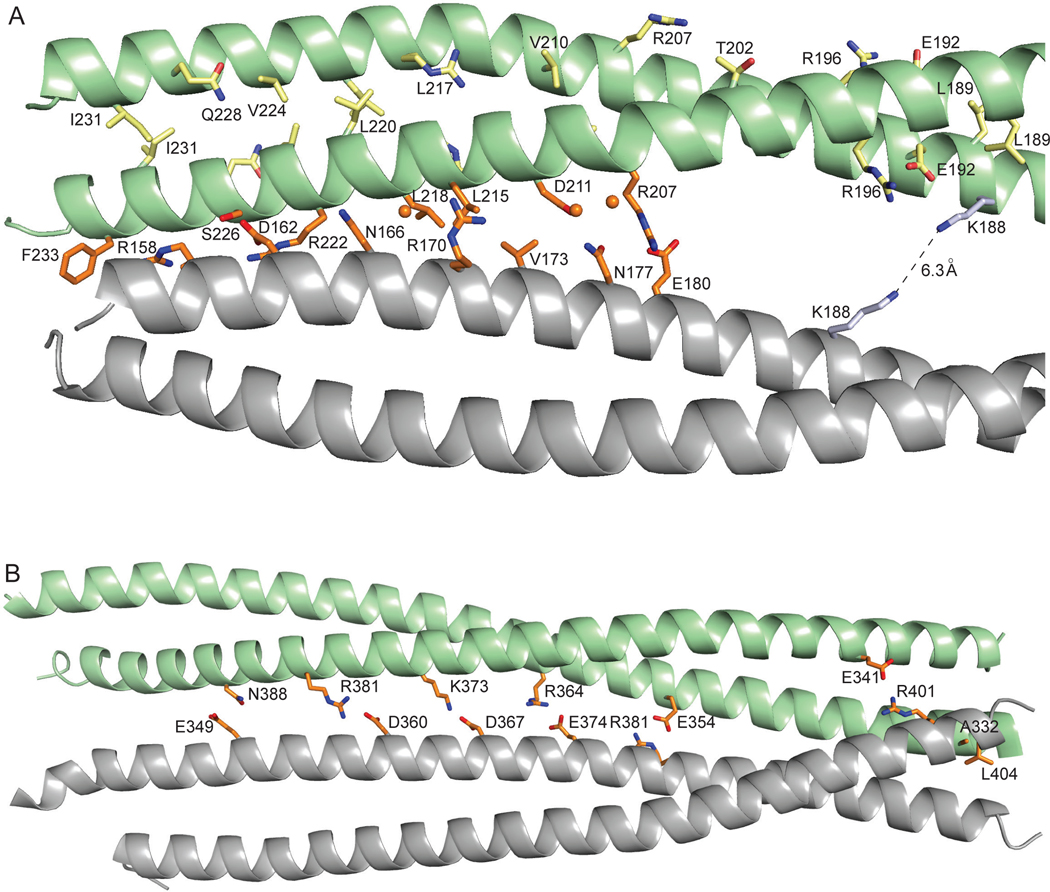
The conformational details of coil 1B in the dimeric and the tetrameric states have been described previously [29, 30]; here we focus on intermolecular interactions. The two parallel coils in a dimer are related by quasi-dyad symmetry. The dimerization interface is largely hydrophobic, composed of interacting hydrophobic residues in positions a and d of a periodic heptad motif that is repeated essentially continuously along the entire rod domain (Fig. 2, Fig. 6A). Frequently, residues at heptad position e and, occasionally, at position g, are also involved. This interface also includes aliphatic regions of charged residues contributing to the nonpolar interface. The charged side chain groups of these residues are almost invariably engaged in intramolecular salt bridges with other charged residues in the same helix. Four salt bridges across the dimerization interface were observed: Glu153-Arg158, Asp167-Lys168, Glu174-Arg175 and Glu192-Arg196.
Figure 6.
Dimerization and tetramerization interfaces of coil 1B and tetramerization interface of coil 2B. A. A zoomed-in view of the dimerization and tetramerization interfaces of coil 1B. The residues involved in the dimerization and tetramerization are shown as sticks in the same color scheme as in Fig. 2. Water molecules are shown as balls. Only a half of the approximately symmetrical interface is shown. B The tetramerization interface of coil 2B (PDB ID: 1GK4). Numerous water-mediated interactions are not shown to simplify the view.
The two coiled coil dimers in a tetramer are antiparallel and related by dyad symmetry, where the dyad axis is perpendicular to the coiled coil axis. The tetramerization interface is less extensive than the dimerization interface, involving the N-terminal one-third and the C-terminal one-third of coil 1B (Fig. 2). The residues in this interface are located at positions c and f of the heptad, with the occasional involvement of positions b, e and g that contain mostly hydrophilic and charged residues. The interactions between the dimers are a combination of hydrophobic contacts, hydrogen bonds (direct and water-mediated) and salt bridges (Fig. 6A). Our structure is consistent with the previously reported intermolecular crosslink between two Lys188 of different molecules of coil 1B, across the tetramerization interface [7]. The Nε- Nε distance of 6.3 Å for the Lys188 side chain rotamers facing each other (Fig. 6A) is consistent with the spacer arm length of 6.4 Å of the bridging bifunctional crosslinking agent disuccinimidyl tartrate used in that study.
We also examined interactions that are involved in the tetramerization of coil 2B (PDB ID: 1GK4 [36]; Fig. 6B). Similarly to the tetramerization interface of coil 1B, this interface predominantly polar, consisting of numerous direct and water-mediated hydrogen bonds. Remarkably, this interface contains several uniformly distributed salt bridges.
Interactions defining the high-order assembly of coil 1B
High-order assembly of IFs beyond the tetrameric state has not been structurally elucidated. In the direction orthogonal to tetramerization, the tetramers are packed against each other in the staggered antiparallel fashion (Fig. 7A). One of the two staggered dimer-dimer interfaces involves interactions between the N-terminal halves of two dimers (Fig. 7B), and the other, more extensive one, involves the C-terminal halves of the other two dimers (Fig. 7C). These high-order interfaces are individually less extensive than the tetramerization interface, due to the involvement of only a portion of a tetramer. The residues that are engaged in these staggered interactions are located in the N- and the C-terminal 15-residue regions of each coil 1B and in the central 20-residue region (Fig. 2). These residues are located at heptad positions b, c, e and f. The hydrophobic contacts between nonpolar residues and the aliphatic portions of polar and charged residues in these interfaces are dominated by hydrogen bonds (direct and water-mediated) involving side chains of polar and charged residues (Fig. 7B and 7C); several salt bridges are also present.
Figure 7.
Staggered packing of coil 1B and interactions involved in high-order assembly. A. A lateral view of the staggered antiparallel packing. B. and C. Zoomed-in views of the pairwise interactions of the coil 1B dimers shown in panel A. The residues involved in these interactions are shown as sticks and colored light blue as in Fig. 2.
Discussion
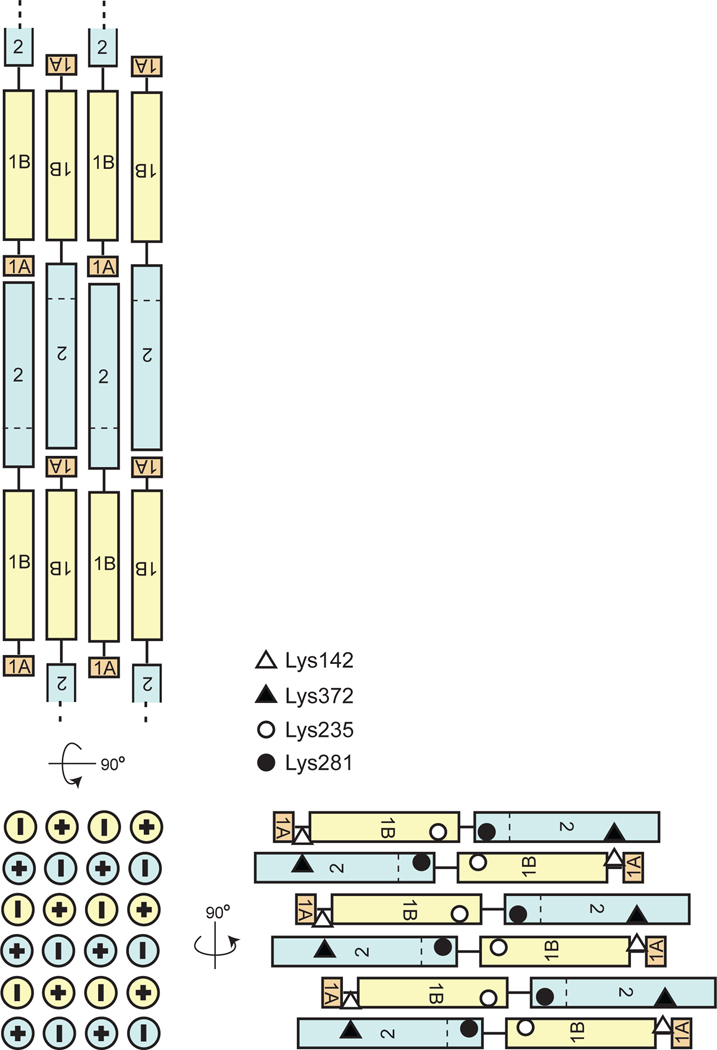
Although prior reports have described the dimeric parallel in-register assembly of individual IF rod domains, our understanding of the high-order assembly of dimers into tetramers and further assemblies into high-order biologically relevant filament structures remains is highly limited. Our crystal structure of vimentin coil 1B demonstrates the lateral packing of dimers of coil 1B in two orthogonal directions to form an infinite-order filament. In one direction, dimers interact in an antiparallel and strictly periodic fashion without a stagger. This mode of packing of coil 1B to itself (termed A11) in the context of a full-length filament was previously proposed based on an elegant crosslinking study [7] and found experimental corroboration in subsequent biochemical studies [37]. For the logical head-to-tail lengthwise assembly of the vimentin into a long filament, this A11 interaction mode naturally results in an antiparallel packing of the coil 2B region of coil 2 (Fig. 8, top view). This mode of coil 2 interactions, known as A22, was also proposed based on studies of vimentin assembly in solution [7, 8, 37, 38]. It is highly possible that the antiparallel dimer-dimer packing that we found in the previous crystal structure of coil 2B (PDB ID: 1GK4 [36]; Figs. 5C and 6B) represents this assembly interface. In the orthogonal direction, tetramers of coil 1B in our structure are packed in an antiparallel, staggered fashion (Fig. 7). These staggered interactions of coil 1B with itself may not be present in a full-length vimentin filament. However, we find that if we replace one of the the tetramers of coil 1B with that of coil 2B in this staggered tertamer-tetramer interface (e.g. in Fig. 7B), the resulting mixed 1B-2B staggered interface is consistent with previouly reported crosslinking of Lys142 to Lys372 and Lys235 to Lys281 [7] that has led to the so called A12 mode of the vimentin assembly (Fig. 8, bottom right view). In this model, Lys235 would interact with coil 2 in the vicinity of Lys281, instead of its interaction with coil 1B shown in Fig. 7C. We propose that, based on the logic of our crystal structure, the A12 mode is an assembly element in an orthogonal direction of the A11, A22 modes, and that all three modes can then be combined into a unified three-dimensional model of a high-order assembly of the entire vimentin rod domain (Fig. 8). Therefore, this model is a generalization of the two-dimensional filament models constructed solely from the crosslinking data [7]. To summarize, we propose that layers of lateral packing, as observed in our crystal and that of coil 2B, are formed by antiparallel, non-staggered continuous packing of coils 1B and 2B (Fig. 8, top view). These layers themselves are packed to each other so that staggered 1B-2B interactions arise (Fig. 8, bottom right view).
Figure 8.
A schematic of the proposed packing of the dimers of the rod domain in a vimentin filament. The panels show three orthogonal views. The opposite orientations of vimentin dimers are designated by symbols “+” and “-”. The horizontal dashed line indicates the N-terminal boundary of the region formerly called coil 2B. The diamonds, triangles and circles designate Lys residues that were previously observed to crosslink to each other, as indicated.
The extent of the intermolecular interactions of coil 1B decreases with its assembly order. Packing of vimentin monomers in a dimeric coiled coil engages residues of coil 1B along its entire length in a tight dimeric interface dominated by hydrophobic interactions. The tetramerization interface is not as extensive, involving only parts of the coils, with a larger relative contributioins of polar or charged interfaces. Finally, packing into higher-order structures is somewhat less extensive than packing of dimers to form a tetramer. The looseness of high-order packing is consistent with the observation of the loose packing of unit-length filaments [9] and further compaction in the late steps of IF formation, which was observed previously [9, 13–17]. This model is likely to be generally relevant for other IFs, due to the conserved domain organization in IFs of various types.
In conclusion, the crystal structure of coil 1B of human vimentin described here, together with previous structural and biochemical studies led us to propose how coiled coil dimers may assemble into a high-order filamentous structure. This model generalizes and refines the previously proposed IF filament models. Future crystallographic and crosslinking experiments will be performed to test the proposed high-order assembly.
Materials and Methods
Cloning, protein expression and purification.
The coding region for vimentin coil 1B (residues 153–238) was amplified by polymerase chain reaction (PCR) from a human cDNA clone (clone ID: 2985712; Dharmacon, Lafayette, CO) with the forward primer 5’-ATCAACCACATATGGAGATGCGGGAGCTGCGCCG-3’ and the reverse primer 5’- ATCGGATCCTCAGTGGAGTTTCTTCAAAAAGG-3’. This region was inserted between NdeI and BamHI sites of a modified pET19b vector [39] encoding an N-terminal decahistidine tag cleavable by PreScission protease (GE Healthcare, Piscataway, NJ, USA). The resulting construct was verified by DNA sequencing (Eurofins, New Orleans, LA, USA) and transformed into E. coli expression strain BL21 (DE3) for protein expression. The bacterial culture was incubated with shaking in Luria-Bertani medium supplemented with 100 μg/mL ampicillin at 15 ºC to an attenuance at 600 nm of 0.6. Protein overexpression was then induced by adding IPTG to the final concentration of 0.4 mM. The culture was incubated for additional 16 hours and harvested by centrifugation for 10 min at 5000 g at 4 ºC. The cell pellet from a 2 L culture was resuspended in 40 mL of buffer A [40 mM Tris–HCl pH 8.0, 400 mM NaCl, 10% of glycerol, 2 mM β-mercaptoethanol]. The cells were disrupted by sonication on ice and clarified by centrifugation at 38000 g for 40 min. The supernatant was filtered through a 0.45 μm Millex-HV PVDF filter (Millipore, Billerica, MA, USA) and applied to a 5 mL Ni2+-IMAC HisTrap FF column (GE Healthcare) equilibrated with buffer A. The column was washed with 100 mL of buffer A containing 50 mM imidazole, and then protein was eluted with 10 mL of buffer A containing 500 mM imidazole. The eluate was dialyzed into PreScission protease cleavage buffer (20 mM Tris–HCl pH 7.0, 150 mM NaCl, 1 mM DTT). After dialysis, vimentin coil 1B was digested overnight at 4 ºC with PreScission protease to cleave the tag, leaving only a GPHM sequence N-terminal to residue 153. Complete digestion was verified on a Coomassie-stained SDS–PAGE gel. Untagged vimentin coil 1B was loaded onto a size-exclusion S-200 HR Sephacryl column (GE Healthcare) equilibrated with gel filtration buffer (10 mM Tris-HCl pH 8.0 and 40 mM NaCl). Peak fractions were checked on SDS–PAGE for purity. Fractions containing pure protein were pooled and concentrated in an Amicon Ultra-15 centrifugal filter unit (Millipore) with a 5 kDa molecular weight cutoff to a final concentration of 10 mg/mL.
Crystallization, data collection and crystal structure determination of vimentin coil 1B.
Initial screening was performed at 22 ºC by the hanging-drop vapor diffusion method using a 48 condition in-house screen. Crystals were grown in 2 μL drops made by mixing 1 μL of the reservoir solution (0.1 M Bis-Tris pH 6.5, 0.2 M magnesium acetate tetrahydrate and 10% PEG 8000) with 1 μL of the concentrated protein and equilibrated against 1 mL of the reservoir solution. Initial crystals were grown with the protein in the presence of 2 mM arylquin, a vimentin binding small molecule [26, 40], but arylquin was abandoned during optimization of crystal growth. Prior to freezing, the crystals were gradually transferred to a cryoprotectant solution, which had the same composition as the crystallization condition with 20% glycerol. X-ray diffraction data were collected at beamline 22-ID of the Advanced Photon Source of the Argonne National Laboratory (Argonne, IL) at 100 K and indexed and integrated with program MOSFLM [41] and scaled with SCALA [42]. Majority of crystals were highly mosaic and all crystals were highly sensitive to radiation damage, ultimately affecting data completeness. Merging data sets collected on multiple crystals did not improve statistics, likely due to the lack of isomorphism or varying degree of radiation damage, or both. The structure of vimentin coil 1B was determined by molecular replacement using MOLREP [43] with a tetramer of coil 1B (PDB ID: 3UF1; residues 144–251 [30]), where in preparation of the search model, the residues not present in our construct were deleted. Initial jelly-body refinement (100 cycles) was carried out using REFMAC5 [44] from the CCP4 program suite [45]. Further refinement was performed using PHENIX refinement [35] (against the maximal likelihood target) iteratively with model building in COOT [46]. The data collection and refinement statistics are given in Table 2. The structure coordinates and structure factor amplitudes were deposited in the Worldwide Protein Data Bank with accession code 5WHF.
Acknowledgements
We thank Dr. David Watt for critical reading of the manuscript and discussions, the staff of sector SER-CAT of the Advanced Photon Source at the Argonne National Laboratory for assistance with remote data collection and Dr. Pavel Afonine for advice on translational noncrystallographic symmetry. This work was supported, in part, by NIH R01 CA60872 (to Dr. Vivek Rangnekar) and by the Center for Pharmaceutical Research and Innovation at the College of Pharmacy, University of Kentucky.
ABBREVIATIONS:
- IF
intermediate filament
- ULF
unit-length filament
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- DTT
1,4-dithiothreitol
Footnotes
Database: structural data are available in the PDB under the accession number 5WHF.
References
- 1.Herrmann H, Bar H, Kreplak L, Strelkov SV & Aebi U (2007) Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol 8, 562–573. [DOI] [PubMed] [Google Scholar]
- 2.Steinert PM & Roop DR (1988) Molecular and cellular biology of intermediate filaments. Annu Rev Biochem 57, 593–625. [DOI] [PubMed] [Google Scholar]
- 3.Albers K & Fuchs E (1992) The molecular biology of intermediate filament proteins. Int Rev Cytol 134, 243–279. [DOI] [PubMed] [Google Scholar]
- 4.Lendahl U, Zimmerman LB & McKay RD (1990) CNS stem cells express a new class of intermediate filament protein. Cell 60, 585–595. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann H, Haner M, Brettel M, Muller SA, Goldie KN, Fedtke B, Lustig A, Franke WW & Aebi U (1996) Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol 264, 933–953. [DOI] [PubMed] [Google Scholar]
- 6.Steinert PM, Marekov LN, Fraser RD & Parry DA (1993) Keratin intermediate filament structure. Crosslinking studies yield quantitative information on molecular dimensions and mechanism of assembly. J Mol Biol 230, 436–452. [DOI] [PubMed] [Google Scholar]
- 7.Steinert PM, Marekov LN & Parry DA (1993) Diversity of intermediate filament structure. Evidence that the alignment of coiled-coil molecules in vimentin is different from that in keratin intermediate filaments. J Biol Chem 268, 24916–24925. [PubMed] [Google Scholar]
- 8.Hess JF, Budamagunta MS, Voss JC & FitzGerald PG (2004) Structural characterization of human vimentin rod 1 and the sequencing of assembly steps in intermediate filament formation in vitro using site-directed spin labeling and electron paramagnetic resonance. J Biol Chem 279, 44841–44846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokolova AV, Kreplak L, Wedig T, Mucke N, Svergun DI, Herrmann H, Aebi U & Strelkov SV (2006) Monitoring intermediate filament assembly by small-angle x-ray scattering reveals the molecular architecture of assembly intermediates. Proc Natl Acad Sci U S A 103, 16206–16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann I, Herrmann H & Franke WW (1991) Assembly and structure of calcium-induced thick vimentin filaments. Eur J Cell Biol 56, 328–341. [PubMed] [Google Scholar]
- 11.Herrmann H, Hofmann I & Franke WW (1992) Identification of a nonapeptide motif in the vimentin head domain involved in intermediate filament assembly. J Mol Biol 223, 637–650. [DOI] [PubMed] [Google Scholar]
- 12.Kirmse R, Portet S, Mucke N, Aebi U, Herrmann H & Langowski J (2007) A quantitative kinetic model for the in vitro assembly of intermediate filaments from tetrameric vimentin. J Biol Chem 282, 18563–18572. [DOI] [PubMed] [Google Scholar]
- 13.Georgakopoulou S, Moller D, Sachs N, Herrmann H & Aebi U (2009) Near-UV circular dichroism reveals structural transitions of vimentin subunits during intermediate filament assembly. J Mol Biol 386, 544–553. [DOI] [PubMed] [Google Scholar]
- 14.Wickert U, Mucke N, Wedig T, Muller SA, Aebi U & Herrmann H (2005) Characterization of the in vitro co-assembly process of the intermediate filament proteins vimentin and desmin: mixed polymers at all stages of assembly. Eur J Cell Biol 84, 379–391. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann H, Haner M, Brettel M, Ku NO & Aebi U (1999) Characterization of distinct early assembly units of different intermediate filament proteins. J Mol Biol 286, 1403–1420. [DOI] [PubMed] [Google Scholar]
- 16.Stromer MH, Ritter MA, Pang YY & Robson RM (1987) Effect of cations and temperature on kinetics of desmin assembly. Biochem J 246, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winheim S, Hieb AR, Silbermann M, Surmann EM, Wedig T, Herrmann H, Langowski J & Mucke N (2011) Deconstructing the late phase of vimentin assembly by total internal reflection fluorescence microscopy (TIRFM). PLoS One 6, e19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckes B, Colucci-Guyon E, Smola H, Nodder S, Babinet C, Krieg T & Martin P (2000) Impaired wound healing in embryonic and adult mice lacking vimentin. J Cell Sci 113, 2455–2462. [DOI] [PubMed] [Google Scholar]
- 19.Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, et al. (2005) Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst 97, 1124–1132. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Yan Q, Long X, Chen X & Wang Y (2008) Vimentin affects the mobility and invasiveness of prostate cancer cells. Cell Biochem Funct 26, 571–577. [DOI] [PubMed] [Google Scholar]
- 21.Moinova H, Leidner RS, Ravi L, Lutterbaugh J, Barnholtz-Sloan JS, Chen Y, Chak A, Markowitz SD & Willis JE (2012) Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer Epidemiol Biomarkers Prev 21, 594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh S, Sadacharan S, Su S, Belldegrun A, Persad S & Singh G (2003) Overexpression of vimentin: role in the invasive phenotype in an androgen-independent model of prostate cancer. Cancer Res 63, 2306–2311. [PubMed] [Google Scholar]
- 23.Hu L, Lau SH, Tzang CH, Wen JM, Wang W, Xie D, Huang M, Wang Y, Wu MC, Huang JF, et al. (2004) Association of vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene 23, 298–302. [DOI] [PubMed] [Google Scholar]
- 24.Shirahata A, Sakata M, Sakuraba K, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y, et al. (2009) Vimentin methylation as a marker for advanced colorectal carcinoma. Anticancer Res 29, 279–281. [PubMed] [Google Scholar]
- 25.Burikhanov R, Sviripa VM, Hebbar N, Zhang W, Layton WJ, Hamza A, Zhan CG, Watt DS, Liu C & Rangnekar VM (2014) Arylquins target vimentin to trigger Par-4 secretion for tumor cell apoptosis. Nat Chem Biol 10, 924–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sviripa VM, Burikhanov R, Obiero JM, Yuan Y, Nickell JR, Dwoskin LP, Zhan CG, Liu C, Tsodikov OV, Rangnekar VM & Watt DS (2016) Par-4 secretion: stoichiometry of 3-arylquinoline binding to vimentin. Org Biomol Chem 14, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suman S, Das TP, Moselhy J, Pal D, Kolluru V, Alatassi H, Ankem MK & Damodaran C (2016) Oral administration of withaferin A inhibits carcinogenesis of prostate in TRAMP model. Oncotarget 7, 53751–53761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chernyatina AA, Hess JF, Guzenko D, Voss JC & Strelkov SV (2016) How to study intermediate filaments in atomic detail. Methods Enzymol 568, 3–33. [DOI] [PubMed] [Google Scholar]
- 29.Chernyatina AA, Nicolet S, Aebi U, Herrmann H & Strelkov SV (2012) Atomic structure of the vimentin central alpha-helical domain and its implications for intermediate filament assembly. Proc Natl Acad Sci U S A 109, 13620–13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aziz A, Hess JF, Budamagunta MS, Voss JC, Kuzin AP, Huang YJ, Xiao R, Montelione GT, FitzGerald PG & Hunt JF (2012) The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography. J Biol Chem 287, 28349–28361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chernyatina AA & Strelkov SV (2012) Stabilization of vimentin coil2 fragment via an engineered disulfide. J Struct Biol 177, 46–53. [DOI] [PubMed] [Google Scholar]
- 32.Nicolet S, Herrmann H, Aebi U & Strelkov SV (2010) Atomic structure of vimentin coil 2. J Struct Biol 170, 369–376. [DOI] [PubMed] [Google Scholar]
- 33.Meier M, Padilla GP, Herrmann H, Wedig T, Hergt M, Patel TR, Stetefeld J, Aebi U & Burkhard P (2009) Vimentin coil 1A-A molecular switch involved in the initiation of filament elongation. J Mol Biol 390, 245–261. [DOI] [PubMed] [Google Scholar]
- 34.Chernyatina AA, Guzenko D & Strelkov SV (2015) Intermediate filament structure: the bottom-up approach. Curr Opin Cell Biol 32, 65–72. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U & Burkhard P (2002) Conserved segments 1A and 2B of the intermediate filament dimer: their atomic structures and role in filament assembly. EMBO J 21, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mucke N, Wedig T, Burer A, Marekov LN, Steinert PM, Langowski J, Aebi U & Herrmann H (2004) Molecular and biophysical characterization of assembly-starter units of human vimentin. J Mol Biol 340, 97–114. [DOI] [PubMed] [Google Scholar]
- 38.Hess JF, Voss JC & FitzGerald PG (2002) Real-time observation of coiled-coil domains and subunit assembly in intermediate filaments. J Biol Chem 277, 35516–35522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biswas T & Tsodikov OV (2008) Hexameric ring structure of the N-terminal domain of Mycobacterium tuberculosis DnaB helicase. FEBS J 275, 3064–3071. [DOI] [PubMed] [Google Scholar]
- 40.Burikhanov R, Sviripa VM, Hebbar N, Zhang W, Layton WJ, Hamza A, Zhan CG, Watt DS, Liu C & Rangnekar VM (2014) Arylquins target vimentin to trigger Par-4 secretion for tumor cell apoptosis. Nat Chem Biol 10, 924–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leslie AG (2006) The integration of macromolecular diffraction data. Acta Cryst D 62, 48–57. [DOI] [PubMed] [Google Scholar]
- 42.Evans P (2006) Scaling and assessment of data quality. Acta Cryst D 62, 72–82. [DOI] [PubMed] [Google Scholar]
- 43.Vagin A & Teplyakov A (2010) Molecular replacement with MOLREP. Acta Cryst D 66, 22–25. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F & Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Cryst D 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011) Overview of the CCP4 suite and current developments. Acta Cryst D 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P & Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Cryst D 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 47.Chen VB, Arendall WB 3rd, Headd JJ, DA Keedy, Immormino RM, Kapral GJ, Murray LW, Richardson JS & Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst D 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou C & Tsodikov OV (2015) Structural basis for dimerization and DNA binding of transcription factor FLI1. Biochemistry 54, 7365–7374. [DOI] [PubMed] [Google Scholar]