Abstract
Background
Machine learning (ML) is increasingly used in research for subtype definition and risk prediction, particularly in cardiovascular diseases. No existing ML models are routinely used for cardiovascular disease management, and their phase of clinical utility is unknown, partly due to a lack of clear criteria. We evaluated ML for subtype definition and risk prediction in heart failure (HF), acute coronary syndromes (ACS) and atrial fibrillation (AF).
Methods
For ML studies of subtype definition and risk prediction, we conducted a systematic review in HF, ACS and AF, using PubMed, MEDLINE and Web of Science from January 2000 until December 2019. By adapting published criteria for diagnostic and prognostic studies, we developed a seven-domain, ML-specific checklist.
Results
Of 5918 studies identified, 97 were included. Across studies for subtype definition (n = 40) and risk prediction (n = 57), there was variation in data source, population size (median 606 and median 6769), clinical setting (outpatient, inpatient, different departments), number of covariates (median 19 and median 48) and ML methods. All studies were single disease, most were North American (n = 61/97) and only 14 studies combined definition and risk prediction. Subtype definition and risk prediction studies respectively had limitations in development (e.g. 15.0% and 78.9% of studies related to patient benefit; 15.0% and 15.8% had low patient selection bias), validation (12.5% and 5.3% externally validated) and impact (32.5% and 91.2% improved outcome prediction; no effectiveness or cost-effectiveness evaluations).
Conclusions
Studies of ML in HF, ACS and AF are limited by number and type of included covariates, ML methods, population size, country, clinical setting and focus on single diseases, not overlap or multimorbidity. Clinical utility and implementation rely on improvements in development, validation and impact, facilitated by simple checklists. We provide clear steps prior to safe implementation of machine learning in clinical practice for cardiovascular diseases and other disease areas.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-021-01940-7.
Keywords: Cardiovascular disease, Machine learning, Subtype, Risk prediction, Informatics, Systematic review
Background
Disease definitions rely on expert consensus, informed by best available evidence [1–3]. Machine learning (ML), the use of algorithms to describe patterns in datasets with (supervised) or without (unsupervised) the need to define them a priori, is increasingly used in research for definition [4] and risk prediction [5]. However, established evaluation frameworks for clinical utility of such models [6, 7] have not been applied.
Better definitions for cardiovascular disease (CVD), the single greatest disease burden in the UK and globally [6], may improve prevention and treatment by characterising target populations, enabling comparability and generalisability across study designs and populations. Heart failure (HF), acute coronary syndromes (ACS) and atrial fibrillation (AF) are among the commonest CVDs globally [8, 9]. Despite frequent changes in disease definitions [3, 10, 11], continued diagnostic and prognostic uncertainties have led to ML research in all three diseases [12–17]. HF, ACS and AF frequently overlap, so phenotyping and risk prediction are relevant across diseases. To date, no existing ML models are routinely used for CVD management, and their phase of clinical utility is unknown, partly due to a lack of clear criteria.
Unlike drugs and devices [18, 19], the pathway to translation from research to routine use is unclear for ML. The AI-TREE criteria (Additional file_Web table 1) were developed for ML healthcare research [20], but consensus is lacking for best practice or regulatory requirements for implementation [21, 22]. Importance of such guidelines is emphasised by systematic reviews showing that ML performs no better than logistic regression or healthcare professionals in prediction across diseases, with high risk of bias and limited external validation in published studies [23, 24].
Evidence for ML in subtype definition and risk prediction in HF, ACS and AF has not been assimilated. Moreover, it is unclear how application of ML in CVD compares with other diseases. Our aims were to (i) develop a simple framework for clinical utility and validity of ML models in healthcare and (ii) conduct a systematic review to evaluate methods and results of ML for subtype definition or risk prediction in HF, AF and ACS.
Methods
We followed the Preferred Reporting Items for Systematic reviews and Meta-Analysis (PRISMA) statement, with a protocol agreed by all authors, without prospectively registering.
Systematic review
Identification of studies
We searched PubMed, MEDLINE and Web of Science databases from 1 January 2000 until 31 December 2019. Our search terms, agreed by co-authors, pertained to machine learning, clustering, cardiovascular disease, heart failure, atrial fibrillation, acute coronary syndromes, subtype and risk prediction. Reference lists and expert recommendations were taken into account, to identify grey literature, including conference reports and proceedings, guidelines, working papers and theses (Additional file_Search strategy).
Selection of studies
All abstracts were independently screened and then full text of selected abstracts were respectively assessed for eligibility by two reviewers (from AB, SC and MZ), and conflicts were resolved by a third reviewer (AB or SC).
Inclusion and exclusion criteria
Studies were eligible if the publication presented:
-
(i)
ML models for disease subtype definition/clustering for HF, ACS and AF; or
-
(ii)
Risk prediction for HF, ACS or AF
Studies were excluded if they:
-
(i)
Were not original empirical data
-
(ii)
Were not English language
-
(iii)
Were not peer-reviewed (e.g. a published dissertation was not eligible)
-
(iv)
Did not concern models developed for humans
-
(v)
Did not have full text available
Data extraction
Tools for extraction were adapted and developed with consensus among co-authors from published frameworks for new risk markers (AHA [6]), diagnostic accuracy (QUADAS-2 [25]), prognostic tools (CHARMS [26], PROGRESS [7], TRIPOD [27]) and ML (AI-TREE [20], Christodoulou [23]) (Additional file_Web table 2). The author, year and country of study, clinical setting, data source, outcome, comparator methods, ML method(s) and covariates were extracted. The final checklist was in the three stages of the translational pathway (“development”, “validation” and “impact”) as per AHA and PROGRESS statements [6, 7]. There were seven domains under the three main stages: clinical relevance, patients, algorithms (“development”), internal validation, external validation (“validation”), and clinical utility and effectiveness (“impact”). The questions for each domain were from published guidelines as above with similar extraction items for subtype definition and risk prediction studies. Data extraction was by two independent reviewers (from AB, MZ and SC), and disagreements were resolved by a third reviewer (AB or SC). Quantitative analysis was beyond the scope of this review and its aims.
Results
Systematic review of HF, AF and ACS
Of 5918 articles identified by our search, 97 met the inclusion criteria (Additional file_Figure S1).
Unsupervised ML for subtype definition
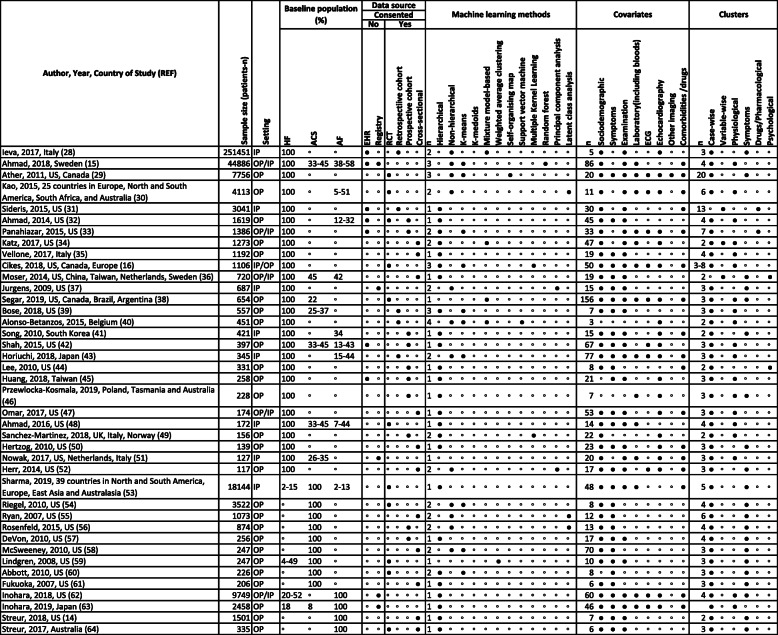
Of the 40 studies of unsupervised ML for subtype definition (included patients: median n = 606; min 117, max 251,451), there were 27 in HF, 9 in ACS and 4 in AF. All studies focused on a single disease, and only 6 (15%) studies included data regarding history of all three diseases. Twenty-three (57.5%) studies involved < 1000 individuals (range 117–874) and 29 (72.5%) were based in North America with no analyses from low- or middle-income countries. Across diseases, 26 (65%) studies were in outpatients (8 inpatient and 6 mixed inpatient and outpatient) and 11 used trial data (10 prospective cohort, 5 retrospective cohort, 11 cross-sectional, 5 registries), with 7 using EHR data. The mean number of covariates was 31 (min 3, max 156), most commonly demographic and symptom variables (Table 1) [28–67].
Table 1.
Systematic review of machine learning studies of subtype definition in heart failure, acute coronary syndromes and atrial fibrillation ( n = 40 studies)
ACS-acute coronary syndrome; AF-atrial fibrillation; CVD-cardiovascular disease; ECG- electrocardiogram; ED-emergency department; EHR-electronic health records; HF-heart failure; IP-hospital inpatient; LV-left ventricular; MI-myocardial infarction; OP- hospital outpatient; RCT- randomised controlled trial; United Kingdom; US-United States.
◦ Negative/No for all columns (except in "Baseline population" column, where it denotes "Unreported")
● Positive/Yes
Most studies used only one ML method (n = 22). Non-hierarchical clustering was used in 12 studies (hierarchical: n = 25). The most commonly used method was K-means (n = 11), but other methods included Gaussian mixture modelling, latent class analysis and random forest (RF). All studies reported disease clusters. Most studies found 3 clusters (n = 17, 42.5%; median 3), and most clusters were based on symptom (n = 20) or physical (n = 17) variables. Clustering was usually case-wise (n = 37), rather than variable-wise (n = 3) and one study used both approaches.
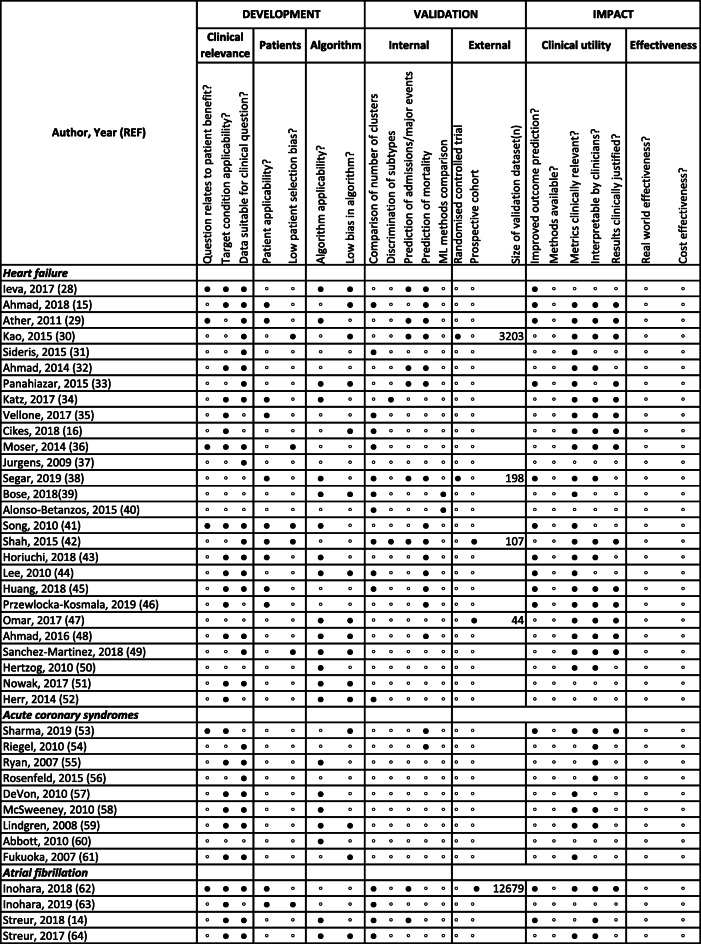
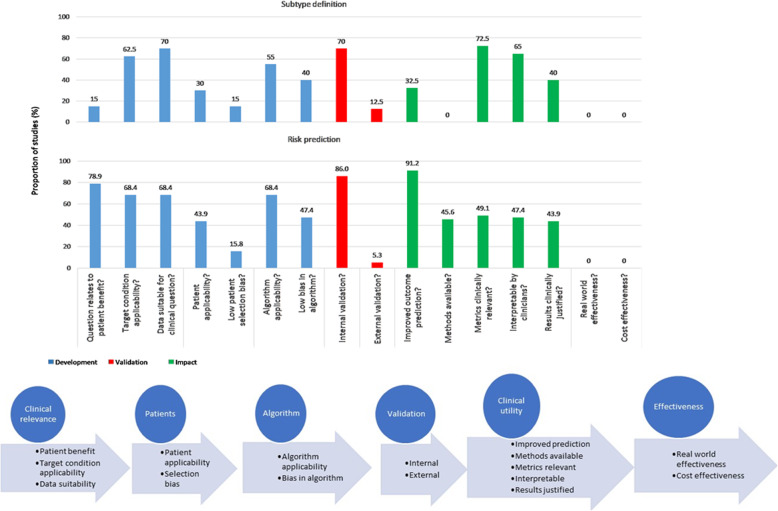
At the development stage, there were limitations at “clinical relevance” (question related to patient benefit 15.0%, target condition applicability 62.5% and data suitable for clinical question 70%), “patient” (patient applicability 30.0%, low patient selection bias 15.0%) and “algorithm” (algorithm applicability 55.0%, low algorithm bias 40.0%) levels. Twelve studies did not validate or replicate findings; 28 (70.0%) had internal validation (most commonly using number of clusters or prediction of mortality/admissions) and only 5 (12.5%) externally validated findings. There were significant deficiencies under “clinical utility” (improved prediction of outcomes 32.5%, methods available 0%, clinically relevant metrics 72.5%, interpretable by clinicians 65%, clinically justified results 40%) and “effectiveness” (no studies showing effectiveness, real-world or cost) domains (Table 2) [28–67].
Table 2.
Quality assessment of machine learning studies of subtype definition for heart failure, acute coronary syndromes and atrial fibrillation ( n = 40)
◦ Negative/No
● Positive/Yes
Supervised ML for risk prediction
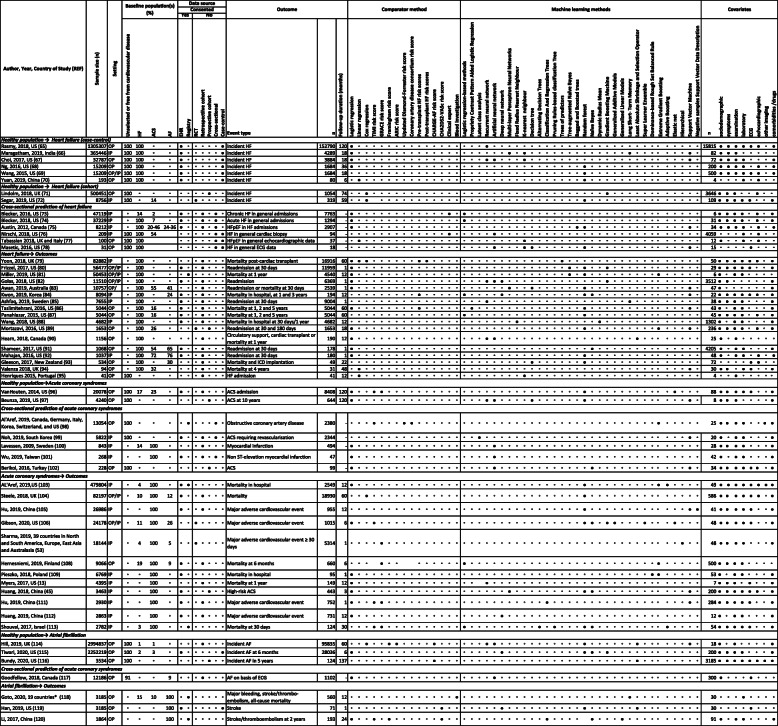
Of the 57 studies of supervised ML for risk prediction (median n = 6769; min 28, max 2,994,837), 31 were in HF, 19 in ACS and 7 in AF. Ten of 57 studies involved < 1000 individuals and most were from North America (n = 32), with one from a low- or middle-income country (n = 1). Risk prediction studies focused on development of (i) HF, ACS or AF in healthy individuals or the general population by case-control, cross-sectional or cohort design (n = 25) and (ii) outcomes in HF, ACS or AF (n = 32) (Table 3) [13, 45, 53, 65–118].
Table 3.
Machine learning risk prediction studies in heart failure, acute coronary syndromes and atrial fibrillation (n = 57)
ACS, acute coronary syndrome; AF, atrial fibrillation; Atherosclerosis Risk in Communities Study; CHARGE Cohorts for Heart and Aging Research in Genomic Epidemiology; CHA2DS2-VASc congestive heart failure, hypertension, age > 75, diabetes mellitus, stroke, vascular disease, sex category; CVD, cardiovascular disease; ECG, electrocardiogram; EHR, electronic health records; GRACE, Global Registry of Acute Coronary Events; HF, heart failure; HFpEF, heart failure with preserved ejection fraction; IP, hospital inpatient; LV, left ventricular; OP, hospital outpatient; RCT, randomised controlled trial; TIMI, thrombolysis in myocardial infarction; UK, United Kingdom; US, United States
*Australia, Austria, Brazil, Canada, China, Denmark, Korea, Finland, France, Germany, Italy, Japan, Mexico, Norway, Poland, Spain, Sweden, Netherlands, and UK
◦ Negative/no for all columns (except in the “Baseline population” column, where it denotes “unreported”)
● Positive/yes
Across diseases, 27 studies were in outpatients (23 inpatient and 7 mixed inpatient and outpatient) and 5 used trial data (6 prospective cohort, 30 retrospective cohort, 6 registry and 11 case-control). Thirty-one studies used EHR data. The mean number of covariates was 723 (median 48; min 6, max 15,815), most commonly demography, symptoms and comorbidities/drugs. The ML methods used were variable with neural networks (n = 19), random forest (n = 23) and support vector machine (n = 16).
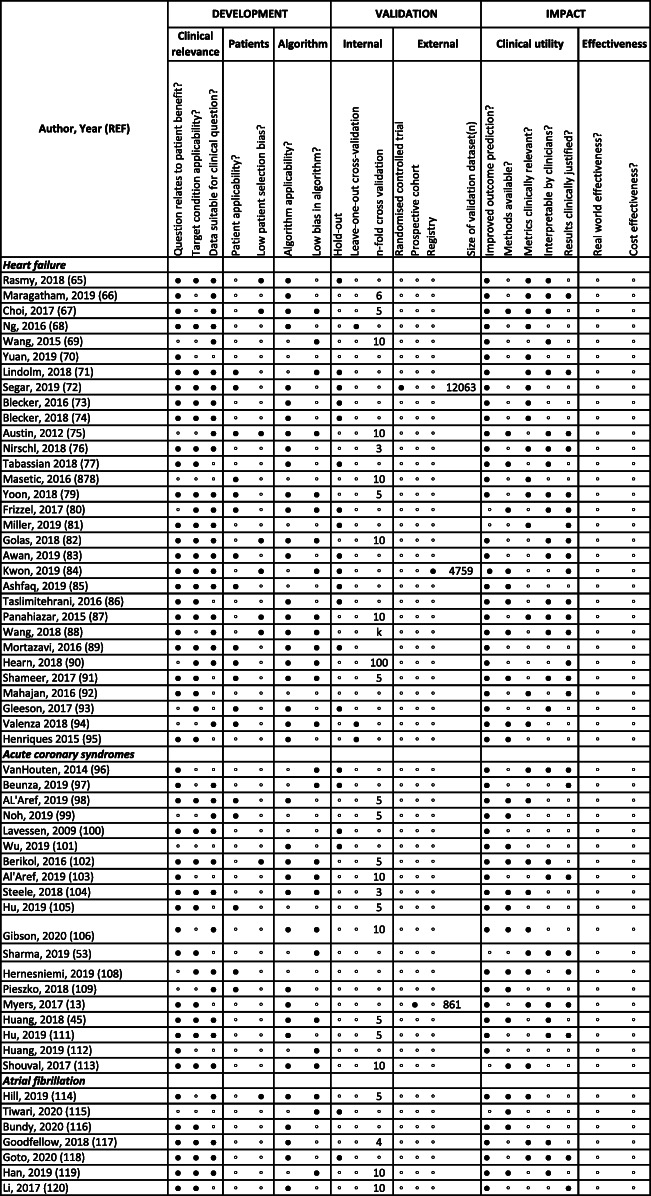
At the development stage, there were concerns at “clinical relevance” (question related to patient benefit 78.9%, target condition applicability 68.4% and data suitable for clinical question 68.4%), “patient” (patient applicability 43.9%, low patient selection bias 15.8%) and “algorithm” (algorithm applicability 68.4%, low algorithm bias 47.4%) levels. Only three studies (5.3%) had external validation in the clinical trial (n = 12,063), prospective cohort (n = 861) and registry (n = 4759) data. There was internal validation in 49/57 (86.0%) by “hold-out”, “leave one out” or k-fold cross-validation. The risk of bias was high in all studies with the commonest causes as patient selection (n = 47), patient applicability to the clinical question (n = 35) and bias in the algorithm(s) (n = 31). Again, there were major limitations under “clinical utility” (improved prediction of outcomes 91.2%, methods available 45.6%, clinically relevant metrics 49.1%, interpretable by clinicians 47.4%, clinically justified results 43.9%) and “effectiveness” (no studies showing effectiveness, real-world or cost) domains (Table 4) [13, 45, 53, 65–118].
Table 4.
Quality assessment of machine learning studies of risk prediction for heart failure, acute coronary syndromes and atrial fibrillation (n = 57)
◦ Negative/No
● Positive/Yes
Discussion
Our study highlighted three key findings with important impact on the use of ML in subtype definition and risk prediction for HF, ACS and AF. First, there is significant variation in methods, data and reporting of results. Second, we provide a pragmatic framework, based on published criteria, for assessing validity and clinical utility of ML studies. Third, there are major limitations at development, validation and impact stages in studies of ML in CVD, and our scoping review (Additional file_Scoping review_Methods and Results, Additional file_Web table 3 [119–129], Additional file_Web table 4 [130–143]) [144, 145] suggests similar problems for non-CVDs. Addressing these issues has potential to improve future data science approaches and enhance value of these methods for clinical decision-making and better patient care.
There is increasing interest in guidelines for development and implementation of ML in healthcare from patients and public, academics and industry, partly due to current uncertainty regarding necessary approval and regulatory procedures [146, 147]. Whether in medicines or medical devices, the “development pipeline” (e.g. phases I to IV of drug development) and guidelines are much clearer, ultimately promoting clinical effectiveness and patient safety. The wide variations in methods, datasets and reporting are not surprising, given the current lack of such guidelines with respect to the use of ML in subtype definition and risk prediction [147]. On the basis of existing checklists, we have developed a straightforward 7-domain checklist (16 points) to capture development, validation and impact. Although there are multiple efforts to standardise reporting guidelines for AI in healthcare [27, 148], the relationship with the “translational pathway” in terms of development, validation and impact has not been emphasised. We show that no published studies have results which are ready to be implemented, again likely to be related to lack of consensus regarding what is required in research and clinical practice to ensure clinical effectiveness and safety of ML.
Problems at the development stage
Studies to date of ML in HF, ACS and AF have been limited by number and type of covariates, population size, geographic location (mainly North American) and clinical setting. As with epidemiologic studies and trials, the generalisability of data and reproducibility of methods [6, 7] are crucial to make findings interpretable and translatable to clinical care. However, the majority of studies to date have not fully considered these factors, resulting in high risk of bias in all studies.
EHR data and advanced data analytics, which have been either under-used or under-reported in studies to date, offer research opportunities across diseases, but ML studies have focused on single diseases, when diseases often co-exist, as for HF, ACS and AF. Outside CVD, perhaps the most promising ML studies to date have been in either large imaging datasets or settings where there is linkage of multimodal clinical data. Whereas the popular concept of ML suggests the use of a wide range of covariates, particularly in clustering, the number and type of variables have been limited. Availability of covariates and quality of recording in EHR determine these limitations rather than research or clinical need. In the future, as more complete and larger EHR datasets become available for research, studies of ML can focus on improved use of limited data in clinical practice, or use of more comprehensive lists of covariates and coexisting diseases for the discovery of new factors in disease definition or prediction. For example, the ESC classification lists 89 causes of HF, which have not been studied together in a single population-based study in EHR or otherwise [3].
The majority of studies have had positive results suggesting publication bias (not formally assessed here), which likely overestimates potential healthcare impact. Lessons must be learned from biomarkers and genomics [149, 150], where lack of standardisation and biased reporting have contributed to lack of translation from science to practice. It is concerning that a significant proportion of studies ML are being developed with questions unrelated to patient benefit, or with data unsuitable to answer them. “Data-driven care” is important in both personalised and precision medicine [151], both of which can benefit from advances in the use of ML. However, a “data-centred” or “data-driven” agenda must remain “patient-centred” rather than “technology-centred” (or in this case, “ML-centred”) [152]. Our framework can be used to plan studies, facilitating clinical relevance and reducing bias.
Problems at the validation stage
In order to improve data-driven characterisation of CVD and influence clinical decision-making, ML studies for subtyping and prediction should be larger-scale, across diseases, with standardised reporting and proven external validity. To date, the degree of external validation across subtype definition and risk prediction ML studies has been disappointing, making ML difficult to implement in routine care. External validation will help to understand which clustering and prediction tools are of greatest use, and greater availability of electronic health record data should facilitate this step in the pathway to implementation of ML.
Problems at the impact stage
There are major gaps at the phases of clinical utility, particularly open availability of methods and interpretability of methods and results by clinicians. A limitation of our study is that we did not conduct meta-analysis for studies of either subtype classification or risk prediction because it was out of scope for our review. However, variation and lack of standardisation in methods and reporting make meta-analysis challenging and potentially unrepresentative. These issues can be addressed at design and implementation phases, but have been relatively neglected. Importantly, in applications of ML in CVD to date, studies of effectiveness and cost-effectiveness are lacking. Without these evaluations, ML cannot be implemented safely or effectively.
Figure 1 shows deficiencies at development, validation and impact stages and that ML research is not necessarily being conducted in a “sequential” manner, e.g. to ensure that the evaluation of impact is in validated ML models, perhaps reflecting lack or under-use of consensus guidelines. Particularly in CVD, with high disease burden globally and in low- and middle-income countries, the impact of ML has to be considered through a global lens. The fact that the majority of the literature regarding ML in CVD is from the North American context does raise concern that AI could broaden research and clinical inequalities both within and across countries, beyond current debates about the inequalities which may be inherent in algorithms. If individuals are not represented in the data feeding into the algorithms, then the AI cannot benefit them. These inequalities still exist in other domains, such as pharmaceutical trials and genomics, but the situation is improving, e.g. the proportion of RCTs recruiting in low- to middle-income countries has increased in recent years. As EHR and digital healthcare become global phenomena, there is scope to use ML in diverse data and settings. Validation of ML applications in clustering and risk prediction across countries and settings, even between high-income countries, are urgently needed to advance this agenda. We have proposed steps to improve the development and validation of ML in clinical datasets (Additional file_Web table 5).
Fig. 1.
Development, validation and impact of machine learning studies in subtype definition and risk prediction for heart failure, acute coronary syndromes and atrial fibrillation
Conclusions
Studies to date of ML in HF, ACS and AF have been limited by number and type of covariates, population size, geographic location (mainly North American), clinical setting, focus on single diseases (where overlap and multimorbidity are common) and ML methods used. Moreover, flaws at stages of development, validation and impact reduce the clinical utility and likelihood of implementation of ML in routine healthcare. To improve the generalisability, applicability and clinical utility of ML in CVD and other diseases, and to influence clinical decision-making internationally, we provide a simple checklist to foster standardised reporting and validation.
Supplementary Information
Additional file 1. Search terms and search strategy.
Additional file 2 Figure S1. PRISMA flow diagram.
Additional file 3 Web Table 1. AI-TREE checklist.
Additional file 4 Web Table 2. Data extraction for included studies.
Additional file 5. Methods and Results for scoping review.
Additional file 6 Web Table 3. Subtype classification studies in other disease areas.
Additional file 7 Web Table 4. Risk prediction studies in other disease areas.
Additional file 8 Web Table 5. Proposed methods for development and validation of ML.
Acknowledgements
Not applicable.
Authors’ contributions
Research question: AB
Funding: AB and HH
Study design and analysis plan: AB and HH
Data extraction: AB, SC and MZ
Drafting initial versions of manuscript: AB
Drafting final versions of manuscript: AB and HH
Critical review of early and final versions of manuscript: all authors
The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. AB is the guarantor. The authors read and approved the final manuscript.
Funding
AB is supported by research funding from NIHR, British Medical Association, Astra-Zeneca and UK Research and Innovation. HH is funded by NIHR (Senior Investigator and University College London Hospitals Biomedical Research Centre). HH is supported by Health Data Research UK (grant No. LOND1), which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome Trust. HH, AB, SD, SG, DK, DFF, JM, GZ, SC and RTL are supported by the BigData@Heart consortium, funded by the Innovative Medicines Initiative-2 Joint Undertaking under grant agreement No. 116074. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA; it is chaired by DE Grobbee and SD Anker partnering with 20 academic and industry partners and ESC. GF is funded by AHA Institutional Data Fellowship programme. SD is supported by an Alan Turing Fellowship. This paper represents independent research [part] funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at University College London Hospital (UCLH BRC). DK reports grants from the National Institute of Health Research (NIHR CDF-2015-08-074 and NIHR HTA-130280), the British Heart Foundation (PG/17/55/33087 and AA/18/2/34218) and IRCCS San Raffaele/Menarini (Beta-blockers in Heart Failure Collaborative Group NCT0083244), in addition to personal fees from Bayer (Advisory Board), AtriCure (Speaker fees), Amomed (Advisory Board), Menarini (Research Grant) and Myokardia (Advisory Board), all outside the submitted work.
Availability of data and materials
The data underlying this article are available in the article and in its online supplementary material.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
AB has received research funding from Astra-Zeneca. DF and JM are employees of Bayer. All other authors declare none.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Otsuka F, Joner M, Prati F, Virmani R, Narula J. Clinical classification of plaque morphology in coronary disease. Nat Rev Cardiol. 2014;11(7):379–389. doi: 10.1038/nrcardio.2014.62. [DOI] [PubMed] [Google Scholar]
- 2.Dunlay SM, Roger VL, Redfield MM. Epidemiology of heart failure with preserved ejection fraction. Nat Rev Cardiol. 2017;14(10):591–602. doi: 10.1038/nrcardio.2017.65. [DOI] [PubMed] [Google Scholar]
- 3.Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JGF, Coats AJS, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J. 2016;37(27):2129–2200. doi: 10.1093/eurheartj/ehw128. [DOI] [PubMed] [Google Scholar]
- 4.Hwang T, Atluri G, Xie M, Dey S, Hong C, Kumar V, et al. Co-clustering phenome-genome for phenotype classification and disease gene discovery. Nucleic Acids Res. 2012;40(19):e146. doi: 10.1093/nar/gks615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beam AL, Kohane IS. Big data and machine learning in health care. JAMA. 2018;319(13):1317–1318. doi: 10.1001/jama.2017.18391. [DOI] [PubMed] [Google Scholar]
- 6.Hlatky MA, Greenland P, Arnett DK, Ballantyne CM, Criqui MH, Elkind MS, Go AS, Harrell FE, Jr, Hong Y, Howard BV, Howard VJ, Hsue PY, Kramer CM, McConnell JP, Normand SL, O’Donnell CJ, Smith SC, Jr, Wilson PW, American Heart Association Expert Panel on Subclinical Atherosclerotic Diseases and Emerging Risk Factors and the Stroke Council Criteria for evaluation of novel markers of cardiovascular risk: a scientific statement from the American Heart Association. Circulation. 2009;119(17):2408–2416. doi: 10.1161/CIRCULATIONAHA.109.192278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steyerberg EW, Moons KG, van der Windt DA, Hayden JA, Perel P, Schroter S, Riley RD, Hemingway H, Altman DG, PROGRESS Group Prognosis Research Strategy (PROGRESS) 3: prognostic model research. PLoS Med. 2013;10(2):e1001381. doi: 10.1371/journal.pmed.1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton JN, Briggs AD, Murray CJ, Dicker D, Foreman KJ, Wang H, et al. Changes in health in England, with analysis by English regions and areas of deprivation, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386(10010):2257–2274. doi: 10.1016/S0140-6736(15)00195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joseph P, Leong D, McKee M, Anand SS, Schwalm JD, Teo K, et al. Reducing the global burden of cardiovascular disease, part 1: the epidemiology and risk factors. Circ Res. 2017;121(6):677–694. doi: 10.1161/CIRCRESAHA.117.308903. [DOI] [PubMed] [Google Scholar]
- 10.Kirchhof P, Curtis AB, Skanes AC, Gillis AM, Samuel Wann L, John CA. Atrial fibrillation guidelines across the Atlantic: a comparison of the current recommendations of the European Society of Cardiology/European Heart Rhythm Association/European Association of Cardiothoracic Surgeons, the American College of Cardiology Foundation/American Heart Association/Heart Rhythm Society, and the Canadian Cardiovascular Society. Eur Heart J. 2013;34(20):1471–1474. doi: 10.1093/eurheartj/ehs446. [DOI] [PubMed] [Google Scholar]
- 11.Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA, et al. Fourth universal definition of myocardial infarction (2018) Circulation. 2018;138(20):e618–ee51. doi: 10.1161/CIR.0000000000000617. [DOI] [PubMed] [Google Scholar]
- 12.Awan SE, Sohel F, Sanfilippo FM, Bennamoun M, Dwivedi G. Machine learning in heart failure: ready for prime time. Curr Opin Cardiol. 2018;33(2):190–195. doi: 10.1097/HCO.0000000000000491. [DOI] [PubMed] [Google Scholar]
- 13.Myers PD, Scirica BM, Stultz CM. Machine learning improves risk stratification after acute coronary syndrome. Sci Rep. 2017;7(1):12692. doi: 10.1038/s41598-017-12951-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Streur M, Ratcliffe SJ, Callans D, Shoemaker MB, Riegel B. Atrial fibrillation symptom clusters and associated clinical characteristics and outcomes: a cross-sectional secondary data analysis. Eur J Cardiovasc Nurs. 2018;17(8):707–716. doi: 10.1177/1474515118778445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad T, Lund LH, Rao P, Ghosh R, Warier P, Vaccaro B, et al. Machine learning methods improve prognostication, identify clinically distinct phenotypes, and detect heterogeneity in response to therapy in a large cohort of heart failure patients. J Am Heart Assoc. 2018;7(8):e008081. [DOI] [PMC free article] [PubMed]
- 16.Cikes M, Sanchez-Martinez S, Claggett B, Duchateau N, Piella G, Butakoff C, et al. Machine learning-based phenogrouping in heart failure to identify responders to cardiac resynchronization therapy. Eur J Heart Fail. 2019;21(1):74–85. doi: 10.1002/ejhf.1333. [DOI] [PubMed] [Google Scholar]
- 17.Alonso-Betanzos A, Bolon-Canedo V. Big-data analysis, cluster analysis, and machine-learning approaches. Adv Exp Med Biol. 2018;1065:607–626. doi: 10.1007/978-3-319-77932-4_37. [DOI] [PubMed] [Google Scholar]
- 18.Schulz KF, Altman DG, Moher D, Group C CONSORT 2010 statement: updated guidelines for reporting parallel group randomised trials. BMJ. 2010;340:c332. doi: 10.1136/bmj.c332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eldridge SM, Chan CL, Campbell MJ, Bond CM, Hopewell S, Thabane L, et al. CONSORT 2010 statement: extension to randomised pilot and feasibility trials. BMJ. 2016;355:i5239. doi: 10.1136/bmj.i5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vollmer S, Mateen BA, Bohner G, Király FJ, Ghani R, Jonsson P, Cumbers S, Jonas A, McAllister KSL, Myles P, Granger D, Birse M, Branson R, Moons KGM, Collins GS, Ioannidis JPA, Holmes C, Hemingway H. Machine learning and artificial intelligence research for patient benefit: 20 critical questions on transparency, replicability, ethics, and effectiveness. BMJ. 2020;368:l6927. [DOI] [PMC free article] [PubMed]
- 21.Department of Health and Social Care. Code of conduct for data-driven health and care technology. 2018.
- 22.Wiens J, Saria S, Sendak M, Ghassemi M, Liu VX, Doshi-Velez F, Jung K, Heller K, Kale D, Saeed M, Ossorio PN, Thadaney-Israni S, Goldenberg A. Do no harm: a roadmap for responsible machine learning for health care. Nat Med. 2019;25(9):1337–40. [DOI] [PubMed]
- 23.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019;110:12–22. [DOI] [PubMed]
- 24.Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, Mahendiran T, Moraes G, Shamdas M, Kern C, Ledsam JR, Schmid MK, Balaskas K, Topol EJ, Bachmann LM, Keane PA, Denniston AK. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digital Health. 2019;1(6):271–297. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 25.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 26.Moons KG, de Groot JA, Bouwmeester W, Vergouwe Y, Mallett S, Altman DG, et al. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 2014;11(10):e1001744. doi: 10.1371/journal.pmed.1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. doi: 10.1136/bmj.g7594. [DOI] [PubMed] [Google Scholar]
- 28.Ieva F, Paganoni AM, Pietrabissa T. Dynamic clustering of hazard functions: an application to disease progression in chronic heart failure. Health Care Manag Sci. 2017;20(3):353–364. doi: 10.1007/s10729-016-9357-3. [DOI] [PubMed] [Google Scholar]
- 29.Ather S, Peterson LE, Divakaran VG, Deswal A, Ramasubbu K, Giorgberidze I, Blaustein A, Wehrens XH, Mann DL, Bozkurt B. Digoxin treatment in heart failure--unveiling risk by cluster analysis of DIG data. Int J Cardiol. 2011;150(3):264–269. doi: 10.1016/j.ijcard.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao DP, Lewsey JD, Anand IS, Massie BM, Zile MR, Carson PE, McKelvie RS, Komajda M, McMurray JJ, Lindenfeld J. Characterization of subgroups of heart failure patients with preserved ejection fraction with possible implications for prognosis and treatment response. Eur J Heart Fail. 2015;17(9):925–935. doi: 10.1002/ejhf.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sideris C, Alshurafa N, Pourhomayoun M, Shahmohammadi F, Samy L, Sarrafzadeh M. A data-driven feature extraction framework for predicting the severity of condition of congestive heart failure patients. Conf Proc IEEE Eng Med Biol Soc. 2015;2015:2534–2537. doi: 10.1109/EMBC.2015.7318908. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad T, Pencina MJ, Schulte PJ, O’Brien E, Whellan DJ, Piña IL, Kitzman DW, Lee KL, O’Connor CM, Felker GM. Clinical implications of chronic heart failure phenotypes defined by cluster analysis. J Am Coll Cardiol. 2014;64(17):1765–1774. doi: 10.1016/j.jacc.2014.07.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panahiazar M, Taslimitehrani V, Pereira NL, Pathak J. Using EHRs for heart failure therapy recommendation using multidimensional patient similarity analytics. Stud Health Technol Inform. 2015;210:369–373. [PMC free article] [PubMed] [Google Scholar]
- 34.Katz DH, Deo RC, Aguilar FG, Selvaraj S, Martinez EE, Beussink-Nelson L, Kim KA, Peng J, Irvin MR, Tiwari H, Rao DC, Arnett DK, Shah SJ. Phenomapping for the identification of hypertensive patients with the myocardial substrate for heart failure with preserved ejection fraction. J Cardiovasc Transl Res. 2017;10(3):275–284. doi: 10.1007/s12265-017-9739-z. [DOI] [PubMed] [Google Scholar]
- 35.Vellone E, Fida R, Ghezzi V, D’Agostino F, Biagioli V, Paturzo M, Strömberg A, Alvaro R, Jaarsma T. Patterns of self-care in adults with heart failure and their associations with sociodemographic and clinical characteristics, quality of life, and hospitalizations: a cluster analysis. J Cardiovasc Nurs. 2017;32(2):180–189. doi: 10.1097/JCN.0000000000000325. [DOI] [PubMed] [Google Scholar]
- 36.Moser DK, Lee KS, Wu JR, Mudd-Martin G, Jaarsma T, Huang TY, Fan XZ, Strömberg A, Lennie TA, Riegel B. Identification of symptom clusters among patients with heart failure: an international observational study. Int J Nurs Stud. 2014;51(10):1366–1372. doi: 10.1016/j.ijnurstu.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurgens CY, Moser DK, Armola R, Carlson B, Sethares K, Riegel B. Heart failure quality of life trialist collaborators. Symptom clusters of heart failure. Res Nurs Health. 2009;32(5):551–560. doi: 10.1002/nur.20343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segar MW, Patel KV, Ayers C, Basit M, Tang WHW, Willett D, Berry J, Grodin JL, Pandey A. Phenomapping of patients with heart failure with preserved ejection fraction using machine learning-based unsupervised cluster analysis. Eur J Heart Fail. 2020;22(1):148–158. doi: 10.1002/ejhf.1621. [DOI] [PubMed] [Google Scholar]
- 39.Bose E, Radhakrishnan K. Using unsupervised machine learning to identify subgroups among home health patients with heart failure using telehealth. Comput Inform Nurs. 2018;36(5):242–248. doi: 10.1097/CIN.0000000000000423. [DOI] [PubMed] [Google Scholar]
- 40.Alonso-Betanzos A, Bolón-Canedo V, Heyndrickx GR, Kerkhof PL. Exploring guidelines for classification of major heart failure subtypes by using machine learning. Clin Med Insights Cardiol. 2015;9(Suppl 1):57–71. doi: 10.4137/CMC.S18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song EK, Moser DK, Rayens MK, Lennie TA. Symptom clusters predict event-free survival in patients with heart failure. J Cardiovasc Nurs. 2010;25(4):284–291. doi: 10.1097/JCN.0b013e3181cfbcbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang CC, Deo RC. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131(3):269–279. doi: 10.1161/CIRCULATIONAHA.114.010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horiuchi Y, Tanimoto S, Latif AHMM, Urayama KY, Aoki J, Yahagi K, Okuno T, Sato Y, Tanaka T, Koseki K, Komiyama K, Nakajima H, Hara K, Tanabe K. Identifying novel phenotypes of acute heart failure using cluster analysis of clinical variables. Int J Cardiol. 2018;262:57–63. doi: 10.1016/j.ijcard.2018.03.098. [DOI] [PubMed] [Google Scholar]
- 44.Lee KS, Song EK, Lennie TA, Frazier SK, Chung ML, Heo S, Wu JR, Rayens MK, Riegel B, Moser DK. Symptom clusters in men and women with heart failure and their impact on cardiac event-free survival. J Cardiovasc Nurs. 2010;25(4):263–272. doi: 10.1097/JCN.0b013e3181cfbb88. [DOI] [PubMed] [Google Scholar]
- 45.Huang Z, Dong W, Duan H, Liu J. A regularized deep learning approach for clinical risk prediction of acute coronary syndrome using electronic health records. IEEE Trans Biomed Eng. 2018;65(5):956–968. doi: 10.1109/TBME.2017.2731158. [DOI] [PubMed] [Google Scholar]
- 46.Przewlocka-Kosmala M, Marwick TH, Dabrowski A, Kosmala W. Contribution of cardiovascular reserve to prognostic categories of heart failure with preserved ejection fraction: a classification based on machine learning. J Am Soc Echocardiogr. 2019;32(5):604–615.e6. doi: 10.1016/j.echo.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Omar AMS, Narula S, Abdel Rahman MA, Pedrizzetti G, Raslan H, Rifaie O, Narula J, Sengupta PP. Precision phenotyping in heart failure and pattern clustering of ultrasound data for the assessment of diastolic dysfunction. JACC Cardiovasc Imaging. 2017;10(11):1291–1303. doi: 10.1016/j.jcmg.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Ahmad T, Desai N, Wilson F, Schulte P, Dunning A, Jacoby D, Allen L, Fiuzat M, Rogers J, Felker GM, O’Connor C, Patel CB. Clinical implications of cluster analysis-based classification of acute decompensated heart failure and correlation with bedside hemodynamic profiles. PLoS One. 2016;11(2):e0145881. doi: 10.1371/journal.pone.0145881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-Martinez S, Duchateau N, Erdei T, Kunszt G, Aakhus S, Degiovanni A, Marino P, Carluccio E, Piella G, Fraser AG, Bijnens BH. Machine learning analysis of left ventricular function to characterize heart failure with preserved ejection fraction. Circ Cardiovasc Imaging. 2018;11(4):e007138. doi: 10.1161/CIRCIMAGING.117.007138. [DOI] [PubMed] [Google Scholar]
- 50.Hertzog MA, Pozehl B, Duncan K. Cluster analysis of symptom occurrence to identify subgroups of heart failure patients: a pilot study. J Cardiovasc Nurs. 2010;25(4):273–283. doi: 10.1097/JCN.0b013e3181cfbb6c. [DOI] [PubMed] [Google Scholar]
- 51.Nowak RM, Reed BP, DiSomma S, Nanayakkara P, Moyer M, Millis S, Levy P. Presenting phenotypes of acute heart failure patients in the ED: identification and implications. Am J Emerg Med. 2017;35(4):536–542. doi: 10.1016/j.ajem.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Herr JK, Salyer J, Flattery M, Goodloe L, Lyon DE, Kabban CS, Clement DG. Heart failure symptom clusters and functional status - a cross-sectional study. J Adv Nurs. 2015;71(6):1274–1287. doi: 10.1111/jan.12596. [DOI] [PubMed] [Google Scholar]
- 53.Sharma A, Sun JL, Lokhnygina Y, Roe MT, Ahmad T, Desai NR, Blazing MA. Patient phenotypes, cardiovascular risk, and ezetimibe treatment in patients after acute coronary syndromes (from IMPROVE-IT) Am J Cardiol. 2019;123(8):1193–1201. doi: 10.1016/j.amjcard.2019.01.034. [DOI] [PubMed] [Google Scholar]
- 54.Riegel B, Hanlon AL, McKinley S, Moser DK, Meischke H, Doering LV, Davidson P, Pelter MM, Dracup K. Differences in mortality in acute coronary syndrome symptom clusters. Am Heart J. 2010;159(3):392–398. doi: 10.1016/j.ahj.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryan CJ, DeVon HA, Horne R, King KB, Milner K, Moser DK, Quinn JR, Rosenfeld A, Hwang SY, Zerwic JJ. Symptom clusters in acute myocardial infarction: a secondary data analysis. Nurs Res. 2007;56(2):72–81. doi: 10.1097/01.NNR.0000263968.01254.d6. [DOI] [PubMed] [Google Scholar]
- 56.Rosenfeld AG, Knight EP, Steffen A, Burke L, Daya M, DeVon HA. Symptom clusters in patients presenting to the emergency department with possible acute coronary syndrome differ by sex, age, and discharge diagnosis. Heart Lung. 2015;44(5):368–375. doi: 10.1016/j.hrtlng.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeVon HA, Ryan CJ, Rankin SH, Cooper BA. Classifying subgroups of patients with symptoms of acute coronary syndromes: a cluster analysis. Res Nurs Health. 2010;33(5):386–397. doi: 10.1002/nur.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McSweeney JC, Cleves MA, Zhao W, Lefler LL, Yang S. Cluster analysis of women’s prodromal and acute myocardial infarction symptoms by race and other characteristics. J Cardiovasc Nurs. 2010;25(4):311–322. doi: 10.1097/JCN.0b013e3181cfba15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindgren TG, Fukuoka Y, Rankin SH, Cooper BA, Carroll D, Munn YL. Cluster analysis of elderly cardiac patients’ prehospital symptomatology. Nurs Res. 2008;57(1):14–23. doi: 10.1097/01.NNR.0000280654.50642.1a. [DOI] [PubMed] [Google Scholar]
- 60.Abbott AA, Barnason S, Zimmerman L. Symptom burden clusters and their impact on psychosocial functioning following coronary artery bypass surgery. J Cardiovasc Nurs. 2010;25(4):301–310. doi: 10.1097/JCN.0b013e3181cfbb46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fukuoka Y, Lindgren TG, Rankin SH, Cooper BA, Carroll DL. Cluster analysis: a useful technique to identify elderly cardiac patients at risk for poor quality of life. Qual Life Res. 2007;16(10):1655–1663. doi: 10.1007/s11136-007-9272-7. [DOI] [PubMed] [Google Scholar]
- 62.Inohara T, Shrader P, Pieper K, Blanco RG, Thomas L, Singer DE, Freeman JV, Allen LA, Fonarow GC, Gersh B, Ezekowitz MD, Kowey PR, Reiffel JA, Naccarelli GV, Chan PS, Steinberg BA, Peterson ED, Piccini JP. Association of of atrial fibrillation clinical phenotypes with treatment patterns and outcomes: a multicenter registry study. JAMA Cardiol. 2018;3(1):54–63. doi: 10.1001/jamacardio.2017.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inohara T, Piccini JP, Mahaffey KW, Kimura T, Katsumata Y, Tanimoto K, Inagawa K, Ikemura N, Ueda I, Fukuda K, Takatsuki S, Kohsaka S. A cluster analysis of the Japanese multicenter outpatient registry of patients with atrial fibrillation. Am J Cardiol. 2019;124(6):871–878. doi: 10.1016/j.amjcard.2019.05.071. [DOI] [PubMed] [Google Scholar]
- 64.Streur M, Ratcliffe SJ, Ball J, Stewart S, Riegel B. Symptom clusters in adults with chronic atrial fibrillation. J Cardiovasc Nurs. 2017;32(3):296–303. doi: 10.1097/JCN.0000000000000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rasmy L, Wu Y, Wang N, Geng X, Zheng WJ, Wang F, Wu H, Xu H, Zhi D. A study of generalizability of recurrent neural network-based predictive models for heart failure onset risk using a large and heterogeneous EHR data set. J Biomed Inform. 2018;84:11–16. doi: 10.1016/j.jbi.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maragatham G, Devi S. LSTM model for prediction of heart failure in big data. J Med Syst. 2019;43(5):111. doi: 10.1007/s10916-019-1243-3. [DOI] [PubMed] [Google Scholar]
- 67.Choi E, Schuetz A, Stewart WF, Sun J. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc. 2017;24(2):361–370. doi: 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ng K, Steinhubl SR, deFilippi C, Dey S, Stewart WF. Early detection of heart failure using electronic health records: practical implications for time before diagnosis, data diversity, data quantity, and data density. Circ Cardiovasc Qual Outcomes. 2016;9(6):649–658. doi: 10.1161/CIRCOUTCOMES.116.002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Ng K, Byrd RJ, Hu J, Ebadollahi S, Daar Z, deFilippi C, Steinhubl SR, Stewart WF. Early detection of heart failure with varying prediction windows by structured and unstructured data in electronic health records. Conf Proc IEEE Eng Med Biol Soc. 2015;2015:2530–2533. doi: 10.1109/EMBC.2015.7318907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan H, Fan XS, Jin Y, He JX, Gui Y, Song LY, Song Y, Sun Q, Chen W. Development of heart failure risk prediction models based on a multi-marker approach using random forest algorithms. Chin Med J. 2019;132(7):819–826. doi: 10.1097/CM9.0000000000000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lindholm D, Fukaya E, Leeper NJ, Ingelsson E. Bioimpedance and new-onset heart failure: a longitudinal study of >500 000 individuals from the general population. J Am Heart Assoc. 2018;7(13):e008970. doi: 10.1161/JAHA.118.008970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Segar MW, Vaduganathan M, Patel KV, McGuire DK, Butler J, Fonarow GC, Basit M, Kannan V, Grodin JL, Everett B, Willett D, Berry J, Pandey A. Machine learning to predict the risk of incident heart failure hospitalization among patients with diabetes: the WATCH-DM risk score. Diabetes Care. 2019;42(12):2298–2306. doi: 10.2337/dc19-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blecker S, Katz SD, Horwitz LI, Kuperman G, Park H, Gold A, Sontag D. Comparison of approaches for heart failure case identification from electronic health record data. JAMA Cardiol. 2016;1(9):1014–1020. doi: 10.1001/jamacardio.2016.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blecker S, Sontag D, Horwitz LI, Kuperman G, Park H, Reyentovich A, Katz SD. Early identification of patients with acute decompensated heart failure. J Card Fail. 2018;24(6):357–362. doi: 10.1016/j.cardfail.2017.08.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Austin PC, Tu JV, Ho JE, Levy D, Lee DS. Using methods from the data-mining and machine-learning literature for disease classification and prediction: a case study examining classification of heart failure subtypes. J Clin Epidemiol. 2013;66(4):398–407. doi: 10.1016/j.jclinepi.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nirschl JJ, Janowczyk A, Peyster EG, Frank R, Margulies KB, Feldman MD, Madabhushi A. A deep-learning classifier identifies patients with clinical heart failure using whole-slide images of H&E tissue. PLoS One. 2018;13(4):e0192726. doi: 10.1371/journal.pone.0192726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tabassian M, Sunderji I, Erdei T, Sanchez-Martinez S, Degiovanni A, Marino P, Fraser AG, D’hooge J. Diagnosis of heart failure with preserved ejection fraction: machine learning of spatiotemporal variations in left ventricular deformation. J Am Soc Echocardiogr. 2018;31(12):1272–1284.e9. doi: 10.1016/j.echo.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 78.Masetic Z, Subasi A. Congestive heart failure detection using random forest classifier. Comput Methods Prog Biomed. 2016;130:54–64. doi: 10.1016/j.cmpb.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 79.Yoon J, Zame WR, Banerjee A, Cadeiras M, Alaa AM, van der Schaar M. Personalized survival predictions via Trees of Predictors: an application to cardiac transplantation. PLoS One. 2018;13(3):e0194985. doi: 10.1371/journal.pone.0194985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frizzell JD, Liang L, Schulte PJ, Yancy CW, Heidenreich PA, Hernandez AF, Bhatt DL, Fonarow GC, Laskey WK. Prediction of 30-day all-cause readmissions in patients hospitalized for heart failure: comparison of machine learning and other statistical approaches. JAMA Cardiol. 2017;2(2):204–209. doi: 10.1001/jamacardio.2016.3956. [DOI] [PubMed] [Google Scholar]
- 81.Miller PE, Pawar S, Vaccaro B, McCullough M, Rao P, Ghosh R, Warier P, Desai NR, Ahmad T. Predictive abilities of machine learning techniques may be limited by dataset characteristics: insights from the UNOS database. J Card Fail. 2019;25(6):479–483. doi: 10.1016/j.cardfail.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 82.Golas SB, Shibahara T, Agboola S, Otaki H, Sato J, Nakae T, Hisamitsu T, Kojima G, Felsted J, Kakarmath S, Kvedar J, Jethwani K. A machine learning model to predict the risk of 30-day readmissions in patients with heart failure: a retrospective analysis of electronic medical records data. BMC Med Inform Decis Mak. 2018;18(1):44. doi: 10.1186/s12911-018-0620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Awan SE, Bennamoun M, Sohel F, Sanfilippo FM, Dwivedi G. Machine learning-based prediction of heart failure readmission or death: implications of choosing the right model and the right metrics. ESC Heart Fail. 2019;6(2):428–435. doi: 10.1002/ehf2.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kwon JM, Kim KH, Jeon KH, Lee SE, Lee HY, Cho HJ, Choi JO, Jeon ES, Kim MS, Kim JJ, Hwang KK, Chae SC, Baek SH, Kang SM, Choi DJ, Yoo BS, Kim KH, Park HY, Cho MC, Oh BH. Artificial intelligence algorithm for predicting mortality of patients with acute heart failure. PLoS One. 2019;14(7):e0219302. doi: 10.1371/journal.pone.0219302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ashfaq A, Sant'Anna A, Lingman M, Nowaczyk S. Readmission prediction using deep learning on electronic health records. J Biomed Inform. 2019;97:103256. doi: 10.1016/j.jbi.2019.103256. [DOI] [PubMed] [Google Scholar]
- 86.Taslimitehrani V, Dong G, Pereira NL, Panahiazar M, Pathak J. Developing EHR-driven heart failure risk prediction models using CPXR (Log) with the probabilistic loss function. J Biomed Inform. 2016;60:260–269. doi: 10.1016/j.jbi.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Panahiazar M, Taslimitehrani V, Pereira N, Pathak J. Using EHRs and machine learning for heart failure survival analysis. Stud Health Technol Inform. 2015;216:40–44. [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Z, Yao L, Li D, Ruan T, Liu M, Gao J. Mortality prediction system for heart failure with orthogonal relief and dynamic radius means. Int J Med Inform. 2018;115:10–17. doi: 10.1016/j.ijmedinf.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 89.Mortazavi BJ, Downing NS, Bucholz EM, Dharmarajan K, Manhapra A, Li SX, Negahban SN, Krumholz HM. Analysis of machine learning techniques for heart failure readmissions. Circ Cardiovasc Qual Outcomes. 2016;9(6):629–640. doi: 10.1161/CIRCOUTCOMES.116.003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hearn J, Ross HJ, Mueller B, Fan CP, Crowdy E, Duhamel J, Walker M, Alba AC, Manlhiot C. Neural networks for prognostication of patients with heart failure. Circ Heart Fail. 2018;11(8):e005193. doi: 10.1161/CIRCHEARTFAILURE.118.005193. [DOI] [PubMed] [Google Scholar]
- 91.Shameer K, Johnson KW, Yahi A, Miotto R, Li LI, Ricks D, Jebakaran J, Kovatch P, Sengupta PP, Gelijns S, Moskovitz A, Darrow B, David DL, Kasarskis A, Tatonetti NP, Pinney S, Dudley JT. Predictive modeling of hospital readmission rates using electronic medical record-wide machine learning: a case-study using Mount Sinai heart failure cohort. Pac Symp Biocomput. 2017;22:276–287. doi: 10.1142/9789813207813_0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mahajan S, Burman P, Hogarth M. Analyzing 30-day readmission rate for heart failure using different predictive models. Stud Health Technol Inform. 2016;225:143–147. [PubMed] [Google Scholar]
- 93.Gleeson S, Liao YW, Dugo C, Cave A, Zhou L, Ayar Z, Christiansen J, Scott T, Dawson L, Gavin A, Schlegel TT, Gladding P. ECG-derived spatial QRS-T angle is associated with ICD implantation, mortality and heart failure admissions in patients with LV systolic dysfunction. PLoS One. 2017;12(3):e0171069. doi: 10.1371/journal.pone.0171069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Valenza G, Wendt H, Kiyono K, Hayano J, Watanabe E, Yamamoto Y, Abry P, Barbieri R. Mortality prediction in severe congestive heart failure patients with multifractal point-process modeling of heartbeat dynamics. IEEE Trans Biomed Eng. 2018;65(10):2345–2354. doi: 10.1109/TBME.2018.2797158. [DOI] [PubMed] [Google Scholar]
- 95.Henriques J, Carvalho P, Paredes S, Rocha T, Habetha J, Antunes M, Morais J. Prediction of heart failure decompensation events by trend analysis of telemonitoring data. IEEE J Biomed Health Inform. 2015;19(5):1757–1769. doi: 10.1109/JBHI.2014.2358715. [DOI] [PubMed] [Google Scholar]
- 96.VanHouten JP, Starmer JM, Lorenzi NM, Maron DJ, Lasko TA. Machine learning for risk prediction of acute coronary syndrome. AMIA Annu Symp Proc. 2014;2014:1940–1949. [PMC free article] [PubMed] [Google Scholar]
- 97.Beunza JJ, Puertas E, García-Ovejero E, Villalba G, Condes E, Koleva G, Hurtado C, Landecho MF. Comparison of machine learning algorithms for clinical event prediction (risk of coronary heart disease) J Biomed Inform. 2019;97:103257. doi: 10.1016/j.jbi.2019.103257. [DOI] [PubMed] [Google Scholar]
- 98.Al'Aref SJ, Maliakal G, Singh G, van Rosendael AR, Ma X, Xu Z, Alawamlh OAH, Lee B, Pandey M, Achenbach S, Al-Mallah MH, Andreini D, Bax JJ, Berman DS, Budoff MJ, Cademartiri F, Callister TQ, Chang HJ, Chinnaiyan K, Chow BJW, Cury RC, DeLago A, Feuchtner G, Hadamitzky M, Hausleiter J, Kaufmann PA, Kim YJ, Leipsic JA, Maffei E, Marques H, Gonçalves PA, Pontone G, Raff GL, Rubinshtein R, Villines TC, Gransar H, Lu Y, Jones EC, Peña JM, Lin FY, Min JK, Shaw LJ. Machine learning of clinical variables and coronary artery calcium scoring for the prediction of obstructive coronary artery disease on coronary computed tomography angiography: analysis from the CONFIRM registry. Eur Heart J. 2020;41(3):359–367. doi: 10.1093/eurheartj/ehz565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Noh YK, Park JY, Choi BG, Kim KE, Rha SW. A machine learning-based approach for the prediction of acute coronary syndrome requiring revascularization. J Med Syst. 2019;43(8):253. doi: 10.1007/s10916-019-1359-5. [DOI] [PubMed] [Google Scholar]
- 100.Lavesson N, Halling A, Freitag M, Odeberg J, Odeberg H, Davidsson P. Classifying the severity of an acute coronary syndrome by mining patient data. 2009. Conference paper: 26th Annual Workshop of the Swedish Artificial Intelligence Society. [Google Scholar]
- 101.Wu CC, Hsu WD, Islam MM, Poly TN, Yang HC, Nguyen PA, Wang YC, Li YJ. An artificial intelligence approach to early predict non-ST-elevation myocardial infarction patients with chest pain. Comput Methods Prog Biomed. 2019;173:109–117. doi: 10.1016/j.cmpb.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 102.Berikol GB, Yildiz O, Özcan IT. Diagnosis of acute coronary syndrome with a support vector machine. J Med Syst. 2016;40(4):84. doi: 10.1007/s10916-016-0432-6. [DOI] [PubMed] [Google Scholar]
- 103.Al'Aref SJ, Singh G, van Rosendael AR, Kolli KK, Ma X, Maliakal G, Pandey M, Lee BC, Wang J, Xu Z, Zhang Y, Min JK, Wong SC, Minutello RM. Determinants of in-hospital mortality after percutaneous coronary intervention: a machine learning approach. J Am Heart Assoc. 2019;8(5):e011160. doi: 10.1161/JAHA.118.011160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Steele AJ, Denaxas SC, Shah AD, Hemingway H, Luscombe NM. Machine learning models in electronic health records can outperform conventional survival models for predicting patient mortality in coronary artery disease. PLoS One. 2018;13(8):e0202344. doi: 10.1371/journal.pone.0202344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu P, Xia E, Li S, Du X, Ma C, Dong J, Chan KCC. Network-based prediction of major adverse cardiac events in acute coronary syndromes from imbalanced EMR data. Stud Health Technol Inform. 2019;264:1480–1481. doi: 10.3233/SHTI190494. [DOI] [PubMed] [Google Scholar]
- 106.Gibson WJ, Nafee T, Travis R, Yee M, Kerneis M, Ohman M, Gibson CM. Machine learning versus traditional risk stratification methods in acute coronary syndrome: a pooled randomized clinical trial analysis. J Thromb Thrombolysis. 2020;49(1):1–9. doi: 10.1007/s11239-019-01940-8. [DOI] [PubMed] [Google Scholar]
- 107.Hernesniemi JA, Mahdiani S, Tynkkynen JA, Lyytikäinen LP, Mishra PP, Lehtimäki T, Eskola M, Nikus K, Antila K, Oksala N. Extensive phenotype data and machine learning in prediction of mortality in acute coronary syndrome - the MADDEC study. Ann Med. 2019;51(2):156–163. doi: 10.1080/07853890.2019.1596302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pieszko K, Hiczkiewicz J, Budzianowski P, Rzeźniczak J, Budzianowski J, Błaszczyński J, Słowiński R, Burchardt P. Machine-learned models using hematological inflammation markers in the prediction of short-term acute coronary syndrome outcomes. J Transl Med. 2018;16(1):334. doi: 10.1186/s12967-018-1702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu D, Dong W, Lu X, Duan H, He K, Huang Z. Evidential MACE prediction of acute coronary syndrome using electronic health records. BMC Med Inform Decis Mak. 2019;19(Suppl 2):61. doi: 10.1186/s12911-019-0754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang Z, Dong W. Adversarial MACE prediction after acute coronary syndrome using electronic health records. IEEE J Biomed Health Inform. 2019;23(5):2117–2126. doi: 10.1109/JBHI.2018.2882518. [DOI] [PubMed] [Google Scholar]
- 111.Shouval R, Hadanny A, Shlomo N, Iakobishvili Z, Unger R, Zahger D, Alcalai R, Atar S, Gottlieb S, Matetzky S, Goldenberg I, Beigel R. Machine learning for prediction of 30-day mortality after ST elevation myocardial infraction: an Acute Coronary Syndrome Israeli Survey data mining study. Int J Cardiol. 2017;246:7–13. doi: 10.1016/j.ijcard.2017.05.067. [DOI] [PubMed] [Google Scholar]
- 112.Hill NR, Ayoubkhani D, McEwan P, Sugrue DM, Farooqui U, Lister S, Lumley M, Bakhai A, Cohen AT, O’Neill M, Clifton D, Gordon J. Predicting atrial fibrillation in primary care using machine learning. PLoS One. 2019;14(11):e0224582. doi: 10.1371/journal.pone.0224582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tiwari P, Colborn KL, Smith DE, Xing F, Ghosh D, Rosenberg MA. Assessment of a machine learning model applied to harmonized electronic health record data for the prediction of incident atrial fibrillation. JAMA Netw Open. 2020;3(1):e1919396. doi: 10.1001/jamanetworkopen.2019.19396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bundy JD, Heckbert SR, Chen LY, Lloyd-Jones DM, Greenland P. Evaluation of risk prediction models of atrial fibrillation (from the Multi-Ethnic Study of Atherosclerosis [MESA]) Am J Cardiol. 2020;125(1):55–62. doi: 10.1016/j.amjcard.2019.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goodfellow SD, Goodwin A, Greer R, Laussen PC, Mazwi M, Eytan D. Atrial fibrillation classification using step-by-step machine learning. Biomed Phys Eng Express. 2018;4:045005. doi: 10.1088/2057-1976/aabef4. [DOI] [Google Scholar]
- 116.Goto S, Goto S, Pieper KS, Bassand JP, Camm AJ, Fitzmaurice DA, Goldhaber SZ, Haas S, Parkhomenko A, Oto A, Misselwitz F, Turpie AGG, Verheugt FWA, Fox KAA, Gersh BJ, Kakkar AK, GARFIELD-AF Investigators. New AI prediction model using serial PT-INR measurements in AF patients on VKAs: GARFIELD-AF. Eur Heart J Cardiovasc Pharmacother. 2019:pvz076. 10.1093/ehjcvp/pvz076 [Epub ahead of print].
- 117.Han L, Askari M, Altman RB, Schmitt SK, Fan J, Bentley JP, Narayan SM, Turakhia MP. Atrial fibrillation burden signature and near-term prediction of stroke: a machine learning analysis. Circ Cardiovasc Qual Outcomes. 2019;12(10):e005595. doi: 10.1161/CIRCOUTCOMES.118.005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li X, Liu H, Du X, Zhang P, Hu G, Xie G, Guo S, Xu M, Xie X. Integrated machine learning approaches for predicting ischemic stroke and thromboembolism in atrial fibrillation. AMIA Annu Symp Proc. 2017;2016:799–807. [PMC free article] [PubMed] [Google Scholar]
- 119.Lynch CM, van Berkel VH, Frieboes HB. Application of unsupervised analysis techniques to lung cancer patient data. PLoS One. 2017;12(9):e0184370. doi: 10.1371/journal.pone.0184370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schuler A, Liu V, Wan J, Callahan A, Udell M, Stark DE, Shah NH. Discovering patient phenotypes using generalized low rank models. Pac Symp Biocomput. 2016;21:144–155. [PMC free article] [PubMed] [Google Scholar]
- 121.Conrad DJ, Bailey BA. Multidimensional clinical phenotyping of an adult cystic fibrosis patient population. PLoS One. 2015;10(3):e0122705. doi: 10.1371/journal.pone.0122705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Niehaus KE, Uhlig HH, Clifton DA. Phenotypic characterisation of Crohn’s disease severity. Conf Proc IEEE Eng Med Biol Soc. 2015;2015:7023–7026. doi: 10.1109/EMBC.2015.7320009. [DOI] [PubMed] [Google Scholar]
- 123.Beaulieu-Jones BK, Greene CS, Pooled Resource Open-Access ALS Clinical Trials Consortium Semi-supervised learning of the electronic health record for phenotype stratification. J Biomed Inform. 2016;64:168–178. doi: 10.1016/j.jbi.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 124.Cho SB, Kim SC, Chung MG. Identification of novel population clusters with different susceptibilities to type 2 diabetes and their impact on the prediction of diabetes. Sci Rep. 2019;9(1):3329. doi: 10.1038/s41598-019-40058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ahlqvist E, Storm P, Käräjämäki A, Martinell M, Dorkhan M, Carlsson A, Vikman P, Prasad RB, Aly DM, Almgren P, Wessman Y, Shaat N, Spégel P, Mulder H, Lindholm E, Melander O, Hansson O, Malmqvist U, Lernmark Å, Lahti K, Forsén T, Tuomi T, Rosengren AH, Groop L. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6(5):361–369. doi: 10.1016/S2213-8587(18)30051-2. [DOI] [PubMed] [Google Scholar]
- 126.Cleret de Langavant L, Bayen E, Yaffe K. Unsupervised machine learning to identify high likelihood of dementia in population-based surveys: development and validation study. J Med Internet Res. 2018;20(7):e10493. doi: 10.2196/10493.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Seymour CW, Kennedy JN, Wang S, Chang CH, Elliott CF, Xu Z, Berry S, Clermont G, Cooper G, Gomez H, Huang DT, Kellum JA, Mi Q, Opal SM, Talisa V, van der Poll T, Visweswaran S, Vodovotz Y, Weiss JC, Yealy DM, Yende S, Angus DC. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA. 2019;321(20):2003–2017. doi: 10.1001/jama.2019.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pikoula M, Quint JK, Nissen F, Hemingway H, Smeeth L, Denaxas S. Identifying clinically important COPD sub-types using data-driven approaches in primary care population based electronic health records. BMC Med Inform Decis Mak. 2019;19(1):86. doi: 10.1186/s12911-019-0805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wojnarski CM, Roselli EE, Idrees JJ, Zhu Y, Carnes TA, Lowry AM, Collier PH, Griffin B, Ehrlinger J, Blackstone EH, Svensson LG, Lytle BW. Machine-learning phenotypic classification of bicuspid aortopathy. J Thorac Cardiovasc Surg. 2018;155(2):461–469.e4. doi: 10.1016/j.jtcvs.2017.08.123. [DOI] [PubMed] [Google Scholar]
- 130.Alaa AM, Bolton T, Di Angelantonio E, Rudd JHF, van der Schaar M. Cardiovascular disease risk prediction using automated machine learning: a prospective study of 423,604 UK Biobank participants. PLoS One. 2019;14(5):e0213653. doi: 10.1371/journal.pone.0213653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weng SF, Reps J, Kai J, Garibaldi JM, Qureshi N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS One. 2017;12(4):e0174944. doi: 10.1371/journal.pone.0174944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhao J, Feng Q, Wu P, Lupu RA, Wilke RA, Wells QS, Denny JC, Wei WQ. Learning from longitudinal data in electronic health record and genetic data to improve cardiovascular event prediction. Sci Rep. 2019;9(1):717. doi: 10.1038/s41598-018-36745-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ambale-Venkatesh B, Yang X, Wu CO, Liu K, Hundley WG, McClelland R, Gomes AS, Folsom AR, Shea S, Guallar E, Bluemke DA, Lima JAC. Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ Res. 2017;121(9):1092–1101. doi: 10.1161/CIRCRESAHA.117.311312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Farran B, Channanath AM, Behbehani K, Thanaraj TA. Predictive models to assess risk of type 2 diabetes, hypertension and comorbidity: machine-learning algorithms and validation using national health data from Kuwait--a cohort study. BMJ Open. 2013;3(5):e002457. doi: 10.1136/bmjopen-2012-002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bellemo V, Lim ZW, Lim G, Nguyen QD, Xie Y, Yip MYT, Hamzah H, Ho J, Lee XQ, Hsu W, Lee ML, Musonda L, Chandran M, Chipalo-Mutati G, Muma M, Tan GSW, Sivaprasad S, Menon G, Wong TY, Ting DSW. Artificial intelligence using deep learning to screen for referable and vision-threatening diabetic retinopathy in Africa: a clinical validation study. Lancet Digital Health. 2019;1(1):35–44. doi: 10.1016/S2589-7500(19)30004-4. [DOI] [PubMed] [Google Scholar]
- 136.Bertsimas D, Dunn J, Pawlowski C, Silberholz J, Weinstein A, Zhuo YD, Chen E, Elfiky AA. Applied informatics decision support tool for mortality predictions in patients with cancer. JCO Clin Cancer Inform. 2018;2:1–11. doi: 10.1200/CCI.18.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Delen D, Walker G, Kadam A. Predicting breast cancer survivability: a comparison of three data mining methods. Artif Intell Med. 2005;34(2):113–127. doi: 10.1016/j.artmed.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 138.Chiew CJ, Liu N, Wong TH, Sim YE, Abdullah HR. Utilizing machine learning methods for preoperative prediction of postsurgical mortality and intensive care unit admission. Ann Surg. 2019. 10.1097/SLA.0000000000003297. [DOI] [PMC free article] [PubMed]
- 139.De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, Askham H, Glorot X, O’Donoghue B, Visentin D, van den Driessche G, Lakshminarayanan B, Meyer C, Mackinder F, Bouton S, Ayoub K, Chopra R, King D, Karthikesalingam A, Hughes CO, Raine R, Hughes J, Sim DA, Egan C, Tufail A, Montgomery H, Hassabis D, Rees G, Back T, Khaw PT, Suleyman M, Cornebise J, Keane PA, Ronneberger O. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 140.Goldstein BA, Pomann GM, Winkelmayer WC, Pencina MJ. A comparison of risk prediction methods using repeated observations: an application to electronic health records for hemodialysis. Stat Med. 2017;36(17):2750–2763. doi: 10.1002/sim.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Taylor RA, Pare JR, Venkatesh AK, Mowafi H, Melnick ER, Fleischman W, Hall MK. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Acad Emerg Med. 2016;23(3):269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tomašev N, Glorot X, Rae JW, Zielinski M, Askham H, Saraiva A, Mottram A, Meyer C, Ravuri S, Protsyuk I, Connell A, Hughes CO, Karthikesalingam A, Cornebise J, Montgomery H, Rees G, Laing C, Baker CR, Peterson K, Reeves R, Hassabis D, King D, Suleyman M, Back T, Nielson C, Ledsam JR, Mohamed S. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572(7767):116–119. doi: 10.1038/s41586-019-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tibble H, Tsanas A, Horne E, Horne R, Mizani M, Simpson CR, Sheikh A. Predicting asthma attacks in primary care: protocol for developing a machine learning-based prediction model. BMJ Open. 2019;9(7):e028375. doi: 10.1136/bmjopen-2018-028375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Moher D, Stewart L, Shekelle P. All in the family: systematic reviews, rapid reviews, scoping reviews, realist reviews, and more. Syst Rev.2015;4:183. [DOI] [PMC free article] [PubMed]
- 145.Peters MD, Godfrey CM, Khalil H, McInerney P, Parker D, Soares CB. Guidance for conducting systematic scoping reviews. Int J Evid Based Healthc. 2015;13(3):141–146. doi: 10.1097/XEB.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 146.Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet. 2019;393(10181):1577–1579. doi: 10.1016/S0140-6736(19)30037-6. [DOI] [PubMed] [Google Scholar]
- 147.Coiera E, Ammenwerth E, Georgiou A, Magrabi F. Does health informatics have a replication crisis? J Am Med Inform Assoc. 2018;25(8):963–968. doi: 10.1093/jamia/ocy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hernandez-Boussard T, Bozkurt S, Ioannidis JPA, Shah NH. MINIMAR (MINimum Information for Medical AI Reporting): developing reporting standards for artificial intelligence in health care. JAMIA. 2020; Published online 28 June 2020. https://academic.oup.com/jamia/article/doi/10.1093/jamia/ocaa088/5864179. [DOI] [PMC free article] [PubMed]
- 149.Sugrue LP, Desikan RS. What are polygenic scores and why are they important? JAMA. 2019;321(18):1820–1821. doi: 10.1001/jama.2019.3893. [DOI] [PubMed] [Google Scholar]
- 150.Gurdasani D, Barroso I, Zeggini E, Sandhu MS. Genomics of disease risk in globally diverse populations. Nat Rev Genet. 2019;20(9):520–535. doi: 10.1038/s41576-019-0144-0. [DOI] [PubMed] [Google Scholar]
- 151.Schussler-Fiorenza Rose SM, Contrepois K, Moneghetti KJ, Zhou W, Mishra T, Mataraso S, et al. A longitudinal big data approach for precision health. Nat Med. 2019;25(5):792–804. doi: 10.1038/s41591-019-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Panch T, Mattie H, Celi LA. The “inconvenient truth” about AI in healthcare. NPJ Digit Med. 2019;2:77. doi: 10.1038/s41746-019-0155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Search terms and search strategy.
Additional file 2 Figure S1. PRISMA flow diagram.
Additional file 3 Web Table 1. AI-TREE checklist.
Additional file 4 Web Table 2. Data extraction for included studies.
Additional file 5. Methods and Results for scoping review.
Additional file 6 Web Table 3. Subtype classification studies in other disease areas.
Additional file 7 Web Table 4. Risk prediction studies in other disease areas.
Additional file 8 Web Table 5. Proposed methods for development and validation of ML.
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.