Graphical abstract
Abbreviation: 2019-nCoV, severe acute respiratory syndrome coronavirus 2; COVID-19, Coronavirus disease; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; RNA, ribonucleic acid; ICU, intensive care units; PCR, polymerase chain reaction; BAL, bronchoalveolar lavage; USFDA, United States Food And Drug Administration; RdRP, RNA-dependent RNA polymerase; ARDS, acute respiratory distress syndrome; G-CSF, (Granulocyte colony-stimulating facto); IP10, 10 Kda interferon gamma-induced protein; MCP1, monocyte chemoattractant protein-1; TNFα, tumor necrosis factor alpha; Th2, T-helper-2; RT-PCR, reverse transcription polymerase chain reaction; cDNA, complementary DNA; LAMP, loop-mediated isothermal amplification; CT, computed tomography; IgM, immunoglobulin M; IgG, immunoglobulin G; AuNP-ab, gold-antibody nanoparticle; HIV, human immunodeficiency virus; CRISPR, clustered regularly interspaced short palindromic repeats; RPA, recombinase polymerase amplification; RCA, rolling circle amplification; ELISA, enzyme-linked immunosorbent assay; MagLev, magnetic levitation; SPION, superparamagnetic iron oxide nanoparticles; HBV, hepatitis B Virus; HCV, hepatitis C Virus; WHO, World Health Organization; FDA, Food And Drug Administration; PICALM, phosphatidylinositol binding clathrin assembly protein; ergic, endoplasmic reticulum-golgi intermediate compartment; NagC, nano-silver colloids; IC, inhibitory concentration; GSH, glutathione; MHV-A59, mouse coronavirus; IFN-γ, interferon gamma; PDT, photodynamic therapy; PS, photosensitizer; MB, methylene blue; TPPS2a, tetraphenyl porphyrin disulphonate; FISH, fluorescent in situ hybridization; MWCNTs, multi-walled carbon nanotubes; EHEC, enterohemorrhagic Escherichia coli; GQD, graphene quantum dot; Ag-MES, silver nanoparticle capped with mercaptoethane sulfonate; HSV-1, herpes Simplex type-1 virus; PDI, photodynamic inactivation; ROS, reactive oxygen species; AMT, 4′-aminomethyl-trioxsalene; PTAF, photocatalytic titanium apatite filter; UV, ultraviolet; EUA, emergency use authorization
Keywords: COVID-19, Diagnosis, Treatment protocols, Nanomedicine, Nanoparticles
Abstract
The 2019 novel coronavirus (2019-nCoV; severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)) has witnessed a rapid and global proliferation since its early identification in patients with severe pneumonia in Wuhan, China. As of 27th May 2020, 2019-nCoV cases have risen to >5 million, with confirmed deaths of 350,000. However, Coronavirus disease (COVID-19) diagnostic and treatment measures are yet to be fully unraveled, given the novelty of this particular coronavirus. Therefore, existing antiviral agents used for severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) were repurposed for COVID-19, taking their biological features into consideration. This study provides a concise review of the current and emerging detection and supervision technologies for SARS-CoV-2, which is the viral etiology of COVID19, and their performance characteristics, with emphasis on the novel Nano-based diagnostic tests (protein corona sensor array and magnetic levitation) and treatment measures (treatment protocols based on nano-silver colloids) for COVID-19.
1. Introduction
Coronaviruses are referred to as enveloped non-segmented positive-sense ribonucleic acid (RNA) viruses, which are members of the Coronaviridae family and the Nidovirales order, with a broad distribution in humans and other mammals [1,2]. Coronaviruses can initiate diseases that are hepatic, respiratory, neurologic and enteric. Although the bulk of coronavirus infections are non-severe, the outbreaks of SARS-CoV [3] and MERS-CoV [4,5] have led to over 10,000 collective cases in the preceding twenty years, with mortality rates of 10 % and 37 %, respectively [6,7]. Nonetheless, the known coronaviruses (SARS-CoV and MERS-CoV) are possibly only a small part of a larger issue, with prospectively more acute zoonotic events to be unraveled. Six (6) coronavirus species that affect human health were recognized. Four viruses — 229E, OC43, NL63, and HKU1—are ubiquitous and normally instigate symptoms of flu in immunocompetent individuals [8].
The COVID-19 was revealed in China in December 2019 [9], where a group of admitted patients displayed symptoms that consist of feverish condition, shortness of breath and cough [10]. SARS-CoV-2 is the viral etiology of COVID19 [11]. Since 2019-nCoV is an RNA virus, it exhibits the intrinsic characteristic of a high mutation rate, although it might be somewhat lesser than other RNA viruses because of its genome encoded exonuclease. This feature supports the adaptability characteristic of this novel zoonotic viral pathogen, allowing it to be effectively transmitted from individual to individual and possibly evolving into a virulent pathogen. The current 2019-nCoV outbreak has indisputable risen the memories of the SARS-CoV epidemic that occurred 17 years ago. Clusters of pneumonia of mysterious cause (now identified as the SARS-CoV) were reported in November, 2002, in Guangdong province. The cases of SARS exponentially increased the following year, subsequently spreading worldwide [12], infecting no less than 8096 persons and resulting in 774 deaths [6]. The global proliferation of SARS-CoV in 2003 was credited to its strong spread under particular conditions and the inadequate preparations and execution of infection control measures. Nonetheless, civic health institutions and scientific competencies of China have significantly evolved since 2003. A competent system was installed for scrutinizing and reacting to epidemics and outbreaks of contagious illness. Accordingly, the 2019-nCoV was swiftly included in the list of infectious diseases and given the maximum priority by China's health authorities.
The 2019-nCoV infection results in a collection of severe respiratory infections resembling SARS coronavirus and is linked to admission into intensive care units (ICU) and high death rate [2]. At the prodromal phase, the symptoms of 2019-nCoV infection include dry cough, fever, and depression; however, they are non-specific [2]. Therefore, several detection and screening protocols for Covid-19 have been introduced. Patients are subjected to computed tomography (CT) scan, which reveals images of lungs with relatively higher density, profusion, and confluent opacity compared to healthy lungs [13]. This discovery initially brought about the prelude diagnosis of viral disease when multiplex real-time was a polymerase chain reaction (PCR) signifying an unfamiliar source of COVID-19 [14]. Subsequently, on the 10th of January, 2020, bronchoalveolar lavage (BAL) fluid samples of the patients were sequenced, resulting in the unraveling of a pathogen with a similar genetic sequence to the betacoronavirus B lineage. This new pathogen had ∼96 %, ∼80 % and ∼ 50 % resemblance to the genomes of bat coronavirus RaTG13, SARS-CoV and MERS-CoV, respectively [13,14]. By 28th of March, 2020, the disease had extended to no less than 202 countries, infecting at any rate 575,444 people, and led to a minimum of 26,654 deaths worldwide. However, it is alleged that the overall cases of COVID-19 infections were underreported since mild or asymptomatic cases can be undetected [15]. For example, approximately 17.9 % of symptomless patients were reported on the Diamond Princess Cruise ship. Given that asymptomatic persons are similarly contagious as symptomatic persons, they can contribute to the disease's proliferation and widespread prevalence [16]. As of 1th of February, 2021, infectious cases have risen to slightly over >100 million, with >2 million recorded deaths [17].
Earlier, when the COVID-19 was spreading globally, clinician made decision to practice treatments of SARS and MERS viruses on COVID-19 infected patients, due to lack of antiviral therapies against the SARS-CoV-2. Corona virous identified as a highly pathogenic into the human population, still there is no effective antiviral treatment. The treatments basically focused on alleviating symptoms and offering respiratory support utilizing oxygen therapy, ventilators, etc. [18,19]. Shen et al. [20] stated that Administration of convalescent plasma derived from a COVID19-recovered patient with high SARS-CoV-2 specific antibody led to the enhance of COVID-19 symptoms. Moreover, the main protease (3 chymotrypsin-like protease (3CLpro) or Mpro) encoded by the viral genome is an attractive drug target because it plays an important role in cleaving viral polyproteins into functional proteins for novel coronavirus. Inhibiting this enzyme is an efficient strategy to block viral replication [21]. Viral papain-like cysteine protease (PLpro, NSP3) indicates promising target for the development of antiviral drugs. Rut et al. [22] performed study on combinatorial substrate library and activity profiling of SARS-CoV-2 PLpro. Elswhere, Lim et al. [23] stated significant reduction of SARS-CoV-2 viral load was reported after treating with protease inhibitor lopinavir and ritonavir in Korean patients diagnosed with COVID-19. Beside, the treatment, accessibility to recognized diagnostic technologies has increased the possibility of studies to plug-and-play in pneumonia diagnostics. Although such approaches require years to optimize scores, they now serve a crucial function in recognizing and managing the proliferation of COVID-19. Thus, the identification and detection of COVID-19 infection were built on lessons garnered from earlier outbreaks. Furthermore, the virus's morphology was determined microscopically, while genome sequencing was utilized for verification, and sequence data was employed to aid the development of PCR primers and probes [24]. These techniques, which were utilized to diagnose SARS-CoV and SARS-CoV-2, present the first line of protection against an outbreak. The subsequent steps being researched include: (a) serological tests, (b) point-of-care tests, in addition to (c) multiplex assays. Furthermore, isothermal amplification, microfluidic technologies and barcoding are being transformed into plug-and-play tests that might be quickly applied into an epidemic. The integration of smartphones with diagnostics can offer better communication and scrutiny [[25], [26], [27]]. Diagnostics plays a significant role in combating epidemics and disease outbreaks because they facilitate the efficient administration of resources and health efforts, which can help control the proliferation of contagious pathogens and lower the rate of mortality. Nonetheless, rapidly developing data on COVID-19 means specific details in this review may need to be altered or modified as recent studies sprout up.
The detection and neutralization of the COVID-19 virus using nanomedicine/nanotechnology is imperative as SARS-CoV-2 is spread via tiny droplets of viral particles, in the course of breathing, talking, sneezing, and coughing. Therefore, nanoparticles can be specialized to fight the causal microbes and disable the viral pathogens before they invade the body [28]. Nanoparticles also enable early diagnosis by interacting with the protein in SARS-CoV-2 to impart a distinct biological identity on the nanoparticle that differs from the pristine nanoparticle [29]. There have been disregarded factors, which are required for effective implementation of nanomedicine [30] such as the implication of the NP protein corona on drug-release profiles [31], disease-specific nanoparticles protein coronas [32], incubating temperature [33], cell division [34], cell age [35,36], cell sex [37], cell shape [38], and plasmonic activation of nanoparticles [39]. These factors limit and cause difficulties for the therapeutic efficacy of nanoparticles. Hence, there is a need for novel concepts such as disease-specific protein corona to ensure nanoparticle protein coronas' efficacy. Therefore, this study provides a concise review of the current and emerging detection and supervised technologies for SARS-CoV-2, which is the viral etiology of COVID19, and their performance characteristics. Also detailed are the reported efficacy as well as pros and cons of therapeutic options for the novel coronavirus.
2. Nanotechnology role
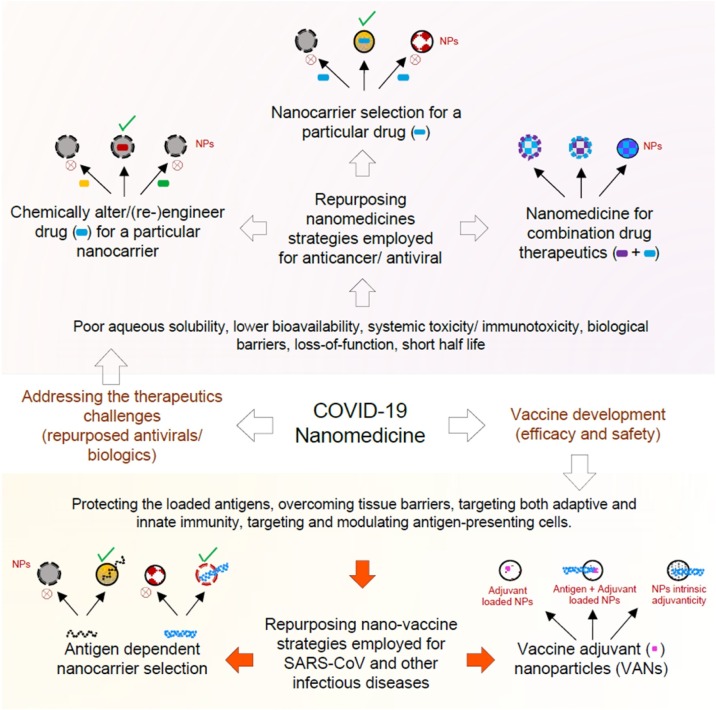
The COVID-19 pandemic also requires a critical assessment of available nanotechnologies. Although nanomedicine techniques are being used to develop vaccine carriers, there remain inadequate nanotechnology approaches to developing vaccines and treatment for the ongoing ravaging epidemic. Therefore, this paper aims to analytically address the existing state of nanotechnology in the production of therapeutics and vaccines. Therapeutic innovation and challenges to SARS-CoV2 infections are not quite isolated from other contagions compared to oncology studies [40]. Additionally, vaccine development has important commonalities with approaches for SARS and MERS [41,42]. It is therefore important to re-examine these therapeutic/vaccine approaches and the novel use of nanotechnology. Thus the goal is to review current studies on the application of nanotechnology in the potential diagnosis and treatment of COVID-19 (Fig. 1 ).
Fig. 1.
Nanomedicine approaches for therapeutics and production of COVID-19 vaccines [43].
3. Biological and clinical features of SARS-CoV-2
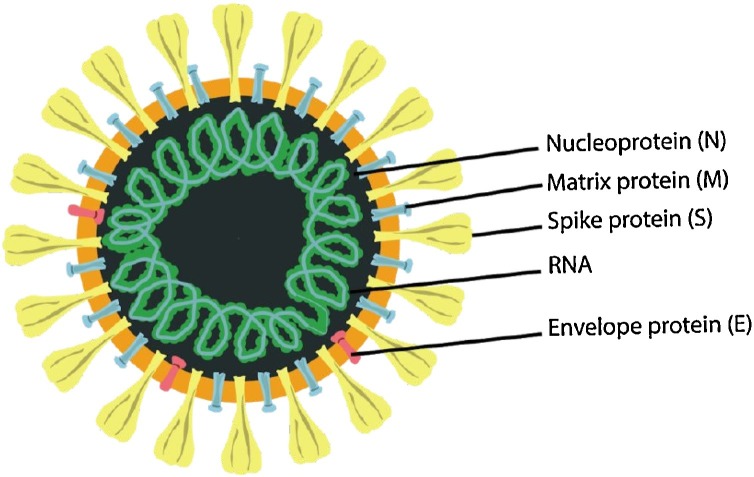
To determine the biological properties of SARS-CoV-2, human airway epithelial cells from patient samples in Wuhan were cultured with the BAL fluid virus taken from patients [44]. The supernatant was obtained from damaged or dead cells and then examined using transmission electron microscopy. Resembling the known coronaviruses (SARS-CoV and MERS-CoV), the diameter of COVID-19 virus ranges from 60 to 140 nm, and has an envelope characterized by protein spikes (thorn like structure) with genetic material [11,44] (Fig. 2 ).
Fig. 2.
The morphology of SARS-CoV-2 : illustration of the structure is presented with its structural viral proteins adapted from [11].
SARS-CoV-2 contains a single-stranded positive sense RNA genome (about 30,000 nucleotides long), which encodes 27 proteins in addition to an RNA-dependent RNA polymerase (RdRP) and 4 structural proteins (spike surface glycoprotein (S), small envelope protein (E), matrix protein (M), and nucleocapsid protein (N)) [45]. RdRP works in combination with nonstructural proteins to ensure genome conformity. The genome sequence of RdRP gene in SARS-CoV-2 was found to be a similarity of 96 % in RaTG13 a RdRP gene of bat [46]. The S gene codes as a target protein facilitates the infection of cells [47] by serving as a mediator between receptor binding as well as membrane fusion that controls host tropism and transmission performance [48]. In SARS-CoV-2, the nucleotide sequence of S gene is only about 75 % similar to all prior SARS-related coronaviruses [14]. The rest of the structural proteins as a target are less active than the spike protein but are essential for overall coronavirus function [24]. 99.9 % sequence homology was noted of the 104 strains sequenced in the period of December 2019 to middle of February 2020, although recent transformations in the viral genome were detailed, confirming greater diversity of sequence [49].
4. Clinical features of patients infected with COVID-19
To recognize clinical features characteristic of COVID-19 patients, Huang et al. [2] performed a cohort study on 41 confirmed 2019-nCoV patients. The patients showed severe, occasionally lethal, pneumonia and were placed in hospital beds of treatment centres selected for COVID-19 treatment in Wuhan, China, by Jan 2, 2020. Clinical manifestations were significantly similar to SARS-CoV. The patients with lethal forms of the infection developed acute respiratory distress syndrome (ARDS), thus needed to be admitted in ICU and subjected to oxygen therapy. The duration from admission to ARDS was an average of 48 h. Within this phase, the 2019-nCoV induced mortality rate rose to15 % (6 out of 41 patients in this cohort expired). The death cases continue to rise exponentially, reaching an estimated 11.1 million confirmed cases and about 525 thousand deaths globally as at 4th of July, 2020.
Furthermore, parallels were noted between the clinical symptoms of 2019-nCoV and preceding betacoronavirus infections. Huang et al. [2] reported most patients of 2019-nCoV infection manifested symptoms that include dry cough, dyspnoea, fever and bilateral ground-glass opacities on CT scans taken of the chest region, which are somewhat comparable to those of SARS-CoV and MERS-CoV infections [50]. Nonetheless, only a small number of 2019-nCoV patients were presented with major upper respiratory tract signs and symptoms such as sore throat and sneezing, signifying that the cells marked by the virus might be sited in the lower airway region. Moreover, unlike MERS-CoV or SARS-CoV infections which had 20–25 % of diarrhea cases [50], COVID-19 infected patients infrequently displayed intestinal issues. Nonetheless, fecal and urine samples have been tested to eliminate a possible substitute course of transmission. Kim et al. [51] detected COVID-19 in urine and faces samples intermittently. However, due to the inability to isolate the virus from the COVID-19 -positive samples, the possibility of viral transmission through stool and urine is anticipated to be low.
The 2019-nCoV patients have relatively higher concentrations of IL1B, IFNγ, IP10, and MCP1 [2], possibly resulting in activated T-helper-1 (Th1) cell responses. In addition, severe ICU cases exhibited higher amounts of (Granulocyte colony-stimulating facto) G-CSF, 10 kDa interferon gamma-induced protein (IP10), monocyte chemoattractant protein-1 (MCP1), MIP1A, and tumor necrosis factor alpha (TNFα) compared to mild or asymptomatic cases, indicating that the cytokine storm is related to infection severity. 2019-nCoV infection also instigated higher secretion of T-helper-2 (Th2) cytokines (e.g., IL4 and IL10) that restrain inflammation, in contrast to the SARS-CoV infection) [52]. Hence, additional research is needed to characterize the Th1 and Th2 responses in 2019-nCoV infection and to explicate their pathogenesis. Moreover, research on autopsy or biopsy of infected patients is crucial to fully elucidate the infection.
5. Current/emerging diagnostic tests for COVID-19
Expressions of symptoms COVID-19 infected patients are random, thus are unable to be employed for precise diagnosis. For instance, less than 45 % of all 1099 COVID-19 patients from China exhibited feverish symptoms, while 89 % caught a fever after being admitted [53]. They further discovered that 68 %, 38 %, 34 % and 19 % of the infected patients displayed fatigue, cough, shortness of breath, and sputum production, respectively. These symptoms are usually linked to other respiratory infections. Hence, emerging procedures that include nucleic acid testing and CT scans are utilized to screen and diagnose COVID-19.
Compared to syndromic testing and CT scans, molecular techniques show more effectiveness for accurate diagnosis because of their capacity to target and recognize particular pathogens. Since SARS-CoV-2 is the viral etiology of COVID19, detection details of SARS-CoV-2 will be presented. Although the genomic and proteomic structures of SARS-CoV-2havebeen recognized, the response of host to the virus remains under stud [11]. The foremost genome sequencing of COVID-19 was performed via RNA sequencing, which is an impartial and high outcome technique of different genome sequencing [54]. The results were released to the public and subsequently included in the GenBank sequence database in January 10, 2020 [55,56]. The Illumina sequencing is a technique that utilizes solid-phase bridge amplification, while nanopore sequencing is used to determine the DNA sequence via the translocation of a DNA molecule through a protein pore and measurement of ensuing voltage variations [57]. Moreover, the genome sequencing is vital to investigate PCR’s sequences by designing of primers.
5.1. Nucleic acid testing
Nucleic acid test is the existing principal technique for diagnosing COVID-19 (Division of Viral Diseases, 2020). Several reverse transcription polymerase chain reaction (RT-PCR) kits were developed for the genetic detection of SARS-CoV-2. RT-PCR entails reversely transcribing the COVID-19 RNA into strands of complementary DNA (cDNA), with a magnification of particular sections of the cDNA [58,59]. The process of designing RT-PCR kits normally takes two key steps: (1) aligning sequences and design of primers, and (2) assay optimization and assessment. A set of primers and probes were designed by aligning and analyzing numerous SARS-related viral genome sequences [60]. Amongst the viral genomes associated with SARS, the study found three regions (the RdRP, E and N genes) that exhibited conserved sequences. RdRP and E genes both possess high diagnostic sensitivity for detection (technical detection limit of 3.6 and 3.9 copies for every reaction) while the N gene showed 8.3 copies for each reaction. Therefore, the design of the assay is a two-target system comprising a primer that commonly detects many coronaviruses (such as SARS-CoV-2) and an additional primer that targets only the detection of SARS-CoV-2.
Based on the probes and primers' design, the assay conditions (e.g., incubation period, reagent type and condition, and temperature) are then optimized, with the next step being PCR testing. RTPCR can be conducted either in a one-step or a two-step assay. For the case of a one-step assay, the PCR amplification and reverse transcription are merged into a solitary reaction. This assay design offers rapid and duplicable data for high-throughput investigation. However, this format shows lower target amplicon generation. Alternatively, the two-step assay reaction is performed successively in different tubes [61]. The sensitivity of the two-step assay format is higher than the one-step assay, although it requires additional time to process and entails the optimization of extra parameters [61,62]. Finally, there is the need to meticulously select controls to guarantee the assay's integrity and recognize experimental inaccuracies.
The use of isothermal amplification in nucleic acid tests is presently being developed to detect SARS-CoV-2. The benefits of isothermal amplification methods include the ability to be deployed for a single temperature, and the non-requirement of dedicated laboratory gear to deliver analytical sensitivities comparable to PCR [63]. The isothermal amplification methods comprise recombinase polymerase amplification, helicase-dependent amplification and loop-mediated isothermal amplification (LAMP). A number of researches have initiated and clinically utilized RT-LAMP tests for detection of COVID-19 [64]. The RT-LAMP involves the binding of DNA polymerase and 4–6 primers to 6 different sections on the target genome. A four-primer system consists of two inner primers (forward and reverse inner primers) and two outer primers. LAMP is exceedingly precise since it employs a more significant amount of primers. When conducting the LAMP diagnosis, the samples are fed into the tube where the amplified DNA is then detected via a number of means that include turbidity (a derivative of the reaction), color (incorporation of a pH-sensitive dye), or fluorescence (inclusion of a fluorescent dye which adsorbs to DNA) [65]. The reaction duration takes below 1 h at 60–65 °C with an analytical detection of approximately 75 copies per μL. The technique is characterized by a simplistic operation, ease of detection visualization, reduced background signal, and non-requirement of a thermocycler. The limitations of LAMP include the optimization problem of primers and reaction parameters. Additional isothermal amplification methods are still in development.
To raise the quantity of data obtained from a solitary test as well as enhance clinical sensitivity and specificity [66], multiplexing of isothermal amplification techniques at the amplification and/or readout stages is achieved using polymeric beads that are encoded with distinctive optical signatures for barcoding. For the detection of multiple analytes from samples of one patient in one reaction tube, barcodes were developed for diverse biomarkers in panels [67]. Agents that emit fluorescent signals are employed to encode distinctive signatures, with each code used to capture DNA or antibody on the bead surface. The presence of a sequence or antigen in a patient’s sample which connects the capture molecule of the bead with a secondary probe denotes positive detection. There exist barcoded-bead multiplex panels that are engineered for the diagnosis of cystic fibrosis and respiratory infections [68], but work are in progress to expand them for the point-of-care. However, this approach is hindered by limitation that can be resolved through the use of inorganic quantum dots for barcoding, which facilitates battery-operated excitation and enables the capture of emission signals using smartphone cameras. Clinical sensitivity and specificity values of 95 %and 91 % were recorded by means of quantum dot barcodes to diagnose hepatitis B infected patients following the isothermal reverse polymerase amplification [69].
Other nucleic acid tests utilized for the detection of COVID-19 virous is SHERLOCK, which is a diagnostic technique that employs Cas13a ribonuclease for the detection of RNA [70]. This approach entails the reverse transcription of viral RNA targets to cDNA, followed by isothermal amplification by means of reverse polymerase amplification. The amplified products are then reversely transcribed into RNA. The Cas13a eventually forms a complex with a RNA guide sequence that is attached/bind to the amplified RNA product [71]. Cas13a is activated following the process of target binding. Afterwards, Cas13a cleaves the adjoining fluorophore-quencher probes to generate a fluorescent signal. Previous research using SHERLOCK detected approximately 2000 copies per mL in serum or urine isolates for viruses such as Zika [72]. A SHERLOCK protocol for the detection of SARS-CoV-2 has been launched, while an additional Cas13a-based detection method was evaluated using SARS-CoV-2 clinical isolates [73].
5.2. Computed tomography (CT)
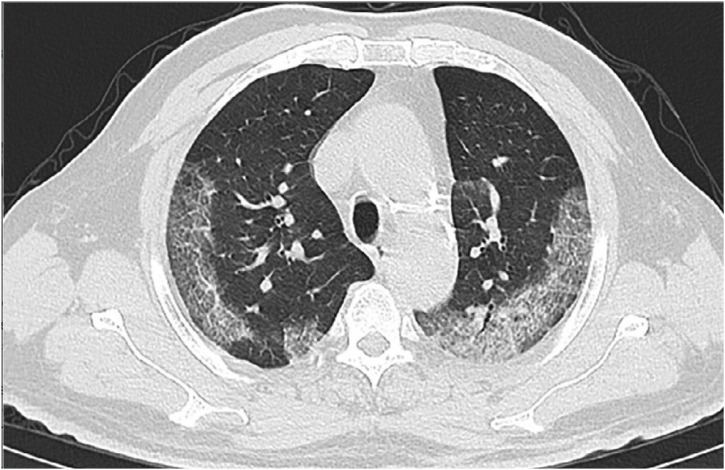
Due to the scarcity of apparatus and the high rate of RT-PCR's false negatives, CT scans were temporarily utilized to clinically diagnose COVID-19 in Hubei Province, China [64]. Chest CT scans are noninvasive and entail the acquisition of several X-ray images at diverse directions that traverses a patient's chest to generate cross-sectional depictions [74]. The goal of generating these images is to search for anomalous textures and features that can bring about a diagnosis [75]. Generally, normal CT findings were reported for 56 % of the cases examined in the preliminary stages of the infection (0–2 days) [76], with the highest lung involvement recorded at approximately 10 days after the inception of symptoms [77].
The prominent characteristic CT features of COVID-19 are (a) bilateral and peripheral ground-glass opacities (regions of indistinct/foggy opacity) and (b) compaction of the lungs (fluid or solid material in compressible lung tissue) [76,77]. The ground-glass opacities are mostly evident 0–4 days following the emergence of symptom. As COVID-19 advances, crazy-paving patterns (i.e. irregular shaped paved stone pattern) begin to emerge, followed by rising consolidation of the lungs [78,79]. Anchored in these CT features of COVID-19, a number of studies demonstrated that CT scans exhibit a higher sensitivity (86–98 %) and lowered false negative rates than RT-PCR. However, the major limitation of CT diagnosis for COVID-19 is its low specificity (25 %) due to the overlap of imaging features with characteristic texture of other viral pneumonia [13].
COVID-19 is currently identified using RT-PCR and subjected to screening with CT scans; however, each method has its limitations. The use of RT-PCR is encumbered by the low availability of PCR reagent kits; lack of PCR infrastructure in community infirmaries on the outskirts of urban cities to hold high sample throughput, and the dependence on the existence of evident SARS-CoV-2 in collected samples. Thus, PCR cannot identify the occurrence of SARS-CoV-2 infection in asymptomatic patients but has since recovered, hence complicating the implementation of control measures. On the other hand, CT systems are costly, involve technical proficiency, and are incapable of specific diagnosis of COVID-19 (Fig. 3 ). Therefore, additional technologies should be modified to resolve these deficiencies in SARS-CoV-2 diagnosis.
Fig. 3.
Typical chest CT scan of the patient who had symptoms of COVID-19 pneumonia [80].
5.3. Protein testing
Viral protein antigens and antibodies, which develop in response to SARS-CoV-2 infection, can be utilized to diagnose COVID-19. However, alteration in viral load across the infection duration may complicate the detection of viral proteins. For instance, high salivary viral loads were observed in the initial few days after the inception of symptoms, accompanied by a gradual decrease over time [81]. On the contrary, antibodies created in reaction to viral proteins possibly will offer a lengthier duration for the indirect detection of SARS-CoV-2. Antibody tests can be mainly valuable for COVID-19 surveillance. A potential limitation that confronts the development of precise serological tests is the possible cross-reaction of SARS-CoV-2 antibodies with antibodies in opposition to other coronaviruses. The possibility of frequent cross-reactivity in COVID-19 patients was confirmed in an earlier report [82].
Serological tests are presently in development [83]. Using the SARS-CoV-2 Rp3 nucleocapsid protein, Zhang et al. [84] was able to detect immunoglobulin G and M from serum of COVID-19 infected cases on an enzyme-linked immunosorbent assay. The presence of anti-SARS-CoV-2 IgG is confirmed if it is packed in the middle of adsorbed nucleoprotein and anti-human IgG probe, leading to a positive signal. Zhang et al. [84] tested 16 infected patients (earlier validated by RTPCR). They discovered that the volume of antibodies rises in the first five days following the emergence of symptoms. Positivity for immunoglobulin M (IgM) and immunoglobulin G (IgG) increased from 50 % and 81 % of patients on day zero to 81 % and 100 % on the fifth day, respectively [80]. Based on the up-to-date findings, other markers (e.g., protein and cellular) can be utilized for D-dimer, in addition to low levels of leukocytes, lymphocytes and number of blood platelets [85]. However, the use of these biomarkers is constrained because they can also be of anomalous levels in other infections. Thus, a multiplex examination that combines antibodies and small molecule markers may enhance the test's specificity.
5.4. Point-of-care testing
Point-of-care tests are utilized for the diagnosis of patients with no need to send samples to central diagnostic centers, thus facilitating the detection of infected patients in places devoid of laboratory equipment and infrastructure. An example of a point-of-care technique being developed for the diagnosis of COVID-19 is the lateral flow antigen detection for SARS-CoV-2 [86]. In the case of lateral flow assays developed for commercial purposes, a membrane is coated with two lines: one for conjugating with gold-antibody nanoparticle (AuNP-ab) conjugates and the other to capture antibodies. The sample is placed on the paper-like strip and the proteins are then spread over the strip via capillary action. As it crosses the first line, the antigens conjugated with the Au nanoparticle-antibody and the complex flow together through the membrane. As they reach the second line, the complex is rendered immobile by the capture antibodies and a red or blue line becomes visible. Individual AuNP is red, although a solution that contains agglomerations of AuNP nanoparticles is blue due to the blend with the plasmon band. The clinical sensitivity of lateral flow assay is 57 %, a specificity of 100 %, and accuracy of 69 % for IgM, in addition to an 81 %, 100 %, and 86 % for IgG, respectively. Clinical sensitivity of 82 % has been reported for the detection of both IgM and IgG [86]. Formerly, a RT-LAMP test was integrated with lateral flow readout for the detection of MERS-CoV [87]. However, these tests can be used just once and are characterized by low analytical sensitivity compared to RT-PCR. Hence, an array of signal amplifying methods was developed (e.g., assembly of multiple Au nanoparticles; thermal imaging) to enhance the assay readout signal [88].
The use of microfluidic devices is another approach for the implementation of point-of-care. Apart from being handy, the main benefits of using microfluidics comprise smallness or miniature scale, sample requirement of small volume, and fast detection periods. These devices are composed of a macro chip with micro-and made from poly-dimethylsulfoxide, glass, or paper. Laksanasopin et al. [89] created a microfluidics-based smartphone accessory to detect antibodies against 3 sexually transferred infections by serially moving reagents pre-stored on a cassette. The platform demonstrated clinical sensitivity and specificity of 100 % and 87 %, respectively, for human immunodeficiency virus (HIV), when tested in 96 patients in Rwanda. These technologies can be modified for the detection SARS-CoV-2 RNA or proteins.
In this review, we provided some emerging diagnostic technologies that have exhibited clinical practicability, which can be modified to detect SARSCoV-2. There are several platforms under development in research labs such as electrochemical sensors, paper-based systems, and surface-modified Raman scattering-based systems. Nonetheless, such approaches are within the preliminary phase of development, hence cannot be deployed to directly diagnose COVID-19. For nucleic acid biomarkers, the platforms include clustered regularly interspaced short palindromic repeats (CRISPR) based on recombinase polymerase amplification (RPA) and RT-RPA technologies, LAMP, RPA, NASBA, rolling circle amplification (RCA), RT-LAMP, quantum dot barcode using barcode technology, and magnetic beads. In the case of protein biomarker, the platforms comprise Smartphone dongle based on Enzyme-linked immunosorbent assay (ELISA) technology, paramagnetic beads based on magnetic sensor technology, ELISA, SIMOA based on digital ELISA, Biobarcode Assay using DNA-assisted Immunoassay, and rapid antigen test based on lateral flow technology. Magnetic bead isolation using magnetic separation technology is also a researched bacteria biomarker-based diagnostic tool. The development of diagnostic technology can be delineated into four phases: conception (i.e., design and verification with synthetic samples); clinical testing (small patient cohorts. i.e., <100 samples); clinical trial (large patient cohorts), and commercialization (launch and patient use). These up-and-coming technologies serve a crucial role in the detection of viral diseases in future pandemics.
5.5. Smartphone surveillance
The control of epidemic demands widespread surveillance, distribution of epidemiological data, and patient monitoring [90]. Health organizations, ranging from neighborhood clinics to the WHO, need gear and equipment that can enhance the rate and easiness of communication to control the proliferation of infections. Smartphones can be purposefully exploited as they have the computational power, connectivity (geospatial contact tracing) and hardware that enable electronic coverage, point-of-care testing, and epidemiological databasing (real-time data upload) (Fig. 4 ). Smartphones are extensively available for the management of responses during pandemics like COVID-19 because of the exponential increase in global smartphone use, counting sub-Saharan Africa [91].
Fig. 4.
Schematic diagram of smartphone diagnosis [92].
Furthermore, smartphones can be harmonized with established diagnostic tests to deliver real-time geospatial data that allows public and international healthcare agencies to execute synchronized control policies. Smartphones can digitize the contact tracing process to acquire more comprehensive and distributable records. Suspected cases of COVID-19 infection can run into communication hurdles with their healthcare providers. Moreover, persons with mild respiratory indications may be cautious about traveling to congested medical centers as they are confronted by a higher risk of getting infected with COVID-19. Smartphones can then be influenced to make available an uninterrupted line of contact between healthcare providers and patients without the risk of contagion on both sides. Smartphone apps can connect mental health counselors to patients to deal with isolation and anxiety experienced during self-quarantine and pandemics [9,83]. Furthermore, patients can personally report symptoms and tendencies, enabling distant screening and observation by health care providers [93]. This mode of reporting also provides epidemiologists with data pertinent to potential transmission mechanisms.
5.6. Nano-based diagnostic tests for COVID-19
In his paper on emerging biomolecular testing to evaluate the risk of mortality from COVID-19 Infection, Mahmoudi [29] suggested a number of potential nanobased technologies (protein corona sensor array and magnetic levitation) that can classify potentially high-risk patients at the early stages of the COVID-19 infection. In biomolecular corona, the intrusion of nanoparticles into a biological environment (e.g., human blood) results in their immediate interaction with various biomolecules, such as proteins. These biomolecules form a coating layer on the nanoparticle surface, i.e., biomolecular corona, thus imprinting a distinctive biological identity on the nanoparticle, which could significantly differ from the original nanoparticle surface [29]. Therefore, the protein corona sensor array technology may define the plasma protein/biomolecule patterns that denote fatal COVID-19 infection at the preliminary phase. Although the bulk of biomolecular corona is engaged by proteins, the other biomolecules (e.g., lipids, metabolomes, and nucleic acids) may have diagnostic capacities that are accessible in the corona structure. Following the identification of a protein pattern to act as a “fingerprint” of high-risk COVID-19 infected patients, colorimetric nanotechnologies (including optoelectronic nose [94] and plasmonic nanoparticle technologies [95]) can also be advanced or modified for point-of-care identification of these susceptible patients.
On the other hand, magnetic levitation (MagLev) is a rapid, uncomplicated, and effective technique for the levitation of nonbiological and biological species, including different types of materials and biogenic cells incubated in paramagnetic fluid [96]. The design of a MagLev device comprises two permanent magnets with two like-poles opposite each other along the vector of gravity. Although proteins are unstable in conventional MagLev paramagnetic solutions, superparamagnetic iron oxide nanoparticles (SPION) can deal with this issue and levitate plasma proteins [92]. Supervised classification analysis confirmed the proof-of-concept that the motion of levitating proteins in MagLev possesses disease detection capacity, with potential use in a point-of-care device by providing important information on the health status of infected individuals [97].
Combining these proposed Nano-based diagnostic approaches with advanced supervised classifiers, the patterns of biomolecules (comprising proteins, nucleic acids, metabolomes, and lipids) characterized by fatal risk diagnostic capacity will be defined [29]. Also, recent studies [98,99] reported a variation of nonprotein biomolecules (e.g., metabolomes) can considerably modify the corona composition of proteins, which can further enhance the diagnostic capacity of the corona sensor array system. In addition, by integrating the protein corona sensor array system with other sensor array elements, its diagnostic accuracy becomes enhanced. However, biomolecular corona and magnetic levitation do not have specific biomarkers or nucleic acid to detect, unlike conventional assays. Therefore, the proposed approaches require the complex efforts of first collecting human plasma and other nonplasma biological fluids from a significant population of COVID-19-infected patients at both fatal and nonfatal stages, followed by evaluation of the collected biological fluids using MagLev and protein corona sensor array platforms, and then analysis using omics techniques and machine learning to classify the library of biomolecular patterns that denote high association with COVID-19 cases that pose the highest risk of mortality.
6. Clinical trials for COVID-19 treatment
Regrettably, drugs or vaccines are yet to be approved for the treatment of human coronaviruses. Nonetheless, several options can be deployed or envisaged to control or prevent the rising 2019-nCoV infection, including vaccines, monoclonal antibodies, oligonucleotide-based therapies, peptides, interferon therapies and small-molecule drugs [100]. However, novel interventions are anticipated to take a lengthy duration (months to years) to develop. Given that treatment measures are urgently required for the ongoing 2019-nCoV outbreak, the urgency of the existing antiviral agents approved or in development for treating infections caused by HIV, hepatitis B virus (HBV), hepatitis C virus (HCV) and influenza [101] are being repurposed to treat SARS-CoV-2 infection, based on therapeutic experiences with SARS and MERS.
Considering the high levels of cytokines induced by SARS-CoV [102] MERS-CoV [103] and 2019-nCoV infections, corticosteroids were regularly used to treat patients with severe illnesses resulting from these infections, with the aim of reducing inflammatory-induced lung injury. However, rather than hinder the effect on mortality, evidence showed that corticosteroids delayed viral clearance [104,105]. Hence, corticosteroids should not be regularly prescribed systemically, according to WHO interim guidance [106]. To confirm their efficacy, Huang et al. [107] administered corticosteroids to very few non-ICU cases among a cohort of 41 cases with 2019-nCoV infection and low-to-moderate dose of corticosteroids to less than half of severely ill patients with ARDS. By Jan 22, 2020, 28 (68 %) of the 41 patients were discharged and six (15 %) patients died. Therefore, further evidence is required without delay to evaluate the supposed efficacy of systematic corticosteroid treatment of patients infected with 2019-nCoV.
6.1. Virally targeted agents
So far, there is no confirmed, effective antiviral treatment for coronavirus, particularly 2019-nCoV infection. Historically, the combined drug therapy of lopinavir and ritonavir for SARS-CoV infection was linked to significant medical benefits (fewer fatal clinical results) [108]. Arabi et al. [109] performed a placebo-controlled interferon beta-1b, lopinavir, and ritonavir trial for MERS infected patients in Saudi Arabia. Preclinical evidence confirmed the potency of remdesivir (a broad-spectrum antiviral nucleotide prodrug) for the treatment of MERS-CoV and SARS-CoV infections [110]. Given that 2019-nCoV is a novel coronavirus infection, there is no effective antiviral treatment. Nonetheless, the availability of the combination of lopinavir and ritonavir in the designated medical centers has facilitated the randomized controlled trial to assess the potency and safe use of a combination of lopinavir and ritonavir for 2019-nCoV infected patients.
Furthermore, both approved and experimental nucleoside analogues may be potentially effective in the treatment of 2019-nCoV [111]. Nucleoside analogues (favipiravir and ribavirin), which are derivatives of adenine or guanine, target the RNA-dependent RNA polymerase and obstruct viral RNA synthesis in a wide range of RNA viruses, as well as human coronaviruses [112]. Favipiravir (T-705) was shown to successfully inhibit the RNA viruses such as yellow fever, influenza, norovirus, Ebola and enterovirus [112]. A recent research [113] reported the activity of Favipiravir against 2019-nCoV (EC50 = 61.88 μM in Vero E6 cells). Therefore, those infected with 2019-nCoV are being enlisted in randomized trials to assess the potency of favipiravir combined with interferon-α (ChiCTR2000029600) as well as favipiravir combined with baloxavir marboxil (ChiCTR2000029544). Ribavirin is a guanine derivative approved for the treatment of Hepatitis C(HCV) and respiratory syncytial virus (RSV) that exist in patients infected with SARS and MERS. However its side effects (e.g., anemia) may be acute if given at high dosages [114], in addition to the uncertainty regarding its sufficient potency against 2019-nCoV.
Experimental nucleoside analogues (remdesivir and galidesivir) have also shown potential. Remdesivir (GS-5734) is a phosphoramidate prodrug of an adenine derivative with a chemical composition close to that of tenofovir alafenamide, which is a standard HIV reverse transcriptase inhibitor. A recent research detailed that remdesivir inhibited 2019-nCoV (EC50 = 0.77 μM in Vero E6 cells) [113] in a US patient who recovered after receiving remdesivir intravenously [53]. The outcome of two phase III trials performed in early February to assess the potency of intravenous remdesivir (200 mg on day 1 and 100 mg on a daily basis for 9 days) in patients infected with 2019-nCoV (NCT04252664 and NCT04257656), are anticipated in April 2020. Galidesivir (BCX4430), an adenosine analogue that was initially created for hepatitis C virus (HCV), is presently undergoing early-stage clinical studies to evaluate its safety in healthy persons and its potency against yellow fever, and has demonstrated antiviral activities against several RNA viruses, counting SARS and MERS [114].
Approved protease inhibitors (e.g., disulfiram, lopinavir and ritonavir) have demonstrated antiviral actions to mitigate SARS and MERS. Disulfiram is approved for treating alcohol dependence/addiction that has an inhibiting effect on the papain-like protease of MERS and SARS in cell cultures. However there is still lack of clinical proof of its effectiveness. Clinical trials (e.g., ChiCTR2000029539) have been launched to analyze HIV protease inhibitors such as lopinavir and ritonavir in patients infected with 2019-nCoV. Lopinavir and ritonavir were initially theorized to hinder the 3-chymotrypsin-like protease of SARS and MERS, and appear to be linked to enhanced clinical outcomes of patients infected with SARS in a non-randomized open-label experiment [114]. Arguably, HIV protease inhibitors could successfully inhibit the 3-chymotrypsin-like and papain-like proteases of 2019-nCoV. This is because HIV protease is a member of the aspartic protease family, while the two coronavirus proteases belong to the cysteine protease family. Moreover, HIV protease inhibitors were specially optimized to fit the C2 symmetry in the catalytic site of the HIV protease dimer, but this C2-symmetric pocket is not present in coronavirus proteases. If HIV protease inhibitors have to modify host pathways to circuitously inhibit coronavirus infections, then their efficacy is still a debatable issue.
Another potential target is spike glycoprotein. Griffithsin is a red-alga-derived lectin that can bind to oligosaccharides on the surface of different viral glycoproteins, such as HIV glycoprotein 120 and SARS-CoV spike glycoprotein [114]. Although Griffithsin was subjected to tests in phase I studies as a gel or an enema to prevent HIV, its efficacy and slow release of spike inhibitors need to be re-assessed to treat or prevent 2019-nCoV.
6.2. Host-targeted agents
PEGylated interferon alfa-2a and -2b, approved for HBV and HCV treatment, could be repurposed to stimulate innate antiviral responses in 2019-nCoV infected patients, and trials relating to interferons have been launched, such as the trial of the approved anti-HCV combination of a PEGylated interferon plus ribavirin (ChiCTR2000029387) [111]. However, the combinatorial action of PEGylated interferon and a nucleoside compound against 2019-nCoV has not been validated. Due to the numerous adverse effects linked to subcutaneous interferon therapies, their assessment should be well scrutinized and dose cutback or therapy termination may be necessary. Other small-molecule agents approved for other infections may serve as a modulator for the virus-host interactions of 2019-nCoV. An approved immune modulator, chloroquine, exhibits inhibitory effects against 2019-nCoV (EC50 = 1.13 μM in Vero E6 cells) [115] and is being assessed as an open-label trial (ChiCTR2000029609), although the trial has been discontinued by the World Health Organization (WHO) due to concerns of associated heart complications. Nitazoxanide, which is approved for diarrhea treatment, has shown inhibitory tendencies toward 2019nCoV (EC50 = 2.12 μM in Vero E6 cells) [113]. However, the antiviral potency of such agents needs to be evaluated using clinical studies. Moreover, despite the numerous attempts to develop host-targeted small molecules against viral infections in the last 5 decades, only maraviroc has been approved by the food and drug administration (FDA), for HIV treatment [101].
7. Nanomedicine against COVID-19
SARS-CoV-2 has a similar size range (60–140 nm) and shape (spherical) [44] compared to the well-researched synthetic nanoparticles [116,117]. Thus, it is probable that one of the mechanisms that explain chloroquine-mediated effects against SARS-CoV-2 is a wide-ranging decline in cells' ability to execute clathrin-mediated endocytosis of nanosized structures, which is attributable to the suppression of phosphatidylinositol binding clathrin assembly protein (PICALM). Chloroquine is a weak base that becomes captured in membrane-enclosed low pH organelles, disrupting their acidification [118]. The implicit chloroquine-induced antiviral effects comprise inhibition of pH-dependent viral fusion/replication, aversion towards viral envelope glycoprotein and host receptor protein glycosylation. Chloroquine may also hinder virion assembly in the endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-like structures. In addition, it is plausible that chloroquine displays host effects, regardless of direct viral action, through attenuation of the expression of pro-inflammatory factors and receptors that can induce acute respiratory distress syndrome, which basically accounts for coronavirus-inflicted deaths [14].
It has been acknowledged that coronaviridae invade the host cells via receptor-mediated endocytosis, with direct fusion with the plasma membrane also noted. For instance, the SARS-CoV virus and human coronavirus NL63 (HCoV-NL63) recognized in 2003 and 2004, respectively, can bind to the angiotensin-converting enzyme 2 (ACE2) receptor, thereby activating endocytosis driven cell entry [119,120]. Both clathrin-mediated and clathrin/caveolae-independent endocytosis mechanisms were described for SARS-CoV entry in human cells [119,121]. SARS-CoV-2 might use comparable ACE2+ mediated mechanisms of cell entry [47]. Furthermore, chloroquine-induced prevention of endosome–lysosome fusion is expected to disrupt general endocytic trafficking, such as membrane receptor recycling, which is considered key for SARS-CoV-2 cellular entry. However, preceding research revealed that though chloroquine has therapeutic activity against SARS-CoV in cell culture, it cannot modify cell-surface levels of ACE2. Besides, therapeutic doses of chloroquine did not considerably alter the biosynthesis or glycosylation of the SARS-CoV spike glycoprotein. Instead, terminal glycosylation of the ACE2 receptor was damaged, affecting viral binding [122]. Chloroquine displays anti SARS-CoV activity in cell culture even when given after the virus's uptake [122], indicating that several intrinsic mechanisms may contribute to its efficacy. Upon invading into cells through endocytosis, the spike protein on the SARS-CoV virion surface is cleaved by local endosomal proteases (e.g., cathepsins), activated by acidification of the endosome. This cleavage transforms the spike protein bringing the viral envelope and the endosomal membrane together to facilitate a fusion. Chloroquine-induced inhibition of endosomal acidification possibly alters this fusion occurrence, thereby halting the virus in endosomes.
Initially, there was guarded optimism that (hydroxy) chloroquine may have prophylactic and/or therapeutic effects against COVID-19, thus elucidating the mechanisms that control these drugs' effect on SARS-CoV-2 is crucial for optimizing and developing preventive and treatment strategies. However, further research by WHO into (hydroxy) chloroquine has been halted due to associated heart complications (irregular heart rates/rhythms).
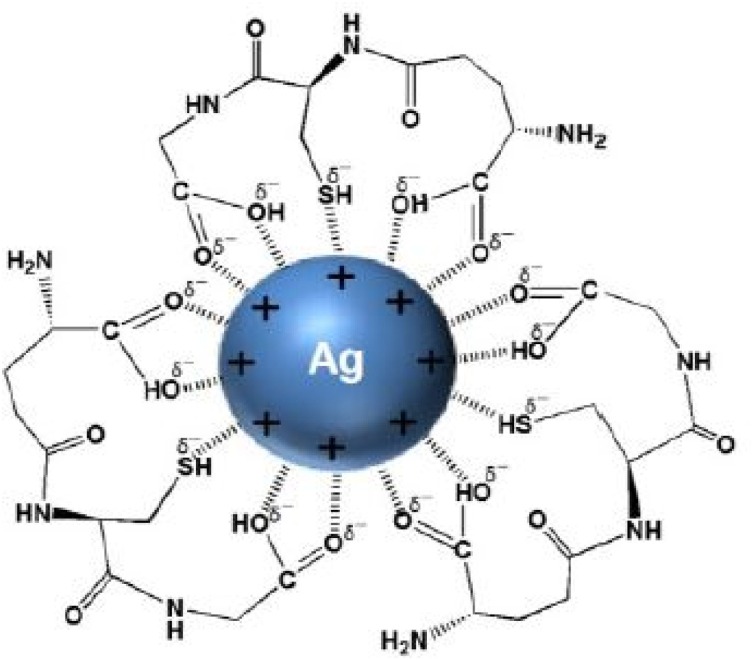
Furthermore, Zachar [123] formulated a treatment serum and protocols based on nano-silver colloids (NagC) to suppress the proliferation of both viral and bacterial respiratory infections. The formulations were delivered via inhalation. The study reported high sensitivity of dosage to the NagC nanoparticles' size, with 3 nm – 7 nm being the optimal size. Effective anti-viral inhibitory concentration (IC) of 10 μg/mL in the mucus of the respiratory system was determined to be the realistic target concentration to attain. The study determined that IC can be achieved in the bronchial tree and alveoli by depositing a total of approximately 0.33cc of a 30 μg/mL NAgC concentration, with 5 nm colloidal silver particles, and delivery inhalation of standard 5 μm diameter droplets aerosol (e.g., using off-the-shelf ultrasonic mesh nebulizers). Under oral breathing of 5 μm aerosol droplets, the tissue deposition fractions include pharynx (30 %), bronchial tree (30 %), and alveoli (25 %). The study recommended that the exhaled fraction of NAgC can be delivered via exhalation through the nose or orally to the nasal cavity by performing nebulized inhalation. Fig. 5 shows the synthesis of silver nanoparticles using GSH [123].
Fig. 5.
Silver nanoparticles prepared by green synthesis method using Glutathione (GSH) [123].
Furthermore, at the prevention phase, both nanofiber-based facial respirators and nanotechnology-enabled highly effectual antimicrobial and antiviral disinfectants are the initial personal protective approach that can avert the proliferation of the virus. In addition, a nano-based vaccine for COVID-19 treatment is currently under development. In diagnostics, nanotechnology has shown significant potential in the design and fabrication of sensors for rapid detection of COVID-19 infection.
8. Attenuated virus vaccines
Vaccines were developed to cure and/or prevent being infected with coronaviruses. For instance, the inclusion of a mutation (Y6398 H) into the Orf1a/b polyprotein (p59/nsp14/ExoN) was demonstrated to fully attenuate virulence of mouse coronavirus (MHV-A59), which is referred to as attenuated virus vaccine. The attenuated MHV virus displayed lowered replication of the virus in mice on the fifth day following intracerebral inoculation [9]. Secondly, antigen-specific CD8 + T cell-mediated responses were developed to fight antigens of the SARS coronavirus. Mice were vaccinated with DNA-coated gold particles against a calreticulin − nucleocapsid fusion protein, using gene gun delivery. The vaccination led to effective nucleocapsid-specific humoral and T cell-mediated immune responses. The vaccinated animals were able to considerably decrease the titer of a complicated vaccinia vector that expresses the N protein of the SARS virus. DNA-based vaccines containing immunogens have also been patented to target MERS-CoV. The vaccines are made up of consensus proteins based on the MERSCoV spike protein. The consensus spike protein considerably stimulates both humoral and cellular immune responses, counting augmented IgG titers and neutralizing antibodies. The stimulated cellular immune response is also associated with augmented CD3+CD8+ and CD3+CD4+ T cell responses that generate IFN-γ, TNF-α, IL-2, or both interferon-gamma (IFN-γ) and TNF-α. On March 3, 2020, Inovio Pharmaceutical, Inc. publicized their design of the DNA vaccine named INO-4800 to be tested on humans in the united states by April [9].
To treat COVID utilizing photodynamic therapy (PDT), the most appropriate method for delivering photosensitizer (PS) to the respiratory system would be by a nebulizer, details of that we are currently exploring. Methylene Blue (MB) influenced PDT in a topical setting is safe, with no expected morbidity. Among those with a tracheostomy, drugs and light are transmitted through a tracheostomy. The light could be transmitted through the cricothyroid membrane using a fine catheter.
As previously discussed, when photo-inactivated viruses behave like a vaccine, they can increase the immune response. The opposite should also be seen as a side effect in which inactivated viruses can reactivate and increase the virus's load [124]. The reversal of SARS-CoV-2 under asymptomatic circumstances may also spread the disease in healthy individuals. Another issue raised in the study is that PS cells' treatment showed internalization and infection of adenoviruses through a photochemical internalization mechanism. In this report, colorectal carcinoma cell lines have been pre-incubated with photosensitizing tetraphenyl porphyrin disulphonate (TPPS2a) or methylene blue derivatives (MBD) accompanied by adenovirus infection and light exposure. Fluorescent in situ hybridization (FISH) and RT-PCR techniques are then used to quantify viral DNA in cellular nuclei.
9. Nanoparticles for treating virus using photodynamic therapy
Multi-walled carbon nanotubes (MWCNTs) were reported as powerful antiviral agents. Visible light-induced excitation of the Porphyrin-conjugated MWCNTs considerably inactivated influenza A virus's ability to infect mammalian cells. This study has inspired others to establish MWCNTs for antiviral therapy. In another document, MWCNT arrays have been used to remove Enterohemorrhagic Escherichia coli (EHEC) serotype O157:H7 using TiO2 nanoparticles [125]. Graphene and graphene oxide sheets were also identified as antimicrobial agents. They were used independently by two different groups for the solubilization of hydrophobic PS hypocrellin A [126] and methylene blue [127]. In another study, an electrochemically produced graphene quantum dot (GQD) was used to kill two bacteria, E. coli and MRSA. The researchers were using a green laser (470 nm, 1 W) for photoactivation. This resulted in ROS, which selectively killed only bacteria and not mouse spleen cells [128].
Ag and Au nanoparticles have shown anti-HIV activity [129,130]. Au nanoparticles were used NPs to prevent T-cell HIV infection in vitro. AuNPs coated with multiple copies of the amphiphilic sulfate ligand bind to glycoprotein gp120 of the HIV envelope, thus affecting the adsorption/fusion process virus. AgNPs capped with mercaptoethane sulfonate (Ag-MES) showed an inhibitory effect of the Herpes Simplex type-1 virus (HSV-1). HSV-1 virus attachment and entry into cells involve interaction between the viral envelope glycoprotein and the heparin sulfate cell surface. Ag-MES NPs block the virus's entry into cells by competing to bind to cellular heparin sulphate through their sulfonate end-groups, thereby preventing infection [131]. AgNPs also demonstrated inhibition of replication in both Hepatitis B and respiratory syncytial viruses [132,133]. They used TiO2/Ag mixture for surface sanitization of buildings in Milan and found it effective against the virus [134].
9.1. Photodynamic therapy and COVID treatment
Depending on its non-invasive nature, PDT or photodynamic inactivation (PDI) overcomes conventional treatment methods such as surgery, radiotherapy and chemotherapy. In addition, there are very few or negligible adverse effects and reduced systemic toxicity [135]. A few researchers have described that PDI can inactivate all known microorganism classes, such as bacteria (Gram-negative and Gram-positive), viruses, protozoa and fungi, etc. [136,137].
PDT could be one of the supplementary therapeutic interventions to target SARS-CoV-2. Reactive oxygen species (ROS) primary sources for PS excitation can target viral membrane, protein, and SARS-CoV-19 RNA, contributing to the virus's inhibition [138,139]. Numerous PSs were tested against a range of viruses and have been shown to be effective in controlling their growth. It is preferable to have PSs that show the inhibition process over a wide range. To prevent the spread of the virus through platelet transfusion, Lin et al. [140] successfully inactivated SARS-CoV utilizing psoralen amotosalen-HCl in a platelet concentrate. Photochemical treatment involved 150 μM/L of amotosalen with UVA light (3 J/cm2) and achieved >5.8 log reduction in viral load. When emitted under UVA light, the psoralen compound 4'-aminomethyl-trioxsalene (AMT) inactivated MERS-CoV [141]. Riboflavin was used in another study conducted to inactivate plasma MERS-CoV. Riboflavin and UV light treatment significantly reduced the titer of the virus below the detection limit. It may help stop the transfusion of a virus via plasma products [142]. Also, there is a study on the use of photocatalytic titanium apatite filter (PTAF) for inactivation of the SARS-CoV UV and non-UV in the titanium photocatalysis section below [143]. In order to enhance the effectiveness of PDT toward COVID-19, the amalgamation of the latest innovations such as nanotechnology can become more successful. Nanoparticles have been extensively investigated for their various uses in diagnosing and treating different microbes and diseases.
10. Conclusion and recommendations
The accessibility to conventional diagnostic tools has empowered the concept of plug-and-play in the design of COVID-19 diagnostics. Although the optimization of these diagnostic tools takes years, they now play vital roles in identifying and managing the transmissivity of COVID-19. The lessons garnered from previous outbreaks of coronavirus have directed the advancements in COVID-19 diagnosis and treatment. Nonetheless, the quick identification of effective interventions in treatment measures and vaccinations against 2019-nCoV remains a significant debacle. Given the available information on the efficacy and safety profiles of interventions against infections closely related to COVID-19, existing antiviral agents are repurposed into an important near-term strategy for the treatment of 2019-nCoV.
The conventional tests emphasize the detection of particular biomolecules to validate or exempt the COVID-19 infection despite their critical risk or errors. The key benefit of the nanotechnology-based approaches, in contrast to the traditional diagnostic tests (e.g., nucleic acid and protein testing), is their ability to develop biomolecular pattern recognition for the various categories of biomolecules. This pattern recognition is vital for rapid and precise diagnosis of fatality-prone COVID-19 infections. The pattern recognition is based on the phenomenon of associating biomolecules with adapted plasma disparity and/or comorbidity.
There are currently few vaccines for nCoV infection such as Comirnaty (BNT162b2), Moderna COVID-19 Vaccine (mRNA-1273), COVID-19 Vaccine Astrazeneca (AZD1222), CoronaVac, and Convaxin. Many other trials are presently underway to test various treatment alternatives such as NVX-coV2373, ZyCov-D and VIR-7831. Besides, there are over 50 obtainable MERS and/or SARS inhibitors, which include galidesivir, compound 3k, protease inhibitors GC813 and helicase inhibitor (SSYA10-00). This variety of antiviral agents, immunotherapies and vaccines are being explored as prospective treatment therapies. Nonetheless, the pandemic has highlighted the critical need for improved and novel efforts to develop broad-spectrum therapies to combat 2019-nCoV infection. This emphasizes the research into nanoparticle treatment protocols such as nano-silver colloids proven to be an antimicrobial and antiviral disinfectant.
To avert wider proliferation of the 2019-nCoV infection in health-care locales, patients with fever and respiratory issues must be given close attention. Respiratory specimens should be immediately tested in cases of suspected diagnosis. Serum antibodies need to be examined among medical personnel prior to and following their contact with suspected 2019-nCoV patients to identify asymptomatic infections. Research should be expanded to cover primary care, outpatient, and community settings to develop a comprehensive perspective of clinical severity scale. Simultaneously, statistical analyses and p values should be thoroughly scrutinized, as non-significant p values do not automatically exempt disparity between ICU and non-ICU patients. Given that the causal pathogen has recently been discovered, the kinetics of viral load and antibody titers can be studied. Inadvertently, 2019-nCoV still requires exhaustive studies since it has evolved into a global health crisis, although reliable data has been generated regarding its transmission, infectivity and treatment. Dependable rapid pathogen tests and practicable range of diagnosis according to medical reports are critical for health workers exposed to suspected cases.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Authors’ contribution
MAD, PMK, MSJ and AAA conceived the study. MAD, PMK, MSJ and AAA, AAO and HAK contributed to acquisition of the data. MAD wrote the manuscript. MAD, PMK, AAA, MSJ, BM, AAO and HAK revised it critically for important intellectual content. All authors agree with the article submission. All authors read and approved the final manuscript.
Funding
This work was funded by the Malaysia Ministry of Higher Education FRGS grant [203/PFIZIK/6711678], and the authors would also like to thank School of Physics, Universiti Sains Malaysia for supporting this research work.
Data availability
All relevant data are made available in the manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgement
Would like to thank the academics, authors and researchers at Universiti Sains Malaysia for their efforts to make this research work readable.
References
- 1.Richman D.D., Whitley R.J., Hayden F.G. John Wiley & Sons; 2016. Clinical Virology. [Google Scholar]
- 2.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A. Commentary: Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.W.H. Organization . 2003. Summary of Probable SARS Cases With Onset of Illness.http://www.who.int/csr/sars/country/table2004_04_21/en/index.html from 1 November 2002 to 31 July 2003. [Google Scholar]
- 7.W.H. Organization . 2019. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) November, [EB/OL].(2019-11)[2020-01-25], in. [Google Scholar]
- 8.Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Publ. 2020 doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.W.H. Organization . 2020. Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (nCoV) Infection Is Suspected: Interim Guidance. Jan 11, 2020, in. [Google Scholar]
- 11.Udugama B., Kadhiresan P., Kozlowski H., Malekjahani A., Osborne M., Li V., Chen H., Mubareka S., Gubbay J., Chan W. 2020. Diagnosing COVID-19: The Disease and Tools for Detection ACS Nano. in, March. [DOI] [PubMed] [Google Scholar]
- 12.Zhong N., Zheng B., Li Y., Poon L., Xie Z., Chan K., Li P., Tan S., Chang Q., Xie J. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T., Jung S.-m., Linton N.M., Kinoshita R., Hayashi K., Miyama T., Anzai A., Yang Y., Yuan B., Akhmetzhanov A.R. Multidisciplinary Digital Publishing Institute; 2020. Communicating the Risk of Death From Novel Coronavirus Disease (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C., Wang Z., Wang G., Lau J.Y.-N., Zhang K., Li W. COVID-19 in early 2021: current status and looking forward. Signal Transduct. Target. Ther. 2021;6:1–14. doi: 10.1038/s41392-021-00527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S., Yang L., Zhang C., Xiang Y.-T., Liu Z., Hu S., Zhang B. Online mental health services in China during the COVID-19 outbreak. Lancet Psychiatry. 2020;7:e17–e18. doi: 10.1016/S2215-0366(20)30077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seth S., Batra J., Srinivasan S. COVID-19: Targeting Proteases in Viral Invasion and Host Immune Response. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q., Kang C. Progress in developing inhibitors of SARS-CoV-2 3C-like protease. Microorganisms. 2020;8:1250. doi: 10.3390/microorganisms8081250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti–COVID-19 drug design. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd4596. eabd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim J., Jeon S., Shin H.Y., Kim M.J., Seong Y.M., Lee W.J., Choe K.W., Kang Y.M., Lee B., Park S.J. Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of Lopinavir/Ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J. Korean Med. Sci. 2020;35 doi: 10.3346/jkms.2020.35.e79. e79-e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 25.Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., de Ramirez S.A.S., Valera E., Cunningham B.T., King W.P., Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y., Chen H., Mubareka S., Gubbay J.B., Chan W.C. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 27.Fozouni P., Son S., de León Derby M.D., Knott G.J., Gray C.N., D’Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dheyab M.A., Aziz A.A., Jameel M.S., Noqta O.A., Khaniabadi P.M., Mehrdel B. Excellent relaxivity and X-ray attenuation combo properties of Fe3O4@ Au CSNPs produced via Rapid sonochemical synthesis for MRI and CT imaging. Mater. Today Commun. 2020;25:101368. [Google Scholar]
- 29.Mahmoudi M. Emerging biomolecular testing to assess risk of mortality from COVID-19 infection. Mol. Pharm. 2020 doi: 10.1021/acs.molpharmaceut.0c00371. [DOI] [PubMed] [Google Scholar]
- 30.Mahmoudi M. Debugging nano–bio interfaces: systematic strategies to accelerate clinical translation of nanotechnologies. Trends Biotechnol. 2018;36:755–769. doi: 10.1016/j.tibtech.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Behzadi S., Serpooshan V., Sakhtianchi R., Müller B., Landfester K., Crespy D., Mahmoudi M. Protein corona change the drug release profile of nanocarriers: the “overlooked” factor at the nanobio interface. Colloids Surf. B Biointerfaces. 2014;123:143–149. doi: 10.1016/j.colsurfb.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Hajipour M.J., Laurent S., Aghaie A., Rezaee F., Mahmoudi M. Personalized protein coronas: a “key” factor at the nanobiointerface. Biomater. Sci. 2014;2:1210–1221. doi: 10.1039/c4bm00131a. [DOI] [PubMed] [Google Scholar]
- 33.Mahmoudi M., Abdelmonem A.M., Behzadi S., Clement J.H., Dutz S., Ejtehadi M.R., Hartmann R., Kantner K., Linne U., Maffre P. Temperature: the “ignored” factor at the nanobio interface. ACS Nano. 2013;7:6555–6562. doi: 10.1021/nn305337c. [DOI] [PubMed] [Google Scholar]
- 34.Rauch J., Kolch W., Mahmoudi M. Cell type-specific activation of AKT and ERK signaling pathways by small negatively-charged magnetic nanoparticles. Sci. Rep. 2012;2:1–9. doi: 10.1038/srep00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foroozandeh P., Aziz A.A., Mahmoudi M. Effect of cell age on uptake and toxicity of nanoparticles: the overlooked factor at the nanobio interface. ACS Appl. Mater. Interfaces. 2019;11:39672–39687. doi: 10.1021/acsami.9b15533. [DOI] [PubMed] [Google Scholar]
- 36.Dheyab M., Aziz A., Jameel M., Khaniabadi P., Mehrdel B., Khaniabadi B. Gold-coated iron oxide nanoparticles as a potential photothermal therapy agent to enhance eradication of breast cancer cells. J. Phys. Conf. Ser. 2020:012003. IOP Publishing. [Google Scholar]
- 37.Serpooshan V., Sheibani S., Pushparaj P., Wojcik M., Jang A.Y., Santoso M.R., Jang J.H., Huang H., Safavi-Sohi R., Haghjoo N. Effect of cell sex on uptake of nanoparticles: the overlooked factor at the nanobio interface. ACS Nano. 2018;12:2253–2266. doi: 10.1021/acsnano.7b06212. [DOI] [PubMed] [Google Scholar]
- 38.Farvadi F., Ghahremani M.H., Hashemi F., Hormozi-Nezhad M.R., Raoufi M., Zanganeh S., Atyabi F., Dinarvand R., Mahmoudi M. Cell shape affects nanoparticle uptake and toxicity: an overlooked factor at the nanobio interfaces. J. Colloid Interface Sci. 2018;531:245–252. doi: 10.1016/j.jcis.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Mahmoudi M., Lohse S.E., Murphy C.J., Fathizadeh A., Montazeri A., Suslick K.S. Variation of protein corona composition of gold nanoparticles following plasmonic heating. Nano Lett. 2014;14:6–12. doi: 10.1021/nl403419e. [DOI] [PubMed] [Google Scholar]
- 40.Kostarelos K. Nature Publishing Group; 2020. Nanoscale Nights of COVID-19. [DOI] [PubMed] [Google Scholar]
- 41.Ahn D.-G., Shin H.-J., Kim M.-H., Lee S., Kim H.-S., Myoung J., Kim B.-T., Kim S.-J. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19) J. Microbiol. Biotechnol. 2020;30:313–324. doi: 10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 43.Chan W.C. Nano research for COVID-19. ACS Nano. 2020 doi: 10.1021/acsnano.0c02540. [DOI] [PubMed] [Google Scholar]
- 44.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu J., Qiao F.L., Wang X.H., et al. A numerical study of transport dynamics and seasonal variability of the Yellow River sediment in the Bohai and Yellow seas. Estuar. Coast. Shelf Sci. 2011;95:39–51. [Google Scholar]
- 47.Berry J.D., Jones S., Drebot M.A., Andonov A., Sabara M., Yuan X.Y., Weingartl H., Fernando L., Marszal P., Gren J. Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J. Virol. Methods. 2004;120:87–96. doi: 10.1016/j.jviromet.2004.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z. On the origin and continuing evolution of SARS-CoV-2. Sci. Rev. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J.-M., Kim H.M., Lee E.J., Jo H.J., Yoon Y., Lee N.-J., Son J., Lee Y.-J., Kim M.S., Lee Y.-P. Detection and isolation of SARS-CoV-2 in serum, urine, and stool specimens of COVID-19 patients from the Republic of Korea. Osong Public Health Res. Perspect. 2020;11:112. doi: 10.24171/j.phrp.2020.11.3.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong C., Lam C., Wu A., Ip W., Lee N., Chan I., Lit L., Hui D., Chan M., Chung S. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller S., Chiu C., Rodino K.G., Miller M.B. Point-counterpoint: should we be performing metagenomic next-generation sequencing for infectious disease diagnosis in the clinical laboratory? J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01739-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheridan C. Coronavirus and the race to distribute reliable diagnostics. Nat. Biotechnol. 2020;38:382. doi: 10.1038/d41587-020-00002-2. [DOI] [PubMed] [Google Scholar]
- 56.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;10 doi: 10.1038/s41586-020-2008-3. [published online ahead of print February 3, 2020] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Freeman W.M., Walker S.J., Vrana K.E. Quantitative RT-PCR: pitfalls and potential. Biotechniques. 1999;26:112–125. doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- 59.Kageyama T., Kojima S., Shinohara M., Uchida K., Fukushi S., Hoshino F.B., Takeda N., Katayama K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corman V., Bleicker T., Brünink S., Drosten C., Zambon M. Vol. 17. World Health Organization; 2020. (Diagnostic Detection of 2019-Ncov by Real-Time RT-PCR). Jan. [Google Scholar]
- 61.Wong M.L., Medrano J.F. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 62.Pfaffl M.W., Bustin S. 2004. AZ of Quantitative PCR, International University Line. La Jolla, CA. [Google Scholar]
- 63.Craw P., Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 64.Yang W., Yan F. Patients with RT-PCR-confirmed COVID-19 and normal chest CT. Radiology. 2020;295 doi: 10.1148/radiol.2020200702. E3-E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 66.Elnifro E.M., Ashshi A.M., Cooper R.J., Klapper P.E. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000;13:559–570. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han H.-S., Kim D.-S. Springer; Netherlands: 2016. Magnetic Levitation. [Google Scholar]
- 68.Dunbar S.A. Applications of Luminex® xMAPTM technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J., Biondi M.J., Feld J.J., Chan W.C. Clinical validation of quantum dot barcode diagnostic technology. ACS Nano. 2016;10:4742–4753. doi: 10.1021/acsnano.6b01254. [DOI] [PubMed] [Google Scholar]
- 70.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Connell M.R. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J. Mol. Biol. 2019;431:66–87. doi: 10.1016/j.jmb.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 72.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y. Development and evaluation of a CRISPR-based diagnostic for 2019-novel coronavirus. medRxiv. 2020 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou C., Gao C., Xie Y., Xu M. COVID-19 with spontaneous pneumomediastinum. Lancet Infect. Dis. 2020;20:510. doi: 10.1016/S1473-3099(20)30156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whiting P., Singatullina N., Rosser J. Computed tomography of the chest: I. Basic principles. Bja Educ. 2015;15:299–304. [Google Scholar]
- 76.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., Diao K., Lin B., Zhu X., Li K. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020:200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020:200370. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lv H., Wu N.C., Tsang O.T.-Y., Yuan M., Perera R.A., Leung W.S., So R.T., Chan J.M.C., Yip G.K., Chik T.S.H. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020:107725. doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., Shen C. Evaluation of enzyme-linked immunoassay and colloidal gold-immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19) MedRxiv. 2020 [Google Scholar]
- 84.Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R., Wei H., Tanner N. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020;2 2020.02. 26.20028373, in. [Google Scholar]
- 85.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D. China medical treatment expert group for covid-19. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiang Y.-T., Yang Y., Li W., Zhang L., Zhang Q., Cheung T., Ng C.H. Timely mental health care for the 2019 novel coronavirus outbreak is urgently needed. Lancet Psychiatry. 2020;7:228–229. doi: 10.1016/S2215-0366(20)30046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang P., Wang H., Cao Z., Jin H., Chi H., Zhao J., Yu B., Yan F., Hu X., Wu F. A rapid and specific assay for the detection of MERS-CoV. Front. Microbiol. 2018;9:1101. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spengler M., Adler M., Niemeyer C.M. Highly sensitive ligand-binding assays in pre-clinical and clinical applications: immuno-PCR and other emerging techniques. Analyst. 2015;140:6175–6194. doi: 10.1039/c5an00822k. [DOI] [PubMed] [Google Scholar]
- 89.Laksanasopin T., Guo T.W., Nayak S., Sridhara A.A., Xie S., Olowookere O.O., Cadinu P., Meng F., Chee N.H., Kim J. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aaa0056. 273re271-273re271. [DOI] [PubMed] [Google Scholar]
- 90.Gates B. Responding to Covid-19—a once-in-a-century pandemic? N. Engl. J. Med. 2020;382:1677–1679. doi: 10.1056/NEJMp2003762. [DOI] [PubMed] [Google Scholar]
- 91.Wood C.S., Thomas M.R., Budd J., Mashamba-Thompson T.P., Herbst K., Pillay D., Peeling R.W., Johnson A.M., McKendry R.A., Stevens M.M. Taking connected mobile-health diagnostics of infectious diseases to the field. Nature. 2019;566:467–474. doi: 10.1038/s41586-019-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ashkarran A.A., Olfatbakhsh T., Ramezankhani M., Crist R.C., Berrettini W.H., Milani A.S., Pakpour S., Mahmoudi M. Evolving magnetically levitated plasma proteins detects opioid use disorder as a model disease. Adv. Healthc. Mater. 2020;9:1901608. doi: 10.1002/adhm.201901608. [DOI] [PubMed] [Google Scholar]
- 93.Karimuribo E.D., Mutagahywa E., Sindato C., Mboera L., Mwabukusi M., Njenga M.K., Teesdale S., Olsen J., Rweyemamu M. A smartphone app (AfyaData) for innovative one health disease surveillance from community to national levels in Africa: intervention in disease surveillance. JMIR Public Health Surveill. 2017;3:e94. doi: 10.2196/publichealth.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Z., Askim J.R., Suslick K.S. The optoelectronic nose: colorimetric and fluorometric sensor arrays. Chem. Rev. 2018;119:231–292. doi: 10.1021/acs.chemrev.8b00226. [DOI] [PubMed] [Google Scholar]
- 95.Saha K., Agasti S., Kim Ch., Li X., Rotello V.M. Chem. Rev. 2012;112:2739. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ashkarran A.A., Suslick K.S., Mahmoudi M. Magnetically levitated plasma proteins. Anal. Chem. 2020 doi: 10.1021/acs.analchem.9b05101. [DOI] [PubMed] [Google Scholar]
- 97.Ashkarran A.A., Dararatana N., Crespy D., Caracciolo G., Mahmoudi M. Mapping the heterogeneity of protein corona by ex vivo magnetic levitation. Nanoscale. 2020;12:2374–2383. doi: 10.1039/c9nr10367h. [DOI] [PubMed] [Google Scholar]
- 98.Tavakol M., Montazeri A., Naghdabadi R., Hajipour M.J., Zanganeh S., Caracciolo G., Mahmoudi M. Disease-related metabolites affect protein–nanoparticle interactions. Nanoscale. 2018;10:7108–7115. doi: 10.1039/c7nr09502c. [DOI] [PubMed] [Google Scholar]
- 99.Palchetti S., Digiacomo L., Pozzi D., Zenezini Chiozzi R., Capriotti A.L., Laganà A., Coppola R., Caputo D., Sharifzadeh M., Mahmoudi M. Effect of glucose on liposome–plasma protein interactions: relevance for the physiological response of clinically approved liposomal formulations. Adv. Biosyst. 2019;3:1800221. doi: 10.1002/adbi.201800221. [DOI] [PubMed] [Google Scholar]
- 100.Liu W., Morse J.S., Lalonde T., Xu S. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem. 2020 doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.He L., Ding Y., Zhang Q., Che X., He Y., Shen H., Wang H., Li Z., Zhao L., Geng J. Expression of elevated levels of pro‐inflammatory cytokines in SARS‐CoV‐infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Faure E., Poissy J., Goffard A., Fournier C., Kipnis E., Titecat M., Bortolotti P., Martinez L., Dubucquoi S., Dessein R. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One. 2014;9 doi: 10.1371/journal.pone.0088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rodrigo C., Leonardi‐Bee J., Nguyen-Van-Tam J., Lim W.S. Corticosteroids as adjunctive therapy in the treatment of influenza. Cochrane Database Syst. Rev. 2016 doi: 10.1002/14651858.CD010406.pub2. [DOI] [PubMed] [Google Scholar]
- 105.Arabi Y.M., Mandourah Y., Al-Hameed F., Sindi A.A., Almekhlafi G.A., Hussein M.A., Jose J., Pinto R., Al-Omari A., Kharaba A. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am. J. Respir. Crit. Care Med. 2018;197:757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 106.Organization W.H. World Health Organization; 2020. Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (nCoV) Infection Is Suspected: Interim Guidance. 25 January 2020, in. [Google Scholar]
- 107.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chu C., Cheng V., Hung I., Wong M., Chan K., Chan K., Kao R., Poon L., Wong C., Guan Y. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arabi Y.M., Alothman A., Balkhy H.H., Al-Dawood A., AlJohani S., Al Harbi S., Kojan S., Al Jeraisy M., Deeb A.M., Assiri A.M. Treatment of Middle East Respiratory Syndrome with a combination of lopinavir-ritonavir and interferon-β1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials. 2018;19:81. doi: 10.1186/s13063-017-2427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li G., De Clercq E. Nature Publishing Group; 2020. Therapeutic Options for the 2019 Novel Coronavirus (2019-nCoV) [DOI] [PubMed] [Google Scholar]
- 112.De Clercq E. New nucleoside analogues for the treatment of hemorrhagic fever virus infections. Chemistry–An Asian Journal. 2019;14:3962–3968. doi: 10.1002/asia.201900841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang L. 2019. Cell Res. in, the press. [Google Scholar]
- 114.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.-Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang M., Cao R., Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019 nCoV) in vitro [published online February 4, 2020] Cell Res. 2021;30 doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wolfram J., Ferrari M. Clinical cancer nanomedicine. Nano Today. 2019 doi: 10.1016/j.nantod.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gentile E., Cilurzo F., Di Marzio L., Carafa M., Anna Ventura C., Wolfram J., Paolino D., Celia C. Liposomal chemotherapeutics. Future Oncol. 2013;9:1849–1859. doi: 10.2217/fon.13.146. [DOI] [PubMed] [Google Scholar]
- 118.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases. Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin-and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ma Y., Chan C.-Y., He M.-L. RNA interference and antiviral therapy. World Journal of Gastroenterology: WJG. 2007;13:5169. doi: 10.3748/wjg.v13.i39.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deleted C.T. Clathrin-dependent entry of severe acute. J. Virol. 2007;81:8722. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zachar O. 2020. Formulations for COVID-19 Treatment Via Silver Nanoparticles Inhalation Delivery. [Google Scholar]
- 124.Wolfsen H.C., Ng C.S. Cutaneous consequences of photodynamic therapy. Cutis. 2002;69:140–142. [PubMed] [Google Scholar]
- 125.Banerjee I., Douaisi M.P., Mondal D., Kane R.S. Light-activated nanotube–porphyrin conjugates as effective antiviral agents. Nanotechnology. 2012;23:105101. doi: 10.1088/0957-4484/23/10/105101. [DOI] [PubMed] [Google Scholar]
- 126.Zhou L., Wei S., Ge X., Zhou J., Jiang H., Li F., Shen J. Combination of chemotherapy and photodynamic therapy using graphene oxide as drug delivery system. J. Photochem. Photobiol. B, Biol. 2014;135:7–16. doi: 10.1016/j.jphotobiol.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 127.Sahu A., Choi W.I., Lee J.H., Tae G. Graphene oxide mediated delivery of methylene blue for combined photodynamic and photothermal therapy. Biomaterials. 2013;34:6239–6248. doi: 10.1016/j.biomaterials.2013.04.066. [DOI] [PubMed] [Google Scholar]
- 128.Ristic B.Z., Milenkovic M.M., Dakic I.R., Todorovic-Markovic B.M., Milosavljevic M.S., Budimir M.D., Paunovic V.G., Dramicanin M.D., Markovic Z.M., Trajkovic V.S. Photodynamic antibacterial effect of graphene quantum dots. Biomaterials. 2014;35:4428–4435. doi: 10.1016/j.biomaterials.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 129.Di Gianvincenzo P., Marradi M., Martínez-Ávila O.M., Bedoya L.M., Alcamí J., Penadés S. Gold nanoparticles capped with sulfate-ended ligands as anti-HIV agents. Bioorg. Med. Chem. Lett. 2010;20:2718–2721. doi: 10.1016/j.bmcl.2010.03.079. [DOI] [PubMed] [Google Scholar]
- 130.Walter J.G., Petersen S., Stahl F., Scheper T., Barcikowski S. Laser ablation-based one-step generation and bio-functionalization of gold nanoparticles conjugated with aptamers. J. Nanobiotechnology. 2010;8:1–11. doi: 10.1186/1477-3155-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Baram-Pinto D., Shukla S., Perkas N., Gedanken A., Sarid R. Inhibition of herpes simplex virus type 1 infection by silver nanoparticles capped with mercaptoethane sulfonate. Bioconjug. Chem. 2009;20:1497–1502. doi: 10.1021/bc900215b. [DOI] [PubMed] [Google Scholar]
- 132.Lu L., Sun R., Chen R., Hui C.-K., Ho C.-M., Luk J.M., Lau G., Che C.-M. Silver nanoparticles inhibit hepatitis B virus replication. Antivir. Ther. 2008;13:253. [PubMed] [Google Scholar]
- 133.Sun L., Singh A.K., Vig K., Pillai S.R., Singh S.R. Silver nanoparticles inhibit replication of respiratory syncytial virus. J. Biomed. Nanotechnol. 2008;4:149–158. [Google Scholar]
- 134.Ruiz‐Hitzky E., Darder M., Wicklein B., Ruiz‐Garcia C., Martín‐Sampedro R., Del Real G., Aranda P. Nanotechnology responses to COVID‐19. Adv. Healthc. Mater. 2020;9:2000979. doi: 10.1002/adhm.202000979. [DOI] [PubMed] [Google Scholar]
- 135.Moore C.M., Pendse D., Emberton M. Photodynamic therapy for prostate cancer—a review of current status and future promise. Nat. Clin. Pract. Urol. 2009;6:18–30. doi: 10.1038/ncpuro1274. [DOI] [PubMed] [Google Scholar]
- 136.Hamblin M.R., Hasan T. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem. Photobiol. Sci. 2004;3:436–450. doi: 10.1039/b311900a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maisch T. Anti-microbial photodynamic therapy: useful in the future? Lasers Med. Sci. 2007;22:83–91. doi: 10.1007/s10103-006-0409-7. [DOI] [PubMed] [Google Scholar]
- 138.Wiehe A., O’Brien J.M., Senge M.O. Trends and targets in antiviral phototherapy. Photochem. Photobiol. Sci. 2019;18:2565–2612. doi: 10.1039/c9pp00211a. [DOI] [PubMed] [Google Scholar]
- 139.Moghissi K., Dixon K., Gibbins S. Does PDT have potential in the treatment of COVID 19 patients? Photodiagn. Photodyn. Ther. 2020;31:101889. doi: 10.1016/j.pdpdt.2020.101889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lin L., Hanson C.V., Alter H.J., Jauvin V., Bernard K.A., Murthy K.K., Metzel P., Corash L. Inactivation of viruses in platelet concentrates by photochemical treatment with amotosalen and long‐wavelength ultraviolet light. Transfusion. 2005;45:580–590. doi: 10.1111/j.0041-1132.2005.04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schneider K., Wronka-Edwards L., Leggett-Embrey M., Walker E., Sun P., Ondov B., Wyman T.H., Rosovitz M., Bohn S.S., Burans J. Psoralen inactivation of viruses: a process for the safe manipulation of viral antigen and nucleic acid. Viruses. 2015;7:5875–5888. doi: 10.3390/v7112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Keil S.D., Bowen R., Marschner S. Inactivation of M iddle E ast respiratory syndrome coronavirus (MERS‐C o V) in plasma products using a riboflavin‐based and ultraviolet light‐based photochemical treatment. Transfusion. 2016;56:2948–2952. doi: 10.1111/trf.13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Han W., Zhang P.H., Cao W.C., Yang D.L., Taira S., Okamoto Y., Arai J.I., Yan X.Y. The inactivation effect of photocatalytic titanium apatite filter on SARS virus. Prog. Biochem. Biophys. 2004;31 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are made available in the manuscript.