Abstract
The zinc finger-associated domain (ZAD) is present in over 90 C2H2 zinc finger (ZNF) proteins. Despite their abundance, only a few ZAD-ZNF genes have been characterized to date. Here, we systematically analyze the function of 68 ZAD-ZNF genes in Drosophila female germ cells by performing an in vivo RNA-interference screen. We identified eight ZAD-ZNF genes required for oogenesis, and based on further characterization of the knockdown phenotypes, we uncovered defects broadly consistent with functions in germ cell specification and/or survival, early differentiation, and egg chamber maturation. These results provide a candidate pool for future studies aimed at functionalization of this large but poorly characterized gene family.
Keywords: multigene family, reproduction, oogenesis, gametogenesis, Zif, odj, M1PB, CG320020, trem, hang, CG4936, CG17802
Introduction
In eukaryotes, C2H2 zinc finger (ZNF) proteins are among the most abundant class of DNA-binding proteins (Stubbs et al. 2011; Fedotova et al. 2017). ZNF proteins are often grouped into families based on the presence of conserved N-terminal domains. In Drosophila, over 90 ZNF proteins belong to the zinc finger-associated domain (ZAD) family. The N-terminal ZAD domain can mediate protein–protein interactions, whereas the ZNF domain can confer DNA-binding specificity (Jauch et al. 2003). Despite their abundance, the function of only a few ZAD-ZNFs has been examined in vivo. These studies suggest essential roles in regulating transcription, silencing transposons, and organizing chromatin architecture during development (Fedotova et al. 2017). Interestingly, the ZAD-ZNF family is mostly restricted to Drosophila and related species (Chung et al. 2002, 2007). Because many of the ZAD-ZNF family members are expressed in ovaries, it has been suggested that the expansion of this gene family was driven by species-specific requirements in the germline (Chung et al. 2002, 2007). However, most of the ZAD-ZNF genes remain uncharacterized and their role in the female germline unknown.
In Drosophila, each adult ovary is subdivided into 16–20 individual ovarioles, which contain oocytes spanning the range of maturity, from germline stem cell (GSC) to fully mature eggs (Hinnant et al. 2020). Two to three GSCs are located at the anterior-most structure of the ovariole, called the germarium. Each GSC divides asymmetrically to produce one daughter cell that retains its GSC identity and a second daughter cell called a cystoblast (CB) that will enter the differentiation pathway. The differentiation program begins with four synchronous incomplete mitotic divisions to give rise to a 16-cell interconnected cyst from which one cell becomes the oocyte and enters meiosis. The other 15 cells, called nurse cells, will provide necessary RNA and protein products to the developing egg. Once the 16-cell cyst is formed, the somatic follicle cells surround the cyst in a monolayer to generate an egg chamber.
In this study, we tested the function of 68 ZAD-ZNF genes in the female germline. Using RNA-interference (RNAi) to knock down expression in germ cells, we identified eight ZAD-ZNF genes whose products are required for egg production. The linear arrangement of oocyte production allows the unequivocal identification of each stage development by location, morphology, and expression of molecular markers. Analysis of the mutant ovaries using these methods uncovered defects consistent with functions in germ cell specification and/or survival, early differentiation, and egg chamber maturation. These results provide a candidate pool for future studies.
Materials and methods
Primary germline-specific RNAi screen
We searched Flybase to identify all genes that encode a ZAD domain (ZNF, AD-type, IPR012934; http://www.ebi.ac.uk/interpro/entry/InterPro/IPR012934/). We identified 93 ZAD-ZNF encoding genes (Table 1). Screening was limited to the 68 ZAD-ZNF genes for which germline-optimized TRiP RNAi lines (in VALIUM 20, 21 or 22) were available at the Bloomington Drosophila Stock Center (BDSC stock numbers listed in Table 1). Expression of the UAS-RNAi transgenic lines was driven by nanos-GAL4 (P{GAL4::VP16-nos.UTR}, BDSC #4937), which is strongly expressed in germ cells. Males from each UAS-RNAi line were crossed to nanos-GAL4 females at 29°C, and the F1 female progeny tested for egg production.
Table 1.
List of annotated ZAD-ZFP genes, the TRiP transgenic lines used for the primary screen and a summary of the results
| FBgn number | Gene name | Symbol or CG number | Germline-optimized TRiP transgenic lines used listed by BDSC stock #s | Numbers of >RNAi at 29 phenotype |
|---|---|---|---|---|
| FBgn0026575 | Hangover | hang | #41870, #35674, #57791 | 2/3 No eggs |
| FBgn0037446 | Zinc-finger protein | Zif | #36641 | No eggs |
| FBgn0037621 | Motif 1-binding protein | M1BP | #32858, #41937 | No eggs |
| FBgn0038549 | – | CG17802 | #43233, #57480 | No eggs |
| FBgn0038551 | Oddjob | Odj | #62184, #61912 | No eggs |
| FBgn0038767 | trade embargo | trem | #40881 | No eggs |
| FBgn0038768 | – | CG4936 | #64550 | No eggs |
| FBgn0050020 | – | CG30020 | #58110 | No eggs |
| FBgn0000520 | Deformed wings (zw5) | dwg | #35666 | Eggs |
| FBgn0001133 | grauzone | grau | #53251, #41940 | Eggs |
| FBgn0001990 | weckle | wek | #35680, #57260 | Eggs |
| FBgn0022699 | D19B | D19B | #51166 | Eggs |
| FBgn0022935 | D19A | D19A | #33371 | Eggs |
| FBgn0024975 | – | CG2712 | #57418 | Eggs |
| FBgn0025874 | Meiotic central spindle | Meics | #50636 | Eggs |
| FBgn0029895 | – | CG14441 | #67271 | Eggs |
| FBgn0029928 | – | CG3032 | #57560 | Eggs |
| FBgn0030206 | – | CG2889 | #77384 | Eggs |
| FBgn0030316 | – | CG11695 | #57246 | Eggs |
| FBgn0030455 | – | CG4318 | #51720 | Eggs |
| FBgn0030659 | – | CG9215 | #57537 | Eggs |
| FBgn0031144 | – | CG1529 | #60094 | Eggs |
| FBgn0031597 | – | CG17612 | #57540 | Eggs |
| FBgn0031608 | – | CG15435 | #65064 | Eggs |
| FBgn0031610 | – | CG15436 | #57867 | Eggs |
| FBgn0032223 | GATAd | GATAd | #33747, #34625, #34640 | Eggs |
| FBgn0032730 | – | CG10431 | #44470 | Eggs |
| FBgn0033581 | – | CG12391 | #40847 | Eggs |
| FBgn0034062 | – | CG8388 | #57545 | Eggs |
| FBgn0034114 | – | CG4282 | #62445 | Eggs |
| FBgn0034878 | pita | pita | #35724, #57732 | Eggs |
| FBgn0035036 | – | CG4707 | #61167, #58229 | Eggs |
| FBgn0035691 | – | CG7386 | #62181 | Eggs |
| FBgn0036395 | – | CG17361 | #57714 | Eggs |
| FBgn0037085 | Neuroectoderm-expressed 2 | Neu2 | #60002 | Eggs |
| FBgn0037584 | – | CG7963 | #60116 | Eggs |
| FBgn0037617 | Numerous disordered muscles | nom | #43551 | Eggs |
| FBgn0037618 | ouija board | ouib | #42531 | Eggs |
| FBgn0037619 | – | CG8159 | #62182 | Eggs |
| FBgn0037620 | ranshi | ranshi | #43156, #58206 | Eggs |
| FBgn0037717 | – | CG8301 | #41643, #62206 | Eggs |
| FBgn0037722 | – | CG8319 | #61173 | Eggs |
| FBgn0037794 | – | CG6254 | #57279 | Eggs |
| FBgn0037876 | – | CG4820 | #60117 | Eggs |
| FBgn0037877 | – | CG6689 | #67282 | Eggs |
| FBgn0037921 | – | CG6808 | #35588 | Eggs |
| FBgn0037922 | – | CG14711 | #57280 | Eggs |
| FBgn0038301 | – | CG6654 | #62183 | Eggs |
| FBgn0038547 | – | CG17803 | #60022 | Eggs |
| FBgn0038548 | – | CG17806 | #77448 | Eggs |
| FBgn0038550 | CG17801 | #60118 | Eggs | |
| FBgn0038765 | – | CG4424 | #43218, #77900 | Eggs |
| FBgn0038766 | – | CG4854 | #42833 | Eggs |
| FBgn0039355 | – | CG4730 | #61169 | Eggs |
| FBgn0039602 | – | CG1647 | #55292 | Eggs |
| FBgn0040467 | Dorsal interacting protein 1 | Dlip1 | #57717 | Eggs |
| FBgn0042205 | – | CG18764 | #64554 | Eggs |
| FBgn0050431 | – | CG30431 | #60023 | Eggs |
| FBgn0051109 | – | CG31109 | #61878 | Eggs |
| FBgn0051365 | – | CG31365 | #57450 | Eggs |
| FBgn0051388 | – | CG31388 | #67288 | Eggs |
| FBgn0051441 | – | CG31441 | #67285 | Eggs |
| FBgn0051457 | – | CG31457 | #63588 | Eggs |
| FBgn0067779 | debra | dbr | #43222 | Eggs |
| FBgn0260741 | – | CG3281 | #44555 | Eggs |
| FBgn0263490 | Molting defective | mld | #43145, #63020 | Eggs |
| FBgn0264744 | – | CG44002 | #57463 | Eggs |
| FBgn0285971 | piragua | prg | #36631 | Eggs |
| FBgn0014931 | – | CG2678 | N/A | N/A |
| FBgn0028647 | – | CG11902 | N/A | N/A |
| FBgn0028895 | – | CG17328 | N/A | N/A |
| FBgn0030240 | – | CG2202 | N/A | N/A |
| FBgn0030314 | – | CG11696 | N/A | N/A |
| FBgn0032150 | – | CG13123 | N/A | N/A |
| FBgn0032763 | – | CG17568 | N/A | N/A |
| FBgn0032814 | – | CG10366 | N/A | N/A |
| FBgn0033569 | – | CG12942 | N/A | N/A |
| FBgn0034379 | – | CG15073 | N/A | N/A |
| FBgn0034643 | – | CG10321 | N/A | N/A |
| FBgn0035690 | – | CG10274 | N/A | N/A |
| FBgn0035702 | – | CG10147 | N/A | N/A |
| FBgn0036294 | – | CG10654 | N/A | N/A |
| FBgn0036396 | – | CG17359 | N/A | N/A |
| FBgn0037317 | – | CG14667 | N/A | N/A |
| FBgn0037920 | – | CG14710 | N/A | N/A |
| FBgn0037923 | – | CG6813 | N/A | N/A |
| FBgn0037931 | – | CG18476 | N/A | N/A |
| FBgn0038418 | poils au dos | pad | N/A | N/A |
| FBgn0039329 | – | CG10669 | N/A | N/A |
| FBgn0039860 | – | CG1792 | N/A | N/A |
| FBgn0043796 | – | CG12219 | N/A | N/A |
| FBgn0260243 | Enhancer of variegation 3-9 | E(var)3-9 | N/A | N/A |
The genes are sorted as follows: (1) genes for which germline-specific knockdown resulted in female sterility (no eggs). (2) genes for which germline-specific knockdown did not disrupt female fertility (eggs), and (3) genes for which we were not able to obtain germline-optimized RNAi lines (N/A).
Immunofluorescence and image analysis
F1 females were raised and aged 3–5 days at 29°C prior to gonad dissection. Ovaries were fixed and stained according to standard procedures with the following primary antibodies: rat anti-HA high affinity (1:500, Roche cat #11867423001, RRID: AB_390919), rat anti-Drosophila Vasa (1:100, Developmental Studies Hybridoma Bank, RRID: AB_760351), and mouse anti-Drosophila Orb 4H8 (1:50, Developmental Studies Hybridoma Bank, RRID: AB_528418) combined with mouse anti-Drosophila Orb 6H4 (1:50, Developmental Studies Hybridoma Bank, RRID: AB_528419). Staining was detected with the following conjugated antibodies: Fluorescein (FITC) AffiniPure Goat anti- rat IgG (H + L) (1:200, Jackson ImmnoResearch Labs, cat #112-095-062, RRID: AB_2338194), or Fluorescein (FITC) AffiniPure Goat Anti-mouse IgG (H + L) (1:200, Jackson ImmnoResearch Labs, cat #115-095-003, RRID: AB_2338589). TO-PRO-3 Iodide monomeric cyanine nucleic acid stain (Thermo Fisher, cat #T3605) was used to stain DNA.
Images were taken on a Leica SP8 confocal with 1024 × 1024 pixel dimensions, a scan speed of 600 Hz, and a frame average of 3. Sequential scanning was done for each channel and three Z-stacks were combined for each image. Processed images were compiled with Gnu Image Manipulation Program and Microsoft PowerPoint.
The HA-tagged bam transgene, P{bamP-Bam::HA}, was used to report on BAM protein expression (Li et al. 2009).
Data availability statement
Drosophila strains are available upon request. The authors affirm that all data necessary for confirming the conclusion of this study are present within the article, figures, and tables.
Results and discussion
Germline depletion screen identifies eight ZAD-ZNF genes necessary for oogenesis
In the primary screen, we knocked down the expression of 68 ZAD-ZNF genes by using the germ cell-specific driver nanos-GAL4 to express the germline-optimized TRiP UAS-RNAi transgenes. Germline knockdown (GLKD) of only eight genes abolished egg production (Table 1). For the remaining 60 genes, knocking down expression in germ cells did not prevent egg production. While the lack of phenotype suggests that the majority of ZAD-ZNF genes are not required intrinsically for germ cell development, we cannot rule out other explanations for the lack of mutant phenotype, such as low RNAi efficiency of the available TRiP lines. Furthermore, our GLKD screen would not have identified genes required in somatic cells that contribute to germ cell development.
Secondary immunofluorescence screen to characterize ovarian defects
For the eight genes whose GLKD abolished egg production, we analyzed the ovarian defects by staining for DNA and Vasa, a germ cell-specific marker. For each GLKD experiment, >120 ovarioles were scored individually, as: (1) agametic, defined by the absence of Vasa-positive cells, (2) a failure to form egg chambers, and (3) late defects, in which egg chambers are formed but fail to progress to the larger, more mature stages. The wild-type ovariole has a stereotypical organization with all stages of oogenesis arranged in developmental order from the GSCs housed at the anterior end (the germarium) to the progressively larger, more developed egg chambers (Figure 1A). We found that GLKD of some of the genes gave rise to more than one ovarian defect, and the “per ovariole” analysis, reported in Figure 2, allowed us to quantify the penetrance of each phenotype. Based on this analysis, we conclude that the GLKD phenotypes fall into two general classes: agametic and the absence of the larger, late-stage egg chambers (late defects). Several of the candidate genes were targeted by more than one UAS-RNAi line. In the following sections, we limit our description to the line that induced the most severe phenotype.
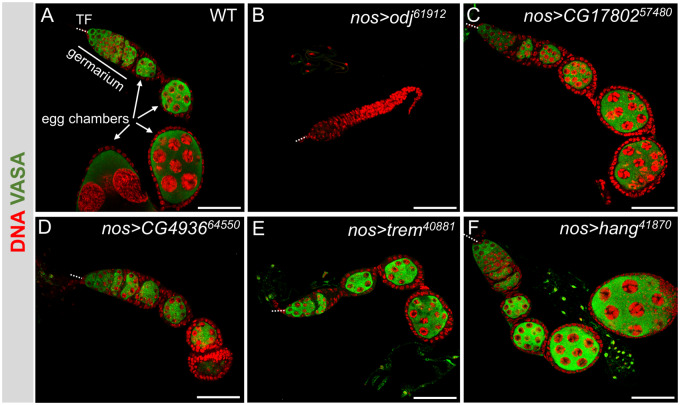
Figure 1.
Loss of 8 ZAD-ZNF genes in germ cells disrupt oogenesis. Representative confocal images of control (A) and mutant ovarioles (B–F) stained for Vasa (green) and DNA (red). All images include the somatic terminal filaments (TF, dotted line), the germarium, and early-stage egg chambers (EC). Scale bar 50 µm. Quantification of phenotypes is presented in Figure 2.
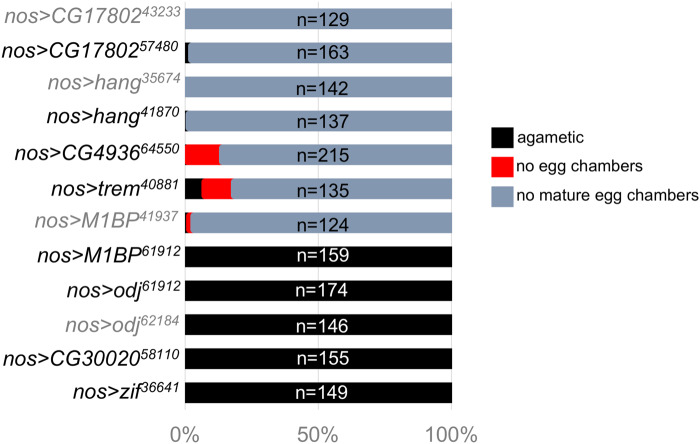
Figure 2.
Range of defects observed in ZAD-ZNF GLKD mutant ovaries. Bar graph showing the percentage of VASA stained ovarioles with no germ cells (agametic), no egg chamber formation, or no mature, vitellogenic, egg chambers. Examples of the most prevalent phenotypes are shown in Figure 1.
Agametic: We found that >90% of the Zif, odj, M1PB, and CG320020 knockdown ovarioles had no germ cells (no Vasa-positive cells, see for example Figure 1B). This agametic phenotype suggests a requirement for germ cell specification, and/or survival. A requirement for odj was confirmed by the second UAS-RNAi line, which recapitulated the agametic phenotype.
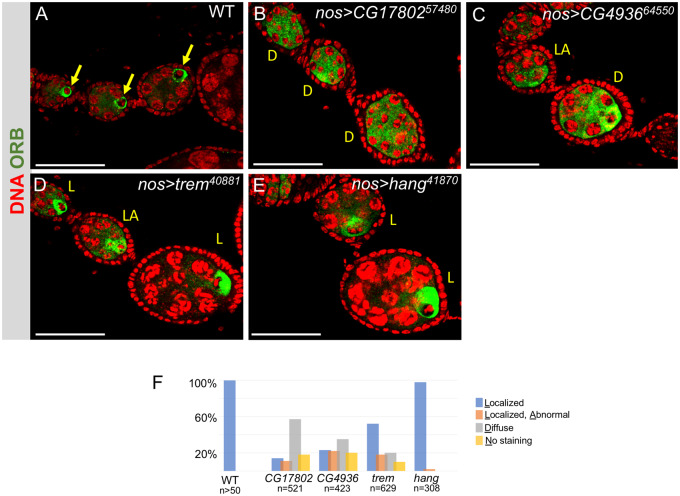
Late defects: We found that >80% of the trem, hang, CG4936 and CG17802 GLKD ovarioles formed only pre-vitellogenic egg chambers (see for example Figure 1, C–F). To better understand the role of these “late defect” genes, we stained the GLKD ovaries with an antibody against Orb, a germline-specific marker for oocyte determination. In the wild-type ovary, the Orb protein concentrates around the cytoplasm of the oocyte (Figure 3A). Major defects were noted in CG17802, CG4936 and trem GLKD egg chambers. Orb staining remained diffuse in the majority (57%) of CG17802 GLKD egg chambers, suggesting a defect in oocyte specification (Figure 3, B and F). On occasion, we observed Orb protein accumulation in the cytoplasm of one (14%) or two (11%) mutant germ cells, or no staining at all (18%). The Orb staining patterns we observed in CG4936 GLKD mutant egg chambers ranged from no staining at all (20%), diffuse staining (35%), localization to more than one germ cell (22%), and localization to a single germ cell (23%; Figure 3, C and 3F). Although some trem GLKD egg chambers displayed defects in Orb localization, Orb was appropriately localized in the majority (52%; Figure 3, D and F). Lastly, Orb protein appeared to be appropriately localized to a single cell in the majority (98%) of hang GLKD egg chambers, suggesting a later defect in egg chamber growth (Figure 3, E and F).
Figure 3.
Impact of ZAD-ZNF germ cell-specific knockdowns on oocyte development. Representative confocal images of egg chambers from control (A) and mutant ovarioles (B–E) stained for Orb (green) and DNA (red). In wild type, the Orb protein only accumulates around the presumptive oocyte (arrow). The range of phenotypes observed in mutants is indicated as follows: localized (L), localized abnormal (LA), and diffuse (D). Scale bar = 50 µm. (F) Quantification of mutant egg chamber phenotypes. Two to three egg chambers per ovariole were scored. N is the number of individual egg chambers scored.
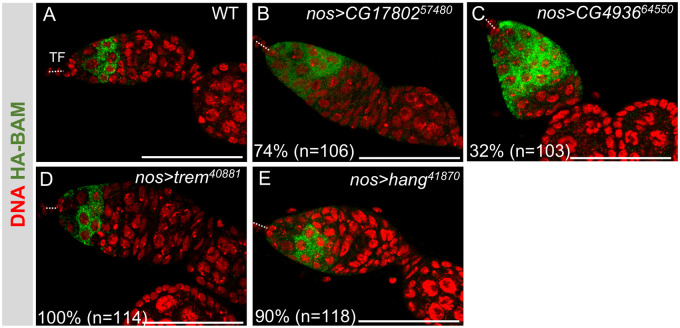
To explore whether any of these mutant phenotypes can be explained by defects that occur prior to oocyte specification, we examined the expression of the cytoplasmic differentiation factor Bag of Marbles (Bam). When GSCs divide, the daughter cell that stays at the tip remains a GSC, while the more posterior daughter cell becomes a CB. Each germarium normally contain a single CB, which goes on to divide four times with incomplete cytokinesis, resulting in a 16-cell germline cyst. When we assessed Bam protein expression with a human influenza hemagglutinin (HA)-tagged bam transgene (Li et al. 2009), we observed that the Bam protein is readily detectable in just a few cells (Figure 4A). Interestingly, we observed an expansion of Bam protein expression in a majority of CG17802 GLKD germaria (74%) and in a significant fraction of CG4936 GLKD germaria (32%), suggesting defects prior to oocyte specification (Figure 4, B and C). On the other hand, Bam protein accumulation is similar to wild type in trem and hang GLKD ovaries, suggesting that these genes do not impact the earliest stages of germ cell differentiation (Figure 4, D and E). These data, together with the observation that Orb was appropriately localized in the majority of trem and hang GLKD egg chambers, suggest that the primary function of trem and hang is after oocyte specification.
Figure 4.
Impact of ZAD-ZNF germ cell-specific knockdowns on expression of the differentiation factor Bam. Representative confocal images of control (A) and mutant ovarioles (B–E) carrying a fully functional copy of a HA-Bam fusion protein, stained for HA (green) and DNA (red). In wild type, Bam protein is only expressed in a few early germ cells. Of note, Bam is not detectable in GSCs, located at the anterior end of the germarium. The location of the somatic terminal filaments (TF) at the anterior end of the germarium is marked by a dashed line. Scale bar = 50 µm. N is the number of germaria scored.
In summary, we have identified eight ZAD-ZNF genes required in germ cells for oogenesis. Several of the ZAD-ZNF genes identified in this study have previously been implicated in germline function, the best characterized of which is trem. Trem is a chromatin-associated protein required for the initiation of meiotic double-strand break formation (Lake et al. 2011). Although not characterized in detail, Zif, CG4936, and hang were identified in an unbiased RNAi screen for factors required to silence transposons (TE) (Czech et al. 2013). However, mutations in genes dedicated to silencing TEs in germ cells induce sterility while maintaining normal ovarian development. Our observations that the loss of Zif, CG4936, and hang disrupts oogenesis suggest a requirement beyond TE silencing.
This analysis also identified ZAD-ZNF genes with no prior reported function in germ cells. For example, we identified a role for the genes encoding the Odj and CG17802 proteins that localize to heterochromatin in somatic cells (Swenson et al. 2016; Kasinathan et al. 2020). Heterochromatin establishment and maintenance is essential for many cellular processes, including chromosome segregation and genome stability (Janssen et al. 2018). A role in germ cells is therefore not entirely unexpected, as mutations in genes that encode crucial heterochromatin functions often disrupt oogenesis (e.g.Giauque and Bickel 2016; Teo et al. 2018; Smolko et al. 2018; Jankovics et al. 2018; Casale et al. 2019; Benner et al. 2019).
We also identified a role for M1BP, a transcription factor that associates with thousands of genes in somatic cells (Li and Gilmour 2013; Zouaz et al. 2017; Barthez et al. 2020). Similarly, CG30020 is also likely to be transcription factor, as in vitro studies have shown that it binds a unique DNA consensus sequence (Krystel and Ayyanathan 2013). Whether CG30020 and M1BP control transcription in a tissue-specific manner remains to be determined.
Overall, the results reported here identify eight members of the large but poorly characterized ZAD-ZNF gene family required in the female germline. The ZAD-ZNF family, which arose in the ancestor of arthropods and vertebrates, expanded to become the most abundant class of transcription factors in Drosophila and related species (Chung et al. 2002, 2007). Our findings suggest that expansion of the ZAD-ZNF family is related to species-specific requirements in the germline. Future work will test these ideas by a more detailed analysis of these ZAD-ZNF genes in Drosophila and other species.
Acknowledgments
We thank the Bloomington Stock Center for providing stocks, the Developmental Studies Hybridoma Bank for antibodies, and Jane Heatwole for fly food. Imaging was performed using equipment purchased through NIH S10OD016164.
All authors contributed to the design and performance of the experiments, analysis and interpretation of the data and writing of the manuscript.
Funding
This work was supported by the National Institutes of Health grant (R01GM129478) to H.K.S.
Conflicts of interest: None declared.
Literature cited
- Barthez M, Poplineau M, Elrefaey M, Caruso N, Graba Y, et al. 2020. Human ZKSCAN3 and Drosophila M1BP are functionally homologous transcription factors in autophagy regulation. Sci Rep. 10:9653–9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner L, Castro EA, Whitworth C, Venken KJT, Yang H, et al. 2019. Drosophila heterochromatin stabilization requires the zinc-finger protein small ovary. Genetics. 213:877–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casale AM, Cappucci U, Fanti L, Piacentini L.. 2019. Heterochromatin protein 1 (HP1) is intrinsically required for post-transcriptional regulation of Drosophila Germline Stem Cell (GSC) maintenance. Sci Rep. 9:4372–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H-R, Löhr U, Jäckle H.. 2007. Lineage-specific expansion of the zinc finger associated domain ZAD. Mol Biol Evol. 24:1934–1943. [DOI] [PubMed] [Google Scholar]
- Chung H-R, Schäfer U, Jäckle H, Böhm S.. 2002. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep. 3:1158–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Preall JB, McGinn J, Hannon GJ.. 2013. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell. 50:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedotova AA, Bonchuk AN, Mogila VA, Georgiev PG.. 2017. C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae. 9:47–58. [PMC free article] [PubMed] [Google Scholar]
- Giauque CC, Bickel SE.. 2016. Heterochromatin-associated proteins HP1a and Piwi collaborate to maintain the association of achiasmate homologs in Drosophila oocytes. Genetics. 203:173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnant TD, Merkle JA, Ables ET.. 2020. Coordinating proliferation, polarity, and cell fate in the Drosophila female germline. Front Cell Dev Biol. 8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovics F, Bence M, Sinka R, Faragó A, Bodai L, et al. 2018. Drosophila small ovary gene is required for transposon silencing and heterochromatin organization, and ensures germline stem cell maintenance and differentiation. Development. 145:dev170639. [DOI] [PubMed] [Google Scholar]
- Janssen A, Colmenares SU, Karpen GH.. 2018. Heterochromatin: guardian of the genome. Annu Rev Cell Dev Biol. 34:265–288. [DOI] [PubMed] [Google Scholar]
- Jauch R, Bourenkov GP, Chung H-R, Urlaub H, Reidt U, et al. 2003. The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein-protein interaction module. Structure. 11:1393–1402. [DOI] [PubMed] [Google Scholar]
- Kasinathan B, Colmenares SU, McConnell H, Young JM, Karpen GH, et al. 2020. Innovation of heterochromatin functions drives rapid evolution of essential ZAD-ZNF genes in Drosophila. eLife. 9:e63368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystel J, Ayyanathan K.. 2013. Global analysis of target genes of 21 members of the ZAD transcription factor family in Drosophila melanogaster. Gene. 512:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake CM, Nielsen RJ, Hawley RS.. 2011. The Drosophila zinc finger protein trade embargo is required for double strand break formation in meiosis. PLoS Genet. 7:e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gilmour DS.. 2013. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J. 32:1829–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, Mckearin DM, Maines JZ.. 2009. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci USA. 106:9304–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolko AE, Shapiro-Kulnane L, Salz HK.. 2018. The H3K9 methyltransferase SETDB1 maintains female identity in Drosophila germ cells. Nat Commun. 9:4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs L, Sun Y, Caetano-Anolles D.. 2011. Function and evolution of C2H2 zinc finger arrays. Subcell Biochem. 52:75–94. [DOI] [PubMed] [Google Scholar]
- Swenson JM, Colmenares SU, Strom AR, Costes SV, Karpen GH.. 2016. The composition and organization of Drosophila heterochromatin are heterogeneous and dynamic. Elife. 5:1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo RYW, Anand A, Sridhar V, Okamura K, Kai T.. 2018. Heterochromatin protein 1a functions for piRNA biogenesis predominantly from pericentric and telomeric regions in Drosophila. Nat Commun. 9:1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouaz A, Auradkar A, Delfini MC, Macchi M, Barthez M, et al. 2017. The Hox proteins Ubx and AbdA collaborate with the transcription pausing factor M1BP to regulate gene transcription. EMBO J. 36:2887–2906., [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Drosophila strains are available upon request. The authors affirm that all data necessary for confirming the conclusion of this study are present within the article, figures, and tables.