Abstract
Angomonas deanei is an endosymbiont-bearing trypanosomatid with several highly fragmented genome assemblies and unknown chromosome number. We present an assembly of the A. deanei nuclear genome based on Oxford Nanopore sequence that resolves into 29 complete or close-to-complete chromosomes. The assembly has several previously unknown special features; it has a supernumerary chromosome, a chromosome with a 340-kb inversion, and there is a translocation between two chromosomes. We also present an updated annotation of the chromosomal genome with 10,365 protein-coding genes, 59 transfer RNAs, 26 ribosomal RNAs, and 62 noncoding RNAs.
Keywords: Angomonas deanei Carvalho (ATCC® PRA-265™), Oxford Nanopore, genome assembly
Introduction
Angomonas deanei is a trypanosomatid that mutually coevolves with an endosymbiont, a β-Proteobacterium of the Alcaligenaceae family that contains a reduced genome when compared to its ancestral prokaryote. The symbiont divides during the host cell cycle such that each new protozoan contains a single bacterium. Trypanosomatid endosymbiosis involves an intense metabolic exchange: the bacterium supplies the protozoan with amino acids, heme, and vitamins, while benefiting from the host’s energy and phospholipid production (de Azevedo-Martins et al. 2007; Motta et al. 2010; Alves et al. 2011, 2013; Klein et al. 2013; Loyola-Machado et al. 2017). Thus, endosymbiosis in trypanosomatids has been used to study cell evolution and the origin of organelles.
Symbiont-harboring trypanosomatids are distributed in four genera: Angomonas, Strigomonas (Teixeira et al. 2011), and Kentomonas (Votýpka et al. 2014), constituting the Strigomonadinae subfamily, and the phylogenetically distant genus Novymonas (Kostygov et al. 2016). They have ultrastructural and biochemical features that distinguish them from other monoxenics and human pathogenic trypanosomatids, such as Trypanosoma cruzi and Leishmania sp., the latter a phylogenetically close genus to symbiont-harboring trypanosomatids. While draft genome assemblies are available for Angomonas and Strigomonas, there are no complete chromosomal assemblies for any of the four genera of symbiont-harboring trypanosomatids.
The first genome sequencing of A. deanei identified the predicted proteins of the protozoan and its symbiont (Motta et al. 2013), and two further sequencing efforts have produced fragmented whole-genome assemblies (Alves et al. 2013; Morales et al. 2016). These assemblies have been used to study the loss, transference, and interference of genes during symbiosis (Alves et al. 2013), as well as to investigate heterologous or endogenous gene and protein expression (Catta-Preta et al. 2016; Morales et al. 2016; Penha et al. 2016). However, the structure and full noncoding regions of the genome have not been resolved yet. Here, we present a new assembly of the A. deanei genome, sequenced using Oxford Nanopore single-molecule technology, which is resolved into 29 chromosomes and reveals several previously unknown special features of the genome. We expect that the new assembly will assist future studies of symbiont-harboring trypanosomatids and other trypanosomatids and monoxenics.
Materials and methods
Supplementary methods and Supplementary file descriptions can be found in Supplementary File S1.
Organism/strain origin and derivation
Crithidia deanei Carvalho (ATCC® PRA-265™) (Carvalho 1973), now A. deanei (Teixeira et al. 2011) was cultivated axenically in Warren’s medium (Warren 1960), supplemented with 10% fetal calf serum for 24 h at 27°C. Cells were concentrated to 108 by centrifugation (×1200 g for 10 min) before DNA extraction.
Sample preparation
DNA was extracted from snap-frozen pellets containing approximately 108 cells using a beta version of the Nanobind CBB Big DNA Kit (Circulomics Inc.), according to the manufacturer’s guidelines, using the HMW protocol for gram-negative bacteria. Briefly, cell pellets were resuspended in 20-μl PBS before addition of equal volumes of proteinase K and kit cell lysis buffer CLE, and incubation at 55°C for 20 min. Samples were then treated with RNase A for 5 min at room temperature, before the addition of kit buffer BL3 and a further 15-min incubation at 55°C. DNA was precipitated with isopropanol, in the presence of the Nanobind disk, washed as per the protocol, and eluted from the disk into Tris elution buffer. DNA was left overnight at 4°C to fully resuspend before further processing.
Sequencing
For high accuracy short-read sequencing, a paired-end library was prepared using the NEBNext Ultra II FS DNA library prep kit for Illumina (New England Biolabs), according to the manufacturer’s instructions, using 100 ng starting DNA, and using four cycles of PCR amplification using NEBNext multiplex oligos for Illumina (unique dual index primers; NEB). The library was then subject to 2 × 150 bp sequencing on an Illumina HiSeq 3000 sequencer, at the University of Leeds Next Generation Sequencing Facility.
Long-fragment DNA sequencing was performed using an Oxford Nanopore Technologies (ONT) MinION sequencer. Approximately 500-ng genomic DNA was subject to shearing using the Covaris g-TUBE™ to a mean fragment size of 20 kb, and mixed with an additional 1 µg of unsheared genomic DNA. The sequencing libraries were generated using the SQK-LSK109 ligation sequencing kit (ONT). Library preparation started with DNA repair/A-tailing using the NEBNext® Ultra™ II End Repair/dA-Tailing Module, with additional NEBNext FFPE repair enzyme (NEB), using sequential incubations for 30 min at 20°C and then 65°C. Following clean up with 0.9× volume AMPure XP beads (Beckman Coulter), adapters were ligated to prepared DNA ends using NEBNext quick T4 DNA ligase, and the ligation buffer provided in the SQK-LSK109 kit. An additional clean up with AMPure XP beads, including two washes using the ONT Long Fragment Buffer, was performed prior to elution into the buffer provided. The total eluted library was then loaded onto an ONT FLO-MIN109 R9.4.1 flow cell, following the manufacturer's guidelines, and run for 48 h using MinKNOW software.
Sequence processing and genome assembly
Oxford Nanopore MinION sequences were base called with Guppy 3.1.5. Adapters were trimmed using Porechop 0.2.3 (https://github.com/rrwick/Porechop). The raw MinION reads were assembled with Canu 1.8 (Koren et al. 2017) (https://canu.readthedocs.io) with option “genomeSize = 23m.” The raw assembly (Supplementary File S2) was manually assessed and edited (see Supplementary File S1 Section 1, File S3, and File S4 for full details). Contig and read alignments for assessments were produced with minimap2 2.17-r941 (Li 2018) (https://github.com/lh3/minimap2) and were inspected using IGV 2.5.3 (Thorvaldsdóttir et al. 2013) (http://software.broadinstitute.org/software/igv/). The genome was edited with seqkit 0.10.0 (Shen et al. 2016) (https://bioinf.shenwei.me/seqkit/).
The filtered assembly was polished with medaka 0.7.1 (https://github.com/nanoporetech/medaka), using a filtered set of reads longer than 20 kb generated with filtlong 0.2.0 (https://github.com/rrwick/Filtlong) using options min_length 20000 and keep_percent 90. The medaka-polished assembly was polished with Illumina data three times using Pilon 1.22 (Walker et al. 2014) (https://github.com/broadinstitute/pilon) (Supplementary File S5 contains the polished assembly). Before polishing, Illumina sequences were adapter trimmed with cutadapt 1.16 (Martin 2011) (https://cutadapt.readthedocs.io) for the Illumina Universal Adapter sequence AGATCGGAAGAG. bwa 0.7.17 (Li 2013) (https://github.com/lh3/bwa) and samtools 1.9 (Li et al. 2009) (http://www.htslib.org) were used to align Illumina reads to the assembly for polishing.
Validation with PCR
PCRs to validate assembly features (Figure 2; see Supplementary File S1 Section 2.2 for further details) were prepared with 10 ng of A. deanei DNA in each reaction (or water for negative controls), mixed with a low ROX SYBR Green master mix and run on a QuantStudio 3, using a 2 step fast PCR with a 2 s denaturing step at 95°C and 30 s anneal and extend step at 60°C, for 32 cycles. Fifteen microliters of each product was run on a 2% agarose gel with an Invitrogen 50-bp DNA ladder.
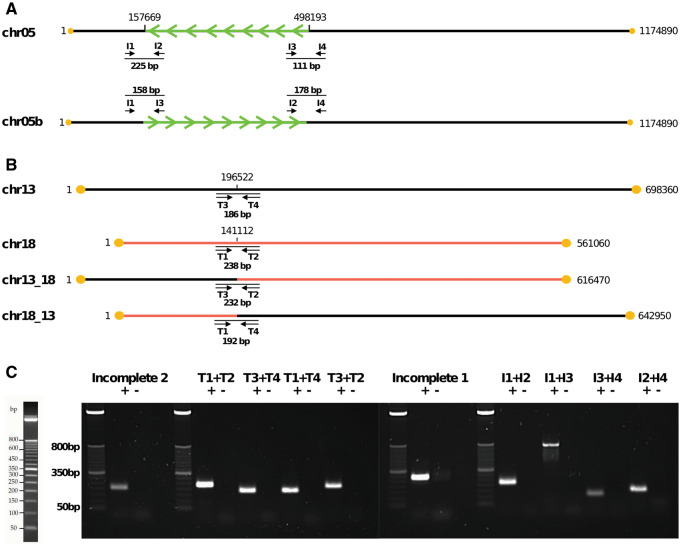
Figure 2.
PCR validation of special features. (A) Chromosome 5 inversion. Inversion shown with arrows in green. Primers I1, I2, I3, and I4 were designed to span the breakpoints of the two inversion haplotypes chr05 and chr05b. Primer products shown as thin black lines (not to scale); expected primer product size is shown for each primer pair. Breakpoint positions in polished genome (Supplementary File S5) given above chr05. Yellow dots are telomeres. (B) Chromosome 13/18 translocation. Primers T1, T2, T3, and T4 were designed to span the breakpoints of the four translocation haplotypes chr13, chr18, chr13_18, and chr18_13. Key as in Figure 2A. (C) PCR products shown via gel electrophoresis against a 50-bp Invitrogen DNA ladder (left) for the inversion (I1–I4), the translocation (T1–T4) and two incomplete chromosomes (Supplementary File S1 Section 2.2, File S5, Table S3 and S4). “+” and “−” lanes show product and negative control (water), respectively.
Annotation
The previous genome annotation (Motta et al. 2013), NCBI accession GCA_000442575.2, was transferred with flo (Pracana et al. 2017). Duplicate annotations and erroneous proteins were fixed with a custom Python script (Supplementary File S6; output in Supplementary File S7), and the genome was also annotated using Companion version 1.0.2 (Steinbiss et al. 2016) (Supplementary Files S8–S12). Full details of the annotation process are in Supplementary File S1 Section 3. Median TriTrypDB statistics were calculated by downloading a table of genome information from https://tritrypdb.org (downloaded on December 11, 2019 via Data Summary → Genomes and Data Types; on January 6, 2021 the same data was available via Data → Organisms: Genome Info & Stats) and restricting to reference genomes only.
Genome analysis
Redundancy of genome assemblies (Figure 1) was assessed by aligning genomes to themselves with minimap2 2.17-r941 (Li 2018) using options -x ava-ont and -a to output SAM format; alignments were then sorted and indexed with samtools 1.9 (Li et al. 2009). Copy numbers were calculated with mosdepth 0.2.5 (Pedersen and Quinlan 2018) (https://github.com/brentp/mosdepth) using option -F 0 to include all alignments. A script (Supplementary File S13) was run to calculate the number of bases assigned to each copy number from the mosdepth output. chr02 was identified as supernumerary (Figure 3) by aligning A. deanei Illumina reads used for polishing to the polished assembly with bwa 0.7.17 (Li 2013) and calling variants with freebayes v1.1.0-3-g961e5f3 (Garrison and Marth 2012) (https://github.com/ekg/freebayes) with option -F 0 to accept variants with any alternate fraction. The freebayes VCF was filtered to heterozygote SNPs only using perl and awk, and filtered to only unique regions of the genome using bcftools 1.9 (Li 2011) (https://www.htslib.org) and the mosdepth BED file from the genome redundancy analysis (see above).
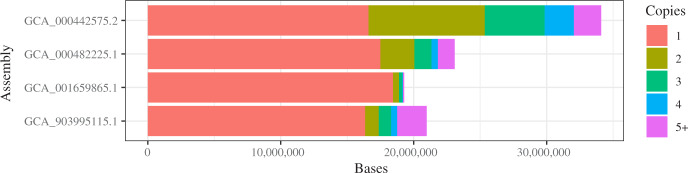
Figure 1.
Redundancy of genome assemblies. Bars show number of bases in assemblies colored by copy number. Unique material has only one copy in the assembly (red). Highly repetitive material has many copies. Large amounts of material with two or three copies suggest haplotypic variation has been retained, although some nonunique material is expected due to common repeats.
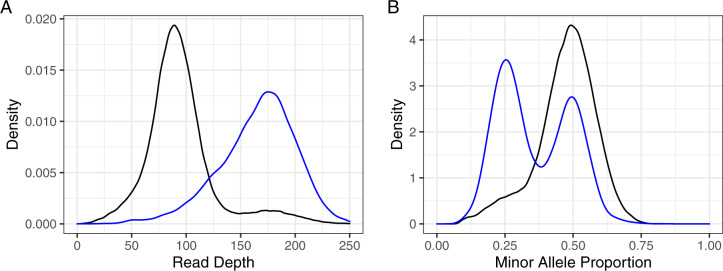
Figure 3.
Chromosome 2 is a supernumerary chromosome. (A) Read depths at SNPs in unique regions across the whole nuclear genome (black) or on chr02 only (blue). Chr02 median depth (170) is roughly double the whole-genome median depth (88), indicating chr02 may have double the copy number of the rest of the genome. (B) Proportion of reads with minor allele for all SNPs in unique regions across the genome (black) or on chr02 only (blue). 0.5 (1:1 ratio) indicates two copies; a mixture of 0.25 (1:3 ratio) and 0.5 indicates four copies.
Assemblies were assessed with BUSCO v4.0.6 (Seppey et al. 2019) (https://busco.ezlab.org) using lineage eukaryota_odb10 and options -m genome and long.
Data availability
Raw reads are available in the European Nucleotide Archive (project, PRJEB36170; study, ERP119328; sample, ERS4235756; Oxford Nanopore reads, ERR3813852; Illumina reads, ERR3813853). The assembly and annotation are available at accession GCA_903995115; chromosome sequences, LR877145-LR877173. All URLs were accessible on 6 January 2021. Supplemental material is available at figshare DOI: https://doi.org/10.25387/g3.13252664.
Results and discussion
The genome size and chromosome number for A. deanei are unknown. Three previous genome assemblies are available (Table 1) (Alves et al. 2013; Motta et al. 2013; Morales et al. 2016). The first (Motta et al. 2013)is a reference-guided assembly aimed at identifying protein-coding gene sequences, using a set of 73.8 thousand protein sequences from TriTrypDB 3.3 as a reference, but also including contigs assembled from reads that did not align to the reference. All three assemblies are fragmented and two contain many gaps. They are also of varying sizes (34.1, 23.1, and 19.3 Mb). However, the first assembly contains only 16.6 Mb of unique material, with a further 13.2 Mb of sequence occurring two or three times in the genome (Figure 1). Nonunique material may be accurate expansions of highly repetitive sequence, but could also be extra haplotypic material that should be removed. Of 129 complete eukaryotic BUSCOs found in this assembly, 88 (68% of complete BUSCOs) are duplicated (Table 1). This suggests the first assembly contains many haplotypic sequences, not found to such an extent in the other assemblies, and so the true genome size is likely to be closer to 20 Mb than 35 Mb.
Table 1.
Summary of A. deanei genome assemblies
| NCBI ID | GCA_000442575.2 | GCA_000482225.1 | GCA_001659865.1 | GCA_903995115.1 |
|---|---|---|---|---|
| Name | Angomonas_deanei_Genome | Adea_1.0 | Angomonas_deanei_v1.0 | Adeanei_nanopore_chromosomes |
| Reference | Motta et al. (2013) | Alves et al. (2013) | Morales et al. (2016) | This paper |
| Scaffolds | 17 339 | 5 616 | 408 | 29 |
| Length (bp) | 34 103 807 | 23 079 371 | 19 282 250 | 20 976 081 |
| Scaffold N50 | 2 497 | 11 595 | 300 798 | 774 942 |
| Gaps (bp) | 30 204 | 197 | 1 728 731 | 0 |
| Complete BUSCOs | 129 (50.6%) | 125 (49.1%) | 127 (49.8%) | 128 (50.2%) |
| Complete, single-copy BUSCOs | 41 (16.1%) | 120 (47.1%) | 124 (48.6%) | 125 (49.0%) |
| Complete, duplicated BUSCOs | 88 (34.5%) | 5 (2.0%) | 3 (1.2%) | 3 (1.2%) |
| Fragmented BUSCOs | 22 (8.6%) | 21 (8.2%) | 20 (7.8%) | 21 (8.2%) |
| Missing BUSCOs | 104 (40.8%) | 109 (42.7%) | 108 (42.4%) | 106 (41.6%) |
BUSCO statistics are from a set of 255 eukaryotic benchmarking universal single-copy orthologs (BUSCOs) (Seppey et al. 2019). Percentages are calculated from all 255 BUSCOs.
We sequenced 2,051,753 Oxford Nanopore MinION reads containing 13,302,088,880 bp of sequence after adapter trimming (665 times coverage of a 20 Mb genome) with a read N50 of 14,610 bp, and 9,775,722 Illumina HiSeq 3000 read pairs totaling 2,952,268,044 bp (read length 150 bp, 148 times coverage of a 20 Mb genome). We assembled the MinION sequence with Canu (Koren et al. 2017) to produce an initial raw genome assembly containing 212 contigs, 27,914,394-bp long (Supplementary File S2), with a contig N50 of 646,966 bp and no gaps, already an improvement on any existing assembly.
To improve the raw Canu assembly (Supplementary File S2), we ran the assembly through Tapestry (Davey et al. 2020) (https://github.com/johnomics/tapestry) to calculate quality information for each contig (Supplementary Table S1, File S3), and then filtered and edited the genome based on this information (Supplementary Table S1, File S1 Section 1, File S4, Figures S1–S13). The assembly contained a symbiont genome in 1 contig (Supplementary File S1 Section 1.1), 127 contigs from the kinetoplast minicircle (which were removed from the assembly; Supplementary File S1 Section 1.2) (Lukeš et al. 2002), and 3 contigs from the kinetoplast maxicircle (which were reduced to one unique copy of the maxicircle) (Supplementary File S1 Section 1.3, Figure S1). As full-length accessory genomes are already publicly available [symbiont: NCBI GenBank GCA_000319225.1 (Motta et al. 2013) and GCF_000340825.1 (Alves et al. 2013), maxicircle: NCBI GenBank KJ778684.1], these have been removed from our public assembly (NCBI GenBank GCA_903995115), but they are available in our polished assembly included with this paper (Supplementary File S5).
This left 81 contigs from the nuclear genome. Of these, 49 contigs were extra repeat copies, subtelomeric, or haplotypic and were removed from the assembly, leaving 32 contigs (see Supplementary Table S1 for details). Manual inspections resolved these contigs to 29 complete or close-to-complete chromosomal sequences, with incomplete contigs explainable due to a translocation (Supplementary Figures S2 and S3), an inversion (Supplementary Figure S4), and several misassemblies (Supplementary Figures S5–S10) (all discussed in detail in Supplementary File S1 Sections 1.4–1.8; genome edits and translocation and inversion haplotypes summarized in Supplementary Table S2). Fifty-six of 58 contig ends have multiple copies of the trypanosome telomere sequence CCCTAA/TTAGGG (Dreesen et al. 2007); although the remaining two contig ends do not contain telomeres, the majority of reads that align to these ends do contain telomeres, so these ends are likely to be almost complete (Supplementary File S1 Section 1.9, Figures S11–S13). The translocation and inversion were validated with read alignments (Supplementary File S1 Section 2.1, Table S2, Figures S14–S21) and with PCR (Figure 2, Materials and methods, Supplementary File S1 Section 2.2, Tables S3 and S4).
We transferred the first A. deanei gene annotation (NCBI genome GCA_000442575.2) to our new nuclear genome assembly, and also predicted new genes and RNAs where possible (see Materials and methods, Supplementary File S1 Section 3, Supplementary Files S6–12). The new annotation has 10,365 protein-coding genes (7199 transferred, 3166 newly predicted), 59 tRNAs covering all 20 standard amino acids and 1 tRNA for selenocysteine, 26 ribosomal RNAs, and 62 noncoding RNAs (45 ncRNA, 14 snoRNA, 3 snRNA). This compares well to other reference genomes in the Kinetoplastid Genomics database TriTrypDB, which have median 8652 protein-coding genes (median absolute deviation 387) and 110 nonprotein coding genes (median absolute deviation 27).
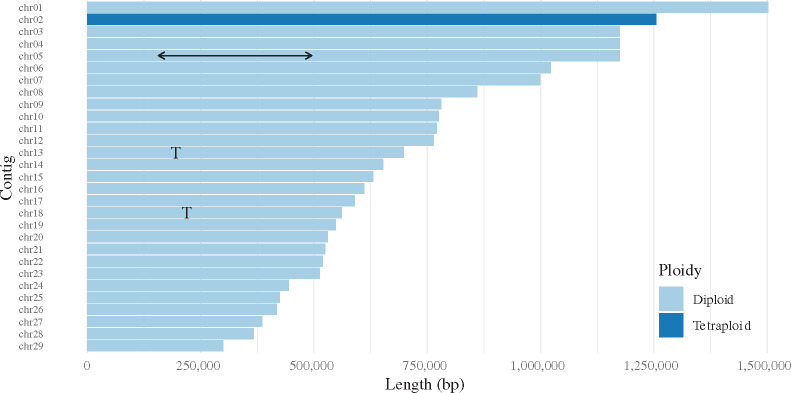
We therefore propose that A. deanei has 29 chromosomes, and have named the remaining 29 contigs chr01 to chr29 in order of size, largest first (Supplementary File S9). These 29 chromosomes make a nuclear genome of 20,976,081 bp, chromosome N50 774,942 bp, with no gaps (Supplementary Table S5). The assembly has a supernumerary chromosome, in common with other trypanosomatids (Downing et al. 2011; Rogers et al. 2011; Reis-Cunha et al. 2018), with chromosome 2 (chr02) having considerably higher read depth than other chromosomes (Figure 3A); the contig has a mixture of 1:1 and 3:1 ratios for SNP calls (Figure 3B), which suggests there are four copies of this chromosome, not two, as for the remaining diploid chromosomes. There is an inversion on chromosome 5 (chr05) between 157.6 and 498.1 kb, 340.5-kb long (1.61% of the nuclear genome), containing 173 genes (1.67% of the protein-coding genes in the nuclear genome). Chromosomes 13 and 18 (chr13, chr18) reciprocally translocate at chromosome 13 196.6 kb and chromosome 18 141.1 kb. Figure 4 shows the genome with these features; the lengths of the contigs are summarized in Supplementary Table S5.
Figure 4.
Chromosome lengths in new A. deanei nuclear genome assembly. chr02 is supernumerary (dark blue), chr05 has a 340-kb inversion (line with arrows), and chromosomes 13 and 18 translocate at the points marked “T.”
All four public A. deanei genome assemblies have very similar BUSCO scores (Table 1), indicating that all four assemblies have similar gene coverage, despite the excess of duplicated genes in the GCA_000442572.2 assembly. The low yet consistent percentages of eukaryotic BUSCO genes across all A. deanei assemblies suggest this eukaryotic gene set is not representative of the A. deanei genome, rather than suggesting a large number of A. deanei genes are missing from all of these assemblies; nevertheless, the BUSCO gene set is useful for comparing the four assemblies. Our new assembly matches the gene coverage of the other assemblies, with slightly higher complete single-copy gene sequences, while greatly improving genome contiguity.
We expect our new high-quality, close-to-complete genome assembly, including full chromosome sequences and many noncoding RNAs and nongenic regions, will be useful for future research. It is the first chromosomal assembly for any endosymbiont-bearing trypanosomatid. MicroRNAs have been reported as important regulators of symbiosis in plants (Hussain et al. 2018; Hossain et al. 2019), an interesting mechanism that can now be investigated in A. deanei, the model of symbiosis in trypanosomatids. Recently, a Brazilian patient presenting symptoms of leishmaniasis was nonresponsive to available treatments and was found to be infected with a new trypanosomatid phylogenetically related to Crithidia fasciculata, a monoxenic trypanosomatid for which only an unpublished draft genome is available (Maruyama et al. 2019). There are few monoxenic genomes that can be used as a reference in such cases, as well as in coinfections of pathogens and the so-called nonpathogens (Pacheco et al. 1998; Srivastava et al. 2010; Ghosh et al. 2012; Kraeva et al. 2015). This new A. deanei assembly can now be used to assist in the identification of new, possibly pathogenic, species. Moreover, a toolkit for reverse genetic studies is being developed for A. deanei, which will illuminate more of the biology of the protozoan and its symbiotic relationship with a prokaryote, and the evolutionary leap from symbiont to organelle. Finally, the assembly provides another example of small genomes being almost completely resolved with single runs of long-read sequencing, with close examination of the sequences revealing special features of the genome never known before.
Acknowledgments
The genome assembly and some other computational tasks were undertaken on the Viking Cluster, which is a high-performance compute facility provided by the University of York. We are grateful for computational support from the University of York High-Performance Computing Service, Viking, and the Research Computing Team.
Funding
This work was funded in part by the Wellcome Trust (grant number 200807/Z/16/Z), by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grant number 305335/2018-9) and by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, grant number E-26/2002.917/2017).
Conflicts of interest: None declared.
Literature cited
- Alves FM, Olifiers N, Bianchi RdC, Duarte AC, Cotias PMT, et al. 2011. Modulating variables of Trypanosoma cruzi and Trypanosoma evansi transmission in free-ranging Coati (Nasua nasua) from the Brazilian Pantanal region. Vector Borne Zoonotic Dis. 11:835–841. [DOI] [PubMed] [Google Scholar]
- Alves JMP, Klein CC, da Silva FM, Costa-Martins AG, Serrano MG, et al. 2013. Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol Biol. 13:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho ALM. 1973. Estudos sobre a posição sistemática, a biologia e a transmissão de tripanosomatídeos encontrados em Zelus leucogrammus (Perty, 1834) (Hemiptera, Reduviidae). Revista Patol Trop. 2:223–274. [Google Scholar]
- Catta-Preta CMC, Dos Santos Pascoalino B, de Souza W, Mottram JC, Motta MCM, et al. 2016. Reduction of tubulin expression in Angomonas deanei by RNAi modifies the ultrastructure of the trypanosomatid protozoan and impairs division of its endosymbiotic bacterium. J Eukaryot Microbiol. 63:794–803. [DOI] [PubMed] [Google Scholar]
- Davey JW, Davis SJ, Mottram JC, Ashton PD.. 2020. Tapestry: validate and edit small eukaryotic genome assemblies with long reads. bioRxiv. doi:10.1101/2020.04.24.059402.
- de Azevedo-Martins AC, Frossard ML, de Souza W, Einicker-Lamas M, Motta MCM.. 2007. Phosphatidylcholine synthesis in Crithidia deanei: the influence of the endosymbiont. FEMS Microbiol Lett. 275:229–236. [DOI] [PubMed] [Google Scholar]
- Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, et al. 2011. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 21:2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesen O, Li B, Cross GAM.. 2007. Telomere structure and function in trypanosomes: a proposal. Nat Rev Microbiol. 5:70–75. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. https://arxiv.org/abs/1303.3997v2.
- Ghosh S, Banerjee P, Sarkar A, Datta S, Chatterjee M.. 2012. Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. J Clin Microbiol. 50:2774–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Hoang NT, Yan Z, Tóth K, Meyers BC, et al. 2019. Characterization of the spatial and temporal expression of two soybean miRNAs identifies SCL6 as a novel regulator of soybean nodulation. Front Plant Sci. 10:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SS, Hussain M, Irfan M, Siddique KHM.. 2018. Legume, microbiome, and regulatory functions of miRNAs in systematic regulation of symbiosis. In:Egamberdieva D, Ahmad P, editors. Plant Microbiome: Stress Response. Singapore: Springer Singapore. p. 255–282. [Google Scholar]
- Klein CC, Alves JMP, Serrano MG, Buck GA, Vasconcelos ATR, et al. 2013. Biosynthesis of vitamins and cofactors in bacterium-harbouring trypanosomatids depends on the symbiotic association as revealed by genomic analyses. PLoS One. 8:e79786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostygov AY, Dobáková E, Grybchuk-Ieremenko A, Váhala D, Maslov DA, et al. 2016. Novel trypanosomatid-bacterium association: evolution of endosymbiosis in action. MBio. 7:e01985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraeva N, Butenko A, Hlaváčová J, Kostygov A, Myškova J, et al. 2015. Leptomonas seymouri: adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani. PLoS Pathog. 11:e1005127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN].
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, 1000 Genome Project Data Processing Subgroup, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola-Machado AC, Azevedo-Martins AC, Catta-Preta CMC, de Souza W, Galina A, et al. 2017. The symbiotic bacterium fuels the energy metabolism of the host trypanosomatid Strigomonas culicis. Protist. 168:253–269. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Lys Guilbride D, Votýpka J, Zíková A, Benne R, et al. 2002. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot Cell. 1:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 17:10–12. [Google Scholar]
- Maruyama SR, de Santana AKM, Takamiya NT, Takahashi TY, Rogerio LA, et al. 2019. Non-Leishmania parasite in fatal visceral Leishmaniasis-like disease, Brazil. Emerg Infect Dis. 25:2088–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J, Kokkori S, Weidauer D, Chapman J, Goltsman E, et al. 2016. Development of a toolbox to dissect host-endosymbiont interactions and protein trafficking in the trypanosomatid Angomonas deanei. BMC Evol Biol. 16:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MCM, Catta-Preta CMC, Schenkman S, de Azevedo Martins AC, Miranda K, et al. 2010. The bacterium endosymbiont of Crithidia deanei undergoes coordinated division with the host cell nucleus. PLoS One. 5:e12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MCM, de Azevedo Martins AC, de Souza SS, Catta-Preta CMC, Silva R, et al. 2013. Predicting the proteins of Angomonas deanei, Strigomonas culicis and their respective endosymbionts reveals new aspects of the trypanosomatidae family. PLoS One. 8:e60209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco RS, Marzochi MC, Pires MQ, Brito CM, de Madeira MF.. 1998. Parasite genotypically related to a monoxenous trypanosomatid of dog’s flea causing opportunistic infection in an HIV positive patient. Mem Inst Oswaldo Cruz. 93:531–537. [DOI] [PubMed] [Google Scholar]
- Pedersen BS, Quinlan AR.. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 34:867–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penha LL, Hoffmann L, de Souza SS, de A, Martins CA, Bottaro T, et al. 2016. Symbiont modulates expression of specific gene categories in Angomonas deanei. Mem Inst Oswaldo Cruz. 111:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracana R, Priyam A, Levantis I, Nichols RA, Wurm Y.. 2017. The fire ant social chromosome supergene variant Sb shows low diversity but high divergence from SB. Mol Ecol. 26:2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Cunha JL, Valdivia HO, Bartholomeu DC.. 2018. Gene and chromosomal copy number variations as an adaptive mechanism towards a parasitic lifestyle in trypanosomatids. Curr Genomics. 19:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, et al. 2011. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 21:2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM.. 2019. BUSCO: assessing genome assembly and annotation completeness. In: Kollmar M, editor. Gene Prediction: Methods and Protocols. New York, New York, NY: Springer. p. 227–245. [DOI] [PubMed] [Google Scholar]
- Shen W, Le S, Li Y, Hu F.. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 11:e0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P, Prajapati VK, Vanaerschot M, Van der Auwera G, Dujardin JC, et al. 2010. Detection of Leptomonas sp. parasites in clinical isolates of Kala-azar patients from India. Infect Genet Evol. 10:1145–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S, Silva-Franco F, Brunk B, Foth B, Hertz-Fowler C, et al. 2016. Companion: a web server for annotation and analysis of parasite genomes. Nucleic Acids Res. 44:W29–W34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MMG, Borghesan TC, Ferreira RC, Santos MA, Takata CSA, et al. 2011. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist. 162:503–524. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votýpka J, Kostygov AY, Kraeva N, Grybchuk-Ieremenko A, Tesařová M, et al. 2014. Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist. 165:825–838. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren LG. 1960. Metabolism of Schizotrypanum cruzi Chagas. I. Effect of culture age and substrate concentration on respiratory rate. J Parasitol. 46:529–539. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw reads are available in the European Nucleotide Archive (project, PRJEB36170; study, ERP119328; sample, ERS4235756; Oxford Nanopore reads, ERR3813852; Illumina reads, ERR3813853). The assembly and annotation are available at accession GCA_903995115; chromosome sequences, LR877145-LR877173. All URLs were accessible on 6 January 2021. Supplemental material is available at figshare DOI: https://doi.org/10.25387/g3.13252664.