Abstract
Genomic selection (GS) is revolutionizing plant breeding since the selection process is done with the help of statistical machine learning methods. A model is trained with a reference population and then it is used for predicting the candidate individuals available in the testing set. However, given that breeding phenotypic values are very noisy, new models must be able to integrate not only genotypic and environmental data but also high-resolution images that have been collected by breeders with advanced image technology. For this reason, this paper explores the use of generalized Poisson regression (GPR) for genome-enabled prediction of count phenotypes using genomic and hyperspectral images. The GPR model allows integrating input information of many sources like environments, genomic data, high resolution data, and interaction terms between these three sources. We found that the best prediction performance was obtained when the three sources of information were taken into account in the predictor, and those measures of high-resolution images close to the harvest day provided the best prediction performance.
Keywords: high-resolution images, genomic data, plant breeding, generalized poisson regression, genomic selection, count data
Introduction
In traditional breeding programs, recognizing the phenotypic appearance of traits is frequently done to obtain the best candidate genotypes. Doing this procedure is costly since all combinations of genotypes must be seen in the field. To solve this problem, a statistical machine learning procedure, known as genomic selection (GS), was introduced. GS became essential since it can find the most desirable genotypes by learning the relationship between the information about the genotype and the phenotype of the training set (Meuwissen et al. 2001). Then, the trained model is used to predict the breeding values or phenotypes of candidate genotypes, based on the available genotypic information. Other variables such as environmental covariates, pedigree information, and their interactions could be included in the model to provide more explanations about the phenotypic variability.
Another way to gain better prediction performance is by applying the current development in high-resolution imaging technology. Modern cameras can provide hundreds of reflectance data at different wavelengths. Continuous examination of this information during the growing season yields much information about the physiological, agronomic, and disease traits of the crops. Moreover, imperfect phenotypic measurements can be generated at very early stages before harvesting (Araus and Cairns 2014). Along with large-scale multi-environmental tests, these conditions yield new opportunities for genetic improvement. Some opportunities are: (i) to increase in the capability of screening large number of genotypes in the field, with nondestructive, repeated, objective observations, without the requirement of an extensive labor force (Rouphael et al. 2018), (ii) to facilitate the study of plants’ responses to various types of environmental stresses (Humplík et al. 2015), (iii) to unraveling complex questions of plant growth, development, responses to environment, as well as selection of appropriate genotypes in molecular breeding strategies (Humplík et al. 2015). The use of high-resolution phenotyping and GS simultaneously can decrease the cost of phenotyping (which often delays the genetic improvement), and allows the expansion of field trials that are logistically and economically viable (Cabrera-Bosquet et al. 2012). However, as one reviewer pointed out temporal phenotype information resulting of hyperspectral images used in GS can mostly give different accuracies depending on the phenotypes belonging to different growing periods of the crop. However, early growth-related gene effects have been overlooked so far because of traditional or low number of phenotyping. For this reason, thanks to using high-resolution phenotyping data, GS can perform better since GS also will use the marker(loci) effects from early generation that has been overlooked so far.
Nowadays, numerous crops with hundreds of genotypes can be examined through high-performance phenotyping platforms at a reasonable cost (Montes et al. 2007). In the beginning, these platforms made use of cameras mounted on tractors or drones and developed for controlled environments (Granier et al. 2006; Montes et al. 2007). Later, the presence of unmanned aerial vehicles promoted the development of high-performance phenotyping for large-scale field trials (Araus and Cairns 2014; Aguate et al. 2017). One of the uses of hyperspectral image data was to determine the vegetative indexes (VI), which are effective algorithms for quantitative and qualitative evaluations of vegetation cover, vigor, and growth dynamics, among other applications (Xue and Su 2017). These VI can be computed using some bands of the available images, for example, Red normalized difference vegetation index (RNDVI), Simple ratio (SRa), Ratio analysis of reflectance spectra chlorophyll a (RARSa), Normalized Green-Red Difference Index (NGRDI) (Gitelson et al. 2002), etc. To see which bands are used for the computation of each index see Table 1 of the paper of Montesinos-López et al. (2017b). The use of a modern camera might give more information that can be used to calculate the VI. Despite these facts, most of the VI are valid for several specific crops.
Table 1.
Proposed models
| Predictor number | Components of the predictor | Type |
|---|---|---|
| P1 | Conventional | |
| P2 | Conventional | |
| P3 | Conventional | |
| P4 | Functional B-splines | |
| P5 | Functional Fourier | |
| P6 | + | Functional B-splines basis |
| P7 | + | Functional Fourier basis |
| P8 | Conventional | |
| P9 | Conventional | |
| P10 | Functional B-splines basis | |
| P11 | Functional Fourier basis | |
| P12 | Functional B-splines basis | |
| P13 | Functional Fourier basis |
Since the existing indexes did not use all available bands, some authors such as Montesinos-López et al. (2017a) and Aguate et al. (2017) introduced the simultaneous use of hundreds of available bands as predictors to increase the model’s predictive capacity. They found that the simultaneous use of all bands yields more accurate predictions than the use of VIs or those bands that presented higher heritability. In the beginning, the bands were used as the only covariates in the predictor of the proposed model. Later, genotype × environment (G × E) interaction and band × environment (band×E) interaction were included in the model using the functional regression method (Montesinos-López et al. 2018), which significantly improve the predictive capacity. Although this method can increase the predictive capacity significantly, this method proposed by Montesinos-López et al. (2018) is only appropriate for a Bayesian framework and continuous response variables.
It is generally accepted that there is no statistical machine learning model that exhibits the best performance for all types of data. Consequently, some types of data should be analyzed using specific models (Wolpert and Macready 1997). For example, logistic regression performs well for binary data with linear patterns, while multinomial regression is suitable for categorical response variables with linear relations (Stroup 2012). Poisson or negative binomial regression performs well when the response variable is a count (Stroup 2012). Also, Poisson or negative binomial regression should be preferred because they guarantee that all predictions are nonnegative (which is not guaranteed with a Gaussian model) (Montesinos-López et al. 2015, 2016, 2017a). When Gaussian regression is used instead of Poisson regression, negative outputs of the Gaussian regression must be truncated to zero, and it is unclear how this affects the optimality of the predictive distribution (Montesinos-López et al. 2015, 2016, 2017a). Apart from those models, empirical studies show that deep learning (DL) and kernel regression can deal with nonlinear patterns in the data (Patterson and Gibson 2017; Chollet and Allaire 2018). These examples illustrate that unfortunately there is no universal statistical machine learning model that works well for all types of data (Wolpert and Macready 1997).
Many count traits can be measured in plant breeding programs, for example, panicles per plant, number of infected spikelets per plant, number of seeds per plant, length of days to maturity, and many more (Montesinos-López et al. 2016, 2017b).Values of nonnegative integers (without a restricted upper limit) could be taken for count traits. Although there is evidence that Poisson or negative binomial regression is suitable for modeling count data, these data are frequently analyzed as if they were continuous response variables. For this reason, we propose using generalized Poisson regression (GPR) as a prediction model for count data in GS. Under a parsimonious framework, this model can integrate genomic information from thousands of markers, high-resolution images from various time points and plants, environmental information, and their interaction effects. Compression of the dimensionality of the high-resolution images would be done using b-spline and Fourier basis functions.
Material and methods
Data
In this study, we only used data from three management practices (call environments in the entire paper)—drought, irrigated, and reduced irrigation—and 976 lines of the original 1,170 wheat lines from the CIMMYT Global Wheat Program (Montesinos-López et al. 2017b). The experimental design used was an alpha-lattice with three replicates and six blocks. The best linear unbiased estimates (BLUEs) were used as response variables after removing the design effect; for more details about how these BLUEs were calculated, the reader should refer to the publication of Montesinos-López et al. (2018). The discretized (converted to discrete values) trait grain yield (GY) was measured in each line, and this was the trait analyzed in this study. Planting dates in the three environments were December 1-5 2014. The bands were measured on nine different dates (January 10, 2015, January 17, 2015, January 30, 2015, February 7, 2015, February 14, 2015, February 19, 2015, February 27, 2015, March 11, 2015, and March 17, 2015), which we call time-points (1, 2, 3, ., 9, respectively), using 250 discrete narrow wavelengths. In each plot for each line and at each time-point, 250 wavelengths from 392 to 851 nm were measured. The th discretized spectrometric curve is given by . We used the notation without subscripts to denote the response of the band measured at 780 wavelengths, to denote the response of the band measured at 670 wavelengths, and so on. The image data was obtained using a Piper PA-16 Clipper flight that was fitted with a hyperspectral camera (Model: A-series, Micro-Hyperspace Airborne sensor, VNIR Headwall Photonics, www.headwallphotonics.com, Fitchburg, Massachusetts, USA) and thermal camera (A600 series Infrared camera, FLIR, www.flir.com, Boston, US). The plane flew at 270 m above the surface (Montesinos-López et al. 2017b). The aerial high-throughput phenotyping (HTP) data was measured around solar noon time every date, aligning the plane to the solar azimuth for the data acquisition. Images of the experimental fields were obtained and formatted to tabular data by calculating the mean value of the pixels inside the center of each individual trial plot represented as a polygon area on a map. The software used to achieve this was ArcMap (ESRI, USA, CA) (Montesinos-López et al. 2017b).
Univariate generalized poisson regression model
We assume that our training set is composed of pairs of inputs (, ) with , for . Also, we assume that the number of independent variables () is larger than the number of observations ; for example, for predictor P2 (See Table 1), the number of independent variables were ; therefore, the penalized loss function for the univariate GPR model is equal to:
where was derived as the negative penalized log likelihood based on a Poisson distribution (Stroup, 2012), represents the inverse link function that is an exponential function and corresponds to a log link function, is a regularization parameter that can be computed using cross-validation and is a parameter that causes Ridge penalization, Lasso penalization or a mixture of both. For example, when the LL corresponds to a univariate Generalized Poisson Ridge Regression (RR); when , the LL corresponds to a univariate Generalized Poisson Lasso Regression (LR), and when the LL corresponds to a univariate Generalized Poisson elastic net regression (ENR). The loss function was optimized with the R package glmnet and the hyper-parameter was performed with 10-fold cross-validations for all regression models. More details of these models can be found in Montesinos-López et al. (2020).
These three models (RR, LR, and ENR) were implemented with the discretized GY response variable. Also, due to the fact that the hyperspectral images (bands) information was measured at 9 time points of the plants, these three models were implemented for each of the 9 time points. Additionally, to be able to integrate in the GPR model the information of environments, lines (genotypes) and high-resolution images with and without interaction terms, we proposed 13 different predictors (P1 to P13) that take into account and all these available information and these were implemented for each of the 27 combinations of 3 models and 9 time points. In Table 1 was described the 13 predictors implemented for each of the 27 combinations of models and time points.
In any predictor that appears in Table 1, represents the design matrix of environments of order , is the vector of beta coefficients of environments of order , is the design matrix of lines with genomic information of order , is the vector of beta coefficients of lines with genomic information of order , is the design matrix of lines with pedigree information of order , is the vector of beta coefficients of lines with pedigree information of order , is the design matrix of the interaction between genotypes (with genomic information) by environment of order , the vector of beta coefficients of genotypes (with genomic information) by environment interaction of order , is the design matrix of the interaction between genotypes (with pedigree information) by environment of order , is the vector of beta coefficients of genotypes (with pedigree information) by environment of order . is the design matrix that contains the information of all the measured bands (hyperspectral images) of order and is the vector of beta coefficients of bands of order . is the compressed design matrix with b-splines basis functions of order , and is the vector of compressed beta coefficients with b-splines basis functions of bands of order . is the compressed design matrix with Fourier basis functions of order , and is the vector of compressed beta coefficients of bands with Fourier basis functions of order . is the design matrix of the interaction term between bands and environments of order and is the beta coefficient of the interaction term between bands and environments of order . is the design matrix of the interaction term between bands with b-splines basis functions and environments of order , and is the beta coefficient of the interaction term between the compressed bands with b-splines basis functions and environments of order . is the compressed design matrix of the interaction term between bands with Fourier basis functions and environments of order , and is the beta coefficient of the interaction term between compressed bands with Fourier basis functions and environments of order . In Table 1 there are two types of predictors: conventional and functional; those called conventional were built directly using the original input information corresponding to the bands, while those called functional were built after compressing the original information of the bands using b-spline basis functions or Fourier basis functions. The code given in Appendix A allows implementing the 13 predictors under the generalized Lasso Poisson regression (LR) for the first time point, T1. By only modifying in the a cv.glmnet() function of the glmnet package used for implementing the 13 predictors, alpha = 1 to alpha = 0 and alpha to a value between 0 and 1, the Ridge regression Poisson (RR) and Elastic net regression Poisson (ENR) models, respectively, can be implemented. However, to implement each of these models for the other 8 time points, the code provided in Appendix A for (o in 1:1) must be changed to any other time point. For example, for time points 2, 3, 4, …, 9 this should be modified as for (o in 2:2), or (o in 3:3), or (o in 4:4), …, (o in 9:9).
Evaluation of prediction performance
We used cross-validation to evaluate the prediction performance in unseen data. Since our data contain the same lines in environments, we used a type of cross-validation that mimics a situation where lines were evaluated in some environments for all traits but where some lines were missing in other environments. We implemented a fivefold cross-validation, where fourfolds were used for training and onefold for testing. We reported the average prediction performance for the test data in terms of average Spearman’s correlation (ASC) and mean arctangent absolute percentage error (MAAPE) for each environment and across environments. These metrics were computed using the observed and predicted response variables in each fold for the testing set and the average of the 5 metrics is reported. It is important to point out that the process for tuning the hyper-parameter () in the implemented univariate GPR (RR, LR, and ENR) was done with 10-fold cross-validation. After selecting in terms of mean square error, the best combination of the hyper-parameter, the model was refitted but using the whole training set (80% of data, since the training and tuning sets were joined) in each fold. Finally, for each testing set, we computed each of the ASC and MAAPE with its corresponding standard error (SE), then the average of the fivefolds and its SE was reported as a measure of prediction performance and variability in each metric. It is important to point out that the fivefold cross-validation strategy was implemented with only 1 replication. We used only one replication to save computational resources.
Results
The results are provided in two sections. The first section provides the results for each model that compares the 13 predictors at each time point under both metrics: (A)ASC, and (B)MAAPE; the second section provides a comparison in terms of prediction performance between the three models (Ridge regression, Lasso regression and Elastic net regression) under the 13 predictors.
Prediction performance of each model
First is given the prediction performance under Ridge regression, then with Lasso regression and finally with Elastic net regression.
Ridge regression
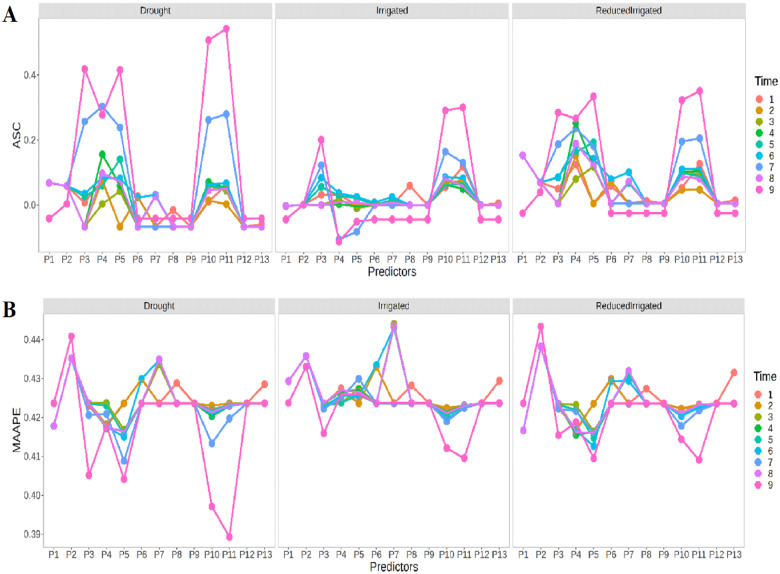
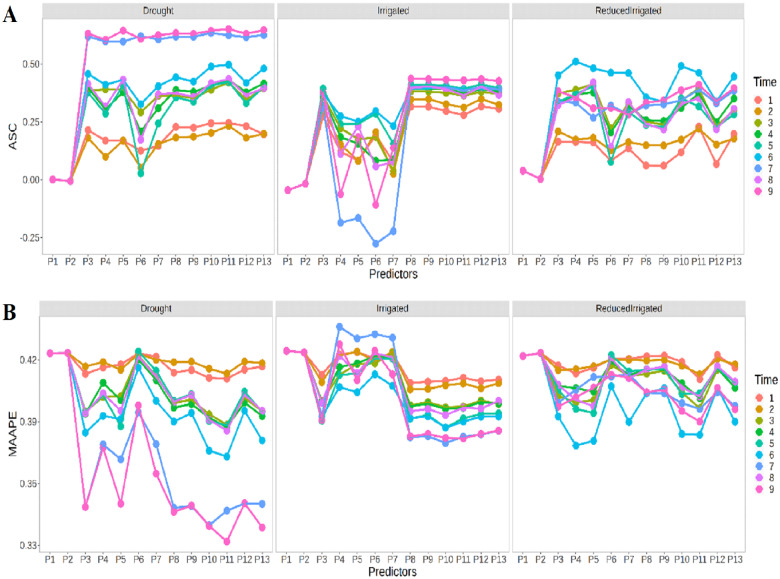
Figure 1A shows in terms of ASC that in general the prediction performance in the three environments was quite similar with the exception being the Drought environment for time points 7 and 9, which presented a better prediction performance than in the other two environments (Irrigated and ReducedIrrigated). Figure 1A also indicates that there are significant differences between the predictors (P1,…,P13), and that predictors P1, P2, P6, P7, P8, and P9 showed the worst prediction performance, while in general predictors P10 and P11 had the best prediction performance. In Figure 1B, we can also observe that in general the best prediction performance was slightly better in the Drought environment. Again, there are significant differences between the prediction performance of the 13 predictors. Now the worst prediction performance was observed with predictor P2 in the Drought and ReducedIrrigation environments, while in the Irrigated environment, the worst prediction performance was observed with predictor P7. Now the best prediction performance was observed under predictors P10 and P11 (in time point 9) in the Drought and Irrigated environments, while in the ReducedIrrigated environment, the best prediction performance was observed under predictors 11 and 5 at the time point 9 (Figure 1B).
Figure 1.
Prediction performance of Ridge regression in each environment in terms of (A) ASC and (B) MAAPE at each time-point under the 13 predictors proposed in Table 1.
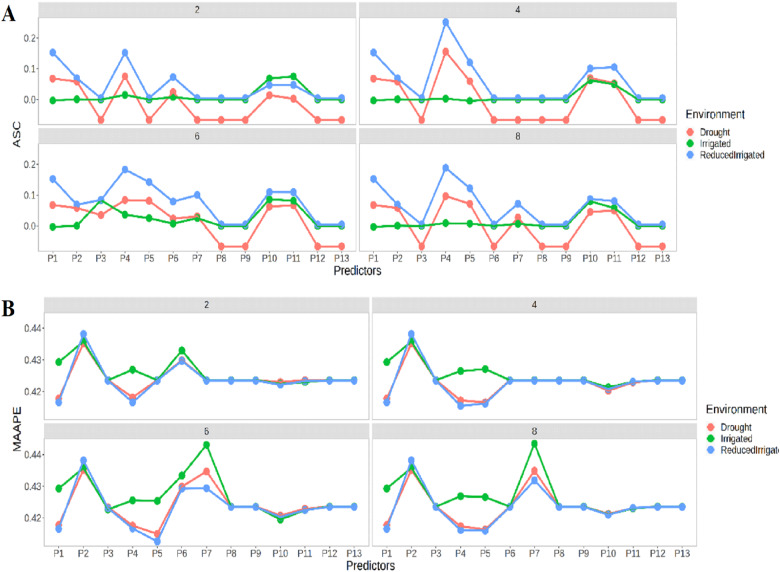
Figure 2A indicates that the in the four time-points (2, 4, 6, and 8), the best predictions in terms of ASC were observed in the ReducedIrrigated environment and the worst in the Drought environment. In general, we can see that the best prediction performance was observed under predictor 4. It is also important to point out that in general we cannot see a significant improvement in the time-points. In terms of MAAPE (Figure 2B), there is a similar performance between environments, but now the best prediction performance was observed in the ReducedIrrigated and Drought environments. Now the worst prediction performances were observed at time points 2 and 4 under predictor P2 and at time points 6 and 8 in predictor P7. On the other hand, the best prediction performances at time points 2, 4, and 6 were observed with predictors P1 and P4 and at time point 8 in predictor P5.
Figure 2.
Prediction performance of Ridge regression at four time points 2, 4, 6, and 8 in terms of (A) ASC and (B) MAAPE in each environment under the 13 predictors proposed in Table 1.
Lasso regression
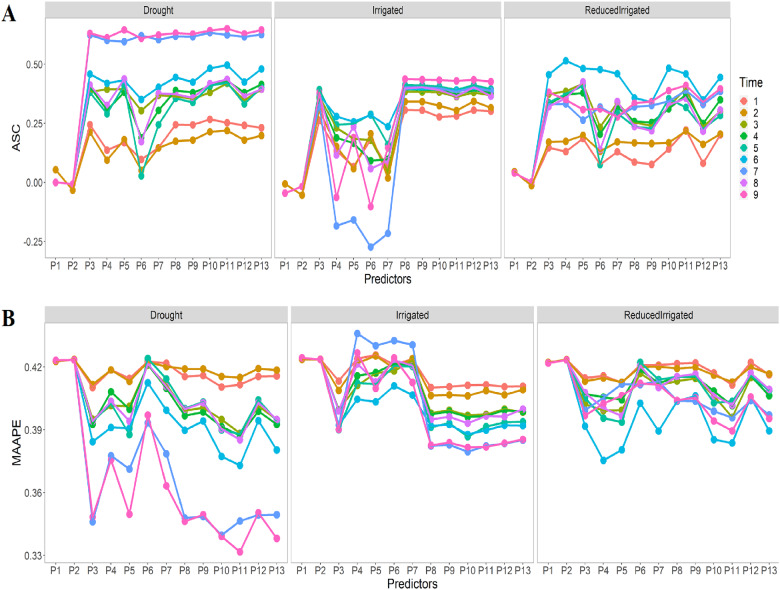
Figure 3A shows that the prediction performances in the three environments were quite similar based on the ASC, with the exception of the Drought environment at time point 7, which showed better prediction performance than the other two environments (Irrigated and ReducedIrrigated environments). Figure 3A also shows that there are significant differences between the predictors (P1,…,P13) and that predictors P1, P2, and P6 show the worst prediction performance, while P5 and P11 shows the best predictions for the Drought, P8 to P13 the best in Irrigated and P3 to P5 had the best prediction for the ReducedIrrigated environment. Figure 3B indicates that in general, the best performance was observed in the Drought environment. Again, we found significant differences between the prediction performances of the 13 predictors. In terms of MAAPE, in general the worst prediction performance was observed with predictor P2 in the Drought and ReducedIrrigated environments, while in the Irrigated environment, the worst prediction performance was found under predictor P4. Now the best prediction performance was observed under predictor P11 (in time-point 9) in the Drought environment, in predictor P10 (time-point 7) in the Irrigated environment and in the predictor P4 (time-point 6) in the ReducedIrrigated environment.
Figure 3.
Prediction performance of Lasso regression in each environment in terms of (A) ASC and (B) MAAPE at each time-point under the 13 predictors proposed in Table 1.
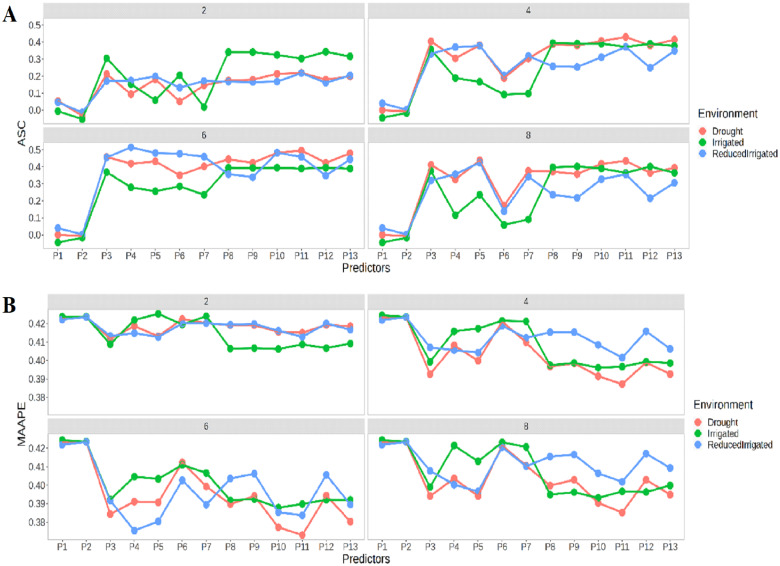
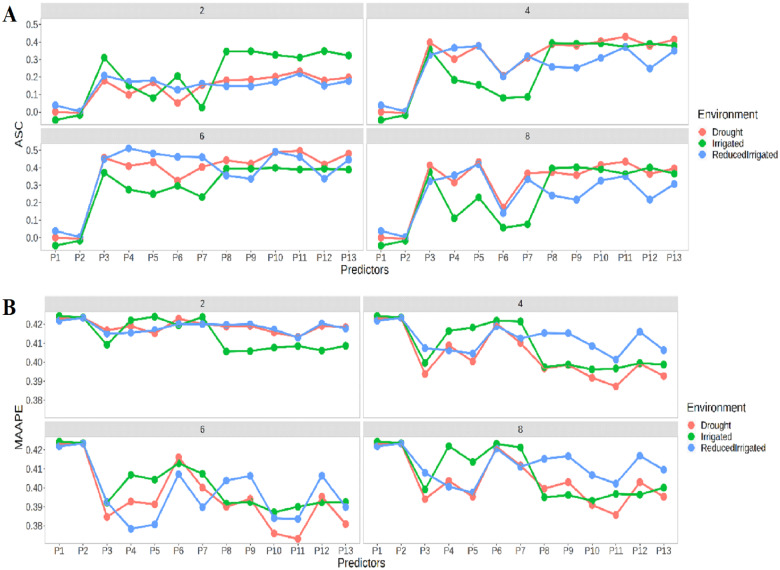
Figure 4A shows that the best prediction in terms of ASC was observed in time-point 2 in the Irrigated environment, in time-points 4 and 8 in the Drought environment, and in time-point 6 in the ReducedIrrigated environment. In general, we found that the best prediction performances were observed under predictors from P8 to P13. In terms of MAAPE (Figure 4B), a similar performance was only observed at time-point 2. The Irrigated environment shows the best prediction performances in the four time-points especially under predictors P1–P7, while the ReducedIrrigated environment stands out under the remaining predictors (P8 until P13). Now, the worst prediction performances in the four time-points were observed under predictors P1 and P2.
Figure 4.
Prediction performance of Lasso regression in four time points 2, 4, 6, and 8 in terms of (A) ASC and (B) MAAPE at each environment under the 13 predictors proposed in Table 1.
Elastic net regression
Figure 5A shows again that the prediction performance in the three environments was quite similar based on the ASC, with the exception of the Drought environment at growth stage 9, which showed better prediction performance than the other two environments (Irrigated and ReducedIrrigated environments). Also, in Figure 5A we can observe that there are significant differences between the predictors (P1, …, P13), and that predictors P1, P2, and P6 show the worst prediction performance, while P3 shows the best predictions in the Drought environment, P8 to P13 in the irrigated environment and P4 shows the best in the ReducedIrrigated environment. We can observe in Figure 5B that the Drought environment produces the best performance in general. Again, we found significant differences between the prediction performances of the 13 predictors. In terms of MAAPE in general the worst prediction performance was observed under predictor P2 in the Drought and ReducedIrrigated environments, while in the Irrigated environment, the worst prediction performance was found under predictor P4. Now the best prediction performance was observed under predictor P11 (in time-point 9) in the Drought environment, under predictor P10 (in time-point 7) in the Irrigated environment and under P4 (in time-point 7) in the ReducedIrrigated environment.
Figure 5.
Prediction performance of Elastic net regression in each environment in terms of (A) ASC and (B) MAAPE at each growth stage (time-point) under the 13 predictors proposed in Table 1.
Figure 6A shows that the best prediction in terms of ASC, again was observed at time-point 2 in the Irrigated environment, at time-points 4 and 8 in the Drought environment, and at time-point 6 in the ReducedIrrigated environment. In general, the best prediction performances were observed under predictors P3 and P11. In terms of MAAPE (Figure 6B), again a similar performance was only observed at time-point 2. The best prediction performances at the four time-points for the Irrigated environment were found under predictors P1 to P7, while for the ReducedIrrigated environment they were found under the remaining predictors (P8 to P13). Again, the worst prediction performances at the four time-points were observed under predictors P1 and P2 in the three environments.
Figure 6.
Prediction performance of Elastic net regression at four time points 2, 4, 6 and 8 in terms of (A) ASC and (B) MAAPE at each environment under the 13 predictors proposed in Table 1.
Comparison between models
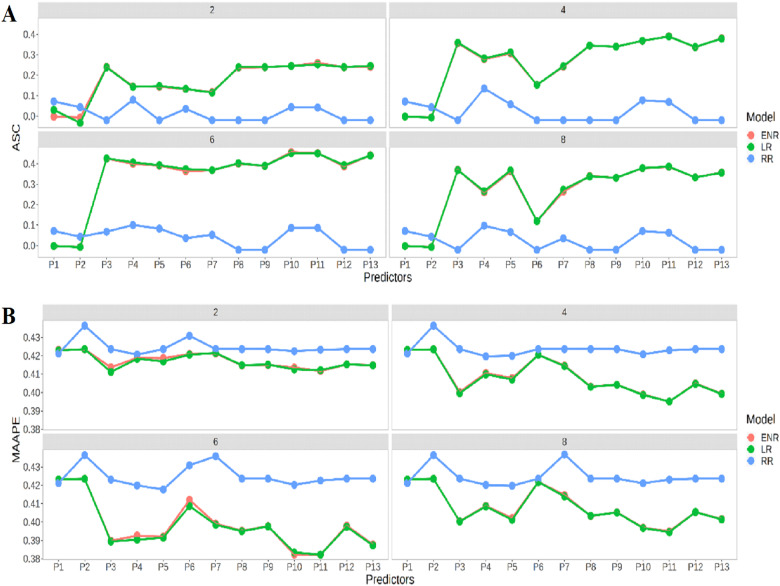
Figure 7A indicates that the Lasso regression and Elastic net regression models have similar prediction performances in terms of ASC, with the best predictions also occurring at the four time-points (2, 4, 6, and 8). In general, the best prediction performance was observed under predictors P2 and P11. Figure 7B shows that the best prediction performance using MAAPE was also found in Lasso regression and Elastic net regression models for the four time-points. We can see that the best performance was observed under P10 and P11. The worst prediction performances were observed at time points 2 and 4 under predictor P2, while in time-points 6 and 8 the worst prediction performances occurred under predictors P2 and P7.
Figure 7.
Comparison of prediction performance between Ridge regression, Lasso regression and Elastic net regression across time points in terms of (A) ASC and (B) MAAPE at each environment under the 13 predictors proposed in Table 1.
Discussion
Phenotyping nowadays uses noninvasive technologies and digital technologies to measure complex traits related to growth, yield, and adaptation to stress, with improved accuracy and precision at different organizational scales, from organs to canopies (Fiorani and Schurr 2013). For this reason, phenotyping is key to understanding complex patterns related to genetics, epigenetics, environmental pressures, and crop management (farming) that can guide selection towards productive plants suitable for their environment (Costa et al. 2019). For these reasons, phenotyping is at the forefront of future plant breeding, but novel statistical machine learning tools are still required to be able to incorporate all these information efficiently in the modeling process.
For this reason, in this study, we propose using Poisson regression when the response variable is count to incorporate, in addition to the information of environments and genotypes (with marker data), the information of hyperspectral images. The Poisson regression framework allows incorporating not only main effects of environments, genotypes and high-resolution images, but also two interaction terms between these three main sources of information. However, due to the fact that by adding more information to the predictor the number of observations was smaller than the number of independent variables, the penalized Poisson regression was implemented. Three penalized versions of Poisson regression were implemented (Ridge regression, Lasso regression and Elastic net regression). In general, we observed that Ridge regression penalization of Poisson regression was the worst in terms of prediction performance, while the other two penalizations (Lasso regression and Elastic net regression) were the best. This can be explained in part by the fact that these two types of penalization not only shrink those large coefficients toward zero, but also make many beta coefficients close to zero exactly zero because they also allow variable selection.
It is very important to point out that when the hyperspectral images were compressed first and then used in the modeling process, the prediction performance was similar than when using the raw high-resolution images (in the original dimensions), with the main advantage that the execution time using the compressed hyperspectral images is considerably low, which is a great advantage since when taking into account the main effects and two-way interaction terms between environment, genotypes and hyperspectral images, the dimension of the prediction is very large, and the larger it is, the more computing resources are needed. For these reasons, the compressed versions of the Poisson regression are very novel since, strictly speaking, they convert the penalized Poisson regression into a Functional penalized Poisson regression model. However, choosing the right number of basis is challenging (in our case we used only 21, which reduced the input of the high resolution of images from 250 to 21). This hyperparameter can be chosen using cross-validation, but this also increases the computation time.
We also observed that the larger the time-point, the better the prediction performance. This is expected since larger time-points are closer to the harvest day of the phenotype. For this reason, in general, time points closer to 9 showed the best prediction performance. Also, under the three models (RR, LR, and ENR), adding the interaction term between environment and genotypes (with markers and pedigree) did not always help to increase the prediction performance as can be observed between predictors P1 (no interaction term) and P2 (with interaction term between genotypes by environment). However, when ignoring the genotype by environment interaction but adding the information of the high-resolution images to the main effects of genotypes (with markers and pedigree), there is a relevant increase in prediction performance which can be appreciated when comparing predictors P1 and P3. This pattern can also be observed when comparing predictors P1 and P4 and P1 and P5; however, in general, using the compressed high-resolution images with Fourier basis does not considerably decrease the prediction performance (see predictor P3 vs. P5), but helps to significantly reduce the execution time since fewer beta coefficients need to be estimated.
In general, predictors P10 and P11 produced the best prediction performance that did not take into account the genotype (with marker and pedigree) by environment interaction terms. These two predictors outperformed predictors P12 and P13 that did take into account the genotype (with marker and pedigree) by environment interaction term, which mean that these two terms in the prediction of genotype by environment interaction did not help to increase the prediction performance and in certain way provided a certain level of overfitting. It is important to point out that predictors P10 and P11 did not use the whole raw hyperspectral images but their compressed versions under b-spline and Fourier basis functions, which provides evidence that a parsimonious version of the penalized Poisson regression model is feasible when compressed hyperspectral images are provided, and strictly speaking, this is a Functional penalized Poisson regression model.
Our results provide evidence that a parsimonious version of the penalized Poisson regression model can be achieved (functional penalized Poisson regression model) when compressed hyperspectral images are used as input. However, this approach requires a two-step process where in the first step, the high-resolution images are compressed, and in the second step, the penalized Poisson regression model is used for the training process with predictors that take into account effects of environments, genotypes and the compressed hyperspectral images. One advantage of the proposed approach for training models with count response variables is that we can use conventional penalized regression software for implementing prediction models with mixed predictors [environment, genotypes (with markers and pedigree) and high-resolution images]. Appendix A provides the R code for implementing the proposed predictors given in Table 1. Finally, since there is no universal model for predicting any type of response variables with any type of input information, the proposed penalized Poisson regression model studied here is an attractive tool for predicting count traits with input from many sources (environments, genotypes with markers and pedigree and hyperspectral images), that is quite efficient in terms of computing resources needed and can accommodate raw and compressed high-resolution images.
Conclusions
In this study, we proposed the Poisson regression model for analyzing hyperspectral images. We found that it is feasible to use this regression model for count data with hyperspectral images combined with environmental and genotype effects in an efficient way. However, in general we found that the best prediction model was the elastic net regression and Lasso regression and the worst was the Ridge regression model. We also found similar prediction performance between using the raw hyperspectral images and the compressed b-splines and Fourier basis functions, with the advantage that the compressed version is more efficient computationally. Also, was quite clear that including the hyperspectral images in the predictor increased the prediction accuracy; however, this was clearer with those hyperspectral images occurring in latter time points. However, other studies need to be performed to be able to generalize our finding for other agronomical traits. Although more research is needed to increase the empirical evidence that we found, in general we found that Poisson regression is a powerful tool for incorporating hyperspectral images and that its efficiency increases when this information is incorporated compressed with b-spline and Fourier basis functions.
Data availability
The data set used in this study (HTP_976_Disc_Data.RData) is available at http://doi.org/10.5281/zenodo.4478247
Acknowledgments
The authors thank CIMMYT for providing the data.
Funding
Financial support for this study was provided by a grant number B/5/UN34.13/HK.06.00/2020 from the Universitas Negeri Yogyakarta, Indonesia: International collaboration research.
Conflict of interest: The authors declare no conflict of interest.
Appendix.
R code for implementing the Generalized Lasso Regression with the 13 predictors for growth state 1 (time point T1).
rm(list=ls())
library(fda)
library(“fda.usc”)
library(glmnet)
library(BMTME)
####################For Getting the name of Wavelengths#########################
load(“HTP_976_Disc_Data.RData”)
Wavelengths=c(Wavelengths)
Wavelengths
LA=t(chol(A976))
LG=t(chol(G976))
########################Selecting the phenotype response-
Yield#########################
X=PhenoD[,-c(1,2,3,4,5)]
###########Information of all
bands##################################################
All.Bands1 = X
All.Bands=All.Bands1
X = X
NIter = 25000
Nburn = 2500
###################Design matrices##############################################
#####Creating the desing matriz of environment ##########################
Z.E=model.matrix(∼0+as.factor(PhenoD$Env))
#####Creating the desing matriz of Lines ##########################
Z.G=model.matrix(∼0+as.factor(PhenoD$Gids))
Z.G1 = Z.G%*%LA
Z.G2 = Z.G%*%LG
KL1 = Z.G1%*%t(Z.G1)
KL11 = Z.G2%*%t(Z.G2)
Z.GE1 = model.matrix(∼0+Z.G1: as.factor(PhenoD$Env))
Z.GE2 = model.matrix(∼0+Z.G2: as.factor(PhenoD$Env))
KL2 = Z.GE1%*%t(Z.GE1)
KL21 = Z.GE2%*%t(Z.GE2)
#############Training-testing partitions#####################
head(PhenoD[ , 1:6])
DataSet=PhenoD[, c(1,4,2)]
colnames(DataSet)=c(“Line”,”Env”,”Response”)
nCV = 5
CrossV<-CV.KFold(DataSet, K =nCV, set_seed = 123)
#length(CrossV$CrossValidation_list$partition3)
Trait_names=colnames(PhenoD[, 2:3])
results_all<-data.frame()
digits = 4
for (t in 1:2){
#t = 1
y1 = c(PhenoD[, t + 1])
y2 = c(y1)
results<-data.frame()
for (o in 1:1)
{
#o = 1
index = 250*(o-1)+250
Data.T=All.Bands[,(((index-250)+1):index)]
X11= as.data.frame(Data.T)
X11 = data.matrix(X11)
for (j in 1: nCV)
{
#j = 1
y1 = y2
tst=c(CrossV$CrossValidation_list[[j]])
y_tr= y1[-tst];
y_tst= y1[tst];
##########Creating the predictor and fitting the model in
BGLR############################
###################Model 1
###############################################################
X1 = cbind(Z.E, Z.G1, Z.G2)
X1_tr=X1[-tst , ];
X1_tst=X1[tst , ]
A1_RR=cv.glmnet(X1_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM1= as.numeric(predict(A1_RR, newx=X1_tst, s="lambda.min",type="response"))
y_pM1 = yhatM1
###Model 2
#####Creating the desing matriz of GenotypexEnviornment
interaction#######################
##########Creating the predictor and fitting the model in
BGLR############################
X2 = cbind(Z.E, Z.G1, Z.G2, Z.GE1, Z.GE2)
X2_tr=X2[-tst , ];
X2_tst=X2[tst , ]
A2_RR=cv.glmnet(X2_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM2= as.numeric(predict(A2_RR, newx=X2_tst, s="lambda.min",type="response"))
y_pM2 = yhatM2
###Model 3- for time point 2
#########Selecting the bands corresponding to point time
2#################################
X3 = cbind(Z.E, Z.G1, Z.G2, data.matrix(X11))
X3_tr=X3[-tst , ];
X3_tst=X3[tst , ]
A3_RR=cv.glmnet(X3_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM3= as.numeric(predict(A3_RR, newx=X3_tst, s="lambda.min",type="response"))
y_pM3 = yhatM3
###Model 4- for time point 2
#########Creating the design matrix for the functional regression part using bspline
basis##############
n.basis = 21
bspl = create.bspline.basis(range(c(Wavelengths)),nbasis=n.basis, breaks = NULL, norder = 4)
n.ind=dim(All.Bands)[1]
X.FDA=matrix(NA, nrow=n.ind, ncol=n.basis)
for (h in 1: n.ind){
#Usando la función directamente
smf=smooth.basisPar(argvals=c(Wavelengths),y=as.numeric(Data.T[h , ]),lambda = 0.1, fdobj=bspl, Lfdobj = 2)
cv_sp_pn = smf$fd$coefs# Coeficientes cj directamente
I_KL = inprod(bspl, bspl)
xt_h = t(I_KL%*%cv_sp_pn)
X.FDA[h , ]=xt_h
}
X.FDA=data.matrix(X.FDA)
X4 = cbind(Z.E, Z.G1, Z.G2, X.FDA)
X4_tr=X4[-tst , ];
X4_tst=X4[tst , ]
A4_RR=cv.glmnet(X4_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM4= as.numeric(predict(A4_RR, newx=X4_tst, s="lambda.min",type="response"))
y_pM4 = yhatM4
###Model 5- for time point 2
#######Creating the design matrix for the functional regression part using Fourier
basis#########
bspF=create.fourier.basis(range(c(Wavelengths)),nbasis=n.basis, period=diff(range(c(Wavelengths))))
X.Fu=matrix(NA, nrow=n.ind, ncol=n.basis)
for (h in 1: n.ind){
#Usando la función directamente
smf=smooth.basisPar(argvals=c(Wavelengths),y=as.numeric(Data.T[h , ]),lambda = 0.1, fdobj=bspF, Lfdobj = 2)
cv_sp_pn = smf$fd$coefs# Coeficientes cj directamente
I_KL = inprod(bspl, bspl)xt_h = t(I_KL%*%cv_sp_pn)
X.Fu[h , ]=xt_h
}
X.Fu=data.matrix(X.Fu)
X5 = cbind(Z.E, Z.G1, Z.G2, X.Fu)
X5_tr=X5[-tst , ];
X5_tst=X5[tst , ]
A5_RR=cv.glmnet(X5_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM5= as.numeric(predict(A5_RR, newx=X5_tst, s="lambda.min",type="response"))
y_pM5 = yhatM5
###Model 6- for time point 2
#######Creating the design matrix for the functional regression part using Fourier
basis#########
X6 = cbind(Z.E, Z.G1, Z.G2, Z.GE1, Z.GE2, X.FDA)
X6_tr=X6[-tst , ];
X6_tst=X6[tst , ]
A6_RR=cv.glmnet(X6_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM6= as.numeric(predict(A6_RR, newx=X6_tst, s="lambda.min",type="response"))
y_pM6 = yhatM6
###Model 7- for time point 2
#######Creating the design matrix for the functional regression part using Fourier
basis#########
X7 = cbind(Z.E, Z.G1, Z.G2, Z.GE1, Z.GE2, X.Fu)
X7_tr=X7[-tst , ];
X7_tst=X7[tst , ]
A7_RR=cv.glmnet(X7_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM7= as.numeric(predict(A7_RR, newx=X7_tst, s="lambda.min",type="response"))
y_pM7 = yhatM7
###Model 8- for time point 2
#######Creating the design matrix for the interaction between Environments and
Bands#########
Z.IT=model.matrix(∼0+Z.E: as.matrix(X11))
X8 = cbind(Z.E, Z.G1, Z.G2, X11, Z.IT)
X8_tr=X8[-tst , ];
X8_tst=X8[tst , ]
A8_RR=cv.glmnet(X8_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM8= as.numeric(predict(A8_RR, newx=X8_tst, s="lambda.min",type="response"))
y_pM8 = yhatM8
###Model 9- for time point 2
X9 = cbind(Z.E, Z.G1, Z.G2, Z.GE1, Z.GE2, X11, Z.IT)
X9_tr=X9[-tst , ];
X9_tst=X9[tst , ];
A9_RR=cv.glmnet(X9_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM9= as.numeric(predict(A9_RR, newx=X9_tst, s="lambda.min",type="response"))
y_pM9 = yhatM9
###Model 10- for time point 2
Z.IF=model.matrix(∼0+Z.E: X.FDA)
X10 = cbind(Z.E, Z.G1, Z.G2, X.FDA, Z.IF)
X10_tr=X10[-tst , ];
X10_tst=X10[tst , ];
A10_RR=cv.glmnet(X10_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM10= as.numeric(predict(A10_RR, newx=X10_tst, s="lambda.min",type="response"))
y_pM10 = yhatM10
###Model 11- for time point 2
Z.IFu=model.matrix(∼0+Z.E: X.Fu)
X11 = cbind(Z.E, Z.G1, Z.G2, X.Fu, Z.IFu)
X11_tr=X11[-tst , ];
X11_tst=X11[tst , ];
A11_RR=cv.glmnet(X11_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM11= as.numeric(predict(A11_RR, newx=X11_tst, s="lambda.min",type="response"))
y_pM11 = yhatM11
###Model 12- for time point 2
X12 = cbind(Z.E, Z.G1, Z.G2, Z.GE1, Z.GE2, X.FDA, Z.IF)
X12_tr=X12[-tst , ];
X12_tst=X12[tst , ];
A12_RR=cv.glmnet(X12_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM12= as.numeric(predict(A12_RR, newx=X12_tst, s="lambda.min",type="response"))
y_pM12 = yhatM12
###Model 13- for time point 2
X13 = cbind(Z.E, Z.G1, Z.G2, Z.GE1, Z.GE2, X.Fu, Z.IFu)
X13_tr=X13[-tst , ];
X13_tst=X13[tst , ];
A13_RR=cv.glmnet(X13_tr, y_tr, family="poisson",
alpha = 1, type.measure=“mse”)
yhatM13= as.numeric(predict(A13_RR, newx=X13_tst, s="lambda.min",type="response"))
y_pM13 = yhatM13
results<-rbind(results, data.frame(Position=tst,
Environment=CrossV$Environments[tst],
Trait =Trait_names[t],
Partition =j,
Observed = round(y2[tst], digits), #$response, digits),
PredictedM1 = round(y_pM1, digits),
PredictedM2 = round(y_pM2, digits),
PredictedM3 = round(y_pM3, digits),
PredictedM4 = round(y_pM4, digits),
PredictedM5 = round(y_pM5, digits),
PredictedM6 = round(y_pM6, digits),
PredictedM7 = round(y_pM7, digits),
PredictedM8 = round(y_pM8, digits),
PredictedM9 = round(y_pM9, digits),
PredictedM10 = round(y_pM10, digits),
PredictedM11 = round(y_pM11, digits),
PredictedM12 = round(y_pM12, digits),
PredictedM13 = round(y_pM13, digits)))
}}results_all<-rbind(results_all, results)
}
results_all
write.csv(results_all, file =“HTP_Count_T1.csv”)
Literature cited
- Aguate FM, Trachsel S, Pérez LG, Burgueño J, Crossa J, et al. 2017. Use of hyperspectral image data outperforms vegetation indices in prediction of maize yield. Crop Sci. 57:2517–2524. [Google Scholar]
- Araus JL, Cairns JE.. 2014. Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci. 19:52–61. [DOI] [PubMed] [Google Scholar]
- Cabrera-Bosquet L, Crossa J, von Zitzewitz J, Serret MD, Luis Araus J.. 2012. High-throughput phenotyping and genomic selection: the frontiers of crop breeding converge. J Integr Plant Biol. 54:312–320. [DOI] [PubMed] [Google Scholar]
- Chollet F, Allaire JJ.. 2018. Deep learning with R. https://www.manning.com/books/deep-learning-with-r.
- Costa C, Schurr U, Loreto F, Menesatti P, Carpentier S.. 2019. Plant phenotyping research trends, a science mapping approach. Front Plant Sci. 9:1933. doi: 10.3389/fpls.2018.01933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorani F, Schurr U.. 2013. Future scenarios for plant phenotyping. Annu Rev Plant Biol. 64:267–291. [DOI] [PubMed] [Google Scholar]
- Gitelson AA, Kaufman YJ, Stark R, Rundquist D.. 2002. Novel algorithms for remote estimation of vegetation fraction. Remote Sens Environ. 80:76–87. [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M,et al. 2006. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 169:623–635. [DOI] [PubMed] [Google Scholar]
- Humplík JF, Lazár D, Husičková A, Spíchal L.. 2015. Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses - A review. Plant Methods 11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME.. 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes JM, Melchinger AE, Reif JC.. 2007. Novel throughput phenotyping platforms in plant genetic studies. Trends Plant Sci. 12:433–436. [DOI] [PubMed] [Google Scholar]
- Montesinos-López OA, Montesinos-López A, Pérez-Rodríguez P, Eskridge K, He X, et al. 2015. Genomic prediction models for count data. JAgri Biol Environ Stat.20:533–554. [Google Scholar]
- Montesinos-López A, Montesinos-López OA, Crossa J, Burgueño J, Eskridge KM, et al. 2016. Genomic Bayesian prediction model for count data with genotype × environment interaction. G3 Genes. G3 (Bethesda). 6:1165–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-López OA, Montesinos-López A, Crossa J, Toledo FH, Montesinos-López JC, et al. 2017a. A Bayesian poisson-lognormal model for count data for multiple-trait multiple-environment genomic-enabled prediction. G3 (Bethesda). 7:1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-López OA, Montesinos-López A, Crossa J, los Campos G, Alvarado G, et al. 2017b. Predicting grain yield using canopy hyperspectral reflectance in wheat breeding data. Plant Methods 13:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-López A, Montesinos-López OA, de los Campos G, Crossa J, Burgueño J,et al. 2018. Bayesian functional regression as an alternative statistical analysis of high-throughput phenotyping data of modern agriculture. Plant Methods 14:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-López OA, Montesinos-López JC, Singh P, Lozano-Ramirez N, Barrón-López A, et al. 2020. A multivariate poisson deep learning model for genomic prediction of count data. G3 (Bethesda). 10:4177–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson J, Gibson A.. 2017. Deep Earning: A Practitioner’sApproach. Sebastopol, CA: O’Reilly [Google Scholar]
- Rouphael Y, Spíchal L, Panzarová K, Casa R, Colla G.. 2018. High-throughput plant phenotyping for developing novel biostimulants: from lab to field or from field to lab? Front Plant Sci. 9:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroup WW. 2012. Generalized Linear Mixed Models: Modern Concepts, Methods and Applications. Boca Raton, FL: CRC Press [Google Scholar]
- Wolpert DH, Macready WG.. 1997. No free lunch theorems for optimization. IEEE Trans Evol Comp. 1:67–82. [Google Scholar]
- Xue J, Su B.. 2017. Significant remote sensing vegetation indices: a review of developments and applications. J Sensors 2017:1–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data set used in this study (HTP_976_Disc_Data.RData) is available at http://doi.org/10.5281/zenodo.4478247