Abstract
The formation of β1,3-linkages on animal glycoconjugates is catalyzed by a subset of β1,3-glycosyltransferases grouped in the Carbohydrate-Active enZYmes family glycosyltransferase-31 (GT31). This family represents an extremely diverse set of β1,3-N-acetylglucosaminyltransferases [B3GNTs and Fringe β1,3-N-acetylglucosaminyltransferases], β1,3-N-acetylgalactosaminyltransferases (B3GALNTs), β1,3-galactosyltransferases [B3GALTs and core 1 β1,3-galactosyltransferases (C1GALTs)], β1,3-glucosyltransferase (B3GLCT) and β1,3-glucuronyl acid transferases (B3GLCATs or CHs). The mammalian enzymes were particularly well studied and shown to use a large variety of sugar donors and acceptor substrates leading to the formation of β1,3-linkages in various glycosylation pathways. In contrast, there are only a few studies related to other metazoan and lower vertebrates GT31 enzymes and the evolutionary relationships of these divergent sequences remain obscure. In this study, we used bioinformatics approaches to identify more than 920 of putative GT31 sequences in Metazoa, Fungi and Choanoflagellata revealing their deep ancestry. Sequence-based analysis shed light on conserved motifs and structural features that are signatures of all the GT31. We leverage pieces of evidence from gene structure, phylogenetic and sequence-based analyses to identify two major subgroups of GT31 named Fringe-related and B3GALT-related and demonstrate the existence of 10 orthologue groups in the Urmetazoa, the hypothetical last common ancestor of all animals. Finally, synteny and paralogy analysis unveiled the existence of 30 subfamilies in vertebrates, among which 5 are new and were named C1GALT2, C1GALT3, B3GALT8, B3GNT10 and B3GNT11. Altogether, these various approaches enabled us to propose the first comprehensive analysis of the metazoan GT31 disentangling their evolutionary relationships.
Keywords: β1,3-glycosyltransferases; evolution; functional genomics; molecular phylogeny; motifs
Introduction
The family glycosyltransferase-31 (GT31) of the Carbohydrate-Active enZYmes (CAZy) classification (Lombard et al. 2014) is a very large family of β1,3-glycosyltransferase (B3GT) including mostly eukaryotic (i.e., animals and plants) and a few bacterial sequences. The eukaryotic subset of B3GT is extremely diverse in terms of sequences and functions. The B3GTs are inverting enzymes that catalyze the transfer of a variety of monosaccharides from activated uridine diphosphate-sugars (i.e., Uridine diphosphate (UDP)-sugars: UDP-Gal, UDP-GlcNAc, UDP-GalNAc, UDP-Glc and UDP-GlcA) to terminal nonreducing positions of oligosaccharides of glycoproteins and glycolipids leading to the formation of a unique β1,3-linkage. Most of the 25 human B3GTs [i.e., 9 β1,3-N-acetylglucosaminyltransferases (B3GNT), 3 Fringe β1,3-N-acetylglucosaminyltransferases (FNG), 2 β1,3-N-acetylgalactosaminyltransferases (B3GALNT), 4 β1,3-galactosyltransferases (B3GALT), 2 core 1 β1, 3-galactosyltransferase (C1GALT), 1 β1,3-glucosyltransferases (B3GLCT) and 4 β1,3-glucuronyl acid transferases involved in chondroitin sulfate synthesis (B3GLCAT or CH)] have been cloned and expressed as recombinant proteins for functional studies (Moremen et al. 2018). Seminal work by Narimatsu and collaborators (Narimatsu 2004; Narimatsu 2006; Togayachi, et al. 2006; Togayachi et al. 2008) and by Clausen and collaborators (Amado et al. 1998) shed light on the substrate specificities of recombinant enzymes and showed that the human B3GTs are implicated in various glycolipids and glycoproteins metabolisms (Narimatsu et al. 2019). As illustrated in Figure 1, the B3GTs are implicated in core extension steps of mucin-type O-glycans (C1GALT/C1GALT1C1 and B3GNT6), of fucosyl-type O-glycans [Radical-, Lunatic-, and Manic-Fringe (RFNG, LFNG, and MFNG) and B3GLCT], of glycosaminoglycan (GAG) linker region (B3GALT6), of mannosyl-type O-glycans (B3GALNT2), of Lacto-, Neolacto-, Ganglio-, Globo- and Neoglobo-series of glycolipids (B3GNT5, B3GALT4; B3GALNT1) biosynthetic pathway and in the specific chondroitin sulfate biosynthetic pathway [chondroitin polymerizing factors (CHPF, CHPF2), and chondroitin sulfate synthases (CHSY1, CHSY3)]. The remaining B3GTs (e.g., B3GALT1, B3GALT2, B3GALT5, B3GNT2, B3GNT3, B3GNT4, B3GNT7, B3GNT8, B3GNT9) are implicated in unspecific glycosylation pathways and poly-N-acetyllactosamine (poly-LacNAc) extension on glycolipids and N- and O-glycans of glycoproteins.
Fig. 1.

Glycan structures and pathways involving B3GTs of the CAZy family GT31. GT31-related enzymes indicated in green letters are involved in various glycosylation pathways: (A) glycolipid core extension, (B) O-glycosylproteins core extension and (C) elongation of glycoconjugates. The glycan structures formed are represented using the symbol nomenclature for graphical representation of glycans (SNFG) (Neelamegham et al. 2019) with DrawGlycan-SNFG (Cheng et al. 2017) and their names are indicated in red.
From a structural point of view, almost nothing is known since only the mouse MFNG structure was solved and shown to adopt a GT-A fold (PDB 2J0A and 2J0B with UDP and Mn) (Jinek et al. 2006). Despite the identification of conserved sequence motifs and cysteine residues in mammalian B3GTs (Heinonen et al. 2006; Hennet et al. 1998; Narimatsu 2006) and more specifically in the B3GALT catalytic domain (Patel and Balaji 2007; Qu et al. 2008; Togayachi et al. 2006), the GT31 members show very limited overall sequence identity (25–28%), which hindered their identification and studies of their structure–function relationships. In an attempt to identify and systematically characterize putative plant GT31, Bacic and collaborators have used phylogenetic approaches (Egelund et al. 2010; Qu et al. 2008). Their studies led to the identification of 11 clades, 4 of which are plant specific (e.g., clades 1, 7, 10 and 11). Several plant sequences from the clades 7 and 10 were biochemically characterized. The Arabidopsis thaliana GALT2, GALT3, GALT4, GALT5, GALT6 as well as GALT31A from clade 7 are involved in the synthesis of β1,3-galactan chains of the arabinogalactan-proteins (AGP) (Basu, Tian et al. 2015a; Basu, Wang et al. 2015b; Geshi et al. 2013), whereas GALT1 is implicated in the synthesis of the Lewisa structure (Fucα1,4[Galβ1,3]GlcNAc-R) found on N-glycans (Strasser et al. 2007). Hydroxyproline-O-galactosyltransferase-1 (HPGT1), HPGT2, HPGT3 and the more recently described KSN4 from clade 10 are three HPGTs (Ogawa-Ohnishi and Matsubayashi 2015; Suzuki et al. 2017). Clades 2–9 group all the remaining Opisthokonta B3GT including B3GNT (clade 8) and Fringe (FNG; clade 3), B3GALNT distributed in clade 8 and clade 9, B3GALNT with B3GALT (clade 8) and C1GALT (clade 5), B3GLCT (clade 2) and CH (clade 4). However, the functional diversification of these GT31 sequences in vertebrates remains poorly understood and the evolutionary history of the metazoan GT31 is still not known.
The objectives of this study were to identify animal GT31-related sequences and to provide a phylogeny-based classification for this GT family. We combined several bioinformatics approaches for GT31-related sequences homology searches with the known human sequences used as seeds and for conserved domains and motifs identification. We extended our analysis to early Metazoa, Choanoflagellata, Fungi and Viridiplantae and we provided evidence of the occurrence of very ancient divergence, which explains the presence of 10 GT31 orthologue groups in the Urmetazoa, the last common ancestor (LCA) of Metazoa. Our synteny and paralogy analyses in vertebrate genomes enabled us to identify five new GT31 vertebrate subfamilies and to predict their donor/acceptor specificity. Altogether, our study clarifies the evolutionary relationships of the 30 vertebrate subfamilies and provides a robust phylogenetic classification of the metazoan B3GT of the CAZy GT31 shedding lights into their evolutionary functional divergence in vertebrates.
Results and discussion
Identification of GT31-related genes in Metazoa and phylogeny analysis
The GT31 CAZy family represents one of the largest multigene CAZy families comprised of a highly diverse set of B3GT enzymes with a remarkable wide range of substrates. Glycosylated products generated such as mucin-type glycoproteins and proteoglycans (Figure 1) are components of the extracellular matrix of utmost importance for multicellular organisms’ life enabling communications between cells. Therefore, GT31 evolutionary history is believed to reflect essential evolutionary steps in cell surface molecule-dependent human biology. To gain insights into the evolution of GT31-related genes in Metazoa, we firstly identified over 900 potential homologues of the human B3GT sequences using the Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1997), the Conserved Domain Architecture Retrieval Tool (Geer et al. 2002) and PFAM database (release 32.0) (El-Gebali et al. 2019) in the metazoan nucleotide divisions of the National Center for Biotechnology Information (NCBI) Entrez Protein Database (Supplemental Data). To ascertain the distribution of metazoan GT31 members, we also investigated the Fungi and the Viridiplantae (plant) divisions of NCBI database as nonmetazoan outgroups taking advantage of the previously described plant GT31 sequences (Egelund et al. 2010). Indeed, several putative homologues of these GT31-related genes could be identified in Viridiplantae species like the flowering plant A. thaliana (Egelund et al. 2010) and the earth moss Physcomitrella patens and in Fungi species like Absidia glauca, Syncephalastrum racemosum, Mortierella verticillata, Basidiobolus meristosporus, Talaromyces marneffei, Aspergillus wentii, Spizellomyces punctatus, Wallemia mellicola, Cytospora leucostoma, Rhodotorula graminis and Rhizopus azygosporus (Supplemental Data 1), further suggesting a very ancient origin of these GT31 genes, which emerged well before the radiation of Metazoa. Focusing on the early branching Metazoa, Porifera and Cnidaria, homologues of the CHPF, CHSY, FNG, B3GLCT and B3GNT/B3GALT could be identified in the sponges Amphimedon queenslandica and Sycon ciliatum, respectively, whereas homologues of the B3GALT6, B3GALNT2, B3GALT/B3GNT, B3GALNT2 and C1GALT were found in Hydra vulgaris. The results of this exploration point to the existence in extant early Metazoa of at least nine distinct orthologue groups of GT31, namely FNG, CHPF, CHSY, B3GLCT, C1GALT/C1GALT1C1, B3GALT/B3GNT, B3GALT6, B3GALNT2 and anc-B3GALT, a group of ancient B3GALT sequences lost in Bilateria. We then further studied the GT31-related sequences evolutionary relationships according to four criteria of analysis.
First criterion: genomic organization of vertebrate GT31 genes
As a first step toward unraveling the evolutionary relationships of these seven GT31 groups, we examined the genomic organization of the coding region of the vertebrate B3GTs genes (Figure 2). The exon/intron organization of these genes was relatively well conserved in each of the orthologue group previously delineated. This analysis revealed two sets of genes sharing the same organization: in the B3GALT/B3GNT, C1GALT1C1 and B3GALT6, the coding sequence is found within a single exon with the notable exception of B3GNT3 that is split into two coding exons in the Amniotes genome, whereas it is distributed in several exons in the B3GALNT2, B3GLCT, C1GALT, FNG, CHPF and CHSY. Focusing on the human B3GTs genes, we noticed their distribution across different human chromosomes. Surprisingly, the vertebrate B3GTs genes encode for proteins with extremely variable lengths ranging from about 350-amino acid (aa) residues for the B3GALTs, B3GNTs, and FNGs to 800-aa residues for the CH (CHPF and CHSY) (Figure 2).
Fig. 2.

Schematic depicting the genomic organization of the 30 GT31 vertebrate genes. The exon/intron organization of the 26 human (capital letters) and 4 additional vertebrate (lower case letters) B3GT genes is represented. The B3GNT10-P is found in the human genome on chromosome 9q33.2 but is likely not translated in an active enzyme. Family name is indicated on the left side. Coding exons are represented by rectangles according to their relative sizes. The gray boxes denote the presence of inserted exons in B3GALNT2 and presence of additional protein domain in B3GLCT (FRINGE-domain) and CH (CHGN domain)-related proteins. The five conserved peptide motifs characteristic of all the GT31-related proteins are represented by black lines above the boxes. The location of nonconserved GT31 signatures is indicated by gray lines above the boxes. Each subfamily name is indicated on the right side. In addition, chromosome location in the human genome and human protein length are given on the right side of the figure; when the human gene and/or protein is absent, its closest vertebrate relative is represented in blue. The inactive proteins, e.g., chaperone C1GALT1C1, CHPF and CHPF2, are indicated in green. A similar genomic organization is found in each orthologue group, and it is conserved in all the vertebrate species with the exception of B3GNT3 genes split into two exons in Amniotes genome.
Second criterion: sequence/structure similarities of GT31 protein sequences
Secondly, we retrieved the aa sequences deduced from these eukaryotic GT31-related genes to further investigate structural similarities. Structure-based analyses performed on the predicted proteins using the Simple Modular Architecture Research Tool (SMART) (Letunic and Bork 2018; Letunic et al. 2015) showed that the eukaryotic GT31 proteins are multidomain proteins containing a combination of either a Galactosyl_T domain (PF01762) or a FRINGE domain (PF02434) and an additional functional domain. Indeed, we identified two major domain architectures of GT31 sequences each associating two domains, e.g., (i) X domain/GALT domain and (ii) FRINGE domain/X domain. In the first X domain/GALT domain architecture, the X domain coupled to the GALT domain can be the domain of unknown function (DUF4094) (PF13334), the galactose-binding domain Gal-bind_lectin (PF00337) or the N-glycosylation protein domain EOS1 (PF12326) mostly found in land plant GT31 sequences of clade 7 (Egelund et al. 2010). Interestingly, the Gal-bind_lectin domain is found alone in other animal proteins (Wang et al. 2017). The X domain is lost in the metazoan B3GALT6 and B3GALNT2 (clade 9), the B3GALT/B3GNT (clade 8) and the plant GALT (clade 10). In the second FRINGE domain/X domain architecture, two Fringe-related (FR) domains (PF02434) named here FRINGE1 and FRINGE2 are encountered in GT31 proteins. FRINGE1 can be associated to an X domain DUF604 (PF04646) found in the plant GT31 clade 1, to another FRINGE1 domain in B3GLCT (clade 2) or the X domain can be lost as in FNG and plant GT31 clade 3. FRINGE2 domain is associated to a CHGN domain (PF05679) as for the metazoan CH sequences of clade 4 or the X domain can be lost as in C1GALT (clade 5). This protein architecture of the vertebrate GT31 corroborates the variable protein length described above in Figure 2.
The 25 human GT31 proteins listed in Figure 2 and a few other representatives of the 30 vertebrate GT31 subfamilies (described in the molecular phylogeny section) were then selected to conduct multiple sequence alignments and investigate the presence of conserved motifs (Supplemental Data) in the GalT and FRINGE domains. Several conserved peptide motifs and cysteine residues have been identified in the catalytic domain of B3GTs (Heinonen et al. 2006; Hennet et al. 1998; Narimatsu 2006) and more specifically in mammalian and land plant B3GALTs (Patel and Balaji 2007; Qu et al. 2008; Togayachi et al. 2006). The functional importance of these motifs and the role of conserved aa residues for enzymatic activity were further investigated in the mouse B3GALT1 through point mutations and enzymatic assays (Malissard et al. 2002). In this present analysis, we retrieved and refined five well-conserved motifs that are signatures of all the GT31 members, and sequence logos were established (Figure 3A). The first motif (motif I) refined here (R/A/L)(R/A)xx(I/V/A)xx(T/S)W corresponds to the RxxxRxT/SW previously described in B3GALTs. It is preceded by a hydrophobic motif that is conserved and specific for each GT31 group (data not shown), which was described using Hydrophobic Cluster Analysis for B3GALTs/B3GNTs (Patel and Balaji 2007; Qu et al. 2008). The tryptophan residue (W) in motif I is highly conserved in each GT31 group (Supplemental Figure 1) and is likely involved in the binding of the sugar nucleotide as suggested for UDP-Gal in the mouse B3GALT1 (Malissard et al. 2002). It is interesting to note that it is not conserved in the vertebrate CHPF and CHPF2 sequences, which support the view that these individual proteins would not be active enzymes but rather chaperones stabilizing CHSY1 and CHSY3 and/or conferring higher activities on CHSY in GAG biosynthesis (Izumikawa et al. 2008; Izumikawa et al. 2007; Ogawa et al. 2010). Similarly, this first motif is not conserved in C1GALT1C1 sequences (Supplemental Figure 1), the well-known cosmc (core 1 βGalT specific molecular chaperone) playing as an essential and private chaperone of the mammalian C1GALT1 for correct protein O-glycosylation (Ju and Cummings 2005; Ju et al. 2006). Intriguingly, this first motif is located far upstream of the other conserved motifs in the B3GALNT2 sequences (Figure 2), which suggests large insertion of exons very early in the evolution of the Opisthokonta sequences. As for the majority of GT-A fold enzymes classified in other CAZy families, B3GT are metal dependent and show a DxD motif in their active site, here included in the second motif (motif II), which helps coordinate the metal ion and nucleotide-sugar (Taujale et al. 2020). This second conserved motif is found in a hydrophobic pocket (Qu et al. 2008) specific of each GT31 group (Figure 3B). A triple point mutation in the DxD motif in the mouse B3GALT1 (DDF residues) led to complete inactivation of the enzyme activity (Malissard et al. 2002). Interestingly, the DxD motif is deleted in the vertebrate C1GALT1C1 sequences and is modified in the CHPF and CHPF2 (Supplemental Figure 1), substantiating the idea that these proteins are not enzymatically active. The third conserved motif (Y/F/W)xG (motif III) is described here for the first time. However, in the 3D structure, the G residue of motif III is located far from the active site. The fourth conserved motif GxxYxxS (motif IV) found in all GT31 (Figure 3) constitutes the C-terminus part of a flexible G-loop, and the conserved glycine residue is involved in donor binding (Taujale et al. 2020), and the first G residue of motif IV is located near the active site. In addition, the conserved aromatic aa residues in GT31 motifs II, III and IV were found to be well aligned in the MFNG structures (PDB 2J0A and 2J0B with UDP and Mn) suggesting their structural importance. The last conserved motif (motif V) (E/D)DVxxGx(W/C) is located at the C-terminus part of the GT31, except for the CH sequences. This conserved motif together with the DxD motif is involved in catalytic functions of GT-A fold inverting GTs (Taujale et al. 2020). The MFNG structure (PDB 2J0B) was solved with UDP but without the substrate. However, motif IV and V seem to form a cavity near the active site where the substrate could be located (Figure 3B). The first two residues of motif V may play a crucial role in substrate binding: (E/D)D. Indeed, the glutamic acid (E) and cysteine (C) residues were shown to be critical for the enzymatic activity of the mouse B3GALT1 (Malissard et al. 2002) and the conserved glycine (G) residue was shown to be critical for the human B3GALNT1 activity (Hellberg et al. 2002).
Fig. 3.

Schematic representation of the 5 conserved motifs of the GT31 sequences. (A) The vertebrate GT31 protein sequences used are those mentioned in Supplemental Figure 2 and Supplemental Figure 3. Briefly, 170 GT31 vertebrate sequences were selected and aligned using the multiple sequence alignment tool Clustal W in MEGA 7.0 (Kumar et al. 2016) (Supplemental Data 2). Identification of the five signature motifs among GT31 homologues and a graphical representation of aa residue conservation at each position of the multiple alignments were obtained using the Berkeley WebLogo tool (Crooks et al. 2004). The letter size is proportional to the degree of aa conservation. The relative position of the 5 motifs noted I, II, III, IV and V is schematized below. (B) Structural representation of known and predicted sequence motifs. Ribbon representation of the mouse MFNG structure (PDB: 2J0A) with sequence motifs shown in colors: I (red), II (blue), III (green), IV (violet) and V (orange). The UDP molecule is shown in black in stick representation. Molecular representation and structural analyses were performed with the UCSF ChimeraX (Goddard et al. 2018).
Third criterion: molecular phylogeny of GT31 sequences
To assess the orthology between the aa sequences of the 25 human GT31 sequences and the ones found in Fungi and Viridiplantae and understand their evolutionary relationships, we conducted phylogenetic analyses. A major limitation of large scale phylogenetic analysis is that it depends on a highly accurate multiple sequence alignment. Therefore, we constructed a first simple dataset (Supplemental Data 1), and for each selected GT31 sequence, we delineated the B3GALT domain (PF01762) strictly assigned to the GT31 CAZy family that was subsequently solely used for multiple sequence alignments (Supplemental Data 2). Using human β1,4-N-acetylgalactosaminyltransferase (B4GALNT2) (CAZy family GT12), β1,4-galactosyltransferase (B4GALT1) and B4GALT2 (CAZy family GT7), α1,4-galactosyltransferase (A4GALT) or Gb3 synthase (CAZy family GT32) and bovine α1,3-galactosyltransferase (GGTA1; CAZy family GT6) sequences as outgroups according to Hennet 2002, a maximum likelihood (ML) procedure was applied (see Materials and Methods section). The resulting phylogenetic tree shown in Figure 4 clearly organizes these GT31 sequences in two deeply rooted branches and several clades. The nine GT31 orthologue groups identified earlier in Metazoa are split into two major subgroups, the FR subgroup encompassing FNG, CHPF, CHSY, B3GLCT, C1GALT and C1GALT1C1 on one hand and the B3GALT-related subgroup (BGR) with the B3GALT/B3GNT, B3GALT6 and B3GALNT2 on the other hand, corresponding, respectively, to the clades 1–5 and clades 6–11 defined by Egelund and collaborators (Egelund et al. 2010). Our phylogeny data indicated that the first FR subgroup is itself divided into FRa grouping FNG and B3GLCT and FRb grouping CHPF, CHSY, C1GALT and C1GALT1C1 sequences, corroborating Egelund views. Interestingly, the introduction of Fungi sequences revealed a new clade restricted to Viridiplantae, sister to clade 1 that was named clade 1b (Figure 4). These Fungi and Viridiplantae sequences are most often annotated as FNG in NCBI database and are sister sequences to metazoan FNG. Of note, an FNG sequence was found in the Fungi Ascomycota C. leucostoma that was not described before (Gazave et al. 2009). The FRb contains Fungi sequences, probably sisters to CH, enriching the clade 4 (Egelund et al. 2010), whereas the C1GALT/C1GALT1C1 (clade 5) appears to be restricted to Metazoa. Similarly, in the second BGR subgroup, the introduction of Fungi sequences revealed a new clade, sister to the plant-specific clade 7 that was named clade 7b (Figure 4). Furthermore, the metazoan B3GALT6 and B3GALNT2 are no longer sister sequences in clade 9 (Egelund et al. 2010) but are associated, respectively, to plant clades 7 and 10. Since the resolution of ancient relationships is a difficult task, we also analyzed the evolution of the domain architectures of the GT31 members (Figure 4). Interestingly, the two major domain architectures of GT31 sequences were found to be split in each FR and BGR subgroup. According to our phylogenetic analysis and studies of domain structure, GT31 sequences form two monophyletic classes, which in turn form several divisions.
Fig. 4.

ML phylogenetic tree of 62 GT31-related sequences from Metazoa, Fungi and Viridiplantae. An ML phylogenetic tree was constructed in MEGA 7.0 based on the JTT + G matrix-based model (Kumar et al. 2016). A discrete gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 1.9932)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 0.00% sites). Sixty-two GT31 sequences limited to their Galactosyl_T domain (PF01762) or FRINGE domain (PF02434) from the Metazoa Bos taurus (Bostau), Homo sapiens (Homsap), L. chalumnae (Latcha), Mus musculus (Musmus), Pan troglodytes (pantro), from the Viridiplantae A. glauca (Absgla), A. thaliana (Aratha), Morus notabilis (Mornot), P. patens (Phymit), and from the fungi A. wentii (Aspwen), B. meristosporus (Basmer), C. leucostoma (Cytleu), M. verticillata (Morver), R. azygosporus (Rhiazy), R. graminis (Rhogra), S. punctatus (Spipun), S. racemosum (Synrac), T. marneffei (Talmar), W. mellicola (Walmel), Xylaria grammica (Xylgram) and the human B4GALNT2 (GT12), B4GALT1 (GT7), B4GALT2 (GT7), A4GALT or Gb3 synthase (GT32) and bovine GGTA1 (GT6) sequences used as the outgroup were selected for multiple sequence alignments performed with MUSCLE in MEGA vers. 7.0 (Kumar et al. 2016) (Supplemental Data 1 for more sequence information and Supplemental Data 2 for multiple sequence alignment). There were a total of 534 positions in the final dataset. Bootstrap values over 50%, obtained from 500 replicates, were indicated on the corresponding branches. The topology of the ML tree indicates two deeply rooted branches corresponding to the subgroups FR and BGR and several clades previously described (Egelund et al. 2010). Four clades, e.g., clades 1, 7, 10 and 11 are plant specific and seven clades (e.g., clades 2–6 and clades 8–9) gather nonplant sequences. The nine metazoan GT31 orthologue groups are split into two subgroups, the FR subgroup comprised of FNG (clade 3), CHPF and CHSY (clade 4), B3GLCT (clade 2) and C1GALT (clade 5) on one hand, and the BGR comprised of the B3GALT/B3GNT (clade 8), B3GALT6 (clade 7) and B3GALNT2 (clade 10), on the other hand. Two new clades restricted to Viridiplantae, e.g., clade 1b and clade 7b are evidenced in this ML tree. The metazoan B3GALT6 and B3GALNT2 are associated, respectively, to plant clades 7 and 10. Representative domain organizations of the eukaryotic GT31 proteins are schematized. SMART (Letunic and Bork 2018; Letunic et al. 2015) showed that the eukaryotic GT31 proteins are multidomain proteins containing a combination of either a Galactosyl_T domain (PF01762, pink box) or a FRINGE domain (PF02434, pink circle and pentagon) and an additional functional X domain. The X domain coupled to the Galactosyl_T domain can be the DUF4094 domain (PF13334, yellow triangle), the galactose-binding domain Gal-bind_lectin (PF00337, blue triangle) or the N-glycosylation protein domain EOS1 (PF12326, light pink diamond). No X domain is found in the metazoan B3GALT6, B3GALNT2, B3GALT/B3GNT and the plant GALT (clade 10). Two FR domains (PF02434) named here FRINGE1 and FRINGE2 are encountered in GT31 domain architecture. FRINGE1 (pink circle) is associated to an X domain DUF604 (PF04646, red triangle) in the plant GT31clade 1, to another FRINGE1 domain in B3GLCT (clade 2, a and b pink circles) or the X domain can be lost as in FNG and plant GT31 clade 3. FRINGE2 domain (pink pentagon) is associated to a CHGN domain (PF05679, green square) in metazoan CH sequences of clade 4 or the X domain can be lost as in C1GALT (clade 5).
To go a step further and get a better understanding of the dating and origin of vertebrate GT31-related gene subfamilies, we constructed two new datasets restricted to Metazoa and their closest sister group, the Choanoflagellata (Supplemental Data) (King et al. 2008). The first dataset is composed of GT31-related sequences of the FR subgroup and was rooted by the human A4GALT and B4GALNT2 and a few sequences of the BGR subgroup. The topology of the ML phylogenetic tree obtained (Supplemental Figure 2) is similar to the one shown in Figure 4, with two major branches corresponding to FRa (FNG and B3GLCT) and FRb (CHPF, CHSY and C1GALT/C1GALT1C1). In the first FRa branch, we identified in the Deuterostomes Branchiostoma floridae, Branchiostoma belcheri tsingtauense and Saccoglossus kowalevski as well as in three Drosophila species, a unique orthologue to the common ancestor present before the split of the vertebrates Fringe sequences (i.e., LFNG, MFNG and RFNG). In early metazoans, we observed at least five copies of this orthologue in the sponge A. queenslandica further suggesting the occurrence of extensive lineage-specific duplications. The FNG sequence found in the choanoflagellate Acanthoeca spectabilis suggests that the Fringe activity was already present before Metazoa emergence, a view supported by FNG-related sequences found in Viridiplantae (clade 1) and Fungi (clade 1b). This further suggests that emergence of the three vertebrate FNG sequences is linked to the two rounds of whole genome duplication events (WGD-2R) that took place at the base of vertebrates evolution (Dehal and Boore 2005). A unique orthologue to the common ancestor of vertebrates B3GLCT is found in early metazoans such as the sponge A. queenslandica, and in the genome of the choanoflagellate Codosiga hollandica, suggesting that orthologues of the B3GLCT gene are also present in unicellular Eukaryotes. In the second FRb branch, an orthologue to the common ancestor present before the split of vertebrate C1GALTs (e.g., c1galt1 and the newly described c1galt2 and c1galt3) and C1GALT1C1 is found in the choanoflagellates Monosiga brevicollis and Salpingoeca rosetta. Since the most basal taxa hosting both C1GALT1C1 and C1GALT are the Deuterostome B. belcheri and the Cnidarians H. vulgaris and Hydra magnifica, it is suggested a post-Spongia and pre-Cnidaria divergence of C1GALT1C1 and C1GALTs (Supplemental Figure 2). Regarding the CHPF and CHSY (previously known as CSS), we identified two series of orthologues to the common ancestor present before the split of vertebrate CHPF (CHPF and CHPF2) and CHSY (CHSY1 and CHSY3). The first divergence of CHPF and CHSY likely dated back before the emergence of Metazoa since an orthologue to the common ancestor of CHSY1 and CHSY3 is already present in the choanoflagellate Salpingoeca urceolata. This view is supported by the presence of CHPF and CHSY sequences in the sponge A. queenslandica (Supplemental Figure 2). Regarding the duplication events that led to CHPF, CHPF2 and CHSY1 and CHSY3, they are likely associated to the WGD-2R events because (i) the Agnathan Lethenteron camtschaticum is linked to the CHPF2 and (ii) there is only one orthologue to the common ancestor of CHPF and CHPF2 in the Deuterostomes Ciona intestinalis and B. belcheri.
The second dataset includes metazoan GT31-related sequences of the BGR subgroup, rooted with P. patens sequences. Phylogenetic analysis using the ML program provided the tree in Supplemental Figure 3. It is organized in two deeply rooted clusters. The first one named BGRa comprises the two enzymes families B3GALT6 and B3GALNT2 and the second one named BGRb groups the remaining B3GALNT1, B3GNTs and B3GALTs. The BGRa cluster is rooted in a P. patens sequence and both enzyme families are present in the Porifera Xestospongia testudinaria, S. ciliatum and Oscarella carmella, highlighting the presence of these two families already in early Metazoa (Supplemental Figure 3). Moreover, a B3GALT6 sequence is found in the choanoflagellate M. brevicollis. The second branch BGRb is a very large cluster rooted by a series of sponge sequences found in S. ciliatum and Sycon raphanus, a series of cnidarian sequences from H. vulgaris and Nematostella vectensis, and two embedded series of mostly bilaterian invertebrate sequences, named invertebrates 1 and 2 in Supplemental Figure 3. Among these sequences, several were identified in Protostome genome of the squids Watasenia scintillans and Octopus bimaculatus and others in the Deuterostome genome of B. floridae and C. intestinalis further suggesting that the last Protostome–Deuterostome common ancestor already had complex β3-glycosylation machinery. However, the ancient invertebrate 1 B3GALT sequences (anc-B3GALT) were lost in vertebrates (Supplemental Figure 3 and Figure 5). The remaining sequences of the BGRb cluster are divided into three subclusters. The first subcluster BGRb1 comprises the vertebrate B3GALT1, B3GALT2, B3GALT5, B4GALNT1 sequences and a newly described vertebrate B3GALT8. It is rooted in two embedded series of bilaterian sequences, named “Invertebrates 3-B3GALT” in Supplemental Figure 3. BGRb1 is rooted in several sequences from the Protostomes Arthropoda Drosophila melanogaster and Apis mellifera and Mollusca (cephalopods) W. scintillans and the Deuterostomes Hemichordata S. kowalevski and Cephalochordata B. floridae, leading us to conclude that emergence of this first subcluster happened in early Bilateria. These B3GALTs sequences are likely responsible for the synthesis of type 1 disaccharide Galβ1,3GlcNAc found on N-glycans recently described in marine Deuterostomes Echinodermata (sea cucumber and brittle star) (Eckmair et al. 2020; Vanbeselaere et al. 2020). Interestingly, B3GALT1 orthologues are found in the Deuterostomes Acanthaster planci (Echinodermata) and S. kowalevski (Hemichordata) further suggesting a prevertebrate emergence of this subfamily. The subcluster BGRb2 gathers the B3GALT4 and the newly described B3GNT10 sequences. BGRb2 is rooted by sequences of Cnidaria N. vectensis and bilaterians, e.g., the Protostome D. melanogaster and the Deuterostome S. kowalevski (noted “invertebrate 4-B3GALT” in Supplemental Figure 3). The subcluster BGRb3 is restricted to B3GNT sequences. The basal B3GNT5 cluster is rooted by a sequence found in C. intestinalis suggesting an ancient emergence of these sequences. The rest of BGRb3 (i.e., B3GNT2, B3GNT8, B3GNT3, B3GNT6, B3GNT7, B3GNT9, B3GNT4 and the newly described vertebrate B3GNT11) is rooted by two C. intestinalis sequences (Supplemental Figure 3). The topology of the ML tree indicates that the B3GNT9, B3GNT4, B3GNT7 and B3GNT11 subfamilies are likely resulting from the WGD-2R events since they are rooted by lamprey sequences of Petromyzon marinus and L. camtschaticum. The evolutionary relationships of B3GNT2, B3GNT8, B3GNT3 and B3GNT6 are less obvious since lamprey sequences were found at the base of the B3GNT2 and of the B3GNT3–B3GNT6 subfamily and required complementary synteny/paralogy-based approaches to disentangle their evolutionary history (see functional divergence of GT31 families in vertebrates section). Anyway, the duplication leading to the two clusters of four subfamilies can be dated before the emergence of vertebrates. In summary, our investigations enabled us to propose a scenario of the evolutionary relationships of the B3GTs in Metazoa illustrated in Figure 5. It points to the existence of 10 GT31 orthologue groups, e.g., B3GALNT2, B3GALT6, B3GALTs, B3GNTs, B3GLCT, FNG, C1GALT, C1GALT1C1, CHPF and CHSY likely present in the ancestral genome of the Urmetazoa, the LCA of Metazoa (Paps and Holland 2018). It suggests the implication of the WGD-2R events at the root of vertebrates, which led to a burst of gene novelties in the B3GALT/B3GNT and the emergence of 16 different vertebrate subfamilies. In addition, extraduplication events during bilaterian and/or deuterostome evolution were observed, before the emergence of vertebrates, as it was already described for the sialyltransferase family ST8Sia (Harduin-Lepers et al. 2008).
Fig. 5.
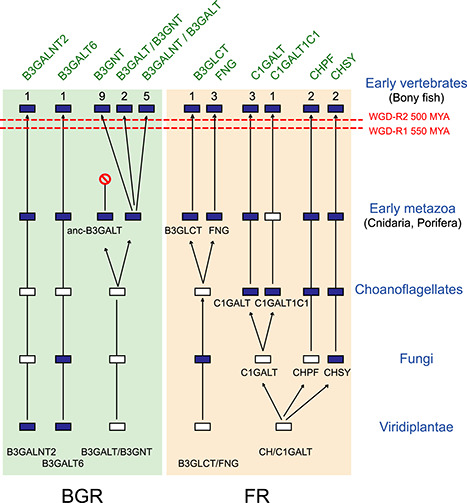
Evolutionary scenario of the metazoan GT31 sequences. This schematic depicts a model for the evolution of the eight orthologues groups (CHPF, CHSY, FNG, C1GALT/C1GALT1C1, B3GALT/B3GNT, B3GALT6, B3GALNT2, B3GLCT) of the BGR and FR clusters in the Viridiplantae, Fungi and early metazoan lineages. This model is based on evidences from phylogenetic analysis and it takes into account the two rounds of WGD-2R (WGD-R1 550 MYA and WGD-R2 500 MYA) that took place early in the vertebrate lineage. It suggests the existence of 10 orthologue groups in early metazoans, 9 of which could be identified in extant early metazoan, e.g., Porifera and Cnidaria. The dark blue-colored boxes indicate the presence of the gene, whereas the white-colored boxes represent gene loss. The numbers of vertebrate subfamilies are indicated above boxes.
Fourth criterion: sequence similarity network
It was desirable to provide a more detailed view of sequence similarity relationships across this diverse GT31 multidomain protein family in a larger set of full-length sequences. Therefore, we also used sequence similarity networks (SSN) (Atkinson et al. 2009) as a complementary approach allowing the rapid generation of an all-versus-all comparison for the analysis of a very large set of related GT31 sequences. A network of 990 GT31 sequences was build using a permissive E-value threshold corresponding to ~ 30% sequence identity. As previously observed in the ML phylogenetic tree (Figure 4), this first SSN analysis highlights the comparatively low degree of similarity between the members of the GT31 family, in particular for the FR-related sequences of the FRa (FNG and B3GLCT) and FRb (CHPF, CHSY and C1GALT/C1GALT1C1) subgroups (Figure 6A). Regardless of the presences of additional protein domains, the GT31 sequences conserve similar relationships. The analysis also shows the close relationship between B3GNT and B3GALT sequences that merge in an impure cluster. The links between B3GALNT2 and B3GALT6 sequences point out the high degree of similarity within BGRa. Additionally, B3GALT6 sequences are connected to an impure cluster of 10 Viridiplantae and Fungi sequences.
Fig. 6.

Protein SSN of metazoan GT31 sequence. The full-length protein sequences listed in supplementary data 1 were used to generate the SSN (Atkinson et al. 2009) visualized with Cytoscape. The sequences are represented as nodes colored according to the subfamily to which they belong. The lines between two nodes are drawn if the BLAST E-value is below a given threshold. (A) The network is composed of 990 GT31-related sequences, the E-value threshold = 1E−29, which corresponds to ~ 30% of sequence identity. B3GALNT2 and B3GALT6 sequences are well separated, FNG and B3GLCT sequences form two distinct clusters, whereas C1GALT1C1 and C1GALT on one hand, and CHPF and CHSY on the other are found in the same cluster. Finally, all the B3GNT (dark pink) and B3GALT (dark green) sequences merge in one cluster. This latter B3GNT/B3GALT cluster boxed with dashed lines was further investigated at more stringent thresholds to visualize subfamilies. (B) A subnetwork composed of 485 sequences of B3GNT and B3GALT at E-value threshold = 1E−55 (~35% sequence identity) and a subnetwork at E-value threshold = 1E−75 (~40% sequence identity) (C) are shown. The “B3GNT” cluster (pink) comprises B3GNT sequences of invertebrates and lampreys, and sequences from the subfamilies B3GNT2, B3GNT3, B3GNT4, B3GNT6, B3GNT7, B3GNT8, B3GNT9, B3GNT11. Interestingly, B3GNT5 sequences form a distinct cluster (yellow), whereas B3GNT10 and B3GALT4 sequences are part of the same connected component where three pure clusters are observed (i.e., B3GNT10, five fish B3GALT4 and the rest of B3GALT4 tetrapod sequences forming a sparse cluster). Besides the B3GALT5 (blue), B3GALT8 (orange) and B3GALNT1 (light blue), the “B3GALT” cluster includes B3GALT1 (light yellow) and B3GALT2 (light green) sequences. B3GALT singletons (green disconnected nodes) correspond to invertebrates, fungi and Viridiplantae sequences.
In a second step, to further investigate the relationships between B3GNT and B3GALT sequences, 485 sequences were used to build a subnetwork with a stringent threshold (E-value = 1E−55), which corresponds to ~ 35% sequence identity (Figure 6B). At this level, the B3GNT5, B3GNT10 and B3GALT4 sequences can be differentiated from the rest of the B3GNT and B3GALT sequences. At a stringent threshold (E-value = 1E−75) corresponding to ~ 40% sequence identity (Figure 6C), the rest B3GNT and B3GALT sequences separated into distinct groups. Sequences of B3GNT10 and B3GALT4 form part of the same connected component where three pure clusters can be distinguished: one of B3GNT10, one of five fish B3GALT4 sequences and one bigger cluster of B3GALT4 tetrapod sequences. Also, B3GALT4 sequences form a sparse cluster, compared with B3GNT10, highlighting the relative high divergence between its members. B3GNT sequences of invertebrates and lampreys, B3GNT2, B3GNT3, B3GNT4, B3GNT6, B3GNT7, B3GNT8, B3GNT9, B3GNT11 (pink cluster on Figure 6C) form a cluster, showing the close relationship between those sequences. B3GALT1, B3GALT2, B3GALT5, B3GALT8 and B3GALNT1 form a connected component.
Functional divergence of GT31 families in vertebrates
To illuminate the origin of the vertebrate GT31 subfamilies and infer which genetic events could account for the emergence of B3GTs functional diversity in vertebrates, we next conducted extensive comparative genomics analyses in vertebrate genomes. It has long been accepted that WGD occurred twice during the evolution of vertebrates (WGD-2R model) around 500 and 555 million years ago (MYA) providing an important source of duplicate genes known as ohnologues (Ohno 1970) with the potential to diverge in various protein functions through neo- or subfunctionalization. We identified ohnologue gene pairs using paralogy and synteny analyses and these findings were confirmed using ancestral genome reconstruction according to the N-model (Nakatani et al. 2007) and the P-model (Putnam et al. 2008) previously described for sialyltransferases (Chang et al. 2019; Harduin-Lepers, 2010; Petit, et al. 2015). A third-round fish-specific genome duplication known as teleost genome duplication (TGD) occurred before Teleosts radiation around 350 MYA (Amores et al. 1998; Meyer and Van de Peer 2005), which generated extra copies of fish genes. For sake of clarity, the details of these various analyses conducted for each vertebrate subfamily are given in Supplementary Data 3. Also, several genetic events of gene deletion or single gene duplication or translocation and extensive chromosomal rearrangements occurred, which have shaped the vertebrate GT31 repertoire (Table I). Altogether, this accurate identification of B3GT homologues (paralogues and orthologues) sharing common ancestry enabled us to predict function evolution of GT31 proteins in vertebrates and the vertebrate β3-glycosylation landscape.
Table I.
Distribution of GT31 in metazoa. This table shows the nine orthologue groups of GT31 found in metazoan genomes and the 30 vertebrate subfamilies. ⨁ the gene is present, ⊚ the gene is absent, /⨁ the gene is present in some species, but not all, ⨁⨁ the gene is duplicated in teleost fish genomes. New subfamilies are described in vertebrates: B3GALT8, which is not found in human genome and is closely related to B3GALT2; B3GNT10 named as it is in HGNC This gene is related to B3GALT4 and should be named B3GALT9 since it is likely a B3GALT. Other newly described vertebrate subfamilies include B3GNT11, C1GALT2 and C1GALT3, which are lost in human genome.

|
In the FR subgroup comprised of B3GLCT and FNG (FRa), and CHPF, CHSY, C1GALT and C1GALT1C1 (FRb), the six metazoan families underwent variously the genome duplication events (WGD-2R and TGD). B3GLCT is the unique member of the vertebrate b3glct subfamily. Despite two rounds of WGD, a single-copy b3glct gene is distributed in all the investigated vertebrate species with the notable exception of Teleosts, which show b3glct co-orthologues. These data and our phylogenetic analysis (Supplemental Figure 2) indicate a highly conserved subfamily in vertebrates and a fundamental role of this β3-glucosyltransferase known to catalyze the attachment of glucose to O-linked fucose via a β1,3-glycosidic linkage on thrombospondin type 1 repeats of extracellular proteins in mammalian tissues (Kozma et al. 2006; Sato et al. 2006). In vertebrates, the FNG expanded into the three subfamilies mfng, rfng and lfng after the WGD-2R events (Supplemental Figure 4). The mammalian β3-N-acetylglucosyltransferases act on O-linked fucose in the epidermal growth factor motifs in Notch (Holdener and Haltiwanger 2019; Rampal et al. 2005), thereby modulating notch signaling Notch/Delta interactions (Brückner et al. 2000; Moloney et al. 2000). Mammalian LFNG, MFNG and RFNG are expressed in a tissue-specific and developmentally regulated manner, further suggesting their subfunctionalization along vertebrate evolution.
The CHPF and CHSY families, which mapped to the root of the Metazoa tree (Figure 4), expanded into four subfamilies in vertebrates CHPF, CHPF2 and CHSY1 and CHSY3 after the WGD-2R. In addition, two chpf-related genes resulting from the TGD were identified in Otocephala fish (Supplemental Data 3). The mammalian CH enzymes possess a dual activity catalyzing the transfer of N-acetylgalactosamine (GalNAc) residue (referred to GalNAcT-II activity) and the transfer of a glucuronic acid residue (called GlcAT-II activity) leading to the formation of (–4GlcAβ1–3GalNAcβ1–) repeat units in chondroitin sulfate (CS) (Mikami and Kitagawa 2013). Although the specific enzymatic activity of each CH enzyme is still a matter of debate, it appears that CS polymerization results from the formation of an enzyme complex involving any of the synthase CHSY1 or CHSY3 with a polymerizing factor CHPF or CHPF2 (Izumikawa et al. 2008; Izumikawa et al. 2007; Ogawa et al. 2010) acting as a chaperone modulating CS biosynthesis. The C1GALT and C1GALT1C1 families are two very ancient families already found in Choanoflagellata (Figure 5). A unique copy of c1galt1c1 gene is found in all the investigated vertebrate species suggesting that the WGD-2R and TGD did not impact this ancient family (Table I). Additional c1galt1c1 homologues identified in the genome of mouse and human likely correspond to two distinct pseudogenes recently translocated in the mammalian genomes (Supplemental Data 3). Our phylogenetic and synteny analyses of the vertebrate c1galt loci illustrated in Figure 7 revealed the existence of three vertebrate C1GALT subfamilies that we named C1GALT1, C1GALT2 and C1GALT3 resulting from the WGD-2R events (Figure 7). The c1galt3 and c1galt1/c1galt2 subfamilies split around 550 MYA (WGD-R1) and c1galt2 and c1galt1 around 500 MYA (WGD-R2). Interestingly, c1galt1 appeared to be lost in Teleost genomes, whereas duplicate c1galt2 genes were found in most ray-finned fish genome, as a result of the TGD. The c1galt2 and c1galt3 were lost in mammalian genomes. The mammalian C1GALT1 catalyzes the transfer of a galactose residue from UDP-Gal onto the mucin-type O-glycan known as Tn antigen (GalNAcα1-Thr/Ser) to form the T antigen (Galβ1,3GalNAcα1-Thr/Ser) (Figure 1B) (Ju, Brewer et al. 2002; Ju, Cummings et al. 2002; Ju et al. 2006). A recent system glycomics analysis of adult zebrafish tissues identified major O-glycans based on core-1 Galβ1,3GalNAc-ol and core-2 Galβ1,3[GlcNAcβ1,6]GalNAc-ol structures (Yamakawa et al. 2018) further suggesting that c1galt2 and/or c1galt3 could share the same function as c1galt1 in fish tissues. Interestingly, the expression of active T-synthase in vertebrates requires a unique molecular chaperone, C1GALT1C1 (cosmc) for correct protein folding, enzyme activity and correct protein O-glycosylation (Ju and Cummings 2005; Ju et al. 2006). A cosmc binding region (CBRT) (LYQKVRILCWVMTGP) was identified within the N-terminal stem domain of the vertebrate C1GALT1, which enables cosmc–C1GALT1 interaction in the endoplasmic reticulum (Aryal et al. 2014). This CBRT region is well conserved in the vertebrate C1GALT2 sequence, but not in the C1GALT3, further suggesting that the C1GALT2 could interact with C1GALT1C1 retaining a T-synthase activity, whereas the C1GALT3 has lost this property. However, this remains to be experimentally checked since the invertebrate C1GALT1 do not require the cosmc chaperone to be correctly folded and fully active (Ju et al. 2006).
Fig. 7.

Evolutionary relationships of vertebrate C1GALT: (A) Molecular phylogenetic analysis by ML method of the C1GALTs. The evolutionary history was inferred by using the ML method based on the JTT matrix-based model in MEGA 7.0 (Kumar et al. 2016). The ML tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 37-aa sequences (sequence information of the eight invertebrate C1GALT, three vertebrate C1GALT3, two lampreys C1GALTs, 10 C1GALT1, six vertebrate C1GALT2, four Teleost C1GALT2-r1 and four C1GALT2-r2 sequences can be found in Supplementary Data). There were a total of 384 positions in the final dataset. (B) Synteny relationships around the c1galt gene loci in vertebrate genomes. The schematic indicates the chromosome localizations of the c1galt1, c1galt2 and c1galt3 in the human (H. sapiens, Hsa), the mouse (M. musculus, Mmu), the chicken (Gallus gallus, Gga), the lizard (Podarcis muralis, Pmur), the coelacanth (L. chalumnae, Lcha), the spotted gar (Lepisosteus oculatus, Locu), the Japanese medaka (O. latipes, Ola) and the reedfish (Erpetoichthys calabaricus, Erpcal). The c1galt genes (e.g., c1galt1, c1galt2 and c1galt3) are indicated in red when present on the chromosome or in gray when lost from the genomic region. The putative orthologues were retrieved from the NCBI and ENSEMBL servers using chromosome walking and reciprocal tblastn and also the latest ENSEMBL dataset (ENS70) at the synteny database site (http://teleost.cs.uoregon.edu/synteny_db/) (Catchen et al. 2009) and they were visualized using the Genomicus 93.01 website (Louis et al. 2012). Conserved neighboring gene loci are indicated in black and those loci in the vicinity of c1galt genes belonging to fish-specific paralogons are indicated in green.
As described above, the BGR subgroup includes three ancient orthologue groups B3GALNT2 and B3GALT6 (BGRa) and B3GALT/B3GNT (BGRb) that individualized before the emergence of early Metazoa (Figure 5). A single-copy b3galnt2 gene is found in all the vertebrate species suggesting that the WGD-2R had no impact on this gene family (Table I). The human B3GALNT2 enzyme catalyzes the transfer of GalNAc residues from UDP-GalNAc leading to the formation of type-1 LacdiNAc (GalNAcβ1,3GlcNAc-R) (Figure 1), a rare structure found onto the O-Man glycans of alpha dystroglycan and in HeLa cells (Hiruma et al. 2004; Nakane et al. 2019). Likewise, a single-copy b3galt6 gene is found in the Metazoa genome and no expansion of this gene family could be evidenced in vertebrates (Table I). The human B3GALT6 enzyme plays essential role in the GAG core synthesis leading to the formation of the Galβ1,3Galβ1,4Xyl-O-Ser linker region (Cole et al. 2001) and mutations in the human B3GALT6 gene cause a spectrum of skeletal and connective tissue disorders (Malfait et al. 2013; Nakajima et al. 2013).
As mentioned, the BGRb is an extremely large cluster that expanded dramatically at the dawn of vertebrates. The BGRb1 subcluster is composed of five vertebrate subfamilies B3GALT1, B3GALT2, B3GALT8, B3GALT5 and B3GALNT1 (Supplemental Figure 3). Our phylogeny and synteny analysis (Supplemental Figure 5A and Supplemental Data 3) indicate that the b3galt1 gene likely emerged before the WGD-2R events. Orthologues were identified in the main groups of Gnathostome vertebrates (Table I) and b3galt1 co-orthologues resulting from the TGD 350 MYA were maintained in Teleosts. Orthologues of the b3galt2 genes were found in all the vertebrate species including Teleosts (Table I). Several copies of the b3galt2 likely resulting from species-specific gene duplications were found in the lampreys P. marinus and L. camtschaticum and the inshore hagfish Eptatretus burgeri genomes. The newly described b3galt2-related genes named b3galt8 represented a novel vertebrate B3GT subfamily found in Squamata (lizards and snakes), in turtles and Archosauria (birds and crocodilian). Orthologues were also identified in Amphibians and ray-finned fishes (Table I). Our data indicate that b3galt2 and b3galt8 are ohnologues likely resulting from the second WGD-2R (WGD-R2) (Figure 8, Supplemental Figure 5A and Supplemental Data 3). Orthologues of the B3GALT5 sequences were found in Tetrapod genomes, but none could be found in ray-finned fishes (Table I). Mammalian B3GALT1, B3GALT2 and B3GALT5 enzymes are known to be involved in the formation of type-1 N-acetyllactosamine (LacNAc) (Galβ1,3GlcNAcβ1,-R) (lacto series) and type 3 Galβ1,3GalNAcβ1,-R (globo-series) disaccharides (Henion et al. 2001; Isshiki et al. 1999; Togayachi et al. 2008). Therefore, B3GALT8 is predicted to catalyze the transfer of a galactose residue from UDP-Gal to form LacNAc disaccharides. The mammalian B3GALT5 enzyme is also known to synthetize the sialylLewisa (sLea) and the stage-specific antigen-3 (CA19.9/SSEA-3-synthase). B3GALT5 is not expressed in fish tissue supporting previous observation that Lea, Leb and type-1 disaccharides were not detected in zebrafish tissues (Yamakawa et al. 2018). Orthologues of the b3galnt1 gene could be identified in Amniotes but was not found in teleost fish (Table I). As illustrated in the ML phylogenetic tree (Supplemental Figure 3), the B3GALNT1 subfamily, previously known as B3GALT3, is evolutionary related to B3GALT1, B3GALT5, B3GALT2 and B3GALT8. These long-branched sequences suggest the occurrence of many substitutions that could explain a change of function at the base of vertebrate evolution. Indeed, the human B3GALNT1 enzyme was shown to use UDP-GalNAc and not UDP-Gal as a donor substrate and the globotriaosylceramide as an acceptor substrate (Gb3, Pk antigen). It is involved exclusively in the synthesis of the P antigen as part of the globoside (GLOB) blood group system to form Gb4 (Figure 1) (Hellberg et al. 2002; Okajima et al. 2000). Teleost fishes lack the b3galnt1 gene, which corroborates with the fact that globosides were not detected in the zebrafish tissues (Yamakawa et al. 2018).
Fig. 8.

Scenario illustrating the evolutionary history of BGRb cluster in vertebrates. For each subcluster (BGRb1: B3GALT2, B3GALT8, B3GALNT1, B3GALT5 and B3GALT1; BGRb2: B3GNT10 and B3GALT4; BGRb3: B3GNT4, B3GNT9, B3GNT7, B3GNT11, B3GNT2, B3GNT8, B3GNT6, B3GNT3 and B3GNT5), the duplication events are localized relatively to the two rounds of WGD-2R that occurred in early vertebrates and the one specific to teleost fishes (TGD). The presence (full dark blue boxes) or loss (full white boxes) of genes in the major vertebrate branches is indicated. In addition, B3GT genes present in some vertebrate species but not all are indicated by dashed light blue boxes. Association of BGRb subfamilies to the ancestral protochromosomes of vertebrates VAC A and VAC F are indicated above in purple letters. In Bilateria and in Deuterostoma, tandem duplications that likely occurred before the WGD-2R events are boxed, whereas the remaining duplication events are of unknown origin.
The BGRb2, rooted by the Deuterostome S. kowalevski, gathers B3GALT4 and the newly described B3GNT10 subfamilies. The b3galt4 orthologues show a patchy distribution in the vertebrate genomes (Table I). Intriguingly, the b3galt4 orthologue appears to be lost in several vertebrate genomes. The mammalian B3GALT4 enzyme determines the synthesis of glycosphingolipids of the ganglio-series, e.g., GD1b, GM1 and GA1 (Figure 1) and therefore is also known as the GM1-synthase (Amado et al. 1998; Daniotti et al. 1999; Miyazaki et al. 1997). Interestingly, it was suggested that it could affect lipid concentrations by modifying lipoprotein receptors (Willer et al. 2008) and more recently it was shown to catalyze the transfer of a Gal residue from UDP-Gal to the β1,4GalNAc side chain of glycosylphosphatdylinositol (GPI)-anchored proteins (Wang et al. 2020). Our data indicate that the B3GALT4 functions are restricted to a few vertebrate species (Supplemental Figure 5B). It remains unclear whether these gangliosides and/or GPI side chain could be synthetized in chicken tissues, for instance (Hirabayashi et al. 1991) and if so, what is the nature of the biosynthetic enzyme. An explanation might come from the existence in the vertebrate genomes of an additional b3galt4-related gene that was annotated B3GNT10 by the HUGO gene nomenclature committee (HGNC ID:53652). This is likely a nonfunctional gene (pseudogene) in the human genome since no start codon could be detected and no encoded product could be demonstrated yet. However, an orthologue was identified in some vertebrate species, which could potentially generate a protein (Table I). Our synteny data indicated that these two genes are ohnologues that emerged from the WGD-2R events (Supplemental Figure 5B and Figure 8). We predict that the B3GNT10 enzyme is a B3GALT catalyzing the transfer of galactose residues from UDP-Gal that could rescue B3GALT4 in b3galt4-defective organisms. Therefore, this enzyme should be named B3GALT9, although this remains to be experimentally assessed.
The last subcluster BGRb3 is extremely large and restricted to B3GNT sequences. The basal B3GNT5 subfamily is rooted by a sequence found in C. intestinalis suggesting its ancient emergence predating the WGD-2R events (Supplemental Figure 3).
Orthologous b3gnt5 genes were found in all the vertebrate species and co-orthologues were identified in some Teleosts like the Cypriniformes Cyprinus carpio and Danio rerio, whereas most of the fish genomes lost one b3gnt5 gene copy (Supplemental Figure 5B). Our comparative genomics data further suggested that TGD impacted this subfamily (Supplementary Data 3). The mammalian B3GNT5 enzymes have a specific core transferase activity catalyzing the entry point for Lacto-/Neolacto-series glycolipids leading to the formation of GlcNAcβ1,3Galβ1,4Glc-Cer structure and therefore are known as the Lc3-synthase (Henion et al. 2001; Togayachi et al. 2001). The rest of BGRb3 subcluster, e.g., B3GNT2, B3GNT8, B3GNT3, B3GNT6, B3GNT7, B3GNT9, B3GNT4 and the newly described B3GNT11 is rooted by two C. intestinalis and one P. marinus sequences (Supplemental Figure 3). One copy of the b3gnt2 gene is found in the Agnathan branch and orthologues of the b3gnt2 gene exist in all the vertebrate genomes (Table I and Figure 8). In addition, b3gnt2 co-orthologues were identified in Teleosts resulting from the TGD (Supplemental Figure 5C). The B3GNT8 subfamily, previously known as B3GALT7 (Huang et al. 2004), represents the closest relative of the B3GNT2 sequences. Interestingly, no b3gnt8 orthologue was found in the in avian branch; it is notably absent from chicken but is found in the Squamata (Lizards and snakes) and Sauria (Crocodilia) branch. Two b3gnt8-related co-orthologues were found in Teleosts indicative of the occurrence of TGD event, although one duplicate was lost in other fish species like in medaka (Supplemental Figure 5C). Our synteny analyses revealed that b3gnt2 and b3gnt8 genes are ohnologues issued from the WGD-2R (Supplemental Data 3 and Figure 8). The mammalian B3GNT2 is the major enzyme involved in polylactosamine synthesis, a carbohydrate structure made of repeats of LacNAc (Galβ1–4GlcNAcβ1–3) unit (Togayachi et al. 2010) carried on glycolipids of the Neolacto-series (GlcNAcβ1,3Galβ1,4GlcNAcβ1,3Galβ1,4Glc-Cer). As illustrated in Figure 1, B3GNT2 is also involved in the initiation and elongation of keratan sulfate-I (KS-I) and KS-II found on N- and O-glycans leading to the formation of GlcNAcβ1,3Galβ1,4GlcNAcβ1,2Man- and GlcNAcβ1,3Gal β1,4GlcNAcβ1,6GalNAcα1-Ser (Sasaki et al. 1997; Shiraishi et al. 2001). The human B3GNT8 is a polylactosamine-synthase acting preferentially on tetra-antennary N-glycans having a β1-6 branch (Ishida et al. 2005). Interestingly, the mammalian B3GNT8 enzyme physically associates with B3GNT2 to increase polylactosamine synthesis (Seko and Yamashita 2005) and its absence in some vertebrate species could have an impact on polylactosamine synthesis in these species. The sister branch encompasses B3GNT3 and B3GNT6 sequences that are rooted by several lamprey sequences found in P. marinus and L. camtschaticum. Interestingly, the open reading frame of the human B3GNT3 gene is split into two coding exons (Figure 2), with the exon/intron junction located between the two conserved motifs I and II. This gene organization was found in all the placental (Eutheria) mammals’ b3gnt3 genes but not into the egg-laying mammals, suggesting that it was acquired in the LCA of Eutheria (Kordiš 2011). Orthologues of the b3gnt3 gene are found in all the vertebrate species (Table I). The human B3GNT3 was shown to have very little activity in the synthesis of poly-LacNAc chains and native acceptors remain to be identified (Shiraishi et al. 2001; Togayachi et al. 2001). Interestingly, we found that the B3GNT6 sequences are restricted to Eutherian mammals (Table I). Our comparative genomics data (Supplemental Figure 5C) suggest two possible evolutionary scenarios for the b3gnt6 gene illustrated in Figure 8: (i) Orthologues could have been lost in birds, amphibians and fish genomes. (ii) Alternatively, the B3GNT6 gene could have a more recent origin in the LCA of Eutheria about 102 MYA. The topology of the ML tree shows that B3GNT6 sequences are embedded in the B3GNT3 cluster (Supplemental Figure 3). This observation favors the idea that they may arise by rearrangement of already existing duplicated B3GNT3 sequences in the stem lineage of Eutheria. The human B3GNT6 gene encodes the core 3 synthase, an enzyme with narrow acceptor specificity that catalyzes the transfer of GlcNAc residue from UDP-GlcNAc to the Tn antigen GalNAc-O-Ser leading to the formation of GlcNAcβ1,3GalNAc-O-Ser on O-glycans (Iwai et al. 2002). Interestingly, core 3 GlcNAcβ1,3GalNAc-ol and core 4 GlcNAcβ1,3[GlcNAcβ1,6]GalNAc-ol are very minor components in zebrafish tissues (Yamakawa et al. 2018). Our data led us to propose that the B3GNT3 enzyme present in the LCA of vertebrates could promiscuously catalyze the synthesis of core 3 O-glycans, an exquisite substrate specificity that was selected by B3GNT6 at the root of placental mammalian evolution. In each of the B3GNT4, B3GNT7, B3GNT9 and the newly described B3GNT11 remaining subfamilies, a shark Rhincodon typus orthologous sequence could be identified. A b3gnt7 gene orthologue was found in the main vertebrate phyla (Table I). We noted the presence B3GNT7-related sequences mostly in sharks, ray-finned fishes, the coelacanth Latimeria chalumnae and frogs Silurana tropicalis, Xenopus laevis, Nanorana parkeri that were named B3GNT11 (Supplemental Figure 3). Our synteny analyses indicated that the B3GNT7 and B3GNT11 were ohnologues issued from the WGD-2R (Supplemental Figure 5C, Supplemental Data 3). The mammalian B3GNT7 enzyme is known to be a B3GNT involved in KS-I and KS-II found on N- and O-glycans leading to the formation of GlcNAcβ1,3Galβ1,4GlcNAcβ1,2Man- and GlcNAcβ1,3Gal β1,4GlcNAcβ1,6GalNAcα1-Ser biosynthesis (Kataoka 2002). Therefore, we suggest that B3GNT11 would be a B3GNT using UDP-GlcNAc as a donor substrate, although its substrate specificities remains to be studied. Orthologues of the b3gnt4 gene show a patchy distribution in vertebrate genomes. It is found in birds and amphibians, in the spotted gar and several Teleost genomes like the Salmoniformes and Esociformes (Table I). However, no b3gnt4 could be identified in Otocephala like the Cypriniforme D. rerio nor in the Neoteleosteii like the Beloniforme Oryzias latipes (Supplementary Figure 5C). As illustrated in Figure 1, the human B3GNT4 uses UDP-GlcNAc to catalyze the initiation and elongation of poly-LacNAc sugar chains and the initiation type-I synthesis leading to the formation of GlcNAcβ1,3Galβ1,4(SO3)GlcNAcβ-R (Shiraishi et al. 2001). Orthologues of the b3gnt9 gene were found in all the vertebrate species (Table I and Supplemental Figure 5C). The enzymatic activity of B3GNT9 is not described yet, but it is likely involved in the formation of GlcNAcβ1,3Gal and polylactosamine biosynthesis.
Altogether, our phylogeny and synteny analyses enabled us to propose a global scenario for the evolutionary history of the BRGb cluster illustrated in Figure 8. Central to this scenario is the observation that the same vertebrate ancestral protochromosome (VAC A) bears the ancestor sequences of b3galt2/8, and b3gnt10/b3galt4 in the subclusters BGRb1 and BGRb2. Each subcluster is rooted by protostomes and deuterostomes sequences, further suggesting that tandem duplication firstly occurred in Bilaterians before the split of Protostoma and Deuterostoma. Similarly, VAC F bears the ancestor sequences of b3gnt5, b3gnt7/11 and b3gnt6 in the BRGb3 subcluster. Since only deuterostomes sequences root this subgroup, it is suggested that two successive tandem duplications occurred in late deuterostomes, giving rise to the ancestors of these genes that were further affected by the WGD-2R events.
Conclusion
In conclusion, among the 10 orthologue groups of GT31 sequences likely present in the Urmetazoa, the LCA of Metazoa, 9 were identified in Porifera and Cnidaria, the still-living earliest clades, testifying of their very ancient divergence. These basal metazoans possess a rich repertoire of GT31 sequences resulting from their own gene duplications and their specificities are still to be investigated. Thirty individual GT31 subfamilies were issued from the 10 GT31 orthologue groups and emerged in vertebrates mainly following the WGD-2R events. Our exploration of the vertebrate genomes evidenced three B3GALT/B3GNT and two C1GALT novel subfamilies that were lost in humans. Also, several GT31 subfamilies were lost in Teleosts constituting evolutionary knock-out (KO) models for human diseases. The activities and substrate specificities of the vertebrate B3GT enzymes have been enlightened by experiments achieved in a few mammalian model organisms (mostly mouse and human) and further experimental is needed to shed light on the similarities and differences in other vertebrate models like the zebrafish and medaka.
Materials and methods
Sequence collection/identification
The vertebrate GT31 CAZyme aa sequences available at URL http://www.cazy.org/ currently (April 2020) contains 110 GT were used as seed sequence to search for homologous sequences in NCBI using BlastP (protein database) or Transcriptome Assembly database to avoid pseudogenes or chimeric sequences. We selected target organisms the Fungi and Viridiplantae, focusing on A. thaliana and P. patens. Among the metazoans, we specially choose Sponges (Porifera), Cnidaria, Ciona, Branchiostoma and most vertebrate models.
Sequence alignment and phylogenetic analysis
Many sequences of GT31 CAZy family include an extradomain that could be identified using the SMART (http://smart.embl-heidelberg.de/) (Letunic and Bork 2018; Letunic et al. 2015) Since these extradomains [e.g., DUF4094 (PF13334), DUF604 (PF04646), the galactose-binding domain Gal-bind_lectin (PF00337), the N-glycosylation protein domain EOS1 (PF12326) and the CHGN domain (PF05679)] prevented correct alignments between sequences as in some cases, there was a loose analogy with the sequences of GTs, we restricted the sequence alignments to the GT31 domain itself, using Muscle program implemented in MEGA vers. 7.0 (Kumar et al. 2016). The parameters corresponding to the best aa substitution model determined by the Akaike criterion implemented in MEGA was chosen (JTT + G) for phylogenetic reconstruction. The phylogenetic topologies were obtained using ML with 500 bootstrap replicates for the evaluation of their branch supports.
Identification of conserved synteny, paralogon detection and ancestral genome reconstruction
To determine the existence of paralogons, i.e., chromosome segments affected by genome duplications within vertebrates, a list of paralogue genes located upstream and downstream each GT31 locus was searched using NCBI and the latest ENSEMBL data set (ENS70). The detection of paralogous blocks was also done at the Synteny Database site (http://teleost.cs.uoregon.edu/synteny_db/, last accessed May 2020) (Catchen et al. 2009) and visualized at the Genomicus site (version 93.01) (http://www.dyogen.ens.fr/genomicus/, last accessed May 2020) (Louis et al. 2012). Synteny (blocks of orthologous genes) between vertebrate GT31-related genes in invertebrates was assessed by manual chromosomal walking and reciprocal BLAST searches of genes adjacent to GT31 gene loci in human (Hsa), mouse (Mmu), chicken (Gga), medaka (Ola) and spotted gar (Loc) genome databases. Search several genes located upstream and downstream each GT31 paralogues in vertebrate genomes through NCBI and ENSEMBL datasets. To firmly establish whether the duplication events involving vertebrate GT31 genes were linked to the two rounds of WGD (WGR-2R), the assignation of each duplicate gene to the protochromosomes of vertebrate ancestral karyotype was searched according to the previously reported scenario of Nakatani suggesting the existence of 10–13 pre-2R protochromosomes (VAC) in the last common vertebrate ancestor (Kasahara et al. 2007; Nakatani et al. 2007) and Putnam suggesting the existence of 17 linkage groups (CLG) in the last common chordate ancestor (Putnam et al. 2008), as previously reported for sialyltransferase genes (Petit et al. 2015). An example of the followed strategy is illustrated for the FNG genes in Supplementary Figure 4.
Detection of conserved domains
The SMART (http://smart.embl-heidelberg.de/) (Letunic and Bork 2018; Letunic, et al. 2015) and database provided domain identification and annotation on the World Wide Web (http://smart.embl.de). The tool compares query sequences with its databases of domain sequences and multiple alignments while concurrently identifying compositionally biased regions such as signal peptide, transmembrane and coiled coil segments. Conserved signature of GT31 sequences were drawn in Logo format using the Berkley WebLogo generator website (Crooks et al. 2004).
Similarity sequence protein network
SSN was constructed using formatdb, from the stand-alone BLAST software and a custom BLAST database was created. The networks provided a graphical overview of interrelationships among and between sets of proteins that are not easily discerned from visual inspection of large trees and multiple alignments. The pairwise relationships between sequences were calculated by a blastall search in the custom database with each individual sequence in the set and the E-value was taken as a measure of similarity between sequences. Three networks were built considering different E-value thresholds: 1E−29, 1E−55 and 1E−75 corresponding to 30, 35 and 40% of sequence identity respectively. The first network includes 990 GT31-related sequences; the two networks at more stringent threshold comprise 485 sequences of B3GNT and B3GALT. Network were visualized using Cytoscape (Shannon et al. 2003), where each sequences was represented as a node and edges were defined between any pair of nodes with an E-value less than threshold, also the default Cytoscape force-directed layout was applied. Nodes were colored according to the subfamily to which the sequence belongs.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the help of Amandine Lecerf-Defer and Anais Barray with the databases screening and Olga Plechakova, who developed and maintained the GT-Database. The contribution of the COST Action CA18103-INNOGLY supported by the European Cooperation in Science and Technology (COST) is greatly acknowledged.
Contributor Information
Daniel Petit, Glycosylation et différenciation cellulaire, EA 7500, Laboratoire PEIRENE, Université de Limoges, 123 Avenue Albert Thomas, 87060 Limoges Cedex, France.
Roxana Elin Teppa, Toulouse Biotechnology Institute, TBI, Université de Toulouse, CNRS, INRA, INSA, 135, Avenue de Rangueil, F-31077 Toulouse Cedex 04, France.
Anne Harduin-Lepers, Université de Lille, CNRS, UMR 8576 - UGSF - Unité de Glycobiologie Structurale et Fonctionnelle, F-59000 Lille, France.
Funding
The Centre National de La Recherche Scientifique (CNRS), the University of Lille (FST) and the University of Limoges.
Conflict of interest statement
None declared.
Abbreviations
aa, amino acid; A4GALT, α1,4-galactosyltransferase or Gb3 synthase; BGR, B3GALT-related; BLAST, Basic Local Alignment Search Tool; B3GALNT, β1,3-N-acetylgalactosaminyltransferase; B3GALT, β1,3-galactosyltransferase; B3GLCAT, β1,3-glucuronyl acid transferases; B3GLCT, β1,3-glucosyltransferase; B3GNT, β1,3-N-acetylglucosaminyltransferase; B3GT, β1,3-glycosyltransferase; B4GALNT2, β1,4-N-acetylgalactosaminyltransferase; B4GALT, β1,4-galactosyltransferase; CAZy, Carbohydrate-Active enZYmes; CBRT, cosmc binding region; CHPF, chondroitin-polymerizing factor; CHSY, chondroitin sulfate synthase (previously known as CSS); Cosmc, core 1βGalT specific molecular chaperone; CS, chondroitin sulfate; C1GALT, core 1 β1,3-galactosyltransferase; DUF, domain of unknown function; FNG, fringe β1,3-N-acetylglucosaminyltransferase; FR, Fringe-related; GAG, glycosaminoglycan; GalNAc, N-acetylgalactosamine; GGTA1, α1,3-galactosyltransferase 1; GLOB, globoside; GPI, glycosylphosphatdylinositol; GT, glycosyltransferase; HGNC, HUGO gene nomenclature committee; HPGT, hydroxyproline-O-galactosyltransferase; KS, keratan sulfate; LacNAc, N-acetyllactosamine; LCA, last common ancestor; LFNG, lunatic FNG; MFNG, manic FNG; ML, maximum likelihood; MYA, million years ago; NCBI, National Center for Biotechnology Information; poly-LacNAc, poly-N-acetyllactosamine; RFNG, radical FNG; SNFG, symbol nomenclature for graphical representation of glycans; TGD, teleost genome duplication; WGD, whole genome duplication.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado M, Almeida R, Carneiro F, Levery SB, Holmes EH, Nomoto M, Hollingsworth MA, Hassan H, Schwientek T, Nielsen PA et al. 1998. A family of human β3-galactosyltransferases: Characterization of four members of a UDP-galactose:β-N-acetyl-glucosamine/β-N-acetyl-galactosamine β-1,3-galactosyltransferase family. J Biol Chem. 273:12770–12778. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science. 282:1711–1714. [DOI] [PubMed] [Google Scholar]
- Aryal RP, Ju T, Cummings RD. 2014. Identification of a novel protein binding motif within the T-synthase for the molecular chaperone Cosmc. J Biol Chem. 289:11630–11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. 2009. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 4:e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Tian L, Wang W, Bobbs S, Herock H, Travers A, Showalter AM. 2015a. A small multigene hydroxyproline-O-galactosyltransferase family functions in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. BMC Plant Biol. 15:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Wang W, Ma S, DeBrosse T, Poirier E, Emch K, Soukup E, Tian L, Showalter AM. 2015b. Two Hydroxyproline galactosyltransferases, GALT5 and GALT2, function in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. PLoS One. 10:e0125624–e0125624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner K, Perez L, Clausen H, Cohen S. 2000. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 406:411–415. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Conery JS, Postlethwait JH. 2009. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 19:1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LY, Teppa E, Noel M, Gilormini PA, Decloquement M, Lion C, Biot C, Mir AM, Cogez V, Delannoy P et al. 2019. Novel zebrafish mono-alpha2,8-sialyltransferase (ST8Sia VIII): An evolutionary perspective of alpha2,8-sialylation. Int J Mol Sci. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Zhou Y, Neelamegham S. 2017. DrawGlycan-SNFG: A robust tool to render glycans and glycopeptides with fragmentation information. Glycobiology. 27:200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SE, Mao MS, Johnston SH, Vogt TF. 2001. Identification, expression analysis, and mapping of B3galt6, a putative galactosyl transferase gene with similarity to Drosophila brainiac. Mamm Genome. 12:177–179. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: A sequence logo generator. Genome Res. 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniotti JL, Martina JA, Zurita AR, Maccioni HJF. 1999. Mouse β 1,3-galactosyltransferase (GA1/GM1/GD1b synthase): Protein characterization, tissue expression, and developmental regulation in neural retina. J Neurosci Res. 58:318–327. [PubMed] [Google Scholar]
- Dehal P, Boore JL. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3:e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmair B, Jin C, Karlsson NG, Abed-Navandi D, Wilson IBH, Paschinger K. 2020. Glycosylation at an evolutionary nexus: The brittle star Ophiactis savignyi expresses both vertebrate and invertebrate N-glycomic features. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J, Ellis M, Doblin M, Qu Y, Bacic A. 2010. Genes and enzymes of the GT31 family: Towards unravelling the function(s) of the plant glycosyltransferase family members. Annu Plant Rev. 41:213–234. [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Lapébie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E. 2009. Origin and evolution of the notch signalling pathway: An overview from eukaryotic genomes. BMC Evol Biol. 9:249–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. 2002. CDART: Protein homology by domain architecture. Genome Res. 12:1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geshi N, Johansen JN, Dilokpimol A, Rolland A, Belcram K, Verger S, Kotake T, Tsumuraya Y, Kaneko S, Tryfona T et al. 2013. A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J. 76:128–137. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2018. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harduin-Lepers A. 2010. Comprehensive analysis of sialyltransferases in vertebrate genomes. Glycobiology Insights. 2:29–61. [Google Scholar]
- Harduin-Lepers A, Petit D, Mollicone R, Delannoy P, Petit JM, Oriol R. 2008. Evolutionary history of the alpha2,8-sialyltransferase (ST8Sia) gene family: Tandem duplications in early deuterostomes explain most of the diversity found in the vertebrate ST8Sia genes. BMC Evol Biol. 8:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen TYK, Pelto-Huikko M, Pasternack L, Mäki M, Kainulainen H. 2006. Murine ortholog of the novel glycosyltransferase, B3GTL: Primary structure, characterization of the gene and transcripts, and expression in tissues. DNA Cell Biol. 25:465–474. [DOI] [PubMed] [Google Scholar]
- Hellberg Å, Poole J, Olsson ML. 2002. Molecular basis of the globoside-deficient Pk blood group phenotype: Identification of four inactivating mutations in the UDP-N-acetylgalactosamine: Globotriaosylceramide 3-β-N-acetylgalactosaminyltransferase gene. J Biol Chem. 277:29455–29459. [DOI] [PubMed] [Google Scholar]
- Henion TR, Zhou D, Wolfer DP, Jungalwala FB, Hennet T. 2001. Cloning of a mouse β1,3N-acetylglucosaminyltransferase GlcNAc(β1,3)gal(β1,4)Glc-ceramide synthase gene encoding the key regulator of lacto-series glycolipid biosynthesis. J Biol Chem. 276:30261–30269. [DOI] [PubMed] [Google Scholar]
- Hennet T. 2002. The galactosyltransferase family. Cell Mol Life Sci. 59:1081–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T, Dinter A, Kuhnert P, Mattu TS, Rudd PM, Berger EG. 1998. Genomic cloning and expression of three murine UDP-galactose: β-N-acetylglucosamine β1,3-galactosyltransferase genes. J Biol Chem. 273:58–65. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Fujita SC, Kon K, Ando S. 1991. Characterization of a novel Le(x)-active ganglioside from chick intestinal tissues recognized by murine monoclonal antibody 188C1. J Biol Chem. 266:10268–10274. [PubMed] [Google Scholar]
- Hiruma T, Togayachi A, Okamura K, Sato T, Kikuchi N, Kwon Y-D, Nakamura A, Fujimura K, Gotoh M, Tachibana K et al. 2004. A novel human beta1,3-N-acetylgalactosaminyltransferase that synthesizes a unique carbohydrate structure, GalNAcbeta1-3GlcNAc. J Biol Chem. 279:14087–14095. [DOI] [PubMed] [Google Scholar]
- Holdener BC, Haltiwanger RS. 2019. Protein O-fucosylation: Structure and function. Curr Opin Struct Biol. 56:78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhou J, Wu S, Shan Y, Teng S, Yu L. 2004. Cloning and tissue distribution of the human B3GALT7 gene, a member of the beta1,3-glycosyltransferase family. Glycoconj J. 21:267–273. [DOI] [PubMed] [Google Scholar]
- Ishida H, Togayachi A, Sakai T, Iwai T, Hiruma T, Sato T, Okubo R, Inaba N, Kudo T, Gotoh M et al. 2005. A novel beta1,3-N-acetylglucosaminyltransferase (beta3Gn-T8), which synthesizes poly-N-acetyllactosamine, is dramatically upregulated in colon cancer. FEBS Lett. 579:71–78. [DOI] [PubMed] [Google Scholar]
- Isshiki S, Togayachi A, Kudo T, Nishihara S, Watanabe M, Kubota T, Kitajima M, Shiraishi N, Sasaki K, Andoh T et al. 1999. Cloning, expression, and characterization of a novel UDP-galactose:β-N-acetylglucosamine β1,3-galactosyltransferase (β3Gal-T5) responsible for synthesis of type 1 chain in colorectal and pancreatic epithelia and tumor cells derived therefrom. J Biol Chem. 274:12499–12507. [DOI] [PubMed] [Google Scholar]
- Iwai T, Inaba N, Naundorf A, Zhang Y, Gotoh M, Iwasaki H, Kudo T, Togayachi A, Ishizuka Y, Nakanishi H et al. 2002. Molecular cloning and characterization of a novel UDP-GlcNAc:GalNAc-peptide β1,3-N-acetylglucosaminyltransferase (β3Gn-T6), an enzyme synthesizing the core 3 structure of O-glycans. J Biol Chem. 277:12802–12809. [DOI] [PubMed] [Google Scholar]
- Izumikawa T, Koike T, Shiozawa S, Sugahara K, Tamura J-i, Kitagawa H. 2008. Identification of chondroitin sulfate glucuronyltransferase as chondroitin synthase-3 involved in chondroitin polymerization: Chondroitin polymerization is achieved by multiple enzyme complexes consisting of chondroitin synthase family members. J Biol Chem. 283:11396–11406. [DOI] [PubMed] [Google Scholar]
- Izumikawa T, Uyama T, Okuura Y, Sugahara K, Kitagawa H. 2007. Involvement of chondroitin sulfate synthase-3 (chondroitin synthase-2) in chondroitin polymerization through its interaction with chondroitin synthase-1 or chondroitin-polymerizing factor. Biochem J. 403:545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chen Y-W, Clausen H, Cohen SM, Conti E. 2006. Structural insights into the notch-modifying glycosyltransferase fringe. Nat Struct Mol Biol. 13:945–946. [DOI] [PubMed] [Google Scholar]
- Ju T, Brewer K, D'Souza A, Cummings RD, Canfield WM. 2002a. Cloning and expression of human core 1 beta1,3-galactosyltransferase. J Biol Chem. 277:178–186. [DOI] [PubMed] [Google Scholar]
- Ju T, Cummings RD. 2005. Chaperone mutation in Tn syndrome. Nature. 437:1252–1252. [DOI] [PubMed] [Google Scholar]
- Ju T, Cummings RD, Canfield WM. 2002b. Purification, characterization, and subunit structure of rat core 1 Beta1,3-galactosyltransferase. J Biol Chem. 277:169–177. [DOI] [PubMed] [Google Scholar]
- Ju T, Zheng Q, Cummings RD. 2006. Identification of core 1 O-glycan T-synthase from Caenorhabditis elegans. Glycobiology. 16:947–958. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature. 447:714–719. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Huh N-h. 2002. A novel β1,3-N-acetylglucosaminyltransferase involved in invasion of cancer cells as assayed in vitro. Biochem Biophys Res Commun. 294:843–848. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I et al. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 451:783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordiš D. 2011. Extensive intron gain in the ancestor of placental mammals. Biol Direct. 6:59–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma K, Keusch JJ, Hegemann B, Luther KB, Klein D, Hess D, Haltiwanger RS, Hofsteenge J. 2006. Identification and characterization of abeta1,3-glucosyltransferase that synthesizes the Glc-beta1,3-Fuc disaccharide on thrombospondin type 1 repeats. J Biol Chem. 281:36742–36751. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. 2015. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 43:D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis A, Muffato M, Roest Crollius H. 2012. Genomicus: Five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 41:D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfait F, Kariminejad A, Van Damme T, Gauche C, Syx D, Merhi-Soussi F, Gulberti S, Symoens S, Vanhauwaert S, Willaert A et al. 2013. Defective initiation of glycosaminoglycan synthesis due to B3GALT6 mutations causes a pleiotropic Ehlers-Danlos-syndrome-like connective tissue disorder. Am J Hum Genet. 92:935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissard M, Dinter A, Berger EG, Hennet T. 2002. Functional assignment of motifs conserved in beta 1,3-glycosyltransferases. Eur J Biochem. 269:233–239. [DOI] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. 2005. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). Bioessays. 27:937–945. [DOI] [PubMed] [Google Scholar]
- Mikami T, Kitagawa H. 2013. Biosynthesis and function of chondroitin sulfate. Biochim Biophys Acta. 1830:4719–4733. [DOI] [PubMed] [Google Scholar]
- Miyazaki H, Fukumoto S, Okada M, Hasegawa T, Furukawa K, Furukawa K. 1997. Expression cloning of rat cDNA encoding UDP-galactose:GD2 β1,3-galactosyltransferase that determines the expression of GD1b/GM1/GA1. J Biol Chem. 272:24794–24799. [DOI] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS et al. 2000. Fringe is a glycosyltransferase that modifies Notch. Nature. 406:369–375. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Ramiah A, Stuart M, Steel J, Meng L, Forouhar F, Moniz HA, Gahlay G, Gao Z, Chapla D et al. 2018. Expression system for structural and functional studies of human glycosylation enzymes. Nat Chem Biol. 14:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Mizumoto S, Miyake N, Kogawa R, Iida A, Ito H, Kitoh H, Hirayama A, Mitsubuchi H, Miyazaki O et al. 2013. Mutations in B3GALT6, which encodes a glycosaminoglycan linker region enzyme, cause a spectrum of skeletal and connective tissue disorders. Am J Hum Genet. 92:927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane T, Angata K, Sato T, Kaji H, Narimatsu H. 2019. Identification of mammalian glycoproteins with type-I LacdiNAc structures syn thesized by the glycosyltransferase B3GALNT2. J Biol Chem. 294:7433–7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S. 2007. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17:1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu H. 2004. Construction of a human glycogene library and comprehensive functional analysis. Glycoconj J. 21:17–24. [DOI] [PubMed] [Google Scholar]
- Narimatsu H. 2006. Human glycogene cloning: Focus on beta 3-glycosyltransferase and beta 4-glycosyltransferase families. Curr Opin Struct Biol. 16:567–575. [DOI] [PubMed] [Google Scholar]
- Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, Ye Z, Chen Y-H, Schjoldager KT, Steentoft C et al. 2019. An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol Cell. 75(e395):394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelamegham S, Aoki-Kinoshita K, Bolton E, Frank M, Lisacek F, Lütteke T, O’Boyle N, Packer NH, Stanley P, Toukach P et al. 2019. Updates to the symbol nomenclature for glycans guidelines. Glycobiology. 29:620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsubayashi Y. 2015. Identification of three potent hydroxyproline O-galactosyltransferases in Arabidopsis. Plant J. 81:736–746. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Shionyu M, Sugiura N, Hatano S, Nagai N, Kubota Y, Nishiwaki K, Sato T, Gotoh M, Narimatsu H et al. 2010. Chondroitin sulfate synthase-2/chondroitin polymerizing factor has two variants with distinct function. J Biol Chem. 285:34155–34167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Heidelberg: Springer. [Google Scholar]
- Okajima T, Chen HH, Ito H, Kiso M, Tai T, Furukawa K, Urano T, Furukawa K. 2000. Molecular cloning and expression of mouse GD1alpha/GT1aalpha/GQ1balpha synthase (ST6GalNAc VI) gene. J Biol Chem. 275:6717–6723. [DOI] [PubMed] [Google Scholar]
- Paps J, Holland PWH. 2018. Reconstruction of the ancestral metazoan genome reveals an increase in genomic novelty. Nat Commun. 9:1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RY, Balaji PV. 2007. Length and composition analysis of the cytoplasmic, transmembrane and stem regions of human Golgi glycosyltransferases. Protein Pept Lett. 14:601–609. [DOI] [PubMed] [Google Scholar]
- Petit D, Teppa E, Mir AM, Vicogne D, Thisse C, Thisse B, Filloux C, Harduin-Lepers A. 2015. Integrative view of alpha2,3-sialyltransferases (ST3Gal) molecular and functional evolution in deuterostomes: Significance of lineage-specific losses. Mol Biol Evol. 32:906–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature. 453:1064–1071. [DOI] [PubMed] [Google Scholar]
- Qu Y, Egelund J, Gilson PR, Houghton F, Gleeson PA, Schultz CJ, Bacic A. 2008. Identification of a novel group of putative Arabidopsis thaliana beta-(1,3)-galactosyltransferases. Plant Mol Biol. 68:43–59. [DOI] [PubMed] [Google Scholar]
- Rampal R, Li ASY, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, Haltiwanger RS. 2005. Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. J Biol Chem. 280:42454–42463. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Kurata-Miura K, Ujita M, Angata K, Nakagawa S, Sekine S, Nishi T, Fukuda M. 1997. Expression cloning of cDNA encoding a human beta-1,3-N-acetylglucosaminyltransferase that is essential for poly-N-acetyllactosamine synthesis. Proc Natl Acad Sci U S A. 94:14294–14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sato M, Kiyohara K, Sogabe M, Shikanai T, Kikuchi N, Togayachi A, Ishida H, Ito H, Kameyama A et al. 2006. Molecular cloning and characterization of a novel human beta1,3-glucosyltransferase, which is localized at the endoplasmic reticulum and glucosylates O-linked fucosylglycan on thrombospondin type 1 repeat domain. Glycobiology. 16:1194–1206. [DOI] [PubMed] [Google Scholar]
- Seko A, Yamashita K. 2005. Characterization of a novel galactose β1,3-N-acetylglucosaminyltransferase (β3Gn-T8): The complex formation of β3Gn-T2 and β3Gn-T8 enhances enzymatic activity. Glycobiology. 15:943–951. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi N, Natsume A, Togayachi A, Endo T, Akashima T, Yamada Y, Imai N, Nakagawa S, Koizumi S, Sekine S et al. 2001. Identification and characterization of three novel β1,3-N-acetylglucosaminyltransferases structurally related to the β1,3-galactosyltransferase family. J Biol Chem. 276:3498–3507. [DOI] [PubMed] [Google Scholar]
- Strasser R, Bondili JS, Vavra U, Schoberer J, Svoboda B, Glössl J, Léonard R, Stadlmann J, Altmann F, Steinkellner H et al. 2007. A unique beta1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell. 19:2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Narciso JO, Zeng W, van de Meene A, Yasutomi M, Takemura S, Lampugnani ER, Doblin MS, Bacic A, Ishiguro S. 2017. KNS4/UPEX1: A type II arabinogalactan β-(1,3)-galactosyltransferase required for pollen exine development. Plant Physiol. 173:183–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taujale R, Venkat A, Huang LC, Zhou Z, Yeung W, Rasheed KM, Li S, Edison AS, Moremen KW, Kannan N. 2020. Deep evolutionary analysis reveals the design principles of fold A glycosyltransferases. Elife. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togayachi A, Akashima T, Ookubo R, Kudo T, Nishihara S, Iwasaki H, Natsume A, Mio H, Inokuchi J-i, Irimura T et al. 2001. Molecular cloning and characterization of UDP-GlcNAc:Lactosylceramide β1,3-N-Acetylglucosaminyltransferase (β3Gn-T5), an essential enzyme for the expression of HNK-1 and Lewis X epitopes on glycolipids. J Biol Chem. 276:22032–22040. [DOI] [PubMed] [Google Scholar]
- Togayachi A, Kozono Y, Kuno A, Ohkura T, Sato T, Hirabayashi J, Ikehara Y, Narimatsu H. 2010. Beta3GnT2 (B3GNT2), a major polylactosamine synthase: Analysis of B3GNT2-deficient mice. Methods Enzymol. 479:185–204. [DOI] [PubMed] [Google Scholar]
- Togayachi A, Sato T, Narimatsu H. 2006. Comprehensive enzymatic characterization of glycosyltransferases with a β3GT or β4GT motif. Methods Enzymol. 416:91–102. [DOI] [PubMed] [Google Scholar]
- Togayachi A, Sato T, Narimatsu H. 2008. β1,3-glycosyltransferase gene family and IGnT gene family. In: Taniguchi N, Suzuki A, Ito Y, Narimatsu H, Kawasaki T, Hase S, editors. Experimental glycoscience: glycobiology. Tokyo, Japan: Springer. p. 24–29. [Google Scholar]
- Vanbeselaere J, Jin C, Eckmair B, Wilson IBH, Paschinger K. 2020. Sulfated and sialylated N-glycans in the echinoderm Holothuria atra reflect its marine habitat and phylogeny. J Biol Chem. 295:3159–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Wang H, Gai J, Tian X, Zhang X, Lv Y, Jian Y. 2017. Evolution of protein N-glycosylation process in Golgi apparatus which shapes diversity of protein N-glycan structures in plants, animals and fungi. Sci Rep. 7:40301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Maeda Y, Liu YS, Takada Y, Ninomiya A, Hirata T, Fujita M, Murakami Y, Kinoshita T. 2020. Cross-talks of glycosylphosphatidylinositol biosynthesis with glycosphingolipid biosynthesis and ER-associated degradation. Nat Commun. 11:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 40:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa N, Vanbeselaere J, Chang LY, Yu SY, Ducrocq L, Harduin-Lepers A, Kurata J, Aoki-Kinoshita KF, Sato C, Khoo KH et al. 2018. Systems glycomics of adult zebrafish identifies organ-specific sialylation and glycosylation patterns. Nat Commun. 9:4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


