Abstract
Phages infecting Salmonella and Escherichia coli are promising agents for therapeutics and biological control of these foodborne pathogens, in particular those strains with resistance to several antibiotics. In an effort to assess the potential of the phage phiC120, a virulent phage isolated from horse feces in Mexico, we characterized its morphology, host range and complete genome. Herein, we showed that phiC120 possesses strong lytic activity against several multidrug-resistant E. coli O157: H7 and Salmonella strains, and its morphology indicated that is a member of Myoviridae family. The phiC120 genome is double-stranded DNA and consists of 186,570 bp in length with a 37.6% G + C content. A total of 281 putative open reading frames (ORFs) and two tRNAs were found, where 150 ORFs encoded hypothetical proteins with unknown function. Comparative analysis showed that phiC120 shared high similarity at nucleotide and protein levels with coliphages RB69 and phiE142. Detailed phiC120 analysis revealed that ORF 94 encodes a putative depolymerase, meanwhile genes encoding factors associated with lysogeny, toxins, and antibiotic resistance were absent; however, ORF 95 encodes a putative protein with potential allergenic and pro-inflammatory properties, making needed further studies to guarantee the safety of phiC120 for human use. The characterization of phiC120 expands our knowledge about the biology of coliphages and provides novel insights supporting its potential for the development of phage-based applications to control unwanted bacteria.
Keywords: Myoviridae, comparative analysis, coliphages, depolymerase
Introduction
Escherichia coli and Salmonella have emerged as zoonotic pathogens with an enormous epidemiological and economic impact as one of the leading causes of foodborne diseases worldwide. These bacteria are broadly distributed in all environments because they may dwell in animals, including wild and livestock, and then delivered to water bodies, soils, and plants through excreta, with the potential to be transmitted to humans (Chlebicz and Sliżewska 2018; Heredia and Garcia 2018). Of note, the overuse of antibiotics has also contributed with the emergence of multidrug-resistant (MDR) strains, which is underlying the urgent need of developing alternative strategies to control bacterial pathogens (Zaman et al. 2017). In this regard, the use of bacteriophages (phages), viruses that naturally infect and lyse bacterial cells, has sparked renewed interest as an alternative to antibiotics for the treatment and biological control of foodborne pathogens (Hagens and Loessner 2010).
Phages have numerous applications in the biotechnology field including the detection of specific bacteria, vehicles for DNA and protein vaccines delivery, phage display systems for proteins and antibodies, as bactericidal agent for food safety and therapeutics, and recently as source of novel genes since genome sequences are available (Haq et al. 2012). Promising results of phage applications have been observed for preventing or treating E. coli and Salmonella infections, as well as reducing contamination in vegetables and other foods (Atenbury 2009; Haq et al. 2012). Despite the potential as antibacterial agents, phage therapy is still limited due to the poor understanding of biology and genetics of phages, in addition to several aspects associated to safety, host resistance, specificity, and effective delivery that remain to be addressed (Roach and Debarbieux 2017).
The progress in the development of phage applications primarily relies on the selection of phages with desirable properties, including criteria regarding the effectivity in preventing or eliminating unwanted bacteria, as well as possessing features that ensure safety for human consumption (Hagens and Loessner 2010). Typically, most of phage-based applications employed strictly lytic phages to guarantee their amplification to kill bacteria, avoiding temperate phages that may integrate into the host genome with the potential to transmit virulence and antibiotic resistance genes by horizontal gene transfer promoting lysogenic conversion (Chen and Novick 2009). With the advances in DNA sequencing technologies and genetic engineering, novel approaches based on synthetic biology have expanded the repertoire of biotechnological applications, including the survey for harmful phage encoded genes, the design of synthetic phages with more predictable and specific prophylactic or therapeutic properties, the engineering of temperate phages to express genes with antimicrobial properties, or even though the development of phage-derived products such as enzybiotics to tackle pathogenic bacteria (Roach and Debarbieux 2017; Sao-José 2018), opening new alternatives to cope against the most menacing pathogens.
The phage phiC120 was isolated from horse feces collected from agricultural settings from Sinaloa, Mexico (Castro del Campo et al. 2011), but still remains to be characterized. Here, we report the morphology, the host range and the complete genome sequence of phage phiC120. A comparative genomic characterization showed phiC120 is highly similar to other phages infecting E. coli and Enterobacteria. Moreover, in silico surveys exposed the absence of genes associated to antibiotic resistance, virulence factors and lysogenic lifestyle, although one putative protein exhibited potential allergenic and inflammatory properties. The detailed characterization of phiC120 provides strong evidence supporting its potential for the development of phage-based applications to control pathogenic bacteria.
Materials and methods
Bacterial strains and bacteriophage
Bacterial strains were obtained from the culture collection maintained by the National Laboratory for Research in Food Safety (LANIIA) and were listed in Table 1 (Amézquita-López et al. 2014; Estrada-Acosta et al. 2014; Jiménez et al. 2014). Phage phiC120 was previously isolated from horse feces collected from agricultural settings in Sinaloa, Mexico (Castro del Campo et al. 2011). E. coli O157: H7 EC-48 (63-Fv18-1) was used for phage propagation and plaque counting. All strains were grown in trypticase soy broth (TSB; BD Bioxon, Mexico) at 37°C for 18–24 h.
Table 1.
Host range spectrum of the bacteriophage phiC120
| Bacterial | Strain | Bacterial lysis |
|---|---|---|
| E. coli O157: H7 | HC14-1 | + |
| E. coli O157: H7 | HE7-1 | + |
| E. coli O157: H7 | HC14-2 | + |
| E. coli O157: H7 | AC6-1 | + |
| E. coli O157: H7 | HE10-1 | − |
| E. coli O157: H7 | AR7-2 | − |
| E. coli O157: H7 | AR17-2 | − |
| E. coli O157: H7 | AC6-1 | − |
| E. coli O157: H7 | AR15-1 | − |
| E. coli O157: H7 | AR17-1 | − |
| E. coli O157: H7 | RM8744 | + |
| E. coli O157: H7 | RM8753 | + |
| E. coli O157: H7 | RM8754 | + |
| E. coli O157: H7 | RM8759 | + |
| E. coli O157: H7 | RM8767 | + |
| E. coli O157: H7 | RM8768 | + |
| E. coli O157: H7 | RM8769 | + |
| E. coli O157: H7 | RM8781 | + |
| E. coli O157: H7 | RM8920 | + |
| E. coli O157: H7 | RM8921 | + |
| E. coli O157: H7 | RM8922 | + |
| E. coli O157: H7 | RM8927 | + |
| E. coli O157: H7 | RM8928 | – |
| E. coli O157: H7 | RM9450 | + |
| E. coli O157: H7 | RM9451 | + |
| E. coli O157: H7 | RM9452 | + |
| E. coli O157: H7 | RM9453 | + |
| E. coli O157: H7 | RM9455 | + |
| E. coli O157: H7 | RM9457 | + |
| E. coli O157: H7 | RM9458 | + |
| E. coli O157: H7 | RM9459 | + |
| E. coli O157: H7 | RM9462 | − |
| E. coli O157: H7 | RM9463 | + |
| Salmonella Weltevreden | AC2-039 | − |
| Salmonella Oranienburg | AC2-041 | − |
| Salmonella Saintpaul | AC2-046 | − |
| Salmonella Minnesota | AC2-070 | + |
| Salmonella Anatum | AC2-079 | − |
| Salmonella Oranienburg | AC2-100 | − |
| Salmonella Montevideo | CM-02 | − |
| Salmonella Saintpaul | AC2-137 | − |
| Salmonella Oranienburg | AC2-142 | − |
| Salmonella Luciana | AC2-240 | + |
| Salmonella Anatum | CM-50 | − |
| Salmonella Minnesota | CM-51 | − |
| Salmonella Montevideo | CM-52 | − |
| Salmonella Agona | AC2-346 | − |
| Salmonella Muenster | CM-08 | − |
| Salmonella Muenster | AC2-366 | − |
| Salmonella Montevideo | AC2-370 | − |
| Salmonella Weltevreden | CM-08 | − |
| Salmonella Poona | CM-18 | − |
| Salmonella Oranienburg | CM-21 | − |
| Salmonella Saintpaul | CM-25 | − |
| Salmonella Give | CM-31 | − |
| Salmonella Saintpaul | AC2-098 | − |
| Salmonella Oranienburg | AC2-026 | + |
| Salmonella Pomona | AC2-248 | − |
| Salmonella Oranienburg | HC2-2 | − |
| Salmonella Oranienburg | HC2-1 | − |
| Salmonella Oranienburg | HC2-3 | − |
| Salmonella Give | HB4-2 | − |
| Salmonella Saintpaul | HE4-1 | − |
| Salmonella Give | HB4-1 | − |
| Salmonella Give | HB4-1 | − |
| Salmonella Weltevreden | HD4-2 | − |
| Salmonella Give | HB4-3 | − |
| Salmonella Saintpaul | HE4-3 | − |
| Salmonella Weltevreden | HD4-3 | − |
| Salmonella Agona | HD5-1 | + |
| Salmonella Give | HD6-3 | − |
| Salmonella Oranienburg | HD5-2 | − |
| Salmonella Oranienburg | HE6-1 | − |
| Salmonella Sandiego | HF6-3 | − |
| Salmonella Montevideo | S-188 | − |
| Salmonella Oranienburg | S-190 | + |
| Salmonella Oranienburg | S-228 | − |
Phage was assessed for host range by double agar overlay technique. (+) indicate positive sensitivity to phage lysis, and (−) indicate negative sensitivity to phage lysis.
Bacteriophage propagation
Phage phiC120 was propagated using the standard double agar layer method (Jamalludeen et al. 2007). Briefly, 1 ml of an overnight culture of E. coli O157: H7 EC-48 was mixed with 3.8 ml of TSB-agarose (0.4%) and then 100 μl of phage phiC120 were added. This mixture was poured onto a tryptic soy agar (TSA) plate, allowed to harden and incubated for 18–24 h at 37°C. Then, 6 ml of Suspension Medium (SM) buffer [50 mM Tris-HCl, pH 7.5; 8 mM MgSO4·7H2O, 100 mM NaCl, 0.002% (w/v) gelatin] were added to recover the top agar from plates and centrifuged for 15 min at 15,000 × g. The supernatant was filtered through 0.45 μm sterile syringe filters (Whatman, UK) and further concentrated at 40,000 × g for 2 h. The supernatant was discarded and the pellet was resuspended in 10 ml of SM buffer and then filtered through 0.22 μm sterile syringe filters of cellulose acetate membrane (GVS, USA) and maintained at 4°C.
Transmission electron microscopy
Electron microscopy was done according to standard method. Briefly, the purified bacteriophage (1010 PFU/ml) was placed on a formvar covered copper grid followed by negative staining with 2% uranyl acetate and dried in a vacuum evaporator (JEE400, JEOL Ltd. Tokyo, Japan). Samples were observed using a transmission electron microscope JEOL JEM-1011 (JEOL Ltd. Tokyo, Japan) at 80 kV accelerated voltage (López-Cuevas et al. 2011). Subsequently, 10 virions of phage were analyzed to calculate average phage dimensions.
Host range analysis
Host range analysis of phiC120 against all strains listed in Table 1 was determined by double agar overlay technique. Three milliliters of TSB-agarose (0.4%) was mixed with 1 ml of bacterial culture (108 CFU/ml) and 100 μl of concentrated phage suspensions (106 PFU/ml), poured onto TSA plates and incubated for 18–24 h at 37°C. The appearance of clear zones indicating bacterial lysis were recorded and compared with a negative control consisting of TSB plus bacteria without phage.
DNA extraction, library preparation, and genome sequencing
Five milliliters of phage suspension (1 × 1012 PFU/ml) were incubated with 10 μl of DNase I/RNase A (10 mg/ml) (Sigma-Aldrich, USA) at 37°C for 30 min, followed by the phage DNA isolation using the SDS-proteinase K protocol previously described (Sambrook and Russell 2001). The DNA library was prepared using an Illumina Nextera DNA library preparation kit, according to the manufacturer’s protocol. The quality of each library was verified using the Agilent Bioanalyzer. Genome sequencing of phage phiC120 was performed using Genome Analyzer HiSeq (Illumina Genome Analyzer, Inc.) (100-bp paired-end reads) technology.
De novo assembly and annotation
Contaminating and trailing sequences with low quality (Q < 25) were trimmed off with Trimmomatic. A total of 54,583,412 reads were generated and assembled de novo into contigs using Geneious R10 (© Biomatters Ltd, New Zealand) (Kearse et al. 2012), resulting in a single contig, with a genome coverage of >100-fold. Potential open reading frames (ORFs) longer than 100 bases were predicted using combined server from GeneMark (http://exon.gatech.edu/) and ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/), with the bacterial, archaeal, and plant plastid code as the translation table. Ribosomal binding sites were confirmed using RBSfinder program online for confirmation of ORF predictions (http://trna.ucsc.edu/tRNAscan-SE/) and the presence of tRNA was determined using tRNAscan-SE (Lowe and Chan 2016) and Aragorn (Laslett and Canback 2004). The deduced amino acid sequences of all the ORFs were analyzed using the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to detect conserved motifs among the proteins. Searching of antibiotic resistance genes were carried out using database ARDB (http://ardb.cbcb.umd.edu/) (Liu and Pop 2009) and integrative-comprehensive database of virulence factors (VFDB, http://www.mgc.ac.cn/VFs/) (Chen et al. 2012), and matched sequences with a minimum of 70% coverage and 30% identity were considered possible homologs. Furthermore, the sequence of virulence genes previously reported in enterobacteriophages (Shiga toxin, enterohemolysin, superoxide dismutase, guanine nucleotide exchange factor SopE, effector protein EspFU, and intimin) were downloaded from NCBI and searched against the phage phiC120 genome by BLASTN. To classify the lifestyle of phage the computational approach phage classification tool set (PHACTS) online prediction program was used (http://www.phantome.org/PHACTS/upload.php). To identify potentially allergenic proteins from phage phiC120 was performed a screening in silico using the FARRP-sliding 80mer FASTA (FAO/WHO >35%) criterion available at the Food Allergy Research and Resource Program (FARRP) allergen database (http://www.allergenonline.com). For proteins with hits, other analyses were performed using AllergenFP (Dimitrov et al. 2014), AllerTOP (Dimitrov et al. 2013), Proinflam (Gupta et al. 2016), SDAP (Ivanciuc 2003), AlgPred (Saha and Raghava 2006), and PREALw (http://lilab.life.sjtu.edu.cn:8080/prealw/). In addition, searches of integrases, transcriptional repressors and other genes which determine temperate development of phage was performed using the BLAST program provided by the NCBI, UniProt, and Pfam databases. Cumulative GC skew analysis was done with GenSkew-genomic nucleotide skew application (http://genskew.csb.univie.ac.at/) using a window size of 1000 bp and a step size of 100 bp.
Comparative genomics and phylogenetic analysis
For comparative genomics, phiC120 genome was compared with highly similar genome phages such as Escherichia phage phiE142 (GenBank no. KU255730.1), Enterobacteria phage RB69 (GenBank no. AY303349.1), E. coli O157 typing phage 3 (GenBank no. NC_041863.1), Escherichia phage vB_EcoM_WFK (GenBank no. MK373775.1) and Shigella phage Shf125875 (GenBank no. KM407600.1), using progressive Mauve alignment to determine homology among phage genomes. Additional analysis was done using CG view (Grant and Stothard 2008) and Easyfig (Sullivan et al. 2011). Comparative analysis of predicted ORFs was conducted with CoreGenes 3.5 using a BLAST-P threshold score of 75 (Turner et al. 2013). Phylogenetic analysis of terminase large (TerL) subunit and major capsid protein were performed using ClustalW for sequence alignment and Neighbor Joining analysis for tree reconstruction with a bootstrap of 1000 replicates, all computed in MEGA X (Kumar et al. 2018).
Data availability
The nucleotide sequence of the phage phiC120 genome was deposited in GenBank under accession number KY703222.1. Supplementary Figure S1 contains a cumulatie GC skew analysis. Supplementary Figure S2 shows a comparative genomic map of phiC120 with similar phages belonging to Mosigvirus using CGView. Supplementary Table S1 contains a detailed description of ORF of phiC120 and homologous proteins. Supplementary Table S2 Showed an analysis of potential allergens of phiC120 proteins using in silico approaches. Supplementary Material available at figshare: https://doi.org/10.25387/g3.13557314.
Results and discussion
Phage morphology and host range
The phage phiC120 was nonenveloped with an isometric and icosahedral capsid with a diameter of 62 ± 2 nm, exhibiting a noncontractile and very long tail (297 ± 5 nm in length by 23 ± 1 nm in width) connected to the head by a narrow neck (Figure 1), suggesting that phiC120 belongs to the Myoviridae family of Caudovirales order. The long tail of phiC120 was significantly larger than the T4 tail (113 nm) (Kulikov et al. 2014), but shorter than the jumbo phage G tail (455 nm), the biggest phage within Myoviridae (Drulis-Kawa et al. 2014). The host range of phiC120 was assayed using 33 E. coli O157: H7 and 44 Salmonella strains, most of them MDR that were previously isolated from domestic animals from rural farms, water samples from rivers and from irrigation channels (Amézquita-López et al. 2014; Estrada-Acosta et al. 2014; Jiménez et al. 2014). Our results revealed that phage phiC120 lysed the 75.7% (25 out of 33) of E. coli O157: H7 strains and 11.6% (5 out of 44) of Salmonella strains from different serotypes (lysed strains: Oranienburg, Agona, Minnesota, Luciana) (Table 1). Hence, phiC120 can be considered as polyvalent phage on its virtue to infect a wide range of host strains.
Figure 1.
Virion morphology of phage phiC120 observed by transmission electron microscopy. The bars represent length in nanometers.
General features of phiC120 genome
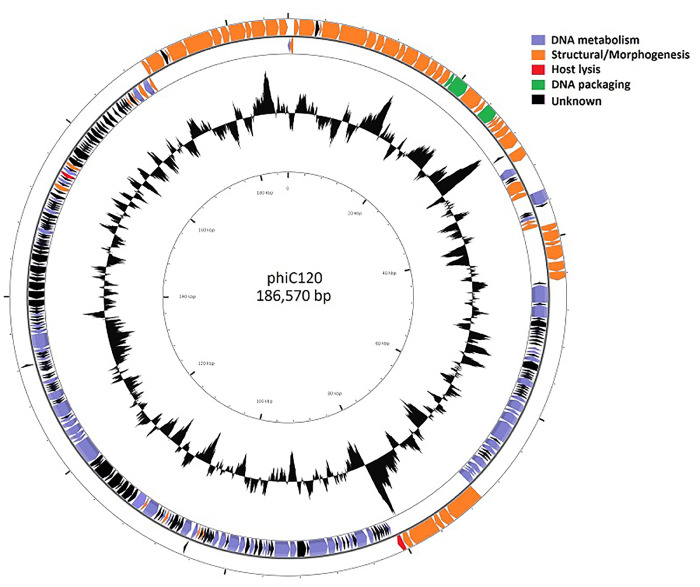
Genome sequencing results indicated that the phiC120 genome is double-stranded and consisted of 186,570 bp with 37.6% G + C content similar to other coliphages such as phiLLS and phiE142 (Amarillas et al. 2016, 2017), but lower than typical E. coli and S. enterica which is around 51% (Wang and Reeves 2000). It is hypothesized that lower G + C content may occur to reduce the metabolic burden to obtain G or C over A and T/U (Rocha and Danchin 2002), but it also reflect the recent evolutionary history with its host suggesting its origin from an ancestor infecting a host with a lower GC content or the acquisition of some elements from other phages (Dalmasso et al. 2016). A total of 281 putative ORFs and two tRNA genes were predicted, where the two tRNA genes were identified on positions 164,441–164,517 and 164,524–164,597 and specific for Arg and Met, respectively.
The circular genome map is depicted in Figure 2 and the gene annotation of predicted ORFs is listed in Supplementary Table S1. About 55 ORFs were encoded on the plus (+) strand while 226 ORFs were located on the minus (−) strand, both showing that 94.5% of total nucleotides were associated to encode putative proteins (coding density), reflecting the compactness of genome organization. Most of predicted ORFs (97.9% ORFs) started with ATG as start codon, although two ORFs (ORF 3 and ORF 269) started with TTG and four ORFs (ORF 69, ORF 152, ORF 188, and ORF 223) started with GTG, representing 0.7% and 1.4% of total ORFs, respectively. The phiC120 genome was organized in functional modules disposing mosaicism that contained gene clusters regarding their biological function: structural/morphogenesis (60 ORFs), DNA packaging (3 ORFs), DNA metabolism (65 ORFs) and host lysis (2 ORFs), suggesting that genome evolution occurred by combining modules from different biological sources (Hendrix et al. 1999). Despite all predicted ORFs had significant matches in NCBI database, 150 ORFs (53.4%) were found to encode hypothetical proteins with unknown function. In addition, 137 ORFs (48.8%) were identified to contain at least one domain, whereas no conserved domains were found in the remaining 144 ORFs (51.2%). Moreover, GC-skew analysis predicted the origin of replication in the position 44,827 nt and the replication terminus at 165,541 nt, indicated by the maximum and minimum cumulative GC-skew (Supplementary Figure S1).
Figure 2.
Graphic representation of genome organization of phage phiC120. Putative ORFs are indicated as arrows, where the orientation indicates the direction of transcription. GC content is indicated in black.
Comparative genomics and phylogenetic analysis
At DNA level, the phiC120 genome showed the highest identity to coliphages RB69 (98.08% identity, 96% coverage) and phiE142 (98.56% identity, 65% coverage) that belong to genus Mosigvirus subfamily Tevenvirinae, by using BLAST-N, findings that were also observed by genome comparison with CGView (Supplementary Figure S2). Despite the high similarity to other phages, progressive Mauve alignment showed phiC120 displayed four blocks with no collinearity (Figure 3A); consistent results were observed using Easyfig CDS, where several genomic regions of phiC120 shared high homology with phages belonging to Mosigvirus, but showed several genome rearrangements (Figure 3B). Moreover, comparison of putative ORFs using CoreGenes revealed that phiC120 shared 64.6% (180) of its proteins with phiE142, 90.39% (254) with RB69, and 89.68% (252) with E. coli O157 typing phage 3, supporting the high relatedness with phage phiC120.
Figure 3.
Comparative genomic analysis of phiC120 genome and other similar phages. (A) Progressive Mauve alignment was used to determine blocks of homolog genomic regions (showed in different colors) and identify genomic rearrangements. (B) Comparison of the genome structure of phiC120 using Easyfig. Lines between genome maps indicate the level of identity in genes sharing orientation (blue/turquoise) or with inverted orientation (red/orange). GenBank accession numbers: phiC120, KY703222.1; RB69, AY303349.1; phiE142, KU255730.1; Escherichia coli O157 typing phage 3 (O157tp3), NC_041863.1; Escherichia phage vB_EcoM_WFK,; Shigella phage Shf125875, MK373775.1.
To investigate the phylogeny of phiC120, we used the gene encoding the major capsid protein (g23) of representative members of some genera within Tevenvirinae subfamily, which confirmed that phiC120 belongs to Mosigvirus genus (Figure 4). Remarkably, phiC120 also contained a predicted large terminase subunit (TerL) almost identical to TerL of RB69 phage (99.84% identity), which is closely related to T4. In an attempt to predict the possible DNA packaging mechanism, a phylogenetic analysis was done with representative members of the distinct DNA packaging mechanisms, revealing that phiC120 is likely a pac-type phage that belongs to T4-like headful cluster (Figure 5). Some phages employing pac-site headful packaging mechanism are known to display generalized transduction in which potentially host DNA may be packaged within the capsid (Penadés et al. 2015). Notwithstanding, T4 uses a pac-type mechanism that involves the degradation of the host DNA during infection by nucleases ensuring the packaging of only DNA phage by headful packaging mechanism (Miller et al. 2003). In this regard, phiC120 genome harbored several ORFs encoding T4 homolog endonucleases II and IV (ORFs: 77, 111, 171, and 174, described below) suggesting that the host DNA degradation process occurs, making phiC120 suitable and promising for further applications.
Figure 4.
Phylogenetic tree using the major capsid protein (g23) genes of phiC120 and representative members of different genera of Tevenvirinae subfamily to investigate the possible genus which phiC120 belongs.
Figure 5.
Phylogenetic tree of TerL subunit protein of phiC120 and representative phages with known packaging mechanisms. Brackets indicate clusters of phages, that follow a similar packaging strategy.
Features of functional modules
Regarding the structural and morphogenesis proteins, most of head, neck and tail structural proteins of phiC120 were organized together in modules, in which DNA packaging, hypothetical and DNA metabolism proteins were intercalated. ORF 23 encoded the major capsid protein exhibiting 99.81% identity to Escherichia phage vB_EcoM_JS09. Besides, ORF 4, ORF 40 and ORF 270 encoding baseplate wedge and hub subunits were found to harbor lysozyme domains (Lyz_like superfamily and GPW_gp25 domains), likely involved in the hydrolysis of bacterial cell wall for further viral DNA ejection (Fernandes and São-José 2018). Likewise, ORF 13 and ORF 279 encoding short tail fibers harbored a S_tail_recep_bd domain (Pfam14928), maybe involved in binding to bacterial LPS (Fernandes and São-José 2018). Strikingly, ORF 94 predicted to encode a long tail fiber distal subunit contained a peptidase_S74 domain (Pfam13884), which has been reported in phage depolymerases that hydrolyze capsular polysaccharides containing sialic acid (Pires et al. 2016). This putative depolymerase shared 84.59% identity to its phage vB_EcoM_IME340 homolog, suggesting the identification of a novel depolymerase. Depolymerases are commonly found in tail spikes or fiber proteins that are capable of degrading capsular polysaccharides, exopolysaccharides and lipopolysaccharides from host bacteria (Knecht et al. 2020). Recombinant depolymerases have shown to disrupt biofilms and enhance antimicrobial activity of antibiotics (Wu et al. 2019), highlighting the potential of the phiC120 depolymerase toward further biotechnological applications.
On the other hand, phage phiC120 encoded several enzymes involved in DNA replication, repair, recombination and transcription, including RNA ligase (ORF 30), DNA helicases (ORF 128, 152), DNA ligase (ORF 51), DNA primase (ORF 142), DNA polymerases (ORF 90, 164), endonuclease V N-glycosylase UV repair enzyme (ORF 236), recombination endonuclease IV (ORF 111), endonuclease VII (ORF 194, resolvase), UvsW helicase (ORF 35), RNA polymerase (ORF 131), and DNA topoisomerase subunits (ORF 105, 118). Additional proteins assisting these functions included UvsX RecA-like recombination protein (ORF 154), ssDNA (ORF 39, 86), and dsDNA (ORF 89) binding proteins, helicase loading protein (ORF 87), clamp loader subunits (ORF 166, 167), and sliding clamp (ORF 168). Interestingly, phiC120 also encoded several transcriptional regulators such as inhibitor of host transcription (ORF 75), promoter transcription accessory protein (ORF 88), putative transcription factors (ORF 125, 165), RNA polymerase binding protein (ORF 169), RNA polymerase sigma factor (ORF 179) and anti-sigma 70 protein (ORF 97), maybe to promote phage transcription. These results clearly indicate that phiC120 encodes the proteins required for DNA replication and associated functions.
Others predicted proteins appear to be involved in nucleotide modification (ORF 49, 69) and degradation by several endonucleases (ORFs: 127, 171, 174, 226, 231), including a homing endonuclease (ORF 117) containing a NUMO_D3 DNA-binding domain. Homing endonucleases are mobile genetic elements that encode a protein with endonuclease activity that mediates their own horizontal transfer (Sitbon and Pietrokovski 2003). Moreover, phiC120 encoded a nucleoid disruption protein (ORF 108) that inhibits host replication through disorganizing the nucleoid architecture (Bouet et al. 1998). Moreover, to preserve the integrity of viral genome, the DNA end protector proteins (ORF 1, 267) may bind to DNA to protect it from host recBCD exonuclease mediated degradation, similar to phage T4 (Petrov et al. 2010).
Phage phiC120 encoded several enzymes for nucleotide metabolism. The ribonucleoside-diphosphate reductase (ORF 78, 79), thymidylate synthase (ORF 81), deoxycytidylate deaminase (ORF 63), deoxynucleoside monophosphate kinase (ORF 265), dihydrofolate reductase (ORF 82), anaerobic NTP reductase subunits (ORF192, 193), thymidine kinase (ORF 219), and dCTP pyrophosphatase (ORF 139) contributed to provide the precursors for DNA synthesis; moreover, glutaredoxin (ORF 188) and thioredoxin (ORF 200) function as hydrogen donor for redox reactions (Gvakharia et al. 1996). Besides, a putative nudix hydrolase (ORF 239) may be involved in NAD+ salvage pathway to support basic processes like DNA synthesis (Lee et al. 2017), although antimutagenic properties for preventing incorporation of oxidized deoxynucleoside triphosphate has also been suggested (Carreras-Puigvert et al. 2017).
The DNA packaging mechanism of phiC120 seems to involve three ORFs encoding the small (ORF 18) and large (ORF19) terminase subunits that translocate the phage dsDNA into an empty capsid, and the portal vertex protein (ORF 22), which multimerizes forming a channel through which DNA is translocated when associates with the terminase complex (Dedeo et al. 2019). In addition, phiC120 genome contains four ORFs encoding homologous endonucleases II and IV maybe involved in host DNA degradation during T4 infection, in which ORF 77 and ORF 111 were highly similar to endonucleases DenA (77.44% identity) and DenB (74.5% identity), respectively, meanwhile ORF 171 and ORF 174 were similar to gp46 (82.83% identity) and gp47 (75.81% identity) products from T4 phage.
For host lysis, phiC120 encoded one putative endolysin, ORF 238 that showed high homology to lysozyme murein hydrolase likely implicated in peptidoglycan degradation of bacterial cell wall during the late stages of phage replication to accomplish host lysis (Young 2014). Moreover, ORF 96 encoded an holin, indicating that the mechanism of host lysis of phage phiC120 may be dependent of hydrolytic activity and holin as occurs in most of phages (Young 2014).
Finally, about 150 hypothetical proteins were identified in phiC120 genome and from these, 116 ORFscontains no identifiable domains. The remaining ORFs contained mostly one domain, but only a few functions were suggested. According to Pfam databases, the ORF 36 harbored a helicase UvsW domain supporting a possible role in recombination; the ORF 158 contained a thymidylate synthase domain indicating a role in nucleotide metabolism; the ORF 210 contained a Nuc-transf superfamily domain, found in nucleotidyl transferases that catalyze the transfer of nucleotides from nucleoside di- or tri-phosphates into dimer or polymer forms; the ORF 230 harbored a RNR_PFL (ribonucleotide reductase and pyruvate formate lyase) superfamily domain, in which RNRs are known to covert nucleotides into deoxynucleotides; the ORFs containing NAD and ADP-ribose (NADAR) and Macro domains (ORF 54 and 223) probably participate as ADP-ribosyl transferases (Aravind et al. 2015). Others, such as ORF 124 and 126, contained one motB domain, generally found in transcriptional factors from Myoviridae. A peptidase activity was also suggested in ORF 159 by the presence of peptidase_U32 domain. Other ORFs contained 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthetase domain related to the biosynthesis of lipopolysaccharide and aromatic amino acid biosynthesis (ORF 64), as well as synthesis and modification of sugars (ORF 161, 163). Lastly, ORF 217 showed no domains and its annotation was not significant suggesting that encoded a unique protein for phage phiC120.
In silico analysis of lifecycle, virulence, and allergenicity
Regarding phage lifecycle, the prediction of PHACTS suggested that phage phiC120 is lytic, which is consistent with the absence of lysogenic genes, and agrees with our observations of clear and nonturbid plaques indicative of strictly lytic phages. Moreover, genes associated with virulence, toxins and resistance to antibiotics were also absent from phiC120 genome, making it suitable for further antibacterial applications (Hagens and Loessner 2010). In addition, from 281 putative ORFs, only ORF 95 predicted as a long tail fiber assembly chaperone showed potential allergenic and proinflammatory properties (Supplementary Table S2), indicating that further toxicological testing in animals and in vivo trials are needed to guarantee the safety for human use, although there is no limitation to prevent pathogen colonization or treat infections for veterinarian use.
Conclusion
In summary, our data revealed that the bacteriophage phiC120 belonging to Myoviridae family and Mosigvirus genus exhibited a broad lytic activity against several pathogenic E. coli O157: H7 and Salmonella strains. Despite phiC120 shared high similarity with other phages belonging to Mosigvirus genus, it is clear that phiC120 displayed a particular genomic structure when compared with those phages. Moreover, a pac-type headful DNA packaging mechanism and the presence of putative endonucleases with homology to T4 host-DNA degrading enzymes support that generalized transduction may not occur during DNA packaging in phiC120. Moreover, a novel putative depolymerase containing the peptidase_S74 domain is encoded by ORF 94. Likewise, functional predictions based on phiC120 genome revealed the absence of undesirable genes, such as those encoding toxins, antibiotic resistance, pathogenicity or elements favoring phage integration into host genome. Nonetheless, only one putative protein exhibited potential allergenic and inflammatory properties, indicating that further toxicological approaches may be done to ensure phage safety for human consumption, although it is not limiting for veterinarian use. Our study provides new insights into the biology of the phage phiC120 as well as strong evidence that poses phiC120 as a promising candidate for developing future technologies directed toward therapeutics and biological control against pathogenic E. coli and Salmonella strains.
Acknowledgments
The authors thank the National Food Safety Research Laboratory (LANIIA) at the Research Center for Food and Development (CIAD, Mexico) for providing laboratory facilities during the research. C.V. is supported by Cátedras CONACYT research project 784: “Functional Genomics of organisms for Food and Agriculture to Mexico.” The authors would like to acknowledge the technical assistance of QFB Lucía Margarita Rubí Rangel and QFB Jesús Héctor Carrillo Yáñez.
Funding
This research was supported by Fundación Produce Sinaloa, Mexico, Grant number: 102/201112 to J.L.F.
Conflicts of interest: None declared.
Literature cited
- Amarillas L, Chaidez C, Gonzalez-Robles A, León-Félix J.. 2016. Complete genome sequence of new bacteriophage phiE142, which causes simultaneously lysis of multidrug-resistant Escherichia coli O157:H7 and Salmonella enterica. Stand Genomic Sci. 11:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarillas L, Rubí-Rangel L, Chaidez C, González-Robles A, Lightbourn-Rojas L, et al. 2017. Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front Microbiol. 8:1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amézquita-López BA, Quiñones B, Lee BG, Chaidez C.. 2014. Virulence profiling of Shiga toxin-producing Escherichia coli recovered from domestic farm animals in Northwestern Mexico. Front Cell Infect Microbiol. 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Zhang D, de Souza RF, Anand S, Iyer LM.. 2015. The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr Top Microbiol Immunol. 384:3–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atenbury RJ. 2009. Bacteriophage biocontrol in animals and meat products. Microb Biotechnol. 2:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet JY, Krisch HM, Louarn JM.. 1998. Ndd, the bacteriophage T4 protein that disrupts the Escherichia coli nucleoid, has a DNA binding activity. J Bacteriol. 180:5227–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras-Puigvert J, Zitnik M, Jemth A-S, Carter M, Unterlass JE, et al. 2017. A comprehensive structural, biochemical and biological profiling of the human NUDIX hydrolase family. Nat Commun. 8:1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro del Campo N, Amarillas-Bueno L, García-Camarena MG, Chaidez-Quiroz C, León-Félix JJ, et al. 2011. Presencia de Salmonella y Escherichia coli O157:H7 en la zona centro del estado de Sinaloa y su control biológico mediante el uso de bacteriófagos. In: Ma. Refugio Torres Vitela and Olga Deli Vázquez Paulino. XIII Congreso Internacional de Inocuidad de Alimentos. Jalisco, Méxi: Prometeo Editores S.A de C.V. p. 165–168. [Google Scholar]
- Chen J, Novick RP.. 2009. Phage-mediated intergeneric transfer of toxin genes. Science. 323:139–141. [DOI] [PubMed] [Google Scholar]
- Chen L, Xiong Z, Sun L, Yang J, Jin Q.. 2012. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40:D641–D645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebicz A, Śliżewska K.. 2018. Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as ZOONOTIC FOODBORNE DISEASES: a review. Int J Environ Res Public Health. 15:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmasso M, Strain R, Neve H, Franz CMAP, Cousin FJ, et al. 2016. Three new Escherichia coli phages from the human gut show promising potential for phage therapy. PLoS One. 11:e0156773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeo CL, Cingolani G, Teschke CM.. 2019. Portal protein: the orchestrator of capsid assembly for the dsDNA tailed bacteriophages and herpesvirus. Annu Rev Virol. 6:141–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I, Flower DR, Doytchinova I.. 2013. AllerTOP-a server for in silico prediction of allergens. BMC Bioinformatics. 14:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I, Naneva L, Doytchinova I, Bangov I.. 2014. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 30:846–851. [DOI] [PubMed] [Google Scholar]
- Drulis-Kawa Z, Olszak T, Danis K, Majkowska-Skrobek G, Ackermann HW.. 2014. A giant Pseudomonas phage from Poland. Arch Virol. 159:567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Acosta M, Jiménez M, Chaidez C, León-Félix J, Castro-del Campo N.. 2014. Irrigation water quality and the benefits of implementing good agricultural practices during tomato (Lycopersicum esculentum) production. Environ Monit Assess. 186:4323–4330. [DOI] [PubMed] [Google Scholar]
- Fernandes S, São-José C.. 2018. Enzymes and mechanisms employed by tailed bacteriophages to breach the bacterial cell barriers. Viruses. 10:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JR, Stothard P.. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36:W181–W184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Madhu MK, Sharma AK, Sharma VK.. 2016. ProInflam: a webserver for the prediction of proinflammatory antigenicity of peptides and proteins. J Transl Med. 14:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gvakharia BO, Hanson E, Koonin EK, Mathews CK.. 1996. Identification of a second functional glutaredoxin encoded by the bacteriophage T4 genome. J Biol Chem. 271:15307–15310. [DOI] [PubMed] [Google Scholar]
- Hagens S, Loessner MJ.. 2010. Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. CPB. 11:58–68. [DOI] [PubMed] [Google Scholar]
- Haq IU, Chaudhry WN, Akhtar MN, Andleeb S, Qadri I.. 2012. Bacteriophages and their implications on future biotechnology: a review. Virol J. 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF.. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 96:2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia N, Garcia S.. 2018. Animals as sources of food-borne pathogens: a review. Anim Nutr. 4:250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanciuc O. 2003. SDAP: database and computational tools for allergenic proteins. Nucleic Acids Res. 31:359–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamalludeen N, Johnson RP, Friendship R, Kropinski AM, Lingohr EJ, et al. 2007. Isolation and characterization of nine bacteriophages that lyse O149 enterotoxigenic Escherichia coli. Vet Microbiol. 124:47–57. [DOI] [PubMed] [Google Scholar]
- Jiménez M, Martinez-Urtaza J, Rodriguez-Alvarez MJ, Leon-Felix J, Chaidez C.. 2014. Prevalence and genetic diversity of Salmonella spp. in a river in a tropical environment in Mexico. J Water Health 12:74–884. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht LE, Veljkovic M, Fieseler L.. 2020. Diversity and function of phage encoded depolymerases. Front Microbiol. 10:2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikov E, Golomidova A, Letarova M, Kostryukova E, Zelenin A, et al. 2014. Genomic sequencing and biological characteristics of a novel Escherichia coli bacteriophage 9g, a putative representative of a new Siphoviridae genus. Viruses. 6:5077–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetic analysis across computing platforms. Mol Biol Ecol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B.. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Li Z, Miller ES.. 2017. Vibrio phage KVP40 encodes a functional NAD+ salvage pathway. J Bacteriol. 199:e00855-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Pop M.. 2009. ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res. 37:D443–D447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Cuevas O, Castro-Del Campo N, León-Félix J, González-Robles A, Chaidez C.. 2011. Characterization of bacteriophages with a lytic effect on various Salmonella serotypes and Escherichia coli O157:H7. Can J Microbiol. 57:1042–1051. [DOI] [PubMed] [Google Scholar]
- Lowe T M, , Chan P P. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57. 10.1093/nar/gkw413 27174935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, et al. 2003. Bacteriophage T4 genome. MMBR. 67:86–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penadés JR, Chen J, Quiles-Puchalt N, Carpena N, Novick RP.. 2015. Bacteriophage-mediated spread of bacterial virulence genes. Curr Opin Microbiol. 23:171–178. [DOI] [PubMed] [Google Scholar]
- Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD.. 2010. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J. 7:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires DP, Oliveira H, Melo LDR, Sillankorva S, Azeredo J.. 2016. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol. 100:2141–2151. [DOI] [PubMed] [Google Scholar]
- Roach DR, Debarbieux L.. 2017. Phage therapy: awakening a sleeping giant. Emerg Top Life Sci. 1:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, Danchin A.. 2002. Base composition bias might result from competition for metabolic resources. Trends Genet. 18:291–294. [DOI] [PubMed] [Google Scholar]
- Saha S, Raghava GPS.. 2006. AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 34:W202–W209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D.. 2001. Molecular Cloning: A Laboratory Manual, 3rd ed.New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sao-José C. 2018. Engineering of phage-derived lytic enzymes: improving their potential as antimicrobials. Antibiotics (Basel). 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon E, Pietrokovski S.. 2003. New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem Sci. 28:473–477. [DOI] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA.. 2011. Easyfig: a genome comparison visualizer. Bioinformatics (Oxford, England). 27:1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D, Reynolds D, Seto D, Mahadevan P.. 2013. CoreGenes3.5: a webserver for the determination of core genes from sets of viral and small bacterial genomes. BMC Res Notes. 6:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Reeves PR.. 2000. The Escherichia coli O111 and Salmonella enterica O35 gene clusters: gene clusters encoding the same colitose-containing O antigen are highly conserved. J Bacteriol. 182:5256–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang R, Xu M, Liu Y, Zhu X, et al. 2019. A novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front Microbiol. 10:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. 2014. Phage lysis: three steps, three choices, one outcome. J Microbiol. 52:243–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman SB, Hussain MA, Nye R, Mehta V, Mamun KT, et al. 2017. A review on antibiotic resistance: alarm bells are ringing. Cureus. 9:e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequence of the phage phiC120 genome was deposited in GenBank under accession number KY703222.1. Supplementary Figure S1 contains a cumulatie GC skew analysis. Supplementary Figure S2 shows a comparative genomic map of phiC120 with similar phages belonging to Mosigvirus using CGView. Supplementary Table S1 contains a detailed description of ORF of phiC120 and homologous proteins. Supplementary Table S2 Showed an analysis of potential allergens of phiC120 proteins using in silico approaches. Supplementary Material available at figshare: https://doi.org/10.25387/g3.13557314.