Abstract
Mucin-type O-glycosylation is one of the most common posttranslational modifications of proteins. The abnormal expression of various polypeptide GalNAc-transferases (GalNAc-Ts) which initiate and define sites of O-glycosylation are linked to many cancers and other diseases. Current O-glycosyation prediction programs utilize O-glycoproteomics data obtained without regard to the transferase isoform (s) responsible for the glycosylation. With 20 different GalNAc-Ts in humans, having an ability to predict and interpret O-glycosylation sites in terms of specific GalNAc-T isoforms is invaluable.
To fill this gap, ISOGlyP (Isoform-Specific O-Glycosylation Prediction) has been developed. Using position-specific enhancement values generated based on GalNAc-T isoform-specific amino acid preferences, ISOGlyP predicts the propensity that a site would be glycosylated by a specific transferase. ISOGlyP gave an overall prediction accuracy of 70% against in vivo data, which is comparable to that of the NetOGlyc4.0 predictor. Additionally, ISOGlyP can identify the known effects of long- and short-range prior glycosylation and can generate potential peptide sequences selectively glycosylated by specific isoforms.
ISOGlyP is freely available for use at ISOGlyP.utep.edu. The code is also available on GitHub (https://github.com/jonmohl/ISOGlyP).
Keywords: GalNAc-T, GALNT, mucin-type O-glycosylation, O-glycosylation prediction, PTM prediction
Introduction
Mucin-type protein O-glycosylation (henceforth O-glycosylation) is one of the most ubiquitous posttranslational modifications (PTMs) of proteins. O-glycosylation is initiated by the transfer of the sugar N-acetylgalactosamine (GalNAc) onto threonine (Thr) and serine (Ser) residues by the UDP-N-acetylgalactosaminyltransferase (GalNAc-Ts) family of enzymes (Bennett et al. 2012). Protein O-glycosylation is critical to development and serves many important biological roles (Joshi et al. 2018). Aberrant O-glycosylation is linked to many disease states, including many cancers which are commonly associated with altered expression and/or mutation of several of the 20 human GalNAc-T isoforms (Hussain et al. 2016). Major difficulties in understanding O-glycosylation’s role in disease include determining which sites may be O-glycosylated by these transferases and identifying which isoform(s) glycosylate the sites. To assist in these efforts several bioinformatics approaches, such as NetOGlyc4.0 (Steentoft et al. 2013), have been developed to predict likely glycosites. However, these approaches are incapable of predicting isoform-specific glycosites or which GalNAc-T isoform(s) are responsible for their glycosylation.
Using random peptide substrates, Gerken et al. (2006, 2008, 2011), showed that the GalNAc-T isoforms possess unique and overlapping peptide substrate specificities, quantified as positional residue specific enhancement values (EVs) spanning +/− 5 residues from the Ser or Thr glycosylation site. Individual EVs are taken as the propensity of a particular amino acid residue being favored (EV > 1) or disfavored (EV < 1) in a peptide sequence glycosylated by a given GalNAc-T. Here, we describe the web-based tool ISOGlyP (Isoform-Specific O-Glycosylation Prediction) that utilizes these EVs to generate predictive enhancement value products (EVP) values for 11 of the 20 human GalNAc-T isoforms (Bennett et al. 2012). Utilizing these multiple EVP predictions, we have validated ISOGlyP’s predictive powers with an in vivo O-glycoproteomics dataset.
ISOGlyP program workflow and implementation
ISOGlyP core program based on EVPs
Written in Python, ISOGlyP is implemented as a web server using the Web.py framework (http://webpy.org/). As seen in Figure 1, the home page contains text areas for inputting protein/peptide sequences in FASTA format and check boxes for selecting parameters allow users to easily customize their analyses. ISOGlyP displays outputs directly onto the webpage with an option to download a CSV file.
Fig. 1.
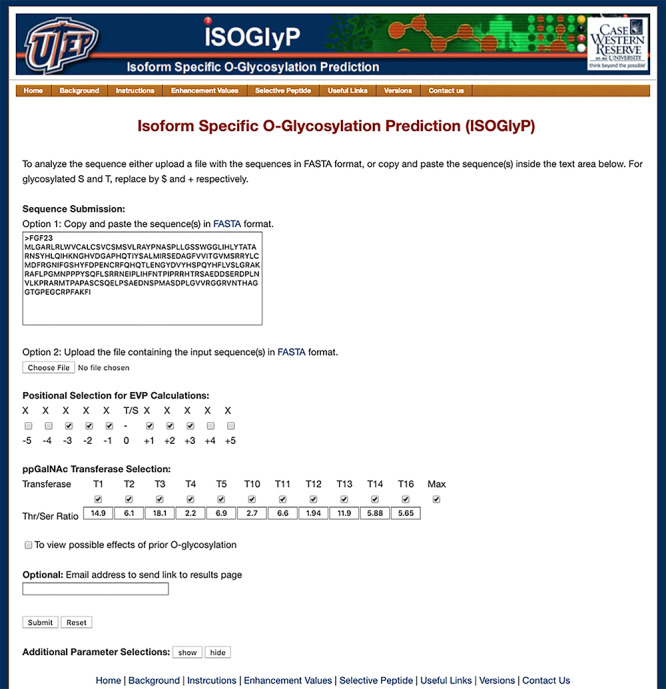
Screenshot of the ISOGlyP website home page. Highlighting options within the main prediction page, which include the choice of entering sequences directly into the text box or uploading a FASTA sequence file, the selection of the flanking residue positions and transferases to use in the EVP calculation, and the possibility to modify the transferase specific Thr/Ser rate ratios. In addition, the user can check a box to examine potential effects of prior O-glycosylation and add an email address to which a link to the results page will be sent. The “Additional Parameter Selections” button permits the user to choose the ISOGlyP EV table version (for repeating previous calculations after EV updates) and to set the EVs of Cys and Thr to the EVs of Ser instead of the default of 1.0.
ISOGlyP generates the EVP by multiplying together the positional EVs of up to +/−5 amino acid residues from the glycosylation site. Currently, EV tables are compiled for 11 GalNAc-Ts (T1–5, T10–14 and T16). Each table contains 10 positional EVs for each amino acid except Cys, Trp and Thr (de las Rivas et al. 2018; Festari et al. 2017; Gerken et al. 2006, 2008, 2011, 2013; Perrine et al. 2009). Due to the preferential glycosylation of Thr over Ser, we have included a correction for the Thr/Ser rate ratios for the GalNAc-T isoforms (Paul Daniel et al. 2020). An EV table (Supplementary Figure S1) and how the final EVPs are calculated are given in Supplemental Section S1. ISOGlyP’s output consists of a table of columns including the peptide sequence and EVP values for the selected GalNAc-T isoforms. We take the EVP values to indicate the likelihood or propensity for a site to be glycosylated. Typically, an EVP greater than 1.0 suggests a potential for glycosylation, with the larger the value the more likely the site is expected to be glycosylated (see Supplemental Section S2). Values less than 1.0 would signify a site unlikely to be glycosylated, again the lower the value the less likely.
Additional features
The activity of a GalNAc-T against a glycopeptide substrate can be significantly increased (or decreased) by neighboring (1–3 residues) and remote (6–17 residues) prior GalNAc glycosylation (Gerken et al. 2013, Revoredo et al. 2016). To inform users of potential prior glycosylation effects, ISOGlyP has an option to add flanking superscripts “+”, “-” or “0” to the EVP to indicate a probable increase, decrease or no change in glycosylation, respectively (see Supplemental Section S3). This functionality is helpful for understanding the glycosylation of heavily O-glycosylated domains and may assist in interpreting experimental data that may be unusually high or low, relative to a prediction, due to prior neighboring/remote glycosylation.
The ability to design peptide sequences preferentially glycosylated by specific GalNAc-T isoform (s) at the exclusion of other GalNAc-Ts has been elusive. This is because the GalNAc-Ts as a class possess both unique and overlapping specificities. Therefore, a Selective Peptide function has been developed utilizing ISOGlyP’s core code to 1) identify highly selective GalNAc-T specific glycosites, and 2) generate peptide sequences that meet such criteria. The latter is performed by ISOGlyP screening a series of randomly generated peptides for selectivity (see Supplemental Section S4).
In vivo prediction accuracy
To validate ISOGlyP’s in vivo predictions the human platelet O-glycoproteome reported by King and coworkers (King et al. 2017) was utilized, as this tissue expresses transferases for which ISOGlyP has EV data tables (GalNAc-T1, T2, T5, T10, T13 and T16). This platelet glycoproteome consisted of 247 glycoproteins with 384 glycosites. Testing sets for various accuracy metrics were built by randomly choosing two-thirds of the glycosites with equal number of unglycosylated sites. ISOGlyP gave an overall average prediction accuracy of 70% compared to NetOGlyc4.0’s accuracy of 75%. Since NetOGlyc4.0 was trained on existing O-proteomics data, the accuracy comparison was repeated using only those unique glycosites within the King et al. (2017) data that had not been previously used in the training of NetOGlyc4.0. This comparison, using the reduced dataset with 125 positive and 10,862 negative sites, gave an overall accuracy of 69% for ISOGlyP and 68% for NetOGlyc4.0. To look at the general overlap of the predictions of ISOGlyP and NetOGlyc4.0, a Venn diagram showing the numbers of predicted positive and negative sites in this unbiased dataset is shown in Figure 2. As seen, NetOGlyc 4.0 correctly predicted 9 (7.2%) more of the glycosites while ISOGlyP correctly predicted 842 (6.9%) more of the negative sites. Further details of the accuracy assessments of the two programs on both the full and unbiased dataset are given in Supplemental Section S5.
Fig. 2.
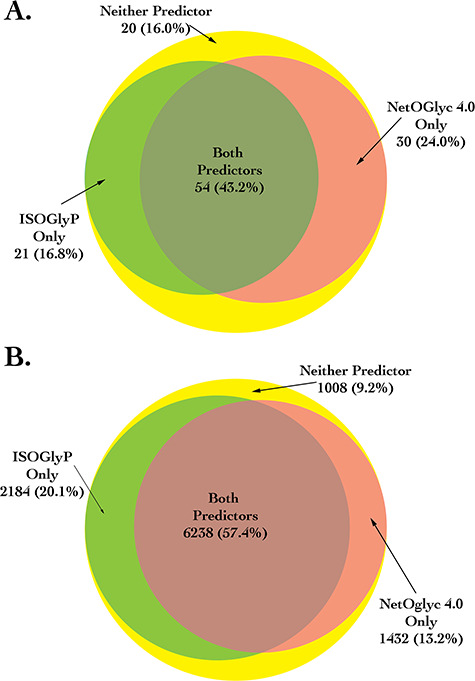
Venn diagram of the comparison of ISOGlyP and NetOGlyc 4.0. The top panel shows the number of positive sites that were correctly predicted by ISOGlyP, NetOGlyc or both using the unbiased dataset of experimentally determined glycosylated sites within platelet data from King et al. (2017). Likewise, the bottom panel shows those numbers for the sites that were not identified as being glycosylated.
Discussion and conclusion
Based solely on the isoform-specific random peptide derived EVs, we have demonstrated that ISOGlyP is equally capable of predicting in vivo sites of O-glycosylation as NetOGlyc4.0 that was trained on O-glycoproteomics data and protein structural data. Unlike NetOGlyc, ISOGlyP’s ability to predict sites of glycosylation based on transferase isoform preferences will be vital for analyzing glycoproteomics data from cell lines and tissues that express different GalNAc-T isoforms, such as in the King et al. (2017) and Steentoft et al. (2013) data. Additionally, this capacity will be helpful in the analysis of the growing number of single cell experiments being collected in databases such as the Expression Atlas (Papatheodorou et al. 2020) to show how variations in O-glycosylation might occur due to cellular differences in GalNAc-T expression in tissues made up of heterogenous cell types. Although the current version of ISOGlyP does not take into account transferases expression levels the ability to incorporate these levels into a final predictive score is being explored.
While the overall accuracies of ISOGlyP and NetOGlyc are similar, the strengths of two programs appear to complement one another, with NetOGlyc being more sensitive and ISOGlyP being more specific. This suggests that an ensemble method taking advantage of the differences of the two predictive programs may increase the overall accuracy of the isoform-specific predictions. Additionally, Mohl et al. (2020) were able to increase the sensitivity of ISOGlyP by extending the features used in the EVP calculations to include structural protein features, such as secondary structure and solvent accessibility, and showed how additional data can readily be applied to the final EVP scores. By incorporating additional isoform-specific experimental data (e.g., more precise EVs and including additional missing isoform EVs) and other computationally derived features, ISOGlyP’s in vivo prediction accuracy is likely to be further improved. It should be noted as with nearly all PTM predictive approaches there is a paucity in our understanding of the competition between different PTMs at the same site (such as between phosphorylation and O-glycosylation), hence additional PTMs may be just as likely be present at the sites of predicted glycosylation which may further affect the accuracy of in vivo predictions.
ISOGlyP provides a unique set of tools for studying the complexity of mucin-type O-glycosylation, in particular identifying the role of neighboring and remote glycosylation and providing the ability to design isoform-specific peptide sequences for introduction into proteins and novel biopharmaceuticals. Presently, we are unable to predict elongated O-glycan structures in a site-specific manner, however, work is currently underway exploring how the initial steps of glycan elongation may be modulated by peptide sequence. If successful, these latter data may be incorporated into future versions of ISOGlyP.
In conclusion, ISOGlyP is a valuable software for interpreting O-glycoproteomic data and understanding the roles of individual GalNAc-Ts in disease. A major advantage of ISOGlyP is its ability to indicate the GalNAc-T isoform (s) that may be responsible for a site’s glycosylation and to identify and predict peptide sequences capable of glycosylation by specific GalNAc-T isoforms.
Supplementary Material
Funding
This work was supported in part by the National Institutes of Health [NIGMS-R01GM113534 to T.A.G. and NIMHD-5U54MD007592 to the Border Biomedical Research Center at UTEP].
Conflict of Interest
None declared.
References
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. 2012. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 22:736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Rivas M, Paul Daniel EJ, Coelho H, Lira-Navarrete E, Raich L, Compañón I, Diniz A, Lagartera L, Jiménez-Barbero J, Clausen H et al. 2018. Structural and mechanistic insights into the catalytic-domain-mediated short-range glycosylation preferences of GalNAc-T4. ACS Central Science. 4:1274–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festari MF, Trajtenberg F, Berois N, Pantano S, Revoredo L, Kong Y, Solari-Saquieres P, Narimatsu Y, Freire T, Bay S et al. 2017. Revisiting the human polypeptide GalNAc-T1 and T13 paralogs. Glycobiology. 27:140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Jamison O, Perrine CL, Collette JC, Moinova H, Ravi L, Markowitz SD, Shen W, Patel H, Tabak LA. 2011. Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J Biol Chem. 286:14493–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Raman J, Fritz TA, Jamison O. 2006. Identification of common and unique peptide substrate preferences for the UDP-GalNAc: Polypeptide alpha-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J Biol Chem. 281:32403–32416. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Revoredo L, Thome JJ, Tabak LA, Vester-Christensen MB, Clausen H, Gahlay GK, Jarvis DL, Johnson RW, Moniz HA et al. 2013. The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J Biol Chem. 288:19900–19914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Ten Hagen KG, Jamison O. 2008. Conservation of peptide acceptor preferences between drosophila and mammalian polypeptide-GalNAc transferase ortholog pairs. Glycobiology. 18:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MR, Hoessli DC, Fang M. 2016. N-acetylgalactosaminyltransferases in cancer. Oncotarget. 7:54067–54081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi HJ, Hansen L, Narimatsu Y, Freeze HH, Henrissat B, Bennett E, Wandall HH, Clausen H, Schjoldager KT. 2018. Glycosyltransferase genes that cause monogenic congenital disorders of glycosylation are distinct from glycosyltransferase genes associated with complex diseases. Glycobiology. 28:284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SL, Joshi HJ, Schjoldager KT, Halim A, Madsen TD, Dziegiel MH, Woetmann A, Vakhrushev SY, Wandall HH. 2017. Characterizing the O-glycosylation landscape of human plasma, platelets, and endothelial cells. Blood Adv. 1:429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl JE, Gerken T, Leung M-Y. 2020. Predicting mucin-type O-glycosylation using enhancement value products from derived protein features. J Theor Comput Chem. 19:2040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatheodorou I, Moreno P, Manning J, Fuentes AM-P, George N, Fexova S, Fonseca NA et al. 2020. Expression atlas update: From tissues to single cells. Nucleic Acids Res. 48:D77–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul Daniel EJ, de las Rivas M, Lira-Navarrete E, García-García A, Hurtado-Guerrero R, Clausen H, Gerken TA. 2020. Ser and Thr acceptor preferences of the GalNAc-Ts vary among isoenzymes to modulate mucin-type O-glycosylation. Glycobiology, cwaa036. Advance Access published [April 18, 2020], doi: 10.1093/glycob/cwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine CL, Ganguli A, Wu P, Bertozzi CR, Fritz TA, Raman J, Tabak LA, Gerken TA. 2009. The glycopeptide preferring polypeptide- GalNAc transferase-10 (ppGalNAc T10), involved in mucin type-O-glycosylation, has a unique GalNAc-O-Ser/Thr binding site in its catalytic domain not found in ppGalNAc T1 or T2. J Biol Chem. 284:20387–20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revoredo L, Wang S, Bennett EP, Clausen H, Moremen KW, Jarvis DL, Ten Hagen KG, Tabak LA, Gerken TA. 2016. Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology. 26:360–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L et al. 2013. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


