Abstract
Certain industrial chemicals accumulate in the environment due to their recalcitrant properties. Bioremediation uses the capability of some environmental bacteria to break down these chemicals and attenuate the pollution. One such bacterial strain, designated Pvy, was isolated from sediment samples from a lagoon in Romania located near an oil refinery due to its capacity to degrade dibenzofuran (DF). The genome sequence of the Pvy strain was obtained using an Oxford Nanopore MiniION platform. According to the consensus 16S rRNA gene sequence that was compiled from six 16S rRNA gene copies contained in the genome and orthologous average nucleotide identity (OrthoANI) calculation, the Pvy strain was identified as Pseudomonas veronii, which confirmed the identification obtained with the aid of MALDI-TOF mass spectrometry and MALDI BioTyper. The genome was analyzed with respect to enzymes responsible for the overall biodegradative versatility of the strain. The Pvy strain was able to derive carbon from naphthalene (NP) and several aromatic compounds of natural origin, including salicylic, protocatechuic, p-hydroxybenzoic, trans-cinnamic, vanillic, and indoleacetic acids or vanillin, and was shown to degrade but not utilize DF. In total seven loci were found in the Pvy genome, which enables the strain to participate in the degradation of these aromatic compounds. Our experimental data also indicate that the transcription of the NP-dioxygenase α-subunit gene (ndoB), carried by the plasmid of the Pvy strain, is inducible by DF. These features make the Pvy strain a potential candidate for various bioremediation applications.
Keywords: Pseudomonas veronii strain Pvy, biodegradation, dioxygenase, dibenzofuran, whole-genome sequencing, nanopore technology, denitrification, organic phosphate mineralization, heavy-metal tolerance
Introduction
The massive production and frequent use of persistent organic pollutants (POPs) in the last century represents a historic environmental burden. Most POPs are halogenated and frequently chlorinated, and their degradation rates are associated with the degree of chlorination. Not only are POPs persistent, but also they are toxic, bio-accumulative, can volatilize and therefore be transported over long distances from the source of emission (Lavric et al. 2004). In humans and other vertebrates, several POPs have proven to be a risk factor for cancer, immune deficiency, nervous system disorders, endocrine disruption, decreased respiratory functions, skin rashes, liver damage, and adverse cardiovascular effects [Wittich et al. 1992; Agency for Toxic Substances and Disease Registry (ATSDR) 1998; Srogi 2008; World Health Organization (WHO) 2014].
The most common approach for the remediation of polluted environments is physico-chemical treatments, with vapor stripping, thermal desorption, soil washing, incineration, pyrolysis, and landfilling being primarily used (Cunningham et al. 1995; Dua et al. 2002). However, these approaches are often economically demanding and difficult to execute. Bioremediation and phytoremediation, which rely on the ability of microorganisms and plants to degrade pollutants, on the other hand, are often a cost-effective and environmental friendly alternative to physico-chemical treatments (Liu and Suflita 1993; Schnoor et al. 1995; Macek et al. 2000; Macková et al. 2006; Macek et al. 2009). One of the major advantages of bioremediation is the possibility of the complete mineralization of contaminants (Tomei et al. 2013; Leewis et al. 2016). Although numerous microbial populations have the metabolic capacity to degrade a broad range of xenobiotics, the biodegradation mechanisms of many of these compounds are not fully understood. Soils and sediments in particular harbor abundant and functionally diverse populations due to numerous microsites and gradients, which can sustain bioremediation (Leigh et al. 2015; Stiborová et al. 2017). Among other taxa, Pseudomonas species have been often associated with the degradation of several anthropogenic compounds, including pesticides, pentachlorophenol, polyaromatic hydrocarbons, polychlorinated biphenyls (PCBs), polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/Fs), and others (Hong et al. 2004; Koubek et al. 2013; Wasi et al. 2013; Wald et al. 2015; Ridl et al. 2018).
Rieske non-heme iron dioxygenases are generally responsible for catalyzing the first reaction in the aerobic degradation pathway of aromatic POPs, which is followed by cleaving dioxygenases involved in the opening of the aromatic ring (Hiraishi 2003; Field and Sierra-Alvarez 2008; Lopez-Echartea et al. 2016). The aim of this study was to analyze the genome of Pseudomonas veronii strain Pvy, which was isolated through its ability to degrade dibenzofuran (DF), with respect to aromatic ring-hydroxylating dioxygenases (ARHD) and other enzymes responsible for the biodegradative versatility of the strain. Furthermore, the induction of Pvy-borne naphthalene (NP) dioxygenase by DF was tested in order to gain deeper insight into the potential of the strain to degrade DF, and potentially PCDD/Fs.
Materials and methods
Strain description and cultivation conditions
The Pvy strain was isolated from sediment samples from a lake in Romania located near an oil refinery (Wald et al. 2015) for its capacity to degrade DF. Briefly, a 10% suspension of the sediment was prepared in 50 mL 1% (wt/vol) sodium pyrophosphate solution, incubated for 2 h at 28°C on a rotary shaker at 130 rpm, followed by a series of 10-fold dilutions to extinction in 0.85% (wt/vol) sodium chloride solution and the plating of 100 µl onto a solid mineral medium (Uhlík et al. 2011). A few crystals of DF (97% purity; Sigma-Aldrich, USA) were provided in the lid to allow for evaporation and the growth of bacterial cultures on the vapors of DF during incubation at 28°C for 72 h. Selected bacterial colonies producing a yellow zone of coloring were reinoculated onto a Plate Count Agar (PCA, Oxoid, UK) for further identification with the aid of MALDI-TOF mass spectrometry and MALDI BioTyper as described previously (Strejček et al. 2018). The DF-degrading bacterial culture, designated Pvy, was preserved over the long term in 25% glycerol stocks at −80°C.
Genome sequencing
For DNA isolation, the culture was grown overnight on PCA at 28°C. Genomic DNA was isolated with a FastDNA SPIN Kit (MPBio, USA) following the manufacturer’s protocol. DNA libraries were prepared with a rapid sequencing kit (Oxford Nanopore) and sequenced on a Nanopore MinION instrument (FLO-MIN106 flow cell). In total, 193 782 Nanopore reads (average read length, 7961 bp; total number of bases, 1 542 749 538) were generated, corresponding approximately to a 200× sequencing depth. Canu software v1.7 (Koren et al. 2017) was used to assemble the reads into continuous genome sequences. The draft assembly was error-corrected by running Racon v1.3.3 twice (Vaser et al. 2017) using minimap2 (Li 2018) to align the input nanopore reads to the draft assembly. Two rounds of nanopolish v0.10.2 (Loman et al. 2015) were performed to further correct the assembly with the fix-homopolymer option turned on and min-candidate-frequency option set to 0.1. The BWA package (Li 2013) was used to map the input nanopore reads against the Racon-corrected assembly. Unless stated otherwise, all the above-mentioned software was run with default parameters, following the authors’ instructions, and using command prompt under a linux operation system. As the resulting assembly contained a large number of frameshifts, a sequence correction was applied as described by Arumugam et al. (2019). Briefly, the assembly (nucleotides) was searched against the bacterial RefSeq protein database (O'Leary et al. 2015) using the DIAMOND v0.9.30 blastx command in frameshift mode with the range-culling argument (Buchfink et al. 2015); the frameshift corrected reads were recovered from the DIAMOND output file using MEGAN Community Edition suite v6.18.5 (Huson et al. 2016). Both the assembly and the frameshift correction workflows are available through complementary scripts deposited in the project GitHub repository (github.com/strejcem/Pseudomonas_veronii_Pvy).
A further manual round of frameshift correction was carried out on proteins that were found to be <0.9 or >1.1 times the length of their closest match to RefSeq protein DB, during which obvious frameshifts located in homopolymer regions were corrected, while substitutions leading to premature STOP codons were left unchanged, as substitution errors are much less likely to occur than indels (Tyler et al. 2018) and naturally occurring pseudogenes were considered a possibility. Both automatic and manual frameshift curation was handled by only inserting one or two ambiguous nucleotides and never deleting a nucleotide, making the corrections trackable. Finally, NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al. 2016) was used to annotate the genome. The genome data were deposited into NCBI repositories as BioProject record PRJNA529862.
For taxonomic purposes, the OrthoANI was calculated between the genomic DNA of the Pvy strain and related strains using the OrthoANI algorithm (Lee et al. 2016) in EzBioCloud (Yoon et al. 2017).
The putative role of genes in the genome of the Pvy strain was assessed through a BLAST search of predicted protein sequences against UniProtKB/Swiss-Prot and Reference Protein (RefSeq) databases, comparison with the literature, and the KEGG database of reference pathways of aromatics degradation. The protein sequences of ARHDs, whose substrate specificities were reported in valid publications, were retrieved from the GenBank database and aligned by Muscle, together with predicted protein sequences of Pvy-borne ARHDs. The phylogenetic tree was constructed using the Maximum Likelihood method and the JTT matrix-based model (Jones et al. 1992) in MEGA X (Kumar et al. 2018).
Utilization/degradation of aromatic substrates
The ability of the Pvy strain to utilize aromatic compounds, since putative genes to degrade them were identified in the genome, was tested as follows: a 0.22-µm filter-sterilized stock solution of DF, NP, biphenyl (BP), carbazole, salicylic acid, protocatechuic acid, indolacetic acid, vanillic acid, vanillin, trans-cinnamic acid, caffeic acid (all purchased from Sigma-Aldrich), and ferulic acid (purchased from Fluka), all at >97% purity and dissolved in molecular biology-grade ethanol were pipetted into 100 ml pre-sterilized flasks and the ethanol was allowed to evaporate. Pvy was inoculated into LB medium and cultivated overnight at 28°C/130 rpm, harvested by centrifugation (10 min at 8000 × g), washed twice and resuspended in an equal volume of mineral salt medium [MSM, composition per liter of deionized water: Na2HPO4·12H2O 11.0 g, KH2PO4 2.7 g, (NH4)2SO4 1 g, MgSO4·7H2O 0.2 g, FeSO4·7H2O 0.02 g, and Ca(NO3)2 0.03 g, pH 7] (Fraraccio et al. 2017) to an initial optical density at 600 nm (OD600) of 0.05. This starting culture was then distributed in aliquots of 15 ml into flasks with individual SPMs, and the cultures were cultivated at 28°C/130 rpm for up to 6 days.
To analyze the depletion of DF by Pvy, a similar modified resting cell assay procedure was employed to that in Garrido-Sanz et al. (2020). Briefly, an overnight grown bacterial culture in LB medium was harvested by centrifugation, washed twice with MSM and diluted with MSM to a final OD600 of 1. The resultant cell suspension was then distributed in 3 ml aliquots into 15 ml glass microcosms, supplemented with 15 µl of DF solution (0.1 M in dimethylsulfoxide, the final concentration of DF in microcosms was 0.5 mM) and sealed. In addition to the Pvy cell suspension and abiotic control (MSM without cells), microcosms containing autoclaved (121°C, 15 min) Pvy cell suspension were prepared as a control of the biomass interference; all samples were prepared in quadruplicates. Microcosms were then incubated at 28°C/130 rpm for 24 h. Upon incubation, 3 ml of methanol was added to the microcosms, cell suspensions were sonicated for 5 min and cells were removed by microfiltration (0.2 µm). The DF content in the samples was then determined with an HPLC NexeraXR system with an SPD-M20A UV/VIS diode array detector (Shimadzu), using the following protocol: 0–2 min—20% methanol, 2–3.5 min—gradient of 20–95% methanol, 3.5–5.5 min—95% methanol, 5.5–7 min—gradient of 95–20% methanol, 7–8 min—20% methanol; mobile phase flow 0.8 ml/min. The separation proceeded in a Luna® Omega Polar C18 column (Phenomenex) at 25°C. DF was monitored at 220 nm; under the conditions applied, DF was eluted at approximately 6.67–6.69 min. To evaluate DF residual concentration in different treatments, average areas under peak values were tested for differences by ANOVA with post hoc Tukey’s test using R project (R Core Team 2017).
Induction of ARHD dioxygenase gene transcription by DF
Induction of the transcription of ARHD large subunit-encoding genes by DF in the Pvy strain was assessed by employing quantitative PCR (qPCR) using cDNA prepared from total RNA from Pvy cells exposed to DF. First, the growth curve of the Pvy strain was determined in MSM amended with 0.5% (wt/vol) sodium acetate (Sigma-Aldrich, USA) as follows: 50 ml of microbial cultures with a starting optical density at 600 nm (OD600 nm) of 0.025 were cultivated in a 250-ml Erlenmeyer flask on a rotary shaker (120 rpm) at 28°C in three independent biological replicates, and the growth was monitored on an hourly basis by measuring OD600. Second, Pvy liquid cultures were cultivated for 17 h corresponding to the mid-log phase as determined from growth curves, harvested by centrifugation (10 min at 8000 × g), washed twice with MSM and resuspended in MSM to OD600 = 1. Stock 0.03 M solution of DF in ethanol was pipetted into an empty 100-ml Erlenmeyer flask and the ethanol was allowed to evaporate, leaving behind the required amount of DF. Subsequently, 10-ml aliquots of prepared cell suspensions were added into the flasks and incubated at 28°C on a rotary shaker (120 rpm). Upon exposure to DF, bacterial cells from 3 ml of the culture were harvested by centrifugation (5 min at 5,000 × g) and stored for subsequent RNA isolation at −80°C. The DF-exposed Pvy cell samples were prepared in quadruplicates, starting from the cultivation of the strain.
RNA isolation was performed using the RNeasy Kit (Qiagen, USA) according to the standard protocol. Concentration of RNA and RNA integrity number (RIN) were determined using Bioanalyzer RNA 6000 Nano assay (Agilent, USA). Only samples with RIN values >7 were subjected to further analyses. Isolated RNA (2000 ng) was treated with 2 U of DNase (Thermo Fisher Scientific, USA) in 50 μl of total reaction volume. After the DNAse treatment, RNA concentration and RIN were measured again, and the absence of leftover contaminating DNA was checked using 16S rRNA gene-targeted qPCR (see further). Finally, a reverse transcription (RT) reaction was performed employing M-MuLV Reverse Transcriptase (200,000 U/ml; New England Biolabs, USA) according to the manufacturer’s instructions starting with 150 ng of RNA per 20 μl of total reaction volume.
All qPCRs were performed in a CFX96 Real-Time System (Bio-Rad, USA) using KAPA SYBR FAST qPCR Master Mix (2×) (Kapa Biosystems, USA). The PCR was performed in a final volume of 15 µl with KAPA SYBR FAST qPCR Master Mix (2×) containing 0.02 U/µl of polymerase, 0.3 µM of each primer (Sigma-Aldrich) and template genomic DNA or cDNA. The 16S rRNA gene (or its cDNA) was amplified using the universal primers 519F 5′-GTGYCAGCMGCNGCGG-3′ (Fraraccio et al. 2017) and 803R 5′-CTACCRGGGTATCTAATCC-3′ (Pei et al. 2010). The following sets of in house-designed primers were used for the quantitation of the transcripts of ARHD large subunits genes found in the Pvy genome, specifically ndoB—465F (5′-CTTCAAGGTATGGCACCCGA-3′) and 737R (5′-ACGACGCCAGGATAAACCTC-3′), ard1—427F (5′-AGCCAGTTCGACAAGTCCAG-3′) and 629R 5′-CACTCAATGGTGGAGCGGAT-3′, and ard2—397F (5′-CTCACCGACGATCAGCTCAA-3′) and 584R (5′-TCGGCGAAGATCCAGTTGAC-3′). The final concentration of each primer was 1.6 µM. The cycling program started with a 5-min denaturation of DNA at 95°C, followed by 30 cycles of 10 s at 95°C, 10 s at the corresponding annealing temperature, 8 s at 72°C, and a final extension of 5 min at 72°C. The annealing temperatures used were 58°C for the 16S rRNA gene, 61°C for ndoB, 66.5°C for ard1, and 63.5°C for ard2. Agarose gel electrophoresis, melting curve analysis, and Sanger sequencing of amplified qPCR products were used to check the specificity of the qPCR. All qPCR measurements were performed in technical triplicates and with a calibration curve constructed using serial dilutions of genomic DNA of the strain. The statistical significance of the induction experiments was tested using R project (R Core Team 2017) and pairwise t-tests, which were performed to compare the control (no inducer present) with DF-induced samples. Multiple P-values were adjusted using the Benjamini and Hochberg method (Benjamini and Hochberg 1995).
Data availability
The strain is available upon request. The supplementary material file contains Supplementary Table S1, which shows the loci in the genome of the Pvy strain that are putatively involved in the metabolism of aromatic compounds including aromatic pollutants, Supplementary Table S2, which summarizes the utilization of aromatic substrates by Pvy strain, and supplementary text on the implications from genome analysis for heavy-metal tolerance and nutrient metabolism. The genome data were deposited into NCBI repositories as BioProject record PRJNA529862. The assembly and frameshift correction workflows are available through complementary scripts deposited in the project GitHub repository at github.com/strejcem/Pseudomonas_veronii_Pvy.
Supplementary material is available at https://gsajournals.figshare.com DOI: https://doi.org/10.25387/g3.13311809.
Results and discussion
Genomic analysis of the Pvy strain
The genome of the Pvy strain is composed of one chromosome and one plasmid, the total size of which is 7 305 203 bp with a total of 6704 genes, of which 6220 are coding genes. There are 94 RNA genes, of which 7 encode for 5S rRNAs, 6 for 16S rRNAs, 6 for 23S rRNAs, 71 for tRNAs, and 4 for ncRNAs. Detailed information on the genome of Pvy is provided in Tables 1 and 2.
Table 1.
Genome project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Rapid sequencing library (SQL-RBK004) |
| MIGS 29 | Sequencing platforms | Oxford Nanopore MiniION |
| MIGS 31.2 | Fold coverage | 200 |
| MIGS 30 | Assemblers | Canu v. 1.7.1 |
| MIGS 32 | Gene calling method | GeneMarkS-2+ |
| Locus Tag | E4167 | |
| Genbank ID | CP039631-CP039632 | |
| GenBank Date of Release | 29-JAN-2020 | |
| GOLD ID | Gs0143776 | |
| BIOPROJECT | PRJNA529862 | |
| Project relevance | Bioremediation, degradation of aromatic compounds |
Table 2.
Genome statistics
| Attribute | Value |
|---|---|
| Genome size (bp) | 7 305 203 |
| GC content (%) | 60.67 |
| DNA scaffolds | 2 |
| Total genes | 6704 |
| Protein coding genes | 6220 |
| RNA genes | 94 |
| Pseudo genes | 390 |
Given that the nanopore technology, which was used for this genome sequencing, is known to be more prone to less randomly distributed errors than other sequencing technologies currently available on the market (Goldstein et al. 2019), two rounds were run of Racon v1.3.3 (Vaser et al. 2017) and nanopolish v0.10.2 (Loman et al. 2015) with the fix-homopolymer option turned on. After such processing, there were a total of 2562 potentially interrupted ORFs. Although some of these ORFs might have been real pseudogenes, in high-quality assembled genomes their numbers typically do not exceed 200 (Stewart et al. 2019), so further manual curation was performed as described in the Materials and methods section. The final number of detected potentially interrupted ORFs was 433; thus, an 83% reduction was achieved. The resulting genome consists of a single 7 110 596-bp-long chromosome and one 194 607-bp-long circular plasmid.
The consensus 16S rRNA gene sequence compiled from six 16S rRNA gene copies contained in the Pvy genome had a 99.79% similarity to the 16S rRNA gene of P. veronii CFML 92-134T, followed by Pseudomonas extremaustralis 14-3T with 99.73% similarity and Pseudomonas grimontii CFML 97-514T with 99.59% similarity, while OrthoANI between the genomic DNA of the Pvy strain and the type strain of P. veronii was 98.16%. OrthoANI with the other closest Pseudomonas species was below 90% (specifically 89.19% with P. extremaustralis 14-3T and 88.22% with P. grimontii CFML 97-514T), thus corroborating the affiliation of the Pvy strain with the species P. veronii, which was also confirmed with the aid of MALDI-TOF mass spectrometry and MALDI BioTyper performed according to Strejček et al. (2018).
The overall characteristics of P. veronii were published by Ajithkumar et al. (2003) and Elomari et al. (1996). It is a ubiquitous bacterial species whose members have been isolated from a wide variety of different environments, including natural mineral waters (Elomari et al. 1996), contaminated soil (Nam et al. 2003; Junca and Pieper 2003), contaminated sludge (Ajithkumar et al. 2003), contaminated sediment (Wald et al. 2015), and biotrickling filter cleaners (Onaca et al. 2007). Finally, P. veronii stains have been isolated from the roots of grapevine plants (Montes et al. 2016), implicating their role as endophytes. As for its common occurrence in contaminated environments, P. veronii has been previously described as a potent degrader of many anthropogenic pollutants, including BTEX (Junca and Pieper 2003; Morales et al. 2016), pentachlorophenol (Nam et al. 2003), NP (Wald et al. 2015), and alkylphenols up to a chain length of six carbons (Ajithkumar et al. 2003). In addition, P. veronii strains are often heavy-metal tolerant and are capable of denitrification and the mineralization of organic phosphate (de Lima-Morales et al. 2013; Montes et al. 2016). With respect to the origin of the Pvy strain as well as its isolation as a DF-degrading bacterium, here we focus on its biodegradative potential, while the implications from the genome analysis in terms of heavy-metal tolerance and nutrient metabolism are provided in the Supplementary material.
Utilization of aromatic substrates by the Pvy strain
To demonstrate its metabolic versatility, the utilization of a range of aromatic substances by the Pvy strain was assessed, the selection of which was based on the composition of the original sludge from which it was isolated, as well as on the genomic context of Pvy (see below). As shown in Table 3, the Pvy strain was able to utilize NP, salicylic acid, protocatechuic acid, p-hydroxybenzoic acid, trans-cinnamic acid, vanillic acid, vanillin, and indoleacetic acid, but not DF, carbazole, BP, ferulic acid, or caffeic acid.
Table 3.
Utilization of aromatic substrates by Pvy strain
| Growth substrate | Growth substrate concentration |
||
|---|---|---|---|
| 5 mM | 10 mM | 15 mM | |
| Dibenzofurana | − | − | − |
| Naphthalene | + | ++ | +++ |
| Carbazole | − | − | − |
| Biphenyl | − | − | − |
| Salicylic acid | ++ | ++ | − |
| Protocatechuic acid | ++ | +++ | +++ |
| p-Hydroxybenzoic acid | ++ | ++ | +++ |
| Vanillic acid | ++ | ++ | ++ |
| Vanillin | + | ++ | ++ |
| Ferulic acid | − | − | − |
| trans-Cinnamic acid | − | − | +++ |
| Caffeic acid | − | − | − |
| Indoleacetic acid | ++ | ++ | +++ |
The maximum growth recorded throughout the cultivation period of 6 days is shown. Legend: (−) no observable growth; (+) maximum OD600 nm < 0.3; (++) 0.3 < OD600 nm < 0.8; (+++) OD600 nm > 0.8; results reported are from two independent experiments. Exact values are reported in Supplementary Table S2.
Dibenzofuran was degraded by Pvy strain as assessed by the resting cell assay.
DF degradation
The Pvy strain was isolated based on its ability to produce yellow coloration zones when grown on plates in the presence of DF; nevertheless, the strain was not able to utilize this substrate as a sole carbon source in liquid cultures (Table 3). Therefore, the ability of the strain to degrade DF was assayed by resting cell assay. Upon co-incubation with DF, the Pvy-bearing microcosms produced a yellow coloration similarly to that observed in the agar plate cultures. As analyzed by HPLC, the final DF concentration in the microcosms with Pvy cells was (0.20 ± 0.07) mM, which corresponded to 42% (significantly lower, P-value <0.001) of the amount in the microcosms with autoclaved Pvy cells [(0.49 ± 0.02) mM]; no significant depletion (P-value >0.1) was observed for autoclaved Pvy cells compared to abiotic controls, showing the matrix effect of Pvy biomass during DF analysis to be negligible.
Implications from genome analysis: degradation of aromatic compounds
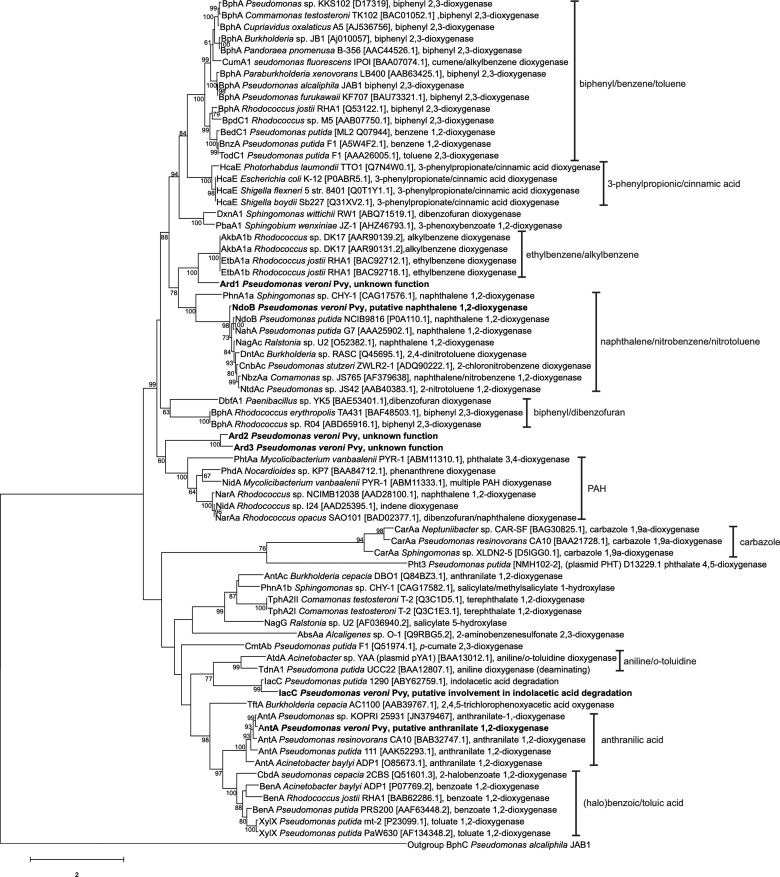
The annotated genome of the Pvy strain was analyzed with respect to the presence of genes potentially involved in the degradation of aromatic compounds, including xenobiotics, with a particular emphasis on genes encoding putative ARHDs. To help elucidate the functions of putative ARHDs found in the Pvy genome, a tree showing their phylogeny was constructed along with ARHDs with assigned substrate specificity (Figure 1).
Figure 1.
Phylogenetic tree showing relationships among ARHDs with assigned substrate specificity (the list might not be complete). The ARHDs found in Pvy strain are shown in bold, and substrates shared in clades are indicated. The analysis involved 73 amino acid sequences, whose Genbank accession numbers are indicated in brackets. Bootstrapping was used to test the tree topology (500 bootstraps, only values >70% are shown). A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories). The tree is drawn to scale, with branch lengths corresponding to the number of substitutions per site. There were a total of 551 positions in the final dataset.
In total, seven loci were found on the chromosome of the Pvy strain for which putative roles in the degradation of aromatic compounds could be assigned based on in silico analyses, as shown in Supplementary Table S1. Coding sequences found within the first locus, here designated as pca I, exhibited high similarity with the coding sequences of enzymes from the protocatechuate branch of the β-ketoadipate pathway, including intradiol-cleaving two-component protocatechuate 3,4-dioxygenase encoded by the genes pcaHG, and enzymes enabling the further utilization of a protocatechuate cleavage product, 3-carboxy-cis,cis-muconate (Zylstra et al. 1989; Harwood and Parales 1996; Shen and Liu 2005). Environmental bacteria use the protocatechuate branch of the β-ketoadipate pathway for the degradation of various aromatic pollutants as well as natural aromatic compounds. Apart from the pca I locus, another copy of pcaHG genes was present elsewhere in the Pvy genome accompanied by the putative regulatory protein gene pcaQ. The ability of the Pvy strain to grow using protocatechuic acid as a sole carbon source was also proven in this study (Table 3).
Within the second region here designated as hmg, three putative enzymes were encoded which mediate the degradation of homogentisate to fumarate and acetoacetate. Homogentisate is an intermediate of the bacterial metabolism of ethylbenzene, styrene, and other pollutants, and at the same time, it represents both the precursor for tyrosine and phenylalanine biosynthesis and a product of their catabolism (Arias-Barrau et al. 2004). For that reason, the potential involvement of hmg-locus-borne genes in the degradation of aromatic xenobiotics is not certain.
ORFs from the kyn/ant/cat region encode putative enzymes enabling the transformation of tryptophan to anthranilate, including tryptophan 2,3-dioxygenase, kynurenine formamidase, and kinureninase (Kurnasov et al. 2003). The further catabolism of anthranilate is likely enabled by anthranilate 1,2-dioxygenase and the catechol branch of the β-ketoadipate pathway (ortho-cleavage) encoded by genes carried within this locus yielding 3-oxoadipate enol-lactone that can be further utilized by in trans-encoded enzymes, e.g. from the pca region.
The dmp/cat region bears putative genes encoding enzymes, which funnel phenol and possibly related phenolic compounds to intermediate metabolism through the catechol meta-cleavage pathway. In addition to this pathway, a short-chain alcohol dehydrogenase family protein is encoded within this locus, which provided no significant hits in databases; therefore, its role in phenol/phenolics degradation remains unknown.
The dmp/ard1 region encodes a putative Rieske-type ARHD of unknown specificity, whose predicted α-subunit exhibits only low sequence similarity with homologous proteins included in the UniProt/Swiss-Prot database (the highest score of 56% similarity was with the BP 2,3-dioxynase subunit α from Rhodococcus jostii RHA1). Moreover, adjacent genes encoding for the unknown putative enzymes aromatic compound-cis-diol dehydrogenase and aromatic ring-opening dioxygenase are present in the dmp/ard1 region. The remaining genes encode the catabolism of 2-hydroxymuconate semialdehyde, an intermediate of catechol meta-cleavage. According to its position in the phylogenetic tree, the ard1-corresponding protein sequence is closely related to ethylbenzene and alkylbenzene dioxygenases from Rhodococcus spp. (Figure 1). Based on these findings, we hypothesized that genes carried by this locus determine the degradation of unknown aromatic substrate(s) in a similar manner as with the model substances DF or BP, i.e. the aromatic ring is first activated by ARHD through dihydroxylation, and then is cleaved and further transformed (Seeger et al. 1995).
The architecture of the iac region resembles the eponymous locus in Pseudomonas putida 1290 enabling the catabolism of the plant auxin hormone and the signaling molecule indole-3-acetic acid (IAA; Scott et al. 2013). Expectably, the Pvy strain was able to utilize IAA as a sole carbon/energy source (Table 3).
The mhp/vanAB/vdh region consists of 22 coding sequences, including three putative regulatory protein- and four transporter protein-encoding genes. The products of genes here designated vdh1 and vanAB exhibit high sequence similarity to vanillin dehydrogenase and two-component vanillate O-demethylase, respectively, from various pseudomonads. These enzymes transform the secondary metabolite vanillin to vanillic acid, which is further converted to protocatechuate. Vanillin dehydrogenases from various bacteria were reported to exhibit broad substrate specificity toward various aromatic aldehydes, such as isovanillin, veratraldehyde, anisaldehyde, protocatechualdehyde, p-hydroxybenzaldehyde, salicylaldehyde, and benzaldehyde (Masai et al. 2007; Mitsui et al. 2010; Graf et al. 2016; Nishimura et al. 2018). The genes fadD and fadB encode for a putative feruloyl-CoA synthase and vanillin synthase, respectively, enabling the catabolism of ferulic acid through its conversion to vanillin, which is further transformed by vanillin dehydrogenase and vanillate O-demethylase (Overhage et al. 1999). The predicted protein encoded by the calA gene exhibited high similarity with coniferyl-alcohol dehydrogenase from Pseudomonas sp. strain HR199 (Achterholt et al. 1998). Furthermore, the mhp/vanAB/vdh locus bears the mhpABCDFE gene cluster, with the mhpA gene encoding for a putative bifunctional 3-(3-hydroxy-phenyl)propionate/3-hydroxycinnamic acid monooxygenase that mediates the metabolism of 3-hydroxycinnamic acid. Moreover, an additional copy of a putative vanillin dehydrogenase gene, vdh2, was present within the mhp/vanAB/vdh locus, the predicted protein sequence of which exhibited a somewhat low relatedness to the vanillin dehydrogenases reported to date, including those from P. putida and P. fluorescens (66% and 57% of sequence similarity, respectively). Therefore, we predicted that the mhp/vanAB/vdh region determines the utilization/degradation of phenylpropanoids, presumably including lignin-derived compounds (Kamimura et al. 2017). The ability of the strain Pvy to utilize trans-cinnamic acid, vanillin, and vanillic acid was demonstrated; however, this was not true of caffeic acid or ferulic acid (Table 3). Predicted proteins encoded by three coding sequences corresponding to locus tags E4167_27585 and E4167_27590 show only low similarity to proteins with annotated functions available in the databases referenced. Nonetheless, domains specific for acyl-CoA dehydrogenase, acyl-CoA acetyltransferase, and putative phenol degradation protein were identified in the predicted amino acid sequences. The substrate specificity of corresponding putative enzymes and their potential involvement in aromatic compound degradation is unclear.
Two other ORFs encoding for putative Rieske-type ARHDs were found within the Pvy chromosome, here designated ard2 and ard3. The amino acid sequences of the corresponding predicted α-subunits (locus tags E4167_29595 and E4167_29825) only exhibited a limited level of relatedness to characterized ARHDs available in databases. In the phylogeny reconstruction, the two predicted proteins formed a separate branch within a cluster belonging to ARHDs catalyzing the transformation of various aromatic substrates including NP, indene, and terephthalate (Figure 1). Surprisingly, at least some of the ORFs surrounding these putative ARHD-encoding genes indicated that these could be part of an aerobic non-specified aromatic compound catabolism pathway through CoA-thioesters, analogously as was reported for the paa-encoded pathway of phenylacetate degradation in both Escherichia coli K12 and Pseudomonas sp. strain Y2 (Teufel et al. 2010). Metabolic pathways that involve the putative ARHDs encoded by the ard2 and ard3 genes remain enigmatic, and discovering their exact function will require more research.
The analysis of the pND plasmid sequence revealed the presence of two ORF clusters here designated ndo/nah and salA/dmp, separated by a DNA span of 26.7 kbp. The predicted amino acid sequences encoded within the above-mentioned region showed a high similarity with proteins from Pseudomonas spp., putatively encoding a two-component NP 1,2-dioxygenase accompanied by a corresponding ferredoxin (genes ndoBC and ndoA, respectively), that initiate NP degradation. In the phylogeny reconstruction, the predicted protein sequence of the corresponding ARHD large subunit NdoB formed a cluster together with other NP, chloronitrobenzene, and nitrotoluene dioxygenases from the genera Pseudomonas, Comamonas, Burkholderia, and Ralstonia (Figure 1). In addition to the ARHD, other enzymes encoded within the ndo/nah cluster are homologous with those that enable subsequent reactions, which yield salicylic acid (genes nahB, nahF, nahC, nahE, nahC). The ndo/nah cluster also includes the pahQ gene, which encodes for a putative outer membrane porin involved in the metabolism of aromatic compounds.
The salA/dmp region is comprised of 12 genes, including those encoding for a putative transcriptional regulator (nahR), salicylate monooxygenase (salA), and the dmp cluster, the combination of which enables the catabolism of salicylate to pyruvate and acetylCoA through the meta-cleavage of the aromatic ring. A putative heme-binding protein of unknown function is encoded within this region (locus tag E4167_34205). Altogether, we hypothesized that the combination of enzymes encoded by the plasmid pND enables the utilization of NP through dihydroxylation with subsequent reactions leading to intermediates of the central metabolism. The ability of the Pvy strain to utilize NP and salicylate as sole carbon sources was confirmed experimentally within this study (Table 3).
Induction of ARHDs by DF in resting cell assay
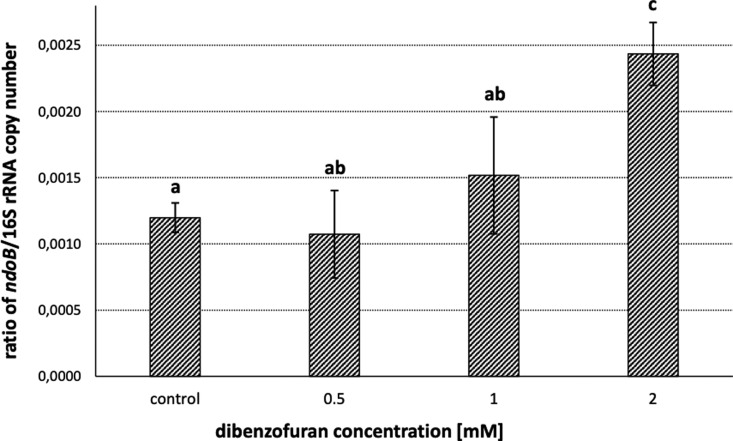
The DF degradation pathway documented in different bacterial strains produces salicylate (Bunz and Cook 1993; Armengaud and Timmis 1997), which is also an intermediate product in the degradation pathway of NP in Pseudomonas strains (Eaton and Chapman 1992). At the same time, the dioxygenation of DF has been reported to be catalyzed by DF 4,4a-dioxygenase (Bunz and Cook 1993; Kasuga et al. 2001), but due to their broad substrate specificity, other ARHDs have been found to catalyze its dioxygenation, such as BP dioxygenase (Seeger et al. 2001; Mohammadi and Sylvestre 2005; L’Abbée et al. 2005) or NP 1,2-dioxygenase (Resnick et al. 1996), which is encoded in the plasmid pND of Pvy. With all of this in mind, and due to the fact that the bacterium Pvy was isolated as a DF-degrading bacterium, we further investigated whether the predicted plasmid-borne NP 1,2-dioxygenase large subunit is involved in DF degradation. For this purpose, the induction of the corresponding large subunit-encoding gene ndoB in the presence of DF was determined. The results of the qPCR with cDNA obtained from DF-exposed Pvy cultures revealed significant differences in the levels of ndoB transcripts (Figure 2). There was no significant difference between the control (no DF added) and 0.5 mM DF, and the difference between the control and 1 mM was marginally insignificant (P = 0.06). Importantly, there was a significant difference between the control and 2 mM DF (P < 0.001). Hence, we assume that the observed DF degradation by the Pvy strain is mediated by the putative NP-dioxygenase encoded by the plasmid-borne gene cluster ndoABC.
Figure 2.
Induction of the transcription of NP-dioxygenase α-subunit (ndoB) gene by DF shown as the ratio of ndoB and 16S rRNA copy number. Error bars indicate standard error of the mean; bars sharing the same letter (a, b, c) are not significantly different (P-value <0.05); control: no DF added.
In addition to ndoB, the induction of the dioxygenases of unknown function ard1, ard2, and ard3 was assessed to determine their possible involvement in DF degradation. Since the deduced aa sequences of ard2 and ard3 exhibited a significant level of similarity (82%), and in the phylogeny reconstruction formed a discrete cluster (Figure 2), only ard2 as a representative of both was followed in this study. Nevertheless, unlike with ndoB, no significant changes in the transcript levels of either ard1 or ard2 were observed, with cp values not differing from the non-template control.
The transcription of NP degradation genes has been reported to be induced by salicylate, NP, or both of them (Schell 1983; Jones et al. 2003; Tomás-Gallardo et al. 2014). DF has been reported to induce ARHDs such as NarA in Rhodococcus opacus (Kimura et al. 2006), DfdA in Terrabacter sp. strain YK3 (Iida et al. 2002), or as we report here, Pvy-borne NdoB. Our results also demonstrated that the induction of biodegradative genes was concentration dependent. Higher concentrations of DF resulted in higher induction (Figure 2), while concentrations of 0.5 mM DF did not induce the gene significantly compared to the control (no DF added). These results are in agreement with the conclusions of other studies; for example Pham et al. (2015) found that the amount of 4-chlorobenzoate (an end metabolite of the Rhodococcus erythropolis U23A BP catabolic pathway) produced by cells co-metabolically grown on BP plus sodium acetate decreased while decreasing BP concentration. A similar result was found by Toussaint et al. (2012), where cells grown on sodium acetate and BP decreased their catabolic activity toward 4-chlorobiphenyl as the concentration of BP decreased.
Conclusions
The analysis of the whole-genome sequence and experiments made with the Pvy strain support the conclusion that this strain is a versatile degrader of aromatic compounds. Introducing the strain or its DNA into the genetic pool of autochthonous microbial populations could potentially contribute to the microbe-assisted bioremediation of sites contaminated with various aromatic hydrocarbons. In addition, several genetic elements, including ARHD-like genes, were identified with low similarity to any known genes/enzymes, requiring their further investigation in order to fully clarify their roles. Furthermore, given the fact that the strain is capable of degrading both pollutants and plant-derived aromatic compounds, it could be employed in the plant-assisted bioremediation (Pham et al. 2015; Musilová et al. 2016; Vergani et al. 2019) of sites with high concentrations of pollutants that the strain is able to tolerate.
Acknowledgment
The authors wish to thank to Dr. Monika Stavelova of AECOM CZ Ltd. for providing the sediment samples.
Funding
The results reported here were obtained with the support of the Ministry of Industry and Trade of the Czech Republic under grant no. FV10471 and the INTER-EXCELLENCE program of the Ministry of Education, Youth and Sports of the Czech Republic under grant no. LTAUSA19013. The whole-genome sequencing was supported by the grant of the Ministry of Defence of the Czech Republic through a long-term organization development plan (no. 907930101413). Further support is acknowledged of the ELIXIR CZ research infrastructure project (Ministry of Education, Youth and Sports of the Czech Republic grant no. LM2015047), specifically for access to computing and storage facilities.
Conflicts of interest: None declared.
Literature cited
- Achterholt S, Priefert H, SteinbüChel A.. 1998. Purification and characterization of the coniferyl aldehyde dehydrogenase from Pseudomonas sp. Strain HR199 and molecular characterization of the gene. J Bacteriol. 180:4387–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR) 1998. Toxicological Profile for Chlorinated Dibenzo-p-Dioxins (CDDs). Atlanta, GA: U.S. Department of Health and Human Services, Public Health Service. [PubMed] [Google Scholar]
- Ajithkumar B, Ajithkumar VP, Iriye R.. 2003. Degradation of 4-amylphenol and 4-hexylphenol by a new activated sludge isolate of Pseudomonas veronii and proposal for a new subspecies status. Res Microbiol. 154:17–23. [DOI] [PubMed] [Google Scholar]
- Arias-Barrau E, Olivera ER, Luengo JM, Fernández C, Galán B, et al. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J Bacteriol. 186:5062–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud J, Timmis KN.. 1997. Molecular characterization of Fdx1, a putidaredoxin-type [2Fe-2S] ferredoxin able to transfer electrons to the dioxin dioxygenase of Sphingomonas sp. RW1. Eur J Biochem. 247:833–842. [DOI] [PubMed] [Google Scholar]
- Arumugam K, Bağcı C, Bessarab I, Beier S, Buchfink B, et al. 2019. Annotated bacterial chromosomes from frame-shift-corrected long-read metagenomic data. Microbiome. 7:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57:289–300. [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12:59–60. [DOI] [PubMed] [Google Scholar]
- Bunz PV, Cook AM.. 1993. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system. J Bacteriol. 175:6467–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham SD, Berti WR, Huang JW.. 1995. Phytoremediation of contaminated soils. Trends Biotechnol. 13:393–397. [Google Scholar]
- de Lima-Morales D, Chaves-Moreno D, Jarek M, Vilchez-Vargas R, Jauregui R, et al. 2013. Draft genome sequence of Pseudomonas veronii strain 1YdBTEX2. Genome Announc. 1:e00258–00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoso R, Leiva-Novoa P, Zuniga A, Timmermann T, Recabarren-Gajardo G, et al. 2016. Biochemical and genetic bases of indole-3-acetic acid (auxin phytohormone) degradation by the plant-growth-promoting rhizobacterium Paraburkholderia phytofirmans PsJN. Appl Environ Microbiol. 83:e01991–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua M, Singh A, Sethunathan N, Johri A.. 2002. Biotechnology and bioremediation: successes and limitations. Appl Microbiol Biotechnol. 59:143–152. [DOI] [PubMed] [Google Scholar]
- Eaton RW, Chapman PJ.. 1992. Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J Bacteriol. 174:7542–7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elomari M, Coroler L, Hoste B, Gillis M, Izard D, et al. 1996. DNA relatedness among Pseudomonas strains isolated from natural mineral waters and proposal of Pseudomonas veronii sp. nov. Int J Syst Evol Microbiol. 46:1138–1144. [DOI] [PubMed] [Google Scholar]
- Field JA, Sierra-Alvarez R.. 2008. Microbial degradation of chlorinated dioxins. Chemosphere. 71:1005–1018. [DOI] [PubMed] [Google Scholar]
- Fraraccio S, Strejček M, Dolinová I, Macek T, Uhlík O.. 2017. Secondary compound hypothesis revisited: selected plant secondary metabolites promote bacterial degradation of cis-1,2-dichloroethylene (cDCE). Sci Rep. 7:8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Sanz D, Sansegundo-Lobato P, Redondo-Nieto M, Šuman J, Cajthaml T, et al. 2020. Analysis of the biodegradative and adaptive potential of the novel polychlorinated biphenyl degrader Rhodococcus sp. WAY2 revealed by its complete genome sequence. Microbial Genomics. 6:000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein S, Beka L, Graf J, Klassen JL.. 2019. Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genomics. 20:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf N, Wenzel M, Altenbuchner J.. 2016. Identification and characterization of the vanillin dehydrogenase YfmT in Bacillus subtilis 3NA. Appl Microbiol Biotechnol. 100:3511–3521. [DOI] [PubMed] [Google Scholar]
- Harwood CS, Parales RE.. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 50:553–590. [DOI] [PubMed] [Google Scholar]
- Hiraishi A. 2003. Biodiversity of dioxin-degrading microorganisms and potential utilization in bioremediation. Microb Environ. 18:105–125. [Google Scholar]
- Hong HB, Nam IH, Murugesan K, Kim YM, Chang YS.. 2004. Biodegradation of dibenzo-p-dioxin, dibenzofuran, and chlorodibenzo-p-dioxins by Pseudomonas veronii PH-03. Biodegradation. 15:303–313. [DOI] [PubMed] [Google Scholar]
- Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, et al. 2016. MEGAN community edition—interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol. 12:e1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Mukouzaka Y, Nakamura K, Kudo T.. 2002. Plasmid-borne genes code for an angular dioxygenase involved in dibenzofuran degradation by Terrabacter sp. strain YK3. AEM. 68:3716–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM.. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 8:275–282. [DOI] [PubMed] [Google Scholar]
- Jones RM, Britt-Compton B, Williams PA.. 2003. The naphthalene catabolic (nag) genes of Ralstonia sp. strain U2 are an operon that is regulated by NagR, a LysR-type transcriptional regulator. J Bacteriol. 185:5847–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junca H, Pieper DH.. 2003. Functional gene diversity analysis in BTEX contaminated soils by means of PCR-SSCP DNA fingerprinting: comparative diversity assessment against bacterial isolates and PCR-DNA clone libraries. Environ Microbiol. 6:95–110. [DOI] [PubMed] [Google Scholar]
- Kamimura N, Takahashi K, Mori K, Araki T, Fujita M, et al. 2017. Bacterial catabolism of lignin-derived aromatics: new findings in a recent decade: update on bacterial lignin catabolism. Environ Microbiol Rep. 9:679–705. [DOI] [PubMed] [Google Scholar]
- Kasuga K, Habe H, Chung J-S, Yoshida T, Nojiri H, et al. 2001. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the Gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem Biophys Res Commun. 283:195–204. [DOI] [PubMed] [Google Scholar]
- Kimura N, Kitagawa W, Mori T, Nakashima N, Tamura T, et al. 2006. Genetic and biochemical characterization of the dioxygenase involved in lateral dioxygenation of dibenzofuran from Rhodococcus opacus strain SAO101. Appl Microbiol Biotechnol. 73:474–484. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubek J, Macková M, Macek T, Uhlík O.. 2013. Diversity of chlorobiphenyl-metabolizing bacteria and their biphenyl dioxygenases in contaminated sediment. Chemosphere. 93:1548–1555. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurnasov O, Jablonski L, Polanuyer B, Dorrestein P, Begley T, et al. 2003. Aerobic tryptophan degradation pathway in bacteria: novel kynurenine formamidase. FEMS Microbiol Lett. 227:219–227. [DOI] [PubMed] [Google Scholar]
- L’Abbée J-B, Barriault D, Sylvestre M.. 2005. Metabolism of dibenzofuran and dibenzo-p-dioxin by the biphenyl dioxygenase of Burkholderia xenovorans LB400 and Comamonas testosteroni B-356. Appl Microbiol Biotechnol. 67:506–514. [DOI] [PubMed] [Google Scholar]
- Lavric ED, Konnov AA, De Ruyck J.. 2004. Dioxin levels in wood combustion—a review. Biomass Bioenerg. 26:115–145. [Google Scholar]
- Lee I, Kim YO, Park S-C, Chun J.. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 66:1100–1103. [DOI] [PubMed] [Google Scholar]
- Leewis M-C, Uhlik O, Fraraccio S, McFarlin K, Kottara A, et al. 2016. Differential impacts of willow and mineral fertilizer on bacterial communities and biodegradation in diesel fuel oil-contaminated soil. Front Microbiol. 7:837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh MB, Wu W-M, Cardenas E, Uhlik O, Carroll S, et al. 2015. Microbial communities biostimulated by ethanol during uranium (VI) bioremediation in contaminated sediment as shown by stable isotope probing. Front Environ Sci Eng. 9:453–464. [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv: 1303.3997.
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Suflita JM.. 1993. Ecology and evolution of microbial populations for bioremediation. Trends Biotechnol. 11:344–352. [DOI] [PubMed] [Google Scholar]
- Loman NJ, Quick J, Simpson JT.. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. bioRxiv: 015552. [DOI] [PubMed]
- Lopez-Echartea E, Macek T, Demnerova K, Uhlik O.. 2016. Bacterial biotransformation of pentachlorophenol and micropollutants formed during its production process. Int J Environ Res Public Health. 13:1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek T, Macková M, Káš J.. 2000. Exploitation of plants for the removal of organics in environmental remediation. Biotechnol Adv. 18:23–34. [DOI] [PubMed] [Google Scholar]
- Macek T, Uhlík O, Ječná K, Nováková M, Lovecká P, et al. 2009. Advances in phytoremediation and rhizoremediation. In: Varma A, editor. Advances in Applied Bioremediation (Soil Biology). Berlin, Germany: Springer. p. 257–277. [Google Scholar]
- Macková M, Dowling D, 2006. Phytoremediation and rhizoremediation. In: Macek T, editor. Focus on Biotechnology. Dordrecht, Netherlands: Springer. p. 300. [Google Scholar]
- Masai E, Yamamoto Y, Inoue T, Takamura K, Hara H, et al. 2007. Characterization of ligV essential for catabolism of vanillin by Sphingomonas paucimobilis SYK-6. Biosci Biotechnol Biochem. 71:2487–2492. [DOI] [PubMed] [Google Scholar]
- Mitsui R, Hirota M, Tsuno T, Tanaka M.. 2010. Purification and characterization of vanillin dehydrogenases from alkaliphile Micrococcus sp. TA1 and neutrophile Burkholderia cepacia TM1. FEMS Microbiol Lett. 303:41–47. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Sylvestre M.. 2005. Resolving the profile of metabolites generated during oxidation of dibenzofuran and chlorodibenzofurans by the biphenyl catabolic pathway enzymes. Chem Biol. 12:835–846. [DOI] [PubMed] [Google Scholar]
- Montes C, Altimira F, Canchignia H, Castro Á, Sánchez E, et al. 2016. A draft genome sequence of Pseudomonas veronii R4: a grapevine (Vitis vinifera L.) root-associated strain with high biocontrol potential. Stand Genomic Sci. 11:76–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales M, Sentchilo V, Bertelli C, Komljenovic A, Kryuchkova-Mostacci N, et al. 2016. The genome of the toluene-degrading Pseudomonas veronii strain 1YdBTEX2 and its differential gene expression in contaminated sand. PLoS One. 11:e0165850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musilová L, Rídl J, Polívková M, Macek T, Uhlík O.. 2016. Effects of secondary plant metabolites on microbial populations: changes in community structure and metabolic activity in contaminated environments. Int J Mol Sci. 17:1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam I-H, Chang Y-S, Hong H-B, Lee Y-E.. 2003. A novel catabolic activity of Pseudomonas veronii in biotransformation of pentachlorophenol. Appl Microbiol Biotechnol. 62:284–290. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Kawakami S, Otsuka H.. 2018. Molecular cloning and characterization of vanillin dehydrogenase from Streptomyces sp. NL15-2K. BMC Microbiol. 18:154–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojiri H, Omori T.. 2002. Molecular bases of aerobic bacterial degradation of dioxins: involvement of angular dioxygenation. Biosci Biotechnol Biochem. 66:2001–2016. [DOI] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, et al. 2015. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaca C, Kieninger M, Engesser K-H, Altenbuchner J.. 2007. Degradation of alkyl methyl ketones by Pseudomonas veronii MEK700. J Bacteriol. 189:3759–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J, Priefert H, Steinbüchel A.. 1999. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. Strain HR199. Appl Environ Microbiol. 65:4837–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei AY, Oberdorf WE, Nossa CW, Agarwal A, Chokshi P, et al. 2010. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol. 76:3886–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TT, Pino Rodriguez NJ, Hijri M, Sylvestre M.. 2015. Optimizing polychlorinated biphenyl degradation by flavonoid-induced cells of the rhizobacterium Rhodococcus erythropolis U23A. PLoS One. 10:e0126033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2017. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Resnick SM, Lee K, Gibson DT.. 1996. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Ind Microbiol Biotechnol. 17:438–457. [Google Scholar]
- Ridl J, Suman J, Fraraccio S, Hradilova M, Strejcek M, et al. 2018. Complete genome sequence of Pseudomonas alcaliphila JAB1 (=DSM 26533), a versatile degrader of organic pollutants. Stand Genomic Sci. 3.13: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato SI, Ouchiyama N, Kimura T, Nojiri H, Yamane H, et al. 1997. Cloning of genes involved in carbazole degradation of Pseudomonas sp. strain CA10: nucleotide sequences of genes and characterization of meta-cleavage enzymes and hydrolase. J Bacteriol. 179:4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA. 1983. Cloning and expression in Escherichia coli of the naphthalene degradation genes from plasmid NAH7. J Bacteriol. 153:822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoor JL, Licht LA, Mccutcheon SC, Wolfe NL, Carreira LH.. 1995. Phytoremediation of organic and nutrient contaminants. Environ Sci Technol. 29:318A–A323. [DOI] [PubMed] [Google Scholar]
- Scott JC, Greenhut IV, Leveau JH.. 2013. Functional characterization of the bacterial iac genes for degradation of the plant hormone indole-3-acetic acid. J Chem Ecol. 39:942–951. [DOI] [PubMed] [Google Scholar]
- Seeger M, Cámara B, Hofer B.. 2001. Dehalogenation, denitration, dehydroxylation, and angular attack on substituted biphenyls and related compounds by a biphenyl dioxygenase. J Bacteriol. 183:3548–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Timmis KN, Hofer B.. 1995. Degradation of chlorobiphenyls catalyzed by the bph-encoded biphenyl-2,3-dioxygenase and biphenyl-2,3-dihydrodiol-2,3-dehydrogenase of Pseudomonas sp. LB400. FEMS Microbiol Lett. 133:259–264. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu S.. 2005. Key enzymes of the protocatechuate branch of the beta-ketoadipate pathway for aromatic degradation in Corynebacterium glutamicum. Sci China Ser C. 48:241–249. [DOI] [PubMed] [Google Scholar]
- Srogi K. 2008. Levels and congener distributions of PCDDs, PCDFs and dioxin-like PCBs in environmental and human samples: a review. Environ Chem Lett. 6:1–28. [Google Scholar]
- Stewart RD, Auffret MD, Warr A, Walker AW, Roehe R, et al. 2019. Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat Biotechnol. 37:953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiborová H, Kolář M, Vrkoslavová J, Pulkrabová J, Hajšlová J, et al. 2017. Linking toxicity profiles to pollutants in sludge and sediments. J Hazard Mater. 321:672–680. [DOI] [PubMed] [Google Scholar]
- Strejček M, Šmrhová T, Junková P, Uhlík O.. 2018. Whole-cell MALDI-TOF MS versus 16S rRNA gene analysis for identification and dereplication of recurrent bacterial isolates. Front Microbiol. 9: 1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44:6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, et al. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci USA. 107:14390–14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás-Gallardo L, Gómez-Álvarez H, Santero E, Floriano B.. 2014. Combination of degradation pathways for naphthalene utilization in Rhodococcus sp. strain TFB. Microb Biotechnol. 7:100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomei MC, Mosca Angelucci D, Annesini MC, Daugulis AJ.. 2013. Ex situ remediation of polluted soils by absorptive polymers, and a comparison of slurry and two-phase partitioning bioreactors for ultimate contaminant degradation. J Hazard Mater. 262:31–37. [DOI] [PubMed] [Google Scholar]
- Toussaint J-P, Pham T, Barriault D, Sylvestre M.. 2012. Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl Microbiol Biotechnol. 95:1589–1603. [DOI] [PubMed] [Google Scholar]
- Tyler AD, Mataseje L, Urfano CJ, Schmidt L, Antonation KS, et al. 2018. Evaluation of Oxford nanopore’s MinION sequencing device for microbial whole. Sci Rep. 8:10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlík O, Strejček M, Junková P, Šanda M, Hroudová M, et al. 2011. Matrix-Assisted Laser Desorption Ionization (MALDI)-Time of Flight Mass Spectrometry- and MALDI Biotyper-based identification of cultured biphenyl-metabolizing bacteria from contaminated horseradish rhizosphere soil. Appl Environ Microbiol. 77:6858–6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Sovic I, Nagarajan N, Sikic M.. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergani L, Mapelli F, Šuman J, Cajthaml T, Uhlík O, et al. 2019. Novel PCB-degrading Rhodococcus strains able to promote plant growth for assisted rhizoremediation of historically polluted soils. PLoS One. 14:e0221253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald J, Hroudova M, Jansa J, Vrchotova B, Macek T, et al. 2015. Pseudomonads rule degradation of polyaromatic hydrocarbons in aerated sediment. Front Microbiol. 6:1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasi S, Tabrez S, Ahmad M.. 2013. Use of Pseudomonas spp. for the bioremediation of environmental pollutants: a review. Environ Monit Assess. 185:8147–8155. [DOI] [PubMed] [Google Scholar]
- Wittich RM, Wilkes H, Sinnwell V, Francke W, Fortnagel P.. 1992. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl Environ Microbiol. 58:1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) 2014. Dioxins and Their Effects on Human Health. Fact sheet No 225. WHO Media Centre.
- Yoon SH, Ha SM, Lim J, Kwon S, Chun J.. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Anton Van Leeuw. 110:1281–1286. [DOI] [PubMed] [Google Scholar]
- Zylstra GJ, Olsen RH, Ballou DP.. 1989. Genetic organization and sequence of the Pseudomonas cepacia genes for the alpha and beta subunits of protocatechuate 3,4-dioxygenase. J Bacteriol. 171:5915–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The strain is available upon request. The supplementary material file contains Supplementary Table S1, which shows the loci in the genome of the Pvy strain that are putatively involved in the metabolism of aromatic compounds including aromatic pollutants, Supplementary Table S2, which summarizes the utilization of aromatic substrates by Pvy strain, and supplementary text on the implications from genome analysis for heavy-metal tolerance and nutrient metabolism. The genome data were deposited into NCBI repositories as BioProject record PRJNA529862. The assembly and frameshift correction workflows are available through complementary scripts deposited in the project GitHub repository at github.com/strejcem/Pseudomonas_veronii_Pvy.
Supplementary material is available at https://gsajournals.figshare.com DOI: https://doi.org/10.25387/g3.13311809.