Abstract
CD33-related Siglecs are often found on innate immune cells and modulate their reactivity by recognition of sialic acid-based “self-associated molecular patterns” and signaling via intracellular tyrosine-based cytosolic motifs. Previous studies have shown that Siglec-11 specifically binds to the brain-enriched polysialic acid (polySia/PSA) and that its microglial expression in the brain is unique to humans. Furthermore, human microglial Siglec-11 exists as an alternate splice form missing the exon encoding the last (fifth) Ig-like C2-set domain of the extracellular portion of the protein, but little is known about the functional consequences of this variation. Here, we report that the recombinant soluble human microglial form of Siglec-11 (hSiglec-11(4D)-Fc) binds endogenous and immobilized polySia better than the tissue macrophage form (hSiglec-11(5D)-Fc) or the chimpanzee form (cSiglec-11(5D)-Fc). The Siglec-11 protein is also prone to aggregation, potentially influencing its ligand-binding ability. Additionally, Siglec-11 protein can be secreted in both intact and proteolytically cleaved forms. The microglial splice variant has reduced proteolytic release and enhanced incorporation into exosomes, a process that appears to be regulated by palmitoylation of cysteines in the cytosolic tail. Taken together, these data demonstrate that human brain specific microglial hSiglec-11(4D) has different molecular properties and can be released on exosomes and/or as proteolytic products, with the potential to affect polySia-mediated brain functions at a distance.
Keywords: exosome, polysialic acid, protein palmitoylation, Siglec-11
Introduction
Sialic acid-binding immunoglobulin superfamily lectins (Siglecs) are a family of single-pass transmembrane cell surface lectins that recognize sialic acid (Sia)-containing ligands (Crocker et al. 2007). Their extracellular domains (ECDs) consist of an amino terminal immunoglobulin V-set-like domain (V-set domain) that recognizes sialoglycans, followed by one or more immunoglobulin-like C2-set domains (C2-set (Varki et al. 2017; Angata 2018).
CD33(Siglec-3)-related Siglecs (CD33rSiglecs) are a subgroup of the Siglecs with sequence similarity that is often found on innate immune cells. Their reactivity is modulated by interacting with sialoglycans via the V-set domain. Siglec-11 is an inhibitory CD33rSiglec that is not found in rodents (Angata et al. 2002; Khan et al. 2020). In humans, Siglec-11 is expressed on tissue macrophages as well as on brain microglia. In contrast, closely related nonhuman primates including chimpanzees showed no expression of Siglec-11 on brain microglia, but rather have an expression on tissue macrophages (Hayakawa et al. 2005; Wang et al. 2011; Wang et al. 2012), i.e., brain microglial expression of Siglec-11 is uniquely human. Genomic sequence data also indicate that in humans SIGLEC11 allele was modified via gene conversion by a nonfunctional SIGLEC16P pseudogene (Wang et al. 2012; Hayakawa et al. 2017). Siglec-11 and Siglec-16 are regarded as paired receptors that have maintained similar ligand-binding preferences via gene conversions (Hayakawa et al. 2017). While Siglec-11 is an inhibitory receptor, Siglec-16 is activating (Schwarz et al. 2017). It is assumed that this conversion event led to selective Siglec-11 expression in human microglia as the converted SIGLEC11 became fixed in human, likely due to the neuroprotective functions it exerts (Wang and Neumann 2010). Notably, human Siglec-11 in brain microglia appears to exist exclusively as an alternate splice form missing the exon encoding the last C2-set domain of extracellular portion of the protein (Wang and Neumann 2010). Previous in vitro analysis demonstrated that Siglec-11 selectively binds to α2-8-linked sialic acid oligomers (oligoSias) and polymers (polySias) (Angata et al. 2002; Hayakawa et al. 2005; Shahraz et al. 2015). PolySia, a ligand of Siglec-11, is known to play important roles in brain development and functions, and expression is mainly observed in embryonic brain as well as in distinct adult brain regions in vertebrates (Angata et al. 2002; Sato and Kitajima 2013; Sato et al. 2016). Other polySia-interactive molecules such as BDNF, FGF2, dopamine and polysialytransferase have been implicated in several neuropsychiatric disorders (Sato and Hane 2018). While the role of Siglec-11 in neuropsychiatric disorders is not well understood, it has been reported to regulate neuroinflammation and microglial neurotoxicity. Indeed, Siglec-11 expression resulted in reduced microglial phagocytosis of apoptotic neuronal material and coculture of Siglec-11-expressing microglia with neurons showed neuronal-surface polySia dependent neuroprotective functions of Siglec-11 (Wang and Neumann 2010). Furthermore, polySia with an average degree of polymerization 20 (DP20) injected into the eyes of SIGLEC11-expressing transgenic mice blocked mononuclear phagocyte reactivity, inhibited complement activation, and gave protection from vascular damage in the retina induced by laser coagulation. (Shahraz et al. 2015; Karlstetter et al. 2017). Despite these reported functions and human-specific expression in the brain, little is known about the molecular properties of Siglec-11.
In this paper, we investigated Siglec-11 molecular properties in detail using in vitro expression systems. We show that the human Siglec-11 microglial form [hSiglec-11(4D)] binds polySia better than the tissue macrophage form [hSiglec-11(5D)] and chimpanzee Siglec-11 [cSiglec-11(5D)], can be processed and secreted differently than the macrophage 5D form and secreted in exosomes. The exosomal hSiglec-11(4D) also binds polySia on human neuroblastoma cells. Mechanistically, cysteine palmitoylation likely plays a role in Siglec-11 distribution. Taken together, our findings suggest that the microglial hSiglec-11(4D) has different molecular properties and have the potential to affect brain functions at a distance.
Results
The human-specific human microglial form Siglec-11(4D)-Fc binds immobilized polySia better than the tissue macrophage form hSiglec-11(5D)-Fc or chimpanzee Siglec-11(5D)-Fc
To evaluate the binding ability of the different Siglec-11forms to polySia, recombinant soluble Ig-Fc chimeras were generated and studied for binding to immobilized E. coli K1-derived polySia chains (Figure 1A). The monoclonal antibody 735 (Mab735) confirmed polySia immobilization (Figure 1B). The microglial form hSiglec-11(4D)-Fc binds this immobilized polySia in a dose-dependent manner (Figure 1C) and binds better to immobilized polySia than the tissue macrophage form hSiglec-11(5D)-Fc (~36%) or the chimpanzee Siglec-11(5D)-Fc (~55%) (Figure 1D). Binding comparison was performed in the linear kinetic range. To evaluate whether hSiglec-11 binding to polySia is dependent on the conserved arginine residue that is known to be important for sialic acid recognition in other Siglecs (Van der Merwe et al. 1996; Vinson et al. 1996), we mutated Arg120 and showed that hSiglec-11(4D)-Fc binding was reduced but not eliminated. Specifically, hSiglec-11(4D; R120A)-Fc showed ~39% binding of wild type binding and hSiglec-11(4D; R120K)-Fc showed ~58% of wild type binding (Figure 1E). Surprisingly, free polySia (up to 5 mg/mL) did not inhibit hSiglec-11(4D)-Fc binding to the immobilized form (Figure 1F), indicating that the immobilized polySia has a distinct conformation that is recognized by Siglec-11.
Fig. 1.
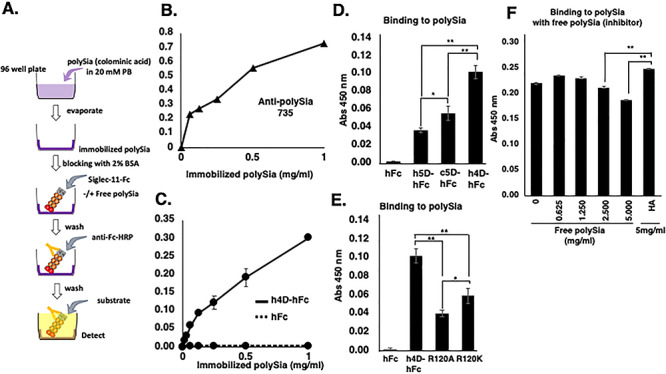
ELISA in vitro binding assay of Siglec-11 to polySia. (A) Experimental scheme for (B–F). PolySia (0~1 mg/mL) was immobilized on the plate and analyzed binding of (B) mab 735, (C) hSiglec-11(4D)-Fc (-) and Fc (…) to the immobilized polySia. PolySia (1 mg/mL) was immobilized on the plate and binding of (D) Fc, hSiglec-11(5D)-Fc, cSiglec-11(5D)-Fc and hSiglec-11(4D)-Fc, (E) Fc, hSiglec-11(4D)-Fc, hSiglec-11(4D;R120A)-Fc and hSiglec-11(4D;R120K)-Fc to immobilized polySia was analyzed. (F) PolySia (1 mg/ml) was immobilized on the plate and binding of hSiglec-11(4D)-Fc was analyzed with the presence of different concentrations of preincubated free polySia (0-5 mg/mL) or HA (5 mg/mL). n = 3, *P < 0.05 **P < 0.005.
The microglial form recognizes polySia on human neuroblastoma SH-SY5Y cells
We next evaluated hSiglec-11(4D)-Fc binding to polySia in a cellular system. Human neuroblastoma SH-SY5Y cells were used as a neuron-like system with cell surface polySia, which could be recognized by the specific Mab 735, and cleaved by the specific enzyme Endo-NF (Figure 2A and B). Indeed, the microglial form hSiglec-11(4D)-Fc bound neuroblastoma cell surface polySia, while control human Fc did not bind (Figure 2C and D). hSiglec-11(4D)-Fc binding was only partially abolished by Endo-NF treatment (Figure 2C and D), likely because Siglec-11 also recognizes the oligo-sialic acids left behind after Endo-NF treatment (Angata et al. 2002). This indicates that hSiglec-11(4D), in addition to binding to immobilized polySia, is also able to bind cell surface polySia.
Fig. 2.
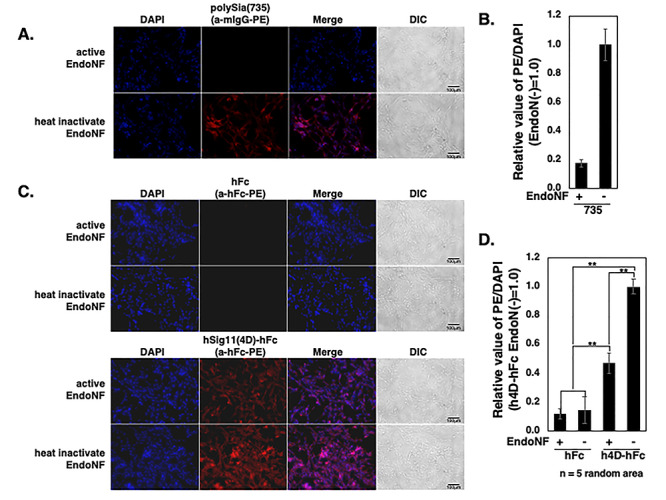
In vitro binding assay of Siglec-11 to cell surface polySia. SH-SY5Y cells were fixed and treated with active/inactive Endo-NF and the cell surface polySia were detected by immunostaining with: (A) mab 735 and PE conjugated antimouse IgG (red), (C) hFc (top two rows) hSiglec-11(4D)-Fc (bottom two rows) and PE conjugated antihuman Fc. Nucleus was visualized with DAPI (blue). All the specimens were observed under the fluoromicroscope. The bars indicate 100 μm. (B) Relative intensity of the PE/DAPI for (A). (D) Relative intensity of the PE/DAPI for (C). Five random areas were measured using ImageJ software, and the data were processed. **P < 0.005.
Siglec-11 is prone to aggregation, independent of disulfide bond formation
Similar to other Siglecs, Siglec-11 is a transmembrane protein that can be expressed on the cell surface. To better understand how Siglec-11 exerts its function, we used Siglec-11 expression vectors with a C-terminal V5 epitope tag and transiently transfected them into HEK293 cells. Western blotting showed that the full-length Siglec11, either hSiglec-11(4D) and hSiglec-11(5D), is prone to aggregation, which occurred even after boiling in SDS and reduction of disulfide bonds by 2-Mercaptoethanol treatment (Figure 3A). Truncated mutants of hSiglec-11 were generated and transiently transfected to evaluate the effect of each domain on the aggregation (Supplementary Figure 1A). Notably, the truncated mutants were still prone to aggregation that was also unaffected by the reduction of disulfide bonds (Figure 3B, C and Supplementary Figure 1B).
Fig. 3.
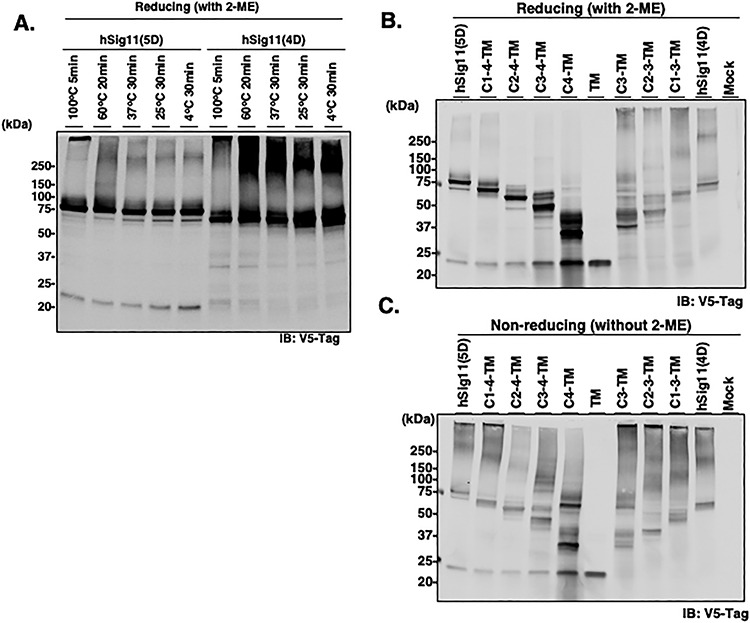
Full length Siglec-11 analysis by western blotting. HEK293 cells were transiently transfected with V5-tagged hSiglec-11 for 3 days and cell lysates were analyzed by western blotting. (A) Denature temperature of cell lysate were changed under the reducing condition with 2-ME and analyzed by western blotting with anti-V5 tag antibody. (B and C) Cell lysates were denatured at 4°C for 30 min under reducing (B) and nonreducing condition (C) and analyzed by Western blotting with anti-V5 tag antibody.
Specific N-glycosylation sites are important for Siglec-11 protein stability
Analysis of the Sigle-11 protein identified 7 and 5 predicted N-glycosylation sites for the 5D and 4 D forms, respectively (Supplementary Figure 1A and Figure 4A). We confirmed Siglec-11 can be glycosylated by using PNGaseF treatment of all three forms of Siglec-11, which resulted in a downward shift in their molecular weight (Figure 4B). We further studied how each N-glycosylation site of hSiglec-11 contributes to its stability by preparing Asn→Gln mutants of Asn-X-Ser/Thr predicted sites and analyzed by western blotting. Transient or stable expression of hSiglec11(5D) with mutations of the sixth and seventh N-glycans sites, which is missing in the 4D microglia form, resulted in protein degradation (Figure 4C and D). These results suggest the importance of sixth and seventh N-glycan for the stability of the 5D macrophage form, and not for the 4D microglial form.
Fig. 4.
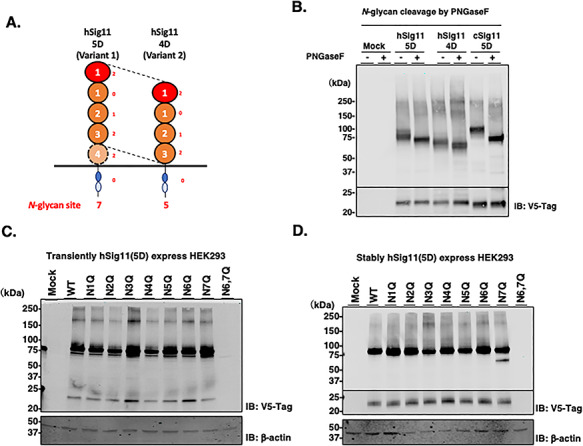
N-glycosylation of Siglec-11. (A) Schematic image of hSiglec-11 and N-glycosylation sites. (B) Cell lysates of V5-tagged Siglec-11 transfected HEK293 cells were treated with PNGaseF and analyzed by western blotting with anti-V5 tag antibody. V5-tagged full length hSiglec-11 and As→Gln mutant of each glycosylation site were transiently (C) or stably (D) transfected into HEK293 cells and cell lysates were analyzed by western blotting with anti-V5 tag antibody. β-actin was used as loading control.
hSiglec-11(4D) and hSiglec-11(5D) are differentially processed and secreted
A small fragment (<25 kDa) was detected in cells transiently transfected with the macrophage hSiglec-11(5D) form and not in the microglial hSig1ec-11(4D) form (Figure 3B and C). This fragment is likely generated by proteolytic cleavage close to the transmembrane domain as an expression vector with just the transmembrane domain yields a similar size fragment (Figure 3B, C and Supplementary Figure 1B). We posited that such a cleavage should also result in production and secretion of a fragment containing the Siglec-11 ECD. To investigate the presence of the ECD, we analyzed the culture medium by western blotting with anti-Siglec-11 (4C4) antibody, which detects the N-terminal domain of Siglec-11 (Figure 5A-C and Supplementary Figure 3). Indeed, Siglec-11 ECD was abundant in the culture medium of cells expressing the 5D form (Figure 5C). Matrix metalloproteinases or β secretases inhibitors did not inhibit the cleavage (Supplementary Figure 2). Although the inhibitors did not alter the total levels of the small fragment in the cell lysate, it is possible that the secreted amount may still be affected by the inhibitors.
Fig. 5.
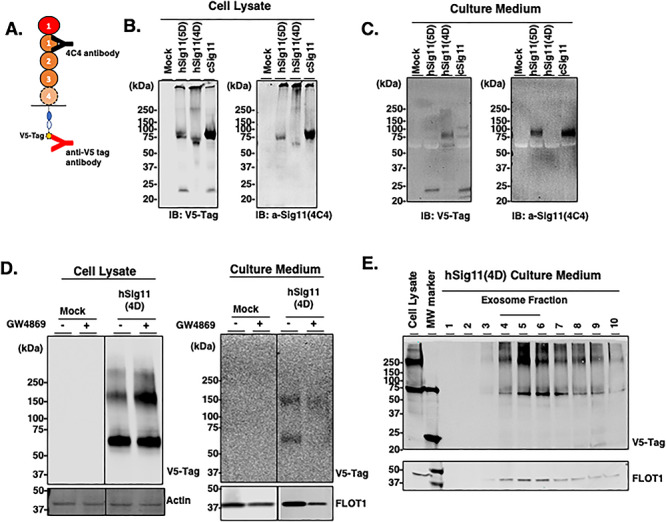
Siglec-11 secretion from transfected cells. HEK293 cells were transiently transfected for 5 days with V5-tagged Siglec-11 and the cell lysates and concentrated culture medium were analyzed. (A) Schematic of anti-V5 tag antibody recognizing C-terminal of Siglec-11 and 4C4 antibody recognizing N-terminal (first C2-set domain) of Siglec-11. Western blotting of cell lysates (B) and culture medium (C) with anti-V5 tag antibody or anti-Siglec11 (4C4) antibody. (D) HEK293 cells were transiently transfected with V5-tagged hSiglec-11(4D) with or without 20 μM of exosome inhibitor GW4869 and the cell lysate and concentrated culture medium were analyzed by western blotting with anti-V5 tag antibody. β-actin was used as loading control and flotillin-1 was used as an exosome marker. (E) Extracellular vesicles were isolated from the culture medium of HEK293 cells transfected with V5-tagged hSiglec-11(4D) and density gradient separation fractions were analyzed by western blotting with anti-V5 tag antibody and anti-flotillin-1 antibody.
In addition to the cleaved products, intact Siglec-11 was also found in the medium by western blotting, especially originating from the 4D microglial form (Figure 5C). We found that this intact Siglec-11 is secreted in exosomes: cells treated with 20 μM of exosome inhibitor GW4869 had reduced levels of intact hSiglec-11(4D) and flotillin-1, an exosome marker, in the culture medium (Figure 5D). Interestingly, 20 μM of GW4869 treatment led to higher levels of hSiglec-11(4D) in the cell lysate (Figure 5D), suggesting that inhibition of exosome production leads to increased production of Siglec-11. Confirming that hSiglec-11(4D) is in exosomes, both intact hSiglec-11(4D) and flotillin-1 are found in the exosome fractions (Figure 5E) following density gradient separation of the extracellular vesicles (de Gassart et al. 2003). These data suggest that microglial Siglec-11 can be processed and secreted differently from macrophage Siglec-11 and that microglial Siglec-11 can be secreted in the exosomes.
Cysteine palmitoylation is likely related to localization on lipid rafts and Siglec-11 secretion on exosomes
To begin to understand how Siglec-11 is secreted in exosomes, we investigated whether S-palmitoylation, a process known to regulate the membrane association of various proteins (Draper et al. 2007) may be involved in Siglec-11-related secretion. Indeed, treating stably transfected HEK293A expressing the three different forms of Siglec-11 with protein acylation inhibitor 2-Bromopalmitate led to a downward molecular weight shift for Siglec-11 (Figure 6A), suggesting acylation modification. Additionally, acyl-PEG exchange analysis resulted in an upward molecular shift of Siglec-11, confirming S-palmitoylation modification (Figure 6B). Furthermore, we noted that in stably transfected HEK293 cells, hSig11(4D) also showed the small <25 kDa fragment, suggesting that, as hSig11(4D) accumulates overtime in stably transfected cells, it can also be proteolytically cleaved. Moreover, density gradient separation of cell lysate showed that Siglec-11 localized in flotillin-1 positive fractions, suggesting their partial localization in cell surface lipid rafts (Figure 7). 2-Bromopalmitate inhibition changed the expression pattern of hSig-11(5D) (Supplementary Figure 4): for example, gradient fraction 3 had detectable hSig-11(5D) after 2BP was added, further suggesting that palmitoylation plays a role in its lipid raft localization.
Fig. 6.
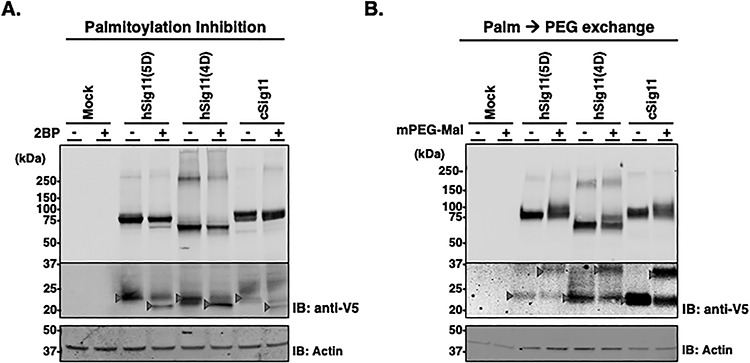
Palmitoylation of Siglec-11. (A) V5-tagged Siglec-11 stably transfected HEK293 cells were cultured with or without protein acylation inhibitor 2BP and the cell lysates were analyzed by western blotting with anti-V5 tag antibody. β-actin was used as loading control. Arrows indicate molecular weight shift as the result of 2BP. (B) Protein S-fatty acylation was tagged by 5 kDa-PEG with cell lysates of V5-tagged Siglec-11 stably transfected HEK293 and analyzed by western blotting with anti-V5 tag antibody. Expression level of full length Siglec-11 (higher molecular weight membrane) and proteolytical fragment (lower molecular weight membrane) were different, thus different exposures were needed. Arrows indicate molecular weight shift in the presence of PEG tag.
Fig. 7.
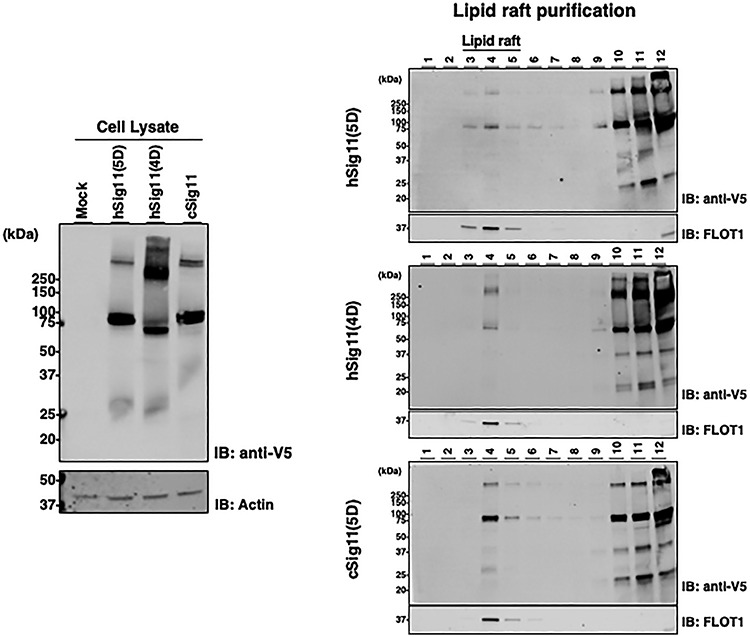
Siglec-11 is localized in lipid rafts. (A) Cell lysates of V5-tagged Siglec-11 stably transfected HEK293 cells were analyzed by western blotting with anti-V5 tag antibody. (B) Same cell lysates were fractionated by density gradient separation and analyzed by western blotting with anti-V5 tag antibody. hSiglec-11(5D) (top panel), hSiglec-11(4D) (middle panel) and cSiglec-11(5D) (lower panel) were analyzed. β-actin was used as loading control and flotillin-1 was used as a lipid raft marker.
The exosome form shows polySia-dependent binding to human neuroblastoma cells
To evaluate the binding ability of hSiglec-11(4D) on exosome with cell surface polySia, we prepared an exosome-labeling construct encoding the N-terminal 183 amino acid of flotillin-1 and an EGFP fused protein (F1-GFP) with or without hSiglec-11(4D) expression (Figure 8A). hSiglec-11(4D), F1-GFP and flotillin-1 were detected in the culture medium of transfected HEK293 cells (Figure 8B), confirming its ability to be secreted. SH-SY5Y cells were used as a cellular model to examine polySia binding as previously described (Figures 2, 9A and B). We found that treating SH-SY5Y cells with the culture media containing the hSiglec-11(4D)-F1-EGFP-labeled exosome (hSig11(4D)-F1-EGFP CM) showed significantly better binding than the culture media with the F1-EGFP-labeled exosome containing culture medium without hSiglec-11(4D) (Mock-F1-EGFP CM) (Figure 9C and D). Endo-NF treatment decreased the binding of EGFP-labeled exosomes with or without hSiglec-11(4D) (Figure 9C and D), suggesting polySia binding. Notably, End-NF treatment lowered the binding of mock-EGFP CM treated cells, suggesting some exosomal binding to polySia.
Fig. 8.
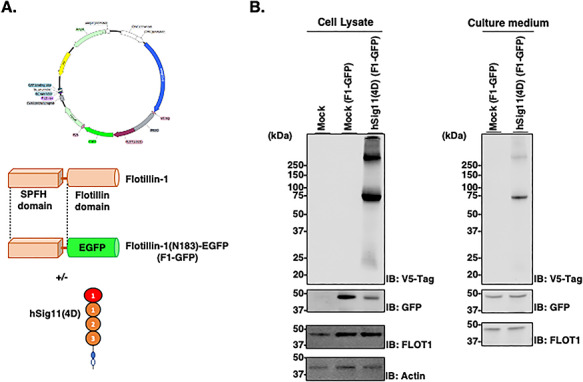
Exosomal Siglec-11 can be secreted. (A) Schematic representation of the F1-GFP protein and expression plasmid. (B) Cell lysates and culture medium of V5-tagged hSiglec-11(4D) and F1-GFP cotransfected HEK293 cells transiently for 5 days were lysed and analyzed by western blotting with anti-V5 tag antibody [hSiglec-11(4D)], anti-GFP (F1-GFP), anti-flotillin-1 (lipid raft/exosome marker) and β-actin (loading control). SH-SY5Y cells cultured with active/inactive Endo-NF.
Fig. 9.
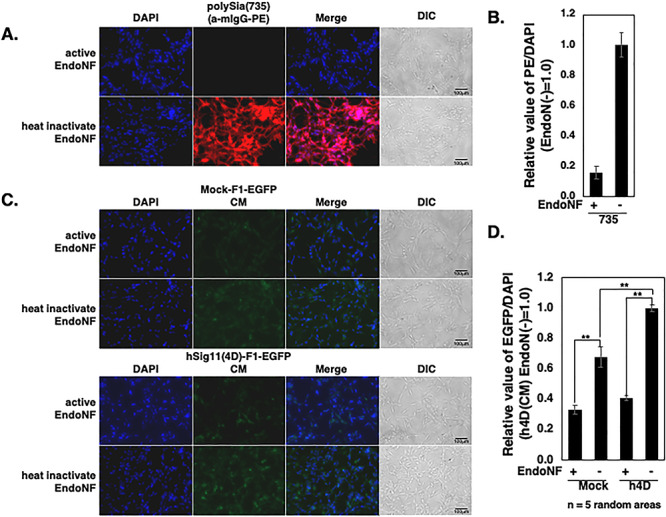
Culture media containing exosomal Siglec-11 binds to polySia on cell surface. (A–D) After fixation, the cell surface polySia were detected by immunostaining with (A) mab 735 and PE conjugated antimouse IgG (red). (C) SH-SY5Y cells were treated with active or inactive endo-NF for 8 h and subjected to culture media without hSiglec-11(4D)-F1-EGFP (Mock-F1-EGFP-CM; upper panel) or with hSig11(4D)-F1-EGFP exosome (hSig11(4D)-F1-EGFP CM; lower panel). Nucleus was visualized with DAPI (blue). All the specimens were imaged under the fluoromicroscope. The bars indicate 100 μm. (B) Relative intensity of the PE/DAPI was analyzed from (A). (D) Relative intensity of the EGFP/DAPI was analyzed from (C). Five random areas were measured using ImageJ software, and the data were processed. **P < 0.005.
Discussion
Siglec-11 is a CD33rSiglec not found in rodents (Angata et al. 2002) and among higher primates is uniquely expressed in microglia only in the human brain (Hayakawa et al. 2005). In the human central nervous system, Siglec-11 in microglia appears to exist exclusively as an alternate splice form missing the exon encoding the last C2-set domain of the extracellular portion of the protein (Wang and Neumann 2010). It is curious that human SIGLEC11 was gene converted from the nonfunctional SIGLEC16P and became fixed in humans (Hayakawa et al. 2005; Wang et al. 2011, 2012), suggesting its importance. In this study, we characterized the molecular properties of the different forms of Siglec-11, including the macrophage and microglial Siglec-11. We showed that the uniquely human microglial form of Siglec-11 binds polySia better and secreted differently from macrophage and chimpanzee forms of Siglec-11. Furthermore, the 4D microglial Siglec-11 is secreted in exosomes that are able to bind cell-surface polySia (Figure 10). We also show that Siglec-11 undergoes palmitoylation that likely affects its localization in lipid raft. Collectively, our results suggest that splicing out of last C2-set domain in microglia changes the proteolytic cleavage and enhances incorporation of the 4D form into exosomes.
Fig. 10.
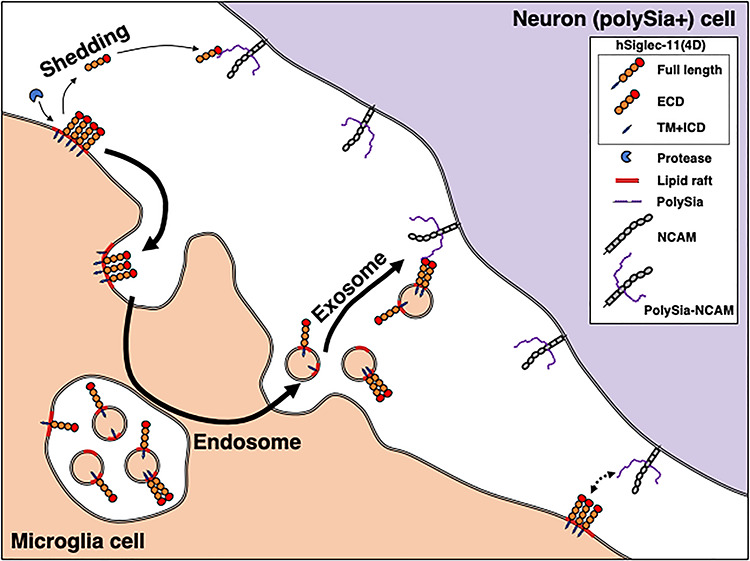
Human Siglec-11 can be secreted and bind cell surface polySia at a distance. Siglec-11 has been considered to exist as a membrane protein on cell surface and interact with oligo/polySia on cell surface. Here, we propose that Siglec-11 can be proteolytically cleaved from cell surface and the ECD exist as a shed soluble protein. Moreover, the microglial form hSiglec-11(4D) can be secreted on exosome. Both soluble ECD and full length of hSiglec-11(4D) showed binding to cell surface polySia. Novel pathways of hSiglec-11(4D)-polySia interaction in human brain are proposed.
Microglial Siglec-11 has been previously demonstrated to alleviate microglia neurotoxicity (Linnartz et al. 2010). It is still unclear how Siglec-11 contribute to the well-documented roles that microglia and polySia have in cellular function including synaptic plasticity, neurogenesis, immune cell recruitment and neuropsychiatric diseases, such as Alzheimer’s disease (Schnaar et al. 2014; Garcez et al. 2017; Keren-Shaul et al. 2017; Sarlus and Heneka 2017; Butovsky and Weiner 2018). Given that Siglec-11 is known to regulate immune responses to a human-specific pathogen E. coli K1 through its interaction with polySia (Schwarz et al. 2017), it is intriguing to speculate that, in the brain, human microglial Siglec-11, through its binding to polySia, may provide a targeted mechanism to regulate neuroinflammation and other uniquely human neurodegenerative conditions. Based on our current data, we propose that microglial Siglec-11 may affect cellular function in the brain both locally and at a distance. However, more work is needed to understand how the modification and secretion are regulated, for example, the contribution of specific cysteine palmitoylation, and how they impact microglial function.
Similar questions remain for the paired receptor of Siglec-11, Siglec-16. Unlike Siglec-11, which is inhibitory, Siglec-16 is activating. Unlike SIGLEC-11, SIGLEC-16 is present in human populations largely as the nonfunctional SIGLEC-16P (Cao et al. 2008). The high prevalence of the nonfunctional SIGLEC16P allele and the fixation of the converted SIGLEC11 in human populations suggest that the dimorphism of Siglec-11/Siglec-16 paired receptors, such as Siglec-11/Siglec-16 and/or Siglec-11/Siglec-16P, have been maintained under some evolutionary constraint in the human lineage. Future work will help to elucidate the functional consequences of the dimorphism and how Siglec-16 and Siglec-11 interaction regulate immune reactivity in the brain.
Materials and methods
Plasmid preparations
All primers were synthesized by Eton Bioscience, Inc. (CA, USA). All polymerase chain reaction (PCR) was performed with Phusion High-Fidelity DNA Polymerase (NEB (MA, USA) with Mastercycler pro 6321 [Eppendorf (Hamburg, Germany)]. All PCR products were purified with QIAquick Gel Extraction Kit [QIAGEN (Venlo, The Netherlands)]. Plasmid miniprep was performed with E.Z.N.A. Plasmid Mini Kit I [OMEGA Bio-tek (GA, USA)] and plasmid maxiprep was performed with HiSpeed Plasmid Maxi Kit (QIAGEN). All plasmids sequences were confirmed by GENEWIZ (CA, USA). IRES2 fragment was amplified from pIRES2-EGFP plasmid with primers CMV-F and IRES2-Puro-R. Puromycin resistant gene fragment was amplified from pSpCas9(BB)-2A-Puro with primers IRES2-Puro-F and Puro-R. pcDNA3.1-V5 plasmid was linearized by PCR with CMV-R and PuroCterm-F. Amplified fragments were combined with NEBuilder HiFi DNA Assembly Master Mix (NEB) and prepared pcDNA3.1-V5-IRES2(PuroR) plasmid. pcDNA3.1-V5-IRES2(PuroR) plasmid was linearized by PCR with Kozak-R and IRES2-F. Human Siglec-11v1 (the tissue macrophage form) and v2 (the brain microglia form) full length gene was amplified from pcDNA3.1(+)-hSiglec11v1 and pcDNA3.1(+)-hSiglec11v2 by PCR with Kzk-hSig11-F and hSig11-IRES2-R. Amplified fragments were combined with NEBuilder HiFi DNA Assembly and prepared pcDNA3.1-hSiglec11v1-V5-IRES2(PuroR) and pcDNA3.1-hSiglec11v2-V5-IRES2(PuroR) plasmid. Chimpanzee Siglec-11 partial ECD was amplified from pcDNA3.1(+)-cSiglec11(3D)-hFc with Kzk-cSig11-F and cSig11(3D)-R and partial ECD, transmembrane and intracellular domain was synthesized by GENEWIZ. Amplified fragments were combined with NEBuilder HiFi DNA Assembly and prepared pcDNA3.1-cSiglec11-V5-IRES2(PuroR). Truncated mutants were prepared by PCR [hSig11 Truncate-R and each Forward primer set, C1-4-TM; C2-1-F, C2-4-TM; C2-2-F, C3-4-TM; C2-3-F, C4-TM; C2-4-F, TM; TM-F with pcDNA3.1-hSiglec11v1-V5-IRES2(PuroR), C1-3-TM; C2-1-F, C2-3-TM; C2-2-F and C3-TM; C2-3-F with pcDNA3.1-hSiglec11v2-V5-IRES2(PuroR)] and digested with DpnI and BamHI, then self-ligation by NEBuilder HiFi DNA Assembly. N-glycan mutants were point mutated from pcDNA3.1-hSiglec11v1-V5-IRES2(PuroR) by PCR (N1Q; N1Q-F and N1Q-R, N2Q; N2Q-F and N2Q-R, N3Q; N3Q-F and N3Q-R, N4Q; N4Q-F and N4Q-R, N5Q; N5Q-F and N5Q-R, N6Q; N6Q-F and N6Q-R, N7Q; N7Q-F and N7Q-R) and following that, pcDNA3.1-hSiglec11v1(N6Q)-V5-IRES2(PuroR) was mutated by PCR (N6,7Q; N7Q-F and N7Q-R), then digested with DpnI and self-ligated with endogenous E. coli ligase; (N1Q, N2Q, N4Q and N5Q), with phosphorylated by T4 Polynucleotide Kinase (NEB) and T4 DNA Ligase (NEB); (N3Q, N6Q, N7Q and N6,7Q). Sequences Human Fc fragment were amplified from pIRES2-hNCAM-hFc plasmid and with primers CH1-F and CH1-R, CH1-2-F and CH2-3-R, and CH2-3-F and CH3-R and combined by overlapping PCR with CH1-F and CH3-R. Following that, hFc fragment was amplified with hFc-F and hFc-IRES2-R. pIRES2-IRES2(EGFP) plasmid was linearized by PCR with pIRES2-hFc-R and IRES2-F. Amplified fragments were combined with NEBuilder HiFi DNA Assembly and prepared pIRES2-hFc-IRES2(EGFP) plasmid. pIRES2-hFc-IRES2(EGFP) plasmid was linearized by PCR with Kozak-R and hFc-F. Transin signal peptide fragment was amplified from pcDNA3.1-pA-hST8SIA2 with kzk-tSP-F and tSP-hFc-R. Amplified fragments were combined with NEBuilder HiFi DNA Assembly and prepared pIRES2-tSP-hFc-IRES2(EGFP) plasmid. Similarly, hSiglec11v1, hSiglec11v2 and cSiglec11 ECD were amplified from pcDNA3.1-hSiglec11v1-V5-IRES2(PuroR), pcDNA3.1-hSiglec11v2-V5-IRES2(PuroR) and pcDNA3.1-cSiglec11-V5-IRES2(PuroR) plasmid by PCR with each primer set [hSiglec11v1; Kzk-hSig11-F and h/cSig11(5D)-hFc-R, hSiglec11v2; Kzk-hSig11-F and hSig11(4D)-hFc-R, cSiglec11; Kzk-cSig11-F and h/cSig11(5D)-hFc-R]. Each fragment was combined with NEBuilder HiFi DNA Assembly and prepared pIRES2-hSiglec11(5D)-hFc-IRES2(EGFP), pIRES2-hSiglec11(4D)-hFc-IRES2(EGFP) and pIRES2-cSiglec11(5D)-hFc-IRES2(EGFP). Essential arginine mutant of pIRES2-hSiglec11(4D)-hFc-IRES2(EGFP) was prepared by point mutation with the following primer sets (R120K; R120K-F and R120K-R, R120A; R120A-F and R120A-R).
The partial FLOT1 gene was cloned from HEK293 cell cDNA with FLOT1-F and FLOT1(183)-R. EGFP fragment was amplified from pIRES2-EGFP plasmid with primers F183-EGFP-F and EGFP-P2A-R. P2A fragment was amplified from pSpCas9(BB)-2A-Puro with EGFP-P2A-F and P2A-Puro-R. pcDNA3.1-IRES2(PuroR) was linearized with kzk-FLOT1-R and PuroR-F and combined all fragments with NEBuilder HiFi DNA Assembly and prepared pcDNA3.1-FLOT183EGFP-P2A-PuroR. pcDNA3.1-FLOT183EGFP-P2A-PuroR was linearized by PCR with Kozak-R and IRES2-F183-F. hSiglec11v2-V5-IRES2 fragment was amplified from pcDNA3.1-hSiglec11v2-V5-IRES2(PuroR) with Kzk-hSig11-F and IRES2-R. Linearized plasmid and the fragment were combined with NEBuilder HiFi DNA Assembly and prepared pcDNA3.1-hSiglec11v2-V5-IRES2(FLOT183EGFP-P2A-PuroR).
Cell culture and transfections
All medium and PBS were purchased from Thermo Fisher Scientific (MA, USA). Human embryonic kidney HEK293 cells and human neuroblastoma SH-SY5Y cells were maintained in Advanced Dulbecco’s modified Eagle’s medium (DMEM) containing 1% fetal bovine serum (FBS) and were transferred to new culture dishes and grown in a 5% CO2 incubator at 37°C until they reached 70% confluency. For purification of Fc-fused protein and culture medium analysis, Basal media [50% RPMI 1640, 50% DMEM containing 1% antibiotic/antimycotic (Invitrogen), 2 mM L-glutamine, 1% Nutridoma and 1 mM sodium pyruvate] were used. For exosome purification and binding experiments, FBS was ultracentrifuged at 100,000 g, 4°C, 70 min by Optima XPN-80 Ultracentrifuge with SW41Ti rotor and 331372 Thinwall Polypropylene Tube [Beckman Coulter (CA, USA)] and Exosome-free FBS was used instead of normal FBS. All plasmids were transfected with Polyethyleneimine (PEI) according to the manufacturer’s instructions. Transient transfected cells were cultured for either 3 or 5 days, and stably transfected gene expression cells were selected with puromycin (1 μg/mL) for a week.
In vitro binding assay (ELISA)
HEK293A cells were cultured for 5 days after the transfection of plasmid for Fc-fused protein. Culture media was removed and incubated with protein A-Sepharose [GE healthcare (IL, USA)] overnight. The incubated resins were packed into Poly-Prep Chromatography Columns [Bio-Rad (CA, USA)] and supernatants were reapplied to the column for binding Fc-fused protein. The resin was washed with 10 mM Tris–HCl (pH 8.0), 150 mM NaCl and exchange buffer to 20 mM HEPES (pH 7.0) with 25 mU of Arthrobacter ureafaciens sialidase and cleaved sialic acid at room temperature (r.t.) for 1 h. The resin was washed again, and the Fc-fused protein was eluted with 0.1 M glycine buffer (pH 3.0). The eluted Fc-fused protein was neutralized with 1 M Tris–HCl (pH 8.0) and exchanged buffer to PBS and concentrated by EMD Millipore Amicon Ultra-15 Centrifugal Filter Units (Thermo Fisher Scientific).
1 mg/mL PolySia (colominic acid from E. coli K1) [EY Laboratories (CA, USA)] dissolved in 20 mM Phosphate buffer (pH 7.4) and applied 50 μL to 96 well polystyrene plate [Corning #3370 (NY, USA)] and evaporated at 37°C overnight. The plate was blocked with 2% BSA/PBS at r.t. for 1 h and applied 0.2 μg in 50 μL of each Fc-fused protein (in 2% BSA/PBS) at r.t. for 1 h. Nonbinding proteins were washed 5 times with PBS containing 0.05% Tween-20 (PBST). Fifty microliter of 1/15,000 diluted Goat Anti-Human (H L) HRP conjugate (#1721050, Bio-Rad) was applied to each well and incubated at r.t. for 1 h and washed five times with PBST, then 100 μL BD OptEIA [Becton, Dickinson and Company (NJ, USA)] was added. Reaction was stopped with 50 μL of 2 M H2SO4. 450 nm absorbance values were measured by EnSpire Alpha Plate Reader [PerkinElmer (MA, USA)]. For binding inhibition assay, Fc-fused proteins was mixed with 0 to 5 mg/mL polySia or as a negative control, 5 mg/mL Hyaluronic acid [Sigma-Aldrich (MO, USA)].
In vitro binding assay (cell staining)
5 × 105 cells were cultured on PEI (25 μg/mL) coated 18 mm × 18 mm slide coverslip. Cells were fixed with 4% paraformaldehyde at r.t. for 8 min and treated with boiled and heat-inactivate/intact active endo-N-acylneuraminidaseF (Endo-NF; gift from Prof. Rita Gerardy-Schahn at Hannover Medical School, Germany). The fixed cells were blocked with 2% bovine serum albumin in PBS, then incubated with anti-polySia antibody (mouse IgG, 20 μg/mL), hFc (100 μg/mL) or hSig11(4D)-hFc (100 μg/mL) at r.t. for 1 h. After washing three times with PBS, the cells were incubated with goat antimouse IgG-PE (2 μg/mL; Thermo Fisher Scientific) for anti-polySia, goat anti-Human IgG Fc-PE (2 μg/mL; Thermo Fisher Scientific) at r.t. for 30 min. The cells were washed three times with PBS and washed with water, then mounted with ProLong Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific). Cells were then observed under a fluoromicroscope [ZEISS Axio Observer d1, Carl Zeiss (Oberkochen, Germany)]. The ratio of anti-polySia/DAPI staining and Fc-fused protein/DAPI for five random areas were measured using ImageJ software, and the data were processed.
SDS-PAGE and western blotting
Cells were incubated with RIPA buffer [Cell Signaling Technology (MA, USA)] and 1 mM PMSF on ice for 1 h. They were then centrifuged, and the protein concentration of the supernatants (cell lysates) were evaluated by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Samples were dissolved in Laemmli buffer with/without (reducing/nonreducing condition) 5% 2-mercaptoethanol and were subsequently incubated at 4°C for 30 min to avoid aggregation. For experiments in Figure 3, samples were incubated at 100°C for 5 min (standard protein denature condition), 60°C for 20 min (standard glycoprotein denature mild condition), 37°C for 30 min, 25°C for 30 min or 4°C for 30 min. The cold denatured samples were then electrophoresed on CosmoPAGE TG Precast Gel 4-15% [Nacalai USA (CA, USA)] and electroblotted onto Immobilon-FL Membrane [Merck Millipore (MA, USA)] using Trans Blot SD (Bio-Rad). Following the transfer, membranes were blocked at r.t. for 1 h with Odyssey Blocking Buffer (PBS) [LI-COR Biosciences (NE, USA)]. The membranes were then incubated at r.t. for 1 h with the primary antibody. Antibodies used included: anti-polySia735 (gift from Prof. Rita Gerardy-Schahn at Hannover Medical School, Germany; 1:3000), 4C4 antibody (BioLegend, 1:1000), mouse V5-tag antibody (Invitrogen, 1:3000), rabbit FLOT1 antibody (Abcam, 1:1000), β-actin antibody (Cell Signaling; 1:1000), anti-GFP antibody (Novus Biologicals, 1:1000). After membranes were washed with PBS three times, 10 min each, the secondary antibody IRDye 800 mouse IgG (LI-COR Biosciences, 1:5000) or IRDye 680 rabbit IgG (LI-COR Biosciences, 1:5000) was incubated at r.t. for 30 min and bands were visualized with LI-COR Odyssey Imaging System (LI-COR Biosciences).
PNGase F treatment
PNGase F was purchased from New England Biolabs (NEB; Ipswich, MA). The experiment was carried out according to manufacturer protocol with nondenaturing reaction condition.
Exosomes and proteolytic products secretion
Cells were cultured in basal media for 5 days, then collected and lysed. The culture medium was collected, and the cell debris were removed. Following the removal, the medium was concentrated with Amicon Ultra 10KD Centrifugal Filter Units (Thermo Fisher Scientific). To inhibit the exosome secretion, cells were cultured in basal media with 20 μM of exosome inhibitor (sphingomyelinase inhibitor) GW4869 (Sigma-Aldrich) for 24 h. To inhibit the proteolytic cleavage, cells were cultured with 1/500 protease inhibitor cocktail (Sigma-Aldrich)/20 μM of matrix metalloproteinases inhibitor; GM6001[Selleckchem (TX, USA)]/100 nM of beta secretase inhibitor; KMI-1303(Wako (Osaka, JAPAN)] for 12 h.
hSigec-11(4D)-F1-EGFP binding assay
pcDNA3.1-FLOT183EGFP-P2A-PuroR or pcDNA3.1-hSiglec11v2-V5-IRES2 (FLOT183EGFP-P2A-PuroR) was transfected into HEK293A cells. Cells were cultured in Exosome-free DMEM for 5 days. The culture medium was collected, and the cell debris were removed. Following the removal, the medium was added to Endo-NF treated 2 × 105 SH-SY5Y cells (cells were cultured with heat-inactivate/intact active Endo-NF and cultured for 8 h.). Cells were fixed with 4% paraformaldehyde at r.t. for 8 min. The cells were washed three times with PBS and washed with water, then mounted and imaged. For Endo-NF control slides, the fixed cells were stained with anti-polySia antibody. The ratio of anti-polySia/DAPI staining and EGFP/DAPI for five random areas were measured using ImageJ software.
Exosome isolation
Cells were cultured in basal media for 5 days, then collected and lysed. The culture media was collected, and the cell debris was removed. Following that, the media was ultracentrifuged (100,000 g, 4°C, 70 min) and extracellular vesicles were isolated as pellets. The pellets were dissolved in 0.5 mL of 2.5 M sucrose/20 mM HEPES (pH 7.4). For further purification, a sucrose gradient [0.25–2.5 M (1.03–1.32 g/mL) sucrose] was prepared over the dissolved pellet and centrifuged at 100,000 g for 5 h. Ten fractions were collected and washed with 20 mM HEPES, and repelleted by ultracentrifugation at 100,000 g for 5 h. The pellets were dissolved in Laemmli buffer and analyzed by western blotting.
Prediction of posttranslational modification of Siglec-11 protein
N-glycosylation sites were predicted using NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc/).
Acyl-PEG exchange analysis
The protocol was carried out similarly to what was previously described by Percher et al. (2017). Briefly, cells were lysed with Lysis Buffer (4% SDS, 5 mM EDTA, 50 mg/mL PMSF, 10 mg/mL leupeptin, 10 mg/mL pepstatin A in PBS) and 8 M urea, the lysate was incubated on ice for 1 h. The centrifuged supernatant was reduced with 25 mM TCEP for 1 h at r.t. and the free cysteine residues were alkylated with 50 mM N-ethylmaleimide (NEM) for 3 h at r.t. After chloroform/methanol precipitation (CM ppt), proteins were resuspended in Resuspend Buffer (4% SDS and 5 mM EDTA in PBS) and incubated with Palm Buffer (1% SDS, 5 mM EDTA, 1 M NH 2OH, pH 7.0) for 1 h at 37°C. After 3 CM ppts, proteins were resuspended in PBS with 4% SDS and PEGylated with 20 mM mPEG-5 kDa for 1 h at r.t. As a negative control, 20 mM NEM was used instead of mPEG-5 kDa. After CM ppt, protein was dissolved in Laemmli buffer with 5% 2-ME and were subsequently incubated at 4°C for 30 min. The samples were analyzed by SDS-PAGE and western blotting as described above.
Funding
This work was support by a grant from Aviceda Therapeutics (D.Y.C.), NIH grants K12HL141956 (D.Y.C.) and R01GM32373 (A.V.).
Conflict of interest statement
There is no conflict of interest.
Supplementary Material
Contributor Information
Masaya Hane, Departments of Medicine and Cellular & Molecular Medicine, Center for Academic Research and Training in Anthropogeny, Glycobiology Research and Training Center, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA.
Dillon Y Chen, Departments of Medicine and Cellular & Molecular Medicine, Center for Academic Research and Training in Anthropogeny, Glycobiology Research and Training Center, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA.
Ajit Varki, Departments of Medicine and Cellular & Molecular Medicine, Center for Academic Research and Training in Anthropogeny, Glycobiology Research and Training Center, University of California San Diego, 9500 Gilman Dr, La Jolla, CA 92093, USA.
References
- Angata T. 2018. Possible influences of endogenous and exogenous ligands on the evolution of human Siglecs. Front Immunol. 9:2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T, Kerr SC, Greaves DR, Varki NM, Crocker PR, Varki A. 2002. Cloning and characterization of human Siglec-11. A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem. 277:24466–24474. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Weiner HL. 2018. Microglial signatures and their role in health and disease. Nat Rev Neurosci. 19:622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Lakner U, de Bono B, Traherne JA, Trowsdale J, Barrow AD. 2008. SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans. Eur J Immunol. 38:2303–2315. [DOI] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. 2007. Siglecs and their roles in the immune system. Nat Rev Immunol. 7:255–266. [DOI] [PubMed] [Google Scholar]
- de Gassart A, Geminard C, Fevrier B, Raposo G, Vidal M. 2003. Lipid raft-associated protein sorting in exosomes. Blood. 102:4336–4344. [DOI] [PubMed] [Google Scholar]
- Draper JM, Xia Z, Smith CD. 2007. Cellular palmitoylation and trafficking of lipidated peptides. J Lipid Res. 48:1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez ML, Mina F, Bellettini-Santos T, Carneiro FG, Luz AP, Schiavo GL, Andrighetti MS, Scheid MG, Bolfe RP, Budni J. 2017. Minocycline reduces inflammatory parameters in the brain structures and serum and reverses memory impairment caused by the administration of amyloid β (1-42) in mice. Prog Neuropsychopharmacol Biol Psychiatry. 77:23–31. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Angata T, Lewis AL, Mikkelsen TS, Varki NM, Varki A. 2005. A human-specific gene in microglia. Science. 309:1693. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Khedri Z, Schwarz F, Landig C, Liang SY, Yu, Chen X, Fujito NT, Satta Y, Varki A et al. 2017. Coevolution of Siglec-11 and Siglec-16 via gene conversion in primates. BMC Evol Biol. 17:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlstetter M, Kopatz J, Aslanidis A, Shahraz A, Caramoy A, Linnartz-Gerlach B, Lin Y, Lückoff A, Fauser S, Düker K et al. 2017. Polysialic acid blocks mononuclear phagocyte reactivity, inhibits complement activation, and protects from vascular damage in the retina. EMBO Mol Med. 9:154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B et al. 2017. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 169:1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- Khan N, Kim SK, Gagneux P, Dugan LL, Varki A. 2020. Maximum reproductive lifespan correlates with CD33rSIGLEC gene number: Implications for NADPH oxidase-derived reactive oxygen species in aging. FASEB J. 34:1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnartz B, Wang Y, Neumann H. 2010. Microglial immunoreceptor tyrosine-based activation and inhibition motif signaling in neuroinflammation. Int J Alzheimers Dis. 2010:587463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percher A, Thinon E, Hang H. 2017. Mass-tag Labeling using acyl-PEG exchange for the determination of endogenous protein S-fatty acylation. Curr Protoc Protein Sci. 89:14.17.1–14.17.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarlus H, Heneka MT. 2017. Microglia in Alzheimer’s disease. J Clin Invest. 127:3240–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Hane M. 2018. Mental disorders and an acidic glycan-from the perspective of polysialic acid (PSA/polySia) and the synthesizing enzyme, ST8SIA2. Glycoconj J. 35:353–373. [DOI] [PubMed] [Google Scholar]
- Sato C, Hane M, Kitajima K. 2016. Relationship between ST8SIA2, polysialic acid and its binding molecules, and psychiatric disorders. Biochim Biophys Acta. 1860:1739–1752. [DOI] [PubMed] [Google Scholar]
- Sato C, Kitajima K. 2013. Impact of structural aberrancy of polysialic acid and its synthetic enzyme ST8SIA2 in schizophrenia. Front Cell Neurosci. 7:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaar RL, Gerardy-Schahn R, Hildebrandt H. 2014. Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol Rev. 94:461–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, Landig CS, Siddiqui S, Secundino I, Olson J, Varki N, Nizet V, Varki A. 2017. Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen. EMBO J. 36:751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahraz A, Kopatz J, Mathy R, Kappler J, Winter D, Kapoor S, Schütza V, Scheper T, Gieselmann V, Neumann H. 2015. Anti-inflammatory activity of low molecular weight polysialic acid on human macrophages. Sci Rep. 5:16800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Merwe PA, Crocker PR, Vinson M, Barclay AN, Schauer R, Kelm S. 1996. Localization of the putative sialic acid-binding site on the immunoglobulin superfamily cell-surface molecule CD22. J Biol Chem. 271:9273–9280. [PubMed] [Google Scholar]
- Varki A, Schnaar RL, Crocker PR. 2017. I-Type Lectins. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH et al., editors. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Vinson M, Van der Merwe PA, Kelm S, May A, Jones EY, Crocker PR. 1996. Characterization of the sialic acid-binding site in sialoadhesin by site-directed mutagenesis. J Biol Chem. 271:9267–9272. [DOI] [PubMed] [Google Scholar]
- Wang X, Chow R, Deng L, Anderson D, Weidner N, Godwin AK, Bewtra C, Zlotnik A, Bui J, Varki A et al. 2011. Expression of Siglec-11 by human and chimpanzee ovarian stromal cells, with uniquely human ligands: Implications for human ovarian physiology and pathology. Glycobiology. 21:1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mitra N, Cruz P, Deng L, NISC CSP, Varki N, Angata T, Green ED, Mullikin J, Hayakawa T et al. 2012. Evolution of siglec-11 and siglec-16 genes in hominins. Mol Biol Evol. 29:2073–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Neumann H. 2010. Alleviation of neurotoxicity by microglial human Siglec-11. J Neurosci. 30:3482–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


