Abstract
Various master key regulators (MKRs) that control a binary switch of sex determination (SD) have been found in fish; these provide an excellent model for the study of vertebrate genetic SD. The SD region in flathead grey mullet has been previously mapped to a 1 Mbp region harboring 27 genes, of which one is follicle-stimulating hormone receptor (fshr). Although this gene is involved in gonad differentiation and function, it has not been considered as an MKR of SD. We systematically investigated polymorphism in mullet fshr using DNA shotgun sequences, and compared them between males and females. Capable of encoding nonconservative amino acid substitutions, c.1732G>A and c.1759T>G exhibited association with sex on a population level (N = 83; P ≤ 6.7 × 10−19). Hence, 1732 A and 1759 G represent a male-specific haplotype of the gene, designated as “fshry.” Additional flanking SNPs showed a weaker degree of association with sex, delimiting the SD critical region to 143 nucleotides on exon 14. Lack of homozygotes for fshry, and the resulting divergence from Hardy–Weinberg equilibrium (N = 170; P ≤ 3.9 × 10−5), were compatible with a male heterogametic model (XY/XX). Capable of replacing a phenylalanine with valine, c.1759T>G alters a conserved position across the sixth transmembrane domain of vertebrate FSHRs. Amino acid substitutions in this position in vertebrates are frequently associated with constant receptor activation and consequently with FSH/FSHR signaling alteration; thus, indicating a potential role of fshr as an MKR of SD.
Keywords: sex determination, XX/XY, bony fishes, Genetics of Sex, gonadotropin receptor
Introduction
Sex dimorphism in gonochoristic species is associated with the differentiation of gonads, but also includes other important behavioral, morphological, and physiological characteristics (Williams and Carroll 2009). Sex determination (SD) is variable and can involve genetic, environmental, and social factors. Studies of species with a genetic SD system, show that in most cases a single gene plays the role of a master key regulator (MKR), initiating the dichotomic division of sexes (males and females). Different MKRs were found in vertebrate and insect taxa including sry (Gubbay et al. 1990; Sinclair et al. 1990), sox3 (Takehana et al. 2014), amh (Eshel et al. 2014; Li et al. 2015), amhr2 (Kamiya et al. 2012), dmrt1 (Raymond et al. 2000; Matsuda et al. 2002; Nanda et al. 2002), gsdf (Myosho et al. 2012), hsd17b1 (Koyama et al. 2019), and sdY (Yano et al. 2012), and in many cases these are found to be associated with gene duplications (Eshel et al. 2014; Curzon et al. 2020). In Table 1, we surveyed the types of reported sequence variation of SD genes in vertebrates. Copy-number variations (CNVs) were the underlying polymorphism in eight instances whereas nonstructural polymorphisms were evident in five SD genes.
Table 1.
Nonstructural versus structural sequence variation of SD genes in vertebrates
| Paralog | SD gene | Organism | Variationa | References |
|---|---|---|---|---|
| sox3 | SRY-related HMG-box 3 (sox3) | Oryzias dancena and other Oryzias species | NSV | Takehana et al. (2014); Myosho et al. (2015) |
| sex determining region Y (sry) | Mammals (most) | MSD | Gubbay et al. (1990) and Sinclair et al. (1990) | |
| amhr2 | Anti-Mullerian hormone receptor type 2 (amhr2) | Takifugu rubripes | NSV | Kamiya et al. (2012) |
| Perca flavescens | MSD | Feron et al. (2020) | ||
| gsdf | gonadal somatic cell derived factor (gsdf) | Oryzias luzonensis | NSV | Myosho et al. (2012) |
| hsd17b | hsd17b1 | Seriola genus | NSV | Koyama et al. (2019) |
| dmrt1 | Y-specific DM-domain (DM-Y) | Oryzias latipes | MSD | Matsuda et al. (2002), Nanda et al. (2002), Yoshimoto et al. (2008), Smith et al. (2009), Chen et al. (2014), Cui et al. (2017), and Mustapha et al. (2018) |
| dmrt1 | Scatophagus argus | |||
| W-linked DM-domain (DM-W) | Xenopus laevis | FSD | ||
| Doublesex and mab-3 related transcription factor 1, (dmrt1) | Gallus gallus, Cynoglossus semilaevis | |||
| irf9 | sexually dimorphic on the Y-chromosome (sdY) | Salmonids | MSD | Yano et al. (2013) |
| gdf6 | gdf6Y | Nothobranchius furzeri | NSV | Reichwald et al. (2015) |
| zk | zinc knuckle on the Y chromosome (zkY) | Gadus morhua | MSD | Kirubakaran et al. (2019) |
| amh | Anti-Mullerian hormone (amh) | Odontesthes hatcheri, Oreochromis niloticus, Ophiodon elongates, Hypoatherina tsurugae, Esox lucius | MSD | Hattori et al. (2012), Eshel et al. (2014), Rondeau et al. (2016), Bej et al. (2019), and Pan et al. (2019) |
Sequence variation types are non-structural (NSV) and structural sequence variation including CNV of male (MSD) or female (FSD) specific gene duplications.
The existence of common MKRs for SD in taxonomically distant species suggests that the number of MKRs is limited to a set of factors, which belong to a conserved pathway (Marshall Graves and Peichel 2010; Graves 2013) that triggers the primary differentiation of the bipotential gonad up to sex hormone synthesis (Kim and Capel 2006). However, the surprising finding of an MKR that had not been previously associated with sex differentiation, the immune-related gene (sdY) in salmonids (Yano et al. 2012, 2013), indicates a more complex regulation of SD (Herpin and Schartl 2015). SD among different fish is considered to be extremely variable, even between families of the same genus for which different MKRs have been found, e.g. Oryzias (Matsuda and Sakaizumi 2016), Cichlidae (Ser et al. 2010), and zebrafish (Kossack and Draper 2019). Thus, fish are an excellent model for the elucidation of the SD pathway and the vertebrate sex differentiation cascade (Baroiller et al. 1999; Schartl 2004; Capel 2017).
The flathead grey mullet (Mugil cephalus) is the most widespread fish of the Mugilidae family and populates both subtropical and temperate coastal waters in the major oceans. Despite its abundance and the fact that it has adapted to a wide range of environments, it is not fully domesticated and fry-production for aquaculture is mainly dependent on collection from river estuaries. The flesh and roe of this mullet are highly prized commodities in Asian and Mediterranean markets (Aizen et al. 2005). The ovaries, processed with salt, are sold as ‘Botarga Caviar’ or as ‘Karasumi’ in Asia, and are considered a delicacy in countries around the Mediterranean, Japan, and Taiwan (Hung and Shaw 2006; Katselis et al. 2006). Thus, on account of a faster growth rate and the high value of their products, production of all-female populations is economically advantageous.
The addition of sex steroids to larva feed can cause sex inversion in many fish species, and is often practiced in commercial production, giving rise to environmental and food safety concerns (Mlalila et al. 2015; Chen et al. 2018). These worries could be allayed by technologies for mono-sex production through parent sex-reversal that do not require the use of steroids. Dor et al (2016) analyzed the sex of M. cephalus progeny in two independent families, and based on genetic markers, showed that the alleles linked with maleness were transmitted from the fathers, indicating that mullets have a genomic XX/XY SD system. Thus, a female mono-sex population can be constructed by female-to-male sex reversal, followed by mating of the XX males with females. As opposed to sex reversing fish that go directly for human consumption, mono-sex production based on such mating presents an environmental friendly and safer alternative. Since sex reversal occurs at an early age prior to the appearance of any external signs of morphological sex differentiation, a genetic marker for SD is required to discriminate between sex-reversed and normal individuals of the flathead grey mullet. The most reliable genetic marker for sex would be the causal variation in the MKR of SD.
Based on synteny with the Nile tilapia (Oreochromis niloticus) genome, the SD region of M. cephalus was initially mapped to a 1.8 Mbp region (Dor et al. 2016). A follow-up study fine-mapped the SD region down to a 1.0 Mbp region on mullet LG9, harboring 27 genes with an enriched set of genes showing sex-biased gene expression (Dor et al. 2020). Although 25 sequence-based markers in 12 of these genes were analyzed, on a population level, none of these polymorphisms exhibited association with sex. One of the genes mapped in the region was Follicle-stimulating hormone receptor (fshr), indicating it as a candidate MKR gene for SD, based on its putative position and relevant function in sex differentiation (Murozumi et al. 2014; Dor et al. 2020). Although the use of the candidate gene approach in the analysis of the genetic control of human diseases has not been effective (Altshuler et al. 2008), the fact that the majority of discovered SD genes in vertebrates have a known function in reproduction, suggests that this approach may be suitable for the search for MKRs of SD (Shirak et al. 2006; Eshel et al. 2014).
In this study, we used our established genomic libraries (Dor et al. 2016, 2020) and resequencing data of fshr in M. cephalus to investigate its role in SD. A panel of wild type (WT) fish and full-sib families was used to test the association of fshr polymorphism with sex. Our findings propose a new MKR in teleost fish, and thus contribute to a broader understanding of the vertebrate SD cascade.
Materials and methods
Fish sampling and sexing
Adult flathead grey mullet specimens from two full-sib families (n = 6 and n = 7) and from a wild population (n = 70) were dissected and sexed based on external observation of gonad morphology (n = 83). The wild-sexed population consisted of fish that have been reared by commercial growers from fry caught in river estuaries along the Israeli Mediterranean coast in years 2016 (n = 46), 2017 (n = 7), and 2018 (n = 12), and of fish reared from fry caught in Ebro River, Spain (n = 5). Of these fish, the gonads of 27 individuals (17 males and 10 females) were also examined microscopically in order to improve confidence in the visual sexing. A caudal fin sample was taken from each of the adult specimens and preserved in ethanol (90–96%). Fin samples were also taken from two groups of 28 dph larvae (Group A, n = 7; Group B, n = 8, Dor et al. 2020), and from 87 nonsexed fingerlings of a wild population caught in year 2019 in river estuaries along the Israeli Mediterranean coast.
Sequence assembly of fshr in M. cephalus using whole-genome sequencing
Mugil cephalus fshr raw reads and previously built SOAPdenovo scaffolds (Dor et al. 2016, 2020) of a male and female were assembled separately. The Gap5 (Staden et al. 2000) wrapper and the BWA program were used for alignment of reads and scaffolds against the masked gene sequence of Dicentrarchus labrax Fshr (GenBank accession no. FQ310507). Masking was performed using CENSOR (Kohany et al. 2006) and Tilapia Repeat Masker version 2 (Shirak et al. 2010). Aligned sequences were then used for de novo assembly using Gap4 (Staden et al. 2000). Raw reads and scaffolds were used again for alignment against the newly assembled sequences using Gap5. A consensus sequence was obtained and used for further alignment. This process was repeated several times until the entire gene assembly was completed. Exon borders were deduced using mRNA sequences that had been previously built using the Trinity software (Dor et al. 2020).
DNA extraction and sequencing procedure
A sample of the caudal fin (100–200 mg) was used for DNA extraction using a commercial kit (MasterPure DNA Purification, Madison). Polymerase chain reaction (PCR) was performed using relevant primers that were designed using Primer3 (Untergasser et al. 2012) (Table 2), and the Bio-X-ACT™ Long kit (Bioline Ltd., London, UK) according to the manufacturers’ instructions under the following conditions: 36 cycles for 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C. Thereafter, the PCR products were excised from the gel and purified with a kit (Montage Gel Extraction, Millipore, Bedford, MA). Sanger sequencing was conducted from both directions of the amplified genomic region. For the purpose of allele-specific PCR, “X” specific and “Y” specific forward primers were designed following the method of Liu et al. (2012), including a mismatch in the third nucleotide position from the 3' end. Annealing temperature was raised to 69°C.
Table 2.
PCR primer pairs
| Pair | Sequence (5ʹ to 3ʹ) | Product size (bp) | Target of amplificationa |
|---|---|---|---|
| 1 |
GGATGTGGAGGATCTGGTGT GAGACGGCGAAGAAGGAGAT |
221 | SD region (Exon 14) |
| 2 |
CGGAATCTAAGGTCCTCCTG GCCTTCTTCTTTGCCTGAGA |
491/481 | Upstream of SD region (Exon 14) |
| 3 |
GCCGGCCCACGCCGACACGCTCG GCCTTCTTCTTTGCCTGAGA |
618/608 | fshry |
| 4 |
GCCGGCCCACGCCGACACGCTCA GCCTTCTTCTTTGCCTGAGA |
618/608 | fshrx |
| 5 |
CGGTGACGATACTGGTGATG CTTCAATGGCTGCAGGACAC |
230 | Non-synonymous SNP (Exon 1) |
fshrx represents the putative non-specific allele of fshr, while fshry represents the male-specific allele.
Characterization of sex-specific alleles for fshr
Analysis of genomic alleles was based on whole- genome sequencing (WGS) of a male and female deposited in the Short Read Archive (SRA) under project IDs PRJEB12265 and PRJEB34342, respectively. Based on sequence variation along the assembled fshr gene and read-pair information, sex-specific alleles were manually separated using Gap5 viewer. The assembly of fshry (ENA accession LR860658) was also deposited in the BAM format, which provided further verification for each read assembly (over 3000 reads, ENA accession ERX4280005). In search for sequence variants, we preferentially targeted the gene regions that are likely to be functional as has been previously described (Dor et al. 2020). Briefly, most variants were excluded by observing their fitness to the XY system model in the male and female WGS data. Variants that fit the model were tested for association with sex by Sanger sequencing of sample of individuals from two families (families A1 and B, n = 17). Variants that passed this test were further examined on the population level by Sanger sequencing (n = 83, using primer pair #1, Table 2).
Fshr gene expression
Expression of previously established RNA-seq libraries (Dor et al. 2020) included two groups of larvae A and B (n = 7 and 8), female and male brains (n = 3 and 2, respectively), female and male gonads (n = 2 and 1, respectively). Existence of fshr variants were examined in these libraries, using the BLASTN algorithm for alignment with 32 bp words: GCATGGCTCAACGCATGGCCGTCCTCATCGTC and GCGTGGCTCAACGCATGGCCGTCCTCATCTTC that represented the male-specific allele of fshr (fshry) and the nonspecific allele of fshr (fshrx), respectively. All available M. cephalus SRA RNA-Seq submissions were screened including 92 runs (ERX3536078-ERX3536169, a total of 1,038,774,806 reads) originating from Israel (Dor et al. 2020) and 4 runs from other sources (SRX3153305 and SRX1817285-8, a total of 140,071,760 reads). No fshr reads were detected in the latter source, whereas 81 reads were found to contain the fshrx probe word, most of them (85%) in adult male testis. The fshr expression values were calculated and normalized in a previous study using Trimmed mean of values (TMM) (Dor et al. 2020).
Statistics
The JMP® statistical package (Pro 13, SAS Institute, Cary, NC) was used for conducting statistical tests. Fisher’s exact Chi-squared test was applied for an association study of individual genetic markers and sex. Pearson Chi-squared test was applied for testing deviation from Hardy–Weinberg equilibrium.
Data availability
DNA- and RNA-seq reads are available at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession numbers PRJEB12265 and PRJEB34342. The M. cephalus assembled and annotated transcriptome is available at http://cowry.agri.huji.ac.il/. Supplementary material includes Figure S1 Comparison of predicted Fshr of M. cephalus with other vertebrate FSHRs (complete alignment) and the annotated mullet fshr sequences (File S2). Supplementary material is available at figshare:
Results
Assembly of fshr in M. cephalus
Our established genomic libraries allowed the assembly of the whole fshr gene in two full-sibs of M. cephalus, representing both sexes. Separation of sequencing fragments based on polymorphism in these paired-read sequences revealed four different fshr alleles, with no evidence of a fifth allele that might indicate a CNV. Annotation based on the RNA-seq data (Dor et al. 2020) was used to characterize the fshr gene structure with its 14 exons, its conserved exon–intron boundaries, and its putative protein sequence, all of which were found similar to those of known orthologous FSHRs (Table 3, Supplementary Figure S1, ENA accession No. LR860658, Additional File 2). Using this predicted Fshr protein and the BLASTP program, we searched GenBank for the reference sequences of mammals, amphibians, birds, and reptiles. This search indicated that the assembled gene was fshr, i.e. 50% identity and 67% similarity to human FSHR, NP_000136; 51 and 66% similarity to clawed frog Fshr, XP_017949625; 50% identity and 66% similarity to duck FSHR, XP_005012152; and 51% identity and 65% similarity to python Fshr, XP_007431488. However, nomenclature for many similar proteins in other fish differed i.e. 77% identity and 84% similarity to Nile tilapia gonadotropin receptor I (gth-ri), NP_001266517; and 49% identity and 64% similarity to Fugu lutropin-choriogonadotropic hormone receptor, XP_029691459. Nevertheless, synteny analysis indicated that, in spite of the different nomenclatures, all these similarities arise from the same ancestral gene, i.e. Nile tilapia gth-ri, which is located on LG8 in a position orthologous to mullet fshr on LG9 (Dor et al. 2016). The mullet exonic fshr SNPs, which were capable of encoding amino acid changes, were marked for analysis, and sex-biased polymorphism was used to assemble the male-specific (single allele) and nonspecific sequences of fshr (three alleles), representing the “Y” and “X” genomic regions, designated as fshry and fshrx, respectively (Table 4; Figure 1). The complete coding sequences (cds) of the predicted transcripts encoded by these alleles were deposited in public database (ENA accession Nos. LR860659, LR860657, LR860660, LR860656).
Table 3.
Genomic organization of the M. cephalus fshr gene, based on the male fshry variant
| Introna | Exon |
Intron |
||
|---|---|---|---|---|
| No. | Size | Size | ||
| ATGATG | 1 | 188 | GTGCTTgtgagtacgc | 3,119 |
| tgcatccacagGGAACT | 2 | 72 | GAGACTgtaagcttta | 120 |
| tccaactttagGAACAT | 3 | 75 | TTACATgtgagtgtgt | 526 |
| gtctgccccagCTTAAT | 4 | 75 | TGAGATgtaagtataa | 107 |
| cctcctttcagAACCAT | 5 | 75 | GTTTATgtgagtactc | 885 |
| tctcgtcacagGACCAT | 6 | 75 | TCAGCTgtgagtctca | 319 |
| ttgacactcagTCACCT | 7 | 81 | GAAGATgtaagggctt | 1,506 |
| gctattgacagATGGCT | 8 | 69 | AAGACTgtgagtcaca | 101 |
| tttccccttagATTTCT | 9 | 75 | AGTGCTgtaagcatga | 380 |
| tgctcccacagTGACAT | 10 | 186 | AAACAAgtgcgcgtct | 115 |
| gtgtgtgctagATCAAG | 11 | 218 | TGTTGAgtatgaaatg | 140 |
| tccattgtcagGCAGCA | 12 | 182 | TTCTCGgtgtgtacgc | 97 |
| gtcacgtgcagGTGTTT | 13 | 192 | AGCAAAgtgggtgtta | 1,046 |
| tgttcgtgcagGTGAGC | 14 | 529 | CGTGCGTAG | |
Intron and exon sizes are given in base pairs, and their sequences are written in lowercase and uppercase letters, respectively. The first and last two bases of the introns are presented in bold type (gt and ag for donor and acceptor splice sites, respectively). The initiation and stop codons are shown in bold and underlined (ATG, TAG). Starting from the initiation codon, the genomic and putative transcript sizes of the fshry gene are 10,553 and 2,118 bp, respectively.
Table 4.
Polymorphism in M. cephalus predicted fshr gene
| Exon 1 | Exon 14a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positionb | 131 | 149 | 1689 | 1732 | 1759 | 1781 | 1875 | 1917 | 2031 | 2047 | 2112 |
| Nucleotide | T/C | T/C | C/A | G/A | T/G | C/A | C/T | C/T | G/A | G/A | C/A |
| AA | L/P | M/T | T/T | V/M | F/V | A/D | I/I | R/R | S/S | A/T | S/S |
| Malesc | 6/2 | 4/4 | 60/26 | 44/42 | 44/42 | 78/8 | 2/2 | 18/6 | 18/6 | 3/21 | 10/14 |
| Femalesc | 7/1 | 3/5 | 80/0 | 77/3 | 77/3 | 78/2 | 4/0 | 9/11 | 6/14 | 5/15 | 15/5 |
Two nonsynonymous SNPs comprising a haplotype that fits an XY model of SD are shown in bold underlined font. The “Y” fshry haplotype is presented on the right side of the divider.
Nucleotide position in the coding DNA sequence (in bp). Nucleotide and amino acid (AA) variation is denoted in both sides of divider.
Number of counts of alternative alleles in each position for samples of males and females, respectively.
Figure 1.
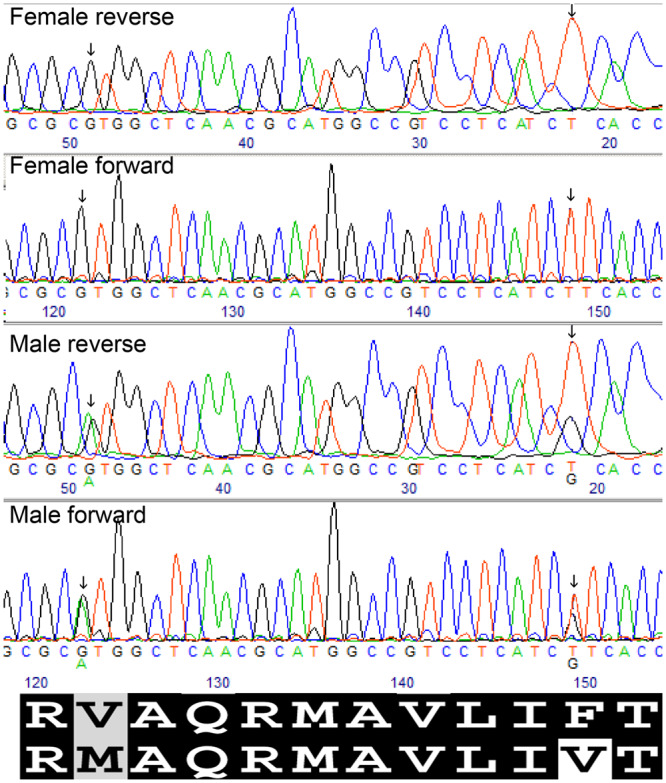
Sanger sequencing of the sex associated region of FSHR in exon 14 after amplification using PCR primers (pair 1, Table 2). SNPs (nt positions 1732 and 1759 of the predicted cds) are marked by an arrow. The fshrx (upper) and fshry (lower) putative translated amino acids are shown below the chromatograms. Partially conserved and highly conserved amino acid residues are indicated by a grey and black background, respectively. The white box indicates the novel male-specific amino acid substitution between the proteins (following the coloration presented in Figure 2).
Analysis of polymorphism in fshr
According to the predicted transcript, two nonsynonymous male-specific SNPs were found in exon 14 at 1732 and 1759 nucleotide positions of the fshr assembled sequence (Table 4). Using PCR primers (pair 1, Table 2), these SNPs were investigated in two full-sib families (n = 6 and n = 7), and were found to be fully associated with sex. Another two nonsynonymous SNPs in exons 1 and 14 (Table 4, nucleotide positions at 131 and 2047 bp, respectively) were found in fshr’s assembly; however, these did not match the expected XY/XX SD model, since in the two established genomic libraries of both sexes used, both SNPs were homozygous for each variant. Using PCR primers (pairs 2 and 5, Table 2), we amplified and sequenced the relevant regions in exons 1 and 14 in a small panel of WT males (n = 4) and females (n = 4), precluding association with sex of these other SNPs. Moreover, they did not conform to any simple genetic sex chromosome model (XY/XX or WZ/ZZ), as homozygous individuals were found for each variant. Apart from these instances, Sanger sequencing revealed another two nonsynonymous SNPs at nucleotide position 149 and 1781 bp, in exons 1 and 14, respectively (Table 4), however, these were not associated with sex.
Two nonsynonymous substitutions in fshr are male-specific
The two male-specific SNPs, c.1732G>A and c.1759T>G representing fshry, found in the full-sib families which are capable of encoding nonconservative amino acid substitutions (Table 4; Figure 1), were further investigated for association with sex in WT samples. All tested samples (n = 83) demonstrated that these two SNPs were a stable haplotype which was significantly associated with sex, with only 4.8% of mismatches (P ≤ 6.7 × 10−19; Table 5). Samples showing discordance between fshry haplotype and sex were all phenotyped for sex without microscope inspection. In addition, neither Sanger sequencing of these samples (n = 83), nor the nonsexed fingerlings (n = 87), were homozygous for the fshry haplotype, even though, based on Hardy–Weinberg equilibrium, their expected frequency was 6.6% (Table 6). Most importantly, on the population level, additional SNPs flanking the fshry haplotype, including other nonsynonymous substitutions (Table 4, positions 131–1689 and 1917–2112), did not show significant association with sex, and thus the observed SD associated region is delimited between positions 1732 and 1875, to 143 nucleotides on exon 14 (Table 4). It should be noted that we have analyzed additional sex-linked polymorphism in candidate genes besides fshr (Dor et al. 2020). In these genes, associations with sex were evident within familial structures only. However, the association with genetic factor affecting sex on the population level was displayed only for exon 14 polymorphism of fshr.
Table 5.
Association between fshr haplotype for SNPs at nt 1732 and 1759 on exon 14, and phenotypic sex of M. cephalus. The homozygous haplotype represents “XX,” while the heterozygous haplotype represents “XY”a
| fshr haplotype | Female | Male | Total count |
|---|---|---|---|
| “XX” | 37 | 1 | 38 |
| “XY” | 3 | 42 | 45 |
| Total count | 40 | 43 | 83 |
Fisher exact test: P < 6.7 × 10−19.
Table 6.
Contingency table comparing observed counts and frequencies of fshr haplotypes with those expected from Hardy–Weinberg equilibrium in the genotyped population (N = 170)a
| fshr haplotype | Actual count (frequency) | Expected count (frequency) |
|---|---|---|
| “XX” | 83 (0.48) | 94 (0.55) |
| “XY” | 87 (0.52) | 65 (0.38) |
| “YY” | 0 (0) | 12 (0.07) |
Pearson Chi-square test for divergence of fshr haplotypes from Hardy–Weinberg equilibrium (P ≤ 3.9 × 10−5).
Analysis of ploidy of fshr by allele specific PCR
As indicated in Introduction, scenarios in which the SD gene is not in diploidic state are common. To confirm that mullet fshr is in a diploidic state, in which each individual carries two alleles for the gene (paternal and maternal), two males and two females were analyzed by allele-specific PCR. Two forward primers were designed to amplify both variants of the c.1732G>A alteration (primer pair #3 and #4, Table 2) and two PCR reactions were performed for each individual. Only males gave a product when using the primer designed for the “A” variant. As opposed to female sequences, the sequenced products of males (608-618 bp), for both variants “A/G,” showed complete homozygosity over multiple SNPs (including positions 1759-2112, Table 4), suggesting no evidence for CNV in males.
Conservation of FSHR protein among vertebrate species
The protein sequence of the fshry variant was used as a template for BLASTP search against GenBank (nonredundant protein sequences). While the V/M substitution (nt 1732 on exon 14) was found in other organisms, the F/V substitution (nt 1759 on exon 14) is novel and causes an alteration in amino acid, which is conserved across vertebrate species (Figures 1 and 2; Supplementary Figure S1).
Figure 2.
Comparison of predicted Fshr of M. cephalus with other vertebrate FSHRs (GenBank: Human Human NP_000136; Mouse NP_038551; Tilapia NP_001266517; Seriola XP_022599280; Labrax AAV48628; Trout NP_001117799; Fugu XP_029691459). Amino acid numbering follows the complete gene alignment presented in supplementary file (Figure S1). The highly conserved phenylalanine (F) residue (nt 1759 in exon 14, Table 4), which was uniquely altered in fshr of M. cephalus male, is marked with an arrow indicating the corresponding human amino acid position. The protein domains are indicated above the amino acid alignment. Identical and similar amino acid residues in at least two of four sequences are indicated by a black and grey background, respectively. White boxes indicate non-conservative amino acid changes between the proteins.
Analysis of full-sib groups for sex-biased expression
In a previous study, two full-sib groups of fry (28 dph) of a mullet family (n = 15), were assigned to different sex groups, designated as A and B, with differential expression of genes that are located on the critical SD chromosomal region (Dor et al. 2020). We genotyped fshr for these groups and revealed that they represent fshr haplotypes “XX” and “XY,” respectively (Table 6). Interestingly, fshry was not found in any expression library, whereas expression of fshrx varied between larvae, adult brains and gonads, with a tendency toward higher values in male tissues (Figure 3). Since the promoter and gene structure of fshrx and fshry were similar, we expected that in males both genes would be similarly expressed. However, as detailed in “Method” section, BLASTN search using a 32 bp word spanning the critical variation sequence revealed only 0, 3, and 68 fshrx reads in male larvae, brain, and testis, respectively, but no fshry reads. This highly significant result (P < 4 × 10−22) and the differentially expressed pattern of fshrx (Figure 3) are compatible with the observation of Dor et al. (2020) that genes in the LG9 SD region are sex-biased differentially expressed and thus may be subjected to partial chromosome silencing.
Figure 3.
Expression data for fshrx in terms of normalized Trimmed Mean of values (TMM) for 28 dph larvae groups: A (“XX”) and B (“XY”), for adult brains and for gonads of both sexes. Standard error bar is shown at the top of columns with biological replicates.
Discussion
Multiple genetic SD factors are considered to be upstream of gonadal hormones, and the SD cascade is believed to be initiated by a single upstream MKR (Nakamura 2010). Many distant vertebrate organisms share common MKRs, suggesting that only a limited number of genes can initiate the fate of the bipotential gonad (Marshall Graves and Peichel 2010; Graves 2013). For example, dmrt1 determines sex in Medaka (Nanda et al. 2002), Spotted scat (Mustapha et al. 2018), African clawed frog (Yoshimoto et al. 2008), Chicken (Smith et al. 2009), and Half-smooth tongue sole (Cui et al. 2017); Anti Mullerian hormone is a MKR of Nile tilapia (Eshel et al. 2014), Patagonian pejerrey (Hattori et al. 2012), lingcod (Rondeau et al. 2016), and silverside (Bej et al. 2019). Moreover, a conserved cascade for initial activation of the gonad is implied by conservation of sex-biased expression genes (cassettes) among different species (Capel 2017). However, the initial low level of MKR’s extra-gonadal expression during early development (the early developmental period), renders it difficult to identify MKRs and their position in the sex cascade (Tilmann and Capel 2002). Furthermore, new models suggest that SD may act as a network, through genes with antagonistic effects on the manifestation of sex, without a conserved order (Capel 2017). The network properties and their degree of conservation require further study (Herpin and Schartl 2015). Fish are an excellent model for SD studies, as different, frequently closely related species exhibit varying MKRs (Herpin and Schartl 2015). Moreover, MKRs in fish include genes that are presumed to be involved in SD by controlling different types of mechanisms at different levels, including differentiation of supporting cells, proliferation of germ cells, and regulation of estrogen and androgen production, allowing a broader understanding of the different mechanisms (Capel 2017). The elucidation of whole MKR sets in different species is important for a wider understanding of the SD cascade (Herpin and Schartl 2015). Here, we have for the first time detected a putative MKR of M. cephalus, fshr, and suggest that this gene is part of the SD pathway. Deciphering the SD mechanism of M. cephalus also has significance for mullet aquaculture, as breeding of all-female populations is expected to increase production.
Our previous study mapped the SD locus to a 1 Mbp region, containing 27 genes including fshr. Although this gene is involved in gonad differentiation, it has not been considered an MKR of SD. In this study, we show a significant association between a haplotype of two nonsynonymous SNPs in fshr, c.1732G>A and c.1759T>G, and sex in M. cephalus. The association, initially traced within familial designs, was further validated in a WT population. Moreover, the fact that the associated male-specific haplotype (c.1732G>A and c.1759T>G, fshry) was flanked by other SNPs that were not male-specific on the population level, delimits the SD critical region to 143 nucleotides on exon 14, and strengthens the possibility that this variation is the causative polymorphism of SD. Seventy-nine individuals (95%) showed concordance between this fshr variation and sex, with only four mismatches (Table 5). M. cephalus is a gonochoristic species; however, when not in the breeding season, its gonads shrink and are absorbed in the surrounding tissues, making sex phenotyping less reliable. It is of interest to note that all four individuals showing discordance between the fshr haplotype and sex, were phenotyped for sex without microscope inspection. Furthermore, the fact that no homozygous individuals were found for the male-specific fshry haplotype, despite its abundancy in the population, may be explained by either mortality of the homozygotes or by the causative nature of the “Y” variant. The latter is a more likely explanation in view of the presumed XX/XY SD system, the localization of fshr haplotype to the SD critical region, and its strong association with sex. Both inheritance models testing a segregating autosomal locus or lethal homozygosity of the GG genotypes were excluded (Table 7).
Table 7.
Goodness of fit of inheritance models with the observed distribution of c.1759T>G genotypes in male population (n = 43)
| TT | GT | GG | ||
|---|---|---|---|---|
| Observed distribution | ||||
| 1 | 42 | 0 | ||
| Inheritance modelsa | Expected distribution | P b | ||
| Autosomal locusc | 11.3 | 21.4 | 10.3 | 2.97 × 10−7 |
| Lethal GG homozygoted | 14.9 | 28.1 | 0 | 2.83 × 10−4 |
| Male determination by Ge | 0 | 43 | 0 | 0.99 |
H0—there is no difference between observed and expected distributions based on the inheritance model with allele frequencies p(T) = 0.512 and q(G) = 0.488.
Fisher exact test: 2 × 3 tool (http://vassarstats.net/fisher2x3.html).
Hardy–Weinberg equilibrium: p2, 2pq, q2.
Elimination of q2: p2/(1 − q2), 2pq/(1 − q2), 0.
Elimination of q2 and p2: 0, 1, 0.
Many SD genes in fish, birds, reptiles, and crocodiles, belong to a gene pathway that drives the balance of sex steroids (Capel 2017). The assumed finding of fshr as an MKR of SD in M. cephalus, together with the knowledge of its involvement in gonad differentiation from other studies, support a general putative role of SD genes through 17β-estradiol (E2) and 11-ketotestosterone (11-KT) equilibrium in fish (Baroiller et al. 1999). The manner in which fshr governs SD could be attributed to its recorded role in SD in amphibians and fish. In mammals, FSH signaling in granulosa cells is known to stimulate the cytochrome P450 aromatase (CYP19A1) expression, affecting estrogen biosynthesis and consequently the conversion of androgens to estrogens (Whitlock 1986; Steinkampf et al. 1987; Fitzpatrick and Richards 1991; Manuel Silva and Price 2000). Different studies in fish and amphibians have shown the involvement of fshr in functions that are related to SD, presumably by controlling cyp19a1 and estrogen biosynthesis (Yamaguchi et al. 2007; Suda et al. 2011; Nakamura 2013; Murozumi et al. 2014). However, findings in Honeycomb grouper (Epinephelus merra), (Kobayashi et al. 2010) and Japanese eel (Anguilla japonica) (Ohta et al. 2007), suggest its role is in upregulating androgen (11KT) biosynthesis, perhaps through the control of P450 11 b-hydoroxylase (P45011β). Indeed, disruption of fshr causes masculinization in medaka and zebrafish whereas in the latter loss of fshr lead sexual reversal to fertile males (Murozumi et al. 2014, Zhang et al., 2015). Thus, FSH/FSHR’s signaling role in sex hormone biosynthesis may possibly influence male determination in M. cephalus. The SD associated haplotype on fshr is capable of encoding two adjacent protein domains: transmembrane α-helices six (TM6), and intracellular loop 3 (IL3). This loop is assumed to be a G protein interaction domain (Inglese et al. 1993; Kosugi et al. 1993), which is conserved in G protein-coupled receptors (GPCRs), such as FSHR, TSHR, and LHR (Kotlar et al. 1997, Figure 4). Many studies have demonstrated that this region, including the two domains, is of crucial importance in maintaining the G protein receptors in an inactive conformation (Tao 2008), furthermore, in humans, mutations in this domain are known to cause alteration of signaling mostly by activation (Kotlar et al. 1997; Tao 2008, Figure 4). The first nucleic acid variation (V/M) is in IL3. However, this substitution is common as it was found in the WT fshr of fugu and trout (Figure 2). In contrast, the second nucleic acid substitution (F/V i.e., phenylalanine/valine) is in a highly conserved position in TM6, and based on comparative analysis, valine is a novel amino acid in this position in vertebrate FSHRs, which is likely to induce FSH/FSHR signaling alteration (Figure 4). A mutation in the same conserved amino acid, was found in thyrotropin receptor gene, and causes cAMP basal activation without induction (Porcellini et al. 1994), which may lead to fshry control of sex via upregulation of androgen biosynthesis. Alternatively, the male fshry may be inactive, thus inducing masculinity by lowering the signal which is needed for female gonad differentiation. Indeed, we were not able to detect the expression of fshry in 28 dph larvae and adult brains and testes. However, our predicted fshry displayed no pseudogene characteristics, as its entire reading frame was intact, which implies that it is functional (Supplementary Figure S1). Thus, it may be postulated that fshry is expressed in a narrow time window, not yet resolved. A more complex mechanism may be postulated based on a hypothesis that LG9 has adopted some primitive characteristics of a sex chromosome, which may involve gene silencing in parts of the chromosome being packaged into transcriptionally inactive structures (heterochromatins). Indeed, of the 107 sex-biased genes that were differentially expressed by M. cephalus larvae, 23 displayed sex-specific inactivation and 22 of these genes were positioned to LG9 (Dor et al. 2020). Moreover, the few genotypes (4 out of 83) that did not fit with the proposed mono factorial genetic model for male determination might be attributed to environmental sex reversal (Baroiller et al. 1999). However, sex phenotyping is prone to errors and thus may be responsible for the occurrence of such observations.
Figure 4.
Different mutations in intracellular loop 3 (IL3) and transmembrane α-helices six (TM6) of human GPCRs (LHR, TSHR and FSHR), their functional outcomes, and the putative positions of the altered M. cephalus male variants for Fshr (following Kotlar et al. 1997).
Previous studies have shown that gene duplications are frequently associated with SD (Eshel et al. 2014; Curzon et al. 2020). Our data from DNA-seq assembly, together with the results from specific PCR and sequencing of the regions representing the “X” and “Y” chromosomes, provide no evidence for duplications or null alleles in M. cephalus SD mechanism. In Oreochromis niloticus, an Anti Müllerian hormone (amh) male-specific duplication, amhy, was suggested as the SD key regulator (Eshel et al. 2014, Curzon et al. 2020). Li et al. (2015) showed that an amhy knockout resulted in male to female sex reversal in XY tilapia, while its over-expression resulted in female to male sex reversal; thus, validating amhy as the sex-determining gene in tilapia. We suggest that in a similar manner, the fshr candidate gene for SD should be further explored by functional validation using advanced methods, such as CRISPR/Cas9, TALEN, or antisense RNA (Ventura and Sagi 2012; Li et al. 2015; Bao et al. 2019; Pan et al. 2019).
Acknowledgments
A. S., H. R., I. M.A., M.R., and E.S. have, together with Kidum—R&D Applications and Technology Transfer, filed an Israeli patent application based on the research described within this article (#275926; dated: 08/07/2020; titled: Nucleic acid sequences for use in sex determination in mullet and methods of using such sequence).
Funding
This work was supported by the Nizan project, the Ministry of Agriculture and Rural Development, Israel. Grant: 30-04-0010.
Conflicts of interest: None declared.
Literature cited
- Aizen J, Meiri I, Tzchori I, Levavi-Sivan B, Rosenfeld H.. 2005. Enhancing spawning in the grey mullet (Mugil cephalus) by removal of dopaminergic inhibition. Gen Comp Endocrinol. 142:212–221. doi: 10.1016/J.YGCEN.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Altshuler D, Daly MJ, Lander ES.. 2008. Genetic mapping in human disease. Science. 322:881–888. doi: 10.1126/science.1156409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Tian C, Liu S, Zhang Y, Elaswad A, et al. 2019. The Y chromosome sequence of the channel catfish suggests novel sex determination mechanisms in teleost fish. BMC Biol. 17:16. doi: 10.1186/s12915-019-0627-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroiller JF, Guiguen Y, Fostier A.. 1999. Endocrine and environmental aspects of sex differentiation in fish. Cell Mol Life Sci. 55:910–931. doi: 10.1007/s000180050344 [DOI] [Google Scholar]
- Bej DK, Miyoshi K, Hattori RS, Strüssmann CA, Yamamoto Y.. 2019. A duplicated, truncated amh gene is involved in male sex determination in an old world silverside. G3 (Bethesda). 7:2489–2495. doi: 10.1534/g3.117.042697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B. 2017. Vertebrate sex determination: Evolutionary plasticity of a fundamental switch. Nat Rev Genet. 18:675–689. doi:10.1038/nrg.2017.60 [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang G, Shao C, Huang Q, Liu G, et al. 2014. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 46:253–260. doi: 10.1038/ng.2890 [DOI] [PubMed] [Google Scholar]
- Chen J, Fan Z, Tan D, Jiang D, Wang D.. 2018. A review of genetic advances related to sex control and manipulation in Tilapia. J World Aquacult Soc. 49:277–291. doi: 10.1111/jwas.12479 [DOI] [Google Scholar]
- Cui Z, Liu Y, Wang W, Wang Q, Zhang N, et al. 2017. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci Rep. 7:1–10. doi:10.1038/srep42213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curzon AY, Shirak A, Dor L, Zak T, Perelberg A, et al. 2020. A conserved truncated male-specific copy of amh is associated. with sex determination of Nile tilapia (Oreochromis niloticus). Heredity. 124:317–327. doi: 10.1038/s41437-020-0340-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor L, Shirak A, Rosenfeld H, Ashkenazi IM, Band MR, et al. 2016. Identification of the sex-determining region in flathead grey mullet (Mugil cephalus). Anim Genet. 47:698–707. doi: 10.1111/age.12486 [DOI] [PubMed] [Google Scholar]
- Dor L, Shirak A, Curzon AY, Rosenfeld H, Ashkenazi IM, et al. 2020. Preferential mapping of sex-biased differentially-expressed genes of larvae to the sex-determining region of flathead grey mullet (Mugil cephalus). Front Genet. 11:839. doi: 10.3389/fgene.2020.00839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel O, Shirak A, Dor L, Band M, Zak T, et al. 2014. Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics. 15:774. doi: 10.1186/1471-2164-15-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feron R, Zahm M, Cabau C, Klopp C, Roques C, et al. 2020. Characterization of a Y-specific duplication/insertion of the anti-Mullerian hormone type II receptor gene based on a chromosome-scale genome assembly of yellow perch, Perca flavescens. Mol Ecol Resour. 20:531–543. doi: 10.1111/1755-0998.13133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick SL, Richards JS.. 1991. Regulation of cytochrome P450 aromatase messenger ribonucleic acid and activity by steroids and gonadotropins in rat granulosa cells. Endocrinology. 129:1452–1462. doi:10.1210/endo-129-3-1452 [DOI] [PubMed] [Google Scholar]
- Graves JAM. 2013. How to evolve new vertebrate sex determining genes. Dev Dyn. 242:354–359. doi: 10.1002/dvdy.23887 [DOI] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, et al. 1990. A gene-mapping to the sex-determining region of the mouse y-chromosome is a member of a novel family of embryonically expressed genes. Nature. 346:245–250. doi: 10.1038/346245a0 [DOI] [PubMed] [Google Scholar]
- Hattori RS, Murai Y, Oura M, Masuda S, Majhi SK, et al. 2012. A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci USA. 109:2955–2959. doi: 10.1073/pnas.1018392109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Schartl M.. 2015. Plasticity of gene regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 16:1260–1274. doi: 10.15252/embr.201540667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CM, Shaw D.. 2006. The impact of upstream catch and global warming on the grey mullet fishery in Taiwan: a non-cooperative game analysis. Mar Resour Econ. 21:285–300. doi: 10.1086/mre.21.3.42629512 [DOI] [Google Scholar]
- Inglese J, Freedman NJ, Koch WJ, Lefkowitz RJ.. 1993. Structure and mechanism of the G protein-coupled receptor kinases. J Biol Chem. 268:23735–23738. [PubMed] [Google Scholar]
- Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, et al. 2012. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger Pufferfish, Takifugu rubripes (Fugu). PLoS Genet. 8:e1002798. doi: 10.1371/journal.pgen.1002798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katselis G, Hotos G, Minos G, Vidalis K.. 2006. Phenotypic affinities on fry of four Mediterranean grey mullet species. Turk J Fish Aquat Sci. 6:49–55. [Google Scholar]
- Kim Y, Capel B.. 2006. Balancing the bipotential gonad between alternative organ fates: a new perspective on an old problem. Dev Dyn. 235:2292–2300. doi: 10.1002/dvdy.20894 [DOI] [PubMed] [Google Scholar]
- Kirubakaran TG, Andersen Ø, De Rosa MC, Andersstuen T, Hallan K, et al. 2019. Characterization of a male specific region containing a candidate sex determining gene in Atlantic cod. Sci Rep. 9:116. doi: 10.1038/s41598-018-36748-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Alam MA, Horiguchi R, Shimizu A, Nakamura M.. 2010. Sexually dimorphic expression of gonadotropin subunits in the pituitary of protogynous Honeycomb Grouper (Epinephelus merra): evidence that follicle-stimulating hormone (FSH) induces gonadal sex change. Biol Reprod. 82:1030–1036. doi: 10.1095/biolreprod.109.080986 [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J.. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 7:474.doi: 10.1186/1471-2105-7-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossack ME, Draper BW.. 2019. Genetic regulation of sex determination and maintenance in zebrafish (Danio rerio). Sex Determin Vertebr. 134:119-149. doi: 10.1016/bs.ctdb.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Okajima F, Ban T, Hidaka A, Shenker A, et al. 1993. Substitutions of different regions of the third cytoplasmic loop of the Thyrotropin (TSH) receptor have selective effects on constitutive, TSH-, and TSH receptor autoantibody-stimulated phosphoinositide and 3’,5’-cyclic adenosine monophosphate signal generation. Mol Endocrinol. 7:1009–1020. doi: 10.1210/mend.7.8.7901757 [DOI] [PubMed] [Google Scholar]
- Kotlar TJ, Young RH, Albanese C, Crowley WF, Scully RE, et al. 1997. A mutation in the follicle-stimulating hormone receptor occurs frequently in human ovarian sex cord tumors. J Clin Endocrinol Metab. 82:1020–1026. doi: 10.1210/jcem.82.4.3870 [DOI] [PubMed] [Google Scholar]
- Koyama T, Nakamoto M, Morishima K, Yamashita R, Yamashita T, et al. 2019. A SNP in a steroidogenic enzyme is associated with phenotypic sex in Seriola fishes. Curr Biol. 29:1901–1909.e8. doi: 10.1016/J.CUB.2019.04.069 [DOI] [PubMed] [Google Scholar]
- Li M, Sun Y, Zhao J, Shi H, Zeng S, et al. 2015. A tandem duplicate of anti-müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile Tilapia. PLoS Genet. 11:e1005678–23. doi: 10.1371/journal.pgen.1005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Huang S, Sun M, Liu S, Liu Y, et al. 2012. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods. 8:34. doi: 10.1186/1746-4811-8-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel Silva J, Price CA.. 2000. Effect of Follicle-stimulating hormone on steroid secretion and messenger ribonucleic acids encoding cytochromes P450 aromatase and cholesterol side-chain cleavage in bovine granulosa cells in vitro. Biol Reprod. 62:186–191. doi: 10.1095/biolreprod62.1.186 [DOI] [PubMed] [Google Scholar]
- Marshall Graves JA, Peichel CL.. 2010. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 11:205.doi: 10.1186/gb-2010-11-4-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, et al. 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 417:559–563. doi: 10.1038/nature751 [DOI] [PubMed] [Google Scholar]
- Matsuda M, Sakaizumi M.. 2016. Evolution of the sex-determining gene in the teleostean genus Oryzias. Gen Comp Endocrinol. 239:80–88. doi: 10.1016/j.ygcen.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Mlalila N, Mahika C, Kalombo L, Swai H, Hilonga A.. 2015. Human food safety and environmental hazards associated with the use of methyltestosterone and other steroids in production of all-male tilapia. Environ Sci Pollut Res. 22:4922–4931. doi: 10.1007/s11356-015-4133-3 [DOI] [PubMed] [Google Scholar]
- Murozumi N, Nakashima R, Hirai T, Kamei Y, Ishikawa-Fujiwara T, et al. 2014. Loss of follicle-stimulating hormone receptor function causes masculinization and suppression of ovarian development in genetically female Medaka. Endocrinology. 155:3136–3145. doi: 10.1210/en.2013-2060 [DOI] [PubMed] [Google Scholar]
- Mustapha UF, Jiang D-N, Liang Z-H, Gu H-T, Yang W, et al. 2018. Male-specific Dmrt1 is a candidate sex determination gene in spotted scat (Scatophagus argus). Aquaculture. 495:351–358., doi: 10.1016/j.aquaculture.2018.06.009 [DOI] [Google Scholar]
- Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, et al. 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics. 191:163–170. doi: 10.1534/genetics.111.137497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myosho T, Takehana Y, Hamaguchi S, Sakaizumi M.. 2015. Turnover of sex chromosomes in celebensis group medaka fishes. G3 (Bethesda). 5:2685–2891. doi: 10.1534/g3.115.021543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M. 2010. The mechanism of sex determination in vertebrates-are sex steroids the key-factor? J Exp Zool. 313A:381–398. doi: 10.1002/jez.616 [DOI] [PubMed] [Google Scholar]
- Nakamura M. 2013. Is a sex-determining gene(s) necessary for sex-determination in amphibians? Steroid hormones may be the key factor. Sex Dev. 7:104–114. doi: 10.1159/000339661 [DOI] [PubMed] [Google Scholar]
- Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, et al. 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci USA. 99:11778–11783. doi: 10.1073/pnas.182314699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Miyake H, Miura C, Kamei H, Aida K, et al. 2007. Follicle-stimulating hormone induces spermatogenesis mediated by androgen production in Japanese Eel, Anguilla japonica1. Biol Reprod. 77:970–977. doi: 10.1095/biolreprod.107.062299 [DOI] [PubMed] [Google Scholar]
- Pan Q, Feron R, Yano A, Guyomard R, Jouanno E, et al. 2019. Identification of the master sex determining gene in Northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLoS Genet. 15:e1008013. doi: 10.1371/journal.pgen.1008013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcellini A, Ciullo I, Laviola L, Amabile G, Fenzi G, et al. 1994. Novel mutations of thyrotropin receptor gene in thyroid hyperfunctioning adenomas. Rapid identification by fine needle aspiration biopsy. J Clin Endocrinol Metab. 79:657–661. doi: 10.1210/jcem.79.2.8045989 [DOI] [PubMed] [Google Scholar]
- Raymond CS, Murphy MW, O'sullivan MG, Bardwell VJ, Zarkower D.. 2000. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14:2587–2595. doi: 10.1101/gad.834100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichwald K, Petzold A, Koch P, Downie BR, Hartmann N, et al. 2015. Insights into sex chromosome evolution and aging from the genome of a short-lived fish. Cell. 163:1527–1538., doi: 10.1016/j.cell.2015.10.071 [DOI] [PubMed] [Google Scholar]
- Rondeau EB, Laurie CV, Johnson SC, Koop BF.. 2016. A PCR assay detects a male-specific duplicated copy of Anti-Müllerian hormone (amh) in the lingcod (Ophiodon elongatus). BMC Res Notes. 9: doi:10.1186/s13104-016-2030-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M. 2004. Sex chromosome evolution in non-mammalian vertebrates. Curr Opin Genet Dev. 14:634–641. doi: 10.1016/J.GDE.2004.09.005 [DOI] [PubMed] [Google Scholar]
- Ser JR, Roberts RB, Kocher TD.. 2010. Multiple interacting loci control sex determination in lake malawi cichlid fish. Evolution (N. Y). 64:486–501. doi: 10.1111/j.1558-5646.2009.00871.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirak A, Grabherr M, Palma FD, Lindblad-Toh K, Hulata G, et al. 2010. Identification of repetitive elements in the genome of Oreochromis niloticus: Tilapia repeat masker. Mar Biotechnol. 12:121–125. doi: 10.1007/s10126-009-9236-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirak A, Seroussi E, Cnaani A, Howe AE, Domokhovsky R, et al. 2006. Amh and Dmrta2 genes map to tilapia (Oreochromis spp.) linkage group 23 within quantitative trait locus regions for sex determination. Genetics. 174:1573–1581. doi: 10.1534/genetics.106.059030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, et al. 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 346:240–244. doi: 10.1038/346240a0 [DOI] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, et al. 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 461:267–271. doi: 10.1038/nature08298 [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK.. 2000. The Staden Package, 1998, Methods Mol Biol. 132: 115–130. doi: 10.1385/1-59259-192-2:115 [DOI] [PubMed] [Google Scholar]
- Steinkampf MP, Mendelson CR, Simpson ER.. 1987. Regulation by follicle-stimulating hormone of the synthesis of aromatase cytochrome P-450 in human granulosa cells. Mol Endocrinol. 1:465–471. doi: 10.1210/mend-1-7-465 [DOI] [PubMed] [Google Scholar]
- Suda M, Kodama M, Oshima Y, Yamamoto K, Nakamura Y, et al. 2011. Up-regulation of FSHR expression during gonadal sex determination in the frog Rana rugosa. Gen Comp Endocrinol. 172:475–486. doi: 10.1016/J.YGCEN.2011.04.011 [DOI] [PubMed] [Google Scholar]
- Takehana Y, Matsuda M, Myosho T, Suster ML, Kawakami K, et al. 2014. Co-option of Sox3 as the male-determining factor on the y chromosome in the fish Oryzias dancena. Nat Commun. 5:1–10. doi:10.1038/ncomms5157 [DOI] [PubMed] [Google Scholar]
- Tao Y-X. 2008. Constitutive activation of G protein-coupled receptors and diseases: insights into mechanisms of activation and therapeutics. Pharmacol Ther. 120:129–148. doi: 10.1016/J.PHARMTHERA.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilmann C, Capel B.. 2002. Cellular and molecular pathways regulating mammalian sex determination. Recent Prog Horm Res. 57:1–18. doi: 10.1210/rp.57.1.1 [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth B C, . et al. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Research. 40:e115–e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura T, Sagi A.. 2012. The insulin-like androgenic gland hormone in crustaceans: From a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnol Adv. 30:1543–1550. doi: 10.1016/j.biotechadv.2012.04.008 [DOI] [PubMed] [Google Scholar]
- Whitlock JP. 1986. The regulation of cytochrome P-450 gene expression. Annu Rev Pharmacol Toxicol. 26:333–369. doi: 10.1146/annurev.pa.26.040186.002001 [DOI] [PubMed] [Google Scholar]
- Williams TM, Carroll SB.. 2009. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat Rev Genet. 10:797–804. doi: 10.1038/nrg2687 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Yamaguchi S, Hirai T, Kitano T.. 2007. Follicle-stimulating hormone signaling and Foxl2 are involved in transcriptional regulation of aromatase gene during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem Biophys Res Commun. 359:935–940. doi: 10.1016/J.BBRC.2007.05.208 [DOI] [PubMed] [Google Scholar]
- Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, et al. 2012. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr Biol. 22:1423–1428. doi: 10.1016/j.cub.2012.05.045 [DOI] [PubMed] [Google Scholar]
- Yano A, Nicol B, Jouanno E, Quillet E, Fostier A, et al. 2013. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol Appl. 6:486–496. doi: 10.1111/eva.12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, et al. 2008. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci USA. 105:2469–2474. doi: 10.1073/pnas.0712244105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lau SW, Zhang L, Ge W.. 2015. Disruption of zebrafish follicle-stimulating hormone receptor (fshr) but not luteinizing hormone receptor (lhcgr) gene by TALEN leads to failed follicle activation in females followed by sexual reversal to males. Endocrinology. 156:3747–3762. doi: 10.1210/en.2015-1039 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DNA- and RNA-seq reads are available at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession numbers PRJEB12265 and PRJEB34342. The M. cephalus assembled and annotated transcriptome is available at http://cowry.agri.huji.ac.il/. Supplementary material includes Figure S1 Comparison of predicted Fshr of M. cephalus with other vertebrate FSHRs (complete alignment) and the annotated mullet fshr sequences (File S2). Supplementary material is available at figshare:





