Abstract
Despite decades of effort, the sensitivity of patient tumors to individual drugs is often not predictable based on molecular markers alone. Therefore, unbiased, high-throughput approaches to match patient tumors to effective drugs—without requiring a priori molecular hypotheses—are critically needed. Here, we improved upon a method we previously reported and developed a method called high-throughput dynamic BH3 profiling (HT-DBP). HT-DBP is a microscopy-based, single-cell–resolution assay that enables chemical screens of hundreds to thousands of candidate drugs on freshy isolated tumor cells. The method identifies chemical inducers of mitochondrial apoptotic signaling, a mechanism of cell death. HT-DBP requires only 24 hours of ex vivo culture, which enables a more immediate study of fresh primary tumor cells and minimizes adaptive changes that occur with prolonged ex vivo culture. Effective compounds identified by HT-DBP induced tumor regression in genetically engineered and patient-derived xenograft (PDX) models of breast cancer. We additionally found that chemical vulnerabilities changed as cancer cells expanded ex vivo. Furthermore, using PDX models of colon cancer and resected tumors from colon cancer patients, our data demonstrated that HT-DBP could be used to generate personalized pharmacotypes. Thus, HT-DBP appears to be an ex vivo functional method with sufficient scale to simultaneously function as a companion diagnostic, therapeutic personalization, and discovery tool.
Introduction
Clinically actionable knowledge can be obtained from the direct study of primary patient tumors. Current research practice on primary samples typically focuses on obtaining large amounts of information about the molecular constituents of tumors (DNA, RNA, proteins, metabolites, and others). These analyses have yielded important successes that have directly improved cancer treatment (1–3). Nonetheless, they necessarily lose information about dynamic interactions of these constituents as they are static measurements of initial conditions(4). Unfortunately, static approaches do not identify active drugs for enough cancer patients, and alternative diagnostic approaches are needed.
To determine how a patient tumor will respond to a drug, it is hard to imagine a more practical way to do this than to put the living cancer cell in contact with that drug. However, inadequate accuracy of prior attempts at ex vivo chemosensitivity determination limited enthusiasm about this approach (5). These older strategies often tested very small numbers of drugs, often in long-term cultures deleterious to maintenance of cancer phenotype and used rudimentary readouts incapable of single cell resolution. The tools were simply not good enough to drive clinical decision making, and enthusiasm waned (5–9). We now have many more potential therapeutics to test, as well as vastly improved cell biological knowledge and better cellular analytics. The potential quality of the information to be garnered from exposing patient cancer cells to drugs has led to a recent re-examination of its application.
A compelling approach for identifying novel or personalized therapies is to perturb living cancer cells with drugs ex vivo and measure clinically relevant cellular phenotypes (4, 10–18). The ideal therapeutic perturbation strategy minimizes ex vivo culture time, maximizes the testable number of therapeutics, and measures effects that are directly relevant to in vivo response. Here, we report a novel strategy called High-Throughput Dynamic BH3 profiling (HT-DBP) that fulfills these criteria. We previously developed BH3 profiling to measure the proximity of a cell to the apoptotic threshold—a property called “apoptotic priming” (fig. S1A) (19, 20). Apoptotic priming is inferred from the degree of mitochondrial outer membrane permeabilization induced by standardized concentrations of synthetic peptides derived from the BH3 domains of pro-apoptotic BCL-2 family proteins. We subsequently developed dynamic BH3 profiling (DBP) which measures how short, 4- to 24-hour ex vivo drug treatments increase apoptotic priming in patient tumor cells (fig. S1, B and C). When a cell exhibits a significant increase in apoptotic priming after drug treatment, that cell is likely to respond to that drug both in vitro and in vivo (11, 21, 22).
A key limitation of DBP is that only a few compounds could be tested on a primary sample at a time (11). This is a small fraction of clinically applicable cancer therapeutics, and an even smaller fraction of pre-clinical or tool compounds (23). With a limitation of 10–20 compounds per sample, we could only perform hypothesis driven drug testing, and could not perform unbiased screening of chemical libraries to discover new therapies or new indications for existing therapies. Therefore, we sought to expand the number of drugs that could be tested with DBP to facilitate unbiased chemical screening of freshly isolated tumor cells. Early technological iterations of DBP required manual transfer of cancer cells from drug treatment vessels into FACS vessels where apoptotic priming is measured (fig. S1D) (11). We therefore miniaturized and automated DBP to facilitate identification of the maximal number of apoptotic sensitizing drugs for any given tumor biopsy specimen.
Results
Our previously reported low-throughput, hypothesis-based use of DBP relied on flow cytometry or well-based fluorimetry (11, 22). To perform unbiased, high-throughput measurements of early induction of apoptotic cell death at single-cell resolution, we implemented automated immunofluorescence microscopy and miniaturized the DBP assay (outlined in Fig. 1A). Single cell suspensions of dissociated tumors were plated in 384-well plates and treated with chemical libraries using pin transfer tools. 24 hours after drug treatment, cells were washed in PBS, and the BH3 profiling buffers and synthetic BH3 peptides were added. To tune the assay for maximal sensitivity to drug-induced apoptotic signaling, 4 hours before completion of drug exposures a single peptide concentration was chosen for each sample such that in the absence of drug 10% of cells lose cytochrome c. (fig. S2). Tumor cells were discriminated from stroma using tumor-discriminating antigens, such as EpCam for epithelial tumors. Drugs that induce apoptotic signaling cause heightened BH3 peptide-induced cytochrome c loss compared to DMSO-treated control wells, measured on a per cell basis using immunofluorescence microscopy. Drug-potentiated peptide-induced loss of cytochrome c was quantified as the difference in percentage of cytochrome c positive cells between DMSO-treated and drug-treated wells (Fig. 1A)—a parameter called “delta priming” (11). In all cases, a concentration of 1μM of drug was used in initial chemical screens which was based on experiments showing good correlation of priming activity at drug concentrations below 1.1 μM (fig. S3).
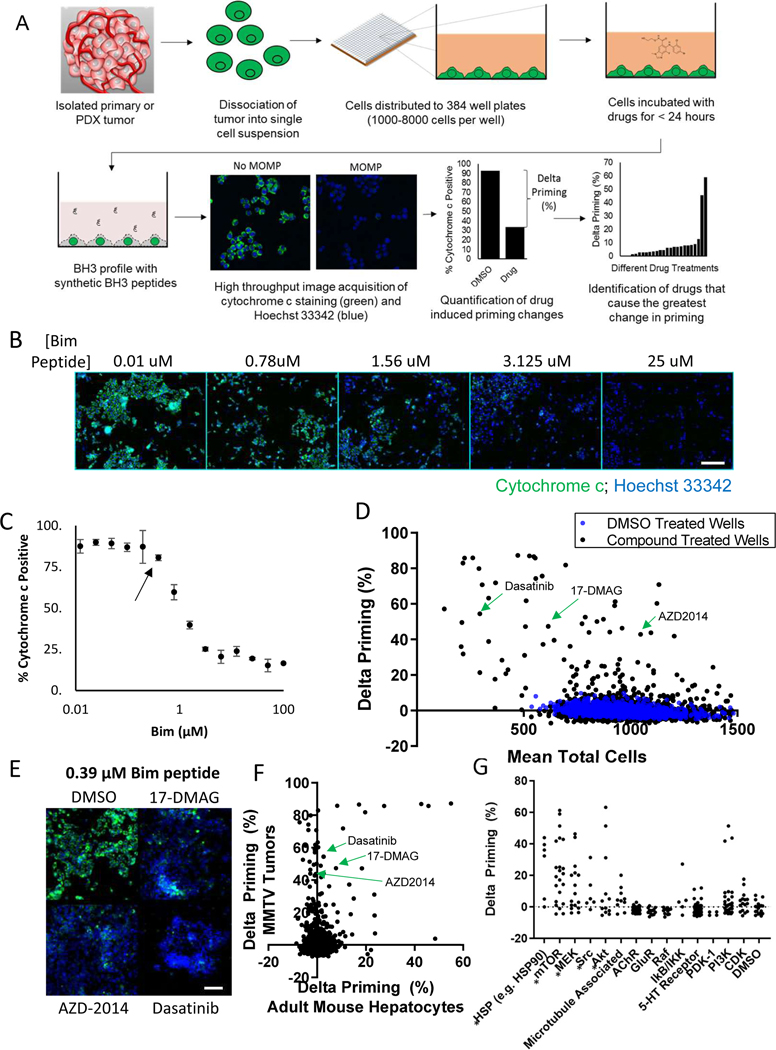
Fig. 1: High-throughput dynamic BH3 profiling screen of 1650 compounds identifies chemicals that sensitize freshly isolated tumor cells, and not healthy cells for apoptosis.
(A) Schematic showing the workflow of the HT-DBP screening platform. Compounds with the largest delta priming cause the largest increase in apoptotic sensitivity. (B) Cytochrome c staining in response to Bim peptide dose in MMTV-PyMT tumors. Scale bars, 100 μm. Images are representative of 2 independent experiments. (C) Quantification of the dose response curve from imaging described in (B). Arrow indicates the peptide concentration chosen for use in the screening. Data are mean ± SD of 6 replicates, representative of 2 independent experiments. (D) HT-DBP on MMTV-PyMT tumors to identify compounds that increase apoptotic sensitivity. DMSO-treated wells shown in blue; compound-treated wells shown in black. Data are mean of 2 independent experiments. (E) Images of selected wells from the chemical screen and the DMSO. Wells were treated with the indicated for 24 hours and subsequently treated with 0.39 μM of the synthetic peptide. Cytochrome c immunofluorescence is shown in green, and Hoechst 33342 staining is shown in blue. Images are representative of 2 independent screens. (F) Comparison of a screen on freshly isolated adult mouse hepatocytes and freshly isolated MMTV-PyMT tumor cells. Data represents mean of 2 independent experiments. (G) Chemical annotation of drug targets from the HT-DBP screen on MMTV-PyMT tumors. Each dot represents a single compound. Asterisks indicate instances where compounds with a similar target increase apoptotic priming (p<0.0001, one-way ANOVA). Data represents mean of 2 independent experiments.
We first verified that HT-DBP could identify compounds that induced apoptotic signaling in the MDA-MB-231 breast cancer cell line. Specifically, we identified the peptide concentration where approximately 10% of cytochrome c was lost from cells (fig S4, A and B), and measured the relative increase in apoptotic signaling (fig S4C). Notably, the change in priming observed in HT-DBP correlated with cytotoxicity 48 hours later (fig. S4, D and E; data files S1 and S2). We next compared an earlier low-throughput FACS based method of dynamic BH3 profiling, with our new high throughput method of dynamic BH3 profiling (fig S5, A and B), and found that these produced similar rankings of chemical sensitivity (fig. S5C).
A rigorous test of our approach is to compare DBP results to treatment response in an in vivo model. Murine models offer the possibility to treat the same tumor in vivo many different ways. Therefore, we next asked if we could assign active therapies to tumors from the MMTV-PyMT mouse autochthonous breast cancer model (24). Single cell suspensions made from freshly isolated tumors were distributed to 384 well plates and treated with a library of 1650 bioactive drugs (described in table S1) at a concentration of 1 μM for 24 hours (further detailed in the Materials and Methods), then subjected to HT-DBP (Fig. 1, B to D, and data file S3). We evaluated drug-induced apoptotic priming only in cells that stained for mouse EpCam (fig. S6). Cells were identified using the multi-wavelength cell scoring module in Metamorph (fig. S7). The screen produced adequate high throughput screening metrics (fig. S8, A and B), and high correlation of replicates in these screens (fig. S8, C and D; data files S4 and S5). Importantly, several chemicals increased apoptotic priming above levels found in DMSO treated wells (Fig. 1, D and E). These results indicate that HT-DBP can identify apoptosis-sensitizing compounds on freshly isolated tumor cells.
We next prioritized compounds with therapeutic potential by excluding broadly toxic compounds. We performed HT-DBP counter screens on non-tumor cells: adult mouse hepatocytes (fig. S9, A to C; data file S6) (25) and human foreskin fibroblasts (19) (fig. S9, D and E; data file S7). We next compared compounds that induced apoptotic priming in freshly isolated MMTV-PyMT tumor cells (Fig 1F, data file S8). Compounds that sensitized non-tumor cells for apoptosis at a concentration of 1 μM were deprioritized for further analysis. Using the multiplicity of unique compounds in the bioactive chemical library with the same nominal target, we sought to infer druggable pathways dependencies of tumor cells and not healthy cells. We determined that several compounds known to target HSP90, mTORC1/2, Src, MEK or AKT preferentially provoked apoptotic signaling in tumor cells (Fig. 1G, data file S9). Note that the three compounds showing highest delta priming in the PI3K column all target mTORC1/2, too. These results indicate that HT-DBP can identify compounds and pathways that selectively prime tumors, and not healthy cells, for apoptosis.
To validate DBP as a predictive tool, we next asked whether drugs identified by HT-DBP cause tumor regression in vivo. We selected 6 drugs for in vivo testing, three predicted to be active and three predictive to be inactive. We prioritized drugs that were inexpensive, readily available, and which had published dosing regimens. Importantly, the dosing regimens of the drugs we chose were previously demonstrated to have on-target in vivo pharmcodynamic effect and were tolerable to the mice (26–31). Treatments based on assay hits with known pharmacokinetic properties [dasatinib (26), 17-DMAG (27), AZD-2014 (28), and a combination of dasatinib and 17-DMAG] showed decreased tumor volume compared to vehicle treated mice at day 14 (Fig. 2, A and B). Drugs that did not score in our assay (navitoclax, lapatinib, and sunitinib(29–31)) did not induce tumor regression (Fig. 2B). Importantly, the magnitude of ex vivo apoptotic sensitization correlated with tumor response in vivo (Fig. 2C). Active drugs 17-DMAG and AZD2014 did not significantly alter cell numbers relative to DMSO-treated wells at 24 hours (Fig. 1D), and would not have been identified by standard screening methods that typically depend upon completion of cell death to register a chemical hit.
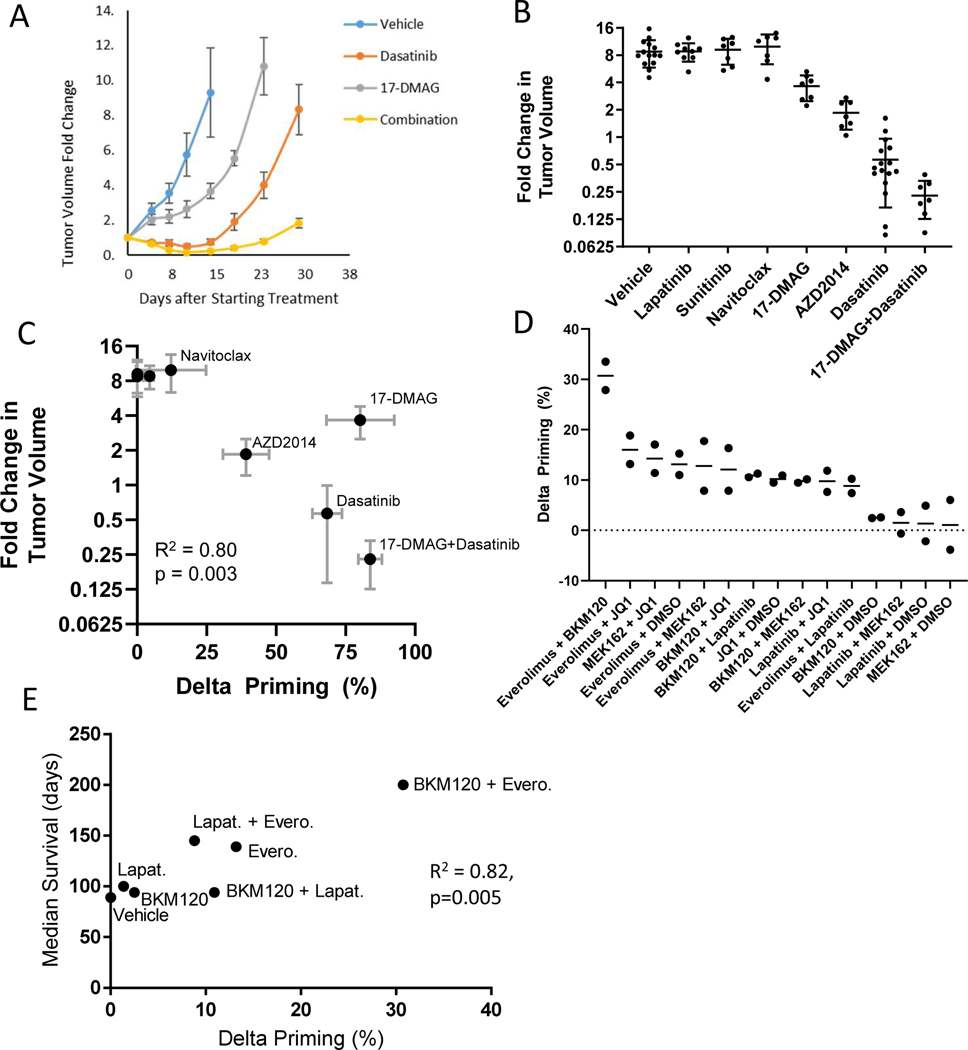
Fig. 2: HT-DBP predicts in vivo response in breast cancer models.
(A) Change in tumor volume relative to the start of treatment over time for select drug treatment in MMTV-PyMT tumors. Data are mean ± SEM of at least 7 mice per group. (B) Fold change in tumor volume at day 14 relative to the start of treatment for MMTV-PyMT tumors. Each dot represents a single mouse; data are mean ± SEM of at least 7 mice per group. Dasatinib (10 mg/kg, i.p), 17-DMAG (10 mg/kg, i.p), AZD2014 (15 mg/kg, p.o), lapatinib (50 mg/kg, p.o), or sunitinib (50 mg/kg, p.o) were dosed daily 5 days a week for 2 weeks. Navitoclax (100mg/kg, p.o) was dosed daily for 2 weeks. (C) Correlation between dynamic BH3 profiling, and MMTV-PyMT tumor response in vivo. (R2 = 0.83; p = 0.004; Pearson). Delta priming data (horizontal) are mean ± SD of n=3 independent experiments at 1 μM of drug. Tumor volume data (vertical) are mean ± SEM from 7–15 mice per group. (D) HT-DBP of select drug combinations in the DF-BM355 breast cancer PDX model. Each point represents an independent experiment. Lines represent mean of N = 2 experiments. (E) Correlation between dynamic BH3 profiling, and median survival of DF-BM355 mice treated with compounds. N = 5–9 mice per group. (R2 = 0.82, p=0.005; Pearson).
We next asked whether we could perform HT-DBP on small volume samples to correlate ex vivo drug sensitivity with known in vivo responses. Using 3×10^5 viable cells from the breast cancer brain metastasis PDX model DF-BM355(32), we used HT-DBP to rank the predicted activity of drug combinations (fig. S10A,B; Fig. 2D). We found good correlation between HT-DBP, and previously reported median survival of drug treated animals (Fig. 2E) (32). The correlation between ex vivo HT-DBP measurements and in vivo response for MMTV-PyMT and DF-BM355 breast cancer models indicate that HT-DBP can potentially assign effective in vivo therapies.
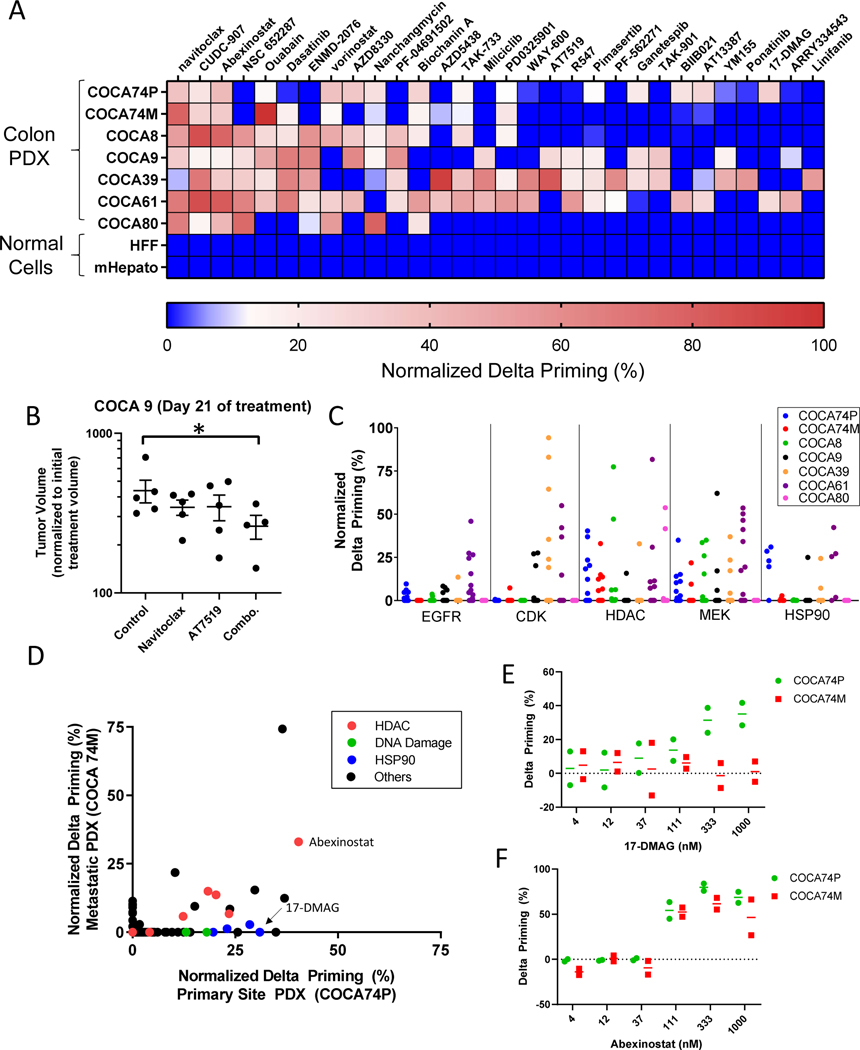
The ability to rapidly pharmacotype human tumors using HT-DBP could enable the use of patient-specific ex vivo drug sensitivity data to guide clinical decision making and drug discovery at a scale heretofore not possible. To investigate the feasibility of this approach, we performed HT-DBP using 1650 compounds at 1μM concentration on 7 different human colorectal tumors isolated from PDX models (33). Freshly isolated tumor cells were identified based on EpCam staining (fig. S11A) and were screened at a single synthetic Bim peptide concentration (fig. S11, B to D). To facilitate comparisons between chemical screens on different tumors, we normalized delta priming between the standard deviation of DMSO treated wells, and the delta priming value of the compound that caused the maximum apoptotic sensitization (fig. S12, A and B). This resulted in a map of chemical sensitivities of the 7 PDX models and 2 non-tumor cells (fig. S13, data file S10). We examined the top 35 hits that did not affect healthy cells from the hepatocyte or human foreskin fibroblast screens (Fig. 3A), and identified chemical vulnerabilities observed across all tumors (such as to navitoclax and abexinostat) as well as more private vulnerabilities observed in a fraction of tumors (such as to vorinostat and milciclib). Based on the screen, we treated the COCA9 PDX model with navitoclax, or the pan-CDK inhibitor AT7519, or both and found that while single agents did not have an effect, the combination treatment delayed tumor growth (Fig. 3B). Ultimately, a resulting matrix of drug sensitivities could form the foundation for development of effective patient-specific combination therapy strategies.
Fig. 3: Identification of apoptotic sensitizing compounds and drug targets in patient derived xenografts of colorectal cancer.
(A) Delta priming measurements of the top 35 hits from HT-DBP on seven PDX models of colorectal cancer and healthy cells Red indicates compounds that cause the highest increase in apoptotic priming. Dark blue indicates compounds that are less than 3 times standard deviation of DMSO treated wells. Data represent mean of 2 replicates. (B) Tumor volume after 21 days of in vivo treatment of COCA9 with navitoclax (100 mg/kg, p.o., daily), AT7519 (15 mg/kg, i.p., daily) or a combination of navitoclax and AT7519. Asterisk indicates a significant difference in tumor volume relative to vehicle treated cells (Mann-Whitney, p=0.03). Each point represents a single mouse; n = 4–5 mice per treatment arm. (C) Delta priming of the different PDX models based on nominal drug targets. Nominal targets include EGFR (31 compounds), pan-CDK (15 compounds), pan-HDAC (20 compounds), MEK (12 compounds), HSP90 (5 compounds). Data represent mean of 2 replicates. (D) Comparison of delta priming in COCA74P and COCA74M. Data represent mean of 2 replicates. (E) Delta priming of 17-DMAG for COCA74P and COCA74M. Each point represents an independent experiment. Lines represent mean of N = 2 experiments. (F) Delta priming of abexinostat for COCA74P and COCA74M. Each point represents an independent experiment. Lines represent mean of N = 2 experiments.
Next, using HT-DBP we asked whether we could use chemical sensitivity results to infer pathway dependencies present in individual colon cancers. While functional genetic tools are frequently used to identify vulnerabilities in cell lines, use of these tools in primary tissue is difficult. By using annotations of nominal small molecule targets of a known bioactive library with sufficient protein target coverage, we asked whether we could identify pathway vulnerabilities of each tumor without extended ex vivo culture (Fig. 3C, fig. S14, data file S11) (34) In deconvoluting protein targets of chemical compounds, we found that broad CDK and not pan-HDAC inhibition was an effective strategy for sensitizing the COCA39 PDX model for apoptosis (fig. S14). Conversely, the COCA74M PDX model were sensitive to pan-HDAC inhibition but not pan-CDK inhibition (fig. S14). Thus, HT-DBP permits chemical biology approaches to identifying mechanism of action in primary tumor cells without extended ex vivo culture.
Notably, in comparing colon cancer PDX models derived from the primary and a metastatic site from the same patient (COCA74P and COCA74M), we identified both common and different chemical vulnerabilities (Fig. 3D). For example, several pan-HDAC inhibitors sensitize both primary and metastatic tumors for apoptosis, however, several HSP90 inhibitors sensitize only the primary tumor for apoptosis. Specifically, the HSP90 inhibitor 17-DMAG shows a preferential sensitization effect on COCA74P relative to COCA74M (Fig 3E), while the pan-HDAC inhibitor abexinostat sensitizes both tumors equivalently for apoptosis (Fig 3F). This data implies that intra-patient heterogeneity in drug response may exist between primary and metastatic sites.
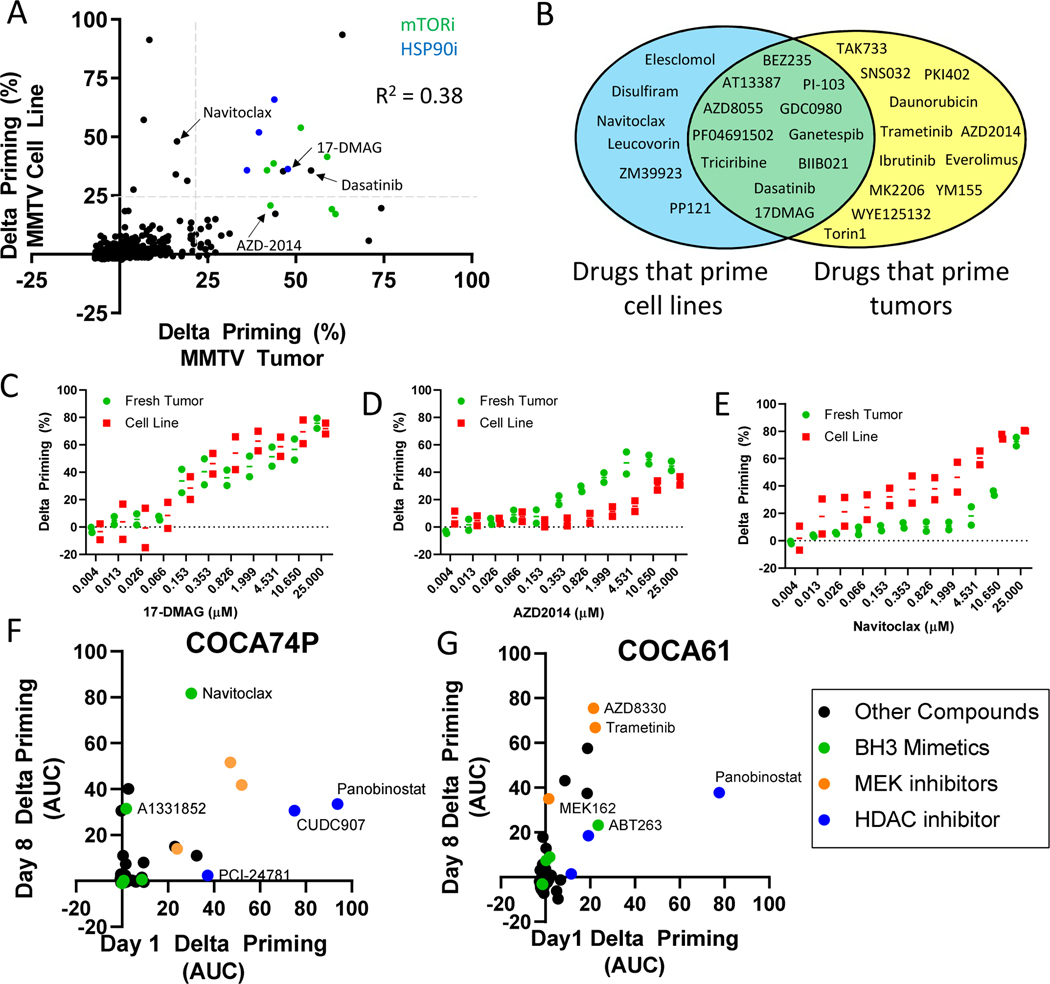
Historically, established cancer cell lines have proven useful but imprecise tools for assigning in vivo therapy (35), driving our concern about ex vivo methods that require prolonged ex vivo culture. We hypothesized that adaptation to long term ex vivo culture might alter chemical sensitivities compared to the fresh tumor. This hypothesis has never, to our knowledge, been directly tested. We realized that since tumors from the MMTV-PyMT model readily form cell lines, we possessed a unique opportunity with HT-DBP to directly test this hypothesis for the first time across a broad panel of compounds. We therefore performed HT-DBP on early passage cancer cell lines, expanded in 2D cell culture for over 1 month, derived from the same pool of primary tumor cells tested above in Fig. 1D (fig. S15, A and B; Fig. 4A; and data file S12). Evaluating compounds that did not prime healthy cells from the hepatocyte or human foreskin fibroblast screen at a concentration of 1 μM, several compounds sensitized both the tumor and the derived cell line for apoptosis, like HSP90 inhibitors. We also identified compounds that primed the cell line but not the tumor, and the tumor but not the cell line (Fig. 4, A and B, and data file S13). Differences between freshly isolated tumor cells and cultured cancer cell lines was greater than assay noise measured in technical replicates or biological replicates (fig. S8, C and D). We used HT-DBP to evaluate drug dose responses of compounds that scored in the screen, and for specific drugs measured both similar or differential activity in cell lines compared to cells from freshly isolated tumors (Fig. 4, C to E; fig S15C; and data file S14). This includes AZD2014 an mTOR inhibitor which preferentially primed the fresh tumor cells for apoptosis relative to the cell line (Fig. 4D), and navitoclax, which preferentially primed the cell line relative to the freshly isolated tumor cells for apoptosis (Fig. 4E). Notably, navitoclax does not noticeably alter tumor growth in vivo, but AZD-2014 delays tumor growth in vivo (Fig. 2, B and C), and may have been missed or deprioritized by performing chemical screens on cancer cell lines.
Fig. 4: Evolution of apoptotic chemical vulnerabilities in cell culture conditions measured by HT-DBP.
(A) Comparison of freshly isolated MMTV-PyMT tumors with cells cultured ex vivo for one month. Compounds that inhibit HSP90 are colored blue. Compounds that inhibit mTOR are colored green. Grey dotted lines indicate 3 standard deviations of DMSO treated wells. R2 = 0.38 by Pearson analysis. Data are means of two independent screens. (B) Identity of compounds that primed the freshly isolated tumor only, the cell line only, or both. This analysis only evaluated compounds that did not prime healthy cells for apoptosis. (C to E) Comparison of delta priming by (C) 17-DMAG, (D) AZD2014, and (E) navitoclax on freshly isolated MMTV-PyMT tumor cells cultured ex vivo for one month. Each point represents an independent experiment. Lines represent mean of N = 2 experiments. (F and G) Comparison of drug-induced delta priming in (F) COCA74P and (G) COCA61 cells at days 1 and 8 after tumor extraction. Data are means of triplicates.
To further evaluate changes in drug induced apoptotic sensitivity during ex vivo culture, we performed HT-DBP on freshly isolated tumor cells from the COCA74P and COCA61 colorectal cancer PDX models, and on cells from each model that were grown ex vivo for 8 days. In COCA74P, we observed that there was enhanced sensitivity to ABT263 and A1331852 after 8 days in culture (Fig. 4F, fig. S16, and data file S15). Conversely, in COCA61, we observed minimal differences in sensitivity to ABT263 and A1331852 after 8 days in culture, however, we found that the MEK inhibitors trametinib and AZD8330 induced greater apoptotic priming in the day 8 cultures relative to the day 1 culture (Fig 4G, fig. S16). Notably several chemical dependencies were preserved as cells remain cultured in vitro (Fig 4, F and G). Ultimately, changes in chemical sensitivity with prolonged ex vivo culture may represent one reason for the mixed translational track record of cancer cell line chemical screens.
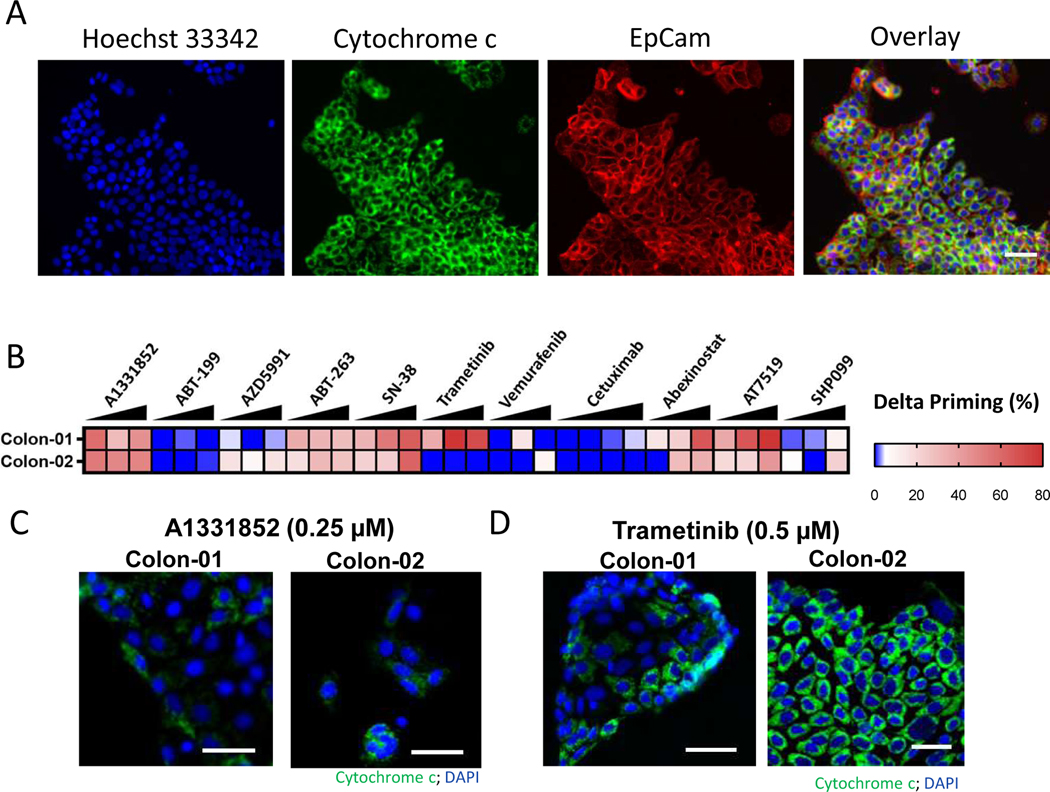
Finally, we sought to determine apoptotic chemical sensitivities on human colon tumors directly obtained from patient tumors without intervening ex vivo culture or propagation in mice. HT-DBP was performed on colon tumors resected from the primary site and using a limited panel of drugs (Fig. 5, A and B, and data file S16). Similar to results in PDX models, we found that Bcl-XL inhibition using A-1331852 increased apoptotic signaling in both tumors (Fig. 5, B and C). However, MEK inhibition using trametinib increased apoptotic signaling in one tumor, but not the other (Fig. 5, B and D). Finally, abexinostat (a pan-HDAC inhibitor) and AT-7519 (a pan-CDK inhibitor) which increased apoptotic signaling in colon PDX models also increased apoptotic signaling in primary human colon tumors (Fig. 5B). Thus HT-DBP enables chemical screening on patient tumors with limited time ex vivo and can identify patient to patient differences in chemical sensitivities.
Fig. 5: Identification of apoptotic sensitizers in primary colon cancer using HT-DBP.
(A) Representative image of Hoechst 33342 (blue), EpCam immunofluorescence (red) and cytochrome c immunofluorescence (green) in a primary colon tumor after 24 hours of ex vivo DMSO treatment and after a BH3 profile. Scale bars, 75 μm. (B) HT-DBP on colon tumors (n=3) using a limited panel of compounds at concentrations of 100 nM, 250 nM and 500 nM. (C) Images of cytochrome c loss in colon tumors using DBP with of the Bcl-XL inhibitor A-1331852. Scale bars, 50 μm. (D) Images of cytochrome c loss in Colon-01 and cytochrome c retention in Colon-02 tumors using DBP of the MEK inhibitor trametinib. Scale bars, 50 μm. Images are representative of 3 replicates.
Discussion
There has not previously been a method available to directly and immediately assess chemical vulnerabilities of a patient’s fresh primary tumor across a large panel of compounds. HT-DBP provides valuable chemical vulnerability information on the actual tumor without intervening model creation. Performing accurate pre-clinical drug screening or determining effective personalized therapy regimens using patient derived models of cancer requires high pharmacologic fidelity of the models to the original tumor. While high genomic fidelity is often observed in patient derived models of cancer (36), an evaluation of pharmacologic fidelity is not typically performed.
Cancer cell lines have been a workhorse of cancer biology for many decades. However, identification of chemical vulnerabilities in cancer cell lines have a mixed record for effective translation to useful clinical treatments. Indeed, even drug screens on nominally identical cell lines cultured by different laboratories return divergent results (37, 38). Here, by developing a high-throughput method to evaluate drug sensitivity of tumors within 24 hours of excision, we provide data that the very process of prolonged ex vivo propagation rapidly alters chemical vulnerabilities compared to the primary tumor. Specifically, dissimilarities that we observed in drug sensitivities of the MMTV-PyMT mouse model of breast cancer at 24 hours ex vivo and after 30 days of ex vivo cell culture demonstrates that drug sensitivity evolves with prolonged ex vivo culture. Importantly AZD-2014 showed minimal drug sensitivity with extended ex vivo culture, however, this drug delayed tumor growth in vivo, indicating the potential to miss promising therapies by screening on cancer cell lines. Applying HT-DBP to other tumor types will help identify drug sensitivity artifacts of extended ex vivo culture. Moreover, HT-DBP could screen for specific culture conditions that minimize ex vivo artifacts of chemical sensitivity.
While the direct identification of clinically relevant apoptotic sensitizing chemicals represents a direct translational application of HT-DBP, identifying signal pathway vulnerabilities in primary tumors could motivate the development of targeted inhibitors (34). Here, using compound libraries with sufficient protein target coverage, we used chemical biology strategies and HT-DBP to evaluate signaling pathways that represent vulnerabilities in freshly isolated tumors from the MMTV-PyMT mouse model and PDX models of colorectal cancer. This was enabled by a 100-fold increase in the number of compounds screened as potential apoptotic sensitizers compared to the original DBP method. Importantly, the identification of signal pathway vulnerabilities using genetic perturbagens with primary samples requires weeks of ex vivo culture. We anticipate that applying clinically relevant compounds and tool compounds in HT-DBP screens will enable the application of chemical biology strategies to identify signal pathway vulnerabilities in primary patient tumors without the need for prolonged ex vivo culture.
A potential use of HT-DBP will be the assembly of combination regimens for typically chemo-refractory tumors. Evaluating combination therapies requires a factorial increase in the number of screening wells, which is not amenable to freshly isolated tumors, or primary tumors. One of the theoretical advantages of DBP over conventional measures of the cell death is the ability to identify compounds that sensitize cells for apoptosis, but may not induce frank cell death, rendering them invisible by most other techniques. Such compounds that sensitize cells for apoptosis, but that do not kill cells could be effective in combination.
Precision medicine in oncology has relied almost exclusively on genomic information. However, most patients do not benefit from such methods, and alternatives must be identified (4). Technologies like HT-DBP that can perform unbiased screening of chemical libraries on human tumors and function as patient biomarkers have theoretical advantages in drug discovery. Identification of effective biomarkers after drug development is non-trivial, and in the absence of accurate biomarkers, drugs that are effective in a small fraction of patients are generally not approved or are not widely adopted (39). We anticipate that the adaptation of functional biomarkers such as HT-DBP into high-throughput screening will identify new therapeutics. In addition, biomarkers like HT-DBP can then be deployed as companion diagnostics to ensure enrollment of appropriate patient populations in clinical trials and clinical practice. This approach would likely decrease the failure rates of drug development programs and provide greater clinical benefit than sequential drug discovery and biomarker identification.
Materials and Methods:
Cell Culture
MDA-MB-231 and SU86.86 cells were cultured in RPMI (Gibco) + 10% FBS (Gibco) and penicillin/streptomycin (Gibco). When extracted, MMTV-PyMT tumor cells were cultured in RPMI + 10% FBS + 1X Pen/Strep. MMTV-PyMT cell lines were derived by culture in the same media for 1 month. Cells were passaged approximately every 3–4 days. Freshly isolated murine hepatocytes were cultured in Advanced DMEM/F12 (Gibco) + 10% FBS (Gibco). Human foreskin fibroblasts were cultured in DMEM/F12 (Gibco) + 10% FBS (Gibco) and penicillin streptomycin (Gibco). Panc8902 cells were cultured in DMEM + 10%FBS + penicillin/streptomycin.
Assessment of Cell Death
Annexin V staining was performed by staining cells in annexin V binding buffer for 15 minutes, fixing cells with paraformaldehyde and glutaraldehyde for 10 minutes, and neutralizing fixatives with Tris and Glycine. Cells were washed and imaged using an IXM XLS high content microscope (Molecular Devices), and the fraction of annexin V positive cells were quantified using the Multi Wavelength Cell Scoring module in Metamorph.
Animal Models
For the MMTV-PyMT model, animals were maintained at the Dana Farber Cancer Institute (Protocol 10–067). For primary screens, tumors were collected from different mammary fat pads, and pooled to get appropriate numbers of cells. For animal treatments, MMTV-PyMT cells that were extracted from mice and not cultured in vivo were injected along with Matrigel (Corning) into the mammary fat pad of syngenic FVB/NJ mice as described previously (40). 7–15 mice were used for each in vivo treatment arm. Dasatinib (ApexBio) and 17-DMAG (ApexBio) were IP dosed at 10 mg/kg. AZD2014 (ApexBio) was dosed via oral gavage once daily at 15 mg/kg. Lapatinib (SelleckChem) was dosed via oral gavage at 50 mg/kg. Sunitinib (Apex Bio) was dosed via oral gavage at 50 mg/kg. Navitoclax (MedChemExpress) was dosed via oral gavage at 100 mg/kg. All treatments were performed daily 5 days a week for 2 weeks.
With regard to breast PDX tumors, DF-BM355 tumors were previously treated with compounds in vivo (32), and were expanded as previously described (32). For colon PDX tumors, tissue to establish PDX models were obtained according to IRB approved research protocols (14–030). Fresh primary colorectal cancer biopsies were first incubated in an antibiotic cocktail of penicillin/streptomycin/amphotericin B/Ciprofloxacin for 1–2 hours and implanted into the flanks of 5 weeks old, female nude mice (Nu/Nu; Taconic). When the xenografts reached ~200 mm, 3 mice were sacrificed, and the tumors were harvested and serially passaged as subcutaneous implants of tumor fragments approximately 2–3 mm in diameter.
Human Samples
Colon tumors were resected from the primary site of patients as a part of standard of care. Patient consent and tumors were obtained according to IRB research protocols (03–189). Tumors were dissociated using collagenase 4 and hyaluronidase for 30 minutes to 1 hour. Cells were plated and drugged with indicated compounds and were BH3-profiled 24 hours after drug treatment.
Tumor and Tissue Dissociation
MMTV-PyMT tumors were dissociated using the GentleMACS and the Miltenyi Mouse dissociation kit (Miltenyi). Colon cancer PDX models were dissociated using the GentleMACS using a mixture of collagenase IV and hyaluronidase (Sigma).
Drug Treatments
HT-DBP and annexin V drug treatments on MDA-MB-231 cells were performed using the Selleck Bioactive library at ICCB at Harvard Medical School consisting of 1902 bioactive compounds. Compounds were screened at a concentration of 1 μM. Screens on freshly isolated tissue from the MMTV-PyMT tumors, murine hepatocytes, and human foreskin fibroblasts were performed at the Broad Institute using a Selleck bioactive library consisting of 1650 compounds all at a concentration of 1 μM. Drug screening libraries are not randomized between technical replicates. Most of the bioactive compounds have been tested in clinical or preclinical models. Permutations of 5 drugs (JQ1, BKM120, Everolimus, lapatinib, and MEK162) were evaluated in the DF-BM355 breast cancer model based on drug treatments in the initial publication. For DF-BM355 treatments, drugs were added using the D300e (Hewlett Packard/Tecan) digital dispenser at a concentration of 1 μM. All screens were at least performed in technical replicate. Screens on MDA-MB-231, MMTV-PyMT primary tissue and cell lines, human foreskin fibroblasts, adult mouse hepatocytes, and DF-BM355 tumors represent the average of biological replicates. Screens on colon PDX models represent the average of technical replicates. Screens on human colon primary samples represent the average of technical triplicates. Compounds from the MMTV-PyMT screen that were subsequently evaluated in vivo, were first evaluated ex vivo using the same stocks newly purchased compound that were also used in vivo. These drugs were added using the D300e digital drug dispenser. Drug treatments where there was a drug dose response were randomized. Cells were plated in either 3712, 3764BC, or 3542 384-well plates (Corning).
High-throughput dynamic BH3 profiling
Upon drug treatment, cells were incubated at 37°C. Subsequently media was washed from plates using the BioTek 406EL plate washer (BioTek). Media was replaced with PBS. A 2X concentrated BH3 profiling buffer was added to cells with appropriate levels of digitonin (0.001% for mouse cells, and 0.002% for human cells), and the appropriate levels of peptide. For peptide titrations, 2X BH3 profiling buffer was added manually. For BH3 profiling of drug treated plates at a single peptide concentration, the 2X buffer was added using the Thermo Multidrop Combi (Thermo Fisher). Cells were fixed in paraformaldehyde. Fixatives were neutralized using a Tris/Glycine buffer, and cells were subsequently stained with antibodies in either a saponin or tween 20 permeabilizing solution. Cells were stained overnight, and prior to imaging, stain solution was washed using the biotek plate washer. A detailed protocol for HT-DBP is provided in the supplemental material (text S1).
With the exception of adult mouse hepatocytes, HT-DBP is performed using the synthetic BH3 peptide modeled after the BIM protein. The synthetic Bim BH3 peptide did not induce cytochrome c release from adult mouse hepatocytes consistent with prior studies (41). Hepatocytes exclusively express the pro-apoptotic effector protein BAK, and not BAX, and more readily undergo mitochondrial outer membrane permeabilization in the presence of the synthetic Bid BH3 peptide. This is consistent with the preferential activation of BAX by BIM and the preferential activation of BAK by BID. We therefore used the synthetic BH3 peptide modeled after the Bid BH3 protein to perform HT-DBP with adult mouse hepatocytes.
Antibodies
Nuclei were stained with Hoechst33342 (Invitrogen). Cytochrome c was measured using the Cytochrome c-Alexa647 antibody (BioLegend). Mouse tumor cells were identified with Mouse EpCam-FITC (BioLegend). Colon tumor cells were identified using human EpCam-Alexa488 (Biolegend)
Imaging
All imaging was performed on the IXM XLS high content widefield microscope (Molecular Devices; at the ICCB at Harvard Medical School or the Broad Institute). A 10X objective was used to perform all imaging. We typically used a DAPI filter cube to measure Hoechst 33342 staining, FITC to measure Alexa488-EpCam staining, and a Cy5 cube to measure Cytochrome c-Alexa647 antibody staining.
Image and Data Analysis
Image analysis was performed in Metamorph using the Multi Wavelength Cell Scoring Module and the Adaptive Background Correction module to segment cells based on an intensity above local background. This results in an approximate single cell segmentation (for example shown in fig S7) and the area of cytochrome c intensity. Cells are scored as being positive or negative based on the area. All subsequent data analysis was performed in Excel or graph pad prism. All statistical analysis was performed in graph pad prism.
Statistical analysis
All chemical screens were performed in technical replicates. Correlation of technical replicates and biological replicates of HT-DBP or annexin staining were evaluated using Pearson two-tailed tests as indicated in figure legends. Correlation of ex vivo and in vivo responses of breast cancer models was performed using Pearson two-tailed tests. Identification of nominal protein/pathway targets that increase apoptotic sensitivity was performed using one-way ANOVA as indicated in the text. Correlation of HT-DBP with annexin staining in breast cancer cells was performed using Pearson two-tailed tests.
Supplementary Material
Text S1: Detailed HT-DBP protocol.
Figure S1: Schematic of dynamic BH3 profiling (DBP).
Figure S2: Selection of informative peptide screening concentrations.
Figure S3: Identification of a drug screening concentration
Figure S4: High-throughput dynamic BH3 profiling enables identification of compounds that sensitize cancer cells for apoptosis.
Figure S5: Similarity between FACS and microscopy dynamic BH3 profiles.
Figure S6: Images of stained MMTV-PyMT tumor cells.
Figure S7: Example of cell masks.
Figure S8: Screening data for freshly isolated MMTV-PyMT tumors.
Figure S9: Counter screens in healthy cells.
Figure S10: Supplemental data for in vivo validation experiments for HT-DBP.
Figure S11: Identification of compounds that sensitize the COCA9 colon cancer patient derived xenograft for apoptosis.
Figure S12: Quantification of apoptotic chemical vulnerabilities in colon cancer PDX models.
Figure S13: Heatmap of apoptotic chemical vulnerabilities in colon cancer PDX models.
Figure S14: Comparison of apoptotic chemical vulnerabilities in colon cancer PDX models.
Figure S15: Supplemental data for comparison of chemical vulnerabilities of freshly isolated and cultured cancer cells.
Figure S16: Supplemental data for comparison of chemical vulnerabilities of freshly isolated and cultured cancer cells.
Table S1: List of compounds used in the chemical screen.
Data file S1: Cell count and delta priming of MDA-MB-231 line.
Data file S2: Apoptosis and delta priming of MDA-MB-231 line.
Data file S3: Cell count and delta priming of MMTV-PyMT tumor.
Data file S10: Normalized delta priming of colorectal PDX models.
Data file S9: Delta priming of freshly isolated MMTV-PyMT tumors, by nominal drug target
Data file S8: Delta priming in adult mouse hepatocytes and MMTV-PyMT tumors
Data file S14: Dose-response by delta priming in MMTV-PyMT tumor and derived cell line.
Data file S15: Delta priming in colorectal PDX models.
Data file S4: Technical replicate of MMTV-PyMT delta priming.
Data file S7: Cell count and delta priming of human foreskin fibroblasts.
Data file S6: Cell count and delta priming of primary mouse hepatocytes.
Data file S12: Delta priming of MMTV-PyMT–derived cell line.
Data file S13: Correlation between MMTV-PyMT tumor and derived cell line.
Data file S16: Delta priming in primary human CRC cells ex vivo.
Data file S5: Biological replicate of MMTV-PyMT delta priming.
Data file S11: Normalized delta priming of colorectal PDX models, by nominal target.
Acknowledgements:
We acknowledge the support from the cDOT group (Broad Institute). We thank Peter Sorger, Laura Mailszewski, Caroline Shamu and Jennifer Smith for support at the LSP, ICCB and HiTS at Harvard Medical School. We acknowledge David Tuveson (Cold Spring Harbor Laboratory) for useful discussions and for coining the term “pharmacotype”.
Funding: AJA acknowledges support from the Doris Duke Charitable Foundation, Pancreatic Cancer Action Network, National Cancer Institute K08 CA218420-01 and P50CA127003. AL acknowledges support from R01 CA205967, R35 CA242427, Ludwig Cancer Research at Harvard and the Starr Cancer Consortium. PB acknowledges support from the Barr Foundation. KN acknowledges support from P50 CA127003, R01 CA205406, and the Project P Fund. JJZ acknowledges support from R35 CA210057, P50 CA168504, DoD W81XWH-18-1-0491 and Breast Cancer Research Foundation.
Footnotes
Competing interests: AL discloses consulting and sponsored research agreements with AbbVie, Novartis, and Astra-Zeneca. He is an equity-holding founder of Flash Therapeutics and is on the SAB of Dialectic Therapeutics. The following are US Patents regarding BH3 profiling, owned by Dana-Farber: 10,393,733; 9,902,759; 9,856,303; 9,540,674; 8,221,966; 7,868,133. AL, JR and PB are inventors on patent applications US20180128813A1, US20180120297A1 held/submitted by Dana-Farber Cancer Institute that covers High-Throughput BH3 Profiling. AJA is a consultant for Oncorus, Inc. KN discloses sponsored research agreements with Pharmavite, Genentech, Gilead Sciences, Celgene, Trovagene, Tarrex Biopharma, Revolution Medicines, and Evergrande Group. KN discloses serving on advisory boards for Genentech, Lilly, Bayer, Seattle Genetics, and Array Biopharma. KN has served as a paid consultant for Tarrexx Biopharma. JN has a consulting relationship with Geode Therapeutics. JJZ is a founder and board director of Crimson Biotech and Geode Therapeutics. JLG is a consultant for Glaxo-Smith Kline (GSK) and Array BioPharma and receives sponsored research support from GSK and Eli Lilly. All other authors declare they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes:
- 1.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O’Dwyer PJ, Lee RJ, Grippo JF, Nolop K, and Chapman PB. 2010. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine 363: 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, and Meyerson M.. 2004. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 3.Aguirre AJ, Nowak JA, Camarda ND, Moffitt RA, Ghazani AA, Hazar-Rethinam M, Raghavan S, Kim J, Brais LK, Ragon D, Welch MW, Reilly E, McCabe D, Marini L, Anderka K, Helvie K, Oliver N, Babic A, Da Silva A, Nadres B, Van Seventer EE, Shahzade HA, St Pierre JP, Burke KP, Clancy T, Cleary JM, Doyle LA, Jajoo K, McCleary NJ, Meyerhardt JA, Murphy JE, Ng K, Patel AK, Perez K, Rosenthal MH, Rubinson DA, Ryou M, Shapiro GI, Sicinska E, Silverman SG, Nagy RJ, Lanman RB, Knoerzer D, Welsch DJ, Yurgelun MB, Fuchs CS, Garraway LA, Getz G, Hornick JL, Johnson BE, Kulke MH, Mayer RJ, Miller JW, Shyn PB, Tuveson DA, Wagle N, Yeh JJ, Hahn WC, Corcoran RB, Carter SL, and Wolpin BM. 2018. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Discov 8: 1096–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Letai A. 2017. Functional precision cancer medicine-moving beyond pure genomics. Nat Med 23: 1028–1035. [DOI] [PubMed] [Google Scholar]
- 5.Burstein HJ, Mangu PB, Somerfield MR, Schrag D, Samson D, Holt L, Zelman D, Ajani JA, and American O. Society of Clinical. 2011. American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29: 3328–3330. [DOI] [PubMed] [Google Scholar]
- 6.Cree IA, Kurbacher CM, Lamont A, Hindley AC, Love S, and T. C. A. O. C. T. Group. 2007. A prospective randomized controlled trial of tumour chemosensitivity assay directed chemotherapy versus physician’s choice in patients with recurrent platinum-resistant ovarian cancer. Anticancer Drugs 18: 1093–1101. [DOI] [PubMed] [Google Scholar]
- 7.Mi Z, Holmes FA, Hellerstedt B, Pippen J, Collea R, Backner A, Bush JE, Gallion HH, Wells A, and O’Shaughnessy JA. 2008. Feasibility assessment of a chemoresponse assay to predict pathologic response in neoadjuvant chemotherapy for breast cancer patients. Anticancer Res 28: 1733–1740. [PubMed] [Google Scholar]
- 8.Ugurel S, Schadendorf D, Pfohler C, Neuber K, Thoelke A, Ulrich J, Hauschild A, Spieth K, Kaatz M, Rittgen W, Delorme S, Tilgen W, Reinhold U, and Dermatologic Cooperative Oncology G.. 2006. In vitro drug sensitivity predicts response and survival after individualized sensitivity-directed chemotherapy in metastatic melanoma: a multicenter phase II trial of the Dermatologic Cooperative Oncology Group. Clin Cancer Res 12: 5454–5463. [DOI] [PubMed] [Google Scholar]
- 9.Wu B, Zhu JS, Zhang Y, Shen WM, and Zhang Q.. 2008. Predictive value of MTT assay as an in vitro chemosensitivity testing for gastric cancer: one institution’s experience. World J Gastroenterol 14: 3064–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevens MM, Maire CL, Chou N, Murakami MA, Knoff DS, Kikuchi Y, Kimmerling RJ, Liu H, Haidar S, Calistri NL, Cermak N, Olcum S, Cordero NA, Idbaih A, Wen PY, Weinstock DM, Ligon KL, and Manalis SR. 2016. Drug sensitivity of single cancer cells is predicted by changes in mass accumulation rate. Nat Biotechnol 34: 1161–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, Piao H, Horowitz NS, Berkowitz RS, Matulonis U, Janne PA, Amrein PC, Cichowski K, Drapkin R, and Letai A.. 2015. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell 160: 977–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, and Clevers H.. 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161: 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sachs N, and Clevers H.. 2014. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev 24: 68–73. [DOI] [PubMed] [Google Scholar]
- 14.Hill SJ, Decker B, Roberts EA, Horowitz NS, Muto MG, Worley MJ Jr., Feltmate CM, Nucci MR, Swisher EM, Nguyen H, Yang C, Morizane R, Kochupurakkal BS, Do KT, Konstantinopoulos PA, Liu JF, Bonventre JV, Matulonis UA, Shapiro GI, Berkowitz RS, Crum CP, and D’Andrea AD. 2018. Prediction of DNA Repair Inhibitor Response in Short-Term Patient-Derived Ovarian Cancer Organoids. Cancer Discov 8: 1404–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich S, Oles M, Lu J, Sellner L, Anders S, Velten B, Wu B, Hullein J, da Silva Liberio M, Walther T, Wagner L, Rabe S, Ghidelli-Disse S, Bantscheff M, Oles AK, Slabicki M, Mock A, Oakes CC, Wang S, Oppermann S, Lukas M, Kim V, Sill M, Benner A, Jauch A, Sutton LA, Young E, Rosenquist R, Liu X, Jethwa A, Lee KS, Lewis J, Putzker K, Lutz C, Rossi D, Mokhir A, Oellerich T, Zirlik K, Herling M, Nguyen-Khac F, Plass C, Andersson E, Mustjoki S, von Kalle C, Ho AD, Hensel M, Durig J, Ringshausen I, Zapatka M, Huber W, and Zenz T.. 2018. Drug-perturbation-based stratification of blood cancer. J Clin Invest 128: 427–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pemovska T, Kontro M, Yadav B, Edgren H, Eldfors S, Szwajda A, Almusa H, Bespalov MM, Ellonen P, Elonen E, Gjertsen BT, Karjalainen R, Kulesskiy E, Lagstrom S, Lehto A, Lepisto M, Lundan T, Majumder MM, Marti JM, Mattila P, Murumagi A, Mustjoki S, Palva A, Parsons A, Pirttinen T, Ramet ME, Suvela M, Turunen L, Vastrik I, Wolf M, Knowles J, Aittokallio T, Heckman CA, Porkka K, Kallioniemi O, and Wennerberg K.. 2013. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov 3: 1416–1429. [DOI] [PubMed] [Google Scholar]
- 17.Schmidl C, Vladimer GI, Rendeiro AF, Schnabl S, Krausgruber T, Taubert C, Krall N, Pemovska T, Araghi M, Snijder B, Hubmann R, Ringler A, Runggatscher K, Demirtas D, de la Fuente OL, Hilgarth M, Skrabs C, Porpaczy E, Gruber M, Hoermann G, Kubicek S, Staber PB, Shehata M, Superti-Furga G, Jager U, and Bock C.. 2019. Combined chemosensitivity and chromatin profiling prioritizes drug combinations in CLL. Nat Chem Biol 15: 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snijder B, Vladimer GI, Krall N, Miura K, Schmolke AS, Kornauth C, Lopez O. de la Fuente, Choi HS, van der Kouwe E, Gultekin S, Kazianka L, Bigenzahn JW, Hoermann G, Prutsch N, Merkel O, Ringler A, Sabler M, Jeryczynski G, Mayerhoefer ME, Simonitsch-Klupp I, Ocko K, Felberbauer F, Mullauer L, Prager GW, Korkmaz B, Kenner L, Sperr WR, Kralovics R, Gisslinger H, Valent P, Kubicek S, Jager U, Staber PB, and Superti-Furga G.. 2017. Image-based ex-vivo drug screening for patients with aggressive haematological malignancies: interim results from a single-arm, open-label, pilot study. Lancet Haematol 4: e595–e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, Moore Vdel G, Deng J, Anderson KC, Richardson P, Tai YT, Mitsiades CS, Matulonis UA, Drapkin R, Stone R, Deangelo DJ, McConkey DJ, Sallan SE, Silverman L, Hirsch MS, Carrasco DR, and Letai A.. 2011. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science 334: 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, and Letai A.. 2006. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 9: 351–365. [DOI] [PubMed] [Google Scholar]
- 21.Townsend EC, Murakami MA, Christodoulou A, Christie AL, Koster J, DeSouza TA, Morgan EA, Kallgren SP, Liu H, Wu SC, Plana O, Montero J, Stevenson KE, Rao P, Vadhi R, Andreeff M, Armand P, Ballen KK, Barzaghi-Rinaudo P, Cahill S, Clark RA, Cooke VG, Davids MS, DeAngelo DJ, Dorfman DM, Eaton H, Ebert BL, Etchin J, Firestone B, Fisher DC, Freedman AS, Galinsky IA, Gao H, Garcia JS, Garnache-Ottou F, Graubert TA, Gutierrez A, Halilovic E, Harris MH, Herbert ZT, Horwitz SM, Inghirami G, Intlekofer AM, Ito M, Izraeli S, Jacobsen ED, Jacobson CA, Jeay S, Jeremias I, Kelliher MA, Koch R, Konopleva M, Kopp N, Kornblau SM, Kung AL, Kupper TS, LeBoeuf NR, LaCasce AS, Lees E, Li LS, Look AT, Murakami M, Muschen M, Neuberg D, Ng SY, Odejide OO, Orkin SH, Paquette RR, Place AE, Roderick JE, Ryan JA, Sallan SE, Shoji B, Silverman LB, Soiffer RJ, Steensma DP, Stegmaier K, Stone RM, Tamburini J, Thorner AR, van Hummelen P, Wadleigh M, Wiesmann M, Weng AP, Wuerthner JU, Williams DA, Wollison BM, Lane AA, Letai A, Bertagnolli MM, Ritz J, Brown M, Long H, Aster JC, Shipp MA, Griffin JD, and Weinstock DM. 2016. The Public Repository of Xenografts Enables Discovery and Randomized Phase II-like Trials in Mice. Cancer Cell 29: 574–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etchin J, Montero J, Berezovskaya A, Le BT, Kentsis A, Christie AL, Conway AS, Chen WC, Reed C, Mansour MR, Ng CE, Adamia S, Rodig SJ, Galinsky IA, Stone RM, Klebanov B, Landesman Y, Kauffman M, Shacham S, Kung AL, Wang JC, Letai A, and Look AT. 2016. Activity of a selective inhibitor of nuclear export, selinexor (KPT-330), against AML-initiating cells engrafted into immunosuppressed NSG mice. Leukemia 30: 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corsello SM, Bittker JA, Liu Z, Gould J, McCarren P, Hirschman JE, Johnston SE, Vrcic A, Wong B, Khan M, Asiedu J, Narayan R, Mader CC, Subramanian A, and Golub TR. 2017. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med 23: 405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, and Pollard JW. 2003. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. The American journal of pathology 163: 2113–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gimenez-Cassina A, Garcia-Haro L, Choi CS, Osundiji MA, Lane EA, Huang H, Yildirim MA, Szlyk B, Fisher JK, Polak K, Patton E, Wiwczar J, Godes M, Lee DH, Robertson K, Kim S, Kulkarni A, Distefano A, Samuel V, Cline G, Kim YB, Shulman GI, and Danial NN. 2014. Regulation of hepatic energy metabolism and gluconeogenesis by BAD. Cell Metab 19: 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, Mestan J, Cowan-Jacob SW, Lee FY, Heinrich MC, Deininger MW, and Druker BJ. 2005. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer research 65: 4500–4505. [DOI] [PubMed] [Google Scholar]
- 27.Ge J, Normant E, Porter JR, Ali JA, Dembski MS, Gao Y, Georges AT, Grenier L, Pak RH, Patterson J, Sydor JR, Tibbitts TT, Tong JK, Adams J, and Palombella VJ. 2006. Design, synthesis, and biological evaluation of hydroquinone derivatives of 17-amino-17-demethoxygeldanamycin as potent, water-soluble inhibitors of Hsp90. J Med Chem 49: 4606–4615. [DOI] [PubMed] [Google Scholar]
- 28.Guichard SM, Curwen J, Bihani T, D’Cruz CM, Yates JW, Grondine M, Howard Z, Davies BR, Bigley G, Klinowska T, Pike KG, Pass M, Chresta CM, Polanska UM, McEwen R, Delpuech O, Green S, and Cosulich SC. 2015. AZD2014, an Inhibitor of mTORC1 and mTORC2, Is Highly Effective in ER+ Breast Cancer When Administered Using Intermittent or Continuous Schedules. Mol Cancer Ther 14: 2508–2518. [DOI] [PubMed] [Google Scholar]
- 29.Rusnak DW, Lackey K, Affleck K, Wood ER, Alligood KJ, Rhodes N, Keith BR, Murray DM, Knight WB, Mullin RJ, and Gilmer TM. 2001. The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol Cancer Ther 1: 85–94. [PubMed] [Google Scholar]
- 30.Sun L, Liang C, Shirazian S, Zhou Y, Miller T, Cui J, Fukuda JY, Chu JY, Nematalla A, Wang X, Chen H, Sistla A, Luu TC, Tang F, Wei J, and Tang C.. 2003. Discovery of 5-[5-fluoro-2-oxo-1,2- dihydroindol-(3Z)-ylidenemethyl]-2,4- dimethyl-1H-pyrrole-3-carboxylic acid (2-diethylaminoethyl)amide, a novel tyrosine kinase inhibitor targeting vascular endothelial and platelet-derived growth factor receptor tyrosine kinase. J Med Chem 46: 1116–1119. [DOI] [PubMed] [Google Scholar]
- 31.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, Roberts L, Tahir SK, Xiao Y, Yang X, Zhang H, Fesik S, Rosenberg SH, and Elmore SW. 2008. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res 68: 3421–3428. [DOI] [PubMed] [Google Scholar]
- 32.Ni J, Ramkissoon SH, Xie S, Goel S, Stover DG, Guo H, Luu V, Marco E, Ramkissoon LA, Kang YJ, Hayashi M, Nguyen QD, Ligon AH, Du R, Claus EB, Alexander BM, Yuan GC, Wang ZC, Iglehart JD, Krop IE, Roberts TM, Winer EP, Lin NU, Ligon KL, and Zhao JJ. 2016. Combination inhibition of PI3K and mTORC1 yields durable remissions in mice bearing orthotopic patient-derived xenografts of HER2-positive breast cancer brain metastases. Nat Med 22: 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon YCS, Fasani R, Aguirre AJ, Ng K, Elez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, and Meyerson M.. 2017. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Root DE, Flaherty SP, Kelley BP, and Stockwell BR. 2003. Biological mechanism profiling using an annotated compound library. Chem Biol 10: 881–892. [DOI] [PubMed] [Google Scholar]
- 35.Tseng YY, and Boehm JS. 2019. From cell lines to living biosensors: new opportunities to prioritize cancer dependencies using ex vivo tumor cultures. Curr Opin Genet Dev 54: 33–40. [DOI] [PubMed] [Google Scholar]
- 36.Lee JK, Liu Z, Sa JK, Shin S, Wang J, Bordyuh M, Cho HJ, Elliott O, Chu T, Choi SW, Rosenbloom DIS, Lee IH, Shin YJ, Kang HJ, Kim D, Kim SY, Sim MH, Kim J, Lee T, Seo YJ, Shin H, Lee M, Kim SH, Kwon YJ, Oh JW, Song M, Kim M, Kong DS, Choi JW, Seol HJ, Lee JI, Kim ST, Park JO, Kim KM, Song SY, Lee JW, Kim HC, Lee JE, Choi MG, Seo SW, Shim YM, Zo JI, Jeong BC, Yoon Y, Ryu GH, Kim NKD, Bae JS, Park WY, Lee J, Verhaak RGW, Iavarone A, Lee J, Rabadan R, and Nam DH. 2018. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat Genet 50: 1399–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ, and Quackenbush J.. 2013. Inconsistency in large pharmacogenomic studies. Nature 504: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, Strathdee CA, Dempster J, Lyons NJ, Burns R, Nag A, Kugener G, Cimini B, Tsvetkov P, Maruvka YE, O’Rourke R, Garrity A, Tubelli AA, Bandopadhayay P, Tsherniak A, Vazquez F, Wong B, Birger C, Ghandi M, Thorner AR, Bittker JA, Meyerson M, Getz G, Beroukhim R, and Golub TR. 2018. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goossens N, Nakagawa S, Sun X, and Hoshida Y.. 2015. Cancer biomarker discovery and validation. Transl Cancer Res 4: 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerriero JL, Sotayo A, Ponichtera HE, Castrillon JA, Pourzia AL, Schad S, Johnson SF, Carrasco RD, Lazo S, Bronson RT, Davis SP, Lobera M, Nolan MA, and Letai A.. 2017. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature 543: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarosiek KA, Chi X, Bachman JA, Sims JJ, Montero J, Patel L, Flanagan A, Andrews DW, Sorger P, and Letai A.. 2013. BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Mol Cell 51: 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Text S1: Detailed HT-DBP protocol.
Figure S1: Schematic of dynamic BH3 profiling (DBP).
Figure S2: Selection of informative peptide screening concentrations.
Figure S3: Identification of a drug screening concentration
Figure S4: High-throughput dynamic BH3 profiling enables identification of compounds that sensitize cancer cells for apoptosis.
Figure S5: Similarity between FACS and microscopy dynamic BH3 profiles.
Figure S6: Images of stained MMTV-PyMT tumor cells.
Figure S7: Example of cell masks.
Figure S8: Screening data for freshly isolated MMTV-PyMT tumors.
Figure S9: Counter screens in healthy cells.
Figure S10: Supplemental data for in vivo validation experiments for HT-DBP.
Figure S11: Identification of compounds that sensitize the COCA9 colon cancer patient derived xenograft for apoptosis.
Figure S12: Quantification of apoptotic chemical vulnerabilities in colon cancer PDX models.
Figure S13: Heatmap of apoptotic chemical vulnerabilities in colon cancer PDX models.
Figure S14: Comparison of apoptotic chemical vulnerabilities in colon cancer PDX models.
Figure S15: Supplemental data for comparison of chemical vulnerabilities of freshly isolated and cultured cancer cells.
Figure S16: Supplemental data for comparison of chemical vulnerabilities of freshly isolated and cultured cancer cells.
Table S1: List of compounds used in the chemical screen.
Data file S1: Cell count and delta priming of MDA-MB-231 line.
Data file S2: Apoptosis and delta priming of MDA-MB-231 line.
Data file S3: Cell count and delta priming of MMTV-PyMT tumor.
Data file S10: Normalized delta priming of colorectal PDX models.
Data file S9: Delta priming of freshly isolated MMTV-PyMT tumors, by nominal drug target
Data file S8: Delta priming in adult mouse hepatocytes and MMTV-PyMT tumors
Data file S14: Dose-response by delta priming in MMTV-PyMT tumor and derived cell line.
Data file S15: Delta priming in colorectal PDX models.
Data file S4: Technical replicate of MMTV-PyMT delta priming.
Data file S7: Cell count and delta priming of human foreskin fibroblasts.
Data file S6: Cell count and delta priming of primary mouse hepatocytes.
Data file S12: Delta priming of MMTV-PyMT–derived cell line.
Data file S13: Correlation between MMTV-PyMT tumor and derived cell line.
Data file S16: Delta priming in primary human CRC cells ex vivo.
Data file S5: Biological replicate of MMTV-PyMT delta priming.
Data file S11: Normalized delta priming of colorectal PDX models, by nominal target.