Abstract
Although previous studies have shown that the host immune response is crucial in determining clinical outcomes in COVID-19 patients, the association between host immune signatures and COVID-19 patient outcomes remains unclear. Based on the enrichment levels of 11 immune signatures (eight immune-inciting and three immune-inhibiting signatures) in leukocytes of 100 COVID-19 patients, we identified three COVID-19 subtypes: Im-C1, Im-C2, and Im-C3, by clustering analysis. Im-C1 had the lowest immune-inciting signatures and high immune-inhibiting signatures. Im-C2 had medium immune-inciting signatures and high immune-inhibiting signatures. Im-C3 had the highest immune-inciting signatures while the lowest immune-inhibiting signatures. Im-C3 and Im-C1 displayed the best and worst clinical outcomes, respectively, suggesting that antiviral immune responses alleviated the severity of COVID-19 patients. We further demonstrated that the adaptive immune response had a stronger impact on COVID-19 outcomes than the innate immune response. The patients in Im-C3 were younger than those in Im-C1, indicating that younger persons have stronger antiviral immune responses than older persons. Nevertheless, we did not observe a significant association between sex and immune responses in COVID-19 patients. In addition, we found that the type II IFN response signature was an adverse prognostic factor for COVID-19. Our identification of COVID-19 immune subtypes has potential clinical implications for the management of COVID-19 patients.
Keywords: COVID-19 subtypes, Antiviral immune response, Gene expression profiles, Clinical outcomes, Clustering analysis
Abbreviations: ssGSEA, single-sample gene-set enrichment analysis; HLA, Human leukocyte antigen; Tfh, T follicular helper cells; Th1 cells, Type 1 T helper cells; Th2 cells, Type 2 T helper cells; NK, Natural killer; pDCs, Plasmacytoid dendritic cells; Treg, Regulatory T cells; ICU, Intensive care unit; MVS, Mechanical ventilatory support; CV, cross validation; RF, Random Forest; APACHE II, Acute Physiology and Chronic Health Evaluation; SOFA, Sequential Organ Failure Assessment; HFD, Hospital-free days at day 45; WGCNA, Weighted gene co-expression network analysis; FDR, False discovery rate; RF, Random forest
1. Introduction
The COVID-19 pandemic caused by SARS-CoV-2 has resulted in more than 122 million cases and 2.7 million deaths as of March 19, 2021 [1]. The host immune defense system and immune response are crucial factors responsible for clinical outcomes in COVID-19 patients [2], [3], [4], [5], [6], [7]. For example, Takahashi et al. demonstrated that sex biases in COVID-19 were associated with differences in immune responses between males and females [2]. The excessive immune response to SARS-CoV-2, known as COVID-19 cytokine storm, may cause severe COVID-19 [8]. In fact, multiple organ dysfunction caused by aberrant immune responses is presented in a substantial number of severe COVID-19 patients [9]. Despite these previous studies, the association between host immune responses and clinical outcomes in COVID-19 patients is worthy of further in-depth investigation.
Previous studies have explored associations between immune signatures and clinical prognosis based on blood transcriptomic profiling in patients [10], [11]. For example, based on whole blood gene expression profiling, Zhang et al. identified two subtypes of sepsis, which displayed different immune responses and clinical outcomes [10]. Shankar et al. identified transcriptomic features in blood to predict paediatric patients with multiple organ dysfunction in need of intensive care [11]. In this study, using a publicly available RNA-Seq gene expression profiles in 100 leukocyte samples from COVID-19 patients [12], we performed an unsupervised learning to identify COVID-19 subtypes based on the enrichment levels of 11 immune signatures. Moreover, we characterized the immunological and clinical features of these COVID-19 subtypes. Furthermore, we investigated associations between immune signatures and clinical features in COVID-19 patients using three publicly available datasets, including GSE157103 [12], GSE156063 [13], and GSE152075 [14] (Table 1 ). This study aimed to provide new insights into the association between immune responses and clinical prognosis in COVID-19 patients.
Table 1.
A summary of the datasets.
| GSE157103 (12) | ||
|---|---|---|
| Tissue resource | Leukocyte from COVID-19 patients | |
| Sample size | n = 100 | |
| Demographic characteristics | ||
| Female sex – No. (%) | 38 (38) | |
| Age Range – No. (%) | ||
| Younger than 49 | 22 (22) | |
| 50–59 | 21 (21) | |
| 60–69 | 21 (21) | |
| 70–79 | 19 (19) | |
| 80 and older | 16 (16) | |
| Missing data | 1 (1) | |
| Clinical characteristics | ||
| ICU admission – No. (%) | 50 (50) | |
| Hospital free days at 45 days | 26 (median) | |
| Mechanical ventilation – No. (%) | 42 (42) | |
| Ventilator-free days | 28 (median) | |
| APACHE II score | 21 (median) | |
| SOFA score | 7 (median) | |
| Laboratory findings | ||
| C-reactive protein (mg/L) | 128.2 (median) | |
| Ferritin (μg/L) | 652 (median) | |
| Procalcitonin (μg/L) | 0.57 (median) | |
| D-dimer (mg/L) | 1.79 (median) | |
| Lactate (mmol/l) | 1.17 (median) | |
| GSE152075 (14) | ||
| Tissue resource | Nasopharyngeal swabs from COVID-19 patients | |
| Sample size | n = 430 | |
| Demographic characteristics | ||
| Female sex – No. (%) | 201 (46.7) | |
| Male sex – No. (%) | 176 (40.9) | |
| Missing data | 53 (12.3) | |
| Age Range – No. (%) | ||
| Younger than 49 | 157 (36.5) | |
| 50–59 | 84 (19.5) | |
| 60–69 | 53 (12.3) | |
| 70–79 | 63 (14.7) | |
| 80 and older | 56 (13.0) | |
| Missing data | 17 (4.0) | |
| GSE156063 (13) | ||
| Tissue resource | Nasopharyngeal swabs from COVID-19 patients | Nasopharyngeal swabs from patients infected with other viruses |
| Sample size | n = 93 | n = 41 |
| Demographic characteristics | ||
| Female sex – No. (%) | 50 (53.8) | 19 (46.3) |
| Age Range – No. (%) | ||
| Younger than 49 | 59 (63.4) | 23 (56.1) |
| 50–59 | 10 (10.8) | 3 (7.3) |
| 60–69 | 16 (17.2) | 6 (14.6) |
| 70–79 | 7 (7.5) | 5 (12.2) |
| 80 and older | 1 (1.1) | 4 (9.8) |
2. Materials and methods
2.1. Datasets
We downloaded the RNA-Seq gene expression profile datasets in leukocyte samples from 100 COVID-19 patients (GSE157103) and in SARS-CoV-2-infected human tissues from nasopharyngeal swabs (GSE152075 and GSE156063) from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). Table 1 summarizes these datasets.
2.2. Quantification of the enrichment levels of immune signatures
We used the single-sample gene-set enrichment analysis (ssGSEA) score [15] to evaluate the enrichment level of an immune signature in a COVID-19 patient based on the gene expression profiles. The ssGSEA score represents the enrichment score of a gene set in a sample based on the degree of the genes in the gene set coordinately up- or down-regulated in the sample. We analyzed 11 immune signatures, including HLA Class II, CD8+ T cells, Tfh, Th1 cells. The gene sets representing these immune signatures are listed in Supplementary Table S2.
2.3. Clustering analysis
We hierarchically clustered 100 leukocyte samples from COVID-19 patients (GSE157103) based on the ssGSEA scores of the 11 immune cell types. We performed the clustering analysis by using the R function “hclust” for hierarchical agglomerative clustering.
2.4. Logistic regression analysis
We used logistic regression with five predictors (age, gender, CD8+ T cell score, NK cell score, and type II IFN response score) to predict intensive care unit (ICU) and mechanical ventilatory support (MVS), respectively. The logistic regression analysis utilized the R function “glm” to fit the binary model and the R function “lm.beta” in the R package “QuantPsyc” to calculate the standardized regression coefficients (β values).
2.5. Identification of gene ontology associated with COVID-19 subtypes
We used WGCNA [16] to identify the gene modules differentially enriched in COVID-19 subtypes and outcomes. The representative gene ontology (GO) terms associated with the gene modules were identified. The WGCNA analysis was performed by using the R package “WGCNA” (version 1.68).
2.6. Pathway analysis
We identified differentially expressed genes between ICU and non-ICU COVID-19 patients using Student’s t test with a threshold of adjusted P-value (false discovery rate (FDR) < 0.05 and fold change (FC) of mean expression levels > 2. Based on the differentially expressed genes, we identified KEGG [17] pathways differentially enriched between ICU and non-ICU COVID-19 patients by GSEA [18] with a threshold of FDR < 0.05. The FDR was calculated by using the Benjamini-Hochberg method [19].
2.7. Class prediction
We predicted ICU versus non-ICU patients based on gene expression profiles in leukocyte samples from 100 COVID-19 patients (GSE157103). We performed 3-fold cross validation (CV) in the 100 samples. Within each loop of the CV, we selected the 100 genes with the largest absolute t-scores in the comparison of ICU versus non-ICU patients in the training set; based on the 100 genes, we trained the Random Forest (RF) classifier and predicted ICU versus non-ICU patients in the test set. We reported the prediction performance (accuracy and the area under the ROC curve (AUC)) as the average of them in the 3-fold CV. We carried out the prediction algorithm in Weka [20] with the number of trees in the RF set to 500.
2.8. Statistical analysis
In comparison of two classes of data, we used Mann–Whitney U test if they were not normally distributed and used Student’s t test if they were normally distributed. We used Spearman’s correlation test to evaluate the correlation between two groups of data on the assumption that they were not normally distributed. We used Fisher’s exact test to evaluate the association between two categorical variables. All statistical analyses were performed in the R programming environment (version 4.0.2).
3. Results
3.1. Identification of COVID-19 subtypes based on immune signature enrichment levels
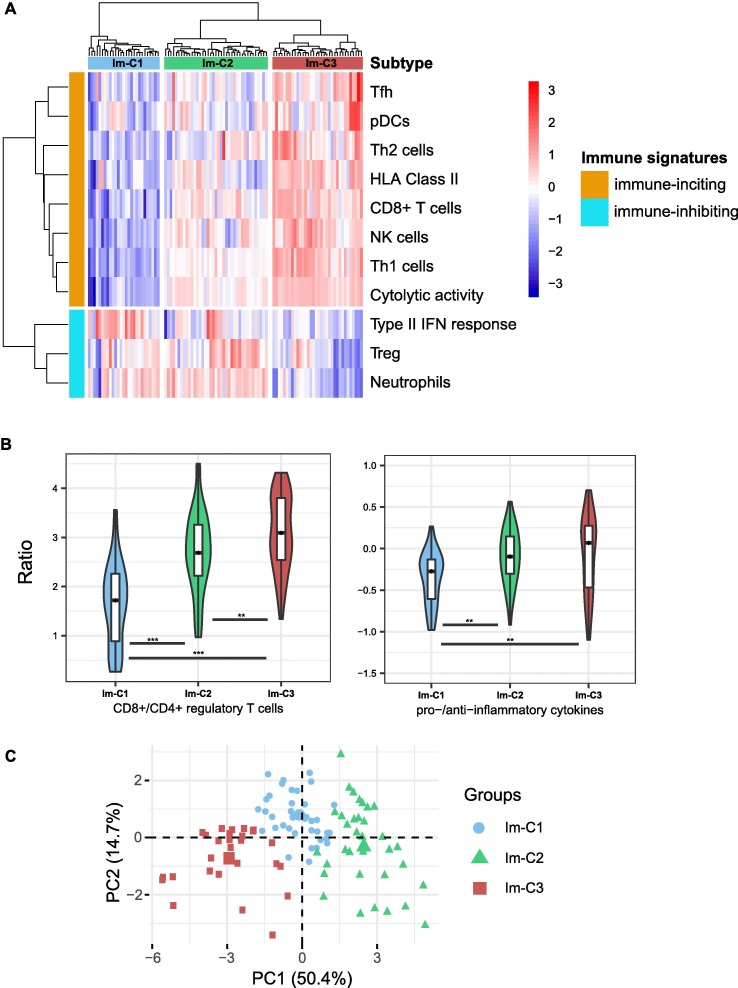
Based on the enrichment levels of 11 immune signatures, using the hierarchical clustering method, we identified three COVID-19 immune subtypes, termed Im-C1, Im-C2, and Im-C3 (Fig. 1 A). Im-C1 had the lowest enrichment levels of HLA Class II, CD8+ T cells, Tfh, Th1 cells, Th2 cells, NK cells, pDCs, and cytolytic activity (termed immune-inciting signatures) while high enrichment levels of type II IFN response, Treg, and neutrophils (termed immune-inhibiting signatures). Im-C2 had medium immune-inciting signatures and high immune-inhibiting signatures. In contrast to Im-C1, Im-C3 had the highest immune-inciting signatures and the lowest immune-inhibiting signatures. Thus, Im-C3 and Im-C1 displayed the strongest and weakest antiviral immune responses, respectively. As expected, the ratios of immune-stimulatory to immune-inhibitory signatures (CD8+/CD4+ regulatory T cells and pro-/anti-inflammatory cytokines) were the highest in Im-C3 and the lowest in Im-C1 (one-tailed Mann–Whitney U test, P < 0.01) (Fig. 1B). Principal component analysis confirmed that these COVID-19 cases could be divided into three subgroups based on the ssGSEA scores of these immune signatures (Fig. 1C).
Fig. 1.
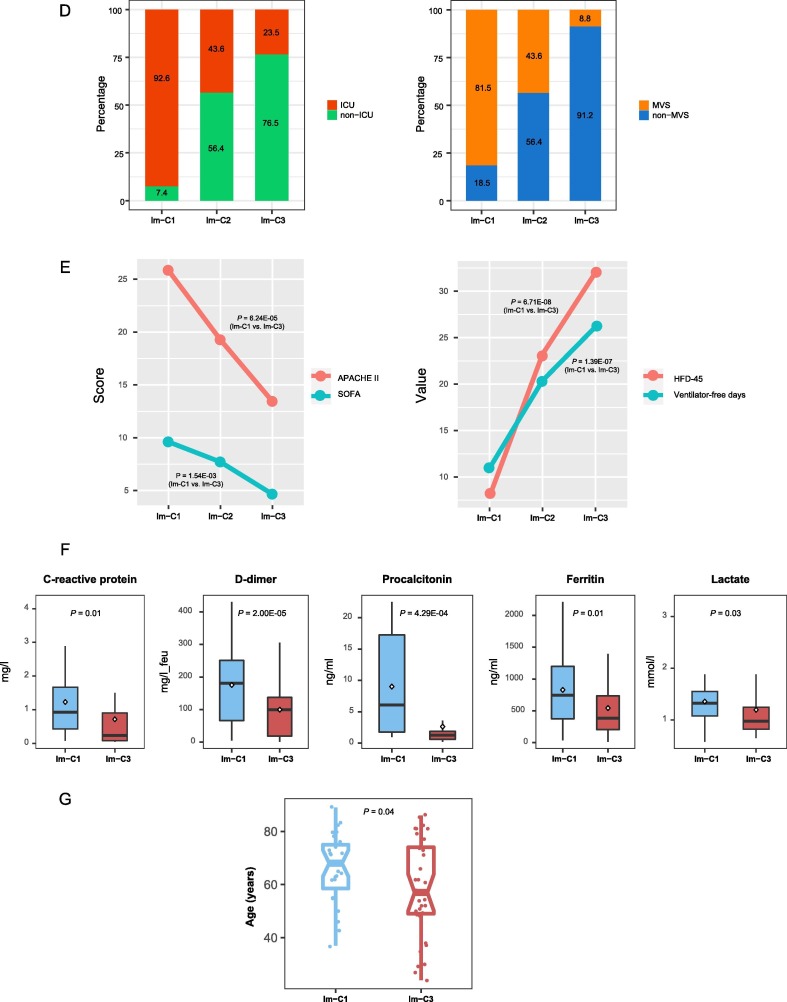
Identification of COVID-19 immune subtypes. A. Hierarchical clustering of 100 COVID-19 patients based on the enrichment scores of 11 immune signatures in leukocytes. B. Comparisons of the ratios of immune-stimulatory to immune-inhibitory signatures between the three COVID-19 subtypes. The ratios were the mean expression levels of the marker genes of immune-stimulatory signatures over those of immune-inhibitory signatures (log2-transformed). The Student’s t test P-values are indicated. * P < 0.05, ** P < 0.01, *** P < 0.001 (This also applies to the following figures). C. Principal component analysis confirming that COVID-19 can be divided into three subgroups based on the enrichment scores of the 11 immune signatures. D. Proportions of COVID-19 patients admitted to intensive care unit (ICU) or requiring mechanical ventilatory support (MVS) in the COVID-19 subtypes. E. Comparisons of the scores of the Acute Physiology and Chronic Health Evaluation (APACHE II) and the Sequential Organ Failure Assessment (SOFA), ventilator-free days, and the hospital-free days at day 45 (HFD-45) values between the three COVID-19 subtypes. F. Comparisons of the COVID-19 severity-associated laboratory measurements between COVID-19 subtypes. G. Comparisons of the age between COVID-19 subtypes. The one-tailed Mann–Whitney U test P-values are shown.
We found that the proportion of the COVID-19 patients admitted to ICU was the lowest in Im-C3 (23.5%) and the highest in Im-C1 (92.6%) (Fisher’s exact test, P < 0.001) (Fig. 1D). Moreover, the proportion of COVID-19 patients requiring MVS was the lowest in Im-C3 (8.8%) and the highest in Im-C1 (81.5%) (Fisher’s exact test, P < 0.001). The scores of the Acute Physiology and Chronic Health Evaluation (APACHE II) and the Sequential Organ Failure Assessment (SOFA), both of which measure the severity of ICU patients [21], were significantly different between the three subtypes: Im-C3 < Im-C2 < Im-C1 (one-tailed Mann–Whitney U test, P < 0.001; mean APACHE II score: 13 versus 19 versus 26; mean SOFA score: 5 versus 8 versus 10) (Fig. 1E). The numbers of ventilator-free days, which is an outcome measure in treatments for acute respiratory distress syndrome [22], and the hospital-free days at day 45 (HFD-45) values, which correlated inversely with disease severity, were significantly different between the three subtypes: Im-C3 > Im-C2 > Im-C1 (one-tailed Mann–Whitney U test, P < 0.001; mean ventilator-free days: 26 versus 20 versus 11; mean HFD-45: 32 versus 23 versus 8) (Fig. 1E). Additionally, some laboratory measurements, such as C-reactive protein, D-dimer, procalcitonin, ferritin, and lactate, whose elevation was associated with COVID-19 severity, tended to display the lowest levels in Im-C3 while the highest levels in Im-C1 (Fig. 1F). Altogether, these results showed that the Im-C3 subtype of COVID-19 had the best outcomes, while the Im-C1 subtype had the worst outcomes. It suggests that antiviral immune responses can reduce COVID-19 disease severity. Furthermore, we found that the patients in Im-C3 were younger than those in Im-C1 (one-tailed Mann–Whitney U test, P = 0.04) (Fig. 1G). It indicates that younger persons tended to have a stronger antiviral immune response than older persons after SARS-CoV-2 infection. However, we did not observe a significant difference in the proportions of female and male patients between Im-C3 and Im-C1 (Fisher’s exact test, P = 0.61).
3.2. Associations between immune signatures and clinical features in COVID-19 patients
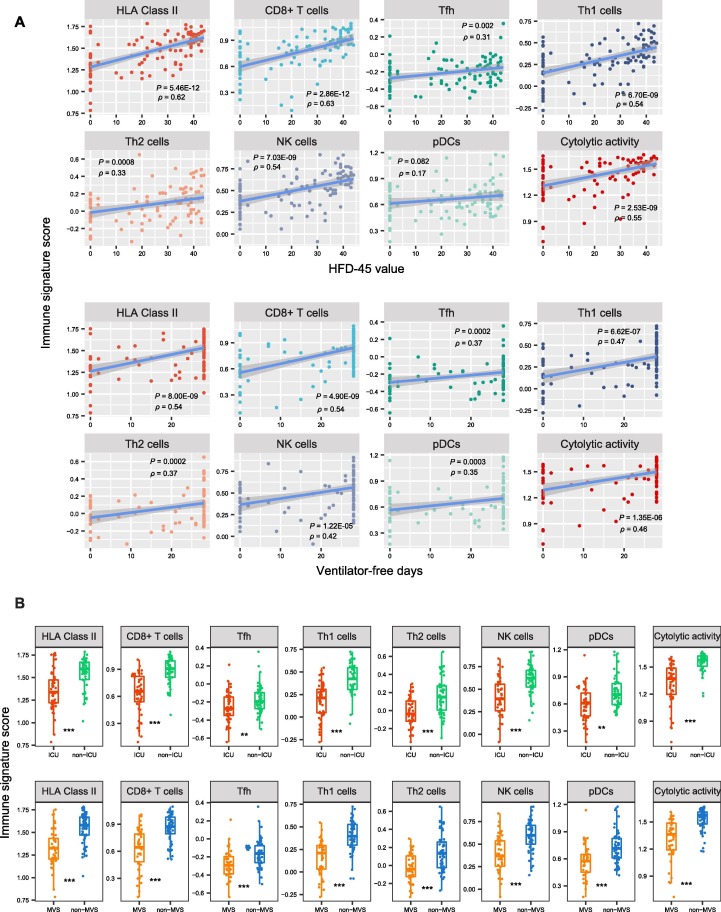
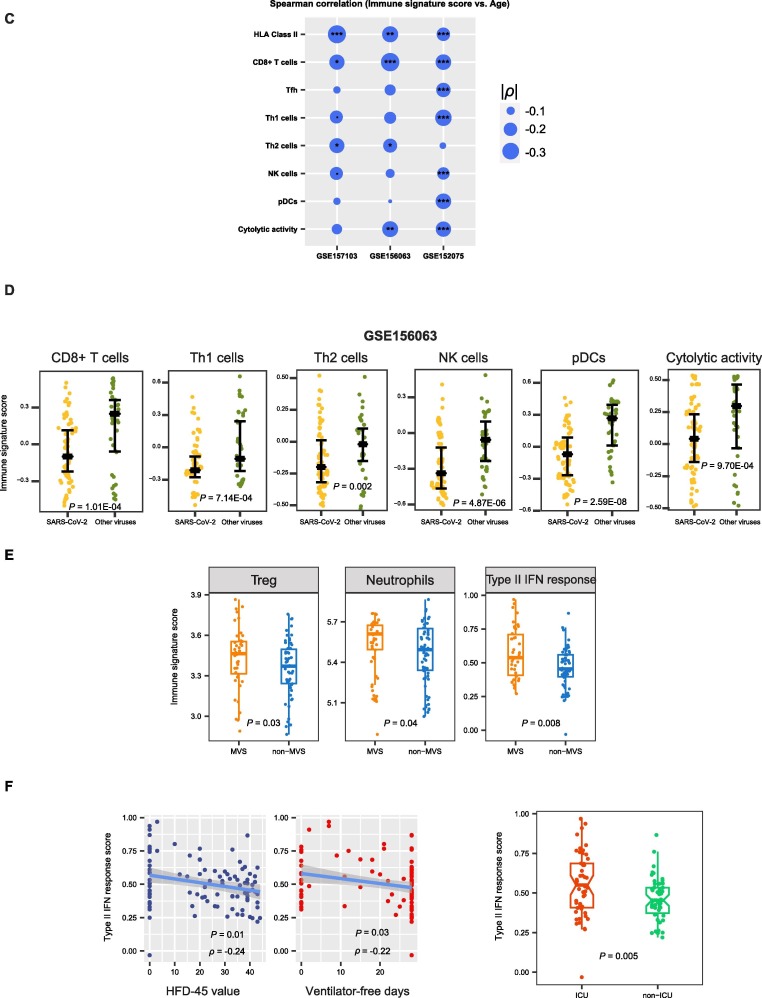
We further analyzed associations between immune signatures and clinical features in COVID-19 patients. As expected, the elevated enrichment of the eight immune-inciting signatures were correlated with higher HFD-45 values and more ventilator-free days (Spearman’s correlation test, P < 0.01, ρ > 0.3) (Fig. 2 A). Moreover, their enrichment levels were significantly higher in non-ICU versus ICU patients and in non-MVS versus MVS patients (one-tailed Mann–Whitney U test, P < 0.01) (Fig. 2B). Collectively, these results indicate that the elevated enrichment of these immune-inciting signatures is associated with better outcomes in COVID-19 patients. In addition, we found five immune-inciting signatures (HLA Class II, CD8+ T cells, Th1 cells, Th2 cells, and NK cells) whose enrichment levels correlated inversely with ages of COVID-19 patients (P < 0.1, ρ < -0.18) (Fig. 2C). However, none of the eight immune-inciting signatures showed significantly different enrichment levels between female and male patients. Again, these results indicate that younger patients have a stronger immune response to SARS-CoV-2 infection than older patients, while the strength of immune response is not different between female and male patients. We further demonstrated the significant negative correlation between the immune-inciting signatures and ages of COVID-19 patients in two other RNA-Seq gene expression profiling datasets for COVID-19 patients (GSE156063 [13] and GSE152075 [14] (Fig. 2C). Likewise, the associations between gender and these immune-inciting signatures were not significant in both datasets. Interestingly, in GSE156063, six immune-inciting signatures displayed significantly lower enrichment levels in COVID-19 patients than in the patients infected with other viruses (one-tailed Mann–Whitney U test, P < 0.01) (Fig. 2D). It suggests that SARS-CoV-2 causes a weaker human host immune response compared to other viruses, a potential explanation for the higher infectivity and pathogenicity of SARS-CoV-2 versus other viruses. In contrast to the immune-inciting signatures, the immune-inhibiting signatures were likely to have a negative correlation with outcomes in COVID-19 patients, as evidenced by that the three immune-inhibiting signatures (type II IFN response, Treg, and neutrophils) displayed significantly higher enrichment levels in MVS than in non-MVS patients (one-tailed Mann–Whitney U test, P < 0.05) (Fig. 2E). In addition, the type II IFN response signature had inverse correlations with ventilator-free days and HFD-45 values (P < 0.05, ρ < -0.21) and was significantly higher in ICU versus non-ICU patients (Fig. 2F).
Fig. 2.
Associations between immune signatures and clinical features in COVID-19 patients. A. Associations between immune signature scores and HFD-45 values and ventilator-free days. The Spearman’s correlation test P-values and correlation coefficients are shown. B. Comparisons of immune signature scores between ICU and non-ICU patients and between MVS and non-MVS patients. The one-tailed Mann–Whitney U test P-values are shown. C. Associations between immune signature scores and age. D. Comparisons of immune signature scores between COVID-19 patients and the patients infected with other viruses. E. Comparisons of the scores of three immune-inhibiting signatures (type II IFN response, Treg, and neutrophils) between non-MVS and MVS patients. F. The scores of type II IFN response have negative correlations with HFD-45 values and ventilator-free days and are higher in ICU than in non-ICU patients.
3.3. Comparison of the contribution of different factors in the prediction of COVID-19 outcomes
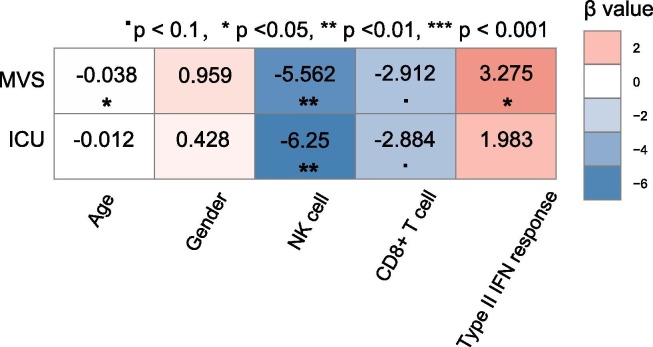
To compare the contribution of different factors in the prediction of outcomes in COVID-19 patients, we used the logistic regression model with five predictors (age, gender, CD8+ T cell score, NK cell score, and type II IFN response score) to predict ICU (=1) versus non-ICU (=0) and MVS (=1) versus non-MVS (=0) patients, respectively. In predicting MVS versus non-MVS, age, CD8+ T cells, and NK cells were significant negative predictors, while type II IFN response was a significant positive predictor (P < 0.05) (Fig. 3 ). In predicting ICU versus non-ICU, CD8+ T cells and NK cells were significant negative predictors (P < 0.1), while type II IFN response was a positive predictor (P = 0.186, β = 1.98). These results indicate that COVID-19 outcomes are correlated positively with immune-inciting signatures and negatively with immune-inhibiting signatures and age, consistent with previous results. Meanwhile, logistic regression analyses indicate that the adaptive immune response (CD8+ T cells) has a stronger impact on COVID-19 outcomes than the innate immune response (NK cells), as evidenced by the larger β values of CD8+ T cells versus NK cells in predicting ICU and MVS.
Fig. 3.
Prediction of ICU versus non-ICU and MVS versus non-MVS patients using five predictors (age, gender, CD8+ T cell score, NK cell score, and type II IFN response score) by logistic regression analyses. The standardized regression coefficients (β values) are shown.
3.4. Identification of gene ontology differentially enriched between COVID-19 subtypes
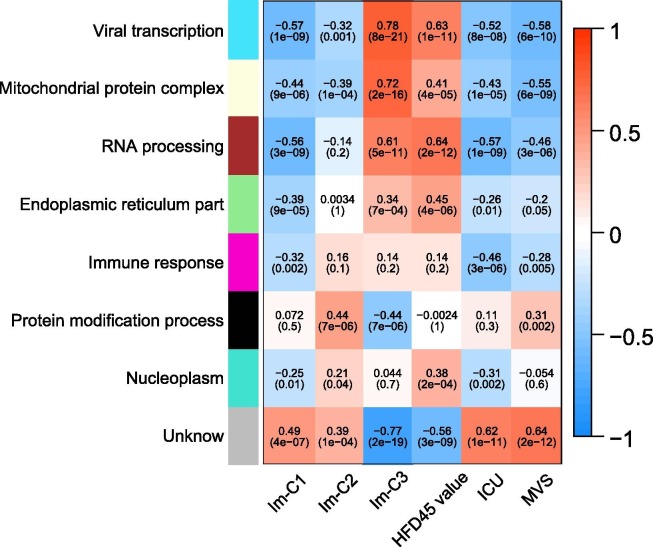
WGCNA identified seven gene modules (indicated in cyan, light yellow, brown, light green, magenta, black, and turquoise color, respectively) that significantly differentiated COVID-19 patients by COVID-19 subtypes (Im-C1, Im-C2, and Im-C3) and outcomes (HFD-45, ICU, and MVS) (Fig. 4 ). The representative GO terms associated with the gene modules highly enriched in Im-C3 while lowly enriched in Im-C1 included viral transcription, mitochondrial protein complex, RNA processing, and endoplasmic reticulum part. Consistently, these modules were associated with better outcomes of COVID-19. Besides, the immune response downregulated in Im-C1 was positively associated with COVID-19 outcomes. In addition, the protein modification process representing the black module, which was downregulated in Im-C3 while upregulated in Im-C2, correlated with worse outcome of MVS.
Fig. 4.
WGCNA identified seven gene modules significantly differentiating COVID-19 patients by COVID-19 subtypes and outcomes. The representative gene ontology terms for gene modules, correlation coefficients, and P-values are shown.
3.5. Identification of genes and pathways differentially expressed between ICU and non-ICU COVID-19 patients
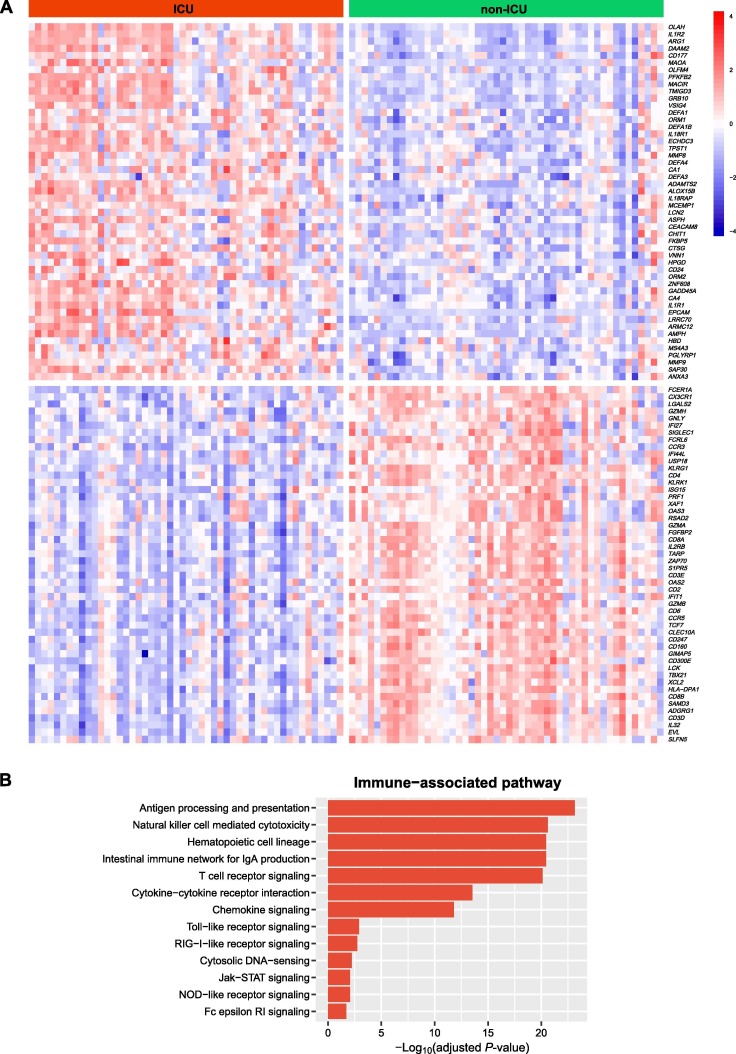
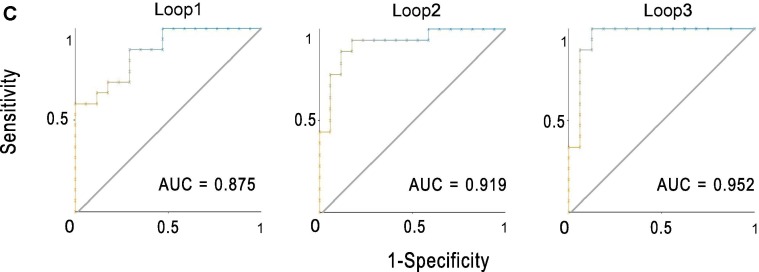
We identified 67 and 309 genes upregulated and downregulated in ICU versus non-ICU COVID-19 patients (Fig. 5 A and Supplementary Table S1). We found a number of immune-related pathways associated with the upregulated genes in non-ICU, including antigen processing and presentation, natural killer cell mediated cytotoxicity, hematopoietic cell lineage, intestinal immune network for IgA production, T cell receptor signaling, cytokine-cytokine receptor interaction, chemokine signaling, Toll-like receptor signaling, RIG-I-like receptor signaling, cytosolic DNA-sensing, Jak-STAT signaling, NOD-like receptor signaling, and Fc epsilon RI signaling (Fig. 5B). Again, these results indicate the stronger immune response in non-ICU versus ICU COVID-19 patients. Furthermore, we performed a prediction of ICU versus non-ICU patients based on gene expression profiles in leukocyte samples from 100 COVID-19 patients (GSE157103). The 3-fold CV accuracy was 83.1%, and the AUC was 91.5% (Fig. 5C). It indicates that the gene expression profiles in leukocytes of COVID-19 patients could be a potentially useful predictor for the severity of COVID-19.
Fig. 5.
Genes and pathways differentially expressed between ICU and non-ICU COVID-19 patients. A. Heatmap for 50 and 50 genes showing the highest increases and decreases of the fold change of mean expression levels in ICU versus non-ICU patients, respectively. B. Immune-related pathways upregulated in ICU versus non-ICU patients. C. Prediction performance of gene expression profiles in leukocyte samples from 100 COVID-19 patients in the prediction of ICU versus non-ICU patients by Random Forest. The area under the ROC curve (AUC) is shown for each loop of the 3-fold cross validation (CV).
4. Discussion
Based on the enrichment levels (ssGSEA scores) of 11 immune signatures in leukocytes of COVID-19 patients, we identified three COVID-19 subtypes: Im-C1, Im-C2, and Im-C3, by clustering analysis. The ssGSEA scores-based clustering method has been shown to be more robust than the gene expression values-based method in identifying subtypes of diseases [23], [24], [25]. Im-C1 had the lowest immune-inciting signatures and high immune-inhibiting signatures. Im-C2 had medium immune-inciting signatures and high immune-inhibiting signatures. Im-C3 had the highest immune-inciting signatures while the lowest immune-inhibiting signatures. Im-C3 and Im-C1 COVID-19 patients had the best and worst clinical outcomes, respectively, suggesting that antiviral immune responses alleviated the severity of COVID-19 patients. We further demonstrated that the adaptive immune response exerted a greater impact on COVID-19 outcomes than the innate immune response. The patients in Im-C3 were younger than those in Im-C1, indicating that younger persons have stronger antiviral immune responses than older persons. Nevertheless, we did not observe a significant association between sex and immune responses in COVID-19 patients. In addition, we found that the type II IFN response signature was an adverse prognostic factor for COVID-19 in the dataset GSE157103. This result appears inconsistent with previous findings [26], [27]. The reason behind this needs to be further investigated.
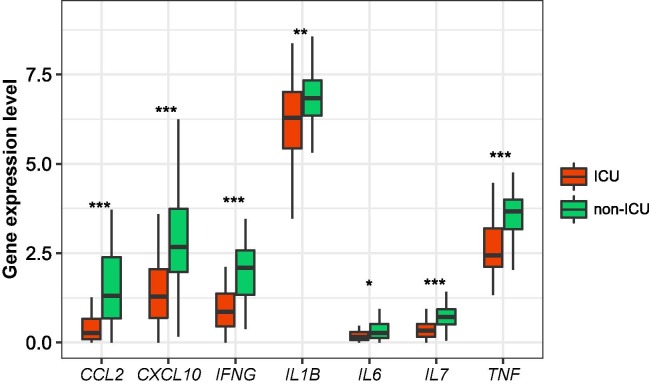
Our data suggest that a strong antiviral immune response can reduce COVID-19 severity. Thus, a strong host immune system is crucial for fighting against COVID-19, as bolstered by a recent study [28]. However, previous studies have revealed that serum inflammatory cytokine levels had an inverse association with clinical outcomes in COVID-19 patients [29], [30], [31], [32]. It suggests that excessive immune response, known as cytokine storm, may cause immunopathological damage in COVID-19 patients [7]. We compared the expression levels of several cytokine genes in leukocytes between ICU and non-ICU COVID-19 patients, including IL-6, IL-1β, TNF, CCL2, CXCL10, IFNG, IL7. We found that these genes displayed significantly higher expression levels in non-ICU than in ICU patients (Fig. 6 ). A potential explanation for these different results could be the different sources of these cytokine. In addition, our findings suggest that weak immune responses are associated with worse prognosis in COVID-19 patients. It appears to conflict with previous indications that strong immune responses may cause severe COVID-19 outcomes [7]. Nevertheless, the present and previous results together may indicate that both inadequate and excessive immune responses will lead to severe COVID-19 cases. Certainly, the association between host immune responses and clinical outcomes in COVID-19 patients needs to be further explored based on large sample size.
Fig. 6.
Comparisons of the expression levels of cytokine genes in leukocytes between ICU and non-ICU COVID-19 patients. The Student’s t test P-values are shown.
Funding
This work was supported by the China Pharmaceutical University (grant number 3150120001) and the Science and Technology Project of Traditional Chinese Medicine in Zhejiang Province (grant number: 2017ZKL003).
CRediT authorship contribution statement
Zuobing Chen: Software, Validation, Formal analysis, Investigation, Data curation, Funding acquisition. Qiushi Feng: Software, Validation, Formal analysis, Investigation, Data curation, Visualization. Tianfang Zhang: Formal analysis, Resources, Investigation. Xiaosheng Wang: Conceptualization, Methodology, Resources, Investigation, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2021.107615.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.COVID-19 Dashboard; https://coronavirus.jhu.edu/map.html. 2020, Johns Hopkins University.
- 2.Takahashi T., et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020 doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khinda J., Janjua N.Z., Cheng S., Heuvel E.R., Bhatti P., Darvishian M. Association between markers of immune response at hospital admission and COVID-19 disease severity and mortality: A meta-analysis and meta-regression. J. Med. Virol. 2021;93(2):1078–1098. doi: 10.1002/jmv.26411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 6.Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.-S., Jeong S.J., Lee H.K., Park S.H., Park S.-H., Choi J.Y., Kim S.-H., Jung I., Shin E.-C. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5(49):eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laing A.G., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020 doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 8.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan R., Zhang Y., Li Y., Xia L.u., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., et al. Deep learning-based clustering robustly identified two classes of sepsis with both prognostic and predictive values. EBioMedicine. 2020;62 doi: 10.1016/j.ebiom.2020.103081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shankar R., et al. Gene expression signatures identify paediatric patients with multiple organ dysfunction who require advanced life support in the intensive care unit. EBioMedicine. 2020;62 doi: 10.1016/j.ebiom.2020.103122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overmyer K.A., et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2020 doi: 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mick E., et al. Upper airway gene expression differentiates COVID-19 from other acute respiratory illnesses and reveals suppression of innate immune responses by SARS-CoV-2. medRxiv. 2020 [Google Scholar]
- 14.N.A.P. Lieberman, V. Peddu, In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age, 18 (9) (2020) e3000849. [DOI] [PMC free article] [PubMed]
- 15.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucl. Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Statist. Soc. B. 1995;57(1):289–300. [Google Scholar]
- 20.I.H. Witten, et al., Data Mining: Practical Machine Learning Tools and Techniques. Fourth ed. 2016: Morgan Kaufmann.
- 21.W.A. Knaus, et al., APACHE II: a severity of disease classification system, Crit. Care Med., 13 (10) (1985) 818–829. [PubMed]
- 22.Schoenfeld D.A., Bernard G.R., Network A. Statistical evaluation of ventilator-free days as an efficacy measure in clinical trials of treatments for acute respiratory distress syndrome. Crit. Care Med. 2002;30(8):1772–1777. doi: 10.1097/00003246-200208000-00016. [DOI] [PubMed] [Google Scholar]
- 23.He Y., Jiang Z., Chen C., Wang X. Classification of triple-negative breast cancers based on Immunogenomic profiling. J. Exp. Clin. Cancer Res. 2018;37(1) doi: 10.1186/s13046-018-1002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu F., Chen J.-X., Yang X.-B., Hong X.-B., Li Z.-X., Lin L., Chen Y.-S. Analysis of Lung Adenocarcinoma Subtypes Based on Immune Signatures Identifies Clinical Implications for Cancer Therapy. Mol. Ther. Oncolyt. 2020;17:241–249. doi: 10.1016/j.omto.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu F., Wang Z.-L., Wang K.-Y., Li G.-Z., Chai R.-C., Liu Y.-Q., Jiang H.-Y., Zhai Y., Feng Y.-M., Zhao Z., Zhang W. Classification of diffuse lower-grade glioma based on immunological profiling. Mol. Oncol. 2020;14(9):2081–2095. doi: 10.1002/1878-0261.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.J. Hadjadj, N. Yatim, Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients, 369 (6504) (2020) 718–724. [DOI] [PMC free article] [PubMed]
- 27.Lagunas‐Rangel F.A., Chávez‐Valencia V. High IL-6/IFN-γ ratio could be associated with severe disease in COVID-19 patients. J. Med. Virol. 2020;92(10):1789–1790. doi: 10.1002/jmv.25900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avanzato V.A., et al. Case Study: Prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised cancer patient. Cell. 2020 doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manjili R.H., Zarei M., Habibi M., Manjili M.H. COVID-19 as an Acute Inflammatory. Disease. 2020;205(1):12–19. doi: 10.4049/jimmunol.2000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burke H., Freeman A., Cellura D.C., Stuart B.L., Brendish N.J., Poole S., Borca F., Phan H.T.T., Sheard N., Williams S., Spalluto C.M., Staples K.J., Clark T.W., Wilkinson T.M.A. Inflammatory phenotyping predicts clinical outcome in COVID-19. Respir. Res. 2020;21(1) doi: 10.1186/s12931-020-01511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangalmurti N., Hunter C.A. Cytokine Storms: Understanding COVID-19. Immunity. 2020;53(1):19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.