Abstract
Background:
The increasing demand for genetic testing for clinical diagnosis and research challenges genetic laboratory capacity to track an increasing number of patient samples through all steps of analysis, from sample collection to report generation. This task is usually performed with the help of a laboratory information management system (LIMS), software that makes it possible to collect, store and retrieve laboratory and sample data. To date there are no open-source options that can manage the entire analytical flow of a genetic laboratory. appMAGI seeks to include all the management aspects of a clinical diagnostic laboratory, making it simpler to process many samples while maintaining the high security and quality standards required in clinical diagnostic practice.
Methods:
appMAGI is written in python using Django. It is a web application that does not require local installation, making development, updates and maintenance a much easier task. appMAGI runs on the Ubuntu server and uses SQLite as engine database.
Results:
In this work we describe an innovative LIMS called appMAGI designed to support all aspects of a clinical diagnostic laboratory. appMAGI can track samples throughout the diagnostic workflow and NGS analysis by virtue of a customizable bioinformatics pipeline. It can handle sample non-compliance, manage laboratory stocks, help generate reports and provide insights into sample data by means of special tools.
Conclusions:
appMAGI is a LIMS endowed with all the features required to manage thousands of samples. Allowing efficient management of patient samples from sample collection to diagnostic report generation. (www.actabiomedica.it)
Keywords: NGS, LIMS, diagnostics, information management
Introduction
Next-generation sequencing (NGS) technologies now play a striking role in the research and diagnosis of genetic diseases. NGS is used to establish gene-disease associations in different fields of medicine. It can analyze many samples per run and investigate thousands of target regions for genetic variations (1-6).
As the number of genetic tests increases, clinics are faced with a growing number of patients (7). The workflow to analyze a patient’s sample is long, complex and involves the contributions of specialized figures, such as clinicians, bioinformaticians, geneticists and quality managers, to cite a few. Since this workflow is also performed in parallel on many samples, it is clear that the complexity of the task increases exponentially. It can therefore be challenging to handle, store and analyze huge amounts of patient data, and generate reports.
To track and manage clinical samples, laboratories normally rely on a laboratory information management system (LIMS), software that stores and retrieves the information belonging to a particular sample. Different open-source LIMS have been developed to deal with various parts of the sample analysis workflow (8-12), but there are unfortunately very few that can manage the entire workflow of NGS clinical diagnosis with a high standard of security, quality and accuracy.
Here we describe appMAGI, a web-based LIMS that can track a sample from acquisition to report generation. It also includes an NGS bioinformatics pipeline to perform sample analysis. Other features are tools for variant confirmation by Sanger sequencing, estimation of sample contamination probability and CNV detection. Our LIMS also has a section dedicated to management of laboratory stocks to keep track of equipment and reagents. A special report-generation section helps the geneticist draft the clinical report.
Materials and methods
Implementation
appMAGI is designed to meet the needs of most clinical genetic laboratories. It is easy to install and configure. appMAGI is written in python using the open-source web framework Django. Being a web application, it does not require local installation, making development, updates and maintenance a much easier task. The user connects to the application through a web client. appMAGI runs on the ubuntu server and uses SQLite as engine database (see Fig. 1 for simplified database architecture). It is composed of different modules for organizing and taking care of all the tasks performed in a genetics laboratory. Fig. 2 gives a brief description of the main sections and functioning.
Figure 1.
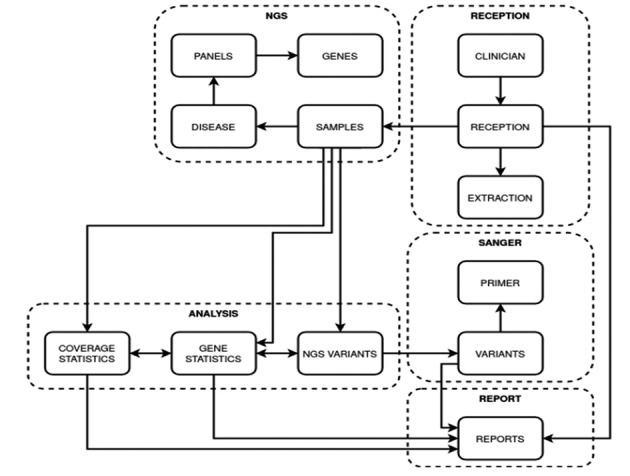
Database Architecture. The Graph show the architecture of the database in SQLlite3. The database is composed by five main tables that are needed to store information on sample clinical, NGS, Sanger, Analysis and Report data
Figure 2.
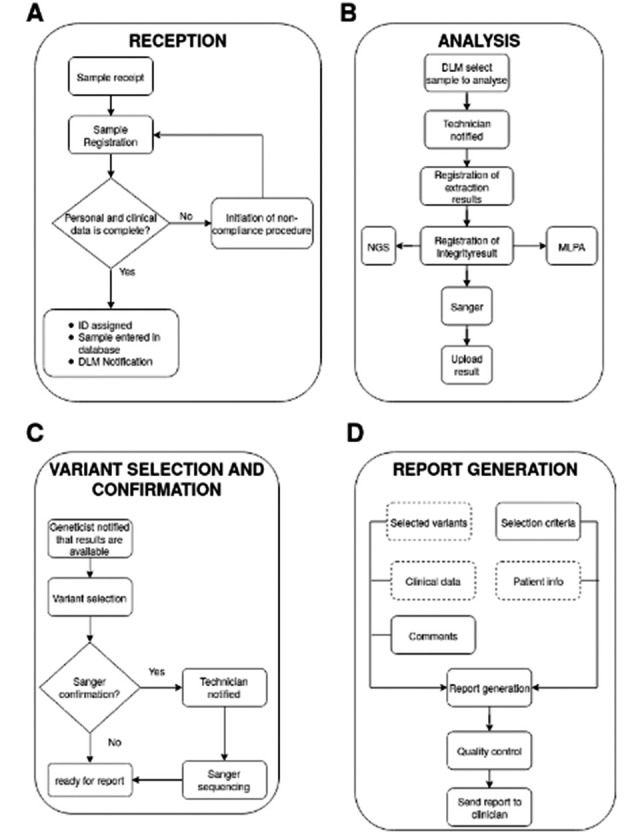
Representation and functioning of appMAGI workflow. The workflow can be divided in 4 main areas. (A) Sample are received from the clinician and get registered in the system. In all the data for a sample are available the sample can be analysed, else a non-compliance instance is open. (B) Samples are analyzed with the technique of choice and results are upload in appMAGI. (C) The geneticist visualize the results and select the variant of interest for a particular sample, if necessary the variants are confirmed with Sanger centrally sequencing. (D) The final report is generated with patient, analysis and variant information and sent back to the clinician
Results
Sample reception
The workflow starts when a sample is received from a clinician. The first step to perform is sample registration by the reception officer (RO) (Fig. 2A), which consists of compiling the clinical and personal data sheet of the patient. Once all the necessary information has been entered, the sample is assigned an identification number so that it can be tracked through all the steps of analysis and so that all information can be retrieved. On registration, the sample is added to the database. The main view allows search and visualization of the current state of the sample. A filtering function can help find a sample through different filtering criteria, such as processing step, date of registration and much more.
A sample may be delivered with incomplete or partial information. When this happens, the RO opens a non-compliance (NC) procedure through a specific LIMS function. The NC is reported to the quality control officer who assigns an ID to the procedure. The sample is kept out of the workflow until the NC is resolved.
Once samples are correctly registered in the system, the RO notifies the diagnostic laboratory manager (DLM) of new samples through the notification function. The DLM assigns the samples to a specific technician for analysis.
Sample analysis
On receipt of notification from the DLM, the technician begins analysis of the samples. NGS analysis has various steps, as shown in Fig. 2B. The first is DNA extraction from the sample, after which the operator enters the concentrations and ratio, along with the lot and kit of reagents used, in the corresponding section of the LIMS. The information becomes visible on the main page of the LIMS. The next step of the workflow is to check the integrity of the extracted DNA by agarose-gel electrophoresis. If the test is positive, a picture of the gel is uploaded to the specific section of the sample entry. After the above quality control, the technician proceeds with the analysis prescribed for the sample. For NGS, the technician runs sequencing and uploads the resulting file to the LIMS database. The LIMS notifies the bioinformatician that the fastq files are ready for bioinformatic analysis (13,14). When the analysis is terminated, VCF files are stored in the LIMS while the intermediate file (e.g. fastq, SAM, BAM) is stored in an external storage device.
Once the analysis is complete, the bioinformatician notifies the geneticists that the results are available. The results are visualized via the NGS results button in the main view of each sample. It is easy to follow the steps of sample analysis, because the main view has a box for each step. The color code of the box indicates the stage of the procedure.
Variant selection and confirmation
Through the NGS results button, the geneticist can see all the variants found in a sample. Each variant is annotated with information from major databases (e.g. dbSNP, HGMD) along with information on homology regions (Fig. 2C). The geneticist selects the variant of interest for the suspected diagnosis indicated. To help the user, the LIMS offers different filters to apply to the list of variants. The user can exclude synonymous variants, filter by MAF and much more. After variant selection, the geneticist specifies those to confirm by Sanger sequencing. The geneticist is helped in this phase by an algorithm that uses machine learning and suggests which variants should be considered for Sanger confirmation (15). Once the variants are selected, the geneticist notifies the operator that Sanger sequencing will be performed. The technician consults the database of primers available in house for the region of interest; if nothing is found the technician designs the primer de-novo and enters it in the database for future use.
Once the primers have been designed the operator sends an order from this section that is automatically submitted to the storage section of appMAGI. After delivery of the primers, the operator confirms the selected variants. The resulting electropherograms are then uploaded to appMAGI in the multimedia section and the technician signals that the analysis is finished so the geneticist can generate the clinical report.
Report generation
Generation of the report is a semi-automated procedure that helps the geneticist at each step (Fig. 2D). The LIMS partially fills in the report with the information provided by the various technicians during analysis, such as the clinical and personal data sheet of the patient, the technology used for analysis and the variants selected by the geneticist. The geneticist comments on the variants, suggests gene segregation and/or writes comments for the clinician. The geneticist also uploads the sheet with the criterion used for variant selection to the LIMS. This classification, along with problematical regions, is kept in the database for future analysis of the same variant/region. Once the report is complete, there is a last quality control step. The geneticist sends the report via the LIMS to another geneticist who reviews the report and suggests modifications or corrections. When this phase is completed, the report can be printed, signed and sent to the clinician.
Storage management
appMAGI contains a section dedicated to the management of laboratory equipment and reagent stocks. While this section is not strictly correlated with the sample analysis workflow, it is important to ensure that the laboratory is always ready and stocked with all necessary materials in order to avoid service interruptions and delays in diagnosis.
Before the system can be used, all the products that need to be monitored must be registered, along with all their providers. Then the stock manager enters the quantities of all the products so that the LIMS has a picture of current stocks. This creates the optimal set up for the stock manager.
The stock manager section has several subsections. The goods subsection allows a check of all registered goods, like reagents and laboratory equipment. The entire inventory can be exported in table format (CSV) or one can search to see whether a particular product is in the inventory, the quantity left and the quantity below which a new order is placed. From this section, it is also possible to make a purchase request.
The notifications section shows automatically generated notifications regarding products in low supply, near their expiry date or open for more than a year, as well as the stage of each purchase request. When a purchase request is placed, a notification is sent for approval to the DLM. Then the purchasing manager places the order and notifies that the order has been sent. For this section to function properly, technicians need to report use of reagents to the LIMS so that the quantity remaining is adjusted accordingly. Whenever products are running low, a notification is sent to the purchase manager who orders replenishment via the LIMS.
Besides management of stocks, this part of the LIMS also manages product expiry dates. A convenient interface can be consulted to check how many days are left before expiry of any product, and warnings can be issued on products that are about to expire.
Conclusions
Efficient management of patient samples is of primary importance in clinical diagnostics. It is time-consuming and requires a considerable amount of work and effort by different specialized figures. The road from sample collection to diagnostic report generation involves many steps, including laboratory, bioinformatic and variant analysis, all of which are repeated for each sample. With an increasing number of genetic tests and samples to analyze, this can become a challenging task. The workflow requires an informatics approach or LIMS to keep track of samples. Open-source LIMS, designed to take care of one or more steps of the workflow, are available, but there is unfortunately no software that can manage the entire workflow of a clinical diagnostic laboratory. This is why we created appMAGI, a LIMS endowed with all the features required to manage thousands of samples. Table 1 summarizes the features offered by appMAGI and a comparison with major open-source LIMS.
Table 1.
Comparison between the features of some of the most common used LIMS
| appMAGI | MendeLIMS | MetaLIMS | Baobab | PASSIM | Galaxy LIMS | |
| Clinical diagnosis | ||||||
| Patients registration | Yes | Yes | No | No | Yes | No |
| NGS analysis | Yes | Yes | No | No | No | Yes |
| Barcode | No | Yes | Yes | No | No | No |
| Quality control | Yes | Yes | No | No | No | No |
| Bionformatics pipeline | Yes | Yes | No | No | No | No |
| Informatics and security | ||||||
| User clas privileges | Yes | Yes | No | Yes | Yes | No |
| Https/ssl | No | Yes | Yes | No | No | No |
| Web-based | Yes | Yes | Yes | Yes | Yes | Yes |
| Technology | Django, Python | Javascript, Ruby | PHP | Python | Java | Javascript |
| Database | SQLite 3 | MySQL | MySQL | ZODB | Oracle | SQL |
| General | ||||||
| Informatics knowledge | Base | Base | Base | Medium | Medium | Base |
| Emails and notifications | Yes | Yes | No | Yes | Yes | No |
| Inventory management | Yes | No | No | Yes | No | No |
| Instruments integration | No | Yes | No | Yes | No | No |
| Report visualization | Yes | No | Yes | No | Yes | No |
appMAGI has a centralized database, from which all information on any sample can be accessed centrally, enabling sample control from clinician to report generation. It also contains a powerful filtering method enabling sample search and visualization based on dozens of features, from working step and personal information to technologies used and much more. appMAGI provides all the modules necessary for sample reception and tracking, including a bioinformatics pipeline for NGS analysis, a primer design section for NGS confirmation and a MLPA management section. It has a module for report generation and a section for managing stocks of reagents, consumables and laboratory equipment. Last but not least, appMAGI implements different tools to improve analysis quality and to draw insights from NGS data, for example an algorithm to estimate contamination probabilities and a tool to detect CNVs from NGS data (16). appMAGI reduces errors in sample analysis by simplifying management of large sample numbers and providing easy access to sample data throughout the workflow.
Conflict of interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article
References
- 1.Su Z, Ning B, Fang H, et al. Next-generation sequencing and its applications in molecular diagnostics. Expert Rev Mol Diagn. 2011;11:333–43. doi: 10.1586/erm.11.3. [DOI] [PubMed] [Google Scholar]
- 2.Sommariva E, Pappone C, Martinelli Boneschi F, et al. Genetics can contribute to the prognosis of Brugada syndrome: a pilot model for risk stratification. Eur J Hum Genet. 2013;21:911–7. doi: 10.1038/ejhg.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daoud H, Luco SM, Li R, et al. Next-generation sequencing for diagnosis of rare diseases in the neonatal intensive care unit. CMAJ. 2016;188:254–60. doi: 10.1503/cmaj.150823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LePichon JB, Saunders CJ, Soden SE. The future of next-generation sequencing in neurology. JAMA Neurol. 2015;72:971–2. doi: 10.1001/jamaneurol.2015.1076. [DOI] [PubMed] [Google Scholar]
- 6.Peters DG, Yatsenko SA, Surti U, Rajkovic A. Recent advances of genomic testing in perinatal medicine. Semin Perinatol. 2015;39:44–54. doi: 10.1053/j.semperi.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips KA, Deverka PA, Hooker GW, Douglas MP. Genetic test availability and spending: Where are we now? Where are we going? Health Aff (Millwood) 2018;37:710–6. doi: 10.1377/hlthaff.2017.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scholtalbers J, Rößler J, Sorn P, et al. Galaxy LIMS for next-generation sequencing. Bioinformatics. 2013;29:1233–4. doi: 10.1093/bioinformatics/btt115. [DOI] [PubMed] [Google Scholar]
- 9.Van Rossum T, Tripp B, Daley D. Slims – A user-friendly sample operations and inventory management system for genotyping labs. Bioinformatics. 2010;26:1808–10. doi: 10.1093/bioinformatics/btq271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bath T, Bozdag S, Afzal V, Crowther D. Limsportal and bonsailims: Development of a lab information management system for translational medicine. Source Code Biol Med. 2011;6:9. doi: 10.1186/1751-0473-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimes S, Ji H. Mendelims: A web-based laboratory information management system for clinical genome sequencing. BMC Bioinformatics. 2014;15:290. doi: 10.1186/1471-2105-15-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viksna J, Celms E, Opmanis M, et al. Passim - An open source software system for managing information in biomedical studies. BMC Bioinformatics. 2007;8:52. doi: 10.1186/1471-2105-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marceddu G, Dallavilla T, Guerri G, Manara E, Bertelli M. PipeMAGI: An integrated and validated workflow for analysis of NGS data for clinical diagnostics. Eur Rev Med Pharmacol Sci. 2019;23:12. doi: 10.26355/eurrev_201908_18566. [DOI] [PubMed] [Google Scholar]
- 14.Maltese PE, Orlova N, Krasikova E, et al. Gene-targeted analysis of clinically diagnosed long QT russian families. Int Heart J. 2017;1:81–7. doi: 10.1536/ihj.16-133. [DOI] [PubMed] [Google Scholar]
- 15.Marceddu G, Dallavilla T, Guerri G, Zulian A, Marinelli C, Bertelli M. Analysis of machine learning algorithms as integrative tools for validation of next generation sequencing data. Eur Rev Med Pharmacol Sci. 2019;23:8139–47. doi: 10.26355/eurrev_201909_19034. [DOI] [PubMed] [Google Scholar]
- 16.Johansson LF, van Dijk F, de Boer EN, et al. CoNVaDING: Single exon variation detection in targeted NGS data. Hum Mutat. 2016;37:457–64. doi: 10.1002/humu.22969. [DOI] [PubMed] [Google Scholar]


