Abstract
The COVID-19 has become of striking interest since the number of deaths is constantly rising all over the globe, and the search for an efficient treatment is more urgent. In light of this worrisome scenario, this opinion review aimed to discuss the current knowledge about the potential role of curcumin and its nanostructured systems on the SARS-CoV-2 targets. From this perspective, this work demonstrated that curcumin urges as a potential antiviral key for the treatment of SARS-CoV-2 based on its relation to the infection pathways. Moreover, the use of curcumin-loaded nanocarriers for increasing its bioavailability and therapeutic efficiency was highlighted. Additionally, the potential of the nanostructured systems by themselves and their synergic action with curcumin on molecular targets for viral infections have been explored. Finally, a viewpoint of the studies that need to be carried out to implant curcumin as a treatment for COVID-19 was addressed.
Keywords: SARS-CoV-2, Curcuma longa, Coronavirus, Nanotechnology, Natural products
Graphical Abstract
1. Introduction
COVID-19 is a viral disease that triggers Severe Acute Respiratory Syndrome (SARS). This disease emerged in December 2019 in Wuhan, China, and spread worldwide, becoming one of the most worrisome pandemic outbreaks of the 21st century [1].
In order to better characterize this infection, the elucidation of the virus’s genome was performed, revealing a similarity of 79% to coronaviruses. Hence, this new strain was named a type 2 coronavirus, known as SARS-CoV-2 [2]. According to the World Health Organization (WHO), the number of COVID-19-confirmed cases has surpassed 120 million, and more than 2.7 million deaths have been reported worldwide (to date) [3]. The absence of an effective pharmacological treatment able to reduce the viral load and minimize the disease progression to the Acute Respiratory Distress Syndrome (ARDS) is one of the main factors that leads to the high mortality rate of this disease [4].
In light of this situation, marketed drugs have been tested to elucidate the COVID-19 biological pathways and identify the SARS-CoV-2 biological target in order to provide a specific and, by consequence, more effective alternative to treat the disease [5]. One of the targets of the drugs selected to treat COVID-19 is the main protease (Mpro also known as 3CLpro), which acts in the coronavirus RNA replication [6]. Besides the aforementioned biological route, another important site is the angiotensin-converting enzyme 2 receptor (ACE II), since it facilitates the entry of the SARS-CoV-2 into the host cell [7]. The latter has encouraged the hypothesis that drugs with suitable ACE II inhibitory activity may become a feasible alternative to be explored.
Indeed, these therapeutic approaches are supported by the SARS-CoV-2 similarity to other viruses from the same family that have infected people before; however, there is still no robust therapeutic evidence for COVID-19 [4]. In this context, some studies have demonstrated the potential of natural product-derived compounds, such as curcumin, against SARS-CoV-2 [8].
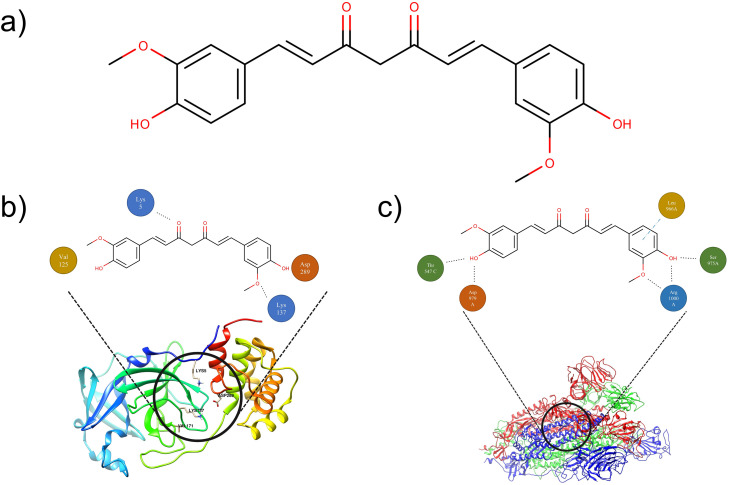
Curcumin ( Fig. 1a), a polyphenol obtained from the Curcuma longa rhizome [9], is the major curcuminoid presented in this plant (77%), along with 17% of demetoxicurcumin (curcumin II) and 3% of bisdemetoxicurcumin (curcumin III) [10].
Fig. 1.
Chemical structure of a) curcumin, and its b) interaction with SARS-CoV-2–3CLpro and c) SARS-CoV-2-S. Dotted lines show hydrogen bonding (black) and pi-sigma interaction (blue). Amino acids are shown according to their side-chain classification: polar (yellow), non-polar (green), acid (orange), and basic (blue). (In silico analysis performed according to the rationale of Dandapat, Jena, Kanungo, Nayak and Chainy [8], Gonzalez-Paz, Lossada, Moncayo, Romero, Paz, Vera-Villalobos, Pérez, San-Blas and Alvarado [11]).
Source: Own authorship.
In this scenario, it is possible to observe that curcumin is one of the most thoroughly investigated and promising dietary natural product-derived molecules. Approximately 3000 preclinical investigations have been carried out, from which the potential beneficial effects and safety (tolerated up to 12 g/day) of curcumin have been reported [10], [12]. Regarding its biological properties, scientific reports have described its use as anti-inflammatory [13], anticancer [14], antioxidant [13], and antidepressant [15]. Additionally, curcumin is an antiviral [16] agent, which also displays potential effects in the treatment of COVID-19, as demonstrated by Zahedipour et al. [17]. These authors suggested that curcumin may act by viral inhibition, inflammatory modulation and/or immunological responses, with the potential to reverse the pulmonary edema and pathways associated to the fibrosis in COVID-19 infection [17].
Indeed, the approach used by Zahedipour and colleagues [17] highlighted curcumin’s promising potential in COVID-19 infection. However, the authors did not properly consider curcumin’s biopharmaceutical limitations and its effect on the biological response. In this perspective, this review discusses not only relevant aspects of the investigation of curcumin against COVID-19 and its mechanisms of antiviral action against SARS-CoV-2, but also highlights the nanotechnological approach as the means to overcome curcumin’s drawbacks and to enable a favorable drug response against SARS-CoV-2.
2. Curcumin as an antiviral alternative against SARS-CoV-2
Curcumin has shown itself as an effective molecule to treat viral infections due to its ability to modulate a great number of molecular targets that contribute to the infection process. Among those, the following could be mentioned: (i) transcription and replication regulation [18], (ii) inhibition of proteases [19], [20], (iii) inhibition of attachment and entry of the virus to the cells [21] and (iv) inactivation and attack of the virus’s structures [22].
Accordingly, curcumin has presented effectiveness against a wide number of viruses, such as the Human Immunodeficiency Virus (HIV), Hepatitis C Virus (HCV), Human Cytomegalovirus (HCMV), Epstein-Barr Virus (EBV), Bovine Herpesvirus 1 (BHV 1), Chikungunya Virus, Ebola Virus, Enterovirus 71 (EV71), Rift Valley Fever Virus (RVFV), Human Norovirus (HuNoV), Respiratory Syncytial Virus (RSV), Fish Viral Hemorrhagic Septicemia Virus (VHSV), and Influenza A Virus (IAV) [23].
Additionally, studies also related curcumin activity to SARS-CoV, a coronavirus, identified in 2003. This molecule inhibited the 3C-like protease (3CLpro) with half-maximum inhibitory concentration (IC50) of 40.0 µM. Such activity represents greater inhibitory potential when compared to other natural product-derived compounds as (i) Hesperetin (Isatis indigotica) IC50 = 60.0 μM, (ii) Quercetin (Allium cepa) IC50 = 52.7 μM, (iii) Broussochalcone A (Broussonetia papyrifera) IC50 = 88.1 μM, (iv) Betulonic acid (Juniperus formosana) IC50 > 100 μM, and (v) Hinokinin (Chamaecyparis obtusa) IC50 > 100 μM [24], [25].
Furthermore, this molecule was also able to inhibit papain-like protease (PLpro), whose IC50 value (5.7 ± 0.3 µM) was more expressive than the natural product-derived compounds, (i) Broussochalcone A (Broussonetia papyrifera) IC50 = 9.2 μM, (ii) Kaempeferol (Zingiber officinale) IC50 = 16.3 μM, and (iii) Quercetin (Allium cepa) IC50 = 16.3 μM [26], [27]. Hence, (i) the large antiviral spectrum due to multiple biological targets, (ii) the expressive in vitro inhibitory activity reported above and (iii) the high in vivo tolerability and, by consequence, the US Food and Drug Administration (FDA) approval [28], make this compound a natural product with high potential to be used against coronaviruses.
Moreover, preliminary computational studies predict curcumin’s ability to inhibit proteases and to interact with S and ACE 2 proteins [28], [29]. Such aspects will be further discussed.
2.1. SARS-CoV-2 protease inhibitor
Proteases play an essential role in viral replication and can represent potential targets for the COVID-19 treatment [30], [31]. Indeed, the protein sequences of the SARS-CoV Mpro and the SARS-CoV-2 Mpro are 96% identical and their active sites, in both proteins, remain free from mutations [32].
In silico preliminary studies have been performed to evaluate different medicinal plant compounds as potential inhibitors of SARS-CoV-2 Mpro [33]. In this perspective, Ibrahim and collaborators [34] evaluated the inhibitory effect of curcumin, quercetin, piperine, kaempferol, capsaicin, carnosol, acetyl eugenol and other natural compounds (32 total) against the main protease of SARS-CoV-2 (Mpro). Curcumin revealed a high potency as Mpro inhibitor, displaying binding energy of - 9.2 kcal/mol. Such affinity is attributed to its ability to form multiple hydrogen bonds, van der Waals interactions, and hydrophobic and pi (π)-based interactions with the key amino acids within the active site [34].
Accordingly, Gonzalez-Paz et al. [11] also performed a molecular docking study to compare capsaicin, piperine, and curcumin with chloroquine and hydroxychloroquine. The authors showed that curcumin was able to promote higher inhibition of 3CLpro of SARS-CoV-2 than the tested control drugs (chloroquine and hydroxychloroquine). The scheme showing the interaction between curcumin and the SARS-CoV-2–3CLpro is shown in Fig. 1b. Based on this rationale, it is expected that curcumin would reduce the viral load in human cells in a shorter treatment time when compared to other drugs with minor inhibitory action, which could prevent the disease from worsening into stages such as ARDS.
2.2. Angiotensin-converting enzyme II as a target for SARS-CoV-2 inhibition
ACE II is a cell receptor extensively present in the human body, mostly in the capillary blood vessels and lung tissue [35]. This receptor is associated to respiratory viral disorders such as SARS [36]. The virus’s ability to bind with ACE II is due to its spike protein (S), which was first described for SARS-CoV [7]. Therefore, since SARS-CoV-S shares ~76% similarity in amino acid sequences with SARS-CoV-2-S, the affinity of the protein S from the new strain to the ACE II was investigated [37]. The study revealed that SARS-CoV-2 displays the most efficient binding to ACE II when compared to the SARS-CoV, which could explain the current increase in human-human transmission ability of this new strain of coronavirus [38].
The virus entry into the cell occurs from a sequence of events. The host cell presents on its membrane the type 2 transmembrane serine protease (TMPRSS2), which cleaves the ACE II and promotes the binding by the SARS-CoV-2-S protein [39]. Subsequently, cellular proteases mediate the virus and cell membrane fusion, leading to the virus replication in the cytosol [37]. Hence, the angiotensin-converting enzyme proved to be a potent in vivo binding site for drugs that acted against SARS-CoV [40]. Recently, Zhou et al. [41] evaluated the infectivity of SARS-CoV-2 on HeLa cells expressing or not the ACE II receptors from humans, Chinese horseshoe bats, civets, pigs, and mice. They observed that SARS-CoV-2 was able to use all ACE II receptors, except for the mice ACE II, and did not use other coronavirus receptors, such as aminopeptidase N and dipeptidyl peptidase 4.
Furthermore, in view of the pursuit of new drug candidates for SARS-CoV-2 inhibition, in silico studies have demonstrated that curcumin has a dual inhibitory action at this target site: (i) inhibition of SARS-CoV-2-S (Fig. 1c) and (ii) inhibition of cell ACE II receptor [8]. Therefore, based on the role of ACE II on the SARS-CoV-2 replication cycle, curcumin emerges as a promising dual-inhibitor (directly and/or indirectly) of this target, which would disrupt the aforementioned viral pathway.
3. Nanotechnological approaches to improve curcumin properties against SARS-CoV-2
Notwithstanding its many biological benefits and potential as a candidate to treat COVID-19 due to its inhibition of SARS-CoV-2 viral pathways, curcumin has physicochemical properties that reduce its bioavailability. Among those, its poor water-solubility (11 ng/ml) and low stability in aqueous media, especially in alkaline pH, are worth to mention [42]. Furthermore, after oral administration, curcumin is rapidly metabolized, i.e., the drug is reduced (Phase I) and conjugated (Phase II) by the gastrointestinal tract and liver metabolism, respectively, before its elimination [42]. To overcome these drawbacks, nanostructured carriers (nanosystems) have been used, since they can protect drugs from chemical and metabolic degradation, enhance their solubility and modify their transport through biological membranes [43].
Different nanotechnological carriers for curcumin have been developed, such as (i) nanoemulsions, (ii) microemulsions, (iii) liposomes, (iv) nanogels, (v) micelles, and (vi) nanoparticles [44]. Table 1 summarizes the main advantages and disadvantages of these nanosystems.
Table 1.
Different nanosystems eligible to be used as curcumin carrier and their advantages and disadvantages.
| Nanosystem | Advantages | Disadvantages | Reference |
|---|---|---|---|
| Nanoemulsions |
|
|
[45] |
| Microemulsions |
|
|
[46] |
| Liposomes |
|
|
[47] |
| Nanogels |
|
|
[48] |
| Micelles |
|
|
[49] |
| Nanoparticles |
|
[50], [51] |
Nanoparticles represent a large group of nanocarrier. Those characteristics may change depending upon the type of particle and its composition material.
Indeed, in light of the current pandemic of COVID-19, it is noteworthy that at least three nanotechnological curcumin-based products are available on the market in the form of polymeric nanoparticles (Nanocurc™), liposomes (Lipocurc™) and nanomicelles (Sinacurcumin®) [52], [53]. To date, only three studies have evaluated the in vivo efficacy of curcumin-loaded nanosystems against COVID-19. Saber-Moghaddam and collaborators [53] conducted an open, non-randomized clinical trial of the effectiveness of an oral curcumin nanosystem (Sinacurcumin®, a soft gel capsules containing 40 mg of curcuminoids in nanomicelles, two capsules twice a day for 2 weeks). The study involved patients hospitalized with COVID-19 classified as mild to moderate. The authors observed that most of the symptoms quickly reduced in the group treated with Sinacurcumin®. They concluded that the curcumin-loaded nanosystem can improve the recovery time in hospitalized patients with COVID-19. However, because these findings are preliminary and could not be statistically representative, the authors recommended other randomized, placebo-controlled clinical trials with larger groups.
Moreover, Valizadeh et al. [54] conducted a randomized, double-blind, placebo-controlled study to evaluate not the antiviral activity, but rather the effects of Sinacurcumin® 40 mg (4 capsules daily for 14 days) on the modulation of inflammatory cytokines in patients with COVID-19. mRNA expression and levels of cytokine secretion were assessed by real-time PCR and ELISA. Sinacurcumin® was able to modulate the increase rate of inflammatory cytokines, especially the expression of IL-1β and IL-6 mRNA, and the secretion of cytokines, in patients with COVID-19, which may increase clinical outcomes and general recovery.
Another study highlighting the anti-inflammatory response of the same formulation in patients with COVID-19 was carried out by Tahmasebi et al. [55]. Indeed, they investigated the therapeutic effects of Sinacurcumin® on the frequency and responses of Th17 cells (T helper cells) in patients with mild and severe COVID-19. They concluded that Sinacurcumin® was able to reduce the frequency of Th17 cells and the related inflammatory factors in patients with mild and severe COVID-19. Therefore, Sinacurcumin® can be considered a potential modulator in improving the patient's inflammatory condition.
In addition to these studies, there is a registered protocol [56] for a prospective placebo-controlled clinical trial with a parallel group, randomized in a single center, in patients with COVID-19. This study will evaluate the effectiveness of nanomicelles containing curcumin and their effects on the patient’s immune responses after treatment.
Despite the positive outcomes from the in vivo studies of Sinacurcumin® in the treatment of COVID-19, this formulation focuses on reducing symptoms and mediating inflammatory responses generated by the disease. However, it is important to emphasize that other nanosystems have advantages (Table 1) that may provide additional features and outcomes in treatment options based on curcumin.
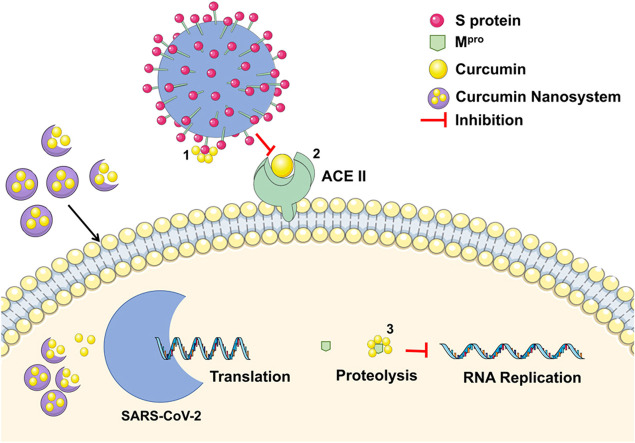
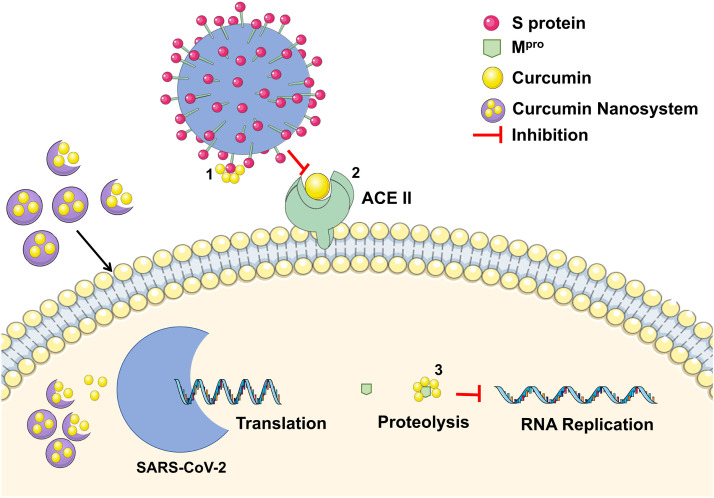
Finally, no in vitro nor in vivo studies have evaluated the response of free and/or entrapped curcumin and its antiviral potential and possible mechanisms against SARS-CoV-2. In view of the potential mechanisms from which curcumin nanosystems could inhibit the activity of SARS-CoV-2, a proposed summary of the aforementioned mechanisms is presented in Fig. 2.
Fig. 2.
Proposed rationale for curcumin nanosystems inhibition mechanisms according to the literature findings. 1) Curcumin inhibition by binding on SARS-CoV-2 S protein. 2) Curcumin inhibition by binding on ACE II receptor on the human cell membrane. 3) Curcumin inhibiting viral RNA replication by binding to SARS-CoV-2 Mpro.
Source: Own authorship.
Beyond the array of advantages of nanosystems on the delivery of burdensome molecules, nanoparticles (NPs) exhibit the advantage of easy customization and functionalization (Table 1) [50]. Moreover, there are a countless number of particles of distinct materials and structural organizations to be explored, such as (i) lipid-based nanoparticles, (ii) polymeric nanoparticles, and (iii) inorganic nanoparticles [57]. On that matter, nanoparticle features can be engineered to strategically target extracellular and intracellular viruses [58], [59]. Indeed, functionalization of NPs may be achieved by linking antibodies [60], carbohydrates [61], heparin [62], polyanionic molecules [63], among other compounds, to their surface. Furthermore, these changes on the particle surface may lead to an extended antiviral spectrum based on the blank-nanoparticle itself [64].
Those nanoparticles mechanisms of action are achieved through different pathways, such as (1) viral inactivation (directly or indirectly), (2) fixation of viruses in host cells, (3) viral penetration, and (4) viral replication, which are mainly dependent on the nature of the nanoparticles used and their functionalization [65]. In this way, nanoparticles are able to block these steps and modify the structure of the capsid protein, reducing viral load, either by physical or chemical means [66].
Therefore, a viral inactivation can occur through interactions between the nanoparticles and the viral surface protein (S-protein) by Kazimir interactions and Van der Waals forces. In fact, this phenomenon was already observed in metallic nanoparticles due to their shape, size, structure, and local field enhancement of action [67]. Since the S-protein can be found on the novel coronavirus surface [68], it is possible that nanoparticles could interact with the SARS-CoV-2 protein.
Additionally, the SARS-CoV-2-S protein is responsible for fixation (subunit 1) and entry (subunit 2) of viruses [37], [69]. The subunit 1 bonds specifically with the receptor of the angiotensin-converting enzyme II (ACE II) on human surface cells. On the other hand, the subunit 2 is related to the fusion of the attached virus to the membrane, which is internalized mostly by endocytosis mechanisms [69], [70], [71]. These mechanisms can also be blocked by electrostatic interactions, expressed by zeta potential readings, between the nanoparticles with different charges and the virus, which neutralizes the effective charge on the virus particles. This leads to viral aggregation, as observed for cationic nanoparticles developed by Ting and colleagues [72].
Furthermore, in a context in which SARS-CoV-2 is already loaded into the cell, the infectant RNA acts as a messenger RNA (mRNA), which is translated by the host’s ribosomes to produce replicative viral enzymes. Subsequently, new RNA and mRNA genomes are produced for the synthesis of necessary components to assemble new viral particles [73].
Briefly, the SARS-CoV-2 replication is a complex process that involves RNA synthesis, proofreading, and capping [73], [74]. In this sense, the SARS-CoV-2 viral replication can be disrupted by nanoparticles, as described by Salleh et al. [71], who affirm that silver nanoparticles (AgNPs) are able to attach to the viral DNA or RNA, thus, inhibiting the replication or propagation of the virus inside the host’s cells.
Considering this view, two approaches can be explored, (a) the increase in bioavailability of curcumin and, by consequence, its antiviral activity by delivery in NPs; and (b) a synergic combination of activities from curcumin and NPs themselves promoting a dual mechanism of action. Accordingly, curcumin could exhibit its inhibition on SARS-CoV-2 targets and the functionalized NPs could further block the virus entrance in the cell or display different mechanisms according to their functionalization.
Motivated by this perspective, some authors have already tested curcumin-nanoparticles against other viruses. Yang, Li and Huang [75] developed curcumin-loaded silver nanoparticles (AgNP) and showed that curcumin-AgNP improved the antiviral activity compared to curcumin and AgNP themselves against a syncytial virus. In another approach, Ting et al. [72] synthesized curcumin-carbon dots and proved their ability to suppress the virus-cell entry and the viral-RNA of porcine epidemic diarrhea virus (Coronaviridae family). These data reassure that nanotechnological approaches to delivery curcumin are viable and should be further investigated.
4. Conclusions
COVID-19 represents a global threat due to its difficulty in treatment once there is no current approved antiviral drug with proven efficacy and minor adverse effects. In this scenario, curcumin becomes a promising drug to be used as an antiviral agent due to its broad-spectrum, low toxicity, and potential pharmacological mechanism against SARS-CoV-2. The latter has been investigated using in silico studies which showed great activity in both target sites of inhibition of SARS-CoV-2. Henceforward, an investigation of the potential of curcumin must be performed to enlarge the possibilities of treatment for the COVID-19. Although this drug has limitations, nanostructured systems can be used as a tool to overcome such drawbacks. Furthermore, curcumin nanocarriers are already available in the market, allowing clinical studies, such as the few already carried out in patients with COVID-19 to reduce the anti-inflammatory process and symptoms associated to this disease. Despite the promising responses of curcumin nanomicelles (Sinacurcumin®) in the symptoms’ management of COVID-19, the in vivo antiviral properties have not yet been investigated. Additionally, other nanosystems, such as nanoparticles, can be investigated due to further possibilities of functionalization and possible antiviral synergy between the particle and the drug. Further, since turmeric (Curcuma longa extract) has a low curcumin concentration, < 7%, its food-grade use should not be encouraged and do not replace proper treatment for COVID-19 once it lacks FDA approval.
5. Perspective
It is noteworthy the aforementioned capability of curcumin as an antiviral agent. Notwithstanding the promising enhancement in curcumin bioavailability by NPs and their in vitro synergistic mechanism against different viruses, studies aiming to verify if these early findings on the Coronaviridae family transpose to SARS-CoV-2 should be further performed. Initially, these studies need to attest curcumin-NPs efficiency to suppress the SARS-CoV-2 viral infection in vitro. Then, since some drugs and nanoparticles fail to ensure in vitro antiviral activity, animal-infected models must be used for in vivo testing. This careful outline would ensure safety in further steps. Finally, after preclinical trials to attest drug efficiency and safety, curcumin nanoparticles, especially the already FDA-approved formulations, may be used on human clinical trials.
Conflict of interest statement
The authors declare no competing financial interest.
Acknowledgments
This study was funded in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brazil (CAPES) - Finance Code 001 and by the Conselho Nacional de Desenvolvimento Científico e Tecnológico - Brazil (CNPQ) through study scholarships. The authors would like to acknowledge the molecular graphics performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO), WHO coronavirus disease (COVID-19) dashboard, (2021). 〈https://www.who.int/emergencies/diseases/novel-coronavirus-2019〉. (Accessed Mar 28th, 2021).
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X.W., Yap Y.L. Old drugs as lead compounds for a new disease? Binding analysis of SARS coronavirus main proteinase with HIV, psychotic and parasite drugs. Bioorg. Med. Chem. 2004;12(10):2517–2521. doi: 10.1016/j.bmc.2004.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J. Dandapat, A.B. Jena, N. Kanungo, V. Nayak, G.B.N. Chainy, Catechin and Curcumin interact with corona (2019-nCoV/SARS-CoV2) viral S protein and ACE2 of human cell membrane: insights from Computational study and implication for intervention, Preprint available at Research Square 10.21203/rs.3.rs-22057/v1 (2020). [DOI]
- 9.Kocaadam B., Sanlier N. Curcumin, an active component of turmeric (Curcuma longa), and its effects on health. Crit. Rev. Food Sci. Nutr. 2017;57(13):2889–2895. doi: 10.1080/10408398.2015.1077195. [DOI] [PubMed] [Google Scholar]
- 10.Goel A., Kunnumakkara A.B., Aggarwal B.B. Curcumin as “Curecumin”: from kitchen to clinic. Biochem. Pharmacol. 2008;75(4):787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 11.L.A. Gonzalez-Paz, C.A. Lossada, L.S. Moncayo, F. Romero, J.L. Paz, J. Vera-Villalobos, A.E. Pérez, E. San-Blas, Y.J. Alvarado, Theoretical molecular docking study of the structural disruption of the viral 3CL-protease of COVID19 induced by binding of Capsaicin, Piperine and Curcumin Part 1: a comparative study with chloroquine and hydrochloroquine two antimalaric drugs, Preprint available at Research Square 10.21203/rs.3.rs-21206/v1 (2020). [DOI]
- 12.Minassi A., Sanchez-Duffhues G., Collado J.A., Munoz E., Appendino G. Dissecting the pharmacophore of curcumin. Which structural element is critical for which action? J. Nat. Prod. 2013;76(6):1105–1112. doi: 10.1021/np400148e. [DOI] [PubMed] [Google Scholar]
- 13.Boroumand N., Samarghandian S., Hashemy S.I. Immunomodulatory, anti-inflammatory, and antioxidant effects of curcumin. J. HerbMed Pharm. 2018;7(4):211–219. [Google Scholar]
- 14.Allegra A., Innao V., Russo S., Gerace D., Alonci A., Musolino C. Anticancer activity of Curcumin and its analogues: preclinical and clinical studies. Cancer Investig. 2017;35(1):1–22. doi: 10.1080/07357907.2016.1247166. [DOI] [PubMed] [Google Scholar]
- 15.Hurley L.L., Akinfiresoye L., Nwulia E., Kamiya A., Kulkarni A.A., Tizabi Y. Antidepressant-like effects of curcumin in WKY rat model of depression is associated with an increase in hippocampal BDNF. Behav. Brain Res. 2013;239:27–30. doi: 10.1016/j.bbr.2012.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Praditya D., Kirchhoff L., Bruning J., Rachmawati H., Steinmann J., Steinmann E. Anti-infective properties of the golden spice Curcumin. Front. Microbiol. 2019;10:912. doi: 10.3389/fmicb.2019.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahedipour F., Hosseini S.A., Sathyapalan T., Majeed M., Jamialahmadi T., Al-Rasadi K., Banach M., Sahebkar A. Potential effects of curcumin in the treatment of COVID-19 infection. Phytother. Res. 2020;34(11):2911–2920. doi: 10.1002/ptr.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayan V., Ravindra K.C., Chiaro C., Cary D., Aggarwal B.B., Henderson A.J., Prabhu K.S. Celastrol inhibits Tat-mediated human immunodeficiency virus (HIV) transcription and replication. J. Mol. Biol. 2011;410(5):972–983. doi: 10.1016/j.jmb.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali A., Banerjea A.C. Curcumin inhibits HIV-1 by promoting Tat protein degradation. Sci. Rep. 2016;6(1):27539. doi: 10.1038/srep27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sui Z., Salto R., Li J., Craik C., Ortiz de Montellano P.R. Inhibition of the HIV-1 and HIV-2 proteases by curcumin and curcumin boron complexes. Bioorg. Med. Chem. 1993;1(6):415–422. doi: 10.1016/s0968-0896(00)82152-5. [DOI] [PubMed] [Google Scholar]
- 21.Mounce B.C., Cesaro T., Carrau L., Vallet T., Vignuzzi M. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antivir. Res. 2017;142:148–157. doi: 10.1016/j.antiviral.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Chen T.-Y., Chen D.-Y., Wen H.-W., Ou J.-L., Chiou S.-S., Chen J.-M., Wong M.-L., Hsu W.-L. Inhibition of enveloped viruses infectivity by Curcumin. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0062482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jennings M.R., Parks R.J. Curcumin as an antiviral agent. Viruses. 2020;12(11):1242. doi: 10.3390/v12111242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G., Lee C.K., Chang S.T., Kuo C.J., Lee S.S., Hou C.C., Hsiao P.W., Chien S.C., Shyur L.F., Yang N.S. Specific plant terpenoids and lignoids possess potent antiviral activities against Severe Acute Respiratory Syndrome Coronavirus. J. Med. Chem. 2007;50:4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 25.Prasansuklab A., Theerasri A., Rangsinth P., Sillapachaiyaporn C., Chuchawankul S., Tencomnao T. Anti-COVID-19 drug candidates: a review on potential biological activities of natural products in the management of new coronavirus infection. J. Tradit. Complement. Med. 2021;11(2):144–157. doi: 10.1016/j.jtcme.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnard D.L., Kumaki Y. Recent developments in anti-severe acute respiratory syndrome coronavirus chemotherapy. Future Virol. 2011;6(5):615–631. doi: 10.2217/fvl.11.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuzimoto A.D., Isidoro C. The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds - additional weapons in the fight against the COVID-19 pandemic? J. Tradit. Complement. Med. 2020;10(4):405–419. doi: 10.1016/j.jtcme.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thimmulappa R.K., Mudnakudu-Nagaraju K.K., Shivamallu C., Subramaniam K.J.T., Radhakrishnan A., Bhojraj S., Kuppusamy G. Antiviral and immunomodulatory activity of curcumin: a case for prophylactic therapy for COVID-19. Heliyon. 2021;7(2) doi: 10.1016/j.heliyon.2021.e06350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandey P., Rane J.S., Chatterjee A., Kumar A., Khan R., Prakash A., Ray S. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: an in silico study for drug development. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1796811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang K.O., Kim Y., Lovell S., Rathnayake A.D., Groutas W.C. Antiviral drug discovery: norovirus proteases and development of inhibitors. Viruses. 2019;11(2):197. doi: 10.3390/v11020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Wang X.J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J. Genet. Genom. 2020;47(2):119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S. Khaerunnisa, H. Kurniawan, R. Awaluddin, S. Suhartati, S. Soetjipto, Potential inhibitor of COVID-19 Main Protease (Mpro) from several medicinal plant compounds by molecular docking study, Preprint available at Preprints 10.20944/preprints202003.0226.v1 (2020). [DOI]
- 34.Ibrahim M.A.A., Abdelrahman A.H.M., Hussien T.A., Badr E.A.A., Mohamed T.A., El-Seedi H.R., Pare P.W., Efferth T., Hegazy M.-E.F. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput. Biol. Med. 2020;126 doi: 10.1016/j.compbiomed.2020.104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuba K., Imai Y., Penninger J.M. Angiotensin-converting enzyme 2 in lung diseases. Curr. Opin. Pharmacol. 2006;6(3):271–276. doi: 10.1016/j.coph.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of Coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 40.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanidad K.Z., Sukamtoh E., Xiao H., McClements D.J., Zhang G. Curcumin: recent advances in the development of strategies to improve oral bioavailability. Annu. Rev. Food Sci. Technol. 2019;10:597–617. doi: 10.1146/annurev-food-032818-121738. [DOI] [PubMed] [Google Scholar]
- 43.Pires P.C., Santos A.O. Nanosystems in nose-to-brain drug delivery: a review of non-clinical brain targeting studies. J. Control Release. 2018;270:89–100. doi: 10.1016/j.jconrel.2017.11.047. [DOI] [PubMed] [Google Scholar]
- 44.Ipar V.S., Dsouza A., Devarajan P.V. Enhancing Curcumin oral bioavailability through nanoformulations. Eur. J. Drug Metab. Pharmacokinet. 2019;44(4):459–480. doi: 10.1007/s13318-019-00545-z. [DOI] [PubMed] [Google Scholar]
- 45.Sabjan K.B., Munawar S.M., Rajendiran D., Vinoji S.K., Kasinathan K. Nanoemulsion as oral drug delivery - a review. Curr. Drug Res. Rev. 2020;12(1):4–15. doi: 10.2174/2589977511666191024173508. [DOI] [PubMed] [Google Scholar]
- 46.Callender S.P., Mathews J.A., Kobernyk K., Wettig S.D. Microemulsion utility in pharmaceuticals: implications for multi-drug delivery. Intern. J. Pharm. 2017;526(1):425–442. doi: 10.1016/j.ijpharm.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 47.Kaul S., Gulati N., Verma D., Mukherjee S., Nagaich U. Role of nanotechnology in cosmeceuticals: a review of recent advances. J. Pharm. 2018;2018:1–19. doi: 10.1155/2018/3420204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah S., Rangaraj N., Laxmikeshav K., Sampathi S. Nanogels as drug carriers – introduction, chemical aspects, release mechanisms and potential applications, Intern. J. Pharm. 2020;581 doi: 10.1016/j.ijpharm.2020.119268. [DOI] [PubMed] [Google Scholar]
- 49.Hanafy N.A.N., El-Kemary M., Leporatti S. Micelles structure development as a strategy to improve smart cancer therapy. Cancers. 2018;10(7):238. doi: 10.3390/cancers10070238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gagliardi M. Biomimetic and bioinspired nanoparticles for targeted drug delivery. Ther. Deliv. 2017;8(5):289–299. doi: 10.4155/tde-2017-0013. [DOI] [PubMed] [Google Scholar]
- 51.Mauricio M.D., Guerra-Ojeda S., Marchio P., Valles S.L., Aldasoro M., Escribano-Lopez I., Herance J.R., Rocha M., Vila J.M., Victor V.M. Nanoparticles in medicine: a focus on vascular oxidative stress. Oxid. Med. Cell. Longev. 2018;2018:1–20. doi: 10.1155/2018/6231482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yallapu M.M., Jaggi M., Chauhan S.C. Curcumin nanoformulations: a future nanomedicine for cancer. Drug Discov. Today. 2012;17(1):71–80. doi: 10.1016/j.drudis.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saber-Moghaddam N., Salari S., Hejazi S., Amini M., Taherzadeh Z., Eslami S., Rezayat S.M., Jaafari M.R., Elyasi S. Oral nano-curcumin formulation efficacy in management of mild to moderate hospitalized coronavirus disease-19 patients: an open label nonrandomized clinical trial. Phytother. Res. 2021:1–8. doi: 10.1002/ptr.7004. [DOI] [PubMed] [Google Scholar]
- 54.Valizadeh H., Abdolmohammadi-Vahid S., Danshina S., Ziya Gencer M., Ammari A., Sadeghi A., Roshangar L., Aslani S., Esmaeilzadeh A., Ghaebi M., Valizadeh S., Ahmadi M. Nano-curcumin therapy, a promising method in modulating inflammatory cytokines in COVID-19 patients. Int. Immunopharmacol. 2020;89 doi: 10.1016/j.intimp.2020.107088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tahmasebi S., El-Esawi M.A., Mahmoud Z.H., Timoshin A., Valizadeh H., Roshangar L., Varshoch M., Vaez A., Aslani S., Navashenaq J.G., Aghebati-Maleki L., Ahmadi M. Immunomodulatory effects of nanocurcumin on Th17 cell responses in mild and severe COVID-19 patients. J. Cell. Physiol. 2020:1–14. doi: 10.1002/jcp.30233. [DOI] [PubMed] [Google Scholar]
- 56.Hassaniazad M., Inchehsablagh B.R., Kamali H., Tousi A., Eftekhar E., Jaafari M.R., Fathalipour M., Nikoofal-Sahlabadi S., Gouklani H., Alizade H., Nikpoor A.R. The clinical effect of Nano micelles containing curcumin as a therapeutic supplement in patients with COVID-19 and the immune responses balance changes following treatment: a structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21(1):876. doi: 10.1186/s13063-020-04824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zazo H., Colino C.I., Lanao J.M. Current applications of nanoparticles in infectious diseases. J. Control Release. 2016;224:86–102. doi: 10.1016/j.jconrel.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 58.Lembo D., Donalisio M., Civra A., Argenziano M., Cavalli R. Nanomedicine formulations for the delivery of antiviral drugs: a promising solution for the treatment of viral infections. Expert Opin. Drug Deliv. 2018;15(1):93–114. doi: 10.1080/17425247.2017.1360863. [DOI] [PubMed] [Google Scholar]
- 59.Abo-zeid Y., Urbanowicz R.A., Thomson B.J., Irving W.L., Tarr A.W., Garnett M.C. Enhanced nanoparticle uptake into virus infected cells: could nanoparticles be useful in antiviral therapy? Int. J. Pharm. 2018;547(1):572–581. doi: 10.1016/j.ijpharm.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 60.Gu J., Al-Bayati K., Ho E.A. Development of antibody-modified chitosan nanoparticles for the targeted delivery of siRNA across the blood-brain barrier as a strategy for inhibiting HIV replication in astrocytes. Drug Deliv. Transl. Res. 2017;7(4):497–506. doi: 10.1007/s13346-017-0368-5. [DOI] [PubMed] [Google Scholar]
- 61.Chiodo F., Marradi M., Calvo J., Yuste E., Penadés S. Glycosystems in nanotechnology: gold glyconanoparticles as carrier for anti-HIV prodrugs. Beilstein J. Org. Chem. 2014;10(1):1339–1346. doi: 10.3762/bjoc.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klimyte E.M., Smith S.E., Oreste P., Lembo D., Dutch R.E. Inhibition of human metapneumovirus binding to heparan sulfate blocks infection in human lung cells and airway tissues. J. Virol. 2016;90(20):9237–9250. doi: 10.1128/JVI.01362-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baram-Pinto D., Shukla S., Gedanken A., Sarid R. Inhibition of HSV-1 attachment, entry, and cell-to-cell spread by functionalized multivalent gold nanoparticles. Small. 2010;6(9):1044–1050. doi: 10.1002/smll.200902384. [DOI] [PubMed] [Google Scholar]
- 64.Cagno V., Andreozzi P., D’Alicarnasso M., Silva P.J., Mueller M., Galloux M., Le Goffic R., Jones S.T., Vallino M., Hodek J. Broad-spectrum non-toxic antiviral nanoparticles with a virucidal inhibition mechanism. Nat. Mater. 2018;17(2):195–203. doi: 10.1038/nmat5053. [DOI] [PubMed] [Google Scholar]
- 65.Chen L., Liang J. An overview of functional nanoparticles as novel emerging antiviral therapeutic agents. Mater. Sci. Eng. C. 2020;112(7) doi: 10.1016/j.msec.2020.110924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gurunathan S., Qasim M., Choi Y., Do J.T., Park C., Hong K., Kim J.H., Song H. Antiviral potential of nanoparticles-can nanoparticles fight against coronaviruses? Nanomaterials. 2020;10(9):1645. doi: 10.3390/nano10091645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Demchenko H., Rusinchuk N. Evaluation of the efficiency of interparticle interactions in nanosystems. J. Nanotechnol. 2019;2019(4):1–8. [Google Scholar]
- 68.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharm. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Medhi R., Srinoi P., Ngo N., Tran H.-V., Lee T.R. Nanoparticle-based strategies to combat COVID-19. ACS Appl. Nano Mater. 2020;3(9):8557–8580. doi: 10.1021/acsanm.0c01978. [DOI] [PubMed] [Google Scholar]
- 70.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(3):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salleh A., Naomi R., Utami N., Mohammad A., Mahmoudi E., Mustafa N., Fauzi M. The potential of silver nanoparticles for antiviral and antibacterial applications: a mechanism of action. Nanomaterials. 2020;10(8):1566. doi: 10.3390/nano10081566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ting D., Dong N., Fang L., Lu J., Bi J., Xiao S., Han H. Multisite inhibitors for enteric coronavirus: antiviral cationic carbon dots based on Curcumin. ACS Appl. Nano Mater. 2018;1(10):5451–5459. doi: 10.1021/acsanm.0c00970. [DOI] [PubMed] [Google Scholar]
- 73.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(10):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang X.X., Li C.M., Huang C.Z. Curcumin modified silver nanoparticles for highly efficient inhibition of respiratory syncytial virus infection. Nanoscale. 2016;8(5):3040–3048. doi: 10.1039/c5nr07918g. [DOI] [PubMed] [Google Scholar]