Abstract
Influenza viruses cause annual epidemics and occasional pandemics of respiratory tract infections that produce a wide spectrum of clinical disease severity in humans. The novel betacoronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December 2019 and has since caused a pandemic. Both viral and host factors determine the extent and severity of virus-induced lung damage. The host’s response to viral infection is necessary for viral clearance but may be deleterious and contribute to severe disease phenotypes. Similarly, tissue repair mechanisms are required for recovery from infection across the spectrum of disease severity; however, dysregulated repair responses may lead to chronic lung dysfunction. Understanding of the mechanisms of immunopathology and tissue repair following viral lower respiratory tract infection may broaden treatment options. In this Review, we discuss the pathogenesis, the contribution of the host response to severe clinical phenotypes and highlight early and late epithelial repair mechanisms following influenza virus infection, each of which has been well characterized. Although we are still learning about SARS-CoV-2 and its disease manifestations in humans, throughout the Review we discuss what is known about SARS-CoV-2 in the context of this broad knowledge of influenza virus, highlighting the similarities and differences between the respiratory viruses.
Subject terms: Immunopathogenesis, Viral host response, Viral pathogenesis, SARS-CoV-2, Influenza virus
In this Review, Schultz-Cherry, Thomas and colleagues discuss the pathogenesis of influenza virus and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the human respiratory tract, the contribution of the host response to severe disease, epithelial repair mechanisms following infection, and current and potential future therapies for influenza virus and SARS-CoV-2 infections.
Introduction
Influenza viruses are enveloped viruses of the Orthomyxoviridae family, which are classified into four genera, which include influenza virus A–D (IAV, IBV, ICV and IDV). With regard to human health, IAVs and IBVs are of main concern; ICVs are endemic and cause only mild disease in humans, and IDVs primarily cause infection in cattle1. IBVs are restricted to humans and are classified into two circulating lineages (B/Victoria and B/Yamagata)2. IAVs, the cause of the majority of annual epidemic and all occasional pandemic human disease, are further subtyped on the basis of two surface glycoproteins located within the host-derived lipid membrane of virions, haemagglutinin (HA) and neuraminidase (NA)3. Sixteen HAs and nine NAs have been described in avian species, the main natural animal reservoir of influenza viruses; two additional HAs and NAs have been described in bats3,4. The IAVs H1N1 and H3N2 currently cause most epidemic disease in humans2. Influenza viruses are in a constant state of evolution within animal and human reservoirs facilitated by high mutation rates due to low-fidelity RNA polymerase proofreading capabilities4. Cumulative changes in sequences encoding HA and NA lead to antigenic drift in IAVs and IBVs, which alters fitness for human infection as the structure of antigenic surfaces recognized by previously protective humoral responses changes and contributes to epidemic disease5. HA (and to a lesser extent NA) gene segments from avian reservoirs may also reassort with contemporaneously circulating human influenza viruses to produce novel strains capable of causing pandemics, a process termed antigenic shift4,5. Since 1889 there have been five IAV pandemics, the most severe of which was in 1918 and the most recent of which was in 2009 (refs4,6). IAV infection may additionally arise as an epizootic infection frequently leading to severe disease. Fortunately, thus far there has been limited human-to-human transmissibility potential demonstrated by these viruses. An example is the highly pathogenic H5N1 IAV that is associated with case fatality rates as high as 60%7.
Coronaviruses are enveloped single-stranded non-segmented RNA viruses of the Coronaviridae family, subfamily Coronavirinae, which are further subdivided into four genera based on phylogenetic analyses: alphacoronaviruses, betacoronaviruses, gammacoronaviruses and deltacoronaviruses8. Similarly to influenza viruses, coronaviruses circulate within non-human reservoirs. Mammalian coronavirus infection is predominantly caused by alphacoronaviruses and betacoronaviruses, which share bats and rodents as natural reservoirs9,10. Avian coronavirus infections are caused by gammacoronaviruses and deltacoronaviruses, which share birds as natural reservoirs9. To date, seven coronaviruses associated with human coronavirus (HCoV) infections have been identified. Four (HCoV-NL63, HCoV-229E, HCoV-OC43 and HCoV-HKU1) tend to cause mild seasonal respiratory infections in otherwise healthy individuals11. The other identified human coronaviruses, including severe acute respiratory syndrome coronavirus (SARS-CoV, which emerged in 2002 and was identified in 2003 (ref.12)), Middle East respiratory syndrome coronavirus (MERS-CoV, which emerged and was identified in 2012 (ref.13)) and the recently identified severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, which emerged in 2019 and was identified in 2020 (ref.14)), cause more severe clinical manifestations. Coronaviruses have the largest genome of any RNA virus11 and use a replication strategy, including ribosomal frameshifting and generation of subgenomic RNAs, that predisposes to recombination events that confer the ability to develop novel host specificity15,16. Indeed, after the emergence of SARS-CoV in 2003, numerous SARS-like coronaviruses were identified in bat populations17. When SARS-CoV-2 was initially sequenced in bronchoalveolar lavage fluid samples from patients identified early in the pandemic in December 2019, the analysis demonstrated 96.2% sequence similarity to a bat coronavirus, Bat CoV RaTG13 (ref.18). Human infection with a coronavirus from its natural reservoir is thought to arise after adaptation in an intermediate host, during which time the ability for efficient human infection and human-to-human transmission develops (for example, camels in the case of MERS-CoV)10. A number of potential intermediate hosts have been proposed for SARS-CoV-2, although none has been definitively proved to date19.
In the following sections, we discuss influenza virus and coronavirus infection in humans, pathogenesis of viral infection, the contribution of the host response to severe disease and late epithelial repair mechanisms following viral infection. Although the mechanisms of SARS-CoV-2 pathogenesis are still being uncovered, throughout the Review we discuss what is known about SARS-CoV-2 and COVID-19 in the context of the wealth of knowledge of influenza virus pathogenesis, highlighting similarities and differences between these respiratory viruses.
Infection in humans
Influenza virus and disease
Human infection with influenza viruses produces a broad spectrum of clinical disease severity, which ranges from asymptomatic infection to death. Adaptive immune memory from prior exposure by either natural infection or immunization can prevent infection or limit the development of symptoms or severe complications (Table 1). Young children without prior exposure who are immunologically naive to influenza virus are at risk of severe disease20. The effectiveness of adaptive immune memory in preventing infection can be subtype specific, and there is evidence suggesting that the first exposure to influenza virus antigen can influence the quality of immune memory acquired over a lifetime21. In individuals who do become infected, complex interactions between viral and host factors influence the site of replication and the corresponding immune response, which determine disease severity. As influenza virus infections are not always medically attended owing to variability in disease manifestations, only estimates are available for yearly infection burden on a global and local basis (Fig. 1a). In a meta-analysis of human volunteer challenge studies, viral shedding is detected in most individuals on the first day after inoculation. Viral titres then peak 2–3 days after inoculation and subsequently fall to undetectable levels by 6–7 days after inoculation. In some individuals, however, viral shedding persists for longer periods of time. Total symptom scores increase on day 1 after inoculation and generally peak by day 2 or 3 with a subsequent return to an asymptomatic baseline by days 8–9 after infection22. Most commonly, upper respiratory tract (URT) signs of tracheobronchitis and pharyngitis coupled with constitutional symptoms, including fever, malaise and myalgia, are reported by symptomatic individuals23. However, severe disease phenotypes, including hospitalization, pneumonia, acute respiratory distress syndrome (ARDS) and death are witnessed more frequently in high-risk patient populations (Fig. 1b; Table 1). Multiple organ dysfunctions have been described in the case of human H5N1 infection with evidence of viral replication outside of lung tissue, although the contribution of direct viral cytopathic effect to these extrapulmonary manifestations is unclear7.
Table 1.
Selected comparisons between influenza virus and SARS-CoV-2
| Parameter | Influenza virus | SARS-CoV-2 |
|---|---|---|
| Receptor usage | Sialic acid | ACE2 |
| Viral surface protein processing | Haemagglutinin processing by trypsin-like proteases | Spike protein processing by host proteases, including TMPRSS2, cathepsin L and furin, neuropilin 1 |
| Cellular tropism | Respiratory epithelial cells: types I and II alveolar epithelial cells; ciliated cells |
Respiratory epithelial cells: type II alveolar epithelial cells, ciliated cells and secretory cells; sustentacular and horizontal basal cells of the olfactory epithelium Intestinal epithelial cells; endothelial cells; renal parenchymal cells |
| Tissues affected and pathology | Upper respiratory tract; lower respiratory tract (severe cases) | Upper respiratory tract; lower respiratory tract; intestinal tract; cardiovascular or endothelial system; kidneys; nervous system |
| Viral recognition in airway epithelial cells | TLR3; RIG-I; ZBP1 | TLR3; RIG-I; MDA5 |
| Site of viral replication | Nuclear | Cytoplasmic |
| Viral evasion of initial host response | NS1; PB2; PB1-F2 | NSP1; ORF6; NSP13; others? (extrapolated from other coronaviruses) |
| Extrapulmonary complications | Limited; cardiac: myocarditis (rare); neurological: encephalitis (rare) | Extensive; olfactory: anosmia; endothelial: thrombosis; neurological: stroke, encephalitis, neuropsychiatric; gastrointestinal: nausea, vomiting, diarrhoea |
| Viral evolution and antigenicity | Antigenic shift; antigenic drift | Antigenic drift? |
| Prior immunity | Previous infection; vaccination; subtype specificity | No specific SARS-CoV-2 immunity prior to late 2019–2020; protective immunity from other human coronaviruses unclear; vaccination started December 2020 |
MDA5, melanoma differentiation-associated 5; NS1, nonstructural protein 1; PB2, polymerase basic protein 2; RIG-I, retinoic acid-inducible gene I; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TLR3, Toll-like receptor 3; TMPRSS2, transmembrane serine protease 2; ZBP1, Z-DNA binding protein 1.
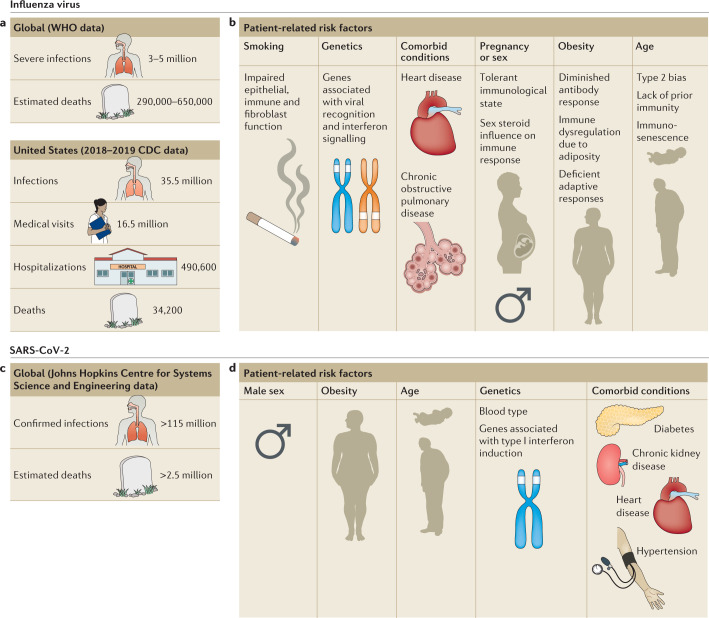
Fig. 1. Patient-related risk factors for severe influenza virus and SARS-CoV-2 infections.
a | Estimates of yearly influenza virus infections worldwide2 and in the United States (2018–2019 season)25. b | Risk factors associated with severe influenza virus infection in epidemiological and genetic studies. c | Global estimated number of cases and deaths from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection46. d | Risk factors identified thus far to be associated with severe coronavirus disease 2019 (COVID-19). Type 2 bias, bias towards type 2 immune responses.
A number of risk factors for complications have been identified through epidemiological studies or genomic analysis (Fig. 1b; reviewed elsewhere24). Individuals >65 years or <6 months of age suffer severe outcomes of influenza virus more often than those not at the extremes of age25. Immunosenescence, characterized by impaired humoral responses26 and reduction in T cell receptor diversity27 and T cell function28 are thought to contribute to the increased disease severity witnessed in elderly populations. In infants, an immature immune system characterized by a bias towards type 2 immunity in response to infection is thought to contribute to increased disease severity early in life29. Individuals with obesity suffer poor outcomes compared with lean individuals when challenged with IAV, an epidemiological observation well recapitulated in murine models30. A predisposition towards severe disease in individuals with obesity is thought to be multifactorial and related to defective adaptive immune responses31, chronic dysfunction of inflammatory signalling related to adiposity32 and insufficient responses to annual epidemic influenza virus vaccination33. Pregnancy increases the risk of hospitalization34, which has been attributed to the development of immune tolerance as a mechanism to prevent the fetus from rejection. As a consequence, there is a shift to type 2 immunity characterized by a cytokine profile that suppresses cytotoxic T lymphocytes and alters antibody class switching to result in less effective responses to viral infections35,36. Men appear to have disproportionate hospitalization rates compared with non-pregnant women37, presumably related to differences in immune responses mediated by sex hormones, which may be age-dependent24,38. Comorbid chronic health conditions, including metabolic syndrome39, heart failure40 and chronic obstructive pulmonary disease41, also increase an individual’s risk of severe disease. Finally, environmental factors, such as cigarette smoke exposure, contribute to disease severity42, potentially as a result of alterations in epithelial and immune cell function43 as well as in fibroblast repair responses to viral infection44.
SARS-CoV-2 and COVID-19
Since the recognition of coronavirus disease 2019 (COVID-19; the disease caused by SARS-CoV-2), a wide range of disease severity has been described, from asymptomatic infection45 (defined by the WHO as an infected individual who never eventually develops clinically apparent symptoms) to severe disease and death. Following its emergence, there have been >115 million cases identified and >2.5 million attributable deaths globally46 (Fig. 1c). Symptoms arise after a median incubation period of 4.8 days, and 95% of symptomatic individuals will display symptoms by 14 days after exposure47. A pre-symptomatic period has been defined by the WHO as an infected individual who does not presently have symptoms but who eventually manifests clinical disease. SARS-CoV-2 RNA is detected by RT–PCR in respiratory tract specimens for weeks after symptom onset48. Peak infectivity has been estimated to occur between 2 days before and 1 day after onset of symptoms in pre-symptomatic people48. The most frequently identified symptom of COVID-19 is cough, followed by fever, myalgia, dyspnoea and headache. Sore throat, diarrhoea and nausea are also reported, although less frequently49,50. Disturbances of smell and/or taste have been increasingly recognized as frequent symptoms of COVID-19 (refs51,52). Although loss of smell (anosmia) following viral URT infection has been recognized previously, anosmia caused by SARS-CoV-2 is potentially due to a different mechanism, which leads to clinical differences in the duration of sensory deficit52,53. As seen in MERS-CoV54 and SARS-CoV55 infection, extrapulmonary manifestations are additionally described with SARS-CoV-2 infection (Table 1). These include acute kidney injury56,57, skin findings58 and thrombosis59.
Several risk factors for severe disease have been identified in early reports detailing the clinical manifestations and outcome of COVID-19 (Fig. 1d). Genome-wide association and epidemiological studies have implicated blood type (A being higher risk than O, and Rhesus factor (Rh)-positive being higher risk than Rh-negative) as a potential heritable trait predisposing to SARS-CoV-2 infection and severe disease60. The mechanism of potential protection afforded by blood type identified in these large population-based studies requires further investigation. Metabolic disorders such as obesity61, diabetes62 and kidney disease63 have been associated with enhanced disease severity. Although the mechanisms remain unknown, obesity as a risk factor could be a consequence of the dampened type I interferon response found in individuals with obesity64; one study that did not include body mass index (BMI) as a factor found individuals with severe disease requiring mechanical ventilation were more likely to have decreased type I interferon levels in the plasma65. Cardiovascular conditions including hypertension, congenital heart disease and coronary artery disease66 have also been linked not only with severe disease but also with exacerbation of pre-existing conditions post-recovery66,67. Age has consistently been identified as an important risk factor for COVID-19 disease severity. Persons of advanced age are more likely to develop severe disease, including ARDS, during SARS-CoV-2 infection68. SARS-CoV-2 has been diagnosed in children and adolescents of all ages, including neonates; however, disease severity is typically milder and outcome is generally more favourable than in adult individuals69,70. Although correlations between disease severity caused by SARS-CoV-2 and certain pre-existing conditions are coming to light, it is important to consider that the novelty of the virus limits our ability to draw conclusions.
Pathological findings in infections
Influenza virus infection and pathology
Pathological analysis of IAV infection in humans is skewed towards a more thorough understanding of severe disease, especially when it occurs during pandemic influenza infection, on examination at autopsy71,72. Owing to the preferential cellular tropism of human influenza viruses, pathological changes of the tracheobronchial epithelium are characteristic (Table 1). With uncomplicated IAV infection in the absence of concurrent bacterial superinfection, the surface of the larynx, trachea and bronchi appear inflamed on visual inspection71. Early in infection, microscopic evaluation of tracheal and bronchial biopsy specimens demonstrates diffuse epithelial sloughing71. Mononuclear cells are the predominant inflammatory cells present73; neutrophils are typically absent early, but may be present with increasing degrees of epithelial necrosis with severe disease or with the presence of a concurrent bacterial superinfection72. More distally, in bronchioles, nearly complete epithelial sloughing may again be present along with bloody exudate in the airway lumen, accompanied by interstitial swelling, mixed inflammatory infiltrates and small-vessel thrombosis in the bronchiolar walls72. Alveolar involvement generates similar general findings of epithelial necrosis and sloughing to more proximal locations but requires different repair mechanisms and leads to severe disease owing to compromised gas exchange. Proteinaceous alveolar fluid, which may be bloody, develops along with cellular debris and impairs efficient diffusion of oxygen and carbon dioxide across the alveolar–capillary interface, leading to severe clinical symptomatology74,75.
SARS-CoV-2 and COVID-19
Lung pathology at autopsy following fatal SARS-CoV-2 infection frequently demonstrates diffuse alveolar damage (DAD) manifest as alveolar septal changes coupled with deposition of proteinaceous, potentially bloody, alveolar fluid, which compromises gas exchange (Table 1). Reactive type II pneumocytes and interstitial oedema are additionally seen76. Neutrophils are common in the inflammatory infiltrate in the lower respiratory tract (LRT)77. Inflammation of the bronchi and bronchioles is also found at autopsy, although less frequently than DAD. Corresponding to known areas of cellular tropism by single-cell gene expression analyses, immunohistochemistry staining for SARS-CoV-2 is seen in alveolar pneumocytes and ciliated epithelial cells. Tracheitis is also witnessed at autopsy with SARS-CoV-2 staining in submucosal glands and lymphocytes77. As described above, COVID-19 is increasingly being recognized as a disease affecting not only the respiratory tract. Intracellular staining by immunohistochemistry for SARS-CoV-2 in renal epithelial cells has been demonstrated in individuals with kidney injury, but it is unclear whether pathological changes are a consequence of pre-existing renal conditions, virus-induced damage or a contribution of both77. Ultrastructural evaluation by electron microscopy has additionally found SARS-CoV-2 particles in the enterocytes of the intestine77. In autopsy specimens, SARS-CoV-2 RNA has been detected in the lung, kidney, large intestine, blood, spleen and heart, with the lowest cycle threshold (Ct) values in pulmonary tissue77. Finally, lung tissue from individuals with COVID-19 exhibits unique vascular findings, including thrombosis, endothelial damage and abnormal angiogenesis patterns, compared with H1N1 infected or control lung tissue, as well as intracellular localization of SARS-CoV-2 particles within endothelial cells76.
Host defence in the respiratory tract
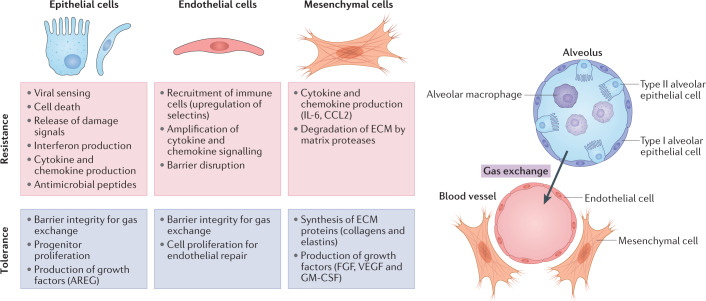
A fundamental challenge to all hosts facing infection is achieving a balance between clearance of the pathogen and maintenance of tissue function. A robust immune response may rapidly clear the pathogen but can also cause extensive collateral damage and compromise tissue function, as described above for influenza virus and SARS-CoV-2 infection. Achieving this balance is particularly important in the respiratory tract as the host cannot survive long without sufficient gas exchange carried out by the lungs. Hosts employ different defence strategies depending on the type of pathogen, chronicity of the infection and the tissue affected. A useful paradigm for characterizing host defence strategies is in terms of disease resistance and tolerance78–80. Disease resistance refers to the set of host processes that actively reduce the burden of the pathogen, which include both innate and adaptive immunity. Disease tolerance refers to a host response that acts to limit the damage in the affected tissue and support its function, thus ‘tolerating’ the pathogen burden. In the respiratory tract, a strategy of tolerance would maintain the essential function of gas exchange and blood oxygenation, and preserve the health of the host. The concepts of resistance and tolerance are often employed at a systems level, considering the host or a specific organ as a system of mechanisms that are connected79. The respiratory system is made up of numerous compartments, including the nasal passages, trachea, bronchioles and alveoli, which carry out distinct functions in supporting respiratory health81. Complex interactions between structural and immune cells ultimately determine the type of tissue environment — resistant or tolerant. The main structural cell types in the respiratory tract — epithelial, endothelial and mesenchymal — have distinct and integral roles in generating these tissue environments (Fig. 2). Each cell type is capable of acquiring distinct activation states that promote resistance or tolerance. Integration of activation signals at both the cellular and tissue level has consequences for innate and adaptive host immune responses, which ultimately determine the outcome of infection.
Fig. 2. Mechanisms of host resistance and tolerance in lung structural cells.
Recent studies have identified structural cells in the lung (epithelial, endothelial and mesenchymal) as crucial regulators of host immune responses. Cells in the lower respiratory tract must effectively communicate to promote effective viral clearance while limiting damage to endothelial cells of the blood vessels (red) and epithelial cells of the alveoli (blue), which together carry out gas exchange. Each cell type, and their phenotypically distinct subsets, contributes to disease resistance and tolerance host strategies through diverse mechanisms indicated in the boxes. The balance of these resistance and tolerance mechanisms ultimately determines the outcome and long-term consequences of respiratory viral infection. AREG, amphiregulin; ECM, extracellular matrix; FGF, fibroblast growth factor; GM-CSF, granulocyte–macrophage colony-stimulating factor; IL-6, interleukin 6.
The effectiveness of the host strategy in response to influenza virus or SARS-CoV-2 infection may depend on the compartment in the respiratory tract. For example, the URT, which is typically the initial site of infection and source of replication for transmission82, may benefit from a resistant host defence that favours a robust immune response to control viral spread. Type I and type III interferons have a crucial role in initially controlling viral replication in the URT and limiting spread to the lower airways83,84. When this resistance is broken, and the virus spreads, the LRT may require a tolerant defence that favours preservation of the crucial alveolar structures that perform gas exchange. This idea is supported by studies in mouse models of influenza virus infection using strains of IAV that preferentially replicate in the URT instead of the LRT85. Numerous survival phenotypes, due to diverse mechanisms, are independent of viral burden, indicating that rapid viral clearance is not necessary to improve disease outcome86–91. In human disease, viral load in respiratory samples is neither a consistent correlate of disease severity nor a reliable predictor of infection outcome92–94. In patients with severe disease who require extended hospitalization, mortality often occurs after viral clearance, indicating that the patient continues to resist a threat that is no longer present, although there are reports of prolonged viral shedding95.
In contrast to viral load, specific cytokines and inflammatory profiles in both serum or plasma and respiratory samples are consistently associated with the severity of infection, including in SARS-CoV-2 infection96,97. High systemic levels of multiple cytokines, and their association with severity, have frequently been referred to as a ‘cytokine storm’98. Although the term cytokine storm has become popular to describe an exuberant immune response during influenza virus infection, few studies have identified specific mechanisms regulating excessive production of inflammatory molecules that could be targeted by therapeutics. Framing the host response as a cytokine storm may be the result of a focus on systemic cytokine levels measured in serum or plasma from humans. Peripheral blood is easier to sample than the LRT and is amenable to longitudinal sampling in patients. However, it is still unclear whether specific inflammatory molecules drive lung pathology or serve as correlates of other tissue-level mechanisms. In studies that have compared respiratory compartment and peripheral blood cytokine levels, poor correlations are often observed94. Interestingly, animal models, which allow for in-depth sampling of respiratory compartments, have identified tissue-resident non-immune and immune cells as crucial regulators of this exuberant response99,100.
The following section will highlight recent studies that have begun to define the compartment-specific mechanisms that prevent or contribute to lung damage following influenza virus infection. Migrating and tissue-resident immune cells certainly have an important role in driving lung pathology, and mechanisms have been reported for many different types of cell, including neutrophils, monocytes/macrophages and T cells. Several reviews have summarized the role of immune-cell-driven lung damage during influenza virus infection80,101. In the following section, we focus on the role of non-immune structural cells that serve as upstream regulators of the host response. Studies of structural cells, including epithelial, endothelial and fibroblast cells (Fig. 2), have primarily reported on their roles in lung function by providing structural support to the tissue or maintaining the barriers required for gas exchange. These studies have underestimated their participation in the inflammatory response. However, it is now becoming clear that these cells have crucial roles in actively coordinating the host immune response to viral infections102. Lung structural cells have many of the same tools as immune cells to respond to infection, including viral sensors, cytokine or chemokine receptors, and antiviral proteins that directly inhibit replication.
Initial response of epithelial cells to influenza virus and SARS-CoV-2
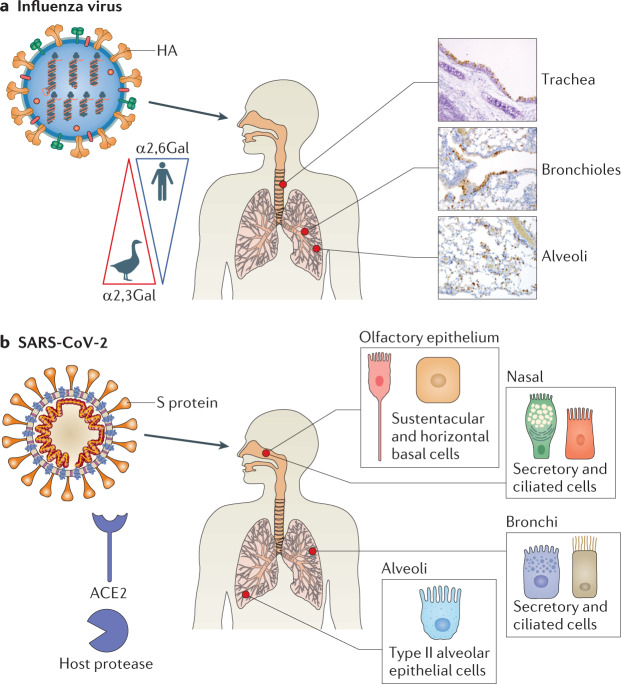
Epithelial cells are the primary targets of influenza virus103 and are the best-studied structural cells in the context of influenza virus infection (Table 1). The type of epithelial cells present and their relative abundance change drastically moving from the trachea to the bronchioles and eventually to the alveoli. As conducting airways, the trachea and bronchioles are rich in mucin-producing goblet cells, mucin and antimicrobial protein-producing submucosal gland cells, secretory club cells and ciliated cells that function to move mucus along the respiratory tract81,104. The alveoli are made up of thin type I pneumocytes that facilitate gas exchange and cuboidal type II pneumocytes that produce surfactant and serve as progenitor cells81,105. Influenza virus cellular tropism, as well as host specificity, is determined by HA and NA, which interact with epithelial cell surface sialosaccharides that contain a sialic acid (SA) residue linked to a galactose (Gal)106 (Fig. 3a; Table 1). Preferences in HA binding to specific SA–Gal linkages in sialosaccharides on epithelial cell surfaces contribute to host species restriction, transmissibility and clinical symptomatology107–109. Whereas avian and equine influenza viruses prefer to bind SAs linked to galactose by α2,3 linkage (SAα2,3Gal), human influenza viruses bind SAs linked to galactose by α(2,6) linkage (SAα2,6Gal)109. SAα2,3Gal and SAα2,6Gal are distributed along a gradient on the surface epithelial cells along the length of the human respiratory tract110. SAα2,6Gal predominates in the nasal mucosa, sinuses, trachea and bronchi; it is also present on epithelial cells of the respiratory and terminal bronchioles. Conversely, SAα2,3Gal molecules are primarily found on the surface of alveolar epithelial cells and rarely in the URT110 (Fig. 3a). Tropism for SAα2,3Gal, which predominates in the LRT, is thought to contribute to the high pathogenicity of epizootic avian IAV infections in humans. Despite the preferential binding noted in the above work, human IAVs are able to infect type I alveolar epithelial cells107.
Fig. 3. Cellular tropism of influenza virus and SARS-CoV-2.
a | The haemagglutinin (HA) protein of influenza viruses preferentially binds sialosaccharides on the surface of pulmonary epithelial cells. Whereas human influenza viruses prefer sialic acids (SAs) linked to galactose by α(2,6) linkage (SAα2,6Gal), avian influenza viruses prefer SAα2,3Gal. These are distributed in a gradient in the human respiratory tract. Immunohistochemistry staining demonstrates intracellular localization of influenza viruses in epithelial cells at three sites from mice challenged with influenza A virus. b | The spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) binds angiotensin-converting enzyme 2 (ACE2) on the surface of certain olfactory and respiratory epithelial cells distributed along the human respiratory tract after activation by a cellular protease, such as transmembrane serine protease 2 (TMPRSS2) (other proteases, including cathepsin L, neuropilin 1 and furin are involved in activation). Histopathological images in part a courtesy of P. Vogel.
Similarly to SARS-CoV, SARS-CoV-2 relies on angiotensin converting enzyme 2 (ACE2) and a host protease for host cell entry (Fig. 3b; Table 1). ACE2 is a transmembrane carboxypeptidase involved in processing angiotensin II, among other signalling proteins. It is constitutively released from the surface of airway epithelial cells as a soluble form (sACE2), which retains proteolytic activity, by the sheddases A disintegrin and metalloproteinase 17 and 10 (ADAM17 and ADAM10)111. The spike (S) protein on the surface of the SARS-CoV-2 particle binds to ACE2 and requires activation by a host protease. Transmembrane serine protease 2 (TMPRSS2) is the most commonly implicated112. Other proteases may also have a role, including cathepsin L113, which is involved in SARS-CoV entry114, neuropilin 1 (ref.115) and furin116. The involvement of multiple host proteases in viral entry may broaden tropism. Single-cell gene expression analyses in both non-human primates and humans have demonstrated overlapping transcription ACE2 and TMPRSS2 throughout the body. In the respiratory tract, overlapping expression is seen in cells of the olfactory, nasal and bronchial epithelial surfaces as well as in alveolar type II epithelial cells52,117–119. For SARS-CoV-2, there is likely a gradient of infectivity along the respiratory tract with susceptible epithelial cells with high ACE2 and TMPRSS2 expression more abundant in the nasal epithelium of the URT than in distal regions of the lung120. Outside of the respiratory tract, sites of ACE2 and TMPRSS2 expression and potential viral replication include superficial cells of the conjunctiva, enterocytes of the ileum and colon, and the gallbladder, among others117–119, which may account for extrapulmonary clinical manifestations of SARS-CoV-2 (Table 1), although further work defining the mechanistic pathophysiology of this association is required.
Despite being the initial and primary cell type infected by respiratory viruses, infected epithelial cells and bystander epithelial cells initiate carefully regulated responses to both cell-intrinsic and extrinsic signals. Epithelial cells are equipped with many of the same host defence tools that professional immune cells are well known for, including viral sensors, direct antiviral molecules and inflammatory mediators, such as type I/III interferons, tumour necrosis factor, interleukin 6 (IL-6) and other chemokines (for example, CXCL10 and IL-8)100,103. Indeed, in vitro models using influenza virus infection of normal human bronchial epithelial cells have demonstrated that epithelial cells are capable of producing inflammatory cytokines in the absence of other professional immune cells103. The epithelial influence on the immune response begins early. Upon infection, the influenza virus HA protein binds to surface sialosaccharides to initiate receptor-mediated endocytosis of the virion, which subsequently fuses with the endosomal membrane and releases viral ribonucleoproteins (vRNPs) into the cytoplasm. vRNPs are trafficked to the nucleus where replication occurs. Pathogen-associated molecular patterns in the form of viral nucleic acids are recognized by the pattern recognition receptors retinoic acid-inducible gene I (RIG-I) and Toll like receptor 3 (TLR3)80,86,121–124 (Table 1). TLR7 is also important for endosomal recognition of IAV in some immune cells125,126. An additional early mediator of the epithelial response to IAV infection is NLR family pyrin domain containing 3 (NLRP3), which forms a complex with other proteins termed the NLRP3 inflammasome that functions to moderate immunopathology86,123. The initial signalling cascades necessary for a coordinated host response to influenza virus entry culminate in the production of hundreds of interferon-stimulated genes (ISGs) and pro-inflammatory cytokines via activation of the transcription factors interferon regulatory factor 3 (IRF3) and nuclear factor κB (NF-κB). Interestingly, influenza viruses encode proteins, such as nonstructural protein (NS1)127 and polymerase basic protein 2 (PB2)128, among others, that interfere with interferon signalling at multiple steps. Although the initial epithelial response to SARS-CoV-2 requires more study, on the basis of recently published data and knowledge extrapolated from other coronaviruses, it is likely that TLR3, RIG-I and RIG-I-like receptor melanoma differentiation-associated 5 (MDA5) also participate in innate sensing of intracellular SARS-CoV-2 and are intrinsic to induction of interferon signalling pathways129–132 (Table 1). Recent evidence suggests that the NLRP3 inflammasome is also activated in response to SARS-CoV-2 infection in CD14+ lung cells obtained from deceased individuals as well as in peripheral blood mononuclear cells133.
Activated interferon pathways and their pleiotropic effects highlight the importance of balancing defence strategies of resistance and tolerance. As two studies recently demonstrated in mice, persistence of antiviral signalling in particular in the LRT during viral inflammation may oppose proliferation of epithelial cells, which are crucial for tissue repair and restoration of lung function90,91. Thus, compartmentalization of interferon and careful regulation of timing are likely crucial to surviving a severe respiratory infection. As in the case of other coronaviruses134, the type I interferon response to SARS-CoV-2 may be crucial. Decreased type I interferon levels in peripheral immune cells was associated with severe disease despite increased interferon-α in the lung65. Loss-of-function variants of TLR7 resulted in a decrease in type I interferon signalling and decreased levels of interferon and ISG mRNA135. Similarly, the presence of neutralizing autoantibodies targeting type I interferons was associated with onset of severe disease130. Finally, compared with individuals with influenza virus, individuals with COVID-19 demonstrate downregulation of interferon signalling pathways in subsets of peripheral blood mononuclear cells136. As in the case of influenza virus, SARS-CoV-2 also produces proteins that interfere with interferon signalling137 (Table 1).
In addition to an interferon and pro-inflammatory response to infection, altruistic programmed cell death (PCD) is an essential component of the initial cellular response to IAV infection. In benefit to the host, PCD serves to limit viral replication and prevent pathological immune responses138,139. However, because there appears to be a threshold of alveolar type I epithelial cell loss of 10%, beyond which gas exchange and survival are impaired85, it is imperative for host survival that PCD responses to prevent continued viral replication are contained139. PCD pathways are intertwined with various levels of crosstalk, which may lead to significantly different inflammatory outcomes with imbalances or preferential activation of one pathway over another139,140. Whereas necroptosis and pyroptosis are inflammatory forms of PCD that result in the release of damage-associated molecular patterns that serve to propagate inflammatory responses, apoptosis is a relatively anti-inflammatory form of PCD140,141. In the context of IAV infection, Z-DNA binding protein 1 (ZBP1), which recognizes Z-RNA, has an integral role in mediating PCD responses124,142–144. Immediately downstream of ZBP1 is receptor interacting protein kinase 3 (RIPK3), which contains a RIP homotypic interaction motif (RHIM) domain complementary to the ZBP1 RHIM124,145. Following ZBP1-dependent RIPK3 activation, necroptosis is mediated by mixed lineage kinase domain-like (MLKL) disruption of the plasma membrane after phosphorylation by RIPK3 (refs146,147) while apoptosis is mediated by RIPK3 interaction with a complex of proteins that includes receptor interacting protein kinase 1 (RIPK1), Fas-associated via death domain (FADD) and caspase 8 (ref.148). It was previously unclear whether both pathways were simultaneously activated in an individual cell or whether they were mutually exclusive. Recently, using a knock-in model in which caspase 8 was engineered to be unable to signal for apoptosis, but still able to carry out its other functions necessary for animal viability, it was demonstrated that necroptosis and apoptosis fates are mutually exclusive events following IAV infection within an individual cell149. Beyond influenza viruses directly activating these PCD pathways, it is important to note that interferons also lead to increased expression of many genes involved in cell death, including ZBP1 and MLKL144,150. Furthermore, proteins produced by influenza viruses themselves, such as PB1-F2, may induce apoptosis151. Early evidence suggests that SARS-CoV-2 results primarily in apoptosis of infected human airway epithelial cells152. Additionally, SARS-CoV-2 encodes the protein ORF3a, which has been shown to induce epithelial cell apoptosis dependent on caspase 8 activity, although to a lesser degree than SARS-CoV ORF3a153.
By releasing inflammatory mediators or other bioactive molecules, either as a result of PCD or by active secretion, epithelial cells help to define the tissue microenvironment. In addition, epithelial cells produce molecules that promote tissue repair and tolerance by interacting with diverse cell types in the LRT. One of these key molecules is granulocyte–macrophage colony-stimulating factor (GM-CSF). During IAV-induced lung injury, alveolar type II epithelial cells are primary producers of GM-CSF in the distal lung154. GM-CSF not only has well-documented roles in promoting inflammatory responses by inducing proliferation and differentiation of myeloid cells, but also helps maintain epithelial barrier integrity by promoting the activity of alveolar macrophages, enhancing adaptive responses through dendritic cell activity, or acting directly on epithelial cells155. Another important molecule is amphiregulin (AREG), a member of the epidermal growth factor family of molecules, which is capable of inducing both cell proliferation and differentiation156. A number of studies in mice have demonstrated that AREG has a protective effect during influenza virus infection and that multiple different cell types, including both immune and parenchymal cells, can produce the molecule87,88,157–159. Although it is unclear which cell type is the primary source of protective AREG during influenza virus infection, epithelial cells are a likely candidate157,159.
Epithelial cells can also alter the tissue microenvironment through regulation of surface molecules and communication with nearby cells. One example is the upregulation of class I major histocompatibility complex (MHC) molecules that present viral peptides to CD8+ T cells. Influenza virus-specific CD8+ T cells recognize the peptide–MHC complex and release cytolytic molecules to kill the infected cell160. This cell killing can further amplify the immune response through the release of inflammatory molecules from the infected cell. Killing of both infected and bystander alveolar type II cells by CD8+ T cells can drive acute lung damage and compromise tissue function161,162.
Epithelium-derived cell adhesion and integrin molecules, which mediate cell–cell and cell–extracellular matrix (ECM) interactions and can activate other extracellular molecules, also regulate the host response during influenza infection. One example is the epithelium-specific αvβ6 integrin, which is upregulated during infection and can modulate the lung microenvironment and collagen deposition through the regulation of transforming growth factor-β and type I and III interferon signalling89,163. β6 integrin-deficient mice are unable to form the functional αVβ6 homodimer and as a consequence have increased interferon signalling, activated alveolar macrophages and enhanced repair, resulting in greater protection from influenza virus and other respiratory pathogens89. This protection extends to high-risk populations164. Although epithelial cells are the most studied structural lung cell involved in host response to influenza virus infection, other structural lung cells have important roles in the response to and, therefore, outcome of infection.
Mesenchymal cells
Mesenchymal cells are a broad group that encompasses fibroblasts, smooth muscle cells, pericytes and other stromal cell types in the respiratory tract. These cells make up the connective tissue in the lung and exhibit diverse functions including regulation of the ECM, production of growth factors, cytokines and chemokines, and the generation of stem cell niches, among others165. As cells of the connective tissue, mesenchymal cells define the tissue environments by generating the ECM and providing signals to nearby cells, which determine their migration, proliferation and differentiation. These functions have been well studied during lung homeostasis but have not been well defined during acute respiratory infection, despite the fact that they communicate with essentially every cell type in the lung either through direct cell–cell interactions or through the ECM. One of the crucial tools that mesenchymal cells use to define the tissue environment is ECM proteases. Mesenchymal cells produce an impressive array of proteases and glycosidases that degrade or modify specific components of the ECM, including structural proteins, cytokines or growth factors, and other proteases166. ECM proteases regulate each stage of lung injury or infection by degrading ECM barriers to facilitate cell migration through the tissue, activating cytokines and growth factors, and modifying the ECM during repair. Bioactive degradation products of the ECM, often referred to as matrikines, can further amplify lung inflammation by stimulating nearby cells167. Several studies in mouse models of influenza virus infection have found that ECM proteases, including members of the matrix metalloproteinase (MMP) and A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family members, have both pathogenic (MMP9, MMP14 and ADAMTS4) and protective functions (ADAMTS5) by degrading diverse ECM proteins168–171. Studies in human cases of influenza virus infection have also established correlations between the levels of ECM proteases and the severity of disease and respiratory failure172. Mechanistic studies in mice have demonstrated that the activity of specific proteases can compromise lung function by directly altering the ECM structure and relatedly by regulating immune cell migration to the site of infection or damage. Although both immune and structural cells produce ECM proteases, recent evidence suggests that mesenchymal cells are the primary producers of proteases that contribute to pathogenesis. One crucial protease is the versican-degrading enzyme ADAMTS4. During influenza virus infection, inflammatory fibroblasts produce ADAMTS4, promoting robust CD8+ T cell infiltration into the lung tissue. In both paediatric and adult cases of influenza virus infection, protein levels of ADAMTS4 in the lower respiratory tract were strongly associated with respiratory failure and mortality173. Thus, fibroblasts serve as gatekeepers of immune cell access to the tissue and their exuberant inflammatory activity can drive lung damage and respiratory failure.
In addition to ECM remodelling activities, mesenchymal cells produce growth factors that directly stimulate epithelial cells to promote tissue repair. In mice, fibroblast growth factor 10 (FGF10), produced by lung mesenchymal cells, has a protective role following severe IAV infection by stimulating a subset of epithelial progenitor cells to undergo proliferation174. Taken together, these studies identify lung mesenchymal cells as crucial regulators of disease resistance and tolerance and of the transition between resolution of inflammation and initiation of tissue repair.
Endothelial cells
Pulmonary endothelial cells make up another barrier that is essential for gas exchange and lung function. The endothelium not only moves blood into the lung for oxygenation but also helps to circulate inflammatory molecules and migratory immune cells. During influenza virus infection, the endothelium becomes activated as cells upregulate surface expression of cell adhesion molecules, including P- and E-selectins74. The endothelium is the entry point for immune cells migrating to the lung tissue. Migrating immune cells interact with cell adhesion molecules on endothelial cells and traverse this barrier on the way to the site of infection. Activation of the endothelium and extravasation of immune cells from blood capillaries into the alveoli can result in vascular permeability and an increase in fluid in the lungs175. Thus, endothelial cells are also important decision-makers in determining resistant and tolerant tissue environments as they balance recruitment of immune cells to clear infected cells with integrity of the vasculature. Although human and avian influenza viruses can infect primary human endothelial cells in vitro176–178, the extent to which they productively infect endothelial cells in vivo remains controversial74,179.
Whether or not endothelial cells become productively infected in vivo, they do respond robustly to infection. In addition to upregulating adhesion molecules to recruit immune cells, endothelial cells have a crucial role in amplifying cytokine and chemokine production, which can lead to lethal immunopathology99. The sphingosine 1-phosphate 1 receptor (S1P1R), which binds to lipid metabolites, regulates this excessive cytokine production, and agonism of S1P1R with small molecules can significantly reduce cytokine levels in the lung99,180. Amplification of inflammation in endothelial cells depends on signalling through the IL-1 receptor and is independent of endosomal or cytosolic sensing of the virus. Dependence on IL-1 receptor signalling suggests that endothelial cells respond to damage signals released from epithelial cells, such as IL-1α, or those produced by professional immune cells, likely IL-1β181.
Like the other major structural cell types in the lung, endothelial cells exhibit heterogeneity based on compartment (microvascular, macrovascular or lymphatic). Single-cell gene expression studies have begun to characterize this heterogeneity during influenza virus infection, identifying groups of cells with distinct transcriptional responses that are highly proliferative and involved in tissue repair182. The tolerogenic potential of pulmonary endothelial cells has been largely unexplored.
As described above, SARS-CoV-2 infection appears to involve the pulmonary endothelium more than other respiratory viruses, such as influenza virus. Observations of severe COVID-19 in humans indicate extensive endothelial cell activation, thrombosis and angiogenesis76,183,184. This endothelial activation and thrombosis has also been observed in a non-human primate model of SARS-CoV-2 infection185. The mechanisms leading to such extensive endothelial involvement are unclear, but possibilities include direct infection of endothelial cells183 and microvascular complement protein deposition186. During infection in humans, endothelial cells express ACE2 (ref.76), and the virus readily infects human blood vessel organoids187.
Lung repair and recovery from infection
Tissue repair in the respiratory tract presents a challenge because of its distinct compartments, the diverse cell types affected and the heterogeneity of infection188. A pair of excellent, recent reviews have detailed the contribution of different subsets of structural cells, including epithelial, endothelial and mesenchymal, to tissue repair and the complex crosstalk between these cells during both pathogen-induced and sterile lung damage188,189.
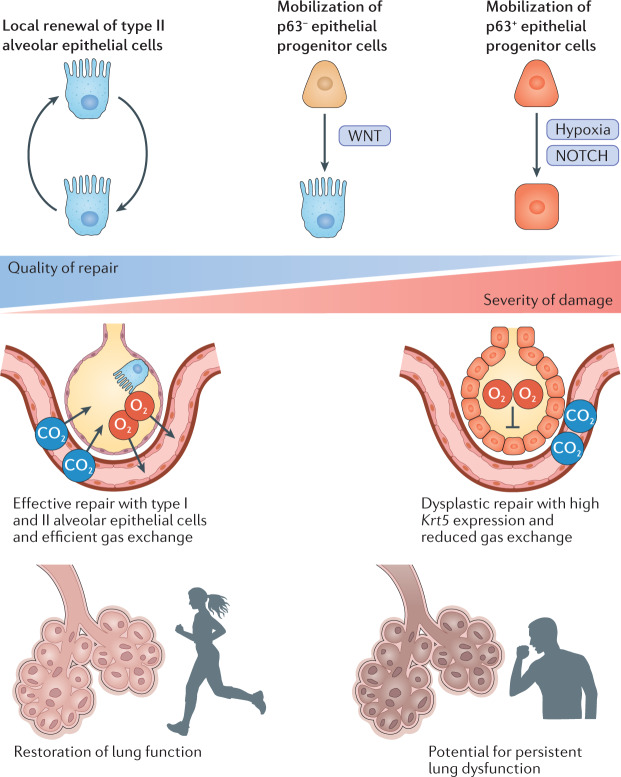
An important theme that has emerged in studies of tissue regeneration during influenza virus infection is that the severity of infection and extent of lung damage determine which progenitor cells mobilize in response to injury and the quality of the repair that they mediate105,190–194 (Fig. 4). Fortunately for the influenza virus field, severe influenza virus infection in mice has become a popular model for lung biologists to study acute lung injury and repair. In general, local proliferation of alveolar epithelial progenitor cells (AEPs), which are alveolar type II cells, mediate rapid and effective repair by differentiating into both alveolar type I and type II cells105,195,196. The stemness of the AEPs is maintained by WNT signalling in a close alveolar niche165,195,196. This type of repair typically occurs when there is localized damage to the alveoli. However, when there is more extensive alveolar damage and severe influenza virus infection ablates alveolar type II cells, including the AEPs, alternative progenitor cell populations fulfill this regenerative role188. In mice, several different alternative progenitor cells have been identified, termed bronchioalveolar stem cells, lineage-negative epithelial progenitors (LNEPs) — also known as distal airway stem cells (DASCs) — and another group of club-like epithelial progenitors174,190,191,194,197–199. Several recent studies indicate that IAVs preferentially infect club-like epithelial progenitors, which express stem cell antigen 1 (SCA1) and elevated levels of MHC class I174,194. Although these cells are able to facilitate alveolar epithelial repair following sterile lung damage, viral infection of the club-like progenitors may limit their ability to participate in effective tissue repair following severe IAV infection, requiring alternative progenitors to take on this role.
Fig. 4. Alveolar epithelial repair along the severity continuum.
The regeneration of alveolar tissue and restoration of function is essential for survival following a severe respiratory infection. Recent studies have demonstrated that the extent and severity of influenza virus infection determines the quality of alveolar epithelial repair. Repair by self-renewing type II alveolar cells occurs during less severe infection and is efficient. During infection with extensive tissue damage when type II alveolar cells are ablated, additional epithelial progenitor cells (p63− and p63+) mobilize to mediate repair. WNT and NOTCH signalling pathways determine localized differentiation of these progenitor cells. In severe damage, mobilization of p63+ progenitors can result in dysplastic alveolar repair characterized by the formation of cyst-like structures with high expression of Krt5 leading to reduced lung function. This dysplastic repair may lead to persistent lung dysfunction.
One of the apparent consequences of mobilizing LNEPs and/or DASCs is that they do not fully differentiate into alveolar type II cells, leaving areas of the lung with cyst-like structures with cytokeratin 5 (KRT5) expression and persistent pathology190,192,193. In these areas of the lung that never fully repair, hypoxia drives NOTCH signalling, which prevented differentiation of Krt5+ cells into alveolar epithelial lineages in mice193. Areas of persistent pathology maintain distinct microenvironments with gene expression signatures of type 2 immunity and the presence of tuft-like cells200,201. The physiological consequences of these signatures and the extent to which this happens in severe influenza virus infection in humans is unknown. It should be noted, however, that ongoing lung dysfunction, evident by both lung imaging and pulmonary function testing, has been described following severe IAV and SARS-CoV-2 infection202–204.
Together, these exciting advances define the mechanistic basis for how the extent and severity of infection determine the quality of tissue repair following influenza virus infection and provide a foundation for developing both cellular and molecular therapies to influence the trajectory of lung regeneration.
Experimental models of SARS-CoV-2
The complexities of the host response to SARS-CoV-2 necessitate in vivo studies, and it is crucial to consider both pathogenesis and transmission when modelling a novel disease (Table 2). Initial reports have described SARS-CoV-2 infection in rodents, ferrets, non-human primates and cats205–207; however, it is unclear which model best recapitulates human infection. Mice are not susceptible to SARS-CoV-2 owing to the inability of murine ACE2 to bind the viral spike protein. Therefore, infection of mice requires either introduction of the human ACE2 receptor or adaptation of SARS-CoV-2 to the murine receptor208. Human ACE2 transgenic mouse lines have been developed with different promoters driving ACE2 expression. The tissue distribution and abundance of human ACE2 expression differ between these transgenic lines and in part determine the severity of disease being modelled208. In mice in which ACE2 expression is driven by the epithelial cytokeratin 18 promoter, unadapted SARS-CoV-2 causes severe pathology and respiratory distress209,210; however, recently reported mouse-adapted strains also cause severe disease in BALB/c mice211,212. Ferrets, the gold-standard model for influenza virus transmission studies, also transmit SARS-CoV-2 by both direct and respiratory contact213. However, disease severity is mild, and viral replication is generally restricted to the URT205,208. Pathogenesis in non-human primates varies by species. African green monkeys develop acute pneumonia and lung injury persisting for >1 month despite viral clearance207, whereas rhesus macaques manifest pulmonary infiltration similar to humans with only mild to moderate disease severity206. Interestingly, hamsters can efficiently transmit SARS-CoV-2 (ref.214) and experience severe disease including weight loss, DAD and high viral load in the alveolar space215. As in the human experience, aged animals, including BALB/c mice211 and Syrian golden hamsters216, demonstrate more severe clinical manifestations of infection. Because human SARS-CoV-2 disease severity is variable, in vivo systems spanning the spectrum of disease could prove invaluable.
Table 2.
Animal models of influenza virus and SARS-CoV-2 infection and pathogenesis
| Animal | Physiology and genetics | Influenza virus | SARS-CoV-2 |
|---|---|---|---|
| Mouse |
Popular model for acute lung injury Key differences from humans in anatomy and respiratory cell distribution Differences in physiological responses (no fever or sneezing or coughing) Not a model for transmission Many genetic tools: knockouts, reporters and other transgenics |
Diverse viral strains that recapitulate spectrum of human disease, including acute respiratory distress syndrome Mouse adaptation not required for some IAVs, including avian viruses Many immunological tools to study host response |
Mice require introduction of human ACE2 receptor (transgenic animal or viral transduction) for infection with unadapted virus Mouse-adapted SARS-CoV-2 does cause severe disease Together, diverse mouse models can recapitulate spectrum of disease in humans |
| Hamster |
Small animal model of contact and airborne transmission for some viruses Limited genetic tools, but some genetic knockouts for key immune genes available |
IAVs typically infect without adaptation Primarily upper respiratory tract infection with limited pathology Model for contact transmission and some evidence of airborne transmission Limited immunological tools to study host response |
Unadapted SARS-CoV-2 infects hamsters Infection causes mild to moderate pathology in respiratory tract Some evidence that the model can recapitulate age- and sex-based differences in disease severity witnessed in humans Model for contact transmission and some evidence of airborne transmission |
| Ferret |
Respiratory anatomy similar to humans Many respiratory viruses do not require adaptation for infection/pathogenesis Similarities in physiological responses (fever and sneezing) Model for transmission Outbred animals Few genetic tools |
IAVs typically infect without adaptation (human and avian IAVs) Reporter viruses available to trace spread (fluorescent and luciferase-based) Limited immunological tools to study host response, but many are in development |
Limited studies on SARS-CoV-2 transmission and pathogenesis thus far Unadapted SARS-CoV-2 infects ferrets Current models do not recapitulate severe disease in humans |
| Non-human primate |
Respiratory anatomy most similar to humans Respiratory viruses do not require adaptation for infection or pathogenesis Similarities in physiological response (fever and respiratory distress) Outbred animals Few genetic tools Often serve as gatekeepers of vaccine candidates and therapeutics |
IAVs infect without adaptation (human and avian viruses) Frequently used to study highly pathogenic IAVs Limited immunological tools to study host response |
SARS-CoV-2 infects NHPs without adaptation of virus or manipulation of host Current models do not recapitulate severe disease in humans Clinical symptoms allow for testing efficacy of therapeutics NHPs exhibit adaptive immune responses to vaccination and infection |
ACE2, angiotensin-converting enzyme 2; IAV, influenza A virus; NHP, non-human primate; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Outlook: current and potential therapies
At both the individual and community level, annual seasonal influenza virus vaccination remains a mainstay of influenza control. However, several considerations make reliance on vaccination alone inadequate. In certain populations at high risk of severe complications from influenza virus infection (that is, elderly individuals and individuals with obesity) vaccine response is less robust. Antigenic drift may reduce vaccine efficacy at population level, placing large communities at risk. Finally, antigenic shifts resulting in pandemic influenza virus as well as epizootic infections with highly pathogenic strains of IAV are not covered by vaccines and may result in severe disease in large numbers of individuals. For these reasons, the availability of effective therapies for influenza virus is crucial. Aside from supportive care to correct physiological derangements, the currently available treatments are all antivirals. Those available are well tolerated and shorten symptom duration, although are most efficacious when given early.
Currently available therapeutics for influenza virus infection are antiviral compounds (Box 1), many of which need to be started in the first 2–3 days of infection (when viral replication is at its peak) to demonstrate benefit. As described above, severe influenza (as well as COVID-19) results in profound disruption of lung integrity, which is only in part driven by viral cytopathic effect. In severe infections, deleterious immune responses and inadequate repair mechanisms contribute to disease outcomes. Given the short therapeutic window for antivirals, targeting host factors that drive excessive inflammation and immunopathology offer a promising alternative treatment strategy. Any host therapeutic aimed at reducing lung injury, however, must consider the potential negative effect of delaying viral clearance and leaving patients with severe disease vulnerable to secondary infections. Broadly immunosuppressive agents, such as corticosteroids, are frequently given to patients in other scenarios to reduce inflammation, but are not recommended as treatment for influenza infection, unless a separate indication for their use exists217. However, in the case of SARS-CoV-2 infection leading to hospitalization, dexamethasone (a corticosteroid) given for a median of 7 days (up to 10 days) has been shown to reduce mortality, which was most evident in patients requiring higher levels of support at randomization218.
As broadly acting immunosuppressants do not appear efficacious in the case of influenza virus infection and have potentially harmful side effects, more targeted therapeutics are needed that modulate specific immunological pathways. Ideally, these agents would target host factors that are active at the site of infection or injury and would avoid systemic effects. Recent work on the role of lung structural cells in determining tissue resistant or tolerant states has identified several promising classes of cellular and molecular targets. Each of the main structural cell types (mesenchymal, endothelial and epithelial) has a distinct role in determining local tissue environments that could be modulated to maintain tissue function. Mesenchymal cells are professional tissue remodellers and produce key lung ECM proteases that drive immunopathology by causing direct damage to the lung structure and regulating the movement of immune cells168,169. Inhibitors that selectively target these proteases may promote tissue tolerance by preserving lung integrity for gas exchange and preventing colonization by opportunistic bacteria. Mesenchymal cells are also primary producers of IL-6, which is consistently associated with severity94,219,220. The IL-6 receptor antagonist tocilizumab is approved to treat chronic inflammatory diseases such as rheumatoid arthritis. Although it is unclear whether IL-6 is causal for influenza virus infection severity, as the cytokine has pleiotropic effects on the host response, there is continued interest in using tocilizumab to dampen excessive inflammation in severe respiratory infection, including influenza virus and SARS-CoV-2 infection97,221, although a recent trial result did not show a benefit of a single dose of tocilizumab over usual care in adult patients hospitalized with COVID-19 (ref.222). As potent amplifiers of cytokine or chemokine production, endothelial cells represent an attractive target to reduce inflammation that is initiated in the respiratory tract. Several agonists of the S1P receptor have been developed to dampen inflammation and could potentially be used for numerous inflammatory diseases180,223. None has yet been approved for respiratory viral infections, but their use in humans, including for COVID-19, has been proposed.
In contrast to targeting the mechanisms driving inflammation and disease resistance, an alternative approach could be to enhance directly the mechanisms of lung repair and disease tolerance. Preclinical studies in animal models of severe IAV infection described above indicate that the growth factors GM-CSF, FGF10 and AREG all have the potential to enhance epithelial cell proliferation in vivo. An added benefit of directly promoting these repair mechanisms to promote recovery from severe infection is that it is possible that the antiviral, or more generally anti-pathogen, host response could be left intact to continue clearance of an ongoing infection or protect against a secondary bacterial infection.
The past decade of research on epithelial cells during lung injury has identified numerous pathways that could be manipulated with therapeutics, from early interferon responses and integrin signalling89, to cell death mechanisms143–145, to repair190,191,193. A better understanding of how these pathways function in severe human infections is needed to identify the most promising targets to advance from preclinical studies. Moreover, it is unclear how these epithelial pathways might be dysregulated in high-risk groups, such as elderly individuals and individuals with obesity. The qualitative differences in epithelial repair from heterogeneous epithelial progenitor populations provide the opportunity to target specific pathways to influence the repair process and improve both short- and long-term outcomes.
Box 1 Antiviral medications for influenza virus and SARS-CoV-2.
Neuraminidase inhibitors (NAIs) were developed on the basis of rational drug design in the early 1990s224 and are influenza virus-specific antiviral medications. Oseltamivir, zanamivir, peramivir and laninamivir are available NAIs, each with different routes of administration. Of the NAIs, oseltamivir is the most studied and used. As their name suggests, these inhibit the function of viral neuraminidase (NA), which serves to limit viral progeny from being released from infected cells, thus reducing the number of cells subsequently infected. In adults with uncomplicated illness, oseltamivir has been shown to reduce the duration of symptomatic illness by ~24 h225. Although there are no randomized controlled trials of oseltamivir for the treatment of severe influenza virus infection, which may call into question the validity of the data226, observational studies have shown that oseltamivir has also been associated with a reduced risk of mortality in hospitalized adults with H3N2 seasonal influenza A virus (IAV)227 and the 2009 pandemic H1N1 IAV228, particularly when given early in hospital admission. Oseltamivir reduces the risk of death in patients hospitalized with highly pathogenic H5N1 IAV infection229. Resistance, conferred by the amino acid substitutions H275Y in N1 viruses or R292K in N2 viruses (among others), has been described230 and may emerge in high-risk groups on prolonged courses of NAI therapy231.
Baloxavir is a newer influenza virus antiviral that inhibits the polymerase acidic (PA) cap-snatching endonuclease activity of the viral RNA-dependent RNA polymerase (RdRP) complex, halting viral replication. It has been shown to reduce the duration of symptoms by ~26 h after influenza virus infection and reduce viral load more quickly than oseltamivir232. No resistance was reported in surveillance efforts in the United States25, although resistance may emerge during treatment with baloxavir233.
Other antiviral compounds, including favipiravir, which inhibits viral RdRPs (not influenza virus specific and with some interest for potential treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2))230, and pimodivir, which inhibits influenza virus polymerase basic protein 2 (PB2)234, have activity against influenza virus and have been studied in human clinical trials, but are not currently widely available (favipiravir is approved in Japan for novel or emerging influenza viruses235). Finally, monoclonal antibodies targeting influenza virus haemagglutinin have been developed and tested in human clinical trials236; however, the role that these compounds have in influenza virus treatment strategies is currently unclear.
Remdesivir (GS-5734) is a RdRP inhibitor with in vivo activity against Ebola virus237, in vitro and in vivo activity against Middle East respiratory syndrome coronavirus (MERS-CoV)238, and in vitro activity against SARS-CoV-2 (ref.239). A large randomized clinical trial using time to recovery as a primary end point demonstrated the benefit of 10 days of remdesivir over placebo in adults hospitalized with lower respiratory tract infection due to SARS-CoV-2 (ref.240). The benefit was most evident if given early in disease and in patients requiring low-flow oxygen at enrolment240. Notably, however, a recent multinational WHO effort to study ‘repurposed’ antiviral medications (including remdesivir) to treat SARS-CoV-2 infection, which used in-hospital mortality as the primary end point, found no apparent benefit of 10 days of remdesivir therapy241. Differences in treatment strategies, patient population and end points may account for variability in trial outcomes, and more study of remdesivir for SARS-CoV-2 infection may be necessary242. As above, favipiravir for the treatment of human infection with SARS-CoV-2 is being studied243. As more is learnt about viral tropism, host response and in vivo replication, additional therapeutic targets throughout the viral life cycle will likely present themselves. Finally, monoclonal antibodies against the spike protein of SARS-CoV-2 have been developed and studied in clinical trials244.
Acknowledgements
This work was funded by the National Institute of Allergy and Infectious Diseases (NIAID) under HHS contract HHSN27220140006C-OPT18I for the St. Jude Center of Excellence for Influenza Research and Surveillance (P.G.T.), NIH grant R01 AI121832 (P.G.T.), Children’s Infection Defense Center (CIDC) Award (St. Jude) (T.F.), HHS contract HHSN27220140006C for the St. Jude Center of Excellence for Influenza (S.S.C.) and ALSAC. The authors thank P. Vogel for providing the histopathological specimens of respiratory tissue from mice experimentally infected with influenza A virus displayed in Fig. 3a.
Glossary
- Tracheobronchitis
Inflammation of the tracheal and bronchial mucosa.
- Pharyngitis
Inflammation of the mucosa between the mouth and nasal cavities and the upper portion of the oesophagus (the throat).
- Myalgia
Muscle ache.
- Acute respiratory distress syndrome
Abrupt onset of respiratory failure characterized by inability to get oxygen into the blood owing to accumulation of lung fluid in the absence of heart failure.
- Chronic obstructive pulmonary disease
A group of diseases that restricts air movement in the lungs.
- Dyspnoea
Shortness of breath.
- Thrombosis
Formation of a blood clot in a blood vessel.
- Interstitial oedema
Fluid build-up in tissues outside of cells.
- Angiogenesis
Development of new blood vessels.
- Extracellular matrix
An intricate supportive collection of proteins and other macromolecules present in all tissues.
Author contributions
T.F., D.F.B. and V.M. researched data for the article. T.F., D.F.B., P.G.T. and S.S.C. contributed substantially to discussion of the content. All authors wrote the article. All authors reviewed and/or edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Microbiology thanks A. Wack and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tim Flerlage, David F. Boyd.
Contributor Information
Paul G. Thomas, Email: paul.thomas@stjude.org
Stacey Schultz-Cherry, Email: stacey.schultz-cherry@stjude.org.
References
- 1.Hause BM, et al. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae family. mBio. 2014;5:e00031–14. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Influenza (Seasonal). https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (2018).
- 3.Houser K, Subbarao K. Influenza vaccines: challenges and solutions. Cell Host Microbe. 2015;17:295–300. doi: 10.1016/j.chom.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster RG, Govorkova EA. Continuing challenges in influenza. Ann. N. Y. Acad. Sci. 2014;1323:115–139. doi: 10.1111/nyas.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treanor J. Influenza vaccine — outmaneuvering antigenic shift and drift. N. Engl. J. Med. 2004;350:218–220. doi: 10.1056/NEJMp038238. [DOI] [PubMed] [Google Scholar]
- 6.Saunders-Hastings PR, Krewski D. Reviewing the history of pandemic influenza: understanding patterns of emergence and transmission. Pathogens. 2016;5:66. doi: 10.3390/pathogens5040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gambotto A, Barratt-Boyes SM, de Jong MD, Neumann G, Kawaoka Y. Human infection with highly pathogenic H5N1 influenza virus. Lancet. 2008;371:1464–1475. doi: 10.1016/S0140-6736(08)60627-3. [DOI] [PubMed] [Google Scholar]
- 8.Payne, S. in Viruses 149–158 (Elsevier, 2017).
- 9.Woo PCY, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rota PA, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 12.Peiris JSM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 14.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo PCY, et al. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 18.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, Cui W, Tian B-P. The potential intermediate hosts for SARS-CoV-2. Front. Microbiol. 2020;11:2400. doi: 10.3389/fmicb.2020.580137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz FM. Influenza virus infection in infancy and early childhood. Paediatr. Respir. Rev. 2003;4:99–104. [PubMed] [Google Scholar]
- 21.Davenport FM, Hennessy AV, Francis T., Jr. Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J. Exp. Med. 1953;98:641–656. doi: 10.1084/jem.98.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrat F, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am. J. Epidemiol. 2008;167:775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 23.Zambon MC. Epidemiology and pathogenesis of influenza. J. Antimicrob. Chemother. 1999;44(Suppl. B):3–9. doi: 10.1093/jac/44.suppl_2.3. [DOI] [PubMed] [Google Scholar]
- 24.Mettelman RC, Thomas PG. Human susceptibility to influenza infection and severe disease. Cold Spring Harb. Perspect. Med. 2020 doi: 10.1101/cshperspect.a038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X, et al. Update: influenza activity in the United States during the 2018-19 season and composition of the 2019-20 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 2019;68:544–551. doi: 10.15585/mmwr.mm6824a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang N, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl Med. 2013;5:171ra19. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egorov ES, et al. The changing landscape of naive T cell receptor repertoire with human aging. Front. Immunol. 2018;9:1618. doi: 10.3389/fimmu.2018.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller RA. The aging immune system: primer and prospectus. Science. 1996;273:70–74. doi: 10.1126/science.273.5271.70. [DOI] [PubMed] [Google Scholar]
- 29.Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proc. Biol. Sci. 2015;282:20143085. doi: 10.1098/rspb.2014.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honce R, Schultz-Cherry S. Impact of obesity on influenza A virus pathogenesis, immune response, and evolution. Front. Immunol. 2019;10:1071. doi: 10.3389/fimmu.2019.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paich HA, et al. Overweight and obese adult humans have a defective cellular immune response to pandemic H1N1 influenza A virus. Obesity. 2013;21:2377–2386. doi: 10.1002/oby.20383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, et al. Adipose tissue dendritic cells enhances inflammation by prompting the generation of Th17 cells. PLoS ONE. 2014;9:e92450. doi: 10.1371/journal.pone.0092450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson EA, et al. Obesity outweighs protection conferred by adjuvanted influenza vaccination. mBio. 2016;7:e01144-16. doi: 10.1128/mBio.01144-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vazquez-Pagan A, Honce R, Schultz-Cherry S. Impact of influenza virus during pregnancy: from disease severity to vaccine efficacy. Future Virol. 2020;15:441–453. [Google Scholar]
- 35.Littauer EQ, et al. H1N1 influenza virus infection results in adverse pregnancy outcomes by disrupting tissue-specific hormonal regulation. PLoS Pathog. 2017;13:e1006757. doi: 10.1371/journal.ppat.1006757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moran TM, Park H, Fernandez-Sesma A, Schulman JL. Th2 responses to inactivated influenza virus can be converted to Th1 responses and facilitate recovery from heterosubtypic virus infection. J. Infect. Dis. 1999;180:579–585. doi: 10.1086/314952. [DOI] [PubMed] [Google Scholar]
- 37.Wang X-L, et al. Age and sex differences in rates of influenza-associated hospitalizations in Hong Kong. Am. J. Epidemiol. 2015;182:335–344. doi: 10.1093/aje/kwv068. [DOI] [PubMed] [Google Scholar]
- 38.Gubbels Bupp MR, Potluri T, Fink AL, Klein SL. The confluence of sex hormones and aging on immunity. Front. Immunol. 2018;9:1269. doi: 10.3389/fimmu.2018.01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith M, Honce R, Schultz-Cherry S. Metabolic syndrome and viral pathogenesis: lessons from influenza and coronaviruses. J. Virol. 2020;94:e00665-20. doi: 10.1128/JVI.00665-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panhwar MS, et al. Effect of influenza on outcomes in patients with heart failure. JACC Heart Fail. 2019;7:112–117. doi: 10.1016/j.jchf.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 41.Mulpuru S, et al. Effectiveness of influenza vaccination on hospitalizations and risk factors for severe outcomes in hospitalized patients with COPD. Chest. 2019;155:69–78. doi: 10.1016/j.chest.2018.10.044. [DOI] [PubMed] [Google Scholar]
- 42.Godoy P, et al. Smoking may increase the risk of hospitalization due to influenza. Eur. J. Public Health. 2016;26:882–887. doi: 10.1093/eurpub/ckw036. [DOI] [PubMed] [Google Scholar]
- 43.Mehta H, Nazzal K, Sadikot RT. Cigarette smoking and innate immunity. Inflamm. Res. 2008;57:497–503. doi: 10.1007/s00011-008-8078-6. [DOI] [PubMed] [Google Scholar]
- 44.Lee SW, et al. Impact of cigarette smoke exposure on the lung fibroblastic response after influenza pneumonia. Am. J. Respir. Cell Mol. Biol. 2018;59:770–781. doi: 10.1165/rcmb.2018-0004OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coronavirus Resource Center. COVID-19 Map - Johns Hopkins Coronavirus Resource Center. https://coronavirus.jhu.edu/map.html (2021).
- 47.Bi Q, et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect. Dis. 2020;20:911–919. doi: 10.1016/S1473-3099(20)30287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 49.Burke RM, et al. Symptom profiles of a convenience sample of patients with COVID-19 - United States, January-April 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:904–908. doi: 10.15585/mmwr.mm6928a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stokes EK, et al. Coronavirus disease 2019 case surveillance - United States, January 22-May 30, 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:759–765. doi: 10.15585/mmwr.mm6924e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butowt R, von Bartheld CS. Anosmia in COVID-19: underlying mechanisms and assessment of an olfactory route to brain infection. Neuroscientist. 2020 doi: 10.1177/1073858420956905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brann DH, et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020;6:eabc5801. doi: 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welge-Lüssen A, Wolfensberger M. Olfactory disorders following upper respiratory tract infections. Adv. Otorhinolaryngol. 2006;63:125–132. doi: 10.1159/000093758. [DOI] [PubMed] [Google Scholar]
- 54.Arabi YM, et al. Middle east respiratory syndrome. N. Engl. J. Med. 2017;376:584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu J, et al. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robbins-Juarez SY, et al. Outcomes for patients with COVID-19 and acute kidney injury: a systematic review and meta-analysis. Kidney Int. Rep. 2020;5:1149–1160. doi: 10.1016/j.ekir.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galván Casas C, et al. Classification of the cutaneous manifestations of COVID-19: a rapid prospective nationwide consensus study in Spain with 375 cases. Br. J. Dermatol. 2020;183:71–77. doi: 10.1111/bjd.19163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poissy J, et al. Pulmonary embolism in patients with COVID-19. Circulation. 2020;142:184–186. doi: 10.1161/CIRCULATIONAHA.120.047430. [DOI] [PubMed] [Google Scholar]
- 60.Ray JG, Schull MJ, Vermeulen MJ, Park AL. Association between ABO and Rh blood groups and SARS-CoV-2 infection or severe COVID-19 illness: a population-based cohort study. Ann. Intern. Med. 2021;174:308–315. doi: 10.7326/M20-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai Q, et al. Obesity and COVID-19 severity in a designated hospital in Shenzhen, China. Diabetes Care. 2020;43:1392–1398. doi: 10.2337/dc20-0576. [DOI] [PubMed] [Google Scholar]
- 62.Kumar A, et al. Is diabetes mellitus associated with mortality and severity of COVID-19? A meta-analysis. Diabetes Metab. Syndr. 2020;14:535–545. doi: 10.1016/j.dsx.2020.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henry BM, Lippi G. Chronic kidney disease is associated with severe coronavirus disease 2019 (COVID-19) infection. Int. Urol. Nephrol. 2020;52:1193–1194. doi: 10.1007/s11255-020-02451-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Honce R, et al. Obesity-related microenvironment promotes emergence of virulent influenza virus strains. mBio. 2020;11:e03341-19. doi: 10.1128/mBio.03341-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hadjadj J, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan W, Aboulhosn J. The cardiovascular burden of coronavirus disease 2019 (COVID-19) with a focus on congenital heart disease. Int. J. Cardiol. 2020;309:70–77. doi: 10.1016/j.ijcard.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu C, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raschetti R, et al. Synthesis and systematic review of reported neonatal SARS-CoV-2 infections. Nat. Commun. 2020;11:5164. doi: 10.1038/s41467-020-18982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mehta NS, et al. SARS-CoV-2 (COVID-19): what do we know about children? A systematic review. Clin. Infect. Dis. 2020;71:2469–2479. doi: 10.1093/cid/ciaa556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walsh JJ, Dietlein LF, Low FN, Burch GE, Mogabgab WJ. Bronchotracheal response in human influenza. Type A, Asian strain, as studied by light and electron microscopic examination of bronchoscopic biopsies. Arch. Intern. Med. 1961;108:376–388. doi: 10.1001/archinte.1961.03620090048006. [DOI] [PubMed] [Google Scholar]
- 72.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu. Rev. Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shieh W-J, et al. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am. J. Pathol. 2010;177:166–175. doi: 10.2353/ajpath.2010.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Short KR, Kroeze EJBV, Fouchier RAM, Kuiken T. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect. Dis. 2014;14:57–69. doi: 10.1016/S1473-3099(13)70286-X. [DOI] [PubMed] [Google Scholar]
- 75.Kuiken T, Taubenberger JK. Pathology of human influenza revisited. Vaccine. 2008;26(Suppl. 4):D59–D66. doi: 10.1016/j.vaccine.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ackermann M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bradley BT, et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ayres JS, Schneider DS. Tolerance of infections. Annu. Rev. Immunol. 2012;30:271–294. doi: 10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- 80.Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyerholz, D. K., Suarez, C. J., Dintzis, S. M. & Frevert, C. W. in Comparative Anatomy and Histology 2nd Edn (eds Treuting, P. M., Dintzis, S. M. & Montine, K. S.) 147–162 (Elsevier, 2018).
- 82.Lakdawala SS, et al. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature. 2015;526:122–125. doi: 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klinkhammer J, et al. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife. 2018;7:e33354. doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galani IE, et al. Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46:875–890.e6. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 85.Sanders CJ, et al. Compromised respiratory function in lethal influenza infection is characterized by the depletion of type I alveolar epithelial cells beyond threshold levels. Am. J Physiol. Lung Cell. Mol. Physiol. 2013;304:L481–L488. doi: 10.1152/ajplung.00343.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Monticelli LA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arpaia N, et al. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meliopoulos VA, et al. An epithelial integrin regulates the amplitude of protective lung interferon responses against multiple respiratory pathogens. PLoS Pathog. 2016;12:e1005804. doi: 10.1371/journal.ppat.1005804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Major J, et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Broggi A, et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Granados A, Peci A, McGeer A, Gubbay JB. Influenza and rhinovirus viral load and disease severity in upper respiratory tract infections. J. Clin. Virol. 2017;86:14–19. doi: 10.1016/j.jcv.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 93.Lee CK, et al. Comparison of pandemic (H1N1) 2009 and seasonal influenza viral loads, Singapore. Emerg. Infect. Dis. 2011;17:287–291. doi: 10.3201/eid1702.100282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oshansky CM, et al. Mucosal immune responses predict clinical outcomes during influenza infection independently of age and viral load. Am. J. Respir. Crit. Care Med. 2014;189:449–462. doi: 10.1164/rccm.201309-1616OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee N, et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J. Infect. Dis. 2009;200:492–500. doi: 10.1086/600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lucas C, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Teijaro, J. R. in Influenza Pathogenesis and ControlVolume II (eds Oldstone, M. B. A. & Compans, R. W.) 3–22 (National Library of Medicine, 2015).
- 98.de Jong MD, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Teijaro JR, et al. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146:980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heaton NS, et al. Long-term survival of influenza virus infected club cells drives immunopathology. J. Exp. Med. 2014;211:1707–1714. doi: 10.1084/jem.20140488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Herold S, Becker C, Ridge KM, Budinger GRS. Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur. Respir. J. 2015;45:1463–1478. doi: 10.1183/09031936.00186214. [DOI] [PubMed] [Google Scholar]
- 102.Krausgruber T, et al. Structural cells are key regulators of organ-specific immune responses. Nature. 2020;583:296–302. doi: 10.1038/s41586-020-2424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sanders CJ, Doherty PC, Thomas PG. Respiratory epithelial cells in innate immunity to influenza virus infection. Cell Tissue Res. 2011;343:13–21. doi: 10.1007/s00441-010-1043-z. [DOI] [PubMed] [Google Scholar]
- 104.Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 2015;16:27–35. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Edinger TO, Pohl MO, Stertz S. Entry of influenza A virus: host factors and antiviral targets. J. Gen. Virol. 2014;95:263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 107.van Riel D, et al. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tumpey TM, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 109.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 110.Shinya K, et al. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 111.Jia HP, et al. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009;297:L84–L96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ou X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Simmons G, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Daly JL, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xia S, et al. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020;5:92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ziegler CGK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zou X, et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sungnak W, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hou YJ, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Crotta S, et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 123.Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thomas PG, Shubina M, Balachandran S. ZBP1/DAI-dependent cell death pathways in influenza a virus immunity and pathogenesis. Curr. Top. Microbiol. Immunol. 2020 doi: 10.1007/82_2019_190. [DOI] [PubMed] [Google Scholar]
- 125.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 126.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl Acad. Sci. USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jia D, et al. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS ONE. 2010;5:e13927. doi: 10.1371/journal.pone.0013927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Graef KM, et al. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J. Virol. 2010;84:8433–8445. doi: 10.1128/JVI.00879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Züst R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bastard P, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang Q, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Frieman M, Heise M, Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133:101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rodrigues TS, et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218:e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Channappanavar R, et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van der Made CI, et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:1–11. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mudd PA, et al. Distinct inflammatory profiles distinguish COVID-19 from influenza with limited contributions from cytokine storm. Sci Adv. 2020;6:eabe3024. doi: 10.1126/sciadv.abe3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lei X, et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yatim N, Albert ML. Dying to replicate: the orchestration of the viral life cycle, cell death pathways, and immunity. Immunity. 2011;35:478–490. doi: 10.1016/j.immuni.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 139.Rodrigue-Gervais IG, et al. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe. 2014;15:23–35. doi: 10.1016/j.chom.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 140.Creagh EM. Caspase crosstalk: integration of apoptotic and innate immune signalling pathways. Trends Immunol. 2014;35:631–640. doi: 10.1016/j.it.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 141.Frank D, Vince JE. Pyroptosis versus necroptosis: similarities, differences, and crosstalk. Cell Death Differ. 2019;26:99–114. doi: 10.1038/s41418-018-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang T, et al. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell. 2020;180:1115–1129.e13. doi: 10.1016/j.cell.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Thapa RJ, et al. DAI senses influenza a virus genomic RNA and activates RIPK3−dependent cell death. Cell Host Microbe. 2016;20:674–681. doi: 10.1016/j.chom.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kuriakose T, et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 2016;1:aag2045. doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nogusa S, et al. RIPK3 activates parallel pathways of MLKL-driven necroptosis and FADD-mediated apoptosis to protect against influenza a virus. Cell Host Microbe. 2016;20:13–24. doi: 10.1016/j.chom.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang J, Yang Y, He W, Sun L. Necrosome core machinery: MLKL. Cell. Mol. Life Sci. 2016;73:2153–2163. doi: 10.1007/s00018-016-2190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 148.Newton K, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343:1357–1360. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- 149.Shubina M, et al. Necroptosis restricts influenza A virus as a stand-alone cell death mechanism. J. Exp. Med. 2020;217:e20191259. doi: 10.1084/jem.20191259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sarhan J, et al. Constitutive interferon signaling maintains critical threshold of MLKL expression to license necroptosis. Cell Death Differ. 2019;26:332–347. doi: 10.1038/s41418-018-0122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hsu AC-Y. Influenza virus: a master tactician in innate immune evasion and novel therapeutic interventions. Front. Immunol. 2018;9:743. doi: 10.3389/fimmu.2018.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhu N, et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ren Y, et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Unkel B, et al. Alveolar epithelial cells orchestrate DC function in murine viral pneumonia. J. Clin. Invest. 2012;122:3652–3664. doi: 10.1172/JCI62139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rösler B, Herold S. Lung epithelial GM-CSF improves host defense function and epithelial repair in influenza virus pneumonia-a new therapeutic strategy? Mol Cell Pediatr. 2016;3:29. doi: 10.1186/s40348-016-0055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zaiss DMW, Gause WC, Osborne LC, Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. 2015;42:216–226. doi: 10.1016/j.immuni.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hall OJ, et al. Progesterone-based therapy protects against influenza by promoting lung repair and recovery in females. PLoS Pathog. 2016;12:e1005840. doi: 10.1371/journal.ppat.1005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guo X-ZJ, et al. Lung γδ T cells mediate protective responses during neonatal influenza infection that are associated with type 2 immunity. Immunity. 2018;49:531–544.e6. doi: 10.1016/j.immuni.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vermillion MS, et al. Production of amphiregulin and recovery from influenza is greater in males than females. Biol. Sex Differ. 2018;9:24. doi: 10.1186/s13293-018-0184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Duan S, Thomas PG. Balancing immune protection and immune pathology by CD8+ T-cell responses to influenza infection. Front. Immunol. 2016;7:25. doi: 10.3389/fimmu.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Moskophidis D, Kioussis D. Contribution of virus-specific CD8+ cytotoxic T cells to virus clearance or pathologic manifestations of influenza virus infection in a T cell receptor transgenic mouse model. J. Exp. Med. 1998;188:223–232. doi: 10.1084/jem.188.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Enelow RI, et al. Structural and functional consequences of alveolar cell recognition by CD8+ T lymphocytes in experimental lung disease. J. Clin. Invest. 1998;102:1653–1661. doi: 10.1172/JCI4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jolly L, et al. Influenza promotes collagen deposition via αvβ6 integrin-mediated transforming growth factor β activation. J. Biol. Chem. 2014;289:35246–35263. doi: 10.1074/jbc.M114.582262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Meliopoulos V, Livingston B, Van de Velde L-A, Honce R, Schultz-Cherry S. Absence of β6 integrin reduces influenza disease severity in highly susceptible obese mice. J. Virol. 2019;93:e01646-18. doi: 10.1128/JVI.01646-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zepp JA, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148.e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gaggar A, Weathington N. Bioactive extracellular matrix fragments in lung health and disease. J. Clin. Invest. 2016;126:3176–3184. doi: 10.1172/JCI83147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bradley LM, Douglass MF, Chatterjee D, Akira S, Baaten BJG. Matrix metalloprotease 9 mediates neutrophil migration into the airways in response to influenza virus-induced toll-like receptor signaling. PLoS Pathog. 2012;8:e1002641. doi: 10.1371/journal.ppat.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Talmi-Frank D, et al. Extracellular matrix proteolysis by MT1-MMP contributes to influenza-related tissue damage and mortality. Cell Host Microbe. 2016;20:458–470. doi: 10.1016/j.chom.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 170.McMahon M, et al. ADAMTS5 is a critical regulator of virus-specific T cell immunity. PLoS Biol. 2016;14:e1002580. doi: 10.1371/journal.pbio.1002580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rojas-Quintero J, et al. Matrix metalloproteinase-9 deficiency protects mice from severe influenza A viral infection. JCI Insight. 2018;3:21. doi: 10.1172/jci.insight.99022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Guan W, et al. Clinical correlations of transcriptional profile in patients infected with avian influenza H7N9 virus. J. Infect. Dis. 2018;218:1238–1248. doi: 10.1093/infdis/jiy317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Boyd DF, et al. Exuberant fibroblast activity compromises lung function via ADAMTS4. Nature. 2020;587:466–471. doi: 10.1038/s41586-020-2877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Quantius J, et al. Influenza virus infects epithelial stem/progenitor cells of the distal lung: impact on Fgfr2b-driven epithelial repair. PLoS Pathog. 2016;12:e1005544. doi: 10.1371/journal.ppat.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Matthay MA, et al. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers. 2019;5:18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chan MCW, et al. Influenza H5N1 virus infection of polarized human alveolar epithelial cells and lung microvascular endothelial cells. Respir. Res. 2009;10:102. doi: 10.1186/1465-9921-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chan LLY, et al. Risk assessment of the tropism and pathogenesis of the highly pathogenic avian influenza A/H7N9 virus using ex vivo and in vitro cultures of human respiratory tract. J. Infect. Dis. 2019;220:578–588. doi: 10.1093/infdis/jiz165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sumikoshi M, et al. Human influenza virus infection and apoptosis induction in human vascular endothelial cells. J. Med. Virol. 2008;80:1072–1078. doi: 10.1002/jmv.21185. [DOI] [PubMed] [Google Scholar]
- 179.Short KR, Kuiken T, Van Riel D. Role of endothelial cells in the pathogenesis of influenza in humans. J. Infect. Dis. 2019;220:1859–1860. doi: 10.1093/infdis/jiz349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Walsh KB, et al. Suppression of cytokine storm with a sphingosine analog provides protection against pathogenic influenza virus. Proc. Natl Acad. Sci. USA. 2011;108:12018–12023. doi: 10.1073/pnas.1107024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Teijaro JR, Walsh KB, Rice S, Rosen H, Oldstone MBA. Mapping the innate signaling cascade essential for cytokine storm during influenza virus infection. Proc. Natl Acad. Sci. USA. 2014;111:3799–3804. doi: 10.1073/pnas.1400593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Niethamer TK, et al. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. eLife. 2020;9:e53072. doi: 10.7554/eLife.53072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Varga Z, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Goshua G, et al. Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. Lancet Haematol. 2020;7:e575–e582. doi: 10.1016/S2352-3026(20)30216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Aid M, et al. Vascular disease and thrombosis in SARS-CoV-2 infected rhesus macaques. Cell. 2020;183:1354–1366. doi: 10.1016/j.cell.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Magro C, et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Monteil V, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Basil MC, et al. The cellular and physiological basis for lung repair and regeneration: past, present, and future. Cell Stem Cell. 2020;26:482–502. doi: 10.1016/j.stem.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zepp JA, Morrisey EE. Cellular crosstalk in the development and regeneration of the respiratory system. Nat. Rev. Mol. Cell Biol. 2019;20:551–566. doi: 10.1038/s41580-019-0141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vaughan AE, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2014;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zuo W, et al. p63+Krt5+ distal airway stem cells are essential for lung regeneration. Nature. 2014;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kanegai CM, et al. Persistent pathology in influenza-infected mouse lungs. Am. J. Respir. Cell Mol. Biol. 2016;55:613–615. doi: 10.1165/rcmb.2015-0387LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xi Y, et al. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat. Cell Biol. 2017;19:904–914. doi: 10.1038/ncb3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kathiriya JJ, Brumwell AN, Jackson JR, Tang X, Chapman HA. Distinct airway epithelial stem cells hide among club cells but mobilize to promote alveolar regeneration. Cell Stem Cell. 2020;26:346–358.e4. doi: 10.1016/j.stem.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359:1118–1123. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zacharias WJ, et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kim CFB, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 198.Kumar PA, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Salwig I, et al. Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. EMBO J. 2019;38:e102099. doi: 10.15252/embj.2019102099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Keeler SP, et al. Influenza a virus infection causes chronic lung disease linked to sites of active viral RNA remnants. J. Immunol. 2018;201:2354–2368. doi: 10.4049/jimmunol.1800671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rane CK, et al. Development of solitary chemosensory cells in the distal lung after severe influenza injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2019;316:L1141–L1149. doi: 10.1152/ajplung.00032.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhao Y-M, et al. Follow-up study of the pulmonary function and related physiological characteristics of COVID-19 survivors three months after recovery. EClinicalMedicine. 2020;25:100463. doi: 10.1016/j.eclinm.2020.100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chen J, et al. Long term outcomes in survivors of epidemic Influenza A (H7N9) virus infection. Sci. Rep. 2017;7:17275. doi: 10.1038/s41598-017-17497-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Huang C, et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shi J, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Munster VJ, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Cross RW, et al. Intranasal exposure of African green monkeys to SARS-CoV-2 results in acute phase pneumonia with shedding and lung injury still present in the early convalescence phase. Virol. J. 2020;17:125. doi: 10.1186/s12985-020-01396-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Muñoz-Fontela C, et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Oladunni FS, et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Winkler ES, et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gu H, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Leist SR, et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;21:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Richard M, et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020;11:3496. doi: 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sia SF, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chan JF-W, et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Imai M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Uyeki TM, et al. Clinical practice guidelines by the Infectious Diseases Society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal influenza. Clin. Infect. Dis. 2019;68:895–902. doi: 10.1093/cid/ciy874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with Covid-19 - Preliminary Report. N. Engl. J. Med. 2020;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hagau N, et al. Clinical aspects and cytokine response in severe H1N1 influenza A virus infection. Crit. Care. 2010;14:R203. doi: 10.1186/cc9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kaiser L, Fritz RS, Straus SE, Gubareva L, Hayden FG. Symptom pathogenesis during acute influenza: interleukin-6 and other cytokine responses. J. Med. Virol. 2001;64:262–268. doi: 10.1002/jmv.1045. [DOI] [PubMed] [Google Scholar]
- 221.Guaraldi G, et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2:e474–e484. doi: 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Stone JH, et al. Efficacy of tocilizumab in patients hospitalized with covid-19. N. Engl. J. Med. 2020;383:2333–2344. doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhao J, et al. Combination of sphingosine-1-phosphate receptor 1 (S1PR1) agonist and antiviral drug: a potential therapy against pathogenic influenza virus. Sci. Rep. 2019;9:5272. doi: 10.1038/s41598-019-41760-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 224.von Itzstein M, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 225.Dobson J, Whitley RJ, Pocock S, Monto AS. Oseltamivir treatment for influenza in adults: a meta-analysis of randomised controlled trials. Lancet. 2015;385:1729–1737. doi: 10.1016/S0140-6736(14)62449-1. [DOI] [PubMed] [Google Scholar]
- 226.Hurt AC, Kelly H. Debate regarding oseltamivir use for seasonal and pandemic influenza. Emerg. Infect. Dis. 2016;22:949–955. doi: 10.3201/eid2206.151037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lytras T, Mouratidou E, Andreopoulou A, Bonovas S, Tsiodras S. Effect of early oseltamivir treatment on mortality in critically Ill patients with different types of influenza: a multiseason cohort study. Clin. Infect. Dis. 2019;69:1896–1902. doi: 10.1093/cid/ciz101. [DOI] [PubMed] [Google Scholar]
- 228.Muthuri SG, et al. Effectiveness of neuraminidase inhibitors in reducing mortality in patients admitted to hospital with influenza A H1N1pdm09 virus infection: a meta-analysis of individual participant data. Lancet Respir Med. 2014;2:395–404. doi: 10.1016/S2213-2600(14)70041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Adisasmito W, et al. Effectiveness of antiviral treatment in human influenza A(H5N1) infections: analysis of a Global Patient Registry. J. Infect. Dis. 2010;202:1154–1160. doi: 10.1086/656316. [DOI] [PubMed] [Google Scholar]
- 230.Yen H-L. Current and novel antiviral strategies for influenza infection. Curr. Opin. Virol. 2016;18:126–134. doi: 10.1016/j.coviro.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 231.Lee N, Hurt AC. Neuraminidase inhibitor resistance in influenza: a clinical perspective. Curr. Opin. Infect. Dis. 2018;31:520–526. doi: 10.1097/QCO.0000000000000498. [DOI] [PubMed] [Google Scholar]
- 232.Hayden FG, et al. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N. Engl. J. Med. 2018;379:913–923. doi: 10.1056/NEJMoa1716197. [DOI] [PubMed] [Google Scholar]
- 233.Uehara T, et al. Treatment-emergent influenza variant viruses with reduced baloxavir susceptibility: impact on clinical and virologic outcomes in uncomplicated influenza. J. Infect. Dis. 2020;221:346–355. doi: 10.1093/infdis/jiz244. [DOI] [PubMed] [Google Scholar]
- 234.Finberg RW, et al. Phase 2b study of pimodivir (JNJ-63623872) as monotherapy or in combination with oseltamivir for treatment of acute uncomplicated seasonal influenza A: TOPAZ trial. J. Infect. Dis. 2019;219:1026–1034. doi: 10.1093/infdis/jiy547. [DOI] [PubMed] [Google Scholar]
- 235.Shiraki K, Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020;209:107512. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ali SO, et al. Evaluation of MEDI8852, an anti-influenza a monoclonal antibody, in treating acute uncomplicated influenza. Antimicrob. Agents Chemother. 2018;62:e00694-18. doi: 10.1128/AAC.00694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Warren TK, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sheahan TP, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wang M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Beigel JH, et al. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.WHO Solidarity Trial Consortium. Repurposed antiviral drugs for covid-19 - interim WHO solidarity trial results. N. Engl. J. Med. 2020;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.McCreary EK, Angus DC. Efficacy of remdesivir in COVID-19. JAMA. 2020;324:1041–1042. doi: 10.1001/jama.2020.16337. [DOI] [PubMed] [Google Scholar]
- 243.Cai Q, et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020;6:1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chen P, et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 2020;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]