Abstract
Tumor progression is associated with dedifferentiated histopathologies concomitant with cancer cell survival within a changing, and often hostile, tumor microenvironment. These processes are enabled by cellular plasticity, whereby intracellular cues and extracellular signals are integrated to enable rapid shifts in cancer cell phenotypes. Cancer cell plasticity, at least in part, fuels tumor heterogeneity and facilitates metastasis and drug resistance. Protein synthesis is frequently dysregulated in cancer, and emerging data suggest that translational reprograming collaborates with epigenetic and metabolic programs to effectuate phenotypic plasticity of neoplasia. Herein, we discuss the potential role of mRNA translation in cancer cell plasticity, highlight emerging histopathological correlates, and deliberate on how this is related to efforts to improve understanding of the complex tumor ecology.
Introduction
Neoplasia is characterized by progression to increasingly less differentiated (i.e., dedifferentiated) states concomitant with disease dissemination, therapy resistance, and poor prognosis. Tumor microenvironments (TMEs) are often hostile, with limited oxygen and nutrient availability. Cellular plasticity (defined as the ability to dynamically alter cell fate through translational and epigenetic alterations; see Glossary) allows cells to survive changing microenvironments by rewiring cellular functions such as metabolism and cytoskeletal dynamics. In tumors, plasticity enables cancer cells to adapt to a plethora of stressors. Accordingly, the molecular regulators of plasticity are attractive therapeutic targets.
John George Adami was one of the first to contemplate the dichotomous nature of plasticity. Based largely on histopathology, he proposed that cell fate changes and proliferation are often mutually exclusive, with plasticity occurring mainly while proliferation is reduced [1]. For example, stem cells are generally less proliferative than their progeny [2–4]. Protein synthesis (or mRNA translation) dictates proliferation rates while playing a central role in differentiation and stress adaptation [5] (Figure 1). To this end, dysregulated protein synthesis promotes drug resistance and metastasis, which are processes driven by cancer cell plasticity [5]. Herein, we review how translational reprograming impacts cancer cell plasticity.
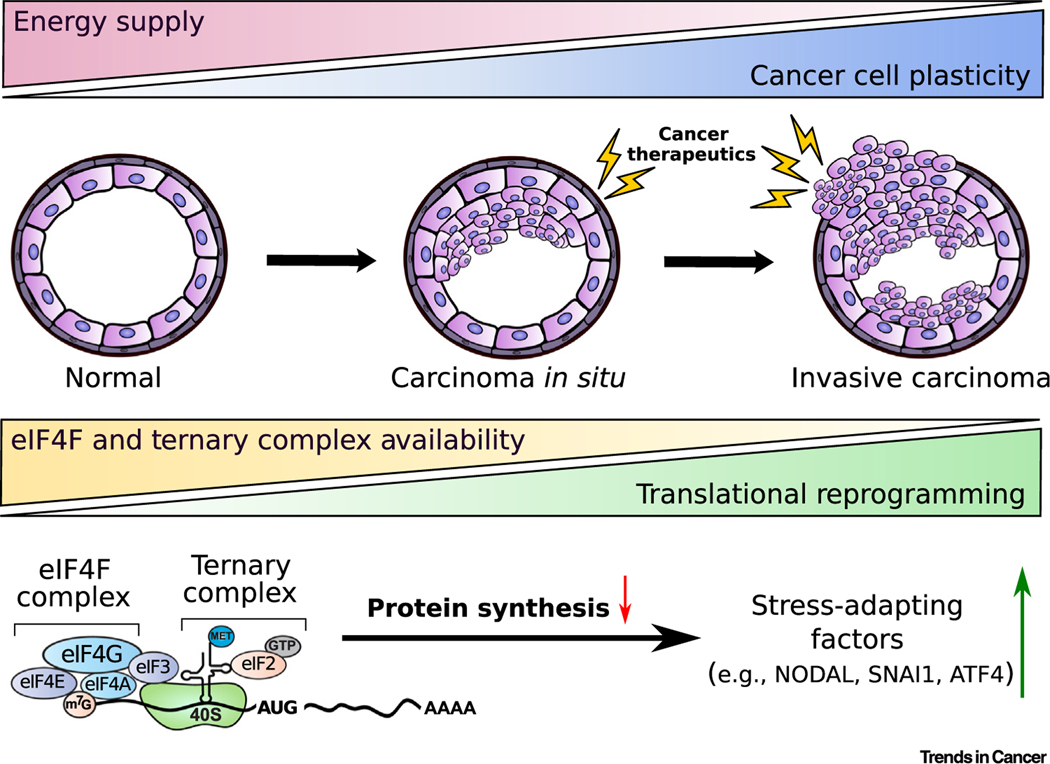
Figure 1. Cancer Progression and Plasticity.
Cancer progression is characterized by increasing cell plasticity. Neoplastic growth is also frequently accompanied by limitations in energy supply due to hypoxia or low nutrient availability. This, in turn, limits eIF4F complex assembly and TC recycling, which are both essential for efficient global translation. Thus, translational reprogramming under limited oxygen and nutrient supply in the TME is characterized by a reduction in global protein synthesis and increased selective translation of mRNAs essential for adaptation and survival, including NODAL, SNAI1, and ATF4. Red arrow indicates a decrease. Green arrow indicates an increase. Abbreviations: ATF4, activating transcription factor 4; eIF, eukaryotic translation initiation factor; GTP, guanosine triphosphate; m7G, 7-methylguanosine; Met, methionine; TC, ternary complex; TME, tumor microenvironment.
Defining Cancer Cell Plasticity
Before genomics and molecular biology became commonplace, histopathology provided early indications of tumor plasticity. Solid tumors can contain cancer cells with vastly different phenotypes, which is suggestive of plasticity. For example, urothelial bladder carcinoma and non-small cell lung cancer can both contain subsets of small cell carcinoma cells, typically heralding poor patient prognosis [6–8]. Other tumors contain mixtures of epithelial and mesenchymal phenotypes, such as carcinosarcomas of the uterus and other tissues of the female genital tract [9]. Some of these lesions with mixed pathology are thought to be collision tumors arising from independent clones that later coalesce [10]. However, genomic sequencing studies show different pathologies arising from a single tumor clone. Indeed, this phenomenon has been observed in acinar and ductal prostate adenocarcinoma, small cell and urothelial bladder cancer, small cell and non-small cell lung cancer, and the epithelial and mesenchymal components of uterine carcinosarcoma [6,8,11–13]. In each case, the distinct pathologies share numerous genomic alterations but they also have divergent changes that are acquired during tumor progression. Overall, the common origins of different pathologies within these tumors indicate that plasticity, at least in part, drives tumor heterogeneity.
In addition to overt differences in pathology, plasticity is evident from changes in tumor differentiation. Epithelial-to-mesenchymal (EMT) and mesenchymal-to-epithelial (MET) transitions are extensively characterized manifestations of cancer plasticity. Both processes resemble transdifferentiation, the reversible transition into different phenotypes [14]. Tumors also undergo dedifferentiation wherein cancer cells revert to a less differentiated state, re-express ‘stem cell’ genes, including NODAL, OCT4, SOX2, and NANOG, and give rise to cancer stem-like cells with a greater ability to metastasize and evade treatment [15–18]. Changes in cellular identity can be driven by epigenetic alterations, rather than genetic changes, in the primary DNA sequence. For example, dedifferentiated endometrial cancer evolves from adjacent, well-differentiated adenocarcinoma through alterations in chromatin remodeling complexes. In this case, tumors with high levels of microsatellite instability that are deficient in ARID1A, a component of a chromatin remodeling complex, progress to a lethal dedifferentiated disease through concurrent loss of either ARID1B or SMARCA4 [19].
Notably, emerging data suggest that transitions in cell differentiation are not regulated by transcriptional changes alone. In contrast to transcription, translation provides cells with the ability to swiftly alter their proteome and adapt to changes in their environment. To this end, translation plays a central role in acute adaptation to stress and, as discussed below, such mechanisms may be co-opted to promote cancer cell plasticity. Indeed, translation of many mRNAs encoding proteins that govern cancer cell plasticity is modulated during adaptation to stressors (Figure 1).
mRNA Translation and Cellular Plasticity
Dysregulation of mRNA translation is a major feature of neoplasia and stem cell maintenance [5,20,21]. Accordingly, stem cells and differentiated adult cells, as well as non-malignant and neoplastic cells, have distinct translational programs [5,21,22]. Recycling of ternary complex (TC) and eukaryotic translation initiation factor 4F (eIF4F) complex assembly are limiting steps in translation that have been most extensively studied in the context of cellular plasticity (Figure 2).
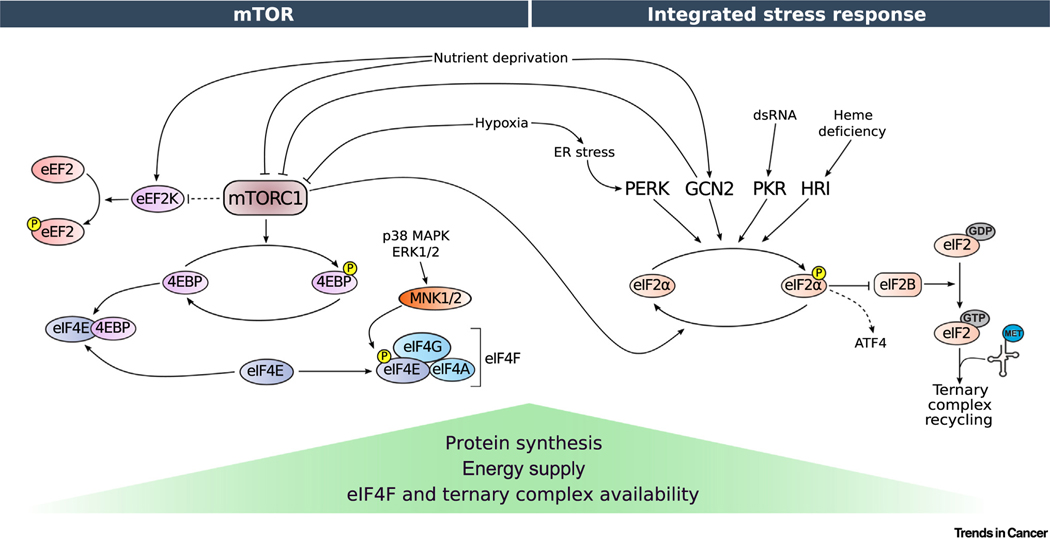
Figure 2. Regulation of Translation Initiation via Mechanistic/Mammalian Target of Rapamycin (mTOR) and the Integrated Stress Response (ISR).
Various stressors act via the mTOR (left) and/or the ISR network (right) that in turn impinge on the translational machinery. mTOR inhibition prevents the phosphorylation of 4E-BPs, which prevents eIF4E:eIF4G binding and interferes with eIF4F complex formation. Protein synthesis is further modulated by the phosphorylation of eIF4E via MNK1/2, although this process is incompletely understood. mTOR inhibition also leads to an activation of eEF2K, resulting in phosphorylation of eEF2 and a reduction in translation elongation (dashed lines denote indirect effects). Induction of ISR is mediated by four different kinases that phosphorylate eIF2α in response to different types of stress. Phosphorylation of eIF2α attenuates GEF activity of eIF2B, thereby decreasing ternary complex (TC; i.e., eIF2: ) levels. However, under these conditions of reduced global translation, select subsets of mRNAs encoding crucial stress response factors, such as ATF4, are translationally upregulated. Elevated eIF4F and TC levels allow high translation rates when energy and oxygen supply are not limiting. Conversely, conditions wherein the levels of eIF4F and TC are low denote translationally repressive states induced by various stressors, resulting in a global reduction in translation. Abbreviations: 4E-BP, 4E-binding proteins; ATF4, activating transcription factor 4; dsRNA, double-stranded RNA; eEF2K, eukaryotic translation elongation factor 2 kinase; eIF, eukaryotic translation initiation factor; ERK, extracellular signal-regulated kinase; GCN2, general control nonderepressible; GDP, guanosine diphosphate; GTP, guanosine triphosphate; HRI, heme-regulated inhibitor; MAPK, mitogen-activated protein kinase; Met, methionine; MNK, MAPK-interacting kinase; mTOR, mammalian/mechanistic target of rapamycin; P, protein phopsphorylation; PERK, PKR-like endoplasmic reticulum kinase; PKR, RNA-activated protein kinase.
TC and Cancer Cell Plasticity
TC, which is composed of initiator tRNA , GTP, and eIF2 (including α, β, and γ subunits), recruits to the 43S preinitiation complex containing the 40S ribosome and associated translation initiation factors [23]. Upon delivery and GTP hydrolysis, the eIF2:GDP complex is recycled by the guanine nucleotide exchange factor (GEF) eIF2B for the next round of initiation [23]. Stressors interfere with TC recycling via eIF2α phosphorylation, which diminishes the GEF activity of eIF2B [24]. Four kinases phosphorylate eIF2α: general control nonderepressible 2 (GCN2) kinase, which senses amino acids; RNA-activated protein kinase (PKR), which is stimulated by double-stranded RNAs; PKR-like endoplasmic reticulum kinase (PERK); and heme-regulated inhibitor (HRI) kinase, which responds to heme deficiency [25]. eIF2α phosphorylation and the subsequent decrease in eIF2B activity and TC levels underpin the integrated stress response (ISR) that plays a central role in stress adaptation [26] (Figure 2). ISR encompasses translational reprogramming, with reduced global protein synthesis and translational activation of a subset of mRNAs containing inhibitory upstream open reading frames (uORFs) [27]. These mRNAs include activating transcription factor 4 (ATF4), which acts as a major coordinator of stress responses, and growth arrest and DNA damage-inducible protein, which recruits protein phosphatase 1 to dephosphorylate eIF2α during the resolution phase of the ISR [27].
ISR promotes or suppresses cancer progression in different contexts [26]. Phospho-eIF2α may suppress initial tumorigenesis by limiting protein synthesis but promotes the survival of cancer cells by inducing adaptive plasticity to low oxygen and nutrient supply or drug treatment. Since nutrient and/or oxygen deprivation lead to eIF2-dependent phenotypic shifts in unicellular organisms such as yeast, ISR may have an evolutionarily conserved role in cell plasticity in response to stress [28]. Indeed, in mouse embryonic stem cells (mESCs), inhibition of eIF2α dephosphorylation by salubrinal induces synthesis of NANOG and c-MYC that are linked to cancer cell plasticity [29]. In leukemic and hematopoietic stem cells, phospho-eIF2α and consequent ATF4 induction are required for stem cell population maintenance [30]. Similarly, in transdifferentiated (EMT) breast cancer cells, overproduction of extracellular matrix components leads to endoplasmic reticulum (ER) stress and activation of the PERK/eIF2α axis, which is paralleled by invasion and metastasis [31]. In the epidermis, an eIF2B5-mediated translational program leads to loss of progenitor self-renewal, which limits tumor initiation and growth [32]. Collectively, these observations suggest a multifaceted role of the ISR in cancer cell phenotypic switching.
eIF4F Complex and Cancer Cell Plasticity
The eIF4F complex consists of the mRNA cap-binding subunit eIF4E, scaffold eIF4G, and DEAD box helicase eIF4A [23]. eIF4E binds to the 5′ mRNA cap, whereas eIF4G associates with eIF3 to recruit mRNA to the 43S preinitiation complex, which is composed of a small ribosomal subunit and associated factors [23]. eIF4A facilitates 5′ untranslated region (UTR) scanning by the 43S preinitiation complex towards the initiation codon [23]. Alterations in eIF4F complex levels lead to selective changes in translation [33]. Increased eIF4F levels are paralleled by the selective upregulation of translation of mRNAs encoding proliferation- and survival-stimulating factors, while translation of mRNAs encoding housekeeping proteins is only marginally sensitive to changes in eIF4F levels [33].
eIF4F assembly is regulated by 4E-binding proteins (4E-BP1–3 in mammals) that interfere with eIF4E:eIF4G binding [33,34]. 4E-BPs are phosphorylated and inactivated by the mechanistic/mammalian target of rapamycin complex 1 (mTORC1) in response to nutrients, hormones (e.g., insulin), and growth factors (e.g., insulin-like growth factors [IGFs]) [35] (Figure 2). mTORC1 increases cellular growth and proliferation by stimulating anabolic pathways, including protein synthesis [36]. mTOR is also a component of mTOR complex 2 (mTORC2), which is involved in cytoskeletal organization, regulation of the AGC-family of kinases (e.g., AKT), and glucose and lipid metabolism [37].
Subunits of the eIF4F complex play a major role in cell fate decisions. For instance, forced eIF4E expression induces the formation of mesoderm in ectodermal explants of Xenopus laevis [38]. In general, stem cells have lower mTOR activity and protein synthesis than differentiated cells [21]. In p53-proficient mouse embryonic fibroblasts, 4E-BP1/2 loss leads to translational induction of p21 that blocks reprogramming into induced pluripotent stem cells (iPSCs). In turn, abrogating 4E-BP expression in p53-deficient fibroblasts increases iPSC induction by stimulating translation of c-MYC and SOX2 mRNAs [39]. Finally, 4E-BPs are implicated in the translational regulation of Yin Yang 2 (YY2) that seem to play a pivotal role in self-renewal and differentiation of mESCs [40]. Altogether, the mTORC1/4E-BP/eIF4F axis is positioned as a central regulator of proteome complexity that underpins cell plasticity. Indeed, in precancerous cells, eIF4E increases self-renewal and proliferation, which simultaneously protects cells with activated RAS or c-MYC from replication catastrophe [41]. Moreover, disruption of eIF4E:eIF4G binding represses clonogenic activity and tumorsphere formation in pancreatic and prostate cancer [42]. Transforming growth factor β (TGFβ), a major stimulator of EMT, induces 4E-BP1 levels thereby reducing CIP2A-BP translation in triple negative breast cancer cells [43]. This in turn activates the oncogene CIP2A, leading to the induction of MMP-2, MMP-9, and SNAIL1 and increased metastasis [43]. Finally, the androgen receptor (AR) has been demonstrated to induce EIF4EBP1. Accordingly, AR-deficient prostate cancers express less EIFE4BP1, leading to increases in eIF4F levels and concomitant prostate cancer progression [44].
eIF4E is also phosphorylated by MAPK-interacting kinases 1 and 2 (MNK1 and 2) [45,46], which leads to translational reprogramming by as of yet incompletely understood mechanisms [33]. Phosphorylation of eIF4E via the TGFβ-MNK1/2 axis is required to establish a translational EMT program, which is signified by upregulation of SNAI1 and MMP3 mRNA translation [47]. In turn, MNK1 inhibition attenuates the progression of ductal carcinoma in situ to a less differentiated invasive ductal carcinoma, which is paralleled by reduced NODAL levels [48]. Accordingly, MNK1/2 inhibitors reduce metastasis in preclinical models [49–51]. Collectively, alterations in eIF4F levels and/or eIF4E phosphorylation have a profound impact on cancer cell plasticity and associated phenotypes.
Extrinsic Drivers of Translationally Induced Plasticity
Plasticity is often a consequence of tumors adapting to the TME, especially hypoxia, nutrient limitation, or cancer therapeutics. The translational machinery has a major function in adaptation to these stressors, which at least in part appears to occur via facilitating cellular plasticity.
Hypoxia
Hypoxia drives cancer cell plasticity [52]. Global protein synthesis is downregulated in hypoxia via mTOR suppression and is mediated chiefly by the induction of regulated in development and DNA damage response 1 [53] and PERK-induced eIF2α phosphorylation [54–56] (Figure 2). Translational downregulation is accompanied by the selective increase in the synthesis of proteins that enable adaptation to oxygen deprivation, such as vascular endothelial growth factor (VEGF) and hypoxia inducible factor 1 α (HIF1α) [57–60]. Precise mechanisms of translational reprogramming under hypoxia are still debated. A number of mRNAs that are preferentially translated under hypoxia, including VEGF and erythropoietin, were reported to harbor uORFs and are thus expected to be induced by ISR [61,62]. In addition, selective translation in hypoxia may be driven by cap-independent mechanisms mediated by internal ribosome entry site (IRES)-like regions in 5′ UTRs. IRES-driven translation was proposed for several mRNAs encoding plasticity-inducing factors such as VEGF [63], c-MYC [64], and HIF1α [65]. However, the extent of cap-independent translation under hypoxia is, at least in some contexts, limited [66]. It also appears that mechanisms of cap-independent translation of mammalian mRNAs are distinct from those engaged by viral IRESs [67].
HIF1 and 2 are transcription factors that regulate adaptation to hypoxia and are linked to the translational machinery. To this end, hypoxia induces the expression of eIF4E1 and its paralog eIF4E2 (4E-HP) in breast cancer cells, whereby EIF4E1, but not EIF4E2, transcription appears to be HIF1α-dependent [68]. This suggests that upregulated eIF4E1 may overcome mTORC1 inhibition in hypoxia and result in cap-dependent induction of translation of a subset of mRNAs, including c-MYC [64] and VEGF [68]. Alternatively, in glioblastoma cells, it was proposed that the alternative complex formed by eIF4E2/4E-HP, HIF2α, and RNA-binding motif protein 4 (RBM4) is responsible for selective cap-dependent translation under hypoxia [69]. The same authors proposed a hypoxia-specific eIF4F complex composed of eIF4E2/4E-HP and eIF4G3 (eIF4G1 paralog) [70]. This model is, however, challenged by findings that eIF4E2/4E-HP is an evolutionarily conserved translational repressor [71–74]. In addition to the proposed effects of hypoxia on the translation initiation machinery, depletion of oxygen induces eukaryotic elongation factor 2 kinase (eEF2K) that inactivates eEF2, thereby decreasing elongation rates [75]. This is mediated by the suppression of the mTORC1/S6 kinase axis and/or the negative regulator of prolyl hydroxylase 2 [75,76]. Finally, a subset of mRNAs, and in particular those induced by HIF1α, may be translationally activated under hypoxia by their subcellular relocalization to the ER due to low numbers of upstream AUGs in 5′ UTRs [75].
Notwithstanding the yet elusive mechanistic underpinnings of reprogramming the translational machinery under hypoxia, oxygen depletion results in rapid alterations in the translatome [77]. Several clues highlight that hypoxia-induced translational reprogramming may promote cancer cell plasticity. For instance, in esophageal squamous cancer cells, hypoxia induces insulin-like growth factor binding protein 3 (IGFBP3), a known promoter of EMT, at least in part by stimulating cap-dependent translation [78]. In turn, IGFBP3 expression mirrors intratumoral enrichment of CD44 high cancer stem cells (CSCs). In breast cancer and glioblastoma models, hypoxia appears to translationally increase the expression of cadherin-22, thereby driving an EMT phenotype [79]. Moreover, hypoxia induces translation of OCT4B, an alternate OCT4 mRNA isoform that promotes EMT and metastasis [80–82]. The enrichment of tumorspheres and CSCs derived from breast cancer cell lines in response to hypoxia may also depend on eIF4E1 upregulation to bolster the translation of c-MYC, VEGF, and cyclin D1 mRNAs [68]. Collectively, this suggests that translation can greatly affect hypoxia-induced cell plasticity.
Nutrient Deprivation and Energy Stress
Nutrient availability can be limited in the TME [83,84]. Even when oxygen and nutrients are not limiting, many cancer subtypes preferably utilize aerobic glycolysis over oxidative phosphorylation (OXPHOS) in order to generate building blocks essential in anabolic processes and to maintain redox balance [85]. Importantly, glycolytic states characterize stem cells and are implicated in the induction of CSCs, EMT, and metastatic phenotypes. For example, in breast cancer, induction of glycolysis through fructose-1,6-biphosphatase (FBP1) inhibition results in CSC enrichment and tumorsphere formation, while SNAIL represses FBP1 during EMT [86]. Conversely, OXPHOS inhibitors reduce CSC populations in some cancer types [87]. Cancer cells also exploit numerous other metabolic pathways that are dependent on non-essential amino acids (NEAAs), such as glutamine [88,89], serine/glycine [90,91], and aspartate [92,93], as well as additional fuels such as fatty acids [94,95] and lactate [96]. Collectively, it is becoming apparent that dynamic metabolic adaptations, not a single metabolic state, signify metabolic plasticity to enable rapid phenotype switching of cancer cells [97].
Translation is highly energy consuming and is tightly linked to energy metabolism [98]. Glucose starvation, for instance, suppresses global translation via ER stress and eIF2α phosphorylation [99]. It also activates adenosine monophosphate-activated protein kinase (AMPK) [100]. AMPK conserves energy by shutting down anabolic processes when nutrients and/or oxygen are limiting [101]. To this end, AMPK downregulates protein synthesis by suppressing mTORC1 [101] and directly activates eEF2K [102], which phosphorylates eEF2 and inhibits translation elongation to promote cell survival when nutrients are limiting [103]. In turn, when cellular energy is not limiting, translation of mRNAs encoding proteins with mitochondrial functions (e.g., TFAM) and components of the electron transport chain (ETC) (e.g., ATP5O and NDUFS6) is stimulated via the mTORC1/4E-BP/eIF4E axis [104]. This increases mitochondrial biogenesis and ATP production to meet high energy demands of protein synthesis [104]. Intriguingly, mRNAs encoding ETC components are enriched in transcripts with extremely short 5′ UTRs (<30 nucleotides) and/or translation initiator of short 5′ UTR elements, which are thought to allow their selective translation [105,106].
In addition to glucose, amino acid supply profoundly affects translation. Amino acid depletion inhibits global protein synthesis by suppressing mTORC1 and activating GCN2 [107,108] (Figure 2). GCN2 upregulates ATF4 and its target metabolic genes, many of which encode amino acid transporters and enzymes involved in NEAA synthesis [109,110]. For example, glutamine deprivation induces ATF4 expression through both transcriptional and translational mechanisms, which subsequently represses microphthalmia-associated transcription factor (MITF) [111]. This results in melanoma phenotype switching characterized by MITF-low/AXL-high cells, which are known to be highly metastatic and drug resistant [111].
Mechanisms that orchestrate translation and metabolic programs are expected to exert a strong impact on cancer cell phenotypes. Indeed, translational dysregulation caused by depletion of 4E-BP1 and 2 increases energy stress tolerance and metabolic plasticity of cancer cells in conjunction with HIF1α [112]. This finding, combined with evidence that 4E-BP1/2 status can determine cell fate decisions [39,40], suggests that translational control of metabolic programs may promote cancer cell plasticity induced by energy stress.
Cancer Therapeutics
Tumor plasticity is linked to therapy resistance. For instance, under the selective pressure of treatment with tyrosine kinase inhibitors, epidermal growth factor receptor (EGFR)-mutant non-small cell lung cancer transforms into EGFR-negative small cell lung cancer [7,113]. Similarly, prostate adenocarcinoma escapes AR-targeted therapies by transforming into small-cell or AR-null phenotypes [114,115]. Enrichment of stem-cell like dormant cancer cells has been described in response to various chemotherapeutics, which limits treatment efficacy [116]. The translational apparatus plays a major role in drug resistance [5] and thus likely participates in therapy-induced cancer plasticity. For instance, high 4E-BP1 levels are associated with resistance to PI3K/Akt/mTOR inhibitors in luminal prostate cancer [117]. In ovarian cancer, drug resistant populations are enriched for CSCs with an activated mTORC1 pathway [118]. Inhibiting mTOR resensitizes cancer cells to carboplatin and is accompanied by a reduced translation of mRNAs supporting cell survival, proliferation, and DNA repair [119,120]. Conversely, in breast cancer cells, chemotherapeutics (e.g., paclitaxel), active-site mTOR inhibitors (e.g., INK128), as well as hypoxia induce translation of NODAL, NANOG, and SNAI1 mRNAs, promoting stem-like phenotypes and drug resistance [121]. Intriguingly, NODAL, NANOG, and SNAI1 mRNAs have multiple 5′ UTR isoforms, some of which are preferentially translated under conditions of limiting eIF4F and TC levels [121]. Accordingly, disruption of adaptive translational programs using an ISR-inhibitor (ISRIB, a small molecule that disrupts ISR translational programs by activating eIF2B) bolsters the efficacy of paclitaxel and mTOR inhibitors [121]. Similarly, ISRIB abolishes the growth and metastasis of prostate cancer patient-derived xenografts [122]. Biguanides are antidiabetics that induce energy stress by inhibiting mitochondrial ATP production, leading to AMPK activation, suppression of mTORC1, and reduced protein synthesis [98]. Intriguingly, CSCs appear to be particularly sensitive to biguanides across a variety of cancers, including glioma [123], gastric [124], pancreatic [125], breast [126], and ovarian cancer [127]. Biguanides exert synergy with HER2, BCR/ABL, or BRAF inhibitors in large part by suppressing the translation of mRNAs encoding enzymes involved in NEAA synthesis via the mTORC1/4E-BP/eIF4E axis [112].
Collectively, these observations suggest that orchestration of the mTOR- and ISR-dependent translational and metabolic programs may play a central, and potentially clinically exploitable, role in cancer cell plasticity that may be targeted to forestall or overcome drug resistance (Figure 3). Importantly, several such crosstalk mechanisms between mTOR and eIF2α phosphorylation have been documented. These mechanisms include mTORC1/CK2-directed phosphorylation of eIF2β, leading to NCK1-dependent eIF2α dephosphorylation [128]; suppression of PERK/GCN2 or PKR via the mTORC2/AKT axis or PTEN, respectively [129]; and protein phosphatase 6-dependent GCN2 activation in response to mTOR inhibition [130].
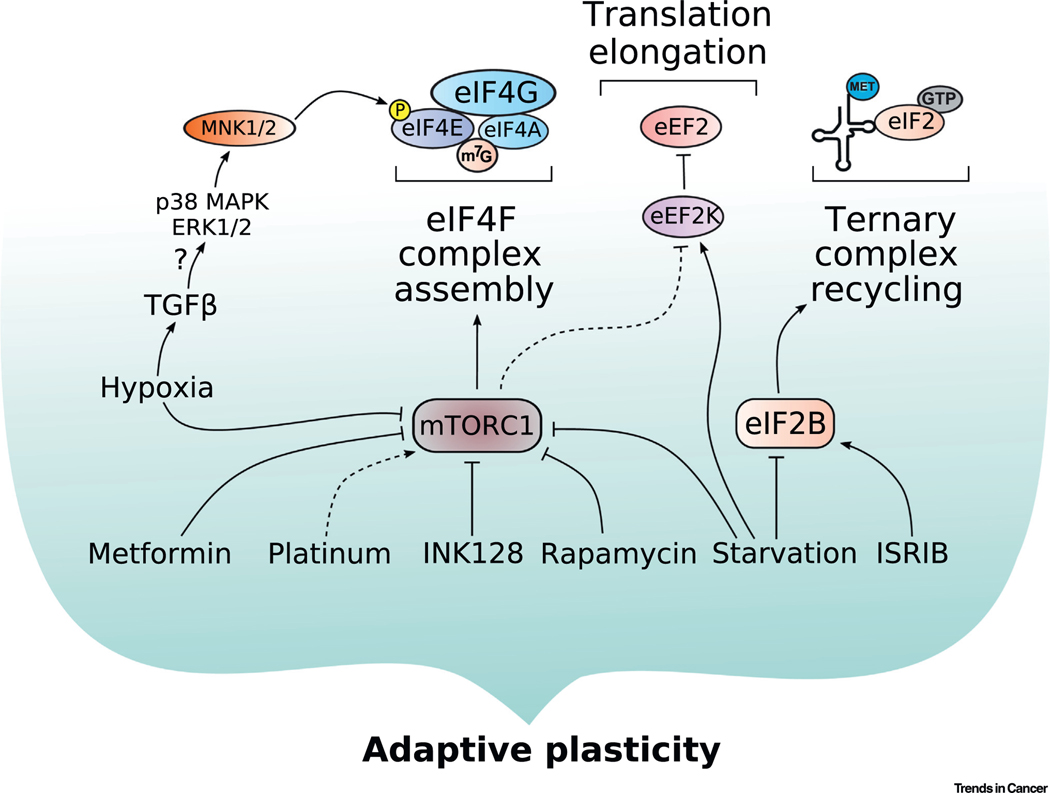
Figure 3. Extrinsic Cues Drive Plasticity through Translational Mechanisms.
Extrinsic cues that drive plastic phenotypes in cancer converge on the mechanistic/mammalian target of rapamycin complex 1 and the integrated stress response, ultimately leading to translational reprogramming. Cellular plasticity is underscored by adaptive processes, and the translational responses to stressors are highly context dependent. Dashed lines indicate new discoveries in the field that, as of yet, have not been robustly verified. Abbreviations: eEF2K, eukaryotic translation elongation factor 2 kinase; eIF, eukaryotic translation initiation factor; ERK, extracellular signal-regulated kinase; GTP, guanosine triphosphate; ISRIB, integrated stress response inhibitor; m7G, 7-methylguanosine; MAPK, mitogen-activated protein kinase; Met, methionine; MNK1/2, MAPK-interacting kinases 1/2; mTORC1, mechanistic/mammalian target of rapamycin complex 1; TGFβ, transforming growth factor β.
Additional Translational Mechanisms Implicated in Cancer Cell Plasticity
Importantly, mechanisms beyond those mediated via eIF4F and/or eIF2/eIF2B engender translational reprogramming that may favor phenotype switching. These include alternative mechanisms of cap-dependent translation centered on DAP5/p97/NAT1 and/or eIF3d [131–133] and cap-independent translation mechanisms mediated by RNA-binding La-protein [134] and Y-box 1 [135]. In addition, miRNA- [136,137] and/or RNA-binding protein (e.g., hnRNP E)-dependent translational regulation have been linked to adaptive plasticity [138]. Abrogation of methyltransferase-like 3 concomitant with decreased m6A mRNA modifications reduces EMT in liver cancer [139]. In turn, ZEB2 mRNA translation, which encodes a protein that promotes EMT, is regulated by stretches of rare UUA (Leu), GGU (Gly), and GUA (Val) codons [140]. This suggests that alterations in tRNA pools and/or tRNA modifications may also contribute to the translational regulation of cancer cell plasticity [140]. Accordingly, U34 tRNA modifications have been linked to metastasis and drug resistance [141]. Finally, ribosome biogenesis represents yet another layer of complexity in the translational regulation of EMT, whereby inhibitors of rRNA synthesis were shown to revert tumors to a more differentiated state [142]. Collectively, these findings suggest multiple, and potentially non-mutually exclusive, mechanisms that link translation to cancer cell plasticity. These findings thus highlight the ability of translational machinery to rapidly alter malignant proteomes to allow cancer cells to evade therapeutic insults and disseminate.
Concluding Remarks
Pathology highlights the breadth of cancer cell plasticity and the ramifications for patient outcomes and treatment; however, it likely underestimates the extent of plasticity within tumors. It can be difficult to detect cells with divergent pathology when they only represent a subpopulation of the tumor [143,144], so the prevalence of plasticity as measured by histopathology may be underappreciated. A more detailed understanding of the molecular drivers of plasticity, including those governed by the translational apparatus, may overcome these limitations. To this end, single cell transcriptomics in conjunction with technological advances towards single cell proteomics hold promise to directly address the diversity and dynamics of cancer cell gene expression programs. Such efforts should help determine the mechanisms of coordination between different steps in the regulation of gene expression that influence phenotypic switching in neoplasia and also establish the full spectrum of cellular plasticity within tumors. In turn, the translational machinery is likely to play a central role in cancer cell plasticity and the corresponding histopathological correlates. Establishing the role of translation in phenotypic switching of cancer cells is thus required to decipher the mechanisms of adaptation of cancer cells to different environmental and therapeutic insults (see Outstanding Questions). Understanding these roles may provide useful biomarkers to monitor and classify cancer cell plasticity and identify new therapeutic targets to attenuate metastasis and resistance.
Outstanding Questions.
How do adaptive responses to stress differ between normal tissue and neoplasia?
How does the translational machinery impact cancer cell plasticity and stress responses?
What are the cellular networks that orchestrate translational, transcriptional, epigenetic, epitranscriptomic, and metabolic programs of cancer cells to enable stress-induced phenotype switching?
Can the translational machinery be targeted to attenuate cancer cell plasticity and thus impede cancer spread and/or alleviate therapy resistance?
Highlights.
Neoplastic progression is characterized by alterations in tissue architecture, such as a loss of structure–function relationships and the emergence of dedifferentiated histopathologies.
To progress, metastasize, and evade therapies, cancer cells must adapt to new and unfavorable microenvironments.
Cancer cell plasticity occurs concomitantly with tumor progression and can limit the effectiveness of anticancer therapies.
Protein synthesis is often dysregulated in neoplasia, with translational reprogramming emerging as a central feature of adaptive plasticity.
Dissecting the mechanisms of translational reprograming in response to changing tumor microenvironments holds a promise to clarify the molecular underpinnings of cancer cell plasticity and its corresponding histopathological manifestations.
Acknowledgements
M.G.L received funding from the National Health and Medical Research Council, Australia, (project grant 1156570) and the Victorian Government acting through the Victorian Cancer Agency (fellowship MCRF18017). S.D.R. is funded by the Canadian Institutes for Health Research (CIHR; grant PJT-162260). D.P. is supported by a CIHR Postdoctoral Fellowship (MFE-171312), and L.J.L is supported by a CIHR Doctoral Research Award (GSD-158587). I.T. is a Senior Scholar of the Fonds de Recherche du Québec-Santé and his laboratory is supported in part by CIHR (MOP-363027), National Institutes of Health (R01 CA 202021–01-A1), and Joint Canada-Israel Health Research Program (108589–001). This work was supported by the CIHR (PLS 9538 and PLS 95381), the Alberta Cancer Foundation, the Women and Children’s Health Research Institute (WCHRI), and Alberta Innovates Health Solutions (AIHS) through grants awarded to L.M.P. L.M.P. was the recipient of the Sawin-Baldwin Chair in Ovarian Cancer, the Dr. Anthony Noujaim Legacy Oncology Chair, and the AIHS Translational Health Chair in Cancer.
Glossary
- Cellular plasticity
defined broadly as the ability of a cell to adjust its phenotype in response to microenvironmental cues. This process can be heritable and involves translational and epigenetic alterations that facilitate the reprogramming of cellular processes such as metabolism, proliferation, cell fate specification, cytoskeletal dynamics, and metastasis
- Epigenetic alterations
mechanisms such as nucleosome repositioning, DNA methylation, and histone modifications that affect DNA accessibility and mediate downstream changes in gene expression and cellular phenotypes
References
- 1.Adami JG (1900) On Growth and Overgrowth and on the Relationship Between Cell Differentiation and Proliferative Capacity: Its Bearing Upon the Regeneration of Tissus and the Development of Tumours, Sherratt & Hughes [Google Scholar]
- 2.Cheshier SH et al. (1999) In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A. 96, 3120–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montgomery RK et al. (2011) Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. U. S. A. 108, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mira H. et al. (2010) Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89 [DOI] [PubMed] [Google Scholar]
- 5.Bhat M. et al. (2015) Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 14, 261–278 [DOI] [PubMed] [Google Scholar]
- 6.Chang MT et al. (2018) Small-cell carcinomas of the bladder and lung are characterized by a convergent but distinct pathogenesis. Clin. Cancer Res. 24, 1965–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oser MG et al. (2015) Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 16, e165–e172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner PL et al. (2009) Combined small cell lung carcinomas: genotypic and immunophenotypic analysis of the separate morphologic components. Am. J. Clin. Pathol. 131, 376–382 [DOI] [PubMed] [Google Scholar]
- 9.Pang A. et al. (2018) Carcinosarcomas and related cancers: tumors caught in the act of epithelial-mesenchymal transition. J. Clin. Oncol. 36, 210–216 [DOI] [PubMed] [Google Scholar]
- 10.Smith SC and Tomlins SA (2014) Prostate cancer SubtyPINg biomarKers and outcome: is clarity emERGing? Clin. Cancer Res. 20, 4733–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillard M. et al. (2019) Integrative genomic analysis of coincident cancer foci implicates CTNNB1 and PTEN alterations in ductal prostate cancer. Eur. Urol. Focus 5, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherniack AD et al. (2017) Integrated molecular characterization of uterine carcinosarcoma. Cancer Cell 31, 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao S. et al. (2016) Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. U. S. A. 113, 12238–12243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W. and Kang Y. (2019) Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 49, 361–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quail DF et al. (2012) Embryonic morphogen nodal promotes breast cancer growth and progression. PLoS One 7, e48237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santini R. et al. (2014) SOX2 regulates self-renewal and tumorigenicity of human melanoma-initiating cells. Oncogene 33, 4697–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiou SH et al. (2010) Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 70, 10433–10444 [DOI] [PubMed] [Google Scholar]
- 18.Keysar SB et al. (2017) Regulation of head and neck squamous cancer stem cells by PI3K and SOX2. J. Natl. Cancer Inst. 109, djw189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coatham M. et al. (2016) Concurrent ARID1A and ARID1B inactivation in endometrial and ovarian dedifferentiated carcinomas. Mod. Pathol. 29, 1586–1593 [DOI] [PubMed] [Google Scholar]
- 20.Sampath P. et al. (2008) A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2, 448–460 [DOI] [PubMed] [Google Scholar]
- 21.Signer RA et al. (2016) The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev. 30, 1698–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingolia NT et al. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelletier J. and Sonenberg N. (2019) The organizing principles of eukaryotic ribosome recruitment. Annu. Rev. Biochem. 88, 307–335 [DOI] [PubMed] [Google Scholar]
- 24.Pavitt GD (2018) Regulation of translation initiation factor eIF2B at the hub of the integrated stress response. Wiley Interdiscip. Rev. RNA 9, e1491. [DOI] [PubMed] [Google Scholar]
- 25.Wek RC (2018) Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 10, a032870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costa-Mattioli M. and Walter P. (2020) The integrated stress response: from mechanism to disease. Science 368, eaat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinnebusch AG et al. (2016) Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Jiménez C. and Goding CR (2019) Starvation and pseudo-starvation as drivers of cancer metastasis through translation reprogramming. Cell Metab. 29, 254–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friend K. et al. (2015) Embryonic stem cell growth factors regulate eIF2α phosphorylation. PLoS One 10, e0139076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Galen P. et al. (2018) Integrated stress response activity marks stem cells in normal hematopoiesis and leukemia. Cell Rep. 25, 1109–1117 e1105 [DOI] [PubMed] [Google Scholar]
- 31.Feng YX et al. (2014) Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 4, 702–715 [DOI] [PubMed] [Google Scholar]
- 32.Cai EY et al. (2020) Selective translation of cell fate regulators mediates tolerance to Broad oncogenic stress. Cell Stem Cell 27, 270–283 e277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roux PP and Topisirovic I. (2018) Signaling pathways involved in the regulation of mRNA translation. Mol. Cell. Biol. 38, e00070–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pause A. et al. (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5’-cap function. Nature 371, 762–767 [DOI] [PubMed] [Google Scholar]
- 35.Brunn GJ et al. (1997) Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277, 99–101 [DOI] [PubMed] [Google Scholar]
- 36.Liu GY and Sabatini DM (2020) mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxton RA and Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 [DOI] [PubMed] [Google Scholar]
- 38.Klein PS and Melton DA (1994) Induction of mesoderm in Xenopus laevis embryos by translation initiation factor 4E. Science 265, 803–806 [DOI] [PubMed] [Google Scholar]
- 39.Tahmasebi S. et al. (2014) Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell 14, 606–616 [DOI] [PubMed] [Google Scholar]
- 40.Tahmasebi S. et al. (2016) Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proc. Natl. Acad. Sci. U. S. A. 113, 12360–12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avdulov S. et al. (2015) eIF4E threshold levels differ in governing normal and neoplastic expansion of mammary stem and luminal progenitor cells. Cancer Res. 75, 687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaiswal PK et al. (2019) Eukaryotic translation initiation factor 4 gamma 1 (EIF4G1): a target for cancer therapeutic intervention? Cancer Cell Int. 19, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo B. et al. (2020) Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J. 39, e102190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y. et al. (2019) The androgen receptor regulates a druggable translational regulon in advanced prostate cancer. Sci. Transl. Med. 11, eaaw4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waskiewicz AJ et al. (1997) Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16, 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukunaga R. and Hunter T. (1997) MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16, 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robichaud N. et al. (2015) Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 34, 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Q. et al. (2019) MNK1/NODAL signaling promotes invasive progression of breast ductal carcinoma. Cancer Res. 79, 1646–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robichaud N. et al. (2018) Translational control in the tumor microenvironment promotes lung metastasis: phosphorylation of eIF4E in neutrophils. Proc. Natl. Acad. Sci. U. S. A. 115, E2202–E2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhan Y. et al. (2017) MNK1/2 inhibition limits oncogenicity and metastasis of KIT-mutant melanoma. J. Clin. Invest. 127, 4179–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y. et al. (2019) Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat. Med. 25, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DF Q. et al. (2012) Microenvironmental regulation of cancer stem cell phenotypes. Curr. Stem Cell Res. Ther. 7, 197–216 [DOI] [PubMed] [Google Scholar]
- 53.Brugarolas J. et al. (2004) Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blais JD et al. (2006) Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol. Cell. Biol. 26, 9517–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L. et al. (2006) Hypoxia induced energy stress regulates mRNA translation and cell growth. Mol. Cell 21, 521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koumenis C. et al. (2002) Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol. Cell. Biol. 22, 7405–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas JD and Johannes GJ (2007) Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA 13, 1116–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koh MY et al. (2011) The hypoxia-associated factor switches cells from HIF-1alpha- to HIF-2alpha-dependent signaling promoting stem cell characteristics, aggressive tumor growth and invasion. Cancer Res. 71, 4015–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang C. et al. (2016) Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A. 113, E2047–E2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Covello KL et al. (2006) HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 20, 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barbosa C. and Romão L. (2014) Translation of the human erythropoietin transcript is regulated by an upstream open reading frame in response to hypoxia. RNA 20, 594–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arcondéguy T. et al. (2013) VEGF-A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res. 41, 7997–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stein I. et al. (1998) Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell. Biol. 18, 3112–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nanbru C. et al. (1997) Alternative translation of the protooncogene c-myc by an internal ribosome entry site. J. Biol. Chem. 272, 32061–32066 [DOI] [PubMed] [Google Scholar]
- 65.Lang KJ et al. (2002) Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell 13, 1792–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young RM et al. (2008) Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J. Biol. Chem. 283, 16309–16319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shatsky IN et al. (2018) Cap-independent translation: what’s in a name? Trends Biochem. Sci. 43, 882–895 [DOI] [PubMed] [Google Scholar]
- 68.Yi T. et al. (2013) Hypoxia-inducible factor-1α (HIF-1α) promotes cap-dependent translation of selective mRNAs through up-regulating initiation factor eIF4E1 in breast cancer cells under hypoxia conditions. J. Biol. Chem. 288, 18732–18742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uniacke J. et al. (2012) An oxygen-regulated switch in the protein synthesis machinery. Nature 486, 126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho JJD et al. (2016) Systemic reprogramming of translation efficiencies on oxygen stimulus. Cell Rep. 14, 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morita M. et al. (2012) A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell. Biol. 32, 3585–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho PF et al. (2005) A new paradigm for translational control: inhibition via 5’−3’ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121, 411–423 [DOI] [PubMed] [Google Scholar]
- 73.Ruscica V. et al. (2019) Direct role for the Drosophila GIGYF protein in 4EHP-mediated mRNA repression. Nucleic Acids Res. 47, 7035–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peter D. et al. (2019) Molecular basis for GIGYF-Me31B complex assembly in 4EHP-mediated translational repression. Genes Dev. 33, 1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moore CE et al. (2015) Elongation factor 2 kinase is regulated by proline hydroxylation and protects cells during hypoxia. Mol. Cell. Biol. 35, 1788–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Connolly E. et al. (2006) Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol. Cell. Biol. 26, 3955–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andreev DE et al. (2015) Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol. 16, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Natsuizaka M. et al. (2012) Hypoxia induces IGFBP3 in esophageal squamous cancer cells through HIF-1α-mediated mRNA transcription and continuous protein synthesis. FASEB J. 26, 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kelly NJ et al. (2018) Hypoxia activates cadherin-22 synthesis via eIF4E2 to drive cancer cell migration, invasion and adhesion. Oncogene 37, 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X. et al. (2009) Alternative translation of OCT4 by an internal ribosome entry site and its novel function in stress response. Stem Cells 27, 1265–1275 [DOI] [PubMed] [Google Scholar]
- 81.Li SW et al. (2015) The differential expression of OCT4 isoforms in cervical carcinoma. PLoS One 10, e0118033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin SC et al. (2019) OCT4B mediates hypoxia-induced cancer dissemination. Oncogene 38, 1093–1105 [DOI] [PubMed] [Google Scholar]
- 83.Kamphorst JJ et al. (2015) Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75, 544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Urasaki Y. et al. (2012) Coupling of glucose deprivation with impaired histone H2B monoubiquitination in tumors. PLoS One 7, e36775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Warburg O. et al. (1927) The metabolism of tumors in the body. J. Gen. Physiol. 8, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dong C. et al. (2013) Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 23, 316–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Janiszewska M. et al. (2012) Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 26, 1926–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.DeBerardinis RJ et al. (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U. S. A. 104, 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hosios AM et al. (2016) Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev. Cell 36, 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Diehl FF et al. (2019) Cellular redox state constrains serine synthesis and nucleotide production to impact cell proliferation. Nat. Metab. 1, 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ducker GS and Rabinowitz JD (2017) One-carbon metabolism in health and disease. Cell Metab. 25, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sullivan LB et al. (2015) Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alkan HF et al. (2018) Cytosolic aspartate availability determines cell survival when glutamine is limiting. Cell Metab. 28, 706–720 e706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zaidi N. et al. (2013) Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 52, 585–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vriens K. et al. (2019) Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature 566, 403–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Faubert B. et al. (2017) Lactate metabolism in human lung tumors. Cell 171, 358–371 e359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McGuirk S. et al. (2020) Metabolic fitness and plasticity in cancer progression. Trends Cancer 6, 49–61 [DOI] [PubMed] [Google Scholar]
- 98.Morita M. et al. (2015) mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 14, 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scheuner D. et al. (2001) Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 100.Zhang CS et al. (2017) Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.González A. et al. (2020) AMPK and TOR: the yin and yang of cellular nutrient sensing and growth control. Cell Metab. 31, 472–492 [DOI] [PubMed] [Google Scholar]
- 102.Proud CG (2015) Regulation and roles of elongation factor 2 kinase. Biochem. Soc. Trans. 43, 328–332 [DOI] [PubMed] [Google Scholar]
- 103.Leprivier G. et al. (2013) The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153, 1064–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morita M. et al. (2013) mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711 [DOI] [PubMed] [Google Scholar]
- 105.Gandin V. et al. (2016) nanoCAGE reveals 5’ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 26, 636–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sinvani H. et al. (2015) Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab. 21, 479–492 [DOI] [PubMed] [Google Scholar]
- 107.Sancak Y. et al. (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang P. et al. (2002) The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell. Biol. 22, 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harding HP et al. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 110.Ye J. et al. (2010) The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 29, 2082–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Falletta P. et al. (2017) Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 31, 18–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hulea L. et al. (2018) Translational and HIF-1α-dependent metabolic reprogramming underpin metabolic plasticity and responses to kinase inhibitors and biguanides. Cell Metab. 28, 817–832 e818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Niederst MJ et al. (2015) RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat. Commun. 6, 6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Beltran H. et al. (2016) Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bluemn EG et al. (2017) Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 32, 474–489 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martins-Neves SR et al. (2018) Therapy-induced enrichment of cancer stem-like cells in solid human tumors: where do we stand? Pharmacol. Res. 137, 193–204 [DOI] [PubMed] [Google Scholar]
- 117.Hsieh AC et al. (2015) Cell type-specific abundance of 4EBP1 primes prostate cancer sensitivity or resistance to PI3K pathway inhibitors. Sci. Signal. 8, ra116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Deng J. et al. (2019) Inhibition of PI3K/Akt/mTOR signaling pathway alleviates ovarian cancer chemoresistance through reversing epithelial-mesenchymal transition and decreasing cancer stem cell marker expression. BMC Cancer 19, 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.David-West G. et al. (2018) mTORC1/2 inhibition re-sensitizes platinum-resistant ovarian cancer by disrupting selective translation of DNA damage and survival mRNAs. Oncotarget 9, 33064–33076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Peng DJ et al. (2010) Role of the Akt/mTOR survival pathway in cisplatin resistance in ovarian cancer cells. Biochem. Biophys. Res. Commun. 394, 600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jewer M. et al. (2020) Translational control of breast cancer plasticity. Nat. Commun. 11, 2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nguyen HG et al. (2018) Development of a stress response therapy targeting aggressive prostate cancer. Sci. Transl. Med. 10, eaar2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sato A. et al. (2012) Glioma-initiating cell elimination by metformin activation of FOXO3 via AMPK. Stem Cells Transl. Med. 1, 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Courtois S. et al. (2017) Metformin targets gastric cancer stem cells. Eur. J. Cancer 84, 193–201 [DOI] [PubMed] [Google Scholar]
- 125.Bao B. et al. (2012) Metformin inhibits cell proliferation, migration and invasion by attenuating CSC function mediated by deregulating miRNAs in pancreatic cancer cells. Cancer Prev. Res. (Phila.) 5, 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hirsch HA et al. (2009) Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 69, 7507–7511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shank JJ et al. (2012) Metformin targets ovarian cancer stem cells in vitro and in vivo. Gynecol. Oncol. 127, 390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gandin V. et al. (2016) mTORC1 and CK2 coordinate ternary and eIF4F complex assembly. Nat. Commun. 7, 11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koromilas AE (2019) M(en)TORship lessons on life and death by the integrated stress response. Biochim. Biophys. Acta Gen. Subj. 1863, 644–649 [DOI] [PubMed] [Google Scholar]
- 130.Wengrod J. et al. (2015) Phosphorylation of eIF2α by mTORC1 inhibition and PP6C activation is required for autophagy and is aberrant in PP6C-mutated melanoma. Sci. Signal. 8, ra27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.de la Parra C. et al. (2018) A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 9, 3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee AS et al. (2016) eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guan BJ et al. (2017) A unique ISR program determines cellular responses to chronic stress. Mol. Cell. 68, 885–900 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Petz M. et al. (2012) PDGF enhances IRES-mediated translation of Laminin B1 by cytoplasmic accumulation of La during epithelial to mesenchymal transition. Nucleic Acids Res. 40, 9738–9749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Evdokimova V. et al. (2009) Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15, 402–415 [DOI] [PubMed] [Google Scholar]
- 136.D’Amato NC et al. (2013) microRNA regulation of epithelial plasticity in cancer. Cancer Lett. 341, 46–55 [DOI] [PubMed] [Google Scholar]
- 137.Syed SN and Brüne B. (2020) microRNAs as emerging regulators of signaling in the tumor microenvironment. Cancers (Basel) 12, 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Grelet S. and Howe PH (2019) hnRNP E1 at the crossroads of translational regulation of epithelial-mesenchymal transition. J. Cancer Metastasis Treat. 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lin X. et al. (2019) RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 10, 2065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Wan Makhtar WR et al. (2017) Short stretches of rare codons regulate translation of the transcription factor ZEB2 in cancer cells. Oncogene 36, 6640–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rapino F. et al. (2017) tRNA modification: is cancer having a wobble? Trends Cancer 3, 249–252 [DOI] [PubMed] [Google Scholar]
- 142.Prakash V. et al. (2019) Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat. Commun. 10, 2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cejalvo JM et al. (2017) Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer. Cancer Res. 77, 2213–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Balanis NG et al. (2019) Pan-cancer convergence to a small-cell neuroendocrine phenotype that shares susceptibilities with hematological malignancies. Cancer Cell 36, 17–34 e17 [DOI] [PMC free article] [PubMed] [Google Scholar]