Abstract
Introduction
The novel Coronavirus disease (COVID-19), caused by the severe acute respiratory syndrome coronavirus—2 (SARS-CoV-2), in Africa is characterised by a more substantial proportion of asymptomatic (or mildly symptomatic) individuals thought to be playing a role in the spread of the infection. The exact proportion and degree of infectiousness of asymptomatic individuals remains unclear. Studies however indicate that their management is crucial for control of SARS-CoV-2 transmission.
Methodology
We developed a simplified deterministic susceptible-exposed-infectious-removed (SEIR) mathematical model to assess the effect of active isolation of SARS-CoV-2 infected but asymptomatic individuals through blanket testing for control of the outbreak in Lusaka Province of Zambia. Here we modelled two scenarios; (1) assuming asymptomatic individuals comprised 70% of all COVID-19 cases and (2) asymptomatic individuals comprised only 50% of the cases. For contrast, the model was assessed first under the assumption that asymptomatic individuals are equally as infectious as symptomatic individuals and then secondly, and more likely, assuming asymptomatic individuals are only half as infectious as symptomatic individuals.
Results
For the model assuming 70% asymptomatic cases, a minimum sustained daily blanket testing rate of ≥ 7911 tests/100000 population was sufficient to control the outbreak if asymptomatic individuals are only half as infectious while if equal infectiousness was assumed then a testing rate of ≥ 10028 tests/ 100000 population would be required. For 50% asymptomatic, minimum blanket testing rates of ≥ 4540 tests/ 100000 population was sufficient to control the outbreak at both assumed levels of infectiousness for asymptomatic individuals relative to symptomatic individuals.
Discussion and conclusion
Our model predicts that active isolation of COVID-19 cases, including asymptomatic individuals, through blanket testing can be used as a possible measure for the control of the SARS-Cov-2 transmission in Lusaka, Zambia, but it would come at a high cost.
Introduction
Since the first reported case on 31 December 2019 in China, the current pandemic of the novel Coronavirus disease (COVID-19), caused by severe acute respiratory syndrome coronavirus—2 (SARS-CoV-2), has killed over 2 million and infected over 100 million people globally by February 2021 [1–4]. The disease is characterised by a more substantial proportion of asymptomatic (or mildly symptomatic) individuals thought to be playing an “in-dismissible” role in the spread of the infection [5–13]. The degree of infectiousness of the asymptomatic individuals remains unclear although recent data from the World Health Organisation (WHO) and others suggest that relative to individuals with symptomatic infections, those with asymptomatic infection are considerably less infectious [14–16]. Nonetheless, various studies, including modelling studies, have suggested that asymptomatic individuals comprise about 50–80% of all COVID-19 cases and may be responsible for as much as 40–73% of new infections [6, 7, 17–19]. Therefore, their management is critical for the control of the disease [6, 7, 12]. To give a striking example, identification and isolation of asymptomatic people through blanket testing (random mass testing) helped eliminate the virus in a completely isolated village of about 3000 people in northern Italy which saw the number of people with COVID-19 symptoms fall by over 90% within ten days [6]. The current study applied a simple mathematical modelling approach to explore the effect of increased blanket testing rates as a possible measure to capture and isolate asymptomatic individuals and control the COVID-19 outbreak in Lusaka Province of Zambia which is the epicentre of the outbreak in Zambia since the first recorded case in the country on 18th March 2020 [20]. Recent studies have modelled the spread and expected burden of the COVID-19 outbreak in Africa and Zambia and explored the effects of various control measures such as applying different levels of physical distancing and shielding in the population [21–24]. Although this provides vital information to guide policy for Zambia, some interventions may not be easy to monitor in practice. Additionally, while such interventions have already been instituted, cases continue to rise in Zambia and other African countries [20, 22, 25, 26]. Assessment of more COVID-19 control options through mathematical modelling based on the known epidemiology of the disease would therefore serve to supplement current information on the possible management of the outbreak in Zambia.
Materials and methods
Study area
Lusaka Province is the smallest and highly urbanised province in Zambia (83.5% urbanisation) with seven districts over an area of about 23,490 km2 [27]. It is one of the most densely populated provinces in the country [28, 29]. It has a total population of about 3,308,438—Density: 140.8/km2 (2019) [27]. The provincial capital, which is also the capital of Zambia, is the highly-populated Lusaka district with the latest population estimated at 2,627,700 –Density: 6,288/km2 (2019) [27]. Lusaka is a busy corporate and commercial hub of Zambia and an outlet to the rest of the world with the busy Kenneth Kaunda International Airport. It is therefore no surprise that the first recorded COVID-19 cases in Zambia occurred in Lusaka district as imported cases [20]. The province also shares borders with neighbouring Zimbabwe and Mozambique [29].
Lusaka province COVID-19 outbreak
On 18th March 2020, Zambia recorded the first confirmed cases of COVID-19 in Lusaka from two residents who had previously travelled to France [20]. By August 2020, Lusaka had recorded about 3900 confirmed cases and 157 deaths [20]. From the onset of the outbreak in Lusaka, the Zambian Ministry of Health and other stakeholders had implemented required preparedness and disease control measures including mandatory physical distancing, surveillance and case notification, heightened sanitation and handwashing in public places, closure of some public institutions as well as general sensitisation [20]. The main laboratory testing approach employed has been targeted testing based on prescribed case definitions and contact tracing to optimise positivity rates (Zambia COVID-19 situation reports No. 1–14) [20]. A few mass testing campaigns have also been conducted in some areas of Lusaka province as by 21st May 2020 e.g., Rhodes park (about 1,190 persons tested), Chirundu, (about 1,000 persons tested), and Kafue (undetermined) (Zambia COVID-19 situation reports No. 29,44–46,62–64) [20].
The model
The spread of SARS-Cov-2 in Lusaka province was modelled through a simplified deterministic susceptible-exposed-infectious-removed (SEIR) compartmental mathematical model as shown in Fig 1.
Fig 1. The SEIR model for the spread of COVID-19 in Lusaka province.
The solid arrows represent flow between compartments and dotted arrows represent the additive contribution relationship to a compartment. Infectious individuals are split into symptomatic and asymptomatic individuals. For symbols used refer to Table 1.

The model both directly and indirectly incorporated the current mitigation measures to attempt to predict the trajectory of the outbreak in Lusaka accurately. We started by denoting the infection states as the total number of susceptible S(t), exposed E(t), infectious I(t), and removed R(t) at any given time (t) in the population of size N. For our analysis, the total population size was assumed to be constant and demographics of natural birth and deaths rates were considered negligible [30, 31]. Table 1 shows the average values of parameters used and equations in set 1 and set 2 give the systems of ordinary differential equations (ODEs) describing the flow of individuals in the model. The ODEs in set-1 describe the spread of infection under the assumption of equal infectiousness for both asymptomatic and symptomatic individuals while the ODEs in set-2 describe the spread of infection assuming asymptomatic individuals are only half as infectious as symptomatic individuals. For the ODEs in set-1, we assume that susceptible individuals are infected at the rate where IT(t) is the total number of active infectious individuals in the population both symptomatic IS(t) and asymptomatic IA(t). On the other hand, for the ODEs in set-2, the susceptible individuals are infected at the rate with the probability of infection from asymptomatic individuals only half that of symptomatic individuals. In both equations, after infection occurs the susceptible individuals are exposed E(t) and enter an incubation period of about 4.8 days on average before they become infectious I(t). Once they have become infectious, they may belong to either one of two classes of infectious individuals; symptomatic infectious (IS), or asymptomatic infectious (IA) determined by the fraction for symptomatic persons ɛ. Based on current interventions in Lusaka, the symptomatic infectious individuals are effectively managed in that once a person becomes symptomatic in the community, they are identified through targeted testing under the current testing criteria and quarantined. However, in our model we assumed an average delay of about 2.5 days in the identification of these symptomatic individuals because sometimes people delay in seeking medical attention when symptoms are unclear, may be in self-denial, plus may have an extra 24hrs turnaround time delay in the laboratory results [20]. The symptomatic infectious individuals are therefore quarantined at a rate π (1/diagnosis delay) and enter the group of quarantined symptomatic infectious individuals ISQ(t). It is taken that during this delay period these symptomatic individuals can infect others but are no longer able to do so once isolated in quarantine. The quarantined infectious individuals recover at rate γ and are removed as ISR(t). On the other hand, in our model the asymptomatic (or mildly symptomatic) infectious individuals (IA) are taken to be generally unnoticed in the community but also recover at a rate γ in which time they can infect susceptible individuals before they become removed as IAR(t) and no longer infectious. Note that some asymptomatic individuals develop symptoms much later in their infection but this does not substantially affect our model because at that time they would still be removed (IAR) if they become diagnosed and quarantined through targeted testing. In this study, we assumed that the current mostly targeted testing for COVID-19 in Lusaka province is restrictive and probably missing some asymptomatic individuals [17]. Therefore, a parameter (θ) was introduced in the model which describes daily blanket testing (random mass testing) applied as tests per 100000 population used to identify and isolate all infectious individuals in the community (symptomatic and asymptomatic) through sustained random mass testing. The total removed individuals for the model R(t) are given as ISR(t)+IAR(t) while the total confirmed cases are given as ISQ(t)+ISR(t) of which a fraction α (Case fatality rate -CFR) are recorded dead D(t) [55].
Table 1. Parameters used for the SEIR model for spread of COVID-19 in Lusaka province.
| PARAMETER | DEFINITION | VALUE | UNITS | COMMENT | REFERENCE |
|---|---|---|---|---|---|
| N | Total population | 3,300,000 | people | Lusaka | [27] |
| R0 | Basic reproduction number | - | Secondary infections/ infectious period (days) | Data fitted | [1, 5, 12, 30, 32–42] |
| β | The average rate of effective contacts per unit time between susceptible people and infectious people | γR0 | Effective contacts per day | Data fitted | - |
| k | The average rate to infectiousness | 1/4.8 | 1/incubation period (days) | Average estimate | [1, 5, 12, 30, 36–39, 43–50] |
| γ | Recovery rate | 1/7.5 | 1/infectious period (days) | Average estimate | [1, 5, 6, 9, 12, 30, 36–38, 40, 43, 45, 47, 51–54] |
| ε | Fraction symptomatic infectious people | 30% - 50% | - | proportion | [6, 7, 9, 12] |
| π | Quarantine rate for symptomatic | 1/2.5 | 1/diagnosis delay (days). | Assumed | [44] |
| θ | Community blanket testing rate | 150/100000–10733/100000 | daily tests per 100000 population | Daily random mass testing | - |
| α | Apparent death rate (CFR) | 0.0206186 | Deaths/confirmed cases | Data fitted | - |
Model optimisation and simulation
Model optimisation and simulation was done using Vensim PLE systems dynamics modelling software for Windows (version-7) [56]. This was done for two scenarios of 70% and a modest 50% assumed proportion of asymptomatic infectious individuals in the population. Data from the first three months of the outbreak in Lusaka (accessed between 18th March - 21st May 2020) as given in the Zambia COVID-19 situation reports No. 1–64 [20] was used to configure the model and optimise parameters. However, due to presence of imported cases in the early days and the considerable variations in recorded cases between some days (probably influenced by variations in availability of testing kits), only data from 10th April 2020 to 16th May 2020 was used. This is because this period had more consistent data and by then community infections had been established [20]. The model initial conditions were estimated from the available data as follows: S (0) = N-92, E (0) = 24, IS (0) = 9, ISQ (0) = 38, ISR (0) = 0, IA (0) = 21, IAR (0) = 0, R (0) = 0, and D (0) = 0. With other parameter values fixed, the model was calibrated to the cumulative number of confirmed cases over time. This was done by manually adjusting values of R0 until the best model fit was achieved (since R0 was expectedly affected by the current mitigation measures). R0 estimates the average number of secondary infections arising from a single infectious individual in a naive population [57, 58]. Model fit was statistically evaluated using Pearson’s correlation at a significance level of 0.01 as well as chi-square goodness of fit test at a significance level of 0.05. For the calibration, θ was approximated at a daily 150 tests/100000 populations–reflective of the mass testing rates achieved in this period [20]. After calibration, the model simulation was extended to 631 days (10th April 2020 – 31st December 2021) to predict the spread of the outbreak in Lusaka under the current transmission rate. To assess which value of θ would sufficiently flatten the curves of both the total number of active infectious individuals and the cumulative number of confirmed cases over time (as a key indicator of control of the outbreak), several iterations of this simulation were then performed using increasingly higher values for θ.
The relation between R0 and θ (daily blanket testing rate)
A simplistic intuition into the relation between θ and R0 can be seen when we consider the use of blanket testing as the only method of diagnosing and isolating all COVID-19 cases in the model system i.e., if we assume no targeted testing is being offered. The relation between θ and R0 can then be easily demonstrated arithmetically using the ODE set-1 as an example as follows:
When an outbreak has established in a community, then we expect the number of new infections to increase in the community. This means that;
Removing the (t) notation for convenience, this statement means that from the ODE set-1 above we have that:
which can be re-written as:
………………….. inequality 1.
Similarly, when the outbreak has established in the community, then we expect the number of total infectious individuals to increase. This means that;
Similarly, removing the (t) notation for convenience, and considering all the terms that are subtracting from I(t), this statement means that from the ODE set-1 above we have that:
Note that here we are considering that there is no contribution from targeted testing which in the model is captured by the automatic isolation of symptomatic cases at rate πIS(t) where π = 1/diagnosis delay from targeted testing (in days). Therefore, in this case, the contribution of the quantity πIS(t) falls off from the model system. Note that by doing so, we are trying to assess the scenario where only blanket testing is used for the diagnosis and isolation of all total infectious individuals in the community. Therefore, in this case the above inequality for total infectious individuals can be written as:
which can be re-written as:
which further simplifies into inequality 2 as:
……………. Inequality 2.
Now considering inequality 1 and inequality 2, we, therefore, have that:
which simplifies into inequality 3 as:
……………. Inequality 3.
Following the definition of R0, inequality 3 is analyzed at t(0) where the entire population is considered completely susceptible except for the index case. At this point therefore N ≈ S(0). This means that at this point inequality 3 is simplified as:
and the quantity on the left-hand side of this inequality gives the estimate of R0 and it is expected to be greater than unity if the infection is to spread in the community at the disease epidemic equilibrium [57–61]. This quantity was therefore also used to assess changes in R0 for increasing values of θ.
Ethical considerations
No ethical issues were encountered as no human or animal subjects were used in this study and the cases were anonymous. Only publicly available data in the form of the total reported counts of COVID-19 daily new confirmed cases and deaths for Lusaka district in the national COVID-19 situation reports compiled by the Zambia National Public Health Institute (ZNPHI available at http://znphi.co.zm/news/situation-reports-new-coronavirus-COVID-19-sitreps/) was used.
Results
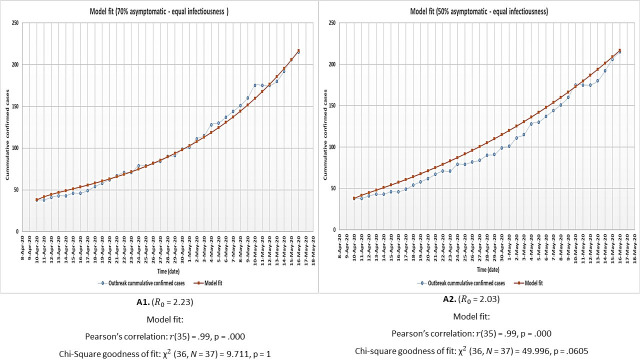
The model had a significant fit to outbreak data under all the assessed conditions and therefore could be used for the general purpose of analysing the outbreak under all these general scenarios. Fig 2 shows the results of model optimisation and fit to outbreak data for both the 70% and 50% asymptomatic scenarios assuming equal infectiousness for asymptomatic and symptomatic individuals (denoted as A1 and A2). Fig 3 shows the model fit results when asymptomatic individuals are only half as infectious as symptomatic individuals (B1 and B2). All R0 values were within the range estimated for COVID-19 [1, 5, 12, 30, 33, 36–39, 41, 42].
Fig 2.
Model optimisation and fit to outbreak data for both the 70% (A1) and 50% (A2) proportion asymptomatic individual’s scenarios under the assumption of equal infectiousness for asymptomatic and symptomatic individuals.
Fig 3.
Model optimisation and fit to outbreak data for both the 70% (B1) and 50% (B2) proportion asymptomatic individual’s scenarios under the assumption that asymptomatic individuals are only half as infectious as symptomatic individuals.
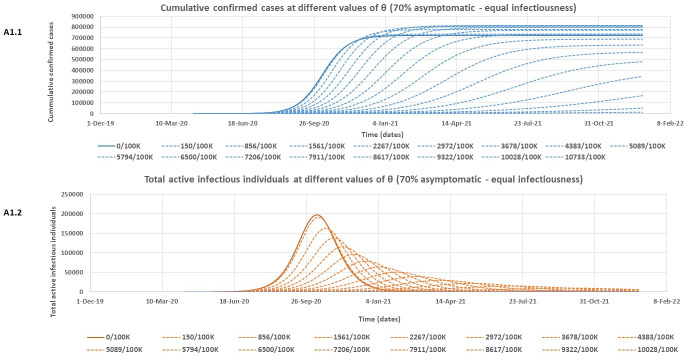
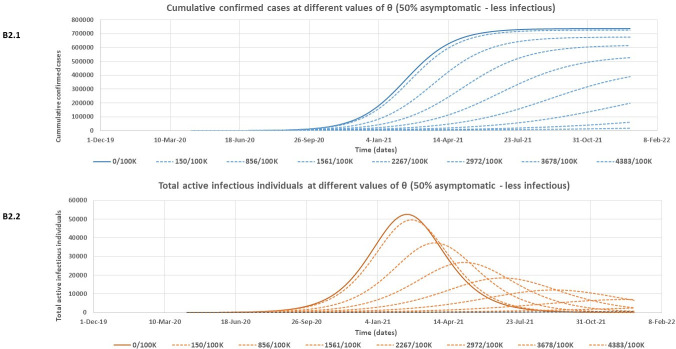
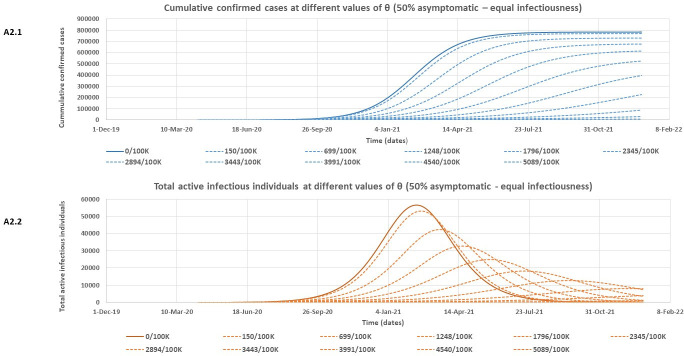
At both assessed levels of infectiousness for asymptomatic individuals, Table 2 shows that the model rightly predicts slightly lower expected peak values for the cumulative number of confirmed cases if there is more (70%) of asymptomatic individuals in the community who are poorly detected. However, this more substantial proportion of asymptomatic individuals (70%) is associated with higher expected peak values of both total active infectious individuals and total quarantined individuals. It is also observed that the outbreak is expected to peak much earlier for all variables in Table 2 for the 70% asymptomatic individual’s scenario compared to only 50% asymptomatic. Note that the model predictions for the Lusaka COVID-19 outbreak given in Table 2 are however subject to the effectiveness of the containment policies in Lusaka province over time. Figs 4 – 7 and Table 3 give the results of the effect of increasing daily blanket testing rates (θ) on control of the outbreak (flattening the curve for the expected cumulative number of confirmed cases and expected total number of active infectious individuals over time) for the assessed model scenarios. If asymptomatic individuals make up 70% of all COVID-19 cases and we assume equal infectiousness for asymptomatic and symptomatic individuals, then a minimum daily blanket testing rate of about ≥ 10028/100000 would be sufficient to flatten the curve for both the expected cumulative number of confirmed cases (Fig 4A1.1) and the total number of active infectious individuals (Fig 4A1.2) by more than 90% (Table 3). However, if the asymptomatic individuals only make up 50% of all cases and are also as infectious as symptomatic individuals, then a minimum daily blanket testing rate of ≥ 4540/100000 would be sufficient to flatten the curve for both the expected cumulative number of confirmed cases (Fig 5A2.1) and the total number of active infectious individuals (Fig 5A2.2) by more than 90% (Table 3). Alternatively, if asymptomatic individuals make up 70% of all COVID-19 cases but are only half as infectious as symptomatic individuals, then a minimum daily blanket testing rate of about ≥ 7911/100000 would be sufficient to flatten the curve for both the expected cumulative number of confirmed cases (Fig 6B1.1) and the total number of active infectious individuals (Fig 6B1.2) by more than 90% (Table 3). However, if the asymptomatic individuals only make up 50% of all cases and are only half as infectious relative to the symptomatic individuals, then a minimum daily blanket testing rate of ≥ 4383/100000 would be sufficient to flatten the curve for both the expected cumulative number of confirmed cases (Fig 7B2.1) and the total number of active infectious individuals (Fig 7B2.2) by more than 90% (Table 3) which is comparable to the result for A2.1 and A2.2.
Table 2. The model predicted key outbreak variables for the Lusaka COVID-19 outbreak for period 10th April 2020 – 31st December 2021 under the different model assumptions.
| Model scenario | Variable | Estimated peak period | Approximated total number at peak |
|---|---|---|---|
| (A1) 70% asymptomatic -equal infectiousness | Cumulative confirmed cases | February 2021 | |
| Total active infectious individuals | October 2020 | ||
| Total quarantined | October 2020 | ||
| Cumulative deaths | February 2021 | 15,146 | |
| (A2) 50% asymptomatic—equal infectiousness | Cumulative confirmed cases | September 2021 | |
| Total active infectious individuals | February 2021 | ||
| Total quarantined | February 2021 | ||
| Cumulative deaths | August 2021 | 15,921 | |
| (B1) 70% asymptomatic—less infectiousness | Cumulative confirmed cases | March–April 2021 | |
| Total active infectious individuals | November 2020 | ||
| Total quarantined | November 2020 | ||
| Cumulative deaths | March–April 2021 | 13,206 | |
| (B2) 50% asymptomatic—less infectiousness | Cumulative confirmed cases | September–October 2021 | |
| Total active infectious individuals | February 2021 | ||
| Total quarantined | February 2021 | ||
| Cumulative deaths | August–September 2021 | 14,994 |
Values are given as single point estimates only.
Fig 4.
The estimated reduction in the cumulative number of confirmed cases (A1.1) and the total number of active infectious individuals (A1.2) at different values of θ (daily blanket testing rate) for the 70% asymptomatic scenario assuming equal infectiousness for asymptomatic and symptomatic individuals.
Fig 7.
The estimated reduction in the cumulative number of confirmed cases (B2.1) and the total number of active infectious individuals (B2.2) at different values of θ (daily blanket testing rate) for the 50% asymptomatic scenario assuming asymptomatic individuals are only half as infectious as symptomatic individuals.
Table 3. Approximated percentage reduction in the predicted cumulative number of confirmed cases and the total number of active infectious individuals for higher values of θ (daily blanket testing rate).
| Model scenario | Variable | θ (daily tests/100000 populations) | Per cent reduction |
|---|---|---|---|
| (A1) 70% asymptomatic -equal infectiousness (R0 = 2.23) | Cumulative confirmed cases (A1.1) | 9322/100000 | ≈ 77.27% |
| 10028/100000 | ≈ 92.98% | ||
| 10733/100000 | ≈ 98.12% | ||
| Total active infectious individuals (A1.2) | 9322/100000 | ≈ 96.92% | |
| 10028/100000 | ≈ 98.99% | ||
| 10733/100000 | ≈ 99.81% | ||
| (A2) 50% asymptomatic -equal infectiousness (R0 = 2.03) | Cumulative confirmed cases (A2.1) | 3991/100000 | ≈ 88.26% |
| 4540/100000 | ≈ 96.175% | ||
| 5089/100000 | ≈ 98.74% | ||
| Total active infectious individuals (A2.2) | 3991/100000 | ≈ 92.76% | |
| 4540/100000 | ≈ 98.04% | ||
| 5089/100000 | ≈ 99.58% | ||
| (B1) 70% asymptomatic -less infectiousness (R0 = 3.53) | Cumulative confirmed cases (B1.1) | 7206/100000 | ≈ 75.42% |
| 7911/100000 | ≈ 92.44% | ||
| 8617/100000 | ≈ 97.998% | ||
| Total active infectious individuals (B1.1) | 7206/100000 | ≈ 95.18% | |
| 7911/100000 | ≈ 98.44% | ||
| 8617/100000 | ≈ 99.72% | ||
| (B2) 50% asymptomatic—less infectiousness (R0 = 3.174) | Cumulative confirmed cases (B2.1) | 3678/100000 | ≈ 72.86% |
| 4383/100000 | ≈ 91.55% | ||
| 5089/100000 | ≈ 97.82% | ||
| Total active infectious individuals (B2.2) | 3678/100000 | ≈ 85.66% | |
| 4383/100000 | ≈ 95.03% | ||
| 5089/100000 | ≈ 99.101% |
Fig 5.
The estimated reduction in the cumulative number of confirmed cases (A2.1) and the total number of active infectious individuals (A2.2) at different values of θ (daily blanket testing rate) for the 50% asymptomatic scenario assuming equal infectiousness for asymptomatic and symptomatic individuals.
Fig 6.
The estimated reduction in the cumulative number of confirmed cases (B1.1) and the total number of active infectious individuals (B1.2) at different values of θ (daily blanket testing rate) for the 70% asymptomatic scenario assuming asymptomatic individuals are only half as infectious as symptomatic individuals.
Effect of θ on R0
Fig 8 shows the results of how increasing θ affects R0 through the relation between R0 and θ given by the quantity as derived from the ODE set-1 when we consider the use of blanket testing as the only method of diagnosing and isolating all COVID-19 cases in the model system for the model scenarios A1 and A2 (Fig 8) as an example.
Fig 8.
The relation between R0 and θ (daily blanket testing rate) for model scenario A1 and A2 when blanket testing alone is applied for diagnosis and isolation of all COVID-19 cases.
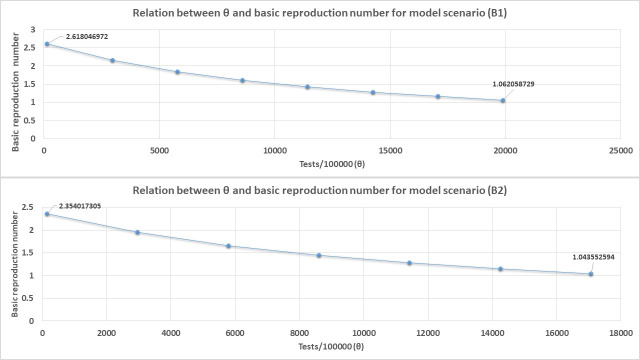
Figs 8 and 9 shows that θ has an inverse relationship with R0 as expected. In fact, our derived equation relating θ to R0 estimates the absolute values of θ required to reduce R0 to exactly one are 16400 test/100000 populations for scenario A1, 13733 tests/100000 populations for scenario A2, 21966/100000 populations for scenario B1, and 18406/100000 populations for scenario B2.
Fig 9.
The relation between R0 and θ (daily blanket testing rate) for model scenario B1 and B2 when blanket testing alone is applied for diagnosis and isolation of all COVID-19 cases. For demonstration purpose only of the direction of the relationship between R0 and θ, an average value of β is used for both scenarios B1 and B2 in Fig 9 given as average β = (β + 0.5β)/2 since these scenarios consist of symptomatic and asymptomatic individuals considered to be transmitting the infection at different rates.
Discussion
In this study, we developed a simple deterministic model to forecast the spread of infection and assess required blanket testing rates for the control of the novel SARS-Cov-2 outbreak in Lusaka province, Zambia, with specific consideration for asymptomatic infectious individuals. We assessed the effect of various levels of blanket testing for control of the COVID-19 outbreak in Lusaka province under different assumed scenarios of the proportion and infectiousness of asymptomatic individuals with results tabulated in Figs 4 – 7 and Table 3. Out of all the assessed scenarios, we nominate scenario B1 to represent the most likely epidemiological dynamics for COVID-19 in the population where asymptomatic individuals make up 70% of cases but are only half as infectious as symptomatic individuals. Therefore, for this scenario, a daily blanket testing rate of about ≥ 7911 tests /100000 (≈ 80/1000) would be sufficient to control the outbreak in Lusaka. It is perceived however that such a testing rate would come at a high cost as it would result in the entire Lusaka population being tested on more than one occasion within a year just to bring the outbreak to some level of control. Therefore, based on these results, we can see that blanket testing is indeed a viable strategy to control the spread of infection but may be costly and is probably less efficient compared to other strategies such as vaccination which would require a usually once-off minimum 1−1/R0 fraction of the population to be vaccinated; assuming vaccination provides full protection [62–65]. Based on the estimated value of R0 for the selected epidemic scenario in B1 in this study, a minimum vaccination coverage of only 70% of the Lusaka population would therefore be required to prevent the spread of the infection if a vaccine for COVID-19 became available and distributed in Zambia. Further, with our results we envisage that random blanket testing as applied in this study would probably be a more feasible and cost effective control measure only in a very small population that is well isolated [6] and where there is enough testing capacity to test 4–10% of the population every day. In their study, a complex model created by Smith and colleagues [66] to assess optimal COVID-19 surveillance strategies in long-term care facilities also found random daily testing to be an inefficient use of resources in the long term. Therefore, overall we suggest that for such large populations as Lusaka, this control strategy would therefore be more realistically applied only besides other control measures such as intensified lock-downs, targeted testing/contact tracing, and vaccinations. Additionally, this study has also demonstrated a relationship of how the identification and isolation of all COVID-19 cases through increased blanket testing negatively affects R0 for the infection (Figs 8 and 9). This is possibly because increased blanket testing results in more cases being physically isolated hence reducing their ability to spread the infection in the population. Estimates by our simple derived equation for the relationship between R0 and θ showed that extensively even higher absolute values of θ would be required to reduce R0 to exactly one as compared to the model simulations which just considered the minimum values of θ required to achieve a certain desirable percentage reduction in projected outbreak outcomes. This showed that when used alone, blanket testing would be an unwieldy and probably less efficient method for reducing R0 to one and therefore other methods would need to be incorporated for this objective e.g vaccinations. Note that the estimated absolute values of θ needed to completely reduce R0 to one are even greater for scenarios B1 and B2 because these scenarios have larger values of R0 as seen in Table 3. One consequence for this is that with such large values of R0, greater than 90% reductions in projected infections through blanket testing would probably not be enough as the outbreak would much easily bounce back later hence the larger required values of θ to completely reduce R0 to one for these scenarios.
In the early stages of novel infectious diseases, and for small populations, stochastic models are used due to uncertainty in parameter values [67–70]. However, deterministic models which require average values can be used after the pandemic has progressed as is the case in this study and other studies that have used deterministic models to study COVID-19 [13, 30]. The deterministic model was also considered appropriate for this study due to its simplicity and also because the modelled population of Lusaka was considered large enough such that stochastic effects could be neglected [69–71]. In the context of Africa, earlier modelling studies have given gloomy predictions of the pandemic for the continent in case of failed early containment [17, 21–23, 26] with Zambia projected to have more than 2.8 million total infections [17] or between 4.8 million and 5.7 million total symptomatic cases [22] in the first year with estimated outbreak peaks as early as July 2020 [17, 21, 22]. Further, the model in [22] also predicted that Zambia would record between 2.5 million– 4.4 million total number of symptomatic cases over 12 months even under various levels of physical distancing and shielding interventions applied at country level. However, early implementation of the outbreak containment measures in Zambia, and most African countries, appears to be substantially averting the given predictions [17, 20]. This may be because Africa was on high alert and prepared for the virus given the weaker health systems on the continent hence early containment of the disease significantly slowed the pandemic. Further, other studies have suggested favorable bio-socioecological factors on the African continent (e.g educated immune systems, young population, climate etc) leading to milder disease and lower rate of transmission in the region with fewer expected cases and deaths compared to other countries such as USA and Italy [13, 17, 72]. With limited incorporation of current containment measures implemented in Zambia, the highest number of cumulative confirmed cases predicted for Lusaka province in our model was 772,161 cases by September 2021 (Table 2). This lower projection is plausible given that it is an estimate for Lusaka province alone and it conforms to the relatively slow progression of the outbreak in Zambia compared to earlier predictions [13, 17, 20, 72]. Further, our model predicts that the outbreak in Lusaka would spread much faster and peak earlier if asymptomatic individuals make up 70% of all COVID-19 cases (earliest peaks October—November 2020 for total active infectious individuals). This is because a higher proportion of poorly detected asymptomatic individuals inflated the population of active infectious individuals in the population (IS+IA = IT) thereby increasing the infection rates in the community; (ODEs in set-1) or (ODEs in set-2), resulting in a faster progression of the outbreak. The higher proportion of asymptomatic individuals was, however, associated with the less overall cumulative number of confirmed cases since asymptomatic individuals are generally less diagnosed under targeted testing approaches such as those employed in Lusaka province [17].
In our model, we refer as asymptomatic all “silent spreaders” of COVID-19 which includes all those that have become infectious but never develop symptoms (asymptomatic or mildly symptomatic) and those that have become infectious but only develop symptoms much later in their infection (pre-symptomatic spreaders) [14, 17–19, 72, 73]. Studies however agree that collectively these asymptomatic individuals (or silent spreaders) are less infectious compared to symptomatic individuals [14–16]. This is most likely because their viral shedding rate (how much virus an infected person releases) may be limited without the symptoms of coughing and sneezing which produces infectious respiratory droplets even if these asymptomatic individuals may have similar viral load, duration of viral shedding, and contact rates in the community as symptomatic individuals [5, 74–78]. Therefore, we suspect that transmission of the infection from asymptomatic individuals may mostly rely on the other possible routes of transmission for COVID-19 including contact with fomites, unwitting close intimate facial contact, and possibly even faecal-oral route [16, 17, 79]. Even with a lowered ability to transmit the infection, lack of detection and awareness of the carrier state may cause the asymptomatic individuals to be playing an important role in the spread of COVID-19 and sustaining the outbreak [5–7, 9, 10, 12]. Active isolation of all infected individuals (symptomatic and asymptomatic) through blanket testing, therefore, offers a possible solution to this challenge [6, 7, 17]. The effect of blanket testing is that it acts to directly reduce the size of the total number of active infectious individuals which leads to a disproportionately higher reduction in the infection rate in the population. This is achieved through active isolation of even the otherwise largely un-detected asymptomatic individuals in the population who possibly participate in the transmission of the infection [5–7, 9]. In the case of Africa, control of asymptomatic infections may be even more pertinent than other regions of the world. This is because Africa has been found to have a younger population and with lower personal vulnerabilities to severe infection compared to Europe, USA, and other regions leading to most of the infections in Africa being asymptomatic [13, 17, 72]. Therefore, more outbreak control strategies with specific consideration of the role of asymptomatic individuals will be required in Africa.
Conclusion
Active isolation of SARS-Cov-2 asymptomatic infectious individuals through blanket testing has been shown to have indeed the potential to control the outbreak in Lusaka province of Zambia. However, we project that this would require a lot of resources to be feasible. For example, we estimate that a minimum sustained daily blanket testing rate of ≥ 7911 tests/100000 population would be sufficient to control the outbreak in Lusaka following the most likely epidemiological dynamics for COVID-19 which may not be very efficient especially for developing nations. Therefore, other control strategies such as vaccinations, not explicitly analyzed in this study, may need to be considered together with blanket testing.
Limitations
As is the case in creating infectious disease models, appropriateness of the model design to incorporate containment measures implemented in Lusaka and quality of data used were some of the challenges faced in this study [29, 80]. Also, limited outbreak data was used due to significant irregularities in the reported outbreak data. Cases were not reported in real-time and there is a possibility of under-reporting due to low testing capacities in the earlier stages of the outbreak in Zambia [20, 81]. Additionally, the model was fitted to cumulative confirmed cases which could still introduce uncertainties [81]. Further modelling studies using more refined models and more outbreak data should, therefore, be conducted to study the COVID-19 outbreak in Lusaka.
Abbreviations
- COVID-19
Novel Coronavirus disease
- SARS-CoV-2
Severe acute respiratory syndrome Coronavirus 2
- CFR
Case fatality rate
- SEIR
susceptible-exposed-infectious-removed compartmental mathematical model
- ODEs
Ordinary differential equations
Data Availability
The data underlying the results presented in the study are available from the Zambia MOH/ZNPH/WHO, | New Coronavirus Disease of 2019 (COVID-19) Situation Reports [1–64]. March-May 2020. Accessed from http://znphi.co.zm/news/situation-reports-new-coronavirus-COVID-19-sitreps/.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Yi Y, Lagniton PN, Ye S, Li E, Xu RH. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int J Biol Sci. 2020;16(10):1753. 10.7150/ijbs.45134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu P, Hao X, Lau EH, Wong JY, Leung KS, Wu JT, et al. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro Surveill. 2020. January 23;25(3):2000044. 10.2807/1560-7917.ES.2020.25.3.2000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlman S. Another decade, another coronavirus. NEJM. 2020. [CrossRef] [PubMed] [DOI] [PMC free article] [PubMed]
- 4.WHO Coronavirus disease (COVID-19) Situation Reports– 123. 2020. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 5.Singhai T. A review of the coronavirus disease-2019. Indian J Pediatr. 2020; 87: 281–6. 10.1007/s12098-020-03263-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day M. Covid-19: identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ (Online). 2020; March 23;368. 10.1136/bmj.m1165 [DOI] [PubMed] [Google Scholar]
- 7.Day M. Covid-19: four fifths of cases are asymptomatic, China figures indicate. BMJ. 2020; 369: m1375 10.1136/bmj.m1375 [DOI] [PubMed] [Google Scholar]
- 8.Du Z, Xu X, Wu Y, Wang L, Cowling BJ, Meyers LA. Serial interval of COVID-19 among publicly reported confirmed cases. Emerg Infect Dis. 2020. June;26(6):1341. 10.3201/eid2606.200357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss P, Murdoch DR. Clinical course and mortality risk of severe COVID-19. The lancet. 2020. March 28;395(10229):1014–5. 10.1016/S0140-6736(20)30633-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai CC, Liu YH, Wang CY, Wang YH, Hsueh SC, Yen MY, et al. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARSCoV-2): facts and myths. J Microbiol Immunol Infect. 2020. March 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis. 2020. March 4. 10.1016/j.ijid.2020.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID‐19) implicate special control measures. J Med Virol. 2020. June;92(6):568–76. 10.1002/jmv.25748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munjita SM, Samutela M, Ndashe K, Munsaka SM. Immunity, parasites, genetics and sex hormones: contributors to mild inflammatory responses in COVID-19?. Pan Afr Med J. 2020. May 15;35(36). 10.11604/pamj.supp.2020.35.2.23267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferretti L, Wymant C, Kendall M, Zhao L, Nurtay A, Abeler-Dörner L, et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020. May 8;368(6491). 10.1126/science.abb6936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schraer R. Coronavirus: Asymptomatic transmission still an ’open question’. BBC NEWS. 9 June 2020. Available from: https://www.bbc.com/news/health-52977940.
- 16.WHO Coronavirus disease (COVID-19) Situation Reports– 73. available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 17.Cabore JW, Karamagi HC, Kipruto H, Asamani JA, Droti B, Seydi AB, et al. The potential effects of widespread community transmission of SARS-CoV-2 infection in the World Health Organization African Region: a predictive model. BMJ Glob Health. 2020. May 1;5(5):e002647. 10.1136/bmjgh-2020-002647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia W, Liao J, Li C, Li Y, Qian X, Sun X, et al. Transmission of corona virus disease 2019 during the incubation period may lead to a quarantine loophole. MedRxiv. 2020. January 1. [Google Scholar]
- 19.Ganyani T, Kremer C, Chen D, Torneri A, Faes C, Wallinga J, et al. Estimating the generation interval for COVID-19 based on symptom onset data. MedRxiv. 2020. January 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MOH/ZNPH/WHO, Situation Reports | New Coronavirus Disease of 2019 (COVID-19) Sitreps [1–141]. March-May 2020. Accessed from http://znphi.co.zm/news/situation-reports-new-coronavirus-COVID-19-sitreps/.
- 21.MOBS Laboratory, Northeastern University, Boston. Center for Inference and Dynamics of Infectious Diseases (CIDID). Fred Hutchinson Cancer Research Center, Seattle.: Modeling local transmission and the burden of the COVID-19 epidemic in African countries. Report 2.0, v1.0. April 12, 2020.
- 22.Nightingale ES, Munday JD, Medley G, Gibbs HP, Abbott S, Houben RMGJ, et al. Modelling projections for COVID-19 epidemic in Zambia. LSHTM CMMID COVID19 Working Group (2020). [Google Scholar]
- 23.Pearson CA, Van Schalkwyk C, Foss AM, O’Reilly KM, Pulliam JR, CMMID COVID-19 working group. Projected early spread of COVID-19 in Africa through 1 June 2020. Euro Surveill. 2020. May 7;25(18):2000543. 10.2807/1560-7917.ES.2020.25.18.2000543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinkala M, Nkhoma P, Zulu M, Kafita D, Tembo R, Daka V. The COVID-19 Pandemic in Africa: Predictions using the SIR Model Indicate the Cases are Falling. medRxiv. 2020. January1. [Google Scholar]
- 25.Ritchie H, Ortiz-Ospina E, Beltekian D, Mathieu E, Hasell J, Macdonald B, et al. Our World in data: Coronavirus Pandemic (COVID-19)–the data. 2020. Available from: https://ourworldindata.org/coronavirus-data [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson CA, Van Schalkwyk C, Foss A, O’Reilly K, Pulliam J. Projection of early spread of COVID-19 in Africa as of 25 March 2020. CMMID. 2020. [DOI] [PMC free article] [PubMed]
- 27.City population. Population statistics for countries, administrative divisions, cities, urban areas and agglomerations–interactive maps and charts. Lusaka Province in Zambia. 2020. Available from: https://www.citypopulation.de/php/zambia-admin.php?adm1id=05.
- 28.ZAMBIA DATA PORTAL. Central Statistical Office. Lusaka. 2020. Available from: https://zambia.opendataforafrica.org/apps/atlas/Lusaka
- 29.Wikipedia. The Free Encyclopedia. Lusaka Province. Available from: https://en.wikipedia.org/wiki/Lusaka_Province
- 30.Prem K, Liu Y, Russell TW, Kucharski AJ, Eggo RM, Davies N, et al. The effect of control strategies to reduce social mixing on outcomes of the COVID-19 epidemic in Wuhan, China: a modelling study. Lancet Public Health. 2020. March 25. 10.1016/S2468-2667(20)30073-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Y Li M. An introduction to mathematical modeling of infectious diseases. Spriger; 2018. [Google Scholar]
- 32.Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. ES. 2020. March;25(10). 10.2807/1560-7917 Euro Surveill.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anastassopoulou C, Russo L, Tsakris A, Siettos C. Data-based analysis, modelling and forecasting of the novel coronavirus (2019-Ncov) outbreak. medRxiv. 2020. January 1. 10.1371/journal.pone.0230405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh R, Adhikari R. Age-structured impact of social distancing on the COVID-19 epidemic in India. arXiv preprint arXiv. 2003;12055. 2020. March 26. [Google Scholar]
- 35.Giordano G, Blanchini F, Bruno R, Colaneri P, Di Filippo A, Di Matteo A, et al. Modelling the COVID-19 epidemic and implementation of population-wide interventions in Italy. Nat Med. 2020. April 22;1–6. 10.1038/s41591-019-0740-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi S, Ki M. Estimating the reproductive number and the outbreak size of COVID-19 in Korea. Epidemiol Health. 2020. March 12;42. 10.4178/epih.e2020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020. February 26:102433. 10.1016/j.jaut.2020.102433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velavan TP, Meyer CG. Mild versus severe COVID-19: laboratory markers. Int J Infect Dis. 2020. April 25. 10.1016/j.ijid.2020.04.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020. February 17:105924. 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuite AR, Fisman DN, Greer AL. Mathematical modelling of COVID-19 transmission and mitigation strategies in the population of Ontario, Canada. CMAJ. 2020. May 11;192(19):E497–505. 10.1503/cmaj.200476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Xu J, Li H, Cao B. A novel coronavirus (COVID-19) outbreak: a call for action. Chest. 2020. April 1;157(4):e99–101. 10.1016/j.chest.2020.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020. March 13. 10.1093/jtm/taaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linton NM, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov AR, Jung SM, et al. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J Clin Med. 2020. February;9(2):538 10.3390/jcm9020538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellewell J, Abbott S, Gimma A, Bosse NI, Jarvis CI, Russell TW, et al. Centre for the Mathematical Modelling of Infectious Diseases COVID-19 Working Group. Funk S, Eggo RM. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health. 2020; 8 (4): e488–e496. 10.1016/S2214-109X(20)30074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Q, Zhao S, Gao D, Lou Y, Yang S, Musa SS, et al. A conceptual model for the outbreak of Coronavirus disease 2019 (COVID-19) in Wuhan, China with individual reaction and governmental action. Int J Infect Dis. 2020. March 4. 10.1016/j.ijid.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamzah FB, Lau C, Nazri H, Ligot DV, Lee G, Tan CL. CoronaTracker: worldwide COVID-19 outbreak data analysis and prediction. Bull World Health Organ. 2020. March;1:32. [Google Scholar]
- 47.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The Lancet. 2020. February 15;395(10223):514–23. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baud D, Qi X, Nielsen-Saines K, Musso D, Pomar L, Favre G. Real estimates of mortality following COVID-19 infection. Lancet Infect Dis. 2020. March 12. 10.1016/S1473-3099(20)30195-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020. May 5;172(9):577–82. 10.7326/M20-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020. February 15;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng L, Yang W, Zhang D, Zhuge C, Hong L. Epidemic analysis of COVID-19 in China by dynamical modeling. arXiv preprint arXiv. 2002;06563. 2020. February 16. [Google Scholar]
- 52.Bastola A, Sah R, Rodriguez-Morales AJ, Lal BK, Jha R, Ojha HC, et al. The first 2019 novel coronavirus case in Nepal. Lancet Infect Dis. 2020. March 1;20(3):279–80. 10.1016/S1473-3099(20)30067-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020. March 17;323(11):1061–9. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The lancet. 2020;395:1054–62. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ritchie H, Roser M. What do we know about the risk of dying from COVID-19? Our World in Data. March 25, 2020. Available from: https://ourworldindata.org/covid-mortality-risk [Google Scholar]
- 56.Vensim. VENTANA systems.inc. 2020. Available from: https://vensim.com/vensim-personal-learning-edition/
- 57.Sichone J, Simuunza MC, Hang’ombe BM, Kikonko M. Estimating the basic reproduction number for the 2015 bubonic plague outbreak in Nyimba district of Eastern Zambia. PLoS Negl Trop Dis. 2020. November 9;14(11):e0008811. 10.1371/journal.pntd.0008811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heffernan JM, Smith RJ, Wahl LM. Perspectives on the basic reproductive ratio. J R Soc Interface. 2005. September 22;2(4):281–93. 10.1098/rsif.2005.0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alshakhoury NS. Mathematical modeling and control of MERS-CoV epidemics (Doctoral dissertation). 2017. Available from: http://hdl.handle.net/11274/9782.
- 60.van den Driessche P. Reproduction numbers of infectious disease models. Infect Dis Model. 2017. August 1;2(3):288–303. 10.1016/j.idm.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pandey A, Mubayi A, Medlock J. Comparing vector–host and SIR models for dengue transmission. Math Biosci. 2013. December 1;246(2):252–9. [PubMed] [Google Scholar]
- 62.Dietz K. The estimation of the basic reproduction number for infectious diseases. Stat Methods Med Res. 1993. March;2(1):23–41. 10.1177/096228029300200103 [DOI] [PubMed] [Google Scholar]
- 63.van den Driessche P. Reproduction numbers of infectious disease models. Infect Dis Model. 2017. August 1;2(3):288–303. 10.1016/j.idm.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mukandavire Z, Liao S, Wang J, Gaff H, Smith DL, Morris JG. Estimating the reproductive numbers for the 2008–2009 cholera outbreaks in Zimbabwe. Proc Natl Acad Sci. 2011. May 24;108(21):8767–72. 10.1073/pnas.1019712108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mukandavire Z, Smith DL, Morris JG Jr. Cholera in Haiti: reproductive numbers and vaccination coverage estimates. Scientific reports. 2013. January 10;3:997. 10.1038/srep00997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith DRM, Duval A, Pouwels KB, Guillemot D, Fernandes J, Huynh B-T, et al. Optimizing COVID-19 surveillance in long-term care facilities: a modelling study. BMC Medicine. 2020. December 8;18(1):386. 10.1186/s12916-020-01866-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang Y, Yang L, Dai H, Tian F, Chen K. Epidemic situation and forecasting of COVID-19 in and outside China. Bull World Health Organ. 2020. March 16;10. [Google Scholar]
- 68.Kucharski AJ, Russell TW, Diamond C, Liu Y, Edmunds J, Funk S, Eggo RM, Sun F, Jit M, Munday JD, Davies N. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis. 2020. March 11. 10.1016/S1473-3099(20)30144-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kretzschmar M, Wallinga J. Mathematical models in infectious disease epidemiology. InModern infectious disease epidemiology 2009. (pp. 209–221). Springer, New York, NY. [Google Scholar]
- 70.Choisy M, Guégan JF, Rohani P. Mathematical modeling of infectious diseases dynamics. Encyclopedia of infectious diseases: modern methodologies. 2007. January;379. [Google Scholar]
- 71.Diekmann O, Heesterbeek H, Britton T. Mathematical tools for understanding infectious disease dynamics. Princeton University Press; 2012. November 18. [Google Scholar]
- 72.Diop BZ, Ngom M, Biyong CP, Biyong JN. The relatively young and rural population may limit the spread and severity of Covid-19 in Africa: a modelling study. BMJ Glob Health. 2020. May 1;5(5):e002699. 10.1136/bmjgh-2020-002699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. NEJM. 2020. April 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, Villamizar-Peña R, Holguin-Rivera Y, Escalera-Antezana JP, et al. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med Infect Dis. 2020. March 13:101623. 10.1016/j.tmaid.2020.101623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Surveillances V. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19)—China, 2020. China CDC Weekly. 2020;2(8):113–22. [PMC free article] [PubMed] [Google Scholar]
- 76.Suxin W, Yi X, Wei F, Yu Z, Boqun L, Yanjun H, et al. Clinical features and treatment of COVID-19 patients in northeast Chongqing. https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/. J Med Virol. 25783. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu N, Zhang D, Wang W. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China. NEJM. 2019. [published January 24, 2020]. [Google Scholar]
- 78.Lavezzo E, Franchin E, Ciavarella C, Cuomo-Dannenburg G, Barzon L, Del Vecchio C, et al. Suppression of COVID-19 outbreak in the municipality of Vo, Italy. medRxiv. 2020. January 1. [Google Scholar]
- 79.Cabore JW, Karamagi HC, Kipruto H, Asamani JA, Droti B, Seydi AB, et al. The potential effects of widespread community transmission of SARS-CoV-2 infection in the World Health Organization African Region: a predictive model. BMJ Glob Health. 2020. May 1;5(5):e002647. 10.1136/bmjgh-2020-002647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fung IC. Cholera transmission dynamic models for public health practitioners. Emerg Themes Epidemiol. 2014. December 1;11(1):1. 10.1186/1742-7622-11-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu FS, Nguyen A, Link N, Santillana M. Estimating the prevalence of COVID-19 in the United States: three complementary approaches. April 23, 2020. (https://www.medrxiv.org/content/10.1101/2020.04.18) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying the results presented in the study are available from the Zambia MOH/ZNPH/WHO, | New Coronavirus Disease of 2019 (COVID-19) Situation Reports [1–64]. March-May 2020. Accessed from http://znphi.co.zm/news/situation-reports-new-coronavirus-COVID-19-sitreps/.