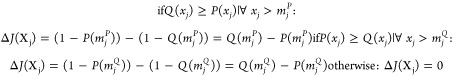Abstract
COVID-19 is still placing a heavy health and financial burden worldwide. Impairment in patient screening and risk management plays a fundamental role on how governments and authorities are directing resources, planning reopening, as well as sanitary countermeasures, especially in regions where poverty is a major component in the equation. An efficient diagnostic method must be highly accurate, while having a cost-effective profile. We combined a machine learning-based algorithm with mass spectrometry to create an expeditious platform that discriminate COVID-19 in plasma samples within minutes, while also providing tools for risk assessment, to assist healthcare professionals in patient management and decision-making. A cross-sectional study enrolled 815 patients (442 COVID-19, 350 controls and 23 COVID-19 suspicious) from three Brazilian epicenters from April to July 2020. We were able to elect and identify 19 molecules related to the disease’s pathophysiology and several discriminating features to patient’s health-related outcomes. The method applied for COVID-19 diagnosis showed specificity >96% and sensitivity >83%, and specificity >80% and sensitivity >85% during risk assessment, both from blinded data. Our method introduced a new approach for COVID-19 screening, providing the indirect detection of infection through metabolites and contextualizing the findings with the disease’s pathophysiology. The pairwise analysis of biomarkers brought robustness to the model developed using machine learning algorithms, transforming this screening approach in a tool with great potential for real-world application.
Coronaviruses (CoVs) are enveloped, single-stranded positive RNA viruses from the Coronaviridae family.1 The recent pandemic, caused by SARS-CoV-2 and denominated COVID-19, opened discussions about measures to control disease spread, such as social distancing and population screening.2 Currently, available tests are based on the direct detection of SARS-CoV-2 virus through antigens or RNA amplification (RT-PCR), serological tests, and the combination of RT-PCR and chest CT (computed-tomography). The development of medical decision-making tools for patient’s risk stratification and management is aligned with COVID-19 testing urgency. Even though the basis for the standard procedures is well-established, there are increased concerns about test’s sensitivity and specificity achieved on the field, time and costs associated with procedures, reagents and trained personnel availability, and the testing window.3−5
Difficulties for an accurate diagnosis of SARS-CoV-2 and patient’s risk categorization are consequences of COVID-19 complexity. SARS-CoV-2 infection pathophysiology reflects in a broad spectrum of patient symptoms, ranging from mild flu-like manifestations to life-threatening acute respiratory distress syndrome (ARDS), vascular dysfunction, and sepsis.2,6 Changes in lipid homeostasis, a common characteristic of viral infections, have been associated with SARS-CoV-2 pathology.7−9 Lipidomic and metabolomic profiling of plasma samples indicate that exosomes enriched with monosialodihexosyl ganglioside (GM3) are associated with the severity of COVID-19.7 Moreover, Fan et al. (2020) proposed the relationship between progressive decrease in serum low-density lipoprotein cholesterol (LDL-C) and cholesterol within deceased patients.8 Individual susceptibility to COVID-19 symptoms is not fully understood, thereby hampering any potential outcome prediction.
Panels of biomarkers that translate disease pathophysiology and sample profiling contribute to SARS-CoV-2 detection and may be proposed through “omics” techniques.7,9−11 The current trend in associating artificial intelligence-explained algorithms and “omics” techniques has yielded platforms involving machine learning (ML) to analyze mass spectrometry (MS) data, aiming at biomarker identification of diseases, including COVID-19 severity assessment.9,11,12 However, applying traditional untargeted mass spectrometry for diagnostic purposes remains laborious.12,13
Considering that the testing tool for COVID-19 introduced in this contribution is based on metabolites from actual patients, it may be considered a new approach for SARS-CoV-2 screening. The proposed end-to-end mass spectrometry and machine learning combination aims at predictively identifying and modeling putative biomarkers for COVID-19 identification, risk assessment and low-risk discrimination from noninfected individuals. The introduced pairwise features approach is critical for effective implementation of untargeted metabolomics on a real-world setting, adding robustness to the model in spite of variations in the input data. Therefore, using the potential of MS-ML techniques in COVID-19 fighting,14 we enrolled a cohort of 815 individuals for the development and testing of this independent system that simultaneously functions as an automated screening test using plasma samples, and provides metabolic information related to the presence and severity risk for the infection.
Experimental Section
Study Design and Patient Recruitment
Participants were recruited from selected sites with proven expertise in research from April to July, 2020 to increase data variability: Hospital das Clinicas, Faculdade de Medicina, Universidade de São Paulo (HCFMUSP), São Paulo, Brazil; Sumaré State Hospital and Paulínia Municipal Hospital localized in São Paulo state inland, Brazil; and Hospital Delphina Rinaldi Abdel Aziz, Manaus, Amazonas State, Brazil. The study was conducted according to principles expressed in Declaration of Helsinki and approved by local Ethics Committees (CAAE 32077020.6.0000.0005, CAAE 31049320.7.1001.5404 and CAAE 30299620.7.0000.0068). All participants provided informed consent before sample collection. Inclusion criteria for COVID-19 group (CV) were adult patients with one or more clinical symptoms of SARS-CoV-2 infection in the last 7 days (fever, dry cough, malaise and/or dyspnea) and positive SARS-CoV-2 RT-PCR in nasopharyngeal samples, following local hospital testing protocols based on Charité and WHO recommendations.15 Control (CT) was formed by symptomatic RT-PCR-negative participants, also discarded by clinical and tomographic picture, and asymptomatic volunteers. Patients with suspicious COVID-19 and negative RT-PCR were separated in a group for posterior assessment.
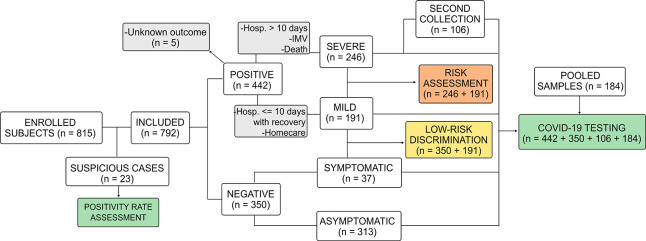
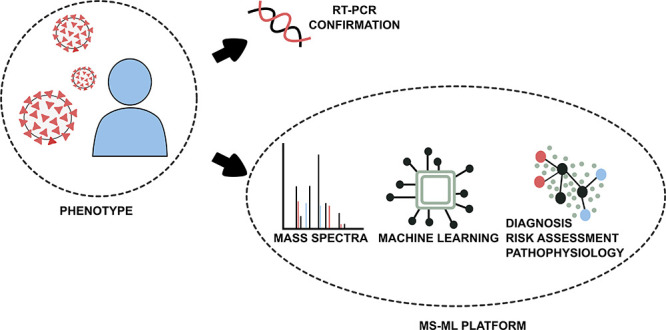
In this study, 815 participants were included. Gender, age, and fasting restrictions were not applied to simulate real-world conditions and to provide results with no patient bias. Table 1 shows detailed COVID-19 and suspicious patients’ demographic information and Figure 1, a flowchart of study design.
Table 1. Characteristics of COVID-19 Confirmed and Suspicious Patients.
| characteristics | CV = 442 | suspicious = 23 |
|---|---|---|
| age, years, mean (SD) | 50 (15.4) | 56 (13.6) |
| female sex, N (%) | 186 (42.1) | 6 (26.1) |
| Severity, N (%) | ||
| homecare | 189 (42.8) | 1 (4.3) |
| hospitalization | 253 (57.2) | 22 (95.7) |
| ≤10 days | 125 (49.4) | 8 (34.8) |
| >10 days | 123 (48.6) | 15 (65.2) |
| transferred | 5 (2.0) | - |
| in-hospital death | 123 (49.6) | 11 (47.8) |
| onset of symptoms to enrolment, days, mean (SD) | 10·6 (6.3)a | 5.5 (3.5) |
| Respiratory Support, N (%) | ||
| no oxygen received | 213 (48.2) | 2 (8.7) |
| oxygen | 76 (17.2) | 4 (17.4) |
| invasive mechanical ventilation | 153 (34.6) | 17 (73.9) |
| Comorbidities, N (%) | ||
| diabetes | 115 (26.4)b | 8 (34.8) |
| hypertension | 176 (40.5)b | 12 (52.2) |
| obesity | 113 (29.9)c | 2 (8.7) |
| cardiomyopathy | 35 (8.1)d | 5 (21.7) |
| respiratory diseases | 37 (8.5)b | 8 (34.8) |
| chronic renal diseases | 13 (3.0)c | 3 (13.0) |
| chronic hepatic diseases | 15 (34.6)c | - |
| HIV | 6 (13.9)d | 1 (4.2) |
N = 431.
N = 435.
N = 378.
N = 432.
N = 433.
Figure 1.
Study design flowchart. Abbreviations: Hosp, hospitalization; IMV, invasive mechanical ventilation.
COVID-19 Diagnostic Modeling (M1)
Diagnosis model (M1) was trained, validated, and tested using CV group composed of 548 plasma samples from 442 symptomatic SARS-CoV-2 confirmed cases upon hospital arrival, and 106 samples representing a second collection from hospitalized patients (mean 9.6 days, SD 3.8). CT group was formed by 37 symptomatic individuals with COVID-19 discarded and 313 asymptomatic controls, totaling 350 individuals, with median age of 50 years-old (IQR 32–72) and 64.9% female. Pooled samples (n = 184) were introduced to the data set to increase method sensitivity: 79 pooled CT, 5 pooled CV, 50 samples with 1:5 (CV:CT) and 50 with 1:10 (CV:CT) dilutions. Positivity rate of 23 suspicious individuals was assessed using this model.
Risk Assessment and Mild Symptoms Discrimination Modeling
Samples from 437 SARS-CoV-2 positive patients with reported outcome were divided into severe cases (n = 191) and mild cases (n = 246). Severe cases were categorized by required hospitalization for more than 10 days with recovery as outcome, or invasive mechanical ventilation, or deceased; mild group consisted of those with moderate (hospitalization lower than 10 day with recovery) and mild symptoms (homecare). Severe cases were compared against mild group for classification and determination of risk biomarkers (M2). The method sensitivity and specificity to discriminate low-risk patients were also accessed comparing controls (CT = 350) against mild group in a third machine learning model (M3).
Mass Spectrometry Sample Preparation
Plasma samples from peripheral venous blood were carefully handled due biohazard, and frozen at −80 °C until analysis. A 20 μL aliquot of plasma was diluted in 200 μL of tetrahydrofuran, followed by homogenization for 30 s at room temperature. Thus, 780 μL of methanol was added followed by homogenization for 30 s and centrifugation for 5 min, 3400 rpm at 4 °C. An aliquot of 5 μL of the supernatant was diluted in 495 μL of methanol and positively ionized by the addition of formic acid (0.1% final concentration) prior analysis.
Mass Spectrometry Analysis and Biomarker Elucidation
All samples were randomized for data acquisition intra- and interdaily and directly infused in a HESI-Q Exactive Orbitrap-MS (Thermo Scientific, Bremen, Germany) with 140 000 FWHM of mass resolution on positive ion mode. MS parameters were set as follows:m/z range 150–1700, 10 mass spectral acquisition per sample, flow rate 10 μL/min, sheath gas flow rate 5 units, capillary temperature 320 °C, aux gas heater temperature 33 °C, spray voltage 3.70 kV, automatic gain control (AGC) at 1 × 106, S-lens RF level 50, and injection time <2 ms. After ML modeling, the presence of each discriminant m/z determined by the algorithm was confirmed in mass spectra using Xcalibur 3.0 software (Thermo, Bremen, Germany). Molecule identity was proposed using METLIN (https://metlin.scripps.edu) database and literature search with mass accuracy ≤5 ppm and confirmed through MSn experiments using an ESI-LTQ XL (Thermos Scientific, Bremen, Germany) with collision energy ranging from 20 to 50 eV (He) and Mass Frontier software (Thermos Scientific, Bremen, Germany) for fragmentation modeling. Biomarkers pathway and meaning were attributed based on Kegg database (https://www.genome.jp/kegg/) and scientific literature.
Machine Learning Data Analysis
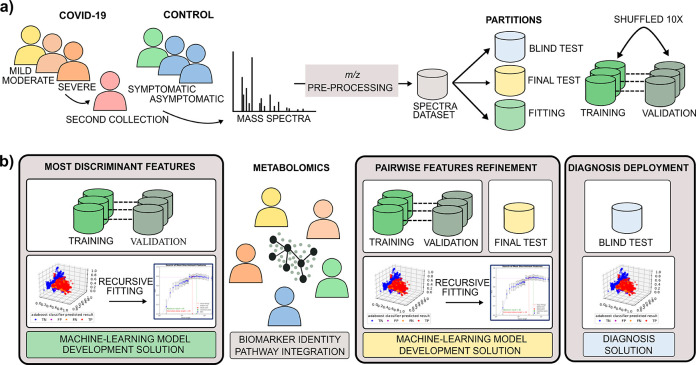
The MS-ML platform16 consists of two primary data analysis phases. The first phase comprises the development of a ML model using a classification algorithm over MS data to determine potential m/z biomarkers. The second phase entails a deployed prediction model for diagnosing and determining risk, used for individuals screening in the field (blind data), as described in Figure 2. First, mass spectrometric data is preprocessed for ion annotation (intensity, width, resolution, and m/z), alignment, normalization, and denoising.17 Three partitions of data are segregated according to the best practices of ML, consisting of fitting (training and validation), test, and blind test partitions. The final classification results are reported using the blind test (see Figure 2a). For the COVID-19 model (M1), a second blind test using suspicious cases was performed to evaluate the positivity rate between SARS-CoV-2 RT-PCR negative patients. The most discriminant features are determined and validated using the ML algorithms (ADA tree boosting (ADA), gradient tree boosting (GDB), random forest (RF), and extreme random forest (XRF), partial least squares (PLS), and support-vector machines (SVM)).18−20 Applied recursive fitting to training and validation data (Figure 2b), with the annotation of averaging and computing the related standard deviation of selected performance metrics is defined in Table 2 for each round of validation (optimized through accuracy, F1score, MCC).
Figure 2.
End to end process for putative biomarkers determination and diagnosis test generation. (a) MS data acquisition and preparation; (b) Sequential steps of ML data analysis and metabolomics biomarkers validation.
Table 2. Statistical Metrics Definition to Evaluate Classification Resultsa.
| metric | formula | |
|---|---|---|
| sensitivity (SEN) | TP/(TP+FN) | |
| specificity (SPE) | TN/(TN+FP) | |
| precision (PRE) | TP/(TP+FP) | |
| accuracy (ACC) | (SEN+SPE)/2 | |
| F1-score (F 1s) | 2·PRE·SEN/(PRE+SEN) | |
| Matthews’ Correlation Coefficient (MCC) | ((TP·TN)-(FP·FN))/sqrt((TP+FP)·(TP+FN)·(TN+FP)·(TN+FN)) | |
| ΔJ |
|
|
| correlation index (r), using Pearson correlation coefficient |
Abbreviations: TP = true positives;
TN = true negatives; FP = false positives; FN = false negatives; sqrt
= square root; A = set of all vectors; xji = value of variable j of vector i in A, xj ∈ 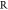 ; yi = label of vector i in A, yi = [0,1]; Xj set of all values of variable j in A; AQ = set of all vectors of
negative samples in A, i.e., labeled yQ = 0; mjQ = median of
values of variable j for all vectors in AQ; Q(xj) = the cumulative probability function (CDF) of values xj ∈ Xj in AQ; AP = set of all vectors of positive samples in A, i.e., labeled yP =
1; mj = median of values of variable j for all vectors
in AP; P(xj) = the cumulative probability function (CDF)
of values xj ∈ Xj in AP; t and u = features.
; yi = label of vector i in A, yi = [0,1]; Xj set of all values of variable j in A; AQ = set of all vectors of
negative samples in A, i.e., labeled yQ = 0; mjQ = median of
values of variable j for all vectors in AQ; Q(xj) = the cumulative probability function (CDF) of values xj ∈ Xj in AQ; AP = set of all vectors of positive samples in A, i.e., labeled yP =
1; mj = median of values of variable j for all vectors
in AP; P(xj) = the cumulative probability function (CDF)
of values xj ∈ Xj in AP; t and u = features.
After the observation of performance metrics, the discriminant features are evaluated through ΔJ importance (see Table 2) and selected for metabolomics biomarkers identification. The marker importance is given by a cumulative distribution function (CDF) analysis: for a specific m/z, a CDF of the feature values for the negative samples (CT group) is compared with the CDF of positive samples (CV group) used in the fitting partition using first the Kolmogorov–Smirnov (KS-test) two samples equality hypothesis test to determine whether those distributions are different (failed on equality hypothesis). Then the ΔJ metric is used to determine if the features contribute negatively ΔJ < 0, or positively ΔJ > 0 for the disease. Features are discarded if CDFs are equal according to KS-test or ΔJ = 0.21 The selected discriminant biomarkers undergo a second round of training and validation with the development software (Figure 2b). As putative biomarkers are validated via metabolomics, they are submitted to pairwise model creation, where the relationship between the biomarker’s intensities are used instead of their relative abundance solely provided in each spectrum, leading to an applied untargeted metabolomics diagnosis software. Features correlated to the selected biomarkers through the correlation index (Table 2) r ≥ 80% were also identified. Detailed information on ML method is displayed in the Supporting Information (SI).
Results and Discussion
COVID-19 Testing Through MS-ML Platform: Modeling and Performance
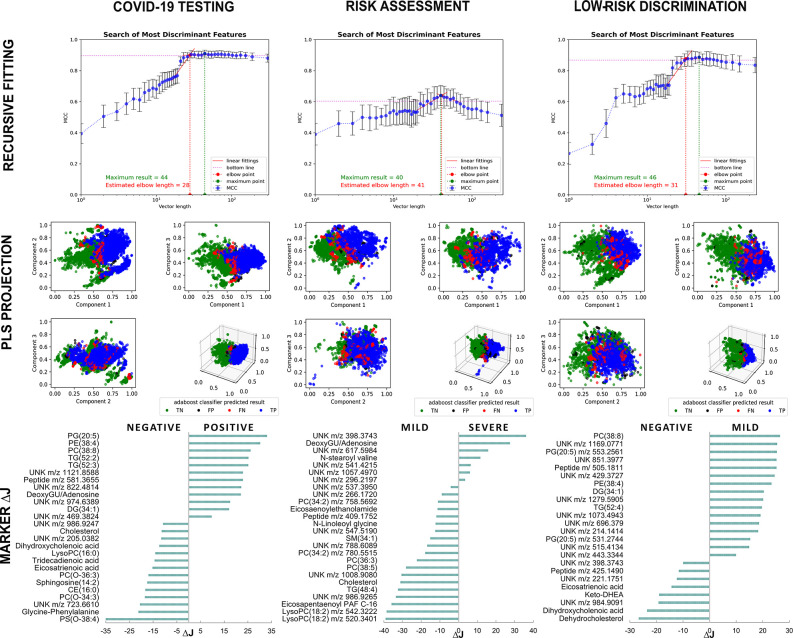
The full data set was segregated as shown in Table 3 for the fitting process (shuffled in 10 rounds of training and validation), and testing. The novel sequential processing of metabolomics data with ML algorithms resulted in a predictive model used for the diagnosis and risk assessment in the field (blind test). Table 4 shows metric results for the predictive models (automated diagnosis (M1), risk assessment (M2), and low-risk discrimination (M3)) using pairwise features. Features selected through recursive fitting using MCC as metric are used to project groups separation (Figure 3). During this process, discriminant features are evaluated through ΔJ importance, with posterior identification into molecules by metabolomics approach. The best final results were obtained with gradient tree boosting (GDB) to COVID-19 automated diagnosis with 96.0% of specificity and 83.1% of sensitivity. COVID-19 suspicious patients with RT-PCR negative results were assessed using the final COVID-19 classifier, resulting in a positivity rate of 78.3%, which may indicate the presence of false negative among RT-PCR results. The best results for risk assessment were obtained with ADA Boosting algorithm with 80.3% of specificity and 85.4% of sensitivity, from blind test.
Table 3. Dataset Subdivisions for Model Fitting (Training and Validation), Testing and Blind Testa.
| model | COVID-19 diagnosis (M1) (n = 1082) |
risk assessment (M2) (n = 437) |
low-risk
discrimination (M3) (n = 595) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| class | positive | negative | subtotal | severe | mild | subtotal - | mild | negative | subtotal |
| training | 260 | 231 | 491 (45) | 94 | 104 | 198 (45) | 113 | 140 | 253 (13) |
| validation | 105 | 95 | 200 (18) | 37 | 43 | 80 (18) | 34 | 42 | 76 (13) |
| testing | 57 | 53 | 110 (10) | 19 | 23 | 42 (10) | 23 | 28 | 51 (9) |
| blind test | 231 | 50 | 281 (26) | 41 | 76 | 117 (27) | 76 | 139 | 215 (36) |
Numbers correspond to individual (N) average and percentages in parentheses.
Table 4. Performance Metrics Using Pairwise Features in 10 Validation Tests, Final Development Testing and Deployed Software Blind Testa.
| Model | COVID-19 diagnosis (M1) | Risk assessment (M2) | Low-risk discrimination (M3) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Algorithm | GDB | ADA | ADA | ||||||
| Data set | Validation | Test | Blind Test | Validation | Test | Blind Test | Validation | Test | Blind Test |
| Vector length | 39 | 39 | 39 | 32 | 32 | 32 | 29 | 29 | 29 |
| # of Estimators | 260 (3) | 256 | 256 | 260 (3) | 256 | 256 | 260 (3) | 256 | 256 |
| TN | 90 (3) | 50 | 48 | 38 (2) | 21 | 61 | 40 (2) | 26 | 121 |
| FP | 5 (2) | 3 | 2 | 5 (2) | 2 | 15 | 2 (1) | 2 | 18 |
| FN | 4 (2) | 3 | 39 | 4 (2) | 4 | 6 | 3 (1) | 2 | 4 |
| TP | 101 (4) | 54 | 192 | 33 (3) | 15 | 35 | 31 (2) | 21 | 72 |
| Accuracy (%) | 95.6 (1.1) | 94.5 | 89.6 | 88.7 (3.2) | 85.1 | 82.8 | 93.4 (1.8) | 92.1 | 90.9 |
| Sensitivity (%) | 95.9 (1.8) | 94.7 | 83.1 | 88.1 (4.6) | 79.0 | 85.4 | 91.8 (3.1) | 91.3 | 94.7 |
| Specificity (%) | 95.2 (2.1) | 94.3 | 96.0 | 89.3 (4.7) | 91.3 | 80.3 | 95.0 (2.4) | 92.9 | 87.1 |
| Precision (%) | 95.3 (1.9) | 94.4 | 95.4 | 89.3 (4.2) | 90.1 | 81.2 | 94.9 (2.3) | 92.7 | 88.0 |
| F1 Score (%) | 95.6 (1.1) | 94.6 | 88.8 | 88.6 (3.2) | 84.2 | 83.2 | 93.3 (1.9) | 92.0 | 91.2 |
| MCC | 0.91 (0.02) | 0.89 | 0.80 | 0.78 (0.06) | 0.71 | 0.66 | 0.87 (0.04) | 0.84 | 0.82 |
Numbers correspond to individual’s classification average and standard deviations in parentheses. Abbreviations: ADA, ADA Boosting; GDB, gradient tree boosting; FN, false negative; FP, false positive; TN, true negative; TP, true positive; MCC, Mathew’s Correlation Coefficient.
Figure 3.
Recursive fitting of mass spectra data followed by model optimization processes allowed the determination of putative biomarkers ranked by ΔJ importance and group contribution. Abbreviations: CE, cholesteryl ester; DG diacylglycerol; DHEA, dehydroepiandrosterone; DeoxyGU, deoxyguanosine; LysoPC, lysophosphatidylcholine; PC, phosphatidylcholine; PE, phosphatidyethanolamine; PG, phosphatidylglycerol; PS, phosphatidylserine; SM, sphingomyelin; TG, triacylglycerol; UNK, unknown.
To assess model specificity and sensitivity, we compared selected moderate and mild symptoms cases with noninfected controls (M3) with ADA boosting, which resulted in the metrics of 92.9% of specificity and 91.3% of sensitivity from blinded data. Supporting data from validation metrics obtained during models’ development with different algorithms are displayed in SI Tables S1 and S2.
Panel of Discriminant Metabolites for COVID-19 Patients Using Untargeted Metabolomics
Twenty-six ions were selected by the ML and used for COVID-19 diagnosis (M1) (Table 4, metrics) and further validated through mass spectrometric data. From those, 19 discriminant biomarkers for COVID-19 infection were proposed, divided into 8 with positive and 11 with a negative contribution to the condition. Out of 19 molecules, seven belong to the glycerophospholipid class, three sterol lipids, three glycerolipids, two fatty acids, one sphingosine, one purine metabolite, and two unknown peptides. The remaining seven molecules have not yet been identified, a common element of nontargeted metabolomics.13 A decrease in lysophosphatidylcholines (LysoPC), cholesterol species and unsaturated fatty acids followed by increased intensities of triacylglycerols (TG), diacylglycerols (DG) and a purine were the main findings that discriminated SARS-CoV-2 infected patients from noninfected individuals. Biomarkers data are displayed in Figure 3 and detailed in SI Table S3.
For risk assessment (M2), 26 ions achieved the metrics displayed in Table 4. Among them, seven markers contributed to the COVID-19 severe condition and 19 contributed to mild group. The main findings shown in Figure 3 pointed to a relative reduction of certain species of LysoPC, phosphatidylcholine (PC), phosphatidylcholine derived plasmalogens, cholesterol, TG, sphingomyelins (SM), and N-acylethanolamines in severe cases in comparison to patients with mild and moderate symptoms (SI Table S4). Severe cases were represented by deoxyguanosine/adenosine, N-stearoyl valine and sterol lipid derivatives. The metrics for low-risk discrimination (M3) (Table 4) were achieved with 24 ions divided in four glycerophospholipid and two glycerolipid markers, one peptide and nine unknown metabolites for mild group, whereas COVID-19 negative patients showed enhanced eicosatrienoic acid, three sterol lipid metabolites, one peptide, and three unknowns (Figure 3, SI Table S5).
Elected Biomarkers and COVID-19 Pathophysiology
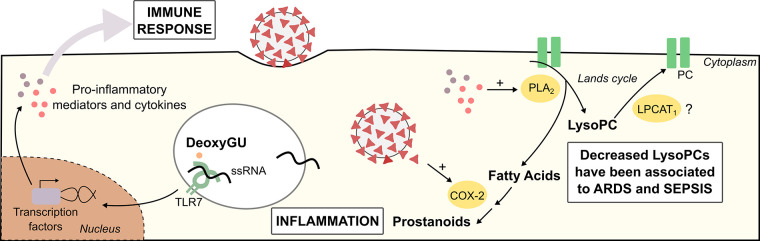
The use of AI-explained algorithms allowed the creation of reliable models that facilitate decision-making in clinics and the investigation of the pathophysiological meaning of distinct biomarker’s levels. Viral recognition is an essential for initial host immune response, and the rapid course and cytokine storm associated with SARS-CoV infection may be involved with the guanosine- and uridine-rich (GU) single-strand RNA potential role as PAMP (pathogen-associated molecular patterns).1 Deoxyguanosine, a metabolite from purine metabolism, triggers the enhanced signaling of TLR7 in the presence of ssRNA, inducing cytokine secretion in macrophages.22 Therefore, further investigations are required to understand the potential role of deoxyguanosine in SARS-CoV-2 immune hyperactivation. On the other hand, N-linoleoyl-glycine and N-acylethanolamines (C20:1 and C22:0), found in this study associated with mild cases, regulate immune response by promoting anti-inflammatory effects.23,24
The main lipidic findings pointed to a remodeling of glycerophospholipid metabolism. We identified enhanced presence of phosphatidylglycerol (PG) [PG(20:5)], phosphatidylethanolamine (PE) [PE(38:4)] and phosphatidylcholine (PC) [PC(38:8)], a diminishment of LysoPCs [LysoPC(16:0), and correlated m/z LysoPC(18:0), LysoPC(18:1), and LysoPC(18:2)] and plasmalogens species25 (PS(O-36:2), PC(O-36:3), PC(O-34:2), PC(O-36:3)) in COVID-19 positive patients; the same PG, PC, and PE markers discriminated low-risk patients from noninfected individuals, as illustrated in Figure 4 by glycerophospholipid pathway recurrence. LysoPC(18:2) were also found as negative contributors in plasma samples from patients with higher risk of death, as well as such PCs markers (PC(34:2), PC(36:3), PC(38:5)) and correlated PC molecules (PC(36:2), PC(36:4), PC(38:3), PC(38:4), PC(38:6)). Cell responses to various stimuli may be mediated by lysophospholipids, which actively participates in inflammation processes.26,27 The relative intensities decrease of LysoPCs and PCs in severely ill patients are in accordance with recent studies of metabolic changes in ARDS and sepsis,26−28 characteristics of COVID-19 severity.2,6 LysoPC is formed through the cleavage of PC by phospho-lipase A2 (PLA2), whose modulation has a crucial role in inflammation processes. PLA2 up-regulation promotes fatty acids formation, precursors of eicosanoids, and LysoPCs.29 Data shows that SARS-CoV nucleocapsid protein stimulates the expression of Cyclooxygenase-2,30 an essential enzyme in the catalysis of prostanoids production from fatty acids, molecules that have been found downregulated in ARDS.31
Figure 4.
Proposed role of identified biomarkers in COVID-19 pathophysiology. Abbreviations: ARDS, acute respiratory distress syndrome; COX-2, cyclooxygenase-2, deoxyGU, deoxyguanosine; LPCAT1, lysophosphatidylcholine acyltransferase 1; LysoPC, lysophosphatidylcholine; PC, phosphatidylcholine; PLA2, phospholipase A2.
The availability of LysoPCs is also finely regulated by the acyltransferase activity of LCAT1 (lysophosphatidylcholine acyltransferase 1), which may promote the restoration of PCs via Lands cycle. The most abundant lipid specie found in alveolar surfactant formed by LCAT1 activity over LysoPC is dipalmitoylphosphatidylcholine (DPPC, PC(16:0/16:0)). This molecule corresponds to 70–80% of surfactant lipid composition, and the dysregulation of surfactant film is directly related to lung injury and ARDS.29 Since DPPC formation is dependent on the availability of lipid substrates and the Lands cycle functioning, interferences in this process may disturb LysoPC and PC availability. Ferrarini et al. (2017) described a decrease in LysoPC species in serum of patients with ARDS derived from influenza infection and sepsis, reinforcing our findings.26
Moreover, COVID-19 pathophysiology seems to impair cholesterol homeostasis.7,8 We found cholesterol and cholesteryl ester (CE (16:0)) diminished in COVID-19 positive patients, and cholesterol decreases within mild/moderate symptoms, which was similarly reported by Song et. al (2020). They demonstrated the correlation between CE abundance and bis (monoacylglycero)phosphate, BMP(38:5), a lipid that influences cellular exportation of cholesterol from endosomes. During recovering progression, it was found increased alveolar macrophages BMP with enhanced CEs.7 Cholesterol and LDL-C (low-density lipoprotein cholesterol) lowering was also observed in clinical practice associated with COVID-19 poor prognosis,8 corroborating to our findings. Herein, based on the proposed m/z markers, we discriminated COVID-19 patients using a diagnostic, risk assessment and low-risk discrimination classifier generated from a MS-ML combination. Although the proposed biomarkers correlates COVID-19 pathophysiology to the mathematical process, a mechanistic biomarker evaluation is needed to better understand their contribution to COVID-19, and identify the unknowns.
Conclusions
The use of machine learning as a mean for the discrimination of diseases from mass spectrometric data aims to develop diagnostic and prognostic tools, treatment targets, and patient management systems.11,12 From published articles to date, mass spectrometry-machine learning approaches employed a MALDI-MS direct method for untargeted analysis of SARS-CoV-2 specimens for diagnosis based on spectra profile,11 or focused on biomarkers evaluation and their significance to severity levels of COVID-19 patients,9 keeping the traditional chromatography–mass spectrometry approach. Our methodology introduced the pairwise m/z analysis, an essential advance in untargeted metabolomics application to provide diagnosis directly from raw data. By combining different m/z, this approach supports the spectra acquired by different mass spectrometers, including the robust use of flow-injection mass spectrometry (FI-MS), on an effort to overcome the ion competition effect.32 Moreover, the proposed MS-ML platform for COVID-19 presented reliable qualitative results, with specificity of 96.0% and sensitivity of 83.1% (in a blind test), similar or even better in performance when compared to available serological and RT-PCR methods.4,5 Our analysis also brings molecular information about the disease pathophysiology that may aid in prognostic markers and treatment targets for COVID-19. Overall, it aggregates, in one solution, an alternative for populational COVID-19 screening and guidance for public health efforts through risk classification. One future work consists in exploring a multiclass model (preliminary results in SI Table S6) for COVID-19 diagnosis and risk assessment. The same approach may be applied to other diseases involved with patient management during the pandemic and contribute to the COVID-19 MS Coalition’s collective effort14 by consolidating the combination of mass spectrometry and artificial intelligence in a real-world setting.
Acknowledgments
We thank the network involved in sample collection, clinical data curation, diagnosis, and technology support (Thermo Scientific, LADETEC (UFRJ)). This work was supported by São Paulo Research Foundation (FAPESP) (2017/12646-3 to A.R.R., 2019/05718-3 to J.D., 2018/10052-1 to W.J.F., 2020/05369-6 to F.T.M.C., 2020/04705-2 to J.C.N., and T.F.D.) Amazonas State Government, Superintendence of the Manaus Free Trade Zone (SUFRAMA), Coordination for the Improvement of Higher Education Personnel (CAPES) (88887.513974/2020-00 to ANO), Department of Science and Technology (DECIT) -Brazilian Ministry of Health (MS), Ministry of Science, Technology and Innovation - National Council for Scientific and Technological Development (CNPq) (403253/2020 to M.V.G.L. and 304497/2018-5 to A.R.R.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c04497.
Detailed data preprocessing and ML modeling method; Tables with model validation with several algorithms, multiclass confusion matrix, and biomarkers elucidation data through mass spectrometry; Preprocessed mass spectrometry17 data available at: 10.5281/zenodo.4329381; Deidentified patient information will be made available from corresponding author upon request (PDF)
Author Contributions
⧖ R.S.F., J.C.N., T.F.D., A.J.B., E.C.S., L.O.R., W.J.F., M.W.P.J., M.V.G.L., G.C.M., W.M.M., F.F.A.V., D.B.-S., V.S.S., J.D., and R.R.C. were involved with study design. R.S.F., T.F.D., A.J.B., R.S., F.G.M.-B., E.C.S., A.E., L.O.R., W.J.F., N.D., L.A.S., J.C.C.A., M.V.G.L., G.C.M., W.M.M., F.F.A.V., R.L.A.-N., D.B.-S., C.C.J., F.T.M.C., and V.S.S. contributed to patient data, clinical support, and network feasibility. L.C.N., J.D., A.R.R., and R.R.C., developed the MS-ML method. J.D., E.N.B.B., G.M.S., A.N.O., and D.N.O. performed mass spectrometry experiments and data interpretation. J.D., L.C.N., R.F.S. wrote the manuscript. E.N.B.B., G.M.S., A.N.O., D.N.O., F.T.M.C., N.D., W.F.J., L.O.R., D.B.-S., F.F.A.V., W.M.M., G.C.M., J.C.N., T.F.D., R.S., F.G.M.-B., A.R.R., and R.R.C. revised the manuscript. R.R.C. idealized the project and managed the research group. All authors have given approval to the final version of the manuscript. ⧖These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Li Y.; Chen M.; Cao H.; Zhu Y.; Zheng J.; Zhou H. Extraor-dinary GU-rich single-strand RNA identified from SARS coronavirus contributes an excessive innate immune response. Microbes Infect. 2013, 15 (2), 88–95. 10.1016/j.micinf.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.; Wang Y.; Li X.; Ren L.; Zhao J.; Hu Y.; Zhang L.; Fan G.; Xu J.; Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395 (10223), 497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Marca A.; Capuzzo M.; Paglia T.; Roli L.; Trenti T.; Nel-son S. M. Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod. BioMed. Online 2020, 41 (3), 483–499. 10.1016/j.rbmo.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhla M.; Boesecke C.; Schulte B.; Diegmann C.; Sib E.; Richter E.; Eschbach-Bludau M.; Aldabbagh S.; Marx B.; Eis-Hübinger A.-M. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health 2020, 182, 170–172. 10.1016/j.puhe.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Yao L.; Li J.; Chen L.; Song Y.; Cai Z.; Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020, 92 (7), 903–908. 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Liu L.; Zhang D.; Xu J.; Dai H.; Tang N.; Su X.; Cao B. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet 2020, 395 (10235), 1517–1520. 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.-W.; Lam S. M.; Fan X.; Cao W.-J.; Wang S.-Y.; Tian H.; Chua G. H.; Zhang C.; Meng F.-P.; Xu Z. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020, 32 (2), 188–202. 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J.; Wang H.; Ye G.; Cao X.; Xu X.; Tan W.; Zhang Y. Low-density lipoprotein is a potential predictor of poor prognosis in patients with coronavirus disease 2019. Metab., Clin. Exp. 2020, 107, 154243. 10.1016/j.metabol.2020.154243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B.; Yi X.; Sun Y.; Bi X.; Du J.; Zhang C.; Quan S.; Zhang F.; Sun R.; Qian L. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 2020, 182 (1), 59–72. 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihling C.; Tänzler D.; Hagemann S.; Kehlen A.; Hüttelmaier S.; Arlt C.; Sinz A. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. J. Proteome Res. 2020, 19 (11), 4389–4392. 10.1021/acs.jproteome.0c00280. [DOI] [PubMed] [Google Scholar]
- Nachtigall F. M.; Pereira A.; Trofymchuk O. S.; Santos L. S. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 2020, 38 (10), 1168–1173. 10.1038/s41587-020-0644-7. [DOI] [PubMed] [Google Scholar]
- Liebal U. W.; Phan A. N.; Sudhakar M.; Raman K.; Blank L. M. Machine Learning Applications for Mass Spectrometry-Based Metabolomics. Metabolites 2020, 10 (6), 243. 10.3390/metabo10060243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz S.; Vallejo M.; García A.; Barbas C. Method validation strategies involved in non-targeted metabolomics. J. Chromatogr. A 2014, 1353, 99–105. 10.1016/j.chroma.2014.04.071. [DOI] [PubMed] [Google Scholar]
- Struwe W.; Emmott E.; Bailey M.; Sharon M.; Sinz A.; Corrales F. J.; Thalassinos K.; Braybrook J.; Mills C.; Barran P. The COVID-19 MS Coalition—accelerating diagnostics, prognostics, and treatment. Lancet 2020, 395 (10239), 1761–1762. 10.1016/S0140-6736(20)31211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.; Bleicker T.; Brünink S.; Drosten C.; Zambon M. Diagnostic detection of 2019-nCoV by real-time RT-PCR. Eurosurveillance. 2020, 25 (3), 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catharino R.; Rocha A.; Navarro L.; Delafiori J.. Automated method for molecular selection. INPI; (Brazil: ). BR1020200159160, 2020. (submitted patent 04/08/2020).
- Delafiori J.; Navarro L. C.; Siciliano R. F.; de Melo G. C.; Busanello E. N. B.; Nicolau J. C.; Sales G. M.; de Oliveira A. N.; Val F. F. A; de Oliveira D. N.; Eguti A.; Dalçóquio T. F.; Bertolin A. J.; Abreu-Netto R. L.; Salsoso R.; Baía-da-Silva D.; Marcondes-Braga F. G.; Sampaio V. S.; Judice C. C.; Costa F. T. M.; Durán N.; Perroud M. W.; Sabino E. C.; de Lacerda M. V. G.; Reis L. O.; Fávaro W. J.; Monteiro W. M.; Rocha A R.; Catharino R. R.. Covid-19 automated diagnosis and risk assessment through Metabolomics and Machine Learning. 2020. (Version 1.0.0) [Data set]. Zenodo. 10.5281/zenodo.4329381. [DOI] [PMC free article] [PubMed]
- Bishop C. M.Pattern recognition and machine learning, 1st ed.; Springer: New York, 2006. [Google Scholar]
- Géron A.Hands-on Machine Learning with Scikit-Learn, Keras, And Tensorflow: Concepts, Tools, aAnd Techniques to Build Intelligent Systems, 2nd ed.; O’Reilly Media: CA, 2019. [Google Scholar]
- Murphy K.Machine Learning: A Probabilistic Perspective, 1st ed.; The MIT Press: Cambridge, 2013. [Google Scholar]
- Lima E. O.; Navarro L. C.; Morishita K. N.; Lamikawa C. M.; Rodrigues R. G. M.; Dabaja M. Z.; de Oliveira D. N.; Delafiori J.; Dias-Audibert F. L.; Ribeiro M. D. S.; Vicentini A. P.; Rocha A.; Catharino R. R. Metabolomics and machine learning approaches combined in pursuit for more accurate paracoccidiodomycosis diagnosis. mSystems. 2020, 5 (3), e00258–20. 10.1128/mSystems.00258-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenne T.; Bridgeman A.; Rigby R. E.; Rehwinkel J. Deoxyguanosine is a TLR7 agonist. Eur. J. Immunol. 2020, 50 (1), 56–62. 10.1002/eji.201948151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein S. H.; Adams J. K.; Bradshaw H. B.; Fraioli C.; Rossetti R. G.; Salmonsen R. A.; Shaw J. W.; Walker J. M.; Zipkin R. E.; Zurier R. B. Potential anti-inflammatory actions of the elmiric (lipoamino) acids. Bioorg. Med. Chem. 2007, 15 (10), 3345–3355. 10.1016/j.bmc.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Carbonare M.; Del Giudice E.; Stecca A.; Colavito D.; Fabris M.; D’Arrigo A.; Bernardini D.; Dam M.; Leon A. A saturated N-acylethanolamine other than N-palmitoyl ethanolamine with anti-inflammatory properties: A neglected story. J. Neuroendocrinol. 2008, 20, 26–34. 10.1111/j.1365-2826.2008.01689.x. [DOI] [PubMed] [Google Scholar]
- Ivanova P. T.; Milne S. B.; Brown H. A. Identification of atypical ether-linked glycerophospholipid species in macrophages by mass spectrometry. J. Lipid Res. 2010, 51 (6), 1581–1590. 10.1194/jlr.D003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarini A.; Righetti L.; Martínez M. P.; Fernández-López M.; Mastrangelo A.; Horcajada J. P.; Betbesé A.; Esteban A.; Ordóñez J.; Gea J. Discriminant biomarkers of acute respiratory distress syndrome associated to H1N1 influenza identified by metabolomics HPLC-QTOF-MS/MS platform. Electrophoresis 2017, 38 (18), 2341–2348. 10.1002/elps.201700112. [DOI] [PubMed] [Google Scholar]
- Drobnik W.; Liebisch G.; Audebert F.-X.; Fröhlich D.; Glück T.; Vogel P.; Rothe G.; Schmitz G. Plasma ceramide and lysophosphatidylcholine inversely correlate with mortality in sepsis patients. J. Lipid Res. 2003, 44 (4), 754–761. 10.1194/jlr.M200401-JLR200. [DOI] [PubMed] [Google Scholar]
- Maile M. D.; Standiford T. J.; Engoren M. C.; Stringer K. A.; Jewell E. S.; Rajendiran T. M.; Soni T.; Burant C. F. Associations of the plasma lipidome with mortality in the acute respiratory distress syndrome: a longitudinal cohort study. Respir. Res. 2018, 19 (1), 1–8. 10.1186/s12931-018-0758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañadas O.; Olmeda B.; Alonso A.; Pérez-Gil J. Lipid–Protein and Protein–Protein Interactions in the Pulmonary Surfactant System and Their Role in Lung Homeostasis. Int. J. Mol. Sci. 2020, 21 (10), 3708. 10.3390/ijms21103708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X.; Hao Q.; Mu Y.; Timani K. A.; Ye L.; Zhu Y.; Wu J. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-kappa B and CCAAT/enhancer binding protein. Int. J. Biochem. Cell Biol. 2006, 38 (8), 1417–1428. 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K. V.; Rao S. M.; Gayani R.; Mohan I. K.; Naidu M. Oxidant stress and essential fatty acids in patients with risk and established ARDS. Clin. Chim. Acta 2000, 298 (1–2), 111–120. 10.1016/S0009-8981(00)00264-3. [DOI] [PubMed] [Google Scholar]
- Sarvin B.; Lagziel S.; Sarvin N.; Mukha D.; Kumar P.; Aizenshtein E.; Shlomi T. Fast and sensitive flow-injection mass spectrometry metabolomics by analyzing sample-specific ion distributions. Nat. Commun. 2020, 11 (1), 1–11. 10.1038/s41467-020-17026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.