Abstract
Motivation
A biomedical relation statement is commonly expressed in multiple sentences and consists of many concepts, including gene, disease, chemical and mutation. To automatically extract information from biomedical literature, existing biomedical text-mining approaches typically formulate the problem as a cross-sentence n-ary relation-extraction task that detects relations among n entities across multiple sentences, and use either a graph neural network (GNN) with long short-term memory (LSTM) or an attention mechanism. Recently, Transformer has been shown to outperform LSTM on many natural language processing (NLP) tasks.
Results
In this work, we propose a novel architecture that combines Bidirectional Encoder Representations from Transformers with Graph Transformer (BERT-GT), through integrating a neighbor–attention mechanism into the BERT architecture. Unlike the original Transformer architecture, which utilizes the whole sentence(s) to calculate the attention of the current token, the neighbor–attention mechanism in our method calculates its attention utilizing only its neighbor tokens. Thus, each token can pay attention to its neighbor information with little noise. We show that this is critically important when the text is very long, as in cross-sentence or abstract-level relation-extraction tasks. Our benchmarking results show improvements of 5.44% and 3.89% in accuracy and F1-measure over the state-of-the-art on n-ary and chemical-protein relation datasets, suggesting BERT-GT is a robust approach that is applicable to other biomedical relation extraction tasks or datasets.
Availability and implementation
the source code of BERT-GT will be made freely available at https://github.com/ncbi/bert_gt upon publication.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The volume of biomedical literature continues to grow rapidly, and it becomes increasingly difficult to maintain and update biomedical knowledge manually. Fortunately, the advance of text-mining techniques is able to greatly help biomedical researchers to retrieve relevant information efficiently in the era of information overload (Fiorini et al., 2018; Sidhoumi et al., 2010). In addition, with the rapid growth of the study on personalized medicine and cancer research, it is necessary to extract and search novel genomic variant information from the biomedical literature (Lee et al., 2020). Biomedical text mining can provide useful data for biomedical curators (Clematide and Rinaldi, 2012) and other researchers in the biomedical communtiy.
Relation extraction (RE) is one of the critical tasks of text mining; it extracts the relation between concepts from the text. It is typically understood as classifying the relation category of a given sentence and an entity pair. Many methods have been proposed for RE. For instance, Lee et al. (2019) and Peng et al. (2019) adapted the Bidirectional Encoder Representation for Transformer (BERT) architecture (Devlin et al., 2019) to biomedical literature and clinical records. Their system achieved state-of-the-art performance on several biomedical and clinical benchmarks against many other machine-learning methods.
No matter whether BERT or other relation extraction methods (Li and Ji, 2019; Peng et al., 2018) are used, they usually focus on classifying a two-entity pair within a single sentence, for example, whether the sentence expresses that the drug ‘anastrozole’ inhibits ‘breast cancer.’ Relationships between entities, however, are often expressed across sentences or may involve more than two entities. Two examples are illustrated inFigure 1. The first example notes that patients with T790M mutations in the epidermal growth factor receptor (EGFR) can be treated with EGFR tyrosine kinase inhibitor (TKI), likes gefitinib. The two sentences collectively express that there is a ternary interaction between the drug (gefitinib), gene/gene product (EGFR tyrosine kinase) and mutation (T790M). Although, in the second example, the disease ‘polyneuropathy’ and the chemical ‘IT-cytosine arabinoside (ara-C)’ do not co-occur in the same sentence, a relation is asserted between the two entities. Extracting such a relation without gathering information across sentences and multiple entities makes it difficult to predict correctly the relation in both cases.
Fig. 1.
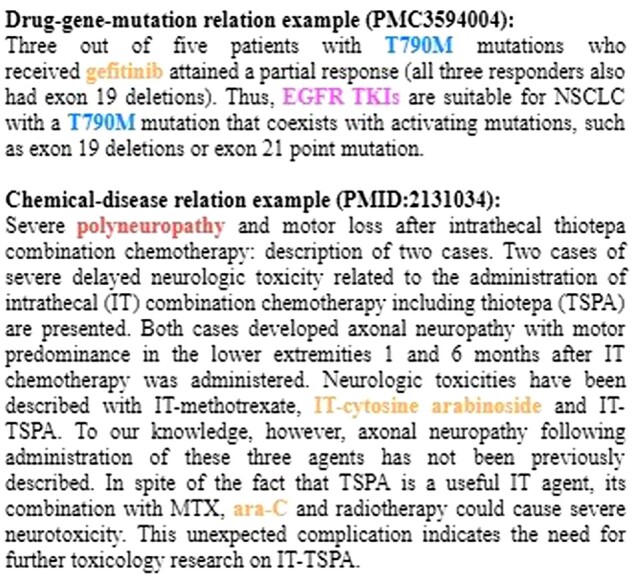
Cross-sentence -ary relation extraction examples
Over time, the above problem has received the attention of the natural language processing (NLP) field. In 2015, an abstract-level relation extraction dataset was created for an NLP task on chemical-disease relation (CDR) extraction (Wei et al., 2016). In the dataset, more than 30% of chemical-induced disease pairs are cross-sentences (Xu et al., 2016). Many cross-sentence methods have been proposed to identify these relations. Verga et al. (2018) proposed a Bi-affine Relation Attention Network (BRAN), which is designed to extract relations between entities in the biological text without requiring explicit mention-level annotation. Their neural network (NN) architecture, however, is limited in terms of binary entities and cannot be adapted to n-ary entities. Peng et al. (2017) developed a cross-sentence n-ary dataset for detecting drug-gene-mutations across sentences. The cross-sentence n-ary relation extraction task is understood as detecting relations among n entities across multiple sentences. Peng et al. (2017) further proposed a graph long short-term memory network (graph LSTM) method.
For a cross-sentence n-ary relation extraction task, previous methods typically utilize dependency information by incorporating long short-term memory (LSTM) or an attention mechanism into a graph neural network (GNN). In recent year, Vaswani et al. (2017) propose an encoder-decoder architecture called Transformer, without the use of LSTM or convolutional neural networks (CNN), and they demonstrate that the Transformer outperforms recurrent neural networks (RNNs) with attention to many sequence-to-sequence tasks. Further, Devlin et al. (2019) used Transformer architecture to develop BERT and proposed two approaches to generate the non-task-specific pre-trained model. They demonstrate that the pre-trained model can be used for different tasks, with transfer-learning approaches, and can outperform state-of-the-art approaches on many NLP tasks.
Because the self-attention mechanism of the Transformer can efficiently utilize the information from the whole input text, we consider that BERT can be used to classify cross-sentence relations as well. We posit, however, that the cross-sentence also may bring noisy information to BERT and can result in difficulties in focusing on explicit information.
In this work, we propose a novel model that adds a Graph Transformer (GT) architecture into BERT (BERT-GT). The Graph Transformer uses a neighbor–attention mechanism, which is a modified version of the self-attention mechanism. In the self-attention mechanism, the whole sentence(s) is used to calculate the attention of the current token. In contrast, the neighbor–attention mechanism calculates its attention based on its neighbor tokens only. Thus, the token can pay attention to its neighbor tokens with limited noise, which is especially important when the text is very long, as in multiple sentences. The main contributions are as follows:
In this work, we focus on classifying the biomedical relations of different text lengths, in particular relations across multiple sentences, an issue that is common in biomedical text but not well studied. In addition, both binary and n-ary relation classification tasks are considered.
While BERT is a robust and state-of-the-art method, its performance is weakened when processing cross-sentence with unrelated relation keywords. In response, we propose a novel architecture that improves BERT by integrating a neighbor–attention mechanism in a Graph Transformer.
BERT-GT is evaluated on two independent biomedical benchmarks and our experimental results demonstrate a significant improvement over other state-of-the-art methods on both n-ary and CDR datasets. This demonstrates the generalizability and robustness of our method.
2 Related work
This section introduces recent works on the cross-sentence relation extraction benchmark and then gives a brief introduction of a recent graph transformer architecture. Peng et al. (2017)’s dataset is the most commonly used dataset for the cross-sentence n-ary relation extraction. The BioCreative CDR dataset (Wei et al., 2016) is also widely used to evaluate abstract-level relation extraction. In what follows, we review the recent work on these benchmarks.
2.1 Cross-sentence n-ary relation extraction
Peng et al. (2017) proposed a graph LSTM architecture. They first use the NLTK dependency parser to parse text input into a directed dependency graph, and then the graph is divided into two acyclic-directed graphs (left-to-right and right-to-left). Then, they respectively apply two LSTM layers to the two graphs to generate the output sequences. Finally, the prediction is calculated by aggregating the hidden states of the entities with a softmax function. Their architecture achieves an accuracy of 80.7% and outperforms the feature-based classifier (Quirk and Poon, 2017) by an accuracy of 3%.
Song et al. (2018) then proposed a graph state LSTM (GS LSTM) architecture for the task. They consider that there are two limitations in Peng et al. (2017)’s architecture. First, Peng et al. (2017) can use only the information from a single direction (either left-to-right or right-to-left) in each LSTM layer. Second, Peng et al. (2017) do not utilize the dependency edge type. Therefore, Song et al. (2018)’s GS LSTM uses a message-passing mechanism where each token can pass the message to itself in the next LSTM layer, and all hidden states of its dependency words can be passed as well. To address the second problem, they proposed a variation of the LSTM layer, which allows inputs to be weighted according to edge types. GS LSTM achieved an accuracy of 83.2%, which outperforms Peng et al. (2017)’s architecture by 2.5%.
Most recently, Guo et al. (2019) introduced an attention-guided graph convolutional networks (AGGCN) model. They consider that the structure of the dependency graph that limits the token can be updated only by its edges. Therefore, they proposed an attention-guided layer to transform the dependency graph, with the word embedding added into the weighted fully connected matrix, where each cell represents the strength of the edge. They, then proposed a densely connected layer to allow each token to receive a message (hidden state) from all tokens of the previous sub-layer. Finally, Guo et al. (2019) aggregate the sentence representation and entity representation to predict the relation type. The AGGCN achieved an accuracy of 87.0% and outperformed Song et al. (2018)’s architecture by 3.8%. However, we also found that Song et al. (2018)’s open-source implementation has a significant improvement over their original results (see our Experiment section).
2.2 Abstract-level chemical-induced disease relation extraction
Xu et al. (2016) proposed a Support Vector Machine (SVM)-based approach to classify CDR that involves two feature-based classifiers: sentence-level and abstract-level. Their system achieved an F1-measure of 50.73%. Zhou et al. (2016) proposed an ensemble approach for the CDR task. They developed three classifiers: tree-kernel-based, feature-based and LSTM-based. Their system achieved an F1-measure of 56%. In addition, they proposed some corpus-specific post-processing rules for boosting performance to 61.31% in F1-measure.
In 2018, Li et al. (2018) introduced recurrent piecewise convolutional neural networks (RPCNN). In their formulation, a candidate means a unique CDR-ID pair, and an instance means a CDR-NE pair. Therefore, one candidate can have multiple instances. They also proposed that the piecewise convolutional neural networks (PCNN) represent the instances of the same candidate as a means to predict the relation type. The recurrent neural network is used to aggregate the hidden states of the instances from the same candidate to predict the relation type. Their architecture achieved an F1-measure of 59.1%.
Recently, Verga et al. (2018) proposed Bi-affine Relation Attention Networks (BRANs). They used the Transformer architecture to encode the input text, and the output sequence of the Transformer is passed into two separate multi-layer perceptrons (MLP): head MLP and tail MLP. The first entity representation is selected from the output of the head MLP, and the second entity representation is selected from the output of the tail MLP. Verga et al. (2018) used a bi-affine function to calculate the correlation between two entity representations and update the hidden sequence. Finally, they aggregate the entity representations of the hidden sequence to calculate the softmax of relation type. Their architecture achieved an F1-measure of 62.1%.
2.3 Graph Transformer
Cai and Lam (2020) proposed a graph transformer architecture for the tasks of the abstract meaning representation and the syntax-based machine translation. They proposed a relation-enhanced global attention mechanism that employed gated recurrent unit (GRU) for learning the relation represent of two nodes, then append the mechanism to the self-attention layer. However, their method does not take generalized pre-trained weights learned with large datasets. Instead, we use the original Transformer and our Graph Transformer (GT) simultaneously. By doing so, the original Transformer part can readily use the pre-trained weights, and only pre-trained word embeddings are needed for the GT part. Therefore, our BERT-GT model is easier to adapt to different languages or text compared with Cai and Lam (2020).
3 Materials and methods
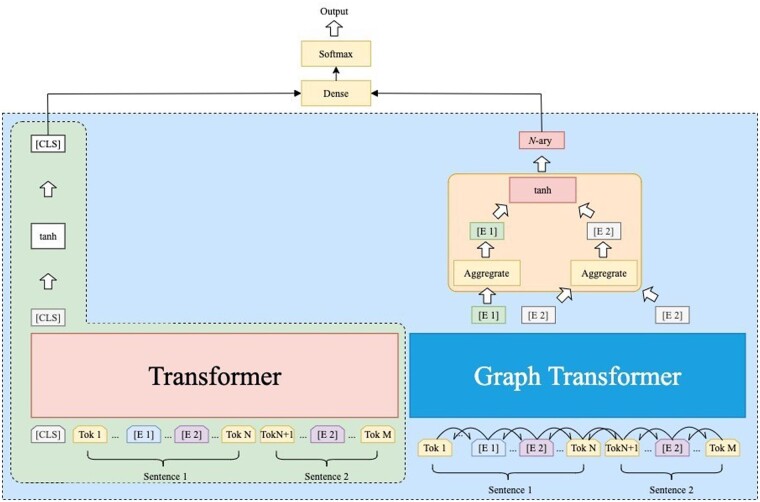
In this section, we introduce BERT-GT and its detailed implementation. The BERT-GT architecture is illustrated in Figure 2, and can be seen as having four parts: (i) input representations for two Transformers, (ii) Transformer, (iii) Graph-Transformer and (iv) layers for aggregating the output states of two Transformers.
Fig. 2.
BERT-GT architecture
The input representations are a tokenized paragraph and its directed graph. The Transformer is the same as BERT’s Transformer, and we take it from BERT, which allows BERT-GT to reuse the pre-trained weights from Lee et al. (2019). GT uses an architecture similar to that of the Transformer but has two modifications. First, the input of GT requires the neighbors’ positions for each token. Second, the self-attention mechanism is replaced with a neighbors–attention mechanism, whereby each token’s output value is calculated by only its neighbor tokens. Finally, we aggregate the hidden states of two Transformers and use the softmax function to calculate the probability of each label. Here, we describe the problem formulation for the cross-sentence n-ary and the CDR tasks, and then we introduce BERT-GT.
3.1 Problem formulation
In our formulation, we assume the text T and entities E are given. Although an input text can be of any arbitrary length, cross-sentence relations are commonly seen within paragraphs instead of across paragraphs or in multiple sections/chapters. Similar to the previous works, we only evaluated BERT-GT on PubMed abstracts. Accordingly, the length of the input text T is within the maximum of an abstract length. Further, an entity can appear more than once in the text and can have more than one ID. Thus, we expand the entity by its IDs. If an entity with many IDs, we will expand the entity into multiple entities, and each has a unique ID. In other words, if an entity of a training/test instance has two IDs, then we will expand it into two training/test instances. Each entity in an instance has a unique ID after the expansion. We follow the problem definition of Guo et al. (2019). The classification problem is defined as whether a relation R holds for the text and an entity subset . In the cross-sentence n-ary task, we assume that given a paragraph that contains a variant/mutation , a gene and a drug . The classification problem checks whether holds for the triple. In the abstract-level chemical-induced disease classification task, a chemical and a disease can appear in a abstract multiple times. Each and can have one or more chemical ID ( and disease ID (). The classification task can be defined as checking whether holds for the pair.
3.2 The graph transformer for the BERT
As noted, BERT-GT is illustrated in Figure 2. The Transformer part is adapted from the BERT model (Peng et al., 2019) and allows us to use the pre-trained model from a large set of biomedical literature. The input of the Transformer is BERT’s preprocessed word embeddings (Devlin et al., 2019) of the text , where denotes the th entity, and the same entity can appear in more than once. For example, in Figure 2, appears twice in the .
The input of GT consists of two representations. One is BERT’s preprocessed word embeddings, which is the same as the Transformer part. Another is the neighbors of each token. For the n-ary dataset, we use the edges provided by the original dataset, and these edges are generated using the NLTK dependency parser and some heuristic rules. These edges are utilized in many of the previous methods. Because CDR does not provide dependency edges, we use ScispaCy (Neumann et al., 2019) to parse the paragraph into dependency trees, and the headword and the adjacent two words of each token are treated as neighbors. Notably, many studies (Miwa and Bansal, 2016; Peng et al., 2018; Xu et al., 2015) show that the use of the shortest path between entities can improve the relation classification. Therefore, for each entity, we also use the headwords between the entity with the other entities as its neighbors.
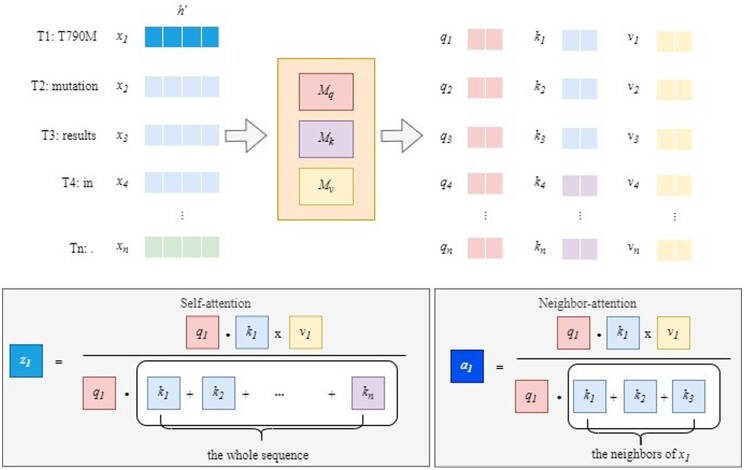
The architectures of the Transformer (Vaswani et al., 2017) and GT are illustrated in Figure 3. GT replaces the self-attention layer of Transformer with the neighbor–attention layer, and and denote the number of gray block layers.
Fig. 3.
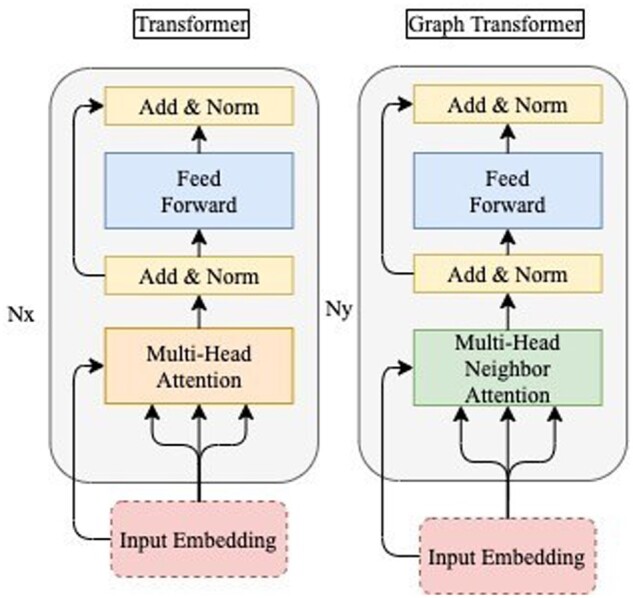
Transformer and Graph Transformer architectures
In the multi-head self-attention mechanism, each input token representation is divided into sub-representations of the input token, and is the number of heads. Assume that the size of the input token representation is , and the size of input token representation for each attention is divided by . Here, we use to denote the size of input token for each attention in Figure 4, which also illustrates that self-attention is neighbors-attention. Each is transformed into , and by multiplying the learnable weighted matrices of , and . The blue vectors of denote the neighbors of . Self-attention calculates by dividing the summation of , multiplying by in the text. Although the text length is longer, may suffer from the noisy message. In contrast, we propose a neighbors–attention mechanism, whereby we use only the neighbors of to calculate the neighbors–attention value . As illustrated in Figure 4, the vectors in blue represent the neighbors of .
Fig. 4.
Two different attention mechanisms. For simplification, we use single cells below to represent the vectors above
3.3 The output layer of the BERT-GT
For the classification task, we obtain the sentence representation of the Transformer and the entity representation of GT. The same entity in different positions is aggregated by using the mean average, as illustrated in Figure 2. We then concatenate the mean averages of different entities to form the entity representation. We also evaluate the effect of using [CLS] for the graph transformer (see our Supplementary Materials). Both sentence representation, for which we choose the output state of the Transformer’s first token, and entity representation are passed into the linear transformation layer and follow the dense and softmax layers for the classification. The dimension of the output layer is the same as the number of labels.
4 Experimental results
4.1 Experimental setup
To test the generalizability of our aproach, we evaluate our method on two independent tasks: (i) Peng et al. (2017)’s n-ary dataset for the cross-sentence n-ary relation extraction task; and (ii) Li et al. (2016) and Wei et al. (2016)’s CDR dataset for the abstract-level relation classification task. We also evaluate our BERT-GT on traditional sentence-level biomedical relation extraction tasks (see our Supplementary Materials).
For both tasks, we compare with four state-of-the-art models: (i) BERT model (Devlin et al., 2019); (ii) BlueBERT model (Peng et al., 2019); (iii) GS LSTM model (Song et al., 2018); and (iv) AGGCN model (Guo et al., 2019), through directly using their online source code. Besides, we evaluate the performances of only using graph transformer (GT) architecture. Note that we do not run the state-of-the-art CDR systems on n-ary due to two main constraints. First, most CDR systems (Li et al., 2018; Xu et al., 2016; Zhou et al., 2016) are not publicly available for retraining on new datasets. Second, the CDR task contains only the relation between two entities. Therefore the features/architectures of these systems cannot be directly used to classify the relation of n entities. In contrast, the n-ary systems can be adapted to classify the relation of two entities in abstract-length text.
4.2 Datasets
The n-ary datasets involve 6987 drug-gene-mutation (ternary) relation instances and 6087 drug-mutation (binary) relation instances, and each instance is categorized into one of the five classes: ‘resistance,’ ‘response,’ ‘resistance or nonresponse,’ ‘sensitivity,’ and ‘none.’ Following Peng et al. (2017)’s experiment settings, we also considered the two-class classification task, whereby the class ‘none’ is treated as ‘no’ and the remaining classes are treated as ‘yes.’ There are, however, two limitations to the n-ary datasets. First, although the n-ary dataset is cross-sentence, the number of sentences is limited to three per instance. Second, the numbers of instances with ‘no’ and ‘yes’ labels are balanced but are not representative of actual biomedical text. Therefore, we also used the BioCreative CDR (Wei et al., 2016) dataset for the evaluation.
The BioCreative CDR dataset consists of 1500 PubMed abstracts and is annotated with 4409 chemicals, 5818 diseases and 3116 chemical-disease relations. The annotations include the spans of entities and their corresponding MeSH IDs. The relations between chemicals and diseases were annotated at the abstract-level, which gives only the chemical-disease MeSH ID pair. In the CDR task, 1000 annotated abstracts are released for participants, and 500 annotated abstracts are used as the test set. Previous work (Xu et al., 2016) shows that approximately 30% of positive CDR instances are cross-sentence relations. The sizes and comparison of the two datasets are summarized in Table 1.
Table 1.
Sizes of the -ary and the CDR datasets. ‘dgm’ means ‘drug-gene-mutation’; ‘dm’ means ‘drug-mutation’
| n-ary | CDR | |
|---|---|---|
| NE types | Drug, gene, mutation | Chemical, disease |
| Normalization | None | MeSH ID |
| Relation type | ‘resistance or non-response’, ‘sensitivity’, ‘response’, ‘resistance,’ and ‘none’ | Positive and negative (no relation) |
| Number of instances |
Resistance or non-response: 1479 dgm and 1259 dm instances;
Sensitivity: 1149 dgm and 1044 dm instances; Response: 488 dgm and 501 dm instances; Resistance: 292 dgm and 327 instances; None: 3582 dgm and 2956 dm instances; |
3116 positive pairs; 13477 negative pairs |
| Size of dataset set | A total of 6987 dgm and 6087 dm instances for the five-fold cross-validation | Training set: 500 abstracts; development set: 500 abstracts; test set: 500 abstracts |
4.3 Evaluation metrics
Previous works used different evaluation metrics for the n-ary nnd CDR tasks. To compare our method with earlier methods, we follow their evaluation metrics for the two tasks. For the n-ary dataset, we use five-fold cross-validation and report the average test accuracy. The partitions of the five-fold cross-validation are the same as those of prior work (Peng et al., 2017). Similar to Song et al. (2018) and Guo et al. (2019), we randomly selected 200 instances from the non-test fold as a held-out set for validation and tuning of hyperparameters. For the CDR dataset, we report the precision, recall and F1-measure on the test set, as the corpus is already divided into the training, development and test sets. We merged training and development sets and randomly selected 200 abstracts for validation and tuning of hyperparameters. Besides, we evaluate the computational costs and effects of different hyper parameters (see our Supplementary Materials).
4.4 Results of the cross-sentence n-ary dataset
The performance of the previous state of the art and all comparing methods is shown in Table 2. ‘Single’ means that we report the accuracy of instances within single sentences, while ‘All’ means the accuracy of all instances. It is of note that the released version of the GS LSTM shows better performance than what was reported in Song et al. (2018). Its performance is also higher than that of AGGCN, especially on the multi-class drug-gene-mutation evaluation metric. Besides, we compare with the pure GT method that removes the Transformer from our BERT-GT. GT scores significantly higher than AGGCN on multi-class, but lower than GS GLSTM. The lower performance might be because GT weights the neighbors by using the neighbor–attention mechanism, and does not utilize the edge type information which is used in GS GLSTM.
Table 2.
Accuracy of different methods on the n-ary test set
| Model | Drug-Mutation |
Drug-Gene-Mutation |
||||
|---|---|---|---|---|---|---|
| Two-class |
Multi-class | Two-class |
Multi-class | |||
| Single | All | All | Single | All | All | |
| Logistic regression (Peng et al., 2017) | 73.90 | 75.20 | – | 74.70 | 77.70 | – |
| Graph LSTM EMBED (Peng et al., 2017) | 74.30 | 76.50 | – | 76.50 | 80.60 | – |
| Graph LSTM FULL (Peng et al., 2017) | 75.60 | 76.70 | – | 77.90 | 80.70 | – |
| GS GLSTM (Song et al., 2018) | 88.15 | 88.56 | 86.92 | 82.86 | 87.60 | 85.91 |
| AGGCN (Guo et al., 2019) | 85.78 | 85.36 | 76.88 | 86.06 | 86.44 | 79.62 |
| GT | 86.04 | 83.69 | 84.62 | 82.85 | 83.67 | 83.65 |
| BERT (Devlin et al., 2019) | 88.59 | 91.01 | 90.03 | 83.75 | 90.80 | 90.73 |
| BlueBERT (Peng et al., 2019) | 89.81 | 92.10 | 92.16 | 85.39 | 91.27 | 91.10 |
| BERT-GT | 91.67 | 93.27 | 93.45 | 85.49 | 91.51 | 91.35 |
Bold indicates that it is the highest score among all models.
Table 2 shows that our BERT-GT method outperforms all of the previous methods. Compared with AGGCN, our method outperforms it by accuracies of 5.07% and 11.73% on two-class and multi-class drug-gene-mutation evaluations, respectively. According to our observation of the error cases that are wrong in the AGGCN but are correct in BERT-GT, the relation information of these cases is less explicit. For example, ‘On the other hand, erlotinibDRUG could not inhibit EGFRGENE phosphorylation in H1975 cells because the T790MMUTATION mutation in EGFR causes a conformation change at the ATP binding pocket, thus decreasing the affinity between erlotinib and EGFR.’ The above case was predicted as ‘None’ because both ‘not’ and ‘inhibit’ indicate a negative relationship. If we take both terms into consideration, however, this indicates a positive relationship: ∼26% of the cases contain negation words, ‘not’ and ‘no.’
In addition, most methods show lower performances on multi-class evaluation. GS GLSTM, BlueBERT and our BERT-GT method, however, show that their multi-class evaluation can retain the same level of performance as do their two-class evaluations. In addition, our method outperforms BlueBERT on the drug-mutation relation classification by 1.17% and 1.29% on two-class and multi-class, respectively.
4.5 Results on the CDR dataset
The performance of our method and recent methods are shown in Table 3. Here, we report only the performance of systems without using domain knowledge in order to compare methods that can be more generalizable. For the same reason, we report their performance that involves no post-processing or ensemble. Table 3 shows that BERT-GT outperforms all of the other methods. In fact, BERT-GT compares favorably to others even when they use additional post-processing methods or ensemble (Gu et al., 2017; Verga et al., 2018; Zhou et al., 2016). These results show that our system has the potential to be adapted to other domains. In addition, our method outperforms BlueBERT by an F1-measure of 2.38%. Considering the size of the CDR test set is small, we implement a statistically significant test to compare BERT-GT and BlueBERT (see our Supplementary Materials).
Table 3.
Performance on the CDR test set in comparison with state-of-the-art systens
| Model | P | R | F |
|---|---|---|---|
| CD-REST (Xu et al., 2016) | 59.60 | 44.00 | 50.73 |
| Feature-TreeK-LSTM (Zhou et al., 2016) | 64.89 | 49.25 | 56.00 |
| + post-processing | 55.56 | 68.39 | 61.31 |
| CNN (Gu et al., 2017) | 60.90 | 59.50 | 60.20 |
| + post-processing | 55.70 | 68.10 | 61.30 |
| RNN-CNN (Li et al., 2018) | 55.20 | 63.60 | 59.10 |
| BRAN (Verga et al., 2018) | 55.60 | 70.8 | 62.10 |
| + ensemble | 63.30 | 67.10 | 65.10 |
| GS LSTM (Song et al., 2018) | 42.31 | 39.21 | 40.70 |
| AGGCN (Guo et al., 2019) | 94.23 | 19.46 | 32.26 |
| GT | 30.04 | 74.67 | 42.84 |
| BERT (Devlin et al., 2019) | 61.41 | 58.82 | 60.09 |
| BlueBERT (Peng et al., 2019) | 62.80 | 64.45 | 63.61 |
| BERT-GT | 64.94 | 67.07 | 65.99 |
Bold indicates that it is the highest score among all models.
GS LSTM and AGGCN, however, show lower recall on the CDR dataset. We observed 25 false-negative cases that are wrong in AGGCN but correct in our proposed method. In ∼65% of these cases, the chemical and disease pairs co-occurred in a sentence at least once. AGGCN, however, cannot identify them correctly. Note that the maximum length of the input text is only three sentences in the n-ary dataset, and each instance in the n-ary dataset consists of only one annotated drug-gene-mutation. In contrast, the CDR dataset is in abstract-length, a chemical can be mentioned in different locations of the article. A chemical-disease can have a positive relation at one location in the abstract but have no relation at the rest of the locations. Hence, it is sometimes challenging to use a graph to distinguish the positive location from the negative ones. As a result, unlike our approach BERT-GT, AGGCN and GS LSTM are not robust and generalizable to another domain or a new dataset without additional effort.
5 Discussion
5.1 The performance improvement as a result of adding the graph transformer
We randomly sampled 50 error cases that were wrong in BERT’s predictions but correct in BERT-GT from the n-ary test set. The most common error cases of BERT are the instances in which ‘None’ labels were assigned to instances with other labels. These cases constitute approximately 56% of the sampled error cases. For example, ‘Interestingly, cells with these mutations also showed greater sensitivity to gefitinib and erlotinibDRUG than cells with the EGFRGENE mutation (exon 19 deletion), which are associated witsh sensitivity to EGFR inhibitors in NSCLC. Mutations in PI3K (H1047RMUTATION) have been shown to enhance HER2 mediated transformation by amplifying the ligand induced signaling output of the HER family of RTKs.’ The BERT model predicts this as ‘resistance or non-response.’ Almost all of these cases are multiple-sentence instances, and it seems that the long instances with some keywords, such as ‘sensitivity,’ ‘associated,’ and ‘enhance,’ which commonly appear in the positive instances, are not used to express the relationship of these entities and can result in the wrong prediction. In addition, we found that 20% of the sampled cases are intra-sentence instances. For example, ‘Consistent with this, we found that the introduction of R1275QVARIANT into EML4-ALKGENE had no negative impact on sensitivity to crizotinibDRUG (IC50 47 ± 8 nm) .’ This is misclassified as ‘resistance’ by the BERT model instead of its ground truth ‘sensitivity.’ It seems that, although BERT can use the key information from cross-sentence, it also is relatively easier to make an incorrect prediction by using that information and that training on cross-sentence also makes it easier to misclassify short text. In contrast, BERT-GT suffered from fewer of these problems, though a GT-only model does not perform well in our observation. Taken together, this suggests the strengths of GT in either focusing on the neighbor information or predicting short-text instances, when built with the default BERT model.
5.2 Error analysis, limitations and future directions
Here, we discuss the error cases of BERT-GT and future research directions. According to our observation of the BERT-GT error cases on the CDR dataset, in 61% of the error cases, either chemical or disease mentions occurred only once. For example, in the PMID : 25986755, the chemical ‘caffeine’ appears only in the first two sentences of the abstract, and the disease ‘dysplasia’ appears only once in the last sentence of the abstract. There is, however, a positive relationship between ‘caffeine’ and ‘dysplasia’ but no explicit information that mentions this relationship; thus, it was misclassified as no relation. Among these error cases, 75% do not have any single sentence that mentions both the target chemical and disease. Therefore, if the entity appears only once in the entire abstract and does not provide explicit information, BERT-GT is likely to generate the wrong predictions.
We also observed that there are some limitations to the n-ary dataset. For example, the n-ary dataset is a balanced dataset in which the numbers of positive and negative instances are balanced, which is not common in biomedical domains, such as CDR. Further, the sentence length of their instances is no more than three, but, in the biomedical domain, the sentence length of the text is usually longer. Therefore, we consider it a research direction to develop a more representative benchmark for evaluating n-ary RE tasks in the future.
6 Conclusion
In the biomedical literature, a biomedical relation usually consists of multiple entities and is represented in multiple sentences. The limitations on the architectures/features of previous methods are that they can perform well only on either cross-sentence n-ary relation extraction or CDR extraction. In this work, we propose a BERT-GT method. We show that BERT can be used to classify a cross-sentence relation, such as a PubMed abstract, because the attention mechanism makes it able to utilize the key information from the whole text and thus make the prediction. Our GT allows the BERT to use the graph information, which provides the neighbors of each token. The neighbors can be specified to focus on specific information for each token. The results demonstrate that BERT-GT achieves the highest performance on both problems and demonstrates its potential to be applied to a more generalizable relation classification problem.
Supplementary Material
Acknowledgements
The authors are grateful to the authors of BERT, BlueBERT, BioBERT, GS LSTM and AGGCN who make their data and source code publicly available. They would like to thank Dr. Zhijiang Guo for helping us to reproduce the results of AGGCN on the n-ary dataset. They thank Dr. Chih-Hsuan Wei for his assistance on revising the manuscript.
Funding
This research was supported by NIH Intramural Research Program, National Library of Medicine.
Conflict of Interest: none declared.
Contributor Information
Po-Ting Lai, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
Zhiyong Lu, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20894, USA.
References
- Cai D., Lam W. (2020) Graph transformer for graph-to-sequence learning. In: Proceedings of Thirty-Fourth Association for the Advancement of Artificial Intelligence (AAAI) Conference on Artificial Intelligence, Vol. 34 No. 05: AAAI-20 Technical Tracks 5, New York, USA. February 2020. pp. 7464–7471. AAAI Press, Palo Alto, CA, USA.
- Clematide S., Rinaldi F. (2012) Ranking relations between diseases, drugs and genes for a curation task. J. Biomed. Semantics, 3, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J. et al. (2019) Bert: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (NAACL-HLT), Vol. 1, June 2019. pp. 4171–4186. Association for Computational Linguistics, Minneapolis, MN, USA.
- Fiorini N. et al. (2018) How user intelligence is improving PubMed. Nat. Biotechnol., 36, 937–945. [DOI] [PubMed] [Google Scholar]
- Gu J. et al. (2017) Chemical-induced disease relation extraction via convolutional neural network. Database, 2017, bax024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z. et al. (2019) Attention guided graph convolutional networks for relation extraction. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics, Florence, Italy. pp. 241–251.
- Lee J. et al. (2019) BioBERT: pre-trained biomedical language representation model for biomedical text mining. Bioinfomatics, 36, 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. et al. (2020) Recent advances of automated methods for searching and extracting genomic variant information from biomedical literature. Brief. Bioinf., [Epub ahead of print, doi: 10.1093/bib/bbaa142, August 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Ji H. (2019) Syntax-aware multi-task graph convolutional networks for biomedical relation extraction. In Proceedings of the Tenth International Workshop on Health Text Mining and Information Analysis (LOUHI 2019), November 2019. pp. 28–33. Association for Computational Linguistics, Hong Kong.
- Li H. et al. (2018) Chemical-induced disease extraction via recurrent piecewise convolutional neural networks. BMC Med. Inf. Decis. Mak., 18, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. (2016) BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database, 2016, baw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M., Bansal M. (2016) End-to-end relation extraction using LSTMS on sequences and tree structures. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics, Berlin, Germany. pp. 1105–1116.
- Neumann M. et al. (2019) ScispaCy: fast and robust models for biomedical natural language processing. In: Proceedings of the 18th BioNLP Workshop and Shared Task. August 2019. pp. 319–327. Association for Computational Linguistics, Florence, Italy. [Google Scholar]
- Peng N. et al. (2017) Cross-sentence n-ary relation extraction with graph lstms. Trans. Assoc. Comput Linguist., 5, 101–115. [Google Scholar]
- Peng Y. et al. (2018) Extracting chemical–protein relations with ensembles of SVM and deep learning models. Database, 2018, bay073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. et al. (2019) Transfer learning in biomedical natural language processing: an evaluation of BERT and ELMo on ten benchmarking datasets. In Proceedings of the 2019 Workshop on Biomedical Natural Language Processing (BioNLP 2019). pp. 58–65, August 2019. Association for Computational Linguistics, Florence, Italy.
- Quirk C., Poon H. (2017) Distant supervision for relation extraction beyond the sentence boundary. In Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics. Vol. 1, Long Papers. Association for Computational Linguistics, Valencia, Spain. pp. 1171–1182.
- Sidhoumi D. et al. (2010) Evaluation of the therapeutic effect of theta burst stimulation on drug-resistant auditory hallucinations in a schizophrenic patient and its impact on cognitive function and neuronal excitability: a case study. Clin. Neurophysiol. Off. J. Int. Federation Clin. Neurophysiol., 121, 802. [DOI] [PubMed] [Google Scholar]
- Song L. et al. (2018) N-ary relation extraction using graph-state LSTM. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, Brussels, Belgium. pp. 2226–2235.
- Vaswani A. et al. (2017) Attention is all you need. In: Advances in Neural Information Processing Systems. vol. 30, pp. 5998–6008. Curran Associates, Inc., Long Beach, California, USA.
- Verga P. et al. (2018) Simultaneously self-attending to all mentions for full-abstract biological relation extraction. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers). Association for Computational Linguistics, New Orleans, Louisiana. pp. 872–884.
- Wei C.-H. et al. (2016) Assessing the state of the art in biomedical relation extraction: overview of the BioCreative V chemical-disease relation (CDR) task. Database, 2016, baw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. et al. (2016) CD-REST: a system for extracting chemical-induced disease relation in literature. Database, 2016, baw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K. et al. (2015) Semantic relation classification via convolutional neural networks with simple negative sampling. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, Lisbon, Portugal. pp. 536–540.
- Zhou H. et al. (2016) Exploiting syntactic and semantics information for chemical–disease relation extraction. Database, 2016, baw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.