Fig. 3.
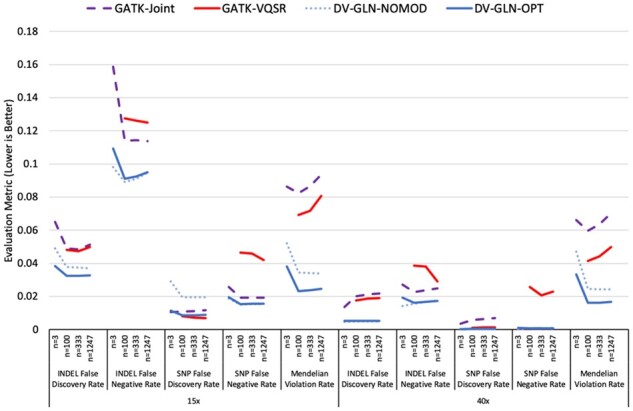
Comparison of four cohort callset creation methods. Four calling and merging pipelines are applied at both 15× and 40× sequence coverage for WGS cohorts of size n = 3, 100, 333 and 1247. Five evaluation metrics are presented: Mendelian Violation Rate, SNP False Discovery Rate (1-Precision), SNP False Negative Rate (1-Recall), indel False Discovery Rate and indel False Negative Rate. In all cases, lower values are better. All evaluation metrics are computed on chr20. See Supplementary Table S5 for the precise values and the variances of each metric
