Abstract
RNA therapeutics have high potential that is yet to be fully realized, largely due to challenges involved in the appropriate delivery to target cells. Extracellular vesicles (EVs) are lipid bound nanoparticles released by cells of all types and possess numerous features that may help overcome this hurdle and have emerged as a promising RNA delivery vehicle candidate. Despite extensive research into the engineering of EVs for RNA delivery, it remains unclear how the intrinsic RNA delivery efficiency of EVs compares to currently used synthetic RNA delivery vehicles. Using a novel CRISPR/Cas9-based RNA transfer reporter system, we compared the delivery efficiency of EVs to clinically approved state-of-the-art DLin-MC3-DMA lipid nanoparticles and several in vitro transfection reagents. We found that EVs delivered RNA several orders of magnitude more efficiently than these synthetic systems. This finding supports the continued research into EVs as potential RNA delivery vehicles.
Keywords: extracellular vesicles, exosomes, LNP, RNA therapeutics, nanomedicine
RNA therapeutics possess great therapeutic potential as they target disease at its genetic source in a highly selective manner.1 In order to function, RNA therapeutics must reach therapeutic concentrations within the cytosol of specific target cells. However, numerous obstacles prevent therapeutic RNA from reaching its site of action. For example, free circulating RNA is subject to renal clearance while extracellular RNases degrade unprotected RNA.2 Even if an RNA molecule is able to reach its target cell it remains unable to cross the plasma membrane due to its large size and charge.3
To bypass these barriers, therapeutic RNA cargo can be delivered inside synthetic nanoparticle (NP) carriers such as lipid nanoparticles (LNPs), which protect the delicate RNA from degradation and facilitate uptake into recipient cells.4 However, synthetic systems are hindered by their own set of challenges. They can be highly immunogenic5 and are subject to uptake and clearance by Kupffer cells of the liver.6 Furthermore, upon cellular uptake, most NPs are destined for lysosomal degradation.7
Extracellular vesicles (EVs) are lipid bound NPs of biological origin. They are released from all cell types, range from 30 to 2000 nm in diameter8 and are involved in intercellular transfer of biological cargo, including RNA.9 Potentially, EVs could avoid the toxicity and immunogenicity that hamper the use and development of clinically effective synthetic NPs.10 In addition, EVs have been shown to be capable of crossing biological barriers and possess endogenous targeting ability.11 These features make EVs an interesting candidate for an RNA delivery vehicle. The concept of hijacking this endogenous RNA transfer system to deliver therapeutic RNAs is an attractive one but despite their advantages, EVs, like synthetic NPs, must bypass cellular barriers in order to release their cargo to the cytosol.
There are numerous examples of EVs functionally delivering RNA in (patho)physiology. A striking example is that of hsa-miR-21, which is relatively highly abundant in EV preparations. The EV-mediated transfer of hsa-miR-21 is strongly implicated in tumor growth and progression.12,13,16 It has been speculated that, to achieve this functional delivery, EVs must be highly efficient at bypassing cellular barriers14 as RNA loading of even the most abundant miRNAs is as low as 1 copy per 100 EVs with other miRNAs present in quantities several orders of magnitude lower.15 For example, despite the suggested pathophysiological function of EV-associated hsa-miR-21, the loading of this RNA into the relevant EVs has been found to be far lower than 1 copy per particle.16 Such analyses of the stoichiometry of functional RNA in EVs are rare, meaning the efficiency of EV-mediated RNA transfer remains unclear.
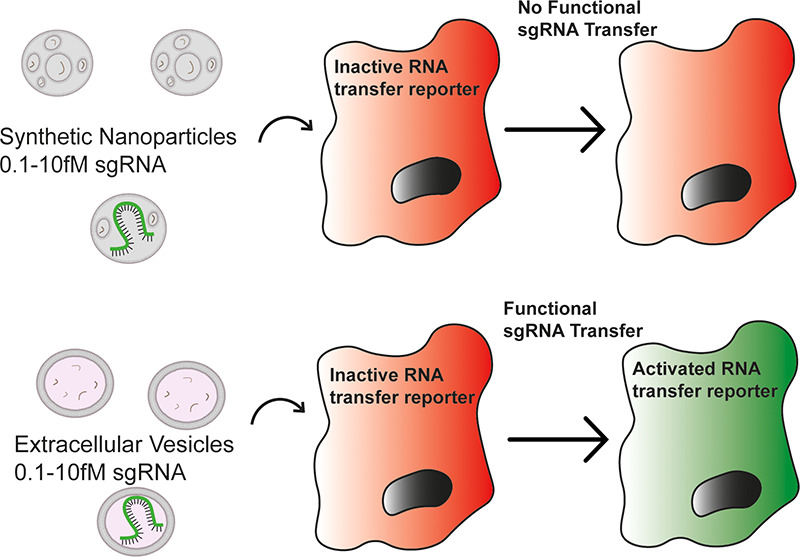
This lack of clarity is partly due to the absence of a suitable assay to allow for the appropriate stoichiometric study of EV-mediated RNA delivery with sufficient sensitivity.17 siRNA or miRNA-mediated gene knockdown as a read-out for RNA delivery is insensitive as it relies on the bulk measurement of cell populations. The Cre recombinase reporter assay used to demonstrate the EV-mediated transfer of mRNA both in vitro and in vivo(18) is able to detect the transfer of mRNA with single-cell resolution. However, it can also be activated by the transfer of miniscule quantities of Cre protein derived from donor cell translation of Cre mRNA,17 the presence of which has been confirmed in Cre mRNA+EVs.19 In addition, experiments assessing the functional transfer of RNA by EVs based on phenotypical changes or expression of endogenous genes can be confounded by the fact that the simple addition of nanoparticles alters cellular behavior.20 To further study the efficiency of EVs, a highly sensitive reporter system with single-cell resolution which can only be activated by RNA transfer is required.
To address this need, we developed the highly sensitive and specific CRISPR Operated Stoplight System for Functional Intercellular RNA Exchange (CROSS-FIRE) reporter system. This system is only activated by the functional transfer of a specific sgRNA and allows the detection of EV-mediated RNA delivery at the single cell level (Figure S1).21 Here we use the CROSS-FIRE system to compare the delivery efficiency of EVs isolated from MDA-MB-231 and A431 cells to in vitro transfection reagents and state-of-the-art DLin-MC3-DMA-LNPs, which are the most advanced therapeutic RNA delivery system available and are used for clinical delivery of siRNAs targeting transthyretin under the name Onpattro (Patisiran).22
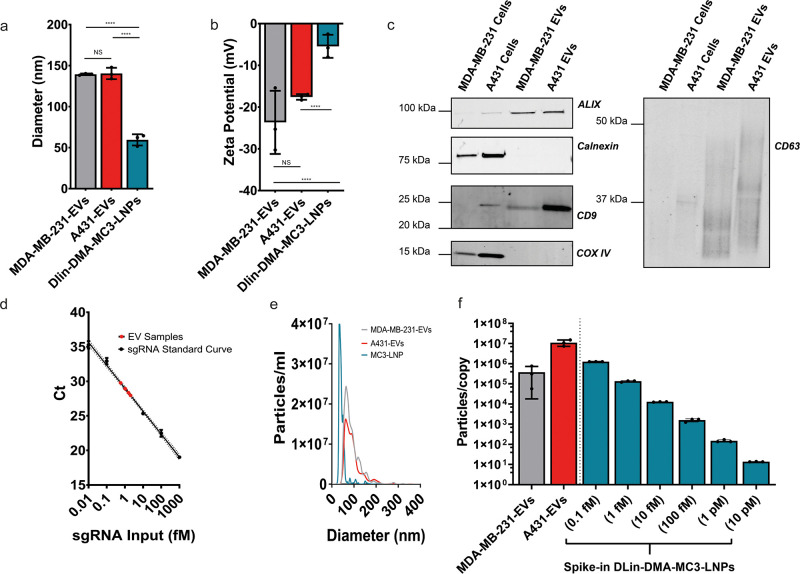
First, EVs were isolated from both MDA-MB-231 and A431 cells by using size exclusion chromatography. DLin-MC3-DMA LNPs were produced by microfluidic mixing. The sizes of these NPs were then determined by using dynamic light scattering (DLS) (Figure 1a). MDA-MB-231-EVs and A431-EVs possessed mean diameters of 139 ± 1 and 140 ± 7 nm, respectively. DLin-MC3-DMA-LNPs were smaller at 59 ± 7 nm, which is comparable to Onpattro which possesses a size of less than 100 nm.23 This may be of significance as endocytosis of nanoparticles can be influenced by particle size, however also by other factors such as shape, rigidity, and the presence of surface ligands.24
Figure 1.
Physical characterization of NPs. DLS analysis of EVs and synthetic nanoparticles (a). ζ potentials of EVs and synthetic nanoparticles (b). Western blot analysis of MDA-MB-231 and A431 cell lysates alongside A431-EVs and MDA-MB-231-EVs for EV markers (ALIX, CD9, CD63) and EV-negative markers (Calnexin and Cox IV) (c). An RT-qPCR interpolation of sgRNA concentration in MDA-MB-231-EV samples from known sgRNA input standard measurements (d). Nanoparticle Tracking Analysis (NTA) size distributions of MDA-MB-231-EVS, A431-EVs, and DLin-MC3-DMA-LNPs (e). Using RT-qPCR and NTA data, we determined the numbers of EVs per single sgRNA and plotted them next to a similar number of particles per single sgRNA of spike-in DLin-DMA-MC3-LNPs used in comparative experiments (f). Means ± SD are displayed, n = 3 biological replicates, Tukey’s multiple comparison test, * = p < 0.05, ** = p < 0.01, **** = p < 0.0001.
To assess the surface charge of particles, ζ-potential analysis was performed (Figure 1b). As is typical of EVs, MDA-MB-231-EVs and A431-EVs possessed negative surface charges of −24 ± 8 and −18 ± 1 mV respectively. In line with previously reported data,23 DLin-MC3-DMA-LNPs were close to neutral charge with a zeta-potential of −5 ± 3 mV. It should be noted that a positive charge is considered important for the endosomal fusion of LNPs and that while DLin-MC3-DMA-LNPs are neutrally charged at physiological pH, they are known to be positively charged at endosomal pH. The mechanisms by which the negatively charged EVs fuse with endosomal membranes remains unclear, but may involve fusogenic proteins or lipids.25
According to MISEV guidelines,26 Western blot analysis (Figure 1c) was used to confirm positive enrichment of EV markers CD9, CD63, and ALIX as compared to cell lysates. The organelle markers calnexin and COX IV were negatively enriched in EVs confirming an absence of cellular contamination.
To determine the concentration of sgRNA in EV samples, RT-qPCR was performed on sgRNA+MDA-MB-231-EVs and A431-EVs alongside an extraction efficiency-corrected sgRNA standard curve. After RT-qPCR analysis, the sgRNA concentrations of EV samples were interpolated (Figure 1d). It should be also noted that we have previously demonstrated that EV associated sgRNA is located within the EV lumen.21 sgRNA was also confirmed to be stable inside all NPs by using RT-qPCR as no reduction in sgRNA signal was observed after 24 h of incubation at 4 °C for any NP. After 24 h of incubation at 37 °C no reduction in sgRNA signal was observed in A431-EVs and DLin-DMA-MC3-LNPs while a slight but significant drop was observed in MDA-MB-231-EVs (Figure S2).
Using the sgRNA concentration values determined by RT-qPCR and particle concentrations measured by NTA (Figure 1e), we could calculate the quantity of sgRNA per EV. This showed that loading was extremely low with 1 sgRNA/3.6 × 105 ± 3.3 × 105 MDA-MB-231-EVs. sgRNA loading into A431 EVs was approximately 30-fold lower, with 1 sgRNA/1.1 × 107 ± 3.9 × 106 EVs (Figure 1f).
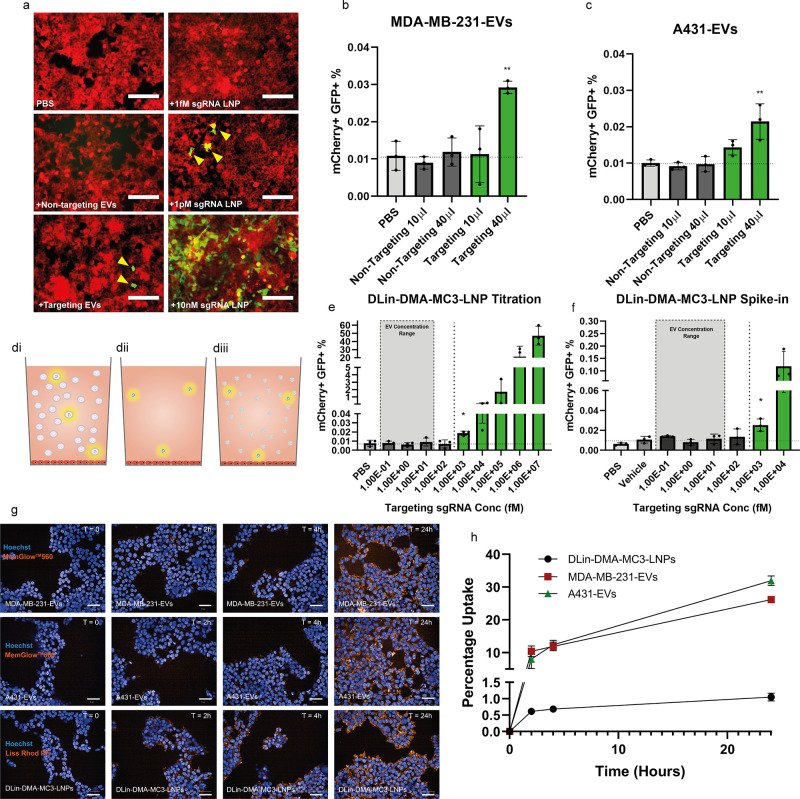
We then tested the ability of EVs to functionally deliver sgRNA. Despite low EV loading, we were able to observe GFP expression in HEK293T CROSS-FIRE reporter cells indicating activation upon 6 daily EV additions (Figure 2a). To demonstrate dose dependence, EVs were applied at two doses. Only the high dose of both EV types was sufficient to induce significant activation as compared to the vehicle control. To rule out nonspecific activation, an equal dose of EVs containing a nontargeting sgRNA was also applied. These nontargeting controls showed no reporter cell activation (Figure 2b,c). In these experiments the average sgRNA high concentration for MDA-MB-231-EV additions was 2.3 ± 2.9 fM while the average A431-EV high concentration was considerably lower at 0.1 ± 0.04 fM. Additional parameters of this experiment can be seen in Table 1.
Figure 2.
EVs functionally deliver RNA at a concentration orders of magnitude lower than those required for synthetic NPs. Fluorescent microscopy images of Stoplight+ spCas9+ HEK293T reporter cells after 6 consecutive daily additions of PBS, nontargeting sgRNA+A431-EVs, targeting sgRNA+A431-EVs or 1 fM, 1 pM, or 10 nM sgRNA DLin-DMA-MC3-LNPs. Scale bars represent 150 μm (a). Flow cytometry analysis of Stoplight+ spCas9+ HEK293T reporter cells after 6 consecutive days of targeting sgRNA+ or nontargeting sgRNA+ MDA-MB-231-EV addition, (b) and 6 consecutive days of targeting sgRNA+ or nontargeting sgRNA+ A431-EV addition (c), n = 3 biological replicates. A graphical representation of the sgRNA composition of samples used for comparative analysis (d): Low abundance of sgRNA copies in a large number of EVs (di), DLin-MC3-DMA-LNPs containing targeting sgRNA only at EV concentration levels (dii), and DLin-MC3-DMA-LNPs containing mainly inert scaffold with targeting sgRNA spiked in to mimic the sgRNA stoichiometry of EV samples (diii). Flow cytometry analysis of Stoplight+ spCas9+ HEK293T reporter cells after 6 consecutive daily doses of DLin-MC3-DMA-LNPs containing targeting sgRNA at a range of 1 × 107 to 1 × 10–1 fM, n = 3 biological replicates (e). Flow cytometry analysis of Stoplight+ spCas9+ HEK293T reporter cells after 6 consecutive daily doses of LNP preparations containing a similar total particle dose to EV samples but with targeting sgRNA spiked in to achieve a targeting sgRNA concentration range of 1 × 104 to 1 × 10–1 fM, n = 3 biological replicates (f). Confocal microscopy images of HEK293T cells at 0, 2, 4, and 24 h after the addition of MemGlow labeled MDA-MB-231-EVs, MemGlow labeled A431-EVs, or Lissamine-Rhodamine PE labeled DLin-DMA-MC3-LNPs (g). Percentage uptake of NPs determined by using a fluorescent plate reader and interpolation from a background corrected standard curve, n = 3 technical replicates (h). Means plus SD are displayed, Tukey’s multiple comparison test was used for statistical analysis of both EV addition experiments and titrations, * = p < 0.05, ** = p < 0.01 versus vehicle.
Table 1. Parameters of Synthetic Nanoparticles and EV Samples Used for Comparative Experimentsa.
| sample | daily sgRNA concentration (fM) | daily sgRNA copy no. | daily particle concn (particles/mL) | daily particle dose | particles/sgRNA |
|---|---|---|---|---|---|
| MDA-MB-231-EVs (high dose) | 2.3E00 (2.9E00) | 3.3E05 (4.2E05) | 1.04E11 (1.3E10) | 2.6E10 (3.2E09) | 3.6E05 (3.3E05) |
| A431-EVs (high dose) | 1.0E-01 (4E-02) | 1.0E04 (5.4E03) | 3.36E11 (9.00E10) | 8.4E10 (2.3E10) | 1.1E07 (3.9E06) |
| sgRNA only- DLin-MC3-DMA-LNPs | 1.0E+07 to 1.0E-01 | 1.51 E12 to 1.51E04 | 2.16E11 (5.4E10) to 2.16E03 (5.4E02) | 5.14E10 (1.3E10) to 5.14E02 (1.3E02) | 3.0E-02 |
| sgRNA spike-in-DLin-MC3-DMA-LNPs (10 000 fM) | 1.0E+04 | 1.5E+09 | 8.0E10 (2.4E09) | 2.0E10 (6.0E08) | 1.4E01 (4.0E-01) |
| sgRNA spike-in-DLin-MC3-DMA-LNPs (1000 fM) | 1.0E+03 | 1.5E+08 | 9.2E10 (1.5E09) | 2.3E10 (3.7E08) | 1.5E02 (2.4E01) |
| sgRNA spike-in-DLin-MC3-DMA-LNPs (100 fM) | 1.0E+02 | 1.5E+07 | 8.8E10 (2.1E09) | 2.2E10 (5.3E08) | 1.5E03 (3.5E02) |
| sgRNA spike-in-DLin-MC3-DMA-LNPs (10 fM) | 1.0E+01 | 1.5E+06 | 7.6E10 (2.5E09) | 1.9E10 (6.2E08) | 1.3E04 (4.1E02) |
| sgRNA spike-in-DLin-MC3-DMA-LNPs (1 fM) | 1.0E+00 | 1.5E+05 | 8.0E10 (8.4E08) | 2.0E10 (2.1E09) | 1.3E05 (1.4E04) |
| sgRNA spike-in-DLin-MC3-DMA-LNPs (0.1 fM) | 1.0E-01 | 1.5E+04 | 7.6E10 (6.4E08) | 1.9E10 (1.6E08) | 1.3E06 (1.1E04) |
| scaffold RNA only-DLin-MC3-DMA-LNPs | 0.0E+00 | 0.0E+00 | 7.2E10 (1.64E09) | 1.8E10 (4.1E08) | NA |
Standard deviations are shown in brackets where applicable.
The fact that EVs caused reporter activation at such low sgRNA concentrations suggests highly efficient delivery. We therefore compared EVs to a state-of-the-art RNA delivery vehicle, the DLin-MC3-DMA-LNP (Onpattro).
DLin-MC3-DMA-LNPs containing targeting sgRNA were produced and a 10-fold serial dilution series from 10 nM to 0.1fM sgRNA was prepared. This covered the average sgRNA concentration delivered per daily dose by both A431-EVs and MDA-MB-231-EVs, which allowed for comparison of CROSS-FIRE activation between EVs and DLin-MC3-DMA-LNPs.
Although this approach allowed the direct comparison of absolute sgRNA concentration between EVs and LNPs, the LNP particle dose was considerably lower at the sgRNA concentrations functionally delivered by EVs. In order to achieve a more comparable sgRNA stoichiometry, DLin-MC3-DMA-LNPs batches were prepared in which the majority of the RNA cargo was composed of an inert scaffold while the sgRNA was spiked in to achieve final targeting sgRNA concentrations ranging from 10 pM to 0.1 fM. This allowed the sgRNA concentration to be titrated while the particle dose remained comparable to EV particle doses (Table 1).
This is visualized in a simplified schematic (Figure 2d) in which sgRNA is delivered by a few highlighted EVs in a background of EVs containing no sgRNA (Figure 2di). An equal amount of sgRNA to that found in EV preparations is delivered by a few highlighted LNP particles delivered from a highly diluted stock LNP preparation (Figure 2dii). To provide a more representative comparison between EVs and LNPs, an equal amount of sgRNA is delivered within a background of empty LNPs (Figure 2diii).
DLin-MC3-DMA-LNPs containing only sgRNA (Figure 2dii) produced a dose dependent response from 10 nM to 1 pM (Figure 2a,e). At 1 pM, activation was 2.5-fold higher than background levels, which is a similar level of activation induced by EVs (Figure 2b,c). Below 1 pM no activation was observed. This is in contrast to EVs, which were able to functionally deliver RNA at these lower concentrations.
When sgRNA was delivered via spike-in DLin-MC3-DMA-LNPs mimicking EV stoichiometry (Figure 2diii) significant activation was observed at 1 pM sgRNA at a level similar to that seen in the direct titration. Again, at concentrations below this, no significant activation was observed (Figure 2f). Taken together, these results indicate that EV-mediated RNA delivery is at least 2 orders of magnitude more efficient than DLin-MC3-DMA-LNP-mediated RNA delivery.
Lastly, in order to allow comparison to commonly used in vitro transfection reagents, we also performed sgRNA titrations by using the transfection reagents lipofectamine RNAiMax, 25 kDa linear polyethylenimine (PEI), and TransIT-2020. The minimal effective dose for lipofectamine RNAiMax and TransIT-2020 was 1 and 10 pM, respectively. PEI induced a clear increase in reporter activation at 100 pM, but a statistically significant minimal effective dose was not reached until 10 nM due to large variation between experiments with this reagent (Figure S3).
These results show that EV-mediated RNA delivery is considerably more efficient than that of synthetic systems. This is demonstrated by the fact that the minimal effective DLin-MC3-DMA-LNP dose (1 pM) was more than 2 orders of magnitude higher than the sgRNA dose required for significant reporter activation when sgRNA was delivered by MDA-MB-231-EVs (2.3 fM). Furthermore, the sgRNA dose delivered by A431-EVs (0.1 fM) was around 4 orders of magnitude lower than the DLin-MC3-DMA-LNP minimal effective dose but was nevertheless capable of inducing significant activation.
There are multiple steps in the uptake and endosomal trafficking process at which EVs could achieve this efficiency. The first step in nanoparticle-mediated RNA delivery is uptake into recipient cells, and there is evidence that EVs may be more efficient at this process than synthetic NPs. For instance, doxorubicin loaded HEK293-EVs have been found to be taken up more rapidly by HEK293 cells and deliver more doxorubicin to the cytosol than a liposome-loaded doxorubicin formulation.27 Furthermore, EV-mimicking liposomes that possessed a lipid composition resembling that of EVs show 3-fold higher rates of cellular uptake as compared to conventional PC-Chol liposomes.28 Therefore, to determine the extent to which uptake efficiency contributed to the differences in RNA delivery efficiency between EVs and DLin-DMA-MC3-LNPs, we compared the uptake of these particles.
To allow visualization and quantification of uptake, MDA-MB-231-EVs and A431-EVs were labeled with MemGlow 560 and DLin-DMA-MC3-LNPs were produced containing 0.2% Lissamine Rhodamine PE. HEK293T cells were then seeded at the same density used in the aforementioned addition experiments. These cells were then treated with a similar particle dose of NPs as used in these experiments. Fluorescent signal was observed inside cells for all NPs after 2 h, and this signal further increased in intensity after 4 and 24 h (Figure 2g), confirming uptake.
To compare the uptake efficiency between these NPs, HEK293T cells were dosed by using the same setup used for the confocal microscopy experiment at 24, 4, and 2 h prior to measurement. The percentage of particles taken up was then determined by comparing the fluorescent intensity in lysed cells to a background corrected standard curve (Figure 2h). It was determined that roughly 10% of both the total MDA-MB-231-EV and A431-EV dose had been taken up after 2 h and after 24 h this had increased to 26% and 32%, respectively. In contrast, after 2 h only 0.6% of the total DLin-DMA-MC3-LNP dose had been taken up rising to 1% after 24 h. It should be noted that the confocal microscopy images show similar signals in cells between EVs and DLin-DMA-MC3-LNPs, due to a higher labeling efficiency of DLin-DMA-MC3-LNPs. These data clearly demonstrate that in this setup, EV uptake is highly more efficient than that of DLin-DMA-MC3-LNPs which may in part explain the observed differences in RNA delivery efficiency.
The physical routes by which EVs are taken up and trafficked postuptake could also differ between EVs and LNPs. Interestingly, in a comparison of EV and LNP uptake, EVs were shown to be rapidly taken up at filopodia active regions while LNPs collected in islands at the cell surface were taken up slowly.29 If the way in which EVs and LNPs are taken up differs, then it is plausible that postuptake trafficking also differs, which may explain their differing delivery efficiencies. The intracellular routes taken by LNPs to deliver RNA to the cytosol are well studied, and endosomes have been identified as the site of escape. This is the rate-limiting step for RNA delivery and occurs at a low efficiency with only 1–2% of LNP cargo escaping into the cytosol.30 In comparison, less is known about the trafficking and endosomal escape efficiency of EVs. Therefore, a study to elucidate the routes EVs and LNPs follow postuptake could help to explain the increased efficiency of EVs observed here.
EVs may also possess features that allow them to fuse with plasma membranes, thereby allowing endolysosomal escape. Evidence to support this is provided by Bonsergent et al., who observed that EVs fuse with plasma membranes and release their cargo in conditions resembling the endolysosome. In an in vitro assay designed to mimic conditions within the endolysosome, they demonstrated that incubation of EVs in a cell-free extract containing purified plasma membrane induced the release of protein cargo. This process is protein dependent as cargo release was abrogated after proteinase pretreatment of either the plasma membrane sheets or EVs.31 In addition, Bhagyashree et al. demonstrated that a proportion of EVs taken up by HEK93T cells fused with the membranes of late endosomes and lysosomes. This process was also dependent on low pH as inhibition of endosome acidification blocked fusion.32 These observations highlight the potential fusogenic properties of EVs that could contribute to their high delivery efficiency.
Furthermore, the results obtained here are in line with those obtained by Reshke et al., who demonstrated that EVs were able to achieve siRNA-mediated target gene knockdown in the liver, intestine, and kidney glomeruli in vivo at a dose at least 10-fold lower than those required for InvivoFectamine 3.0 or C12-200 LNP-mediated knockdown.33
Conversely, Stremersch et al. found that anionic liposomes were able to functionally deliver siRNA while EVs coated with cholesterol anchored siRNA were not.34 This difference could be explained by the way in which EVs were loaded with RNA. In contrast to the cholesterol anchored siRNA used by Stremersch et al., the sgRNA used here is located within the EV lumen.21 Similarly, the siRNA used by Reshke et al. was loaded by insertion into a pre-miR-451 backbone, which is abundant in the EV lumen.33 It is possible that siRNA anchoring prevented escape of the siRNA postuptake, meaning that it was unable to function. This may explain the difference in results between Stremersch et al. and those presented here and by Reshke et al.
It is also possible that EVs of different origins could possess inherently different delivery efficiencies. Although A431-EVs and MDA-MB-231-EVs appear capable of efficient delivery, it is possible that EVs from different sources are not. Furthermore, EV-delivery efficiency may also differ depending on the recipient cell.
Although these results suggest EVs are a highly efficient delivery vehicle, they also demonstrate that passively loaded EVs are unlikely to be utilized as therapeutics for RNA delivery. To induce reporter activation, multiple doses of EVs were isolated and concentrated from a large volume of donor-cell conditioned medium. Even with this EV enrichment, reporter activation was only induced in a small percentage of cells, albeit at concentrations much lower than those required for similar activation levels by DLin-MC3-DMA-LNPs. This minimal response is most likely the result of low sgRNA loading. Therefore, to fully harness the delivery efficiency of EVs, a suitable RNA loading strategy must be found.
In conclusion, this analysis demonstrates that EVs possess a higher RNA delivery efficiency than the synthetic RNA delivery systems tested here. Further research is required to determine the features of EVs that confer this efficiency in order to utilize EVs as RNA delivery vehicles.
Acknowledgments
We thank Dr. Emilia Nagyova, Jerney Gitz Francois, and Omnia M. Elsharkasy for their technical assistance. In addition, we thank Dr. Maria Laura Tognoli for her critical reading of the manuscript and Dr. Sander van der Laan for his assistance with statistical analysis. The work of D.E.M., M.J.W.E., R.M.S., and P.V. is supported by the European Union’s Horizon 2020 Research and Innovation program in the project B-SMART (to P.V. and R.M.S.) under grant agreement No. 721058. O.G.d.J. is supported by a VENI Fellowship (VI.Veni.192.174) from the Dutch Research Council (NWO).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.1c00094.
Methods used in these experiments, a schematic of the CROSS-FIRE reporter system (Figure S1), the results of an sgRNA stability study (Figure S2), the results of an in vitro transfection reagent titration (Figure S3), a schematic of the FACS gating strategy used to count GFP+ reporter cells (Figure S4) and RNA and primer sequences (PDF)
The authors declare the following competing financial interest(s): P.V. serves on the scientific advisory board of Evox Therapeutics.
Supplementary Material
References
- Dowdy S. F. Overcoming Cellular Barriers for RNA Therapeutics. Nat. Biotechnol. 2017, 35 (3), 222–229. 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Hong J.; Zheng S.; Ding Y.; Guo S. Elimination Pathways of Systemically Delivered SiRNA. Mol. Ther. 2011, 19 (2), 381–385. 10.1038/mt.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer A. D.; Dowdy S. F. GalNAc-SiRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid Ther. 2018, 28 (3), 109–118. 10.1089/nat.2018.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers M. J. W.; Kulkarni J. A.; Meel R.; Van Der Cullis P. R.; Vader P.; Schiffelers R. M. State-of-the-Art Design and Rapid-Mixing Production Techniques of Lipid Nanoparticles for Nucleic Acid Delivery. Small Methods 2018, 2, 1700375. 10.1002/smtd.201700375. [DOI] [Google Scholar]
- Engin A. B.; Hayes A. W. The Impact of Immunotoxicity in Evaluation of the Nanomaterials Safety. Toxicol. Res. Appl. 2018, 10.1177/2397847318755579. [DOI] [Google Scholar]
- Sadauskas E.; Wallin H.; Stoltenberg M.; Vogel U.; Doering P.; Larsen A.; Danscher G. Kupffer Cells Are Central in the Removal of Nanoparticles from the Organism. Part. Fibre Toxicol. 2007, 4, 10. 10.1186/1743-8977-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay G.; Alakhova D. Y.; Kabanov A. V. Endocytosis of Nanomedicines. J. Controlled Release 2010, 145 (3), 182–195. 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andaloussi S.; Mäger I.; Breakefield X. O.; Wood M. J. A. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat. Rev. Drug Discovery 2013, 12 (5), 347–357. 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Yáñez-Mó M.; Siljander P. R.; Andreu Z.; Bedina A.; Borràs F. E.; Buzas E. I.; Buzas K.; Casal E.; Cappello F.; Carvalho J.; Colás E.; Cordeiro-Da A.; Fais S.; Falcon-Perez J. M.; Ghobrial I. M.; Giebel B.; Gimona M.; Graner M.; Gursel I.; Gursel M.; Niels H. H.; Hendrix A.; Kierulf P.; Kokubun K.; Kosanovic M.; Kralj-Iglic V.; Laitinen S.; Lässer C.; Lener T.; Ligeti E.; Line̅ A.; Lipps G.; Llorente A.; Manček-Keber M.; Marcilla A.; Mittelbrunn M.; Hoen E. N. M. N.; Nyman T. A.; Driscoll L. O.; Olivan M.; Oliveira C.; Pállinger É.; Portillo H. A.; Rigau M.; Rohde E.; Sammar M.; Sánchez F.; Santarém N.; Schallmoser K.; Ostenfeld M. S.; Stoorvogel W.; Stukelj R.; Grein S. G.; Van Der Helena M.; Wauben M. H. M.; Wever O. De Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader P.; Mol E. A.; Pasterkamp G.; Schiffelers R. M. Extracellular Vesicles for Drug Delivery. Adv. Drug Delivery Rev. 2016, 106, 148–156. 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Murphy D. E.; de Jong O. G.; Brouwer M.; Wood M. J.; Lavieu G.; Schiffelers R. M.; Vader P. Extracellular Vesicle-Based Therapeutics: Natural versus Engineered Targeting and Trafficking. Exp. Mol. Med. 2019, 51, 1–12. 10.1038/s12276-019-0223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abels E. R.; Maas S. L. N.; Nieland L.; Krichevsky A. M.; Breakefield X. O.; Abels E. R.; Maas S. L. N.; Nieland L.; Wei Z.; Cheah P. S.; Tai E.; Kolsteeg C. Glioblastoma-Associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular Article Glioblastoma-Associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular MiR-21. Cell Rep. 2019, 28 (12), 3105–3119. 10.1016/j.celrep.2019.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J.; Liu R. A. N.; Shi Y.; Yin L.; Pu Y. Exosome-Shuttling MicroRNA-21 Promotes Cell Migration and Invasion-Targeting PDCD4 in Esophageal Cancer. Int. J. Oncol. 2016, 48 (87), 2567–2579. 10.3892/ijo.2016.3453. [DOI] [PubMed] [Google Scholar]
- Margolis L.; Sadovsky Y. The Biology of Extracellular Vesicles: The Known Unknowns. PLoS Biol. 2019, 17 (7), e3000363. 10.1371/journal.pbio.3000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillet J. R.; Kang Q.; Ruf I. K.; Briggs H. A.; Vojtech L. N.; Hughes S. M.; Cheng H. H.; Arroyo J. D.; Meredith E. K.; Gallichotte E. N.; Pogosova-Agadjanyan E. L. Quantitative and Stoichiometric Analysis of the MicroRNA Content of Exosomes. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (41), 14888–14893. 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D.; Wang H.; Ho S.; Chan H.; Hai L.; He X.; Wang K. Total Internal Reflection-Based Single-Vesicle in Situ Quantitative and Stoichiometric Analysis of Tumor-Derived Exosomal MicroRNAs for Diagnosis and Treatment Monitoring. Theranostics 2019, 9 (15), 4494–4507. 10.7150/thno.33683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somiya M. Where Does the Cargo Go?: Solutions to Provide Experimental Support for the “Extracellular Vesicle Cargo Transfer Hypothesis.. J. Cell Commun. Signal. 2020, 14 (2), 135–146. 10.1007/s12079-020-00552-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A.; Maynard C.; Verweij F. J.; Kamermans A.; Schäfer R.; Beerling E.; Schiffelers R. M.; De Wit E.; Berenguer J.; Ellenbroek S. I. J.; Wurdinger T.; Pegtel D. M.; Van Rheenen J. In Vivo Imaging Reveals Extracellular Vesicle-Mediated Phenocopying of Metastatic Behavior. Cell 2015, 161 (5), 1046–1057. 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci F.; Garris C.; Lai C. P.; Newton A.; Pfirschke C.; Engblom C.; Alvarez D.; Sprachman M.; Evavold C.; Magnuson A.; von Andrian U. H.; Glatz K.; Breakefield X. O.; Mempel T. R.; Weissleder R.; Pittet M. J. SCS Macrophages Suppress Melanoma by Restricting Tumor-Derived Vesicle-B Cell Interactions. Science 2016, 352 (6282), 242–246. 10.1126/science.aaf1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K.; Somiya M.; Hinuma S. Biochemical and Biophysical Research Communications Induction of Lipid Droplets in Non-Macrophage Cells as Well as Macrophages by Liposomes and Exosomes. Biochem. Biophys. Res. Commun. 2019, 510 (1), 184–190. 10.1016/j.bbrc.2019.01.078. [DOI] [PubMed] [Google Scholar]
- de Jong O. G.; Murphy D. E.; Mäger I.; Willms E.; Garcia-Guerra A.; Gitz-Francois J. J.; Lefferts J.; Gupta D.; Steenbeek S. C.; van Rheenen J.; El Andaloussi S.; Schiffelers R. M.; Wood M. J. A.; Vader P. A CRISPR-Cas9-Based Reporter System for Single-Cell Detection of Extracellular Vesicle-Mediated Functional Transfer of RNA. Nat. Commun. 2020, 11 (1), 1113. 10.1038/s41467-020-14977-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy S. M. Patisiran: First Global Approval. Drugs 2018, 78 (15), 1625–1631. 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- Akinc A.; Maier M. A.; Manoharan M.; Fitzgerald K.; Jayaraman M.; Barros S.; Ansell S.; Du X.; Hope M. J.; Madden T. D.; Mui B. L.; Semple S. C.; Tam Y. K.; Ciufolini M.; Witzigmann D.; Kulkarni J. A.; van der Meel R.; Cullis P. R. The Onpattro Story and the Clinical Translation of Nanomedicines Containing Nucleic Acid-Based Drugs. Nat. Nanotechnol. 2019, 14 (12), 1084–1087. 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- Manzanares D.; Cena V. Endocytosis: The Nanoparticle and Submicron Nanocompounds Gateway into the Cell. Pharmaceutics 2020, 12, 371. 10.3390/pharmaceutics12040371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maugeri M.; Nawaz M.; Papadimitriou A.; Angerfors A.; Camponeschi A.; Na M.; Hölttä M.; Skantze P.; Johansson S.; Sundqvist M.; Lindquist J.; Kjellman T.; Mårtensson I.-L.; Jin T.; Sunnerhagen P.; Östman S.; Lindfors L.; Valadi H. Linkage between Endosomal Escape of LNP-MRNA and Loading into EVs for Transport to Other Cells. Nat. Commun. 2019, 10 (1), 4333. 10.1038/s41467-019-12275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C.; Witwer K. W.; Aikawa E.; Alcaraz M. J.; Anderson J. D.; Andriantsitohaina R.; Antoniou A.; Arab T.; Archer F.; Atkin-Smith G. K.; Ayre D. C.; Bach J.-M.; Bachurski D.; Baharvand H.; Balaj L.; Baldacchino S.; Bauer N. N.; Baxter A. A.; Bebawy M.; Beckham C.; Bedina Zavec A.; Benmoussa A.; Berardi A. C.; Bergese P.; Bielska E.; Blenkiron C.; Bobis-Wozowicz S.; Boilard E.; Boireau W.; Bongiovanni A.; Borràs F. E.; Bosch S.; Boulanger C. M.; Breakefield X.; Breglio A. M.; Brennan M. Á.; Brigstock D. R.; Brisson A.; Broekman M. L. D.; Bromberg J. F.; Bryl-Górecka P.; Buch S.; Buck A. H.; Burger D.; Busatto S.; Buschmann D.; Bussolati B.; Buzás E. I.; Byrd J. B.; Camussi G.; Carter D. R. F.; Caruso S.; Chamley L. W.; Chang Y.-T.; Chen C.; Chen S.; Cheng L.; Chin A. R.; Clayton A.; Clerici S. P.; Cocks A.; Cocucci E.; Coffey R. J.; Cordeiro-Da-Silva A.; Couch Y.; Coumans F. A. W.; Coyle B.; Crescitelli R.; Criado M. F.; D’Souza-Schorey C.; Das S.; Datta Chaudhuri A.; de Candia P.; De Santana E. F.; De Wever O.; del Portillo H. A.; Demaret T.; Deville S.; Devitt A.; Dhondt B.; Di Vizio D.; Dieterich L. C.; Dolo V.; Dominguez Rubio A. P.; Dominici M.; Dourado M. R.; Driedonks T. A. P.; Duarte F. V.; Duncan H. M.; Eichenberger R. M.; Ekström K.; El Andaloussi S.; Elie-Caille C.; Erdbrügger U.; Falcón-Pérez J. M.; Fatima F.; Fish J. E.; Flores-Bellver M.; Försönits A.; Frelet-Barrand A.; Fricke F.; Fuhrmann G.; Gabrielsson S.; Gámez-Valero A.; Gardiner C.; Gärtner K.; Gaudin R.; Gho Y. S.; Giebel B.; Gilbert C.; Gimona M.; Giusti I.; Goberdhan D. C. I.; Görgens A.; Gorski S. M.; Greening D. W.; Gross J. C.; Gualerzi A.; Gupta G. N.; Gustafson D.; Handberg A.; Haraszti R. A.; Harrison P.; Hegyesi H.; Hendrix A.; Hill A. F.; Hochberg F. H.; Hoffmann K. F.; Holder B.; Holthofer H.; Hosseinkhani B.; Hu G.; Huang Y.; Huber V.; Hunt S.; Ibrahim A. G.-E.; Ikezu T.; Inal J. M.; Isin M.; Ivanova A.; Jackson H. K.; Jacobsen S.; Jay S. M.; Jayachandran M.; Jenster G.; Jiang L.; Johnson S. M.; Jones J. C.; Jong A.; Jovanovic-Talisman T.; Jung S.; Kalluri R.; Kano S.; Kaur S.; Kawamura Y.; Keller E. T.; Khamari D.; Khomyakova E.; Khvorova A.; Kierulf P.; Kim K. P.; Kislinger T.; Klingeborn M.; Klinke D. J.; Kornek M.; Kosanović M. M.; Kovács Á. F.; Krämer-Albers E.-M.; Krasemann S.; Krause M.; Kurochkin I. V.; Kusuma G. D.; Kuypers S.; Laitinen S.; Langevin S. M.; Languino L. R.; Lannigan J.; Lässer C.; Laurent L. C.; Lavieu G.; Lázaro-Ibáñez E.; Le Lay S.; Lee M.-S.; Lee Y. X. F.; Lemos D. S.; Lenassi M.; Leszczynska A.; Li I. T. S.; Liao K.; Libregts S. F.; Ligeti E.; Lim R.; Lim S. K.; Line̅ A.; Linnemannstöns K.; Llorente A.; Lombard C. A.; Lorenowicz M. J.; Lörincz Á. M.; Lötvall J.; Lovett J.; Lowry M. C.; Loyer X.; Lu Q.; Lukomska B.; Lunavat T. R.; Maas S. L. N.; Malhi H.; Marcilla A.; Mariani J.; Mariscal J.; Martens-Uzunova E. S.; Martin-Jaular L.; Martinez M. C.; Martins V. R.; Mathieu M.; Mathivanan S.; Maugeri M.; McGinnis L. K.; McVey M. J.; Meckes D. G.; Meehan K. L.; Mertens I.; Minciacchi V. R.; Möller A.; Møller Jørgensen M.; Morales-Kastresana A.; Morhayim J.; Mullier F.; Muraca M.; Musante L.; Mussack V.; Muth D. C.; Myburgh K. H.; Najrana T.; Nawaz M.; Nazarenko I.; Nejsum P.; Neri C.; Neri T.; Nieuwland R.; Nimrichter L.; Nolan J. P.; Nolte-’t Hoen E. N. M.; Noren Hooten N.; O’Driscoll L.; O’Grady T.; O’Loghlen A.; Ochiya T.; Olivier M.; Ortiz A.; Ortiz L. A.; Osteikoetxea X.; Østergaard O.; Ostrowski M.; Park J.; Pegtel D. M.; Peinado H.; Perut F.; Pfaffl M. W.; Phinney D. G.; Pieters B. C. H.; Pink R. C.; Pisetsky D. S.; Pogge von Strandmann E.; Polakovicova I.; Poon I. K. H.; Powell B. H.; Prada I.; Pulliam L.; Quesenberry P.; Radeghieri A.; Raffai R. L.; Raimondo S.; Rak J.; Ramirez M. I.; Raposo G.; Rayyan M. S.; Regev-Rudzki N.; Ricklefs F. L.; Robbins P. D.; Roberts D. D.; Rodrigues S. C.; Rohde E.; Rome S.; Rouschop K. M. A.; Rughetti A.; Russell A. E.; Saá P.; Sahoo S.; Salas-Huenuleo E.; Sánchez C.; Saugstad J. A.; Saul M. J.; Schiffelers R. M.; Schneider R.; Schøyen T. H.; Scott A.; Shahaj E.; Sharma S.; Shatnyeva O.; Shekari F.; Shelke G. V.; Shetty A. K.; Shiba K.; Siljander P. R.-M.; Silva A. M.; Skowronek A.; Snyder O. L.; Soares R. P.; Sódar B. W.; Soekmadji C.; Sotillo J.; Stahl P. D.; Stoorvogel W.; Stott S. L.; Strasser E. F.; Swift S.; Tahara H.; Tewari M.; Timms K.; Tiwari S.; Tixeira R.; Tkach M.; Toh W. S.; Tomasini R.; Torrecilhas A. C.; Tosar J. P.; Toxavidis V.; Urbanelli L.; Vader P.; van Balkom B. W. M.; van der Grein S. G.; Van Deun J.; van Herwijnen M. J. C.; Van Keuren-Jensen K.; van Niel G.; van Royen M. E.; van Wijnen A. J.; Vasconcelos M. H.; Vechetti I. J.; Veit T. D.; Vella L. J.; Velot É.; Verweij F. J.; Vestad B.; Viñas J. L.; Visnovitz T.; Vukman K. V.; Wahlgren J.; Watson D. C.; Wauben M. H. M.; Weaver A.; Webber J. P.; Weber V.; Wehman A. M.; Weiss D. J.; Welsh J. A.; Wendt S.; Wheelock A. M.; Wiener Z.; Witte L.; Wolfram J.; Xagorari A.; Xander P.; Xu J.; Yan X.; Yáñez-Mó M.; Yin H.; Yuana Y.; Zappulli V.; Zarubova J.; Žėkas V.; Zhang J.; Zhao Z.; Zheng L.; Zheutlin A. R.; Zickler A. M.; Zimmermann P.; Zivkovic A. M.; Zocco D.; Zuba-Surma E. K. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7 (1), 1535750. 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christina S.; Collinson A.; Matthews C.; Pointon A.; Jenkinson L.; Minter R. R.; Vaughan T. J.; Tigue N. J. Exosomal Delivery of Doxorubicin Enables Rapid Cell Entry and Enhanced in Vitro Potency. PLoS One 2019, 14 (3), e0214545. 10.1371/journal.pone.0214545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M.; Zhao X.; Xing H.; Xun Z.; Zhu S.; Lang L.; Yang T.; Cai C.; Wang D.; Ding P. Comparison of Exosome-Mimicking Liposomes with Conventional Liposomes for Intracellular Delivery of SiRNA. Int. J. Pharm. 2018, 550 (1–2), 100–113. 10.1016/j.ijpharm.2018.08.040. [DOI] [PubMed] [Google Scholar]
- Heusermann W.; Hean J.; Trojer D.; Steib E.; von Bueren S.; Graff-Meyer A.; Genoud C.; Martin K.; Pizzato N.; Voshol J.; Morrissey D. V.; Andaloussi S. E. L.; Wood M. J.; Meisner-Kober N. C. Exosomes Surf on Filopodia to Enter Cells at Endocytic Hot Spots, Traffic within Endosomes, and Are Targeted to the ER. J. Cell Biol. 2016, 213 (2), 173–184. 10.1083/jcb.201506084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron J.; Querbes W.; Zeigerer A.; Borodovsky A.; Marsico G.; Schubert U.; Manygoats K.; Seifert S.; Andree C.; Stöter M.; Epstein-Barash H.; Zhang L.; Koteliansky V.; Fitzgerald K.; Fava E.; Bickle M.; Kalaidzidis Y.; Akinc A.; Maier M.; Zerial M. Image-Based Analysis of Lipid Nanoparticle-Mediated SiRNA Delivery, Intracellular Trafficking and Endosomal Escape. Nat. Biotechnol. 2013, 31 (7), 638–646. 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- Bonsergent E.; Lavieu G. Content Release of Extracellular Vesicles in a Cell-Free Extract. FEBS Lett. 2019, 593 (15), 1983–1992. 10.1002/1873-3468.13472. [DOI] [PubMed] [Google Scholar]
- Joshi B. S.; de Beer M. A.; Giepmans B. N. G.; Zuhorn I. S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14 (4), 4444–4455. 10.1021/acsnano.9b10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshke R.; Taylor J. A.; Savard A.; Guo H.; Rhym L. H.; Kowalski P. S.; Trung M. T.; Campbell C.; Little W.; Anderson D. G.; Gibbings D. Reduction of the Therapeutic Dose of Silencing RNA by Packaging It in Extracellular Vesicles via a Pre-MicroRNA Backbone. Nat. Biomed. Eng. 2020, 4 (1), 52–68. 10.1038/s41551-019-0502-4. [DOI] [PubMed] [Google Scholar]
- Stremersch S.; Vandenbroucke R. E.; Wonterghem E.; Van Hendrix A.; Smedt S. C.; De Raemdonck K. Comparing Exosome-like Vesicles with Liposomes for the Functional Cellular Delivery of Small RNAs. J. Controlled Release 2016, 232, 51–61. 10.1016/j.jconrel.2016.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.