Abstract
InterMetalDB is a free-of-charge database and browser of intermolecular metal binding sites that are present on the interfaces of macromolecules forming larger assemblies based on structural information deposited in Protein Data Bank (PDB). It can be found and freely used at https://intermetaldb.biotech.uni.wroc.pl/. InterMetalDB collects the interfacial binding sites with involvement of metal ions and clusters them on the basis of 50% sequence similarity and the nearest metal environment (5 Å radius). The data are available through the web interface where they can be queried, viewed, and downloaded. Complexity of the query depends on the user, because the questions in the query are connected with each other by a logical AND. InterMetalDB offers several useful options for filtering records including searching for structures by particular parameters such as structure resolution, structure description, and date of deposition. Records can be filtered by coordinated metal ion, number of bound amino acid residues, coordination sphere, and other features. InterMetalDB is regularly updated and will continue to be regularly updated with new content in the future. InterMetalDB is a useful tool for all researchers interested in metalloproteins, protein engineering, and metal-driven oligomerization.
Keywords: metalloprotein, protein−protein interaction, interprotein site, protein assembly, interfacial metal
Introduction
Nearly every macromolecule in living organisms needs to interact either in a transient or permanent way with another macromolecule to fulfill its function. Taking into account that metal ions are associated with an estimated 30–40% of all proteins,1,2 often performing essential structural or functional roles, it is no wonder that the areas of macromolecule–macromolecule interaction and metal–macromolecule interaction overlap.3 What is more surprising is the fact that this area of research remains almost unexplored and our knowledge is only fragmentary. With the growth of identified macromolecules containing metal ions, efforts have begun to identify and differentiate specific characteristics of binding sites that determine the affinity of the metal ion to the site, and its function in the binding sites. Among the first features described were the metal ion-binding ligands and the distinction whether the bonded metal ion has a catalytic or structural function.4−6 For the most part, the concept of binding metal ions on the interface has escaped researchers’ attention. Although it was described in an extensive review paper in 2014,3 few preceding reviews mentioned intermolecular zinc binding.7,8 It is possible that the presence of metal ions on macromolecules’ interfaces has not attracted much attention because of the rarity or instability of this type of interaction, but it might also be due to the great difficulty in testing and investigating intermolecularly bound metal ions, especially with transient character. In addition to developing our knowledge of intermolecular metal ion binding, it is worth noting that the tool we provide can be used for the construction or improvement of existing models that predict metal ion binding by macromolecules. The aggregation of intermolecular metal binding sites in the form of a database, combined with coordination chemistry and statistical models, may facilitate the engineering of artificial macromolecular interfaces involving metal binding.9 We believe that our very recent contribution in the field of interfacial metal binding together with the presented resource will help researchers to expand knowledge about factors determining interfacial metal binding and its role in biological systems.10
It seems that, so far, the best-explored and described d-block metal ion found in macromolecules is the zinc ion (formally Zn2+). This is fully understandable, given the prevalence of Zn2+ in the living world—Zn2+ is estimated to occur in about 10% of all human proteins—so it will be used as a background for the comparison of intermolecular ion binding.11 However, it is important to mention that estimated zinc protein number is based on already known fingerprints found in proteins encoded in the human genome, and this number does not take into account interprotein sites due to the lack of available bioinformatic tools facilitating identification of such sites.5,10 The first systematic attempt to describe all Zn2+-binding sites in protein structures appeared at the end of 1990. The description of the structures deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB)12 was rerun several more times, usually without leaving the data deposited in electronic form.13−16 In comparison, the first review of the biological sites of intermolecular metal binding appeared only in 2014,3 although two important contributions related to interprotein zinc sites were published earlier, and ours appeared just recently.7,8,10 Although several electronic resources have made searching for PDBs (of metal containing proteins) possible—MESPEUS,17 ZifBase,18 MetalPDB,19,20 ZincBind21—none of them allow for filtering of intermolecular metal binding sites. In order to allow the scientific community to investigate this obscure area and simultaneously efficiently explore the vast amount of structural information, we have aggregated intermolecular binding sites in the entire RCSB PDB database and stored the results in our freely and publicly accessible InterMetalDB database. InterMetalDB is also a browser for deposited structures and offers several useful options for filtering records such as searching for structures by particular parameters, e.g., structure resolution, structure description, and date of deposition. Identified intermolecular binding sites can be filtered by coordinated metal ion, number of coordinating amino acids, coordination sphere, and other features. Nevertheless, records stored in InterMetalDB should be considered with caution. As discussed in our recent review article,10 interprotein Zn2+-binding sites that are not physiological are quite common. Out of around 600 structures containing interfacial Zn2+ (after redundancy removal and preselection via a Python script), we manually selected around 170 structural complexes that we believe contain intermolecular Zn2+ of physiological importance.10 Because currently, we do not have any algorithms or tools that allow for precise artifact prediction, we do not filter in any way metal binding sites, thus leaving it to the user’s experience to judge whether a bound metal has a physiological function. The goal of InterMetalDB is to collect and present all intermolecular metal binding sites in the RCSB PDB and allow the user to easily filter and access useful information regarding them. In order to allow this, it contains the newest possible data set of all known intermolecular metal binding sites deposited in the RCSB PDB.22 InterMetalDB has a user-friendly querying interface and is automatically and regularly updated at https://intermetaldb.biotech.uni.wroc.pl/. The source code for InterMetalDB can be found at https://github.com/jzftran/InterMetalDB/, where it can be viewed, downloaded, and modified under MIT license.
Methods
Acquiring Intermolecular Metal Binding Sites
The RCSB PDB search application programming interface (API) allows one to run queries across RCSB PDB Search Services and retrieve a list of accordant identifiers (e.g., PDB ID) (https://search.rcsb.org/). Structures containing metal elements were acquired from the RCSB PDB,22 querying for the relevant metal element via RCSB PDB Search API using the chemical component identifier rcsb_chem_comp_container_identifiers.comp_id, which is an exact search attribute, and returns only structures that contain a standalone metal ion, not metal bound by any kind of molecule; i.e., structures containing iron in heme or iron–sulfur clusters are not returned. In the future we hope to broaden our search to molecules like this as well. During database construction, no constraints regarding structure resolution were applied. After acquiring corresponding structural file identifiers, each file was received and processed using the Python parser library atomium, which allows for processing of structural files deposited in the RCSB PDB.22
When working on PDB files, the coordinate for biological assembly and asymmetric unit are often the same. Nevertheless, for some files there is a difference and some space operations are needed to analyze the biological assembly. The asymmetric unit is the nonreducible (smallest) model of the crystal which, when duplicated and moved by crystal symmetry operation, will produce the unit cell of the crystal, i.e., part of the crystal that is repeated (https://dictionary.iucr.org). The asymmetric unit should not be confused with the biological functional unit, which is the tertiary or quaternary protein structure that is believed to be a functional macromolecule in an organism. Biological assembly is constructed from an asymmetric unit after selecting a subset of the deposited coordinates (biological assembly will be a portion of the asymmetric unit) or selecting a subset of the deposited coordinates and duplicating or applying symmetry operations (e.g., translation, rotation, and their combination).
In order to deal with biological assemblies, using assembly instructions given in a structural file, the biological assembly containing the metal element of interest and having the lowest energy (if given in assembly instruction) was chosen for further examination. If no macromolecular binding energy was given in a structural file, the first assembly containing the metal has been selected. Sometimes structural files contain duplicated atoms. This is especially often true for atoms lying on a point of symmetry rotation. In order to deal with this redundancy, duplicated atoms are removed, considering as duplicated atoms those that are within a radius of 1 Å or less than the original atom. Each metal ion from the biological assembly is examined for the surrounding environment in a radius of 3 Å (center-to-center) of the metal ion, and a coordination environment is assumed to include all noncarbon, non-hydrogen atoms. PDB structures are considered to contain an intermolecular metal binding site if the metal ion is bound by at least two amino acid residues or nucleotide residues from at least two different macromolecular chains. For example, if a metal ion is coordinated by three amino acid residues from chain A, and a chlorine ion assigned to chain B, such a metal binding site is not considered as intermolecularly bound. For each coordinating atom a one letter abbreviation of the corresponding residue is used to construct a coordination identifier (e.g., a metal ion coordinated by three cysteinyl residues and one histidinyl residue will have C3H1 as the coordination identifier). A group identifier is constructed in a similar way, but for a radius of 5 Å and without restrictions for atom type. Coordination identifier can be understood as a description of the coordination environment of a metal ion, while a group identifier is a description of all amino acid residues located in a radius of 5 Å of the metal ion. The first identifier allows the user to query for a specific coordination environment, while the latter is used for the purposes of clustering. If a metal ion is coordinated by two or more chains in the way described above, the metal site record is added to the SQLite database (https://www.sqlite.com), together with the oxidation state, coordination identifier, group identifier, number of coordinating amino acid residues, number of all ligands, number of coordinating chains and other information. Metal oxidation state is read directly from the file; no additional steps are taken to determine the oxidation state. Oxidation state should be taken with caution as there is no separate identifier for metals with uncertain oxidation state.
Redundancy Removal, Representative Sites
The RCSB PDB as a worldwide repository for macromolecular structures contains structures of the same macromolecules or highly similar macromolecules. This structure redundancy is caused by representation of different variants of the same macromolecule (various bound ligands or small mutations in structure) or existence of highly homologous macromolecules. Because the RCSB PDB holds a body of data that contains considerable redundancy of structures, the next step for database construction was to identify redundancy and select representative intermolecular metal binding sites. In order to account for this redundancy, a similar approach to MetalPDB19 has been used. MMseqs2,23 chain clustering with 50% sequence identity for both query and target, has been used, ensuring that the clusters have the same fold.24 Metal binding sites may not be unique in structure and may appear many times—an extreme example of this is the structure of rotavirus inner capsid particle (PDB ID: 3KZ4) containing 240 Zn2+-binding sites.25 In order to group similar binding sites and deal with metal binding sites’ redundancy, in each sequence cluster the binding sites are then themselves clustered based on the group identifier (described above). The first unique metal site of the best-resolution structure is chosen as a representative metal site.
Web Interface
The InterMetalDB database is integrated into a Django-based web application (https://www.djangoproject.com). Metal binding sites and structures are visualized in the web front-end molecular viewer NGL Viewer.26 The user can filter results with various specific parameters: PDB ID, structure title, keywords, etc. Additionally, one can search for interfacial metal binding sites by coordinating residues, number of coordinating chains, and other parameters. Filtered results can be exported as a CSV or JSON file for further analysis. Statistics for the whole database and for a specific metal can be viewed with the help of the JavaScript library for data visualization Chart.js (www.chartjs.org).
Results and Discussion
User Interface
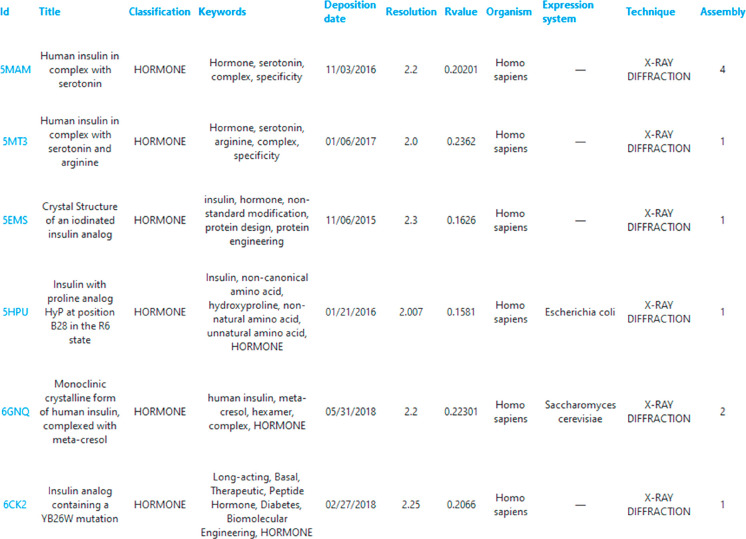
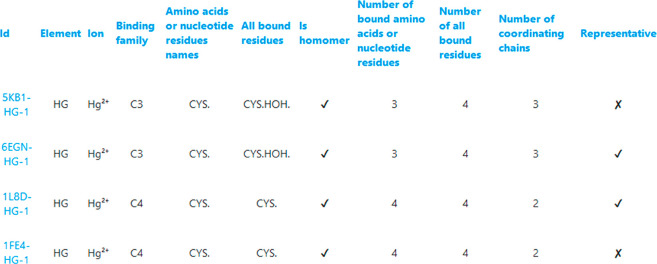
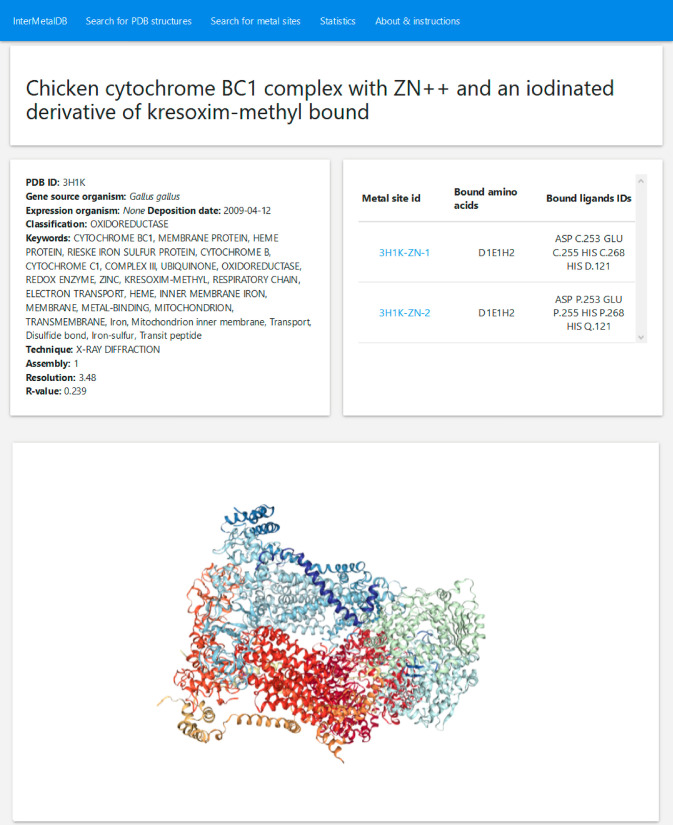
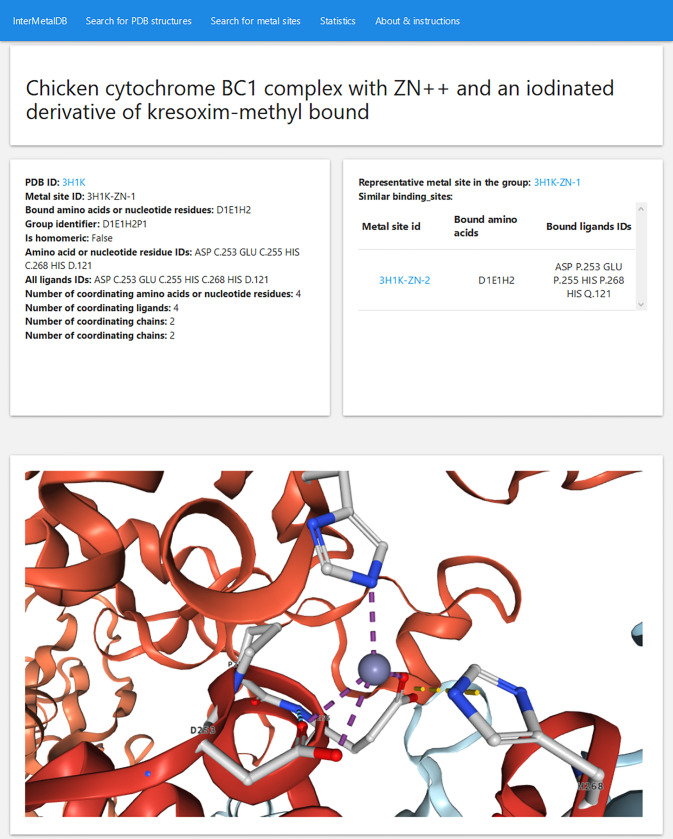
InterMetalDB can be queried with the web interface at https://intermetaldb.biotech.uni.wroc.pl/ via the Django web application. It allows the user to search for the data by multiple criteria from the PDB and metal sites search sites. PDB files can be queried by title, keywords, resolution, source organism, etc. (Figure 1). The database contains all PDB files containing a metal-involved macromolecule–macromolecule interface published in the RCSB PDB so far. Metal sites can be queried by coordinated metal element, types of bound residues, number of bound residues chains, etc. (Figure 2). The complexity of the query depends on the user as querying conditions are connected by a logical AND operator. In both cases searches will return a list of records that can be downloaded to a CSV or JSON file, allowing for further analysis. From the list the user can select one PDB or metal site to view more details (Figure 3, Figure 4), PDB records and metal sites records are associated via id, and allow browsing one based on another. After selecting a record, the user can view the visualized structure using NGL Viewer.26
Figure 1.
Querying InterMetalDB for Protein Data Bank deposited structures of macromolecules. Query fields are connected with logical AND. Every field in the query contains a placeholder that helps the user to fill in the appropriate term. In this case InterMetalDB is queried for PDB title containing “insulin”, gene source organism “Homo sapiens”, PDB classification “hormone”, resolution better than 2.0 A, deposition date between 2015–01–01 and 2020–06–29. Results can be sorted by clicking a title table and downloaded to the file of interest.
Figure 2.
Querying of InterMetalDB for metal binding sites. Query fields are connected with logical AND. Every field in the query contains a placeholder that helps the user to fill in the appropriate term. Each result can be viewed separately by clicking on the metal site ID. Obtained records can be sorted by clicking on table title and downloaded to the file of interest.
Figure 3.
PDB structure details can be found in top left card. In top right card are links to interfacial metal binding sites in the structure. PDB visualization (bottom) is made with help of NGL Viewer.26 From this page the user can choose one of the metal binding sites to be viewed in detail.
Figure 4.
Interfacial metal site details can be found in top-left card. Representative site and similar sites (if available) can be found in top-right card. From this card the user can choose to view another metal binding site. Visualization of metal site is achieved with NGL Viewer.26
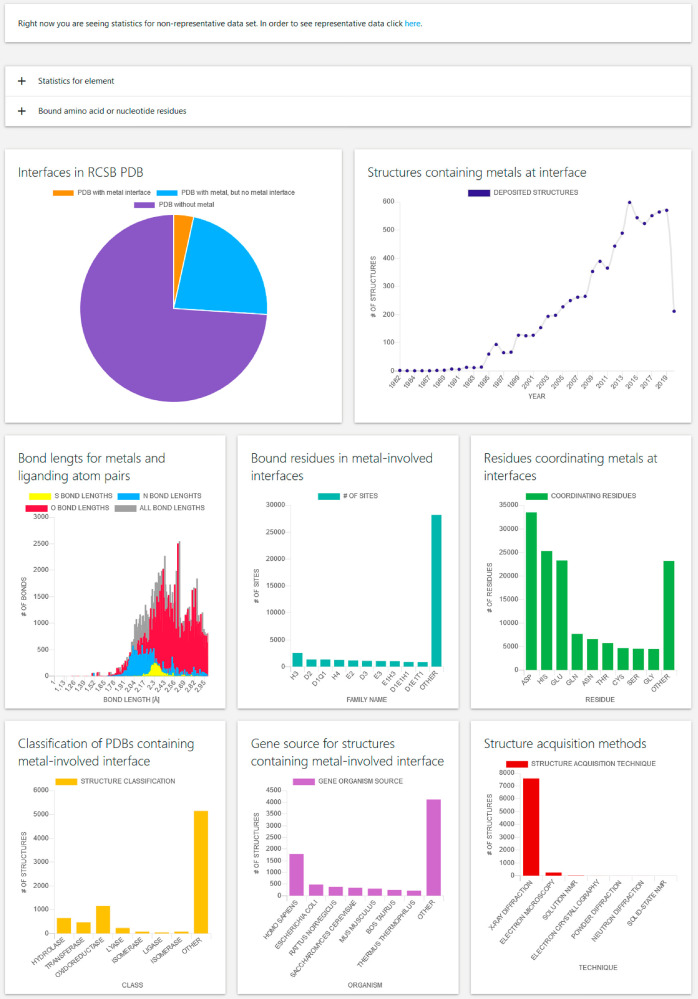
Statistics
The Statistics page contains basic information about bond lengths in metal sites between heteroatoms and metal ion, residues creating metal sites, amount of records, types of protein gathered in the database, gene source of observed macromolecules and other information (Figure 5). By clicking on the Statistics panel in the header, the user is redirected to nonrepresentative (for the whole data set) data records statistics. Whether representative statistics or statistics for the whole data set are displayed can be changed by clicking at the very top of the web page and choosing the preferred option. Below are placed two drop-down panels, the first of which allows one to choose statistics for a certain metal. The other panel shows coordination identifiers for metals in the database. From the first drop-down panel the user can choose a metal element for which statistics are displayed. In the second panel the user sees the most common coordination identifiers and performs a search of metal sites. The first graph presented on a nonrepresentative data set statistics web page allows one to see how many of all structures deposited in the RCSB PDB contain the metal and how many of them contain the intermolecular metal binding site. These data do not currently include metals bound in any kind of compounds (e.g., iron in heme or iron–sulfur clusters). In the future the database will also be extended in order to contain such structures as well. A web page showing statistics for a representative data set instead of the number of interface-containing PDB files shows the number of representative versus nonrepresentative structures gathered in the database. When viewing statistics for a single metal instead of all metals, a pie chart showing the number of particular metal binding sites versus the number of other binding sites is displayed. The rest of the statistics are the same type. Next to the pie chart is placed a graph that shows the number of structures containing intermolecular metal-binding sites published per year. This graph shows the upward trend reflecting the number of published structures in the RCSB PDB. Below on the left is placed a histogram of bond lengths between heteroatoms and metals in binding sites. This type of evaluation of bond length is best done for the whole data set, because in this case having a representative data set is not important for the precise determination of geometric factors, while a large number of observations and high resolution are important.27 Structures that were taken into account in order to prepare this graph have a resolution better than 3 Å. The graph shows that nitrogen and sulfur form distinct groups with clearly defined median and narrow distribution, while in the case of oxygen donors, the length distribution is not so compact. This is due to the large variety of metal-binding oxygen donors. The groups that coordinate metal ions through oxygen donors may be different, such as carboxylates derived from asparaginate residues or glutaminate, carboxylates of the protein C-terminus, but also different low molecular weight ligands such as water, organic acids, etc. An additional factor increasing the variation in the case of bond lengths between oxygen donors and metals is the type of metal; for different metals, different bond lengths with the same metals will be observed. This effect is not so well visible in the case of sulfur and nitrogen donors because these are donors for a narrower group of metals. Next to the distribution of bond lengths there is the number of the most frequent coordination identifiers. Both in the case of representative and nonrepresentative data, records with a small number of bound amino acid or nucleotide residues (two or three) will be frequent. In some cases, the coordination sphere in the structure will be filled by low molecular weight ligands, while in other cases they will not be described in the PDB structure for various reasons, including low resolution. Note that there is a high probability that these types of structures will not be physiological. The last graph describing metal binding sites presents information about the occurrence of a certain residue in metal-binding sites. Generally, occurrence of residues in metal-binding sites follows the HSAB (hard and soft acids and bases) concept; thus residues that can coordinate metal via carboxylates will be most present in metal sites containing Ca2+, Mg2+, Na+, K+. Higher occurrence of histidyl residues and acidic residues in Zn2+ coordination may reflect moderate binding affinity and stability of intermolecular binding sites. Next to the chart representing residues in the metal binding site a bar graph showing classification of PDB files deposited in the RCSB PDB is placed. Because of the huge variation of classifiers, making classification and data presentation almost impossible to do, and because enzymes are the most common group in gathered records, we decided to classify PDB files based on enzyme classification. The succeeding graph shows the gene source for structures containing intermolecular metal binding site, roughly reflecting the gene source distribution in the RCSB PDB, meaning that structures containing intermolecular metal binding sites are not particularly represented in a specific organismal group, but rather follow the trend in RCSB. The last graph presented on the Statistics web page informs the user about techniques that have been used to acquire the structural model, which again is consistent with the trend in the RCSB PDB.
Figure 5.
General statistics page for whole MPPI database. The database statistics can be viewed depending on whether they are displayed for a representative data set or not; this can be chosen on top of the Web site. Below the option of data set selection there are two drop-down panels, which allow one to select the metal for which data are displayed and to select the coordination identifier for a given data set. By clicking on a specific coordination identifier the user is redirected to the search option. Below there is a set of graphs described in more detail in the text.
Prevalence of Interfacial Metal Binding Sites
Of the 227 854 structures deposited in the RCSB PDB at the time of the last database update (October 30, 2020), 50 565 contain a metal as a standalone ion, while 7854 of them were found to contain a metal-involved interface as nonrepresentative sites. Among 6345 representative metal binding sites gathered in the InterMetalDB database, Ca2+ binding sites are the most common, represented by 1403 sites, followed by Zn2+ (1357 sites) and Mg2+ (1110 sites) (Table 1). These three elements are also the most common metal ions in the entire RCSB PDB, and it is no wonder that they will be profoundly represented in InterMetalDB. A lower, but still high, number of protein complex structures contain monovalent Na+ and K+ at the interfaces. Their role is in most examples linked with protein or nucleotide charge compensation and structure stabilization. As a result of the stabilization metal-mediated macromolecule-macromolecule complexes are formed. Interestingly, the high content of iron ions (both Fe2+ and Fe3+) in the RCSB PDB does not correspond to the number in the InterMetalDB. While Fe3+ is present in only 118 representative sites, the Fe2+ ion was found at 23 unique interfaces. It is probably caused by increased likelihood of oxidation at interfaces, but also the fact that iron ions usually do not play a structural role in proteins, but rather catalytic.28 One additional reason why iron ion representation on the macromolecular interfaces is low, and does not correspond to abundance in RCSB PDB, may be due to querying only for the chemical component identifier, which means that only structures that contain standalone ion metals are returned, i.e., structures containing iron–sulfur clusters, heme, or other similar iron-containing particles are not analyzed.
Table 1. Number of Metal Binding Sites in InterMetalDB for Particular Elementsa.
| metal ion | representative | nonrepresentative |
|---|---|---|
| Ca2+ | 1434 | 13991 |
| Zn2+ | 1350 | 6128 |
| Mg2+ | 1104 | 4248 |
| Na+ | 774 | 4148 |
| K+ | 413 | 2703 |
| Cd2+ | 220 | 2565 |
| Mn2+ | 302 | 1625 |
| Cu2+ | 168 | 1193 |
| Fe2+ | 58 | 1035 |
| Fe3+ | 112 | 965 |
| Ni2+ | 173 | 682 |
| Co2+ | 85 | 519 |
| Au+ | 5 | 271 |
| Ba2+ | 23 | 120 |
| Pd2+ | 4 | 99 |
| Ag+ | 38 | 82 |
| Hg2+ | 31 | 54 |
| Rb+ | 5 | 53 |
| Tl+ | 11 | 52 |
| Cs+ | 11 | 48 |
| Cu+ | 22 | 45 |
| Pt2+ | 12 | 41 |
| Sr2+ | 18 | 28 |
| Tb3+ | 1 | 22 |
| La3+ | 8 | 20 |
| Li+ | 9 | 18 |
| Sm3+ | 11 | 16 |
| Pb2+ | 6 | 13 |
| Mn3+ | 0 | 12 |
| Gd | 2 | 6 |
| Ho | 2 | 4 |
| Lu3+ | 3 | 3 |
| Au3+ | 0 | 2 |
| Cr3+ | 1 | 2 |
| Eu3+ | 1 | 2 |
| Re | 1 | 2 |
| Pr3+ | 2 | 2 |
| Yb3+ | 1 | 2 |
| Eu2+ | 1 | 1 |
| Gd3+ | 1 | 1 |
| total | 6423 | 40823 |
The most common interfacial metal binding sites contain Ca2+, Zn2+, and Mg2+.
Very similar to Fe2+, the presence of Cu+ (22 representative sites) on protein interfaces is rather rare due to its capability for oxidation and lack of structural properties. However, it is worth underlining that Cu+ is cellularly transported between chaperone proteins through the formation of interfacial sites, and therefore the list of interfacial copper sites contains such transport-active complexes.29 Manganese is present in metalloproteins as Mn2+ and Mn3+ where it serves catalytically and structurally, but interfacial sites contain only Mn2+, and this state is recognized as a structural one. Metal ions such as Cd2+, Hg2+, Co2+, Ni2+, and Ag+ are frequently used as metal probes for Zn2+ or Cu+ and therefore are frequently investigated by structural methods. The question how found interfacial sites probe native sites is rather an individual example and requires solution studies. It was shown that interfacial Hg2+ or Cd2+ in the Rad50 homodimer very well mimics the Zn2+ complex, and they have been used for characterization of the complex.30,31 The presence of other metal ions in structurally characterized macromolecule-macromolecule complexes is more likely to be linked with a particular interest and can be explored individually by searching in original reports, a list of which can be easily downloaded using InterMetalDB.
Ligands of Interprotein Metal Binding Sites
The most common residue coordinating metal ions in interfaces identified by the InterMetalDB database is an aspartyl residue followed by histidyl residue (Table 2). The first one is usually found in sites containing Ca2+, Mg2+ but also Zn2+, Na+ and Mn2+, while the second is more common for zinc sites. Acidic residues, glutaminyl and asparaginyl with histidinyl and threonyl residues account for 72.5% of all amino acid residues in all metal binding sites and 66.4% in the nonredundant data set where in order to remove bias to more often studied macromolecules only representative metal-binding sites are analyzed. It means that binding sites in the nonrepresentative data set are characterized to some degree by smaller variation than a more representative set. Although a cysteinyl residue is found in many physiologically confirmed Zn2+-involved protein–protein complexes, in the whole database it accounts for only 3.73%. One reason why acidic and histidyl residues are frequent in interprotein metal sites is the fact that Ca2+, Mg2+, Na+, K+ are hard acids according to the HSAB concept and Zn2+ demonstrates moderate character, and therefore they prefer coordination of oxygen and nitrogen donors, respectively.32 Another explanation is linked with the fact that those residues are flexible and have a large size, which allows main chains of interacting protein subunits to have a longer distance without or with minimal conformational change of protein molecules. Moreover, in the case of Zn2+ those residues guarantee moderate stability, which is required for transient sites.10 This is in contrast to cysteinyl residues, which are closer to each other at metal interfaces and require a more significant change of protein structure upon metal binding, increasing the thermodynamic stability of such a site.33,34
Table 2. Most Common Amino Acid Residues Found in the Metal Sites Located at Macromolecular Interfacesa.
| representative |
nonrepresentative |
||
|---|---|---|---|
| residue | count | residue | count |
| Asp | 4576 | Asp | 33468 |
| His | 3558 | His | 25297 |
| Glu | 3442 | Glu | 23250 |
| Asn | 922 | Gln | 7685 |
| Gly | 891 | Asn | 6587 |
| dG | 819 | Thr | 5755 |
| other | 7183 | other | 36877 |
Residue is considered to be bound to metal if any heteroatom (e.g., oxygen, nitrogen) is in radius 3 Å or less from metal. In order to see the detailed distribution of the residues based on bound metal, please visit the statistics web page of InterMetalDB.
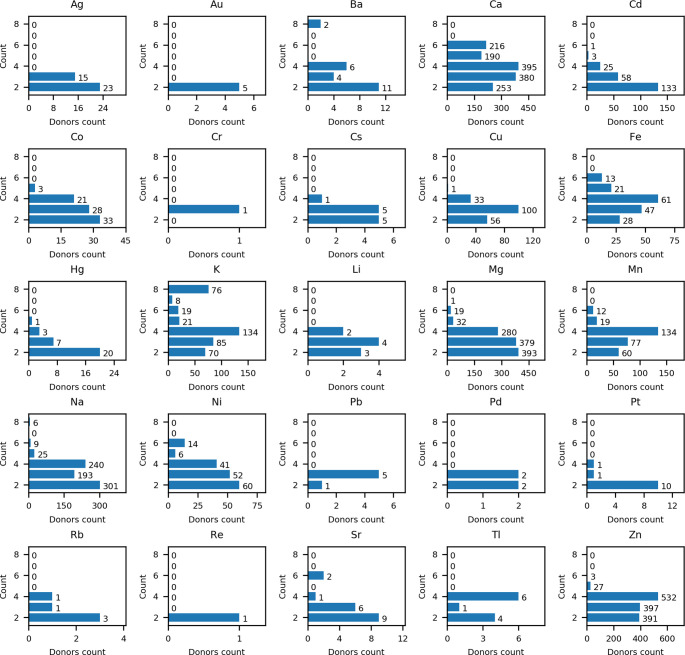
In the nonrepresentative set the most common number of bound ligand donors is three, followed by four and two. In the representative set, the most common number of bound ligand donors is two, followed by four and three, corresponding to 30.1, 29.4, and 29.0% of all sites (Table 3). Interestingly, the higher coordination number in intermolecular sites is relatively low and accounts for 5.3, 4.8, 0.1, and 1.2% in the case of five, six, seven, and eight donors, respectively. The largest number of sites with six donors was identified for Ca2+ while K+ demonstrates the largest tendency to form sites with eight donors. Detailed information on the number of ligand donors of a particular metal ion is presented in Figure S1 and Figure 6, for nonrepresentative and representative data sets, respectively. It is worth underlining that the number of donors bound to various metal ions depends on their chemical features according to bioinorganic rules, but metal sites in X-ray structures may differ from those present in the solution.35 The high representation of such a number of low-filled coordination spheres can be explained by unresolved crystal structures of low molecular weight ligands such as water molecules and others. While probably most of these sites are not physiological, or metal binding affinities to such sites are extremely weak, we have not decided to remove such sites from InterMetalDB, since there may be sites that are physiologically important. An example of this is the structure of P. furiosus Rad50’s zinc hook domain (PDB ID: 6ZFF), with a not fully resolved Zn2+-coordination sphere.10,36
Table 3. Number of Metal Binding Sites Containing a Specific Number of Residues Coordinating Metala.
| no. of donors | representative | nonrepresentative |
|---|---|---|
| 2 | 1883 | 9560 |
| 3 | 1865 | 13794 |
| 4 | 1924 | 12266 |
| 5 | 349 | 3166 |
| 6 | 309 | 1358 |
| 7 | 9 | 57 |
| 8 | 84 | 622 |
Precise distribution of donors over metal can be found in Figure S3.
Figure 6.
Number of donors in a metal site depending on a metal ion plotted for a representative data set. The most common places are those that have the number of donors in the range between 2 and 4. Data for lanthanides are presented in Supporting Information (Figure S1).
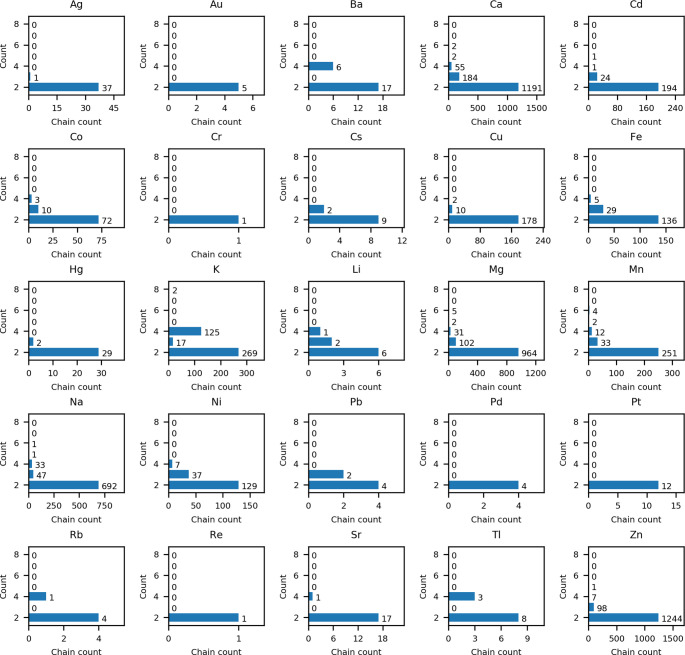
The most common assembly in InterMetalDB is the association of the smallest possible number of macromolecules at the metal interface, that is two, accounting for 86% of all representative interfaces. Subsequent numbers of metal-bound chains, that is three and four, correspond to 9.2% and 4.4% of all representative interfaces, respectively (Table 4). Detailed information on the number of chain ligands depending on metal ions is presented in Figure S2 and Figure 7, for nonrepresentative and representative data sets, respectively. The macromolecular interfaces gathered in InterMetalDB involving metal binding occur in nucleic acid molecules, proteins and also between nucleic acid and protein (e.g., the binding of catalytic Ca2+ by Hinc II restriction endonuclease (PDB ID 1TW8).37 The largest number of chains to be bound is the complex of K+ with nucleic acid creating the i-motif (PDB ID 1V3P).38 In the case of nucleic acids, metal ions participate in the stabilization of G-quadruplexes and i-motif DNA structures. While different types of cations will promote the formation of G-quadruplex structures, starting with bivalent ions such as Ba2+ (PDB ID: 4U92)39 or Pb (PDB ID: 6A85),40 the physiologically relevant G-quadruplex structures will be Na+ and K+.41 Although the coordination number correlates with the number of ligands to a certain degree, the most important factor deciding on the quantity of macromolecules at the interface is the number of donors coordinated to the metal ion from a particular ligand (chain).
Table 4. Number of Metal Sites Containing a Certain Number of Chainsa.
| no. of metal sites | representative | nonrepresentative |
|---|---|---|
| 2 | 5501 | 31549 |
| 3 | 605 | 5789 |
| 4 | 294 | 3290 |
| 5 | 9 | 174 |
| 6 | 12 | 15 |
| 8 | 2 | 6 |
| total | 6423 | 40823 |
The most binding sites are created by two chains.
Figure 7.
Number of chains creating metal sites, depending on a bound metal ion. Graphs are prepared for representative data set. Formation of an intermolecular metal ion binding site occurs between two macromolecule chains. Data for lanthanides are presented in Supporting Information (Figure S2).
Comparison with Other Databases
Integration of structural information about metalloproteins provides the basis for utilization of metal ions and their roles in proteins. It is no wonder that in recent years several databases aggregating metalloproteins have been provided. Nevertheless, some of them are no longer maintained or even accessible, e.g., MDB (Metalloprotein Database and Browser),42 Mespeus,17 or dbTEU.43 Unfortunately, available electronic resources that are regularly updated (MetalPDB,19,20 ZincBind21), although providing a user-friendly interface, do not allow for filtering for metal ions that are bound at macromolecular interfaces. Furthermore, MetalPDB records are based mostly on asymmetric units and ZincBind provides only information of proteins that bind zinc. While ZincBind seems to be updated weekly or monthly, MetalPDB is not updated so often; the last update, as of the time of writing, was 2019–09–18. Both resources are a good resource of knowledge about metalloproteins. MetalPDB contains structures with intermolecularly bound metal but does not have a function to query for such records.
An additional obstacle that makes MetalPDB not suitable to find intermolecularly bound metals is the fact that records in MetalPDB are mostly based on asymmetry units. In the case of examining intermolecularly bound metal ions, this is extremely important, as the metal ions bound in this way will often be bound on the surface of the chains, which will only create an interface after constructing a biological assembly, as exemplified by the human rhinovirus 16 coat protein structure (PDB ID: 1AYM),44 in which the zinc is located on the interface created by the five chains, but this is only visible in the biological assembly. ZincBind overcomes this obstacle by aggregating data that are based on the biological assembly. In addition, ZincBind offers a much friendlier record search interface and a GraphQL application programming interface that allows programmatic access to the aggregated data. MetalPDB allows for downloading only partial information from its database, and download of a 5 Å-radius cut-out of the structure around the metal ion. Currently, in the case of InterMetalDB, data can be retrieved from the site after prior filtering. All updated resources allow one to view directly the structure of the selected record, although by using different front end viewers, JSmol in the case of MetalPDB, and NGL Viewer in the case of both ZincBind and InterMetalDB. Both InterMetalDB and other available resources contain web pages allowing quick insight into general statistics of records contained in the database. All these statistics relate to the interaction of metal ions with proteins and nucleic acids, although each database gives an insight into a slightly different part of this field, because ZincBind focuses only on the interaction of macromolecules with zinc ions, MetalPDB aggregates all records containing the metal, while InterMetalDB focuses only on structures that contain intermolecularly bound metal, which is why the statistics provided may differ. This difference may be mainly seen in terms of what residues will be involved in the metal ion binding and what the coordination identifier will be, and this seems to be related to the fact that the amino acid residues forming intermolecular metal binding sites must have slightly different properties. As it has been discussed, in the case of Zn2+ residues, creating an intermolecular binding site will provide the moderate stability needed for transient binding and enabling association and dissociation.10 InterMetalDB allows for advanced search of structures and metal binding sites in a very similar way to the databases discussed here, except for one function. The function, which is not yet implemented in InterMetalDB, is searching for structures by a sequence. In the future InterMetalDB will also be extended with this feature as well. So rather than replacing those existing databases, InterMetalDB aims to complement existing resources, providing the possibility for advanced searching of intermolecular interfaces.
Conclusions
InterMetalDB has been created in order to provide a resource that identifies and aggregates all metal ions involved in macromolecular interfaces from the RCSB PDB. Although other databases also contain this type of interaction, none of them allows for filtering of such records. InterMetalDB is the first database strictly focused on aggregating and searching for this type of metal binding sites. The database is updated on a regular basis and allows for the retrieval of searched results in different forms. The InterMetalDB clusters intermolecular metal binding sites in accordance with 50% sequential similarity of a given molecule and the nearest metal environment, then the representative site is selected on the basis of the best resolution of the examined structure. No restraints on structure resolution were applied during data acquisition, and structures included in the InterMetalDB are based on biological assemblies (described in PDB files). The web interface allows for searching, browsing and downloading the data. Query filters allow for filtering based on structure quality, deposition date, as well as other parameters such as number of ligands, number of coordinating chains, etc. InterMetalDB gives insight into interfacial metal binding, additionally serving as a useful resource for researchers willing to develop machine learning models predicting macromolecular interactions and involvement of metal ions in such processes. We believe that the data set contained in InterMetalDB will be helpful to other researchers interested in interfacial metal binding, metal-induced protein polymerization, aggregation, nanoparticle creation, and metalloprotein engineering and will boost research in those fields. In the future the resource as well as the web interface will be expanded as needed.
InterMetalDB can be accessed at https://intermetaldb.biotech.uni.wroc.pl and the source code can be viewed and downloaded at https://github.com/jzftran/InterMetalDB.
Acknowledgments
This research and authors were supported by the National Science Centre of Poland (NCN) under the Opus grant no. 2016/21/B/NZ1/02847 (to A.K.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00906.
Figures S1: Number of donors in a metal site depending on a lanthanide metal ion; Figure S2: Number of donors in a metal site depending on a metal ion; Figure S3: Number of donors in a metal site depending on a lanthanide metal ion; Figure S4: Number of chains creating metal sites, depending on a bound metal ion; Figure S5: Number of chains creating metal sites, depending on a bound metal ion; Figure S6: Number of chains creating metal sites, depending on a bound lanthanide metal ion (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Andreini C.; Bertini I.; Cavallaro G.; Holliday G. L.; Thornton J. M. Metal ions in biological catalysis: From enzyme databases to general principles. JBIC, J. Biol. Inorg. Chem. 2008, 13, 1205–1218. 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- Waldron K. J.; Rutherford J. C.; Ford D.; Robinson N. J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- Song W. J.; Sontz P. A.; Ambroggio X. I.; Tezcan F. A. Metals in protein–protein interfaces. Annu. Rev. Biophys. 2014, 43, 409–431. 10.1146/annurev-biophys-051013-023038. [DOI] [PubMed] [Google Scholar]
- Vallee B. L.; Auld D. S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 1990, 29, 5647–5659. 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- Maret W.Zinc and the zinc proteome. In Metallomics and the Cell; Banci L., Ed.; Springer Netherlands: Dordrecht, 2013; pp 479–501. [Google Scholar]
- Kochańczyk T.; Drozd A.; Krężel A. Relationship between the architecture of zinc coordination and zinc binding affinity in proteins – insights into zinc regulation. Metallomics 2015, 7, 244–257. 10.1039/C4MT00094C. [DOI] [PubMed] [Google Scholar]
- Auld D. S. Zinc coordination sphere in biochemical zinc sites. BioMetals 2001, 14, 271–313. 10.1023/A:1012976615056. [DOI] [PubMed] [Google Scholar]
- Maret W.Protein interface zinc sites: The role of zinc in the supramolecular assembly of proteins and in transient protein-protein interactions. In Handbook of Metalloproteins; John Wiley & Sons, Ltd.: Chichester, UK, 2006; pp 1–10. [Google Scholar]
- Sontz P. A.; Song W. J.; Tezcan F. A. Interfacial metal coordination in engineered protein and peptide assemblies. Curr. Opin. Chem. Biol. 2014, 19, 42–49. 10.1016/j.cbpa.2013.12.013. [DOI] [PubMed] [Google Scholar]
- Kocyła A.; Tran J. B.; Krężel A. Galvanization of protein–protein interactions in a dynamic zinc interactome. Trends Biochem. Sci. 2021, 46, 64. 10.1016/j.tibs.2020.08.011. [DOI] [PubMed] [Google Scholar]
- Andreini C.; Banci L.; Bertini I.; Rosato A. Counting the zinc-proteins encoded in the human genome. J. Proteome Res. 2006, 5, 196–201. 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- Berman H.; Henrick K.; Nakamura H.; Markley J. L. The Worldwide Protein Data Bank (WwPDB): Ensuring a Single, Uniform Archive of PDB Data. Nucleic Acids Res. 2007, 35, D301–D303. 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.; Chruszcz M.; Lasota P.; Lebioda L.; Minor W. Data mining of metal ion environments present in protein structures. J. Inorg. Biochem. 2008, 102, 1765–1776. 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C.; Bertini I.; Cavallaro G. Minimal functional sites allow a classification of zinc sites in proteins. PLoS One 2011, 6, e26325. 10.1371/journal.pone.0026325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai K.; Nagata N. Metal–ligand interactions: an analysis of zinc binding groups using the Protein Data Bank. Eur. J. Med. Chem. 2012, 51, 271–276. 10.1016/j.ejmech.2012.02.028. [DOI] [PubMed] [Google Scholar]
- Laitaoja M.; Valjakka J.; Jänis J. Zinc coordination spheres in protein structures. Inorg. Chem. 2013, 52, 10983–10991. 10.1021/ic401072d. [DOI] [PubMed] [Google Scholar]
- Hsin K.; Sheng Y.; Harding M. M.; Taylor P.; Walkinshaw M. D. MESPEUS: A database of the geometry of metal sites in proteins. J. Appl. Crystallogr. 2008, 41, 963–968. 10.1107/S002188980802476X. [DOI] [Google Scholar]
- Jayakanthan M.; Muthukumaran J.; Chandrasekar S.; Chawla K.; Punetha A.; Sundar D. ZifBASE: a database of zinc finger proteins and associated resources. BMC Genomics 2009, 10, 421. 10.1186/1471-2164-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C.; Cavallaro G.; Lorenzini S.; Rosato A. MetalPDB: a database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2012, 41, D312–D319. 10.1093/nar/gks1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putignano V.; Rosato A.; Banci L.; Andreini C. MetalPDB in 2018: a database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2018, 46 (D1), D459–D464. 10.1093/nar/gkx989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland S. M.; Martin A. C. R. ZincBind - the database of zinc binding sites. Database 2019, 2019, baz006. 10.1093/database/baz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland S. M.; Martin A. C. R. Atomium - a python structure parser. Bioinformatics 2020, 36, 2750–2754. 10.1093/bioinformatics/btaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M.; Steinegger M.; Söding J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 2019, 35, 2856–2858. 10.1093/bioinformatics/bty1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí-Renom M. A.; Stuart A. C.; Fiser A.; Sánchez R.; Melo F.; Šali A. comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- McClain B.; Settembre E.; Temple B. R. S.; Bellamy A. R.; Harrison S. C. X-Ray crystal structure of the rotavirus inner capsid particle at 3.8 Å resolution. J. Mol. Biol. 2010, 397, 587–599. 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A. S.; Bradley A. R.; Valasatava Y.; Duarte J. M.; Prlić A.; Rose P. W. NGL Viewer: web-based molecular graphics for large complexes. Bioinformatics 2018, 34, 3755–3758. 10.1093/bioinformatics/bty419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokmanić I.; Šikić M.; Tomić S. Metals in proteins: Correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008, 64, 257–263. 10.1107/S090744490706595X. [DOI] [PubMed] [Google Scholar]
- Andreini C.; Putignano V.; Rosato A.; Banci L. The human iron-proteome. Metallomics 2018, 10, 1223–1231. 10.1039/C8MT00146D. [DOI] [PubMed] [Google Scholar]
- Banci L.; Bertini I.; Cantini F.; Felli I. C.; Gonnelli L.; Hadjiliadis N.; Pierattelli R.; Rosato A.; Voulgaris P. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat. Chem. Biol. 2006, 2, 367–368. 10.1038/nchembio797. [DOI] [PubMed] [Google Scholar]
- Hopfner K.; Craig L.; Moncalian G.; Zinkel R. A.; Usui T.; Owen B. A. L.; Karcher A.; Henderson B.; Bodmer J.-L.; McMurray C. T.; Carney J. P.; Petrini J. H. J.; Tainer J. A. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 2002, 418, 562–566. 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Padjasek M.; Maciejczyk M.; Nowakowski M.; Kerber O.; Pyrka M.; Koźmiński W.; Krężel A. Metal exchange in the interprotein ZnII binding site of the Rad50 hook domain: Structural insights into CdII -induced DNA-repair inhibition. Chem. - Eur. J. 2020, 26, 3297–3313. 10.1002/chem.201904942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R. G. Hard and soft acids and bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. 10.1021/ja00905a001. [DOI] [Google Scholar]
- Sikorska M.; Krężel A.; Otlewski J. Femtomolar Zn2+ Affinity of LIM domain of PDLIM1 protein uncovers crucial contribution of protein–protein interactions to protein stability. J. Inorg. Biochem. 2012, 115, 28–35. 10.1016/j.jinorgbio.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Kochańczyk T.; Nowakowski M.; Wojewska D.; Kocyła A.; Ejchart A.; Koźmiński W.; Krężel A. Metal-coupled folding as the driving force for the extreme stability of Rad50 zinc hook dimer assembly. Sci. Rep. 2016, 6, 36346. 10.1038/srep36346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I.; Gray H. B.; Stiefel E. I.; Valentine J. S. Biological inorganic chemistry: Structure and reactivity. Choice Rev. Online 2007, 44, 6242. 10.5860/CHOICE.44-6242. [DOI] [Google Scholar]
- Soh Y.; Basquin J.; Gruber S. A Rod conformation of the Pyrococcus furiosus Rad50 coiled coil. Proteins: Struct., Funct., Genet. 2021, 89, 251. 10.1002/prot.26005. [DOI] [PubMed] [Google Scholar]
- Etzkorn C.; Horton N. C. Ca2+ binding in the active site of HincII: Implications for the catalytic mechanism. Biochemistry 2004, 43, 13256–13270. 10.1021/bi0490082. [DOI] [PubMed] [Google Scholar]
- Kondo J. Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats. Nucleic Acids Res. 2004, 32, 2541–2549. 10.1093/nar/gkh575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.; Huang T.; Lukeman P. S.; Paukstelis P. J. Crystal structure of a DNA/Ba2+ G-quadruplex containing a water-mediated C-tetrad. Nucleic Acids Res. 2014, 42, 13422–13429. 10.1093/nar/gku1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Wang R.; Yu X.; Shen F.; Lan W.; Haruehanroengra P.; Yao Q.; Zhang J.; Chen Y.; Li S.; Wu B.; Zheng L.; Ma J.; Lin J.; Cao C.; Li J.; Sheng J.; Gan J. High-resolution DNA quadruplex structure containing all the A-, G-, C-, T-tetrads. Nucleic Acids Res. 2018, 46, 11627–11638. 10.1093/nar/gky902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D.; Mirihana Arachchilage G.; Basu S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 38. 10.3389/fchem.2016.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnetto J. M. MDB: The Metalloprotein database and browser at the Scripps Research Institute. Nucleic Acids Res. 2002, 30, 379–382. 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Gladyshev V. N. DbTEU: a protein database of trace element utilization. Bioinformatics 2010, 26, 700–702. 10.1093/bioinformatics/btp705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield A. T.; Lee W.; Zhao R.; Oliveira M. A.; Minor I.; Rueckert R. R.; Rossmann M. G. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure 1997, 5, 427–441. 10.1016/S0969-2126(97)00199-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.