Abstract
Background
We examined the prognostic significance of circulating tumor cell (CTC) dynamics during treatment in metastatic breast cancer (MBC) patients receiving first-line chemotherapy.
Methods
Serial CTC data from 469 patients (2202 samples) were used to build a novel latent mixture model to identify groups with similar CTC trajectory (tCTC) patterns during the course of treatment. Cox regression was used to estimate hazard ratios for progression-free survival (PFS) and overall survival (OS) in groups based on baseline CTCs, combined CTC status at baseline to the end of cycle 1, and tCTC. Akaike information criterion was used to select the model that best predicted PFS and OS.
Results
Latent mixture modeling revealed 4 distinct tCTC patterns: undetectable CTCs (56.9% ), low (23.7%), intermediate (14.5%), or high (4.9%). Patients with low, intermediate, and high tCTC patterns had statistically significant inferior PFS and OS compared with those with undetectable CTCs (P < .001). Akaike Information Criterion indicated that the tCTC model best predicted PFS and OS compared with baseline CTCs and combined CTC status at baseline to the end of cycle 1 models. Validation studies in an independent cohort of 1856 MBC patients confirmed these findings. Further validation using only a single pretreatment CTC measurement confirmed prognostic performance of the tCTC model.
Conclusions
We identified 4 novel prognostic groups in MBC based on similarities in tCTC patterns during chemotherapy. Prognostic groups included patients with very poor outcome (intermediate + high CTCs, 19.4%) who could benefit from more effective treatment. Our novel prognostic classification approach may be used for fine-tuning of CTC-based risk stratification strategies to guide future prospective clinical trials in MBC.
Analysis of circulating tumor cells (CTCs) in blood offers a minimally invasive approach for evaluating prognosis in patients with metastatic breast cancer (MBC) (1,2). Accumulating evidence from clinical studies conducted over the past decade and a half have demonstrated that CTCs are strong predictors of outcome in MBC (1,2). In 2004, Cristofanilli and colleagues (3) showed that pretreatment detection of 5 or more CTCs in 7.5 mL of blood using CellSearch was strongly associated with increased risk of disease progression and death. A follow-up study by Hayes and colleagues (4) showed that, in addition to baseline, increased levels of CTCs at any time point during treatment were also highly prognostic. Subsequent validation studies, including a pooled analysis by Bidard and colleagues (2)—involving 20 studies and approximately 1900 MBC patients (European Pooled Analysis of individual CTC [EPAC])—confirmed the prognostic significance of CTCs. More recently, a study involving 2436 patients with MBC conducted by a team of international experts unequivocally showed that patients with baseline levels of greater than or equal to 5 CTCs per 7.5 mL of blood (stage IVaggressive) had inferior overall survival (OS) compared with those with less than 5 CTCs (stage IVindolent) (5). Based on these confirmatory results, the authors recommended considering these 2 prognostic groups (stage IVindolent vs stage IVaggressive) as stratification factors in prospective clinical trials in MBC (5) .
CTCs offer a great advantage over tissue-based markers because they are amenable to repeat measurements with minimal harm to patients. To date, little is known about CTC dynamics during treatment and its clinical significance. We hypothesized that analysis of serial CTCs can aid in further stratification of poor-prognosis patients into distinct prognostic subgroups. The current CTC-based risk stratification approach (2-4) defines poor-prognosis patients as having greater than or equal to 5 CTCs per 7.5 mL of blood at baseline [CTC-positive or stage IVaggressive (5)]. Given that the stage IVaggressive subset represents approximately 50% of the MBC patients (2-4), its utility for risk stratification may be limited because of the potential clinical heterogeneity in this large group of patients. We posit that serial assessment of CTCs could reveal prognostic groups based on CTC trajectory (tCTC) patterns (ie, changes in CTC levels across timepoints) and therefore help refine CTC-based risk stratification in MBC.
To assess CTC dynamics over the course of therapy, we performed serial testing in 783 patients with MBC patients enrolled in Cancer and Leukemia Group B (CALGB) 40502/NCCTG N063H, a randomized phase III trial that compared the efficacy of first-line nab-paclitaxel or ixabepilone with paclitaxel (6). A novel latent mixture model was developed to classify patients into groups according to tCTC patterns. We assessed the prognostic impact of the tCTC model and compared it with previously clinically validated prognostic models (ie, baseline CTCs [bCTC] and combined CTC status at baseline and end of first cycle of therapy [cCTC]), then validated the model in an independent dataset.
Methods
Patients and Samples
This planned retrospective correlative study was designed to evaluate associations between CTCs and clinical outcomes in patients enrolled in the CALGB 40502 trial (NCT00785291; Supplementary Table 1; Supplementary Methods, available online) (6). The institutional review boards at the National Cancer Institute and at each site approved the study. All participants provided written informed consent.
CTCs were enumerated using CellSearch at pretreatment and end of cycles 1 and 2 (approximately 28 and approximately 56 days after initiation of treatment), and then at every 3 cycles of chemotherapy until the patient went off study treatment because of consent withdrawal, toxicity, or disease progression. Samples with at least 5 CTCs per 7.5 mL of blood were considered CTC positive.
Statistical Analysis
Serial CTC data were used to build a novel latent mixture model to identify groups with similar tCTC patterns during the course of treatment (Supplementary Methods, available online). The Akaike information criterion (AIC) scores were used to evaluate model quality, with a lower value indicating a higher quality model (7). Parameters from the final best model of tCTC built using CALBG 40502 data (Supplementary Table 2, available online) were then used to predict trajectory groups (8) in an independent dataset of 1920 MBC patients with serial CTC measurements (EPAC) (2). Using the coefficients in Supplementary Table 2 (available online) and the EPAC data, we predicted the probability of class membership for each patient based on formulas developed by Nielsen and colleagues (8).
Clinical endpoints were progression-free survival (PFS) and OS. The median follow-up for this subset was 26 months. Multivariable Cox regression adjusted for known prognostic factors was used to estimate hazard ratios (HR) and 95% confidence intervals (CI), with tCTC models weighted according to trajectory group membership probabilities. The abilities of each model (bCTC vs cCTC vs tCTC) to fit the data were compared using AIC (7).
Log-rank test was performed to compare survival curves. Associations between 2 variables were examined using Pearson χ2 test. A P less than .05 was considered significant. All statistical tests were 2-sided.
Results
Patient Characteristics
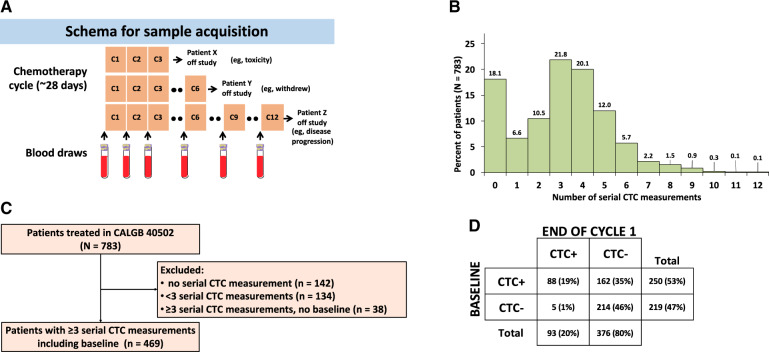
Serial blood samples were collected from 783 patients receiving first-line chemotherapy for MBC (6) (Figure 1, A). CTCs were successfully enumerated in 2418 blood samples with a median of 3 CTC serial measurements per patient (range = 0-12) (Figure 1, B). Of the 783 patients, 314 had no or incomplete CTC data and were excluded from the analysis (Figure 1, C). The final analytic cohort consisted of 469 patients with 3 or more serial CTC data, including baseline. The total number of informative blood samples from these patients was 2202.
Figure 1.
Serial circulating tumor cell (CTC) enumeration in metastatic breast cancer receiving first-line chemotherapy. A) Schema depicting schedule of blood collection in 3 hypothetical patients who came off study because of various reasons. B) Percentage of patients with corresponding number of serial CTC measurements (N = 783 patients and 2202 samples). C) Flowchart showing selection of samples for CTC trajectory analysis. D) CTC status at baseline and at the end of cycle 1.
Clinical and demographic characteristics of patients are summarized in Supplementary Table 1 (available online). Of the 469 patients, 347 (74.0%) were hormone receptor positive (HR+) and 122 (26.0%) were triple negative; 375 (80.1%) had visceral disease, 329 (70.3%) had soft tissue involvement, and 298 (63.7%) had bone metastasis; 182 (38.9%) had received prior taxanes, and 223 (47.5%) had received or were undergoing hormone treatment. Clinicopathologic characteristics and bCTC-positivity rates were balanced across all 3 arms of the original trial (χ2P = .14).
Baseline CTCs
A total of 250 patients (53.3%) had at least 5 CTCs per 7.5 mL of blood and were considered CTC positive (Figure 1, D). A statistically significant association was observed between CTC status and HR+ subtype (P < .001) and the presence of visceral, soft tissue, and bone metastases (P = .03, P = .04, and P < .001, respectively), and the number of prior hormone treatments received (P < .001) (Supplementary Table 3, available online).
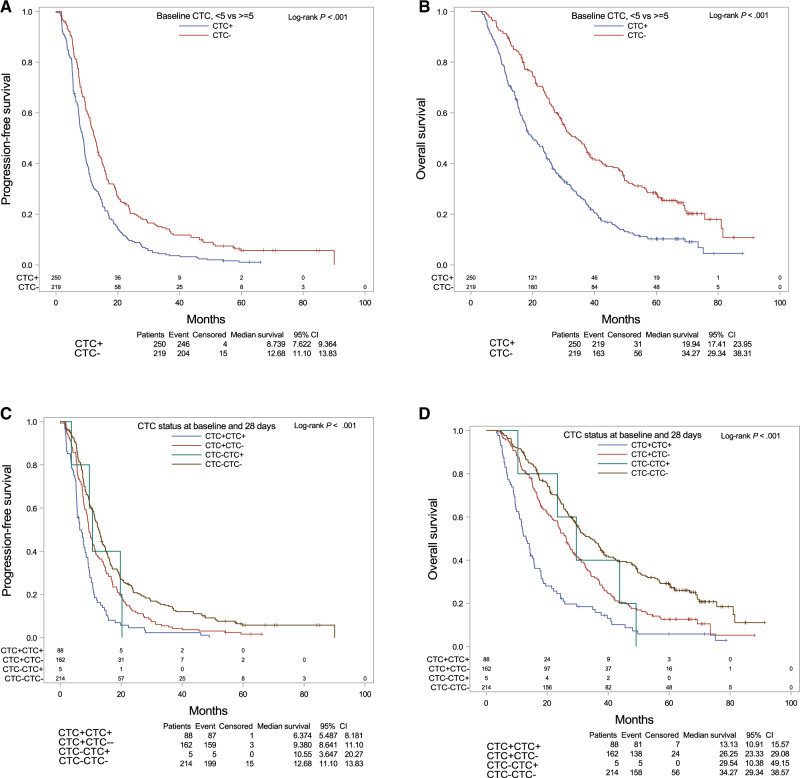
Survival analysis revealed that the PFS (median 8.7 vs 12.7 months) and OS (median = 19.9 vs 34.3 months) were statistically significantly shorter in CTC-positive patients compared with CTC-negative patients (PFS and OS log-rank P < .001) (Figure 2, A and B).
Figure 2.
Prognostic impact of baseline circulating tumor cells (CTCs) and combined CTC status at baseline and early treatment. A) Progression-free survival (PFS) and B) overall survival (OS) based on CTC status at baseline. C) PFS and D) OS based on CTC status at baseline and at the end of cycle 1 in Cancer and Leukemia Group B 40502. Log-rank test was performed to compare survival curves. A 2-sided P < .05 was considered significant.
In multivariable Cox regression models that adjusted for potential confounders, CTCs remained statistically significant predictors of PFS (HR = 1.58, 95% CI = 1.28 to 1.96) and OS (HR = 1.90, 95% CI =1.51 to 2.39) (Table 1;Supplementary Table 4 available online). In addition, triple-negative disease (HR = 1.74, 95% CI = 1.32 to 2.27), prior taxane (HR = 1.31, 95% CI = 1.02 to 1.69), and hormone treatments (1 [HR = 1.39, 95% CI = 1.02 to 1.89] or more [HR = 1.42, 95% CI = 1.06 to 1.90]) were statistically significantly associated with inferior PFS (P = .04 to >.001). Triple-negative disease (HR = 2.03, 95% CI = 1.51 to 2.72), prior taxanes (HR = 1.41, 95% CI = 1.07 to 1.86), and visceral metastatic involvement (HR = 1.45, 95% CI = 1.07 to 1.97) were statistically significantly associated with inferior OS (P = .02 to >.001, respectively).
Table 1.
Association between survival and CTC status and trajectory
| Datasets and modelsa | No. (%)b | PFS |
OS |
||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| CALGB 40502 | |||||
| Baseline CTCs (n = 469) | |||||
| CTC− | 219 (46.7) | Reference | Reference | ||
| CTC+ | 250 (53.3) | 1.58 (1.28 to 1.6) | <.001 | 1.90 (1.51 to 2.39) | <.001 |
| Change in CTC status (baseline to end of cycle 1) (n = 469) | |||||
| CTC−CTC− | 214 (45.6) | Reference | Reference | ||
| CTC+CTC− | 162 (34.5) | 1.34 (1.05 to 1.69) | .02 | 1.51 (1.16 to 1.95) | .002 |
| CTC−CTC+ | 5 (1.1) | 2.15 (0.77 to 6.01) | .14 | 2.88 (1.04 to 7.96) | .04 |
| CTC+CTC+ | 88 (18.8) | 2.48(1.86 to 3.32) | <.001 | 3.65 (2.68 to 4.98) | <.001 |
| tCTC (n = 469) | |||||
| tCTCneg | 267 (56.9) | Reference | Reference | ||
| tCTClo | 111 (23.7) | 1.89 (1.46 to 2.45) | <.001 | 2.25 (1.71 to 2.98) | <.001 |
| tCTCmid | 68 (14.5) | 2.48 (1.79 to 3.43) | <.001 | 3.91 (2.76 to 5.53) | <.001 |
| tCTChi | 23 (4.9) | 3.04 (1.85 to 4.98) | <.001 | 7.54( 4.45 to 12.79) | <.001 |
| EPAC | |||||
| tCTC (n = 1856)c | |||||
| tCTCneg | 892 (48.1) | Reference | Reference | ||
| tCTClo | 627 (33.8) | 1.57 (1.38 to 1.79) | <.001 | 2.49 (2.11 to 2.95) | <.001 |
| tCTCmid | 179 (9.6) | 1.94 (1.59 to 2.36) | <.001 | 3.02 (2.38 to 3.83) | <.001 |
| tCTChi | 158 (8.5) | 3.41 (2.78 to 4.17) | <.001 | 6.55( 5.10 to 8.40) | <.001 |
| tCTC, first line only (n = 776)d | |||||
| tCTCneg | 380 (49.0) | Reference | Reference | ||
| tCTClo | 281 (36.2) | 1.73 (1.40 to 2.12) | <.001 | 3.43 (2.53 to 4.64) | <.001 |
| tCTCmid | 64 (8.2) | 2.43 (1.77 to 3.33) | <.001 | 3.61 (2.34 to 5.57) | <.001 |
| tCTChi | 51 (6.6) | 3.66 (2.58 to 5.18) | <.001 | 6.94 (4.32 to 11.15) | <.001 |
| tCTC, single bCTC only (n = 916) | |||||
| tCTCneg | 487 (53.2) | Reference | Reference | ||
| tCTClo | 251 (27.4) | 1.67 (1.36 to 2.05) | <.001 | 2.50 (2.0 to 3.2) | <.001 |
| tCTCmid | 85 (9.3) | 2.02 (1.49 to 2.73) | <.001 | 2.6 (1.8 to 3.7) | <.001 |
| tCTChi | 93 (10.2) | 3.61 (2.70 to 4.82) | <.001 | 6.2 (4.4 to 8.7) | <.001 |
A summary of the multivariable Cox regression analysis to determine correlation between survival in the following CTC models: CTC status at baseline (see Supplementary Table 4, available online), combined CTC status at baseline and end of cycle 1 (see Supplementary Table 5, available online), tCTC groups in CALGB 40502 (see Supplementary Table 6, available online), and the Bidard et al. pooled analysis (EPAC) (2) (see Supplementary Tables 8-10, available online). bCTC = baseline circulating tumor cell; CALGB = Cancer and Leukemia Group B; CI = confidence interval; CTC = circulating tumor cell; EPAC = European Pooled Analysis of individual CTC; HR = hazard ratio; OS = overall survival; PFS = progression-free survival; tCTC = circulating tumor cell trajectory; tCTChi = high trajectory for circulating tumor cells; tCTClo = low trajectory for circulating tumor cells; tCTCmid = intermediate trajectory for circulating tumor cells; tCTCneg = negative trajectory for circulating tumor cells.
Percentages may not total 100 because of rounding.
n = 1748 for PFS analysis as 108 patients had missing data: tCTCneg = 847 (48.5%); tCTClo = 592 (33.9%); tCTCmid = 165 (9.4%); tCTChi = 144 (8.2%).
n = 773 for PFS analysis as 3 patients had missing data: tCTCneg = 379 (49.0%); tCTClo = 280 (36.2%); tCTCmid = 63 (8.2%); tCTChi = 51 (6.6%).
cCTC Status Early During Treatment
Of the 469 patients, 162 (34.5%) converted to CTC-negative (CTC+CTC−) after the first chemotherapy cycle, 214 (45.6%) remained CTC-negative (CTC−CTC−), 88 (18.8%) remained CTC-positive (CTC+CTC+), and 5 patients converted from negative to positive (CTC−CTC+) (1.1%) (Figure 1, D). Change in CTC status was statistically significantly associated with bone metastases (P < .001) and number of hormone treatments (P < .001) (Supplementary Table 3, available online). The proportion of patients who did not clear CTCs (CTC+CTC+) was statistically significantly higher in HR+ patients compared with triple negatives (21.3% vs 12.4%, P = .002).
Survival analysis showed that PFS was worst in the CTC+CTC+ group (median = 6.4 months) compared with the CTC+CTC− (median = 9.4 months), CTC−CTC+ (median = 10.6 months), and CTC−CTC− (median = 12.7 months) groups (log-rank P < .001). Similar results were observed for OS, with the shortest median OS time for the CTC+CTC+ group (median = 13.1 months) compared with the CTC+CTC− (median = 26.3 months), CTC−CTC+ (median = 29.5 months), and CTC−CTC− (median = 34.3 months) groups (log-rank P < .001) (Figure 2, C and D).
In multivariable models with the CTC−CTC− group as reference, CTC+CTC+ and CTC+CTC− patients had statistically significantly increased risk of progression (HR = 2.48, 95% CI = 1.86 to 3.32, P < .001 and HR = 1.34, 95% CI = 1.05 to 1.69, P = .02, respectively). All 3 cCTC groups were statistically significantly associated with increased risk of death, with the highest hazard ratio for CTC+CTC+ (HR = 3.65, 95% CI = 2.68 to 4.98), followed by CTC−CTC+ (HR = 2.88, 95% CI = 1.04 to 7.96) and CTC+CTC− (HR = 1.51, 95% CI = 1.16 to 1.95) (P = .04 to >.001, respectively) (Table 1; Supplementary Table 5, available online).
CTC Trajectory
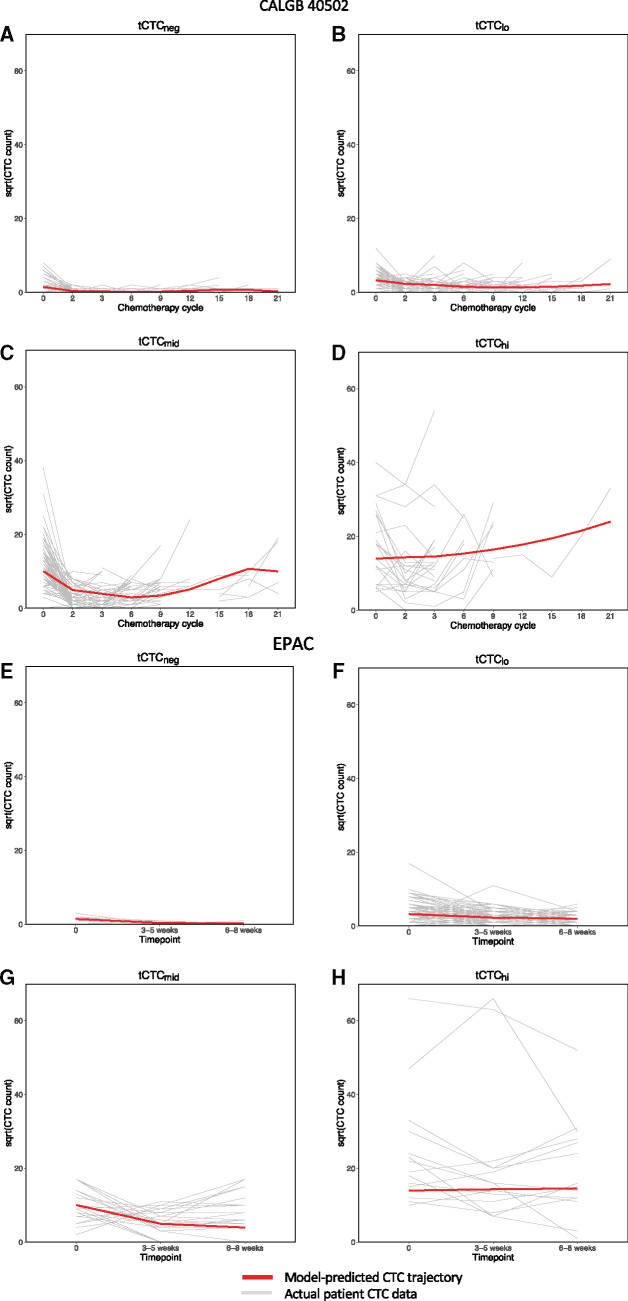
The best-fit tCTC model (adjusted for subtype and treatment arm) consisted of 4 groups of patients with distinct trajectory patterns: consistently negative for CTCs (tCTCneg, n = 267, 56.9%), low (tCTClo, n = 111, 23.7%), intermediate (tCTCmid, n = 68, 14.5%), and high (tCTChi, n = 23, 4.9%) (Figure 3, A-D). Details of the tCTC model parameters can be found in Supplementary Table 2 (available online). In univariate analysis, tCTC was statistically significantly associated with bone metastases (P < .001) (Supplementary Table 3, available online). The total percentage of tCTCmid and tCTChi was statistically significantly higher in HR+ patients compared with those with triple-negative disease (23.4% vs 8.2%, P < .001).
Figure 3.
Circulating tumor cell (CTC) trajectory patterns during therapy. CTCs were counted in individual metastatic breast cancer patients (gray lines) before treatment (time point 0 in the x-axis) and subsequent time points during therapy. A latent mixture model was then built to group patients according to similarities in CTC trajectory (tCTC) patterns over time. This analysis, first performed on serial CTC data from Cancer and Leukemia Group B (CALGB) 40502 patients (A-D), revealed 4 different tCTC patterns (red lines): A) high trajectory for CTCs (tCTCneg), B) low trajectory for CTCs (tCTClo), C) intermediate trajectory for CTCs (tCTCmid), and D) high trajectory for CTCs (tCTChi) groups. For validation, the model was then applied to the European Pooled Analysis of individual CTC (EPAC) dataset (E-H), which also grouped patients into 4 tCTC patterns: E) tCTCneg, F) tCTClo, G) tCTCmid, and H) tCTChi. The y-axis represents the levels of CTCs (square root of CTC per 7.5 mL of blood) in individual patients (gray lines) vs model-predicted CTC trajectories (red lines). The x-axis indicates different time points during therapy: cycles of chemotherapy for CALGB 40502 or time after initiation of treatment for EPAC.
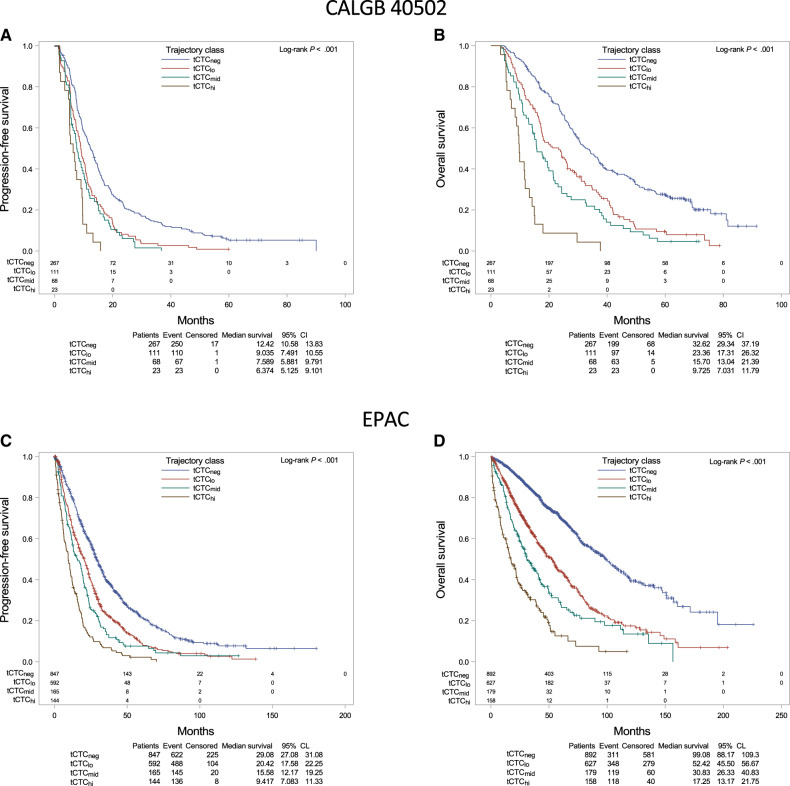
Kaplan-Meier analysis showed that patients in the tCTChi group had the shortest PFS (median = 6.4 months) compared with the tCTCmid (median = 7.6 months), tCTClo (median = 9.0 months), and tCTCneg (median = 12.4 months) groups (log-rank P < .001) (Figure 4, A). Similarly, the tCTChi group had the shortest OS (median = 9.7 months) compared with the tCTCmid (median = 15.7 months), tCTClo (median = 23.4 months), and tCTCneg groups (median = 32.6 months) (log-rank P < .001) (Figure 4, B).
Figure 4.
Prognostic impact of circulating tumor cells (CTCs) based on trajectory patterns during chemotherapy. A) Progression-free survival (PFS) and B) overall survival (OS) based on CTC trajectory (tCTC) in patients enrolled in the Cancer and Leukemia Group B 40502 trial. C) PFS and D) OS based on tCTC in patients in the Bidard et al. pooled analysis (European Pooled Analysis of individual CTC). Log-rank test was performed to compare survival curves. A 2-sided P less than .05 was considered significant.
In multivariable Cox regression analysis, patients in the tCTChi (HR = 3.04, 95% CI = 1.85 to 4.98), tCTCmid (HR = 2.48, 95% CI = 1.79 to 3.43), and tCTClo (HR = 1.89, 95% CI = 1.46 to 2.45) groups had consistently increased risk of progression compared with tCTCneg patients (Table 1; Supplementary Table 6, available online). Similarly, patients in the tCTChi (HR = 7.54, 95% CI = 4.45 to 12.79), tCTCmid (HR = 3.91, 95% CI = 2.76 to 5.53), and tCTClo (HR = 2.25, 95% CI = 1.71 to 2.98) groups had consistently increased risk of death compared with tCTCneg patients (Table 1;Supplementary Table 7, available online).
Comparison of AICs from the bCTC, cCTC, and tCTC models indicated that the tCTC model provided the best fit to the data for PFS (AIC = 4048.8) compared with bCTC (AIC = 4073.2) and cCTC (AIC = 4059.6). The tCTC model also provided the best fit for OS (AIC = 3446.5) compared with the 2 other models (bCTC, AIC = 3502.9; and cCTC, AIC = 3475.0) (Table 2).
Table 2.
Model selection by AIC analysis
| Dataset and endpoints | AIC scores of CTC modelsa |
||
|---|---|---|---|
| bCTC |
cCTC |
tCTC |
|
| Baseline CTC | Change in CTC | tCTC | |
| CALGB 40502 (n = 469) | |||
| Progression-free survival | 4073.2 | 4059.6 | 4048.8b |
| Overall survival | 3502.9 | 3475.0 | 3446.5b |
| EPAC | |||
| ≥1 CTC time point (n = 1856) | |||
| Progression-free survival | 11 108.0 | 11 098.9 | 11 069.5b |
| Overall survival | 6742.9 | 6734.8 | 6679.3b |
| First line patients only (n = 776) | |||
| Progression-free survival | 4167.1 | 4166.5 | 4157.8b |
| Overall survival | 2017.8 | 2018.6 | 1988.2b |
| Baseline CTC only (n = 916) | |||
| Progression-free survival | 2933.1 | n/a | 2907.1b |
| Overall survival | 4332.0 | n/a | 4314.2b |
The model with the best fit, that is, with the lowest AIC score (b), indicates best performance for predicting survival. AIC = Akaike information criterion; CALGB = Cancer and Leukemia Group B; cCTC = combined circulating tumor cell status at baseline and end of first cycle of therapy; CTC = circulating tumor cell; EPAC = European Pooled Analysis of individual CTC; n/a = not applicable; tCTC= circulating tumor cell trajectory.
Lowest AIC score.
Model Validation
The EPAC dataset was used for model validation studies, which consisted of 1920 patients with MBC (2). Because the initial model adjusted for subtype, 64 patients with missing subtype data were excluded. The resulting analytic cohort for the trajectory analysis included 1856 patients who had at least 1 CTC measure (baseline, n = 916), and the rest had 2 (n = 448) or 3 (n = 492) measures. Applying the parameters from the original trajectory model (Supplementary Table 2, available online), the 1856 patients were classified into groups as follows: 892 (48.1%) tCTCneg, 627 (33.7%) tCTClo, 179 (9.6%) tCTCmid, and 158 (8.5%) tCTChi (Figure 3, E-H).
Similar to the initial model, tCTC was statistically significantly associated with hormone receptor status (P = .009) and bone metastases in univariate analysis (P < .001) (Supplementary Table 7, available online). Moreover, patients in the tCTChi group had the shortest PFS (median = 9.4 months, log-rank P < .001) and OS (median = 17.3 months, log-rank P < .001) compared with other trajectory groups (Figure 4, C and D). In multivariable Cox regression analysis, the assigned tCTC groups were strongly associated with outcome, with the worst prognosis for patients in the tCTChi group (PFS HR = 3.41, 95% CI = 2.78 to 4.17; OS HR = 6.55, 95% CI = 5.10 to 8.40) (Table 1; Supplementary Table 8, available online).
Comparison of AICs from the bCTC, cCTC, and tCTC models indicated that the tCTC model provided the best fit to the data for PFS (AIC = 11069.5) and OS (AIC = 6679.3) compared with the bCTC and cCTC models (Table 2). The CALGB 40502 study involved first-line treatment in MBC; hence, we also applied the model to a subset of EPAC patients who received only first-line treatment and observed similar results (Table 1; Table 2;Supplementary Figure 1; Supplementary Table 9, available online).
Applying the Model to Single-Sample Prediction
To evaluate the clinical significance of the tCTC model when CTC measures are limited, we applied the model to a subset of patients with only a single bCTC measure and compared the distributions of model-predicted group probabilities with those obtained from models including patients with additional measures. Supplementary Figure 2 (available online) shows that we can use 1 CTC measure (ie, only bCTCs) to achieve adequate probability class assignment.
In multivariable Cox regression analysis, the assigned tCTC groups were strongly associated with increased risk of progression or death, with the worst prognosis among tCTChi (PFS HR = 3.61, 95% CI = 2.70 to 4.82; OS HR = 6.17, 95% CI = 4.40 to 8.66) (Table 1;Supplementary Table 10, available online). Comparison of AIC scores revealed that the tCTC model was a better fit to the data than the bCTC model, an observation that is consistent with findings from previous subset analyses (Table 2).
Discussion
CTCs are a promising biomarker for monitoring treatment response and predicting outcome in patients with MBC (2,5). In this study, we found that CTC positivity at baseline and persistent detection of CTCs at baseline and at first follow-up were predictive of reduced PFS and OS (Figure 2; Table 1). These findings are consistent with those from previous research (2-4,9-13), including a recent large pooled analysis by Bidard and colleagues (EPAC study) (2). Here, we build on this previous knowledge by demonstrating that additional prognostic information can be gleaned from comprehensive serial analysis of CTCs during treatment. To our knowledge, our report—which includes 2202 samples from 783 patients—represents the largest single study on serial analysis of CTCs to date.
The current and widely accepted CTC prognostic model uses bCTC counts to predict PFS and OS (2-5). In this model, patients are classified into approximately 50% good prognosis (CTC-negative or stage IVindolent, <5 CTC per 7.5 mL) and approximately 50% poor prognosis (CTC-positive or stage IVaggressive, at least 5 CTC per 7.5 mL). We improved on this model by analyzing tCTC patterns (ie, changes in the levels of CTCs over time) in serial blood during treatment. Modeling of CTC dynamics during treatment revealed groups of patients with distinct tCTC patterns (tCTCneg, tCTClo, tCTCmid, and tCTChi) (Figure 3). We showed that these tCTC groups were highly prognostic of PFS and OS (Figure 4).
Our tCTC model is similar to the bCTC model (2-5) in that it identified a group of patients—representing approximately 50% of the cohort (tCTCneg)—with relatively better prognosis compared with other groups (Figure 4). But unlike the bCTC model, our model stratified the remaining approximately 50% poor-prognosis patients into 3 risk groups: tCTClo (approximately 30%), tCTCmid (approximately 15%), and tCTChi (approximately 5%), each with increasing risk of progression and death.
We also demonstrated that a single CTC measurement at baseline can predict patients’ trajectory pattern and provide statistically significant prognostic information (Table 2). Comparative analysis showed that the tCTC model outperformed previously demonstrated CTC-based prognostic models (ie, CTCs at baseline and early change in CTCs status) for prediction of OS and PFS. This finding has major implications in risk stratification strategies in MBC (5) because it demonstrates that further refinement of CTC-based prognostic classification (eg, stage IVindolent vs stage IVaggressive) (5) is feasible using our tCTC model. In addition, our novel prognostic classification developed in CALGB 40502 and its subsequent validation in the EPAC dataset suggest the generalizability of our approach across different therapeutic strategies.
We envisage several applications of the findings of this study including guiding treatment and reducing—at study initiation—heterogeneity of patients across treatment arms in clinical trials. We found that approximately 20% of patients (tCTCmid and tCTChi groups) responded poorly to first-line chemotherapy. These patients can in principle forgo first-line chemotherapy and instead receive a more personalized and targeted treatment. Furthermore, we recommend the utilization of our novel CTC-based risk assessment approach in future clinical trials to guide stratification and randomization of patients. This in turn might limit biases and could magnify differences in the efficacy of the therapeutic agents being investigated. Indeed, by taking into account the differences in clinical outcomes of MBC patients at study onset, a more systematic and rational pipeline for drug development can be implemented.
In addition to CTC analysis, other liquid biopsy approaches may also help improve risk stratification in MBC (14). For example, clinical studies have recently shown that the detection of circulating tumor DNA (ctDNA) in blood is strongly associated with poor prognosis (15,16). Contemporaneous analysis of CTCs and ctDNA in MBC have been reported (17-21). These studies have shown the nonoverlapping prognostic value of these liquid biopsy-based biomarkers (17-21). In this study, paired CTC and ctDNA data were not available. Analysis of ctDNA has also enabled the detection of actionable mutations, which could potentially help match patients to appropriate targeted therapies (22-24). Collectively, these advancements indicate the potential for combined use of CTC and ctDNA for prognostication and biomarker-directed treatment in MBC.
Observational studies that involve CTC enumeration and CTC-based risk stratification can be complemented with genotypic and phenotypic characterization of CTCs as surrogate representatives of systemic disease (25-27). Molecular analysis of CTCs would be most feasible and may be more informative in patients where these cells are more numerous and outcomes are dismal (eg, CTCmid and tCTChi groups) (25). Real-time information from molecular testing of CTCs may also aid in personalization of treatment to improve survival in poor-prognosis patients (28). Therefore, clinical studies that examine CTCs as companion diagnostics are warranted to fully demonstrate the utility of these cells.
Clinical trials have been conducted to investigate the potential utility of CTCs (5,29,30). A prospectively randomized study by Smerage and colleagues (SWOG0500) (29) investigated whether changing treatment in patients with persistently high CTCs could improve OS. The study, however, failed to show that early changes in treatment based on high CTC counts could lead to improvements in patient outcomes (29). The STIC CTC trial examined the potential role of CTCs in first-line treatment selection (ie, hormone therapy or chemotherapy) for HR+HER2-negative MBC (30). Results of the trial showed that switching to chemotherapy in patients with high bCTC counts (≥5 CTC per 7.5 mL) resulted in statistically significant improvements in PFS compared with those who received single-agent hormone therapy (30). Application of the novel tCTC-based risk stratification approach in HR+HER2-negative MBC may identify patients who are at high risk of early disease progression (tCTCmid and tCTChi) despite more aggressive and toxic treatment (ie, chemotherapy over hormone therapy). These patients might be preferentially enrolled in clinical trials aimed at addressing mechanisms of resistance and improving outcome, and additional work may help to further clarify risk factors that could optimize therapy.
We found that in our survival models, other factors in addition to CTCs were also statistically significant predictors of PFS and OS. These included breast cancer subtype (triple-negative disease) and prior therapies. Our latent mixture model did account for the effect of subtype, and our validation studies provided evidence of generalizability across different therapeutic strategies. Testing for interactions between CTCs and other statistically significant covariables was beyond the scope of this study and thus represents a limitation. Understanding these interactions may provide valuable insights into the building of robust and integrated clinicopathologic- and CTC-based prognostic models for MBC.
We posit that screening of CTCs and identification of prognostic groups (tCTCneg, tCTClo, tCTCmid, or tCTChi) in MBC patients enrolling in prospective trials could aid in balancing of arms to facilitate accurate measurement of treatment efficacy and help develop appropriate and effective therapies for MBC. This is a critical step as we add other liquid biopsy-based biomarkers (eg, ctDNA) to our risk stratification to enable better assessment of novel therapies or new combinations.
In summary, accurate prognostic biomarkers are needed to guide therapeutic decisions in MBC (31). Analysis of tCTC patterns identified 4 prognostic groups. More important, performance of the trajectory model for predicting outcomes was consistent even when single bCTC information was analyzed. Our risk stratification approach based on tCTC and subtype may yield better discriminatory power to identify poor-prognosis patients (ie, tCTCmid + tCTChi, approximately 20% poor prognosis) compared with the current model (CTC-positive, cutoff ≥5 CTC per 7.5 mL, approximately 50% poor prognosis). The information derived from our approach may be used to fine-tune risk stratification in clinical trials, facilitate accurate measurement of treatment efficacy, and help identify appropriate and effective therapies for MBC.
Funding
This work was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821, U10CA180882 (to the Alliance for Clinical Trials in Oncology), UG1CA233180, UG1CA233290, UG1CA233339, UG1CA233373, and U10CA180820 (ECOG-ACRIN). https://acknowledgments.alliancefound.org. Also supported in part by funds from Bristol-Myers, Celgene, P30CA014236 (TH and LHH), and the Breast Cancer Research Foundation (HSR and MJMM).
Notes
Role of the funders: The sponsor, National Cancer Institute of the National Institutes of Health, assisted in design of the study and review of the manuscript. The nonfederal sponsors did not contribute to design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Disclosures: J-YP, FCB, JWP, HSR received research grants from Menarini Silicon Biosystems (formerly Veridex); TF declares membership in board of directors or advisory committees for Daichu' Saukyo, Roche, and Novartis; JAG-S declares consultancy/speaker fees from Novartis, Celgene, Eli Lilly, EISAI AstraZeneca, Research funding from AstraZeneca, Travel support from Novartis, Roche, Pfizer; JS declares being the Editor-in-Chief of Oncogene, sat on scientific advisory boards for Celltrion, Singapore Biotech, Vor Biopharma, TLC Biopharmaceuticals and Benevolent AI, consulted with Lansdowne partners, Vitruvian and Social Impact Capital, and Chairs the Board of Directors for BB Biotech Healthcare Trust and Xerion Healthcare; LM declares employment with Hosp. Univ. 12 de Octubre, consultant or advisory role with Lilly, Tesaro, Astra-Zeneca, Roche, Novartis, Pfizer, and Celgene, research funding from Tesaro, speaking for Lilly, Roche, Astra-Zeneca, and Novartis, Pfizer; CC declares being member of AstraZeneca iMed External Science Panel, Illumina Scientific Advisory Board member, recipient of research grants (administered by the University of Cambridge) from AstraZeneca, Genentech, Roche and Servier; EM declares consulting or advisory role for Genomic Health, Pierre Fabre, and Eisai; CM declares consulting and research support from Pfizer, consulting fee from Lilly, Novartis, and Seattle Genetics, research support from Puma; The rest of the authors declare no conflict of interest.
Role of the authors: LHH: Formal analysis. TH: Formal analysis. WTB: Formal analysis. EPW: Resources. CH: Resources. DT: Resources. LAC: Resources. AHP: Resources. J-YP: Resources. TF: Resources. JV-M: Resources. DM: Resources. JAG-S: Resources. JS: Resources. PG: Resources. LM: Resources. RZ: Resources. MLA: Resources. LDM-A: Resources. DG: Resources. CC: Resources. EM: Resources. LD: Resources. ALD: Writing—review & editing. HB: Resources. MQ: Resources. CM: Resources. JHS: Data curation; Formal analysis. F-CB: Resources; Validation. JWP: Conceptualization; Supervision. HSR: Conceptualization; Funding acquisition; Supervision.
Acknowledgments: We thank the Alliance for Clinical Trials in Oncology for funding the Cancer and Leukemia Group B (CALGB) 40502 study (https://acknowledgments.alliancefound.org); Alliance Statistics and Data Center and the Breast Correlative Science Committee for critical review of the manuscript. CALGB is now part of the Alliance for Clinical Trials in Oncology.
Disclaimer: The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Data availability
The data underlying this article were provided by the Alliance for Clinical Trials in Oncology by permission. Data will be shared on request to the Alliance for Clinical Trials in Oncology. De-identified patient data may be requested from Alliance for Clinical Trials in Oncology via concepts@alliancenctn.org if data is not publicly available. A formal review process includes verifying the availability of data, conducting a review of any existing agreements that may have implications for the project, and ensuring that any transfer is in compliance with the IRB. The investigator will be required to sign a data release form before transfer.
Supplementary Material
References
- 1. Lee JS, Magbanua MJ, Park JW.. Circulating tumor cells in breast cancer: applications in personalized medicine. Breast Cancer Res Treat. 2016;160(3):411–424. [DOI] [PubMed] [Google Scholar]
- 2. Bidard FC, Peeters DJ, Fehm T, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15(4):406–414. [DOI] [PubMed] [Google Scholar]
- 3. Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. [DOI] [PubMed] [Google Scholar]
- 4. Hayes DF, Cristofanilli M, Budd GT, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12(14):4218–4224. [DOI] [PubMed] [Google Scholar]
- 5. Cristofanilli M, Pierga JY, Reuben J, et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit Rev Oncol Hematol. 2019;134:39–45. [DOI] [PubMed] [Google Scholar]
- 6. Rugo HS, Barry WT, Moreno-Aspitia A, et al. Randomized phase III trial of paclitaxel once per week compared with nanoparticle albumin-bound nab-paclitaxel once per week or ixabepilone with bevacizumab as first-line chemotherapy for locally recurrent or metastatic breast cancer: CALGB 40502/NCCTG N063H (Alliance). J Clin Oncol. 2015;33(21):2361–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Akaike H. Information theory and an extension of the maximum likelihood principle. In: Parzen E, Tanabe K, Kitagawa G, eds. Selected Papers of Hirotugu Akaike. New York: Springer New York; 1998:199–213. [Google Scholar]
- 8. Nielsen JD, Rosenthal JS, Sun Y, et al. Group-based criminal trajectory analysis using cross-validation criteria. Communic Stat-Theory Methods. 2014;43(20):4337–4356. [Google Scholar]
- 9. Liu MC, Shields PG, Warren RD, et al. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol. 2009;27(31):5153–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartkopf AD, Wagner P, Wallwiener D, et al. Changing levels of circulating tumor cells in monitoring chemotherapy response in patients with metastatic breast cancer. Anticancer Res. 2011;31(3):979–984. [PubMed] [Google Scholar]
- 11. Wallwiener M, Riethdorf S, Hartkopf AD, et al. Serial enumeration of circulating tumor cells predicts treatment response and prognosis in metastatic breast cancer: a prospective study in 393 patients. BMC Cancer. 2014;14(1):512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Magbanua MJ, Carey LA, DeLuca A; for the Translational Breast Cancer Research Consortium, et al. Circulating tumor cell analysis in metastatic triple-negative breast cancers. Clin Cancer Res. 2015;21(5):1098–1105. [DOI] [PubMed] [Google Scholar]
- 13. Magbanua MJM, Yau C, Wolf DM, et al. Synchronous detection of circulating tumor cells in blood and disseminated tumor cells in bone marrow predict adverse outcome in early breast cancer. Clin Cancer Res. 2019;25(17):5388–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36(16):1631–1641. [DOI] [PubMed] [Google Scholar]
- 15. Coombes RC, Page K, Salari R, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res. 2019;25(14):4255–4263. [DOI] [PubMed] [Google Scholar]
- 16. Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. [DOI] [PubMed] [Google Scholar]
- 17. Ye Z, Wang C, Wan S, et al. Association of clinical outcomes in metastatic breast cancer patients with circulating tumour cell and circulating cell-free DNA. Eur J Cancer. 2019;106:133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bidard F-C. Abstract SY31-02: clinical utility trials for CTC and ctDNA in ER+ advanced breast cancer. Cancer Res. 2019;79(suppl 13):SY31-02. [Google Scholar]
- 19. Tzanikou E, Markou A, Politaki E, et al. PIK3CA hotspot mutations in circulating tumor cells and paired circulating tumor DNA in breast cancer: a direct comparison study. Mol Oncol. 2019;13(12):2515–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davis AA, Zhang Q, Gerratana L, et al. Association of a novel circulating tumor DNA next-generating sequencing platform with circulating tumor cells (CTCs) and CTC clusters in metastatic breast cancer. Breast Cancer Res. 2019;21(1):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pierga J-Y, Silveira A, Tredan O, et al. Multimodality liquid biopsy for early monitoring and outcome prediction in first-line metastatic HER2-negative breast cancer: final results of the prospective cohort from the French Breast Cancer InterGroup Unicancer (UCBG)—COMET study. J Clin Oncol. 2019;37(15_suppl):3019. [Google Scholar]
- 22. Rothwell DG, Ayub M, Cook N, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25(5):738–743. [DOI] [PubMed] [Google Scholar]
- 23. Murtaza M, Dawson SJ, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6(1):8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25(9):1729–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Magbanua MJM, Rugo HS, Wolf DM, et al. Expanded genomic profiling of circulating tumor cells in metastatic breast cancer patients to assess biomarker status and biology over time (CALGB 40502 and CALGB 40503, Alliance). Clin Cancer Res. 2018;24(6):1486–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossi G, Ignatiadis M.. Promises and pitfalls of using liquid biopsy for precision medicine. Cancer Res. 2019;79(11):2798–2804. [DOI] [PubMed] [Google Scholar]
- 27. Paoletti C, Schiavon G, Dolce EM, et al. Circulating biomarkers and resistance to endocrine therapy in metastatic breast cancers: correlative results from AZD9496 Oral SERD phase I trial. Clin Cancer Res. 2018;24(23):5860–5872. [DOI] [PubMed] [Google Scholar]
- 28. Gulbahce N, Magbanua MJM, Chin R, et al. Quantitative whole genome sequencing of circulating tumor cells enables personalized combination therapy of metastatic cancer. Cancer Res. 2017;77(16):4530–4541. [DOI] [PubMed] [Google Scholar]
- 29. Smerage JB, Barlow WE, Hortobagyi GN, et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol. 2014;32(31):3483–3489. 10.1200/J Clin Oncol.2014.56.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bidard F-C, Jacot W, Dureau S, et al. Abstract GS3-07: clinical utility of circulating tumor cell count as a tool to choose between first line hormone therapy and chemotherapy for ER+ HER2- metastatic breast cancer: results of the phase III STIC CTC trial. Cancer Res. 2019;79(suppl 4):GS3-07–GS3-07. [Google Scholar]
- 31. Van Poznak C, Somerfield MR, Bast RC, et al. Use of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2015;33(24):2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article were provided by the Alliance for Clinical Trials in Oncology by permission. Data will be shared on request to the Alliance for Clinical Trials in Oncology. De-identified patient data may be requested from Alliance for Clinical Trials in Oncology via concepts@alliancenctn.org if data is not publicly available. A formal review process includes verifying the availability of data, conducting a review of any existing agreements that may have implications for the project, and ensuring that any transfer is in compliance with the IRB. The investigator will be required to sign a data release form before transfer.