Abstract
One measure of research productivity within the University of Kansas Cancer Center (KU Cancer Center) is peer-reviewed publications. Considerable effort goes into searching, capturing, reviewing, storing and reporting cancer- relevant publications. Traditionally, the method of gathering relevant information to the publications is done manually. This manuscript describes the efforts to transition KU Cancer Center’s publication gathering process from a heavily manual to a more automated and efficient process. To achieve this transition in the most customized and cost-effective manner, a homegrown, automated system was developed using open source API among other software. When comparing the automated and the manual processes over several years of data, publication search and retrieval time dropped from an average of 59 hours to 35 minutes, which would amount to a cost savings of several thousand dollars per year. The development and adoption of an automated publications search process can offer research centers great potential for less-error prone results with a savings in time and cost.
Introduction:
All NCI designated cancer centers must contend with publications’ search strategies1. A 2018 poll of cancer center administrators and IT professionals found that some centers spend as many as 85 hours a month on publications data management2. Most cancer centers in the poll spend an average of 13 hours a month, while the more technically sophisticated cancer centers spend between one to three hours a month searching, retrieving, and verifying publications. Of the cancer centers polled, 34% were using a homegrown system, 17% NIH systems, and 13% vendor systems.
Homegrown systems are any set of tools and processes developed, deployed, and managed directly by cancer centers for the capture, storage, verification, and reporting of publications3. These can include Application Programming Interface (API) that can automatically interact with Entrez, the NIH information retrieval system containing PubMed. Additionally, homegrown systems can include an ensemble of locally managed databases for storage and retrieval, as well as custom-built web tools to allow for publication review and verification.
According to a 2018 survey4 of nearly 300 managers at various businesses, 62% responded that there are processes in their work that could be automated. From that same survey, 55% of managers spend about eight hours a week on “administrative tasks rather than strategic, high-value initiatives.” This response can also be heard throughout academic institutions as there is usually a great amount of time spent to integrate such a system5. To better understand and appreciate the opportunities and benefits of a homegrown, automated publication process, it is helpful to detail the manual publication process.
The KU Cancer Center is a matrix cancer center with consortium members spread across four different campuses. The current publications search process requires manual time and labor and uses a combination of NIH and locally managed tools. At the start of each calendar year, saved searches are set up for all KU Cancer Center members using MyNCBI, an NIH search tool and notification system within PubMed. MyNCBI can be scheduled to deliver email notifications for new publications with any level of frequency the user requires. KU Cancer Center receives MyNCBI notifications on a weekly basis.
Each publication is then reviewed by querying the PMID number against the locally managed KU Cancer Center SQL Server database. A query match indicates that the publication already exists in the database. Thus, the MyNCBI notice is disregarded. A query that returns no results indicates that the publication does not currently exist in the database. At this point, a review is conducted to determine if the publication is a correct match by PI name and institutional affiliation.
This vetting occurs by opening the hyperlink within the MyNCBI email and viewing the PubMed citation. When more information is required, the citation is then imported into EndNote where citation meta data can be viewed to verify that a publication belongs to a PI affiliated with the KU Cancer Center. Vetted publications are then entered into a local SQL Server database. This manual process requires several hours per work throughout the calendar year.
Clearly there is a real administrative need for efficiency in publications searches. Several studies suggest that there is promise in piloting an automated search process. Choong et al described an internally developed automatic citation method that, when compared to a manual search process, was accurate in identifying and retrieving citations6. According to Ongenaert and Dehaspe, automated publications searches and text mining can bring results with increased speed and selectivity7. While initial implementation costs for an automated search system may be high, Padilla et al found that an automated search method, targeting cancer related publications authored by educational trainees, retrieved 20% more publications than a manual search8. This paper examines the KU Cancer Center’s effort to leverage the obvious advantages of an automated publications search system.
METHODS AND MATERIAL:
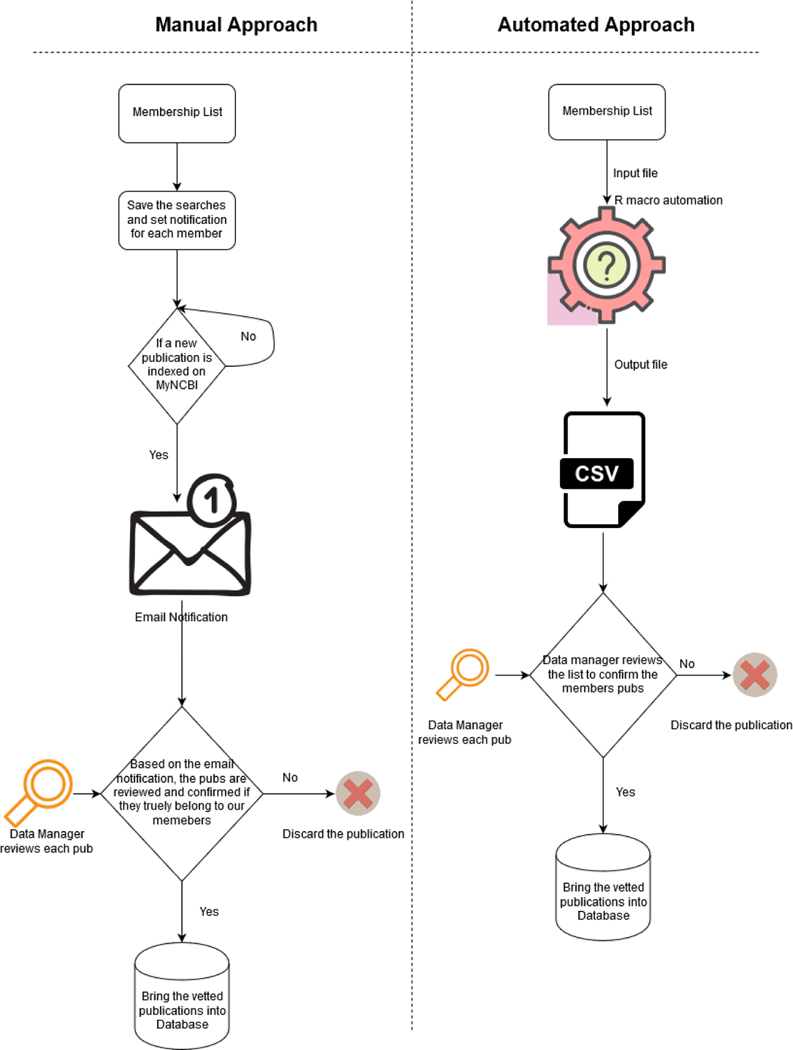
Fig 01 describes both the process the manual and automated and how they vary. There is a paradigm shift for the data manager, instead of manually keeping track of every publications and weeding the false positives he or she now can just focus on reviewing a single list or source of truth that is generated by the automated R script.
Figure 01:
Comparison of Manual Approach Versus Automated Approach
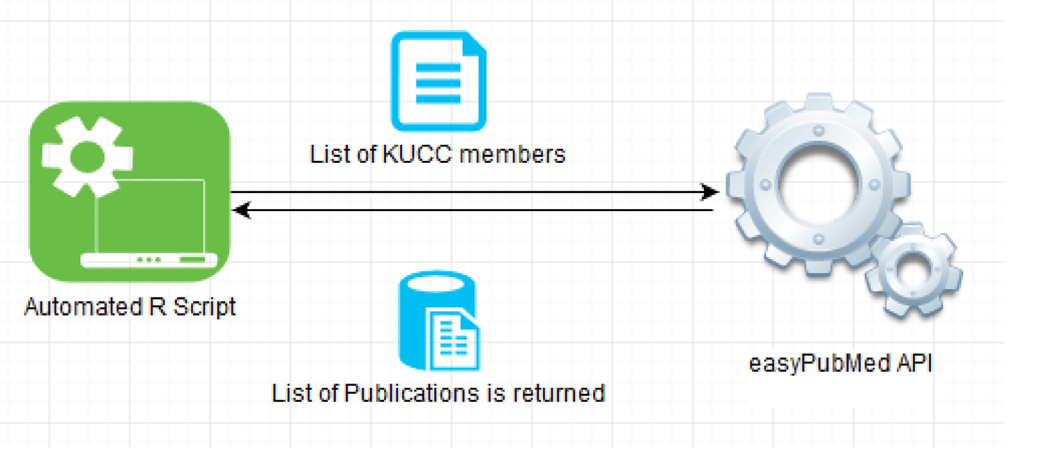
In response to this heavy workload, Scientific Citation Data Mining Search (SCiDS) was developed. SCiDS was developed using R9, which utilizes the “easyPubMed” package10 to call the APIs from PubMed to retrieve the publications. Figure 02 demonstrates the pseudo code as well as the architecture of SCiDS. A membership list for a calendar year is supplied as the input file. The program then searches PubMed for all the publications for the corresponding member.
Figure 02:
Architecture of SCiDS
As a validation check, the program makes sure the full name matches along with the affiliation of the member. This process is repeated for every member included on the membership list. Once the entire list of members has been searched, all the publications found are documented to a file, which is then reviewed and validated before finalization. A pilot test was run on the research membership lists for calendar years 2016 through 2019. The PMIDs from the automated results were then compared to PMIDs from the manually gathered list to see what was and was not caught. Both methods were limited to full published papers. Initially, parameters included variables such as title, authors, the author’s affiliation, and the PMID. Any duplicate articles from the automated list were checked and removed.
Pseudo code:

As described in Fig 02, the custom R macro is automated, to run at a fixed frequency utilizing the dynamic KU cancer center members list as the input. Based on the authors full name all the publications that have been indexed are retrieved and housed into a excel file which is then imported into a SQL database8. Our data scientist would use the newly populated information to perform the final vetting process before freezing and generating the list of publications for a given calendar year.
RESULTS:
After optimizing the R code to quickly search for publications and related information such as publisher and impact factor, the process was run for the member lists in each year from January 2016 to August 2019. This was done to verify the publications found for those years and comparing them to an already established list.
A significant reduction in time spent searching was seen. On average it took roughly 35 minutes, across four different runs, to generate the list of publications that would take at least 60 hours on an average manually. A cost analysis was done to examine the differences in cost of creating these lists using time and median yearly salary for a data analyst. Not only is there significant time being saved but cost as well. Across cancer centers nationally that use a manual method, the potential for cost saving is substantial.
In many cases, SCiDS was able to capture more publications than the manual approach. From this, new publications could be added to the already established list. The automated list tends to capture about three-fourths, on average, of the manually created lists, using PubMed alone. There are publications that can be found on other scientific publication search engines such as Web of Science and Google Scholar, but not on PubMed. Sensitivity was measured by comparing the percentage of articles from the automated lists that were also on the manually gathered lists.
DISCUSSION:
Certain metrics come with difficulties to obtain with an automated process. It is common that the Journal Impact Factor (JIF) cannot be obtained until sometime after research is published since JIFs are calculated and updated every year. One future goal is to develop an improved and automated way of obtaining JIFs for each publication. When it comes to common names, having their middle name when applicable is helpful in narrowing down specific members. Ideally, having researchers submit their ORCIDs, a unique identifier for scientific researchers, for their work would increase the process greatly since there would not have to be a need to rely solely on searching names. Future mandating of ORCIDs by NIH will help this process11. Integrating a new system always does not come quickly and usually has some form of challenges, but an automated process like this is a step forward for a more efficient work flow12.
Since SCiDS mainly utilizes PubMed, it misses out on publications that can only be found on other search engines such as Web of Science and Google Scholar. Adding these to SCiDS is a future goal. Additionally, SCiDS would be utilized more often to keep up with the most up to date publications, additionally the list that is produced by SCiDS would be more precise in that there would be less of a chance of adding duplicate publications. By filtering the data on an annual basis, this would also reduce the chance of adding publications that are not relevant to the cancer members. This would make verifying the list go faster since the data analyst would not have to sift anywhere from 500 to even 800 publications. Even in the early stages, SCiDS has shown that it can drastically reduce the time spent searching for member publications in an efficient manner. Also, because SCiDS uses API and homegrown coding, additional modules can be built that could improve publication verification and incorporated value-added information such as JIFs.
Table 01:
Cost Saving Between Automated and Manual Method
| 2019 | 2018 | 2017 | 2016 | |||||
|---|---|---|---|---|---|---|---|---|
| Method | Auto | Manual | Auto | Manual | Auto | Manual | Auto | Manual |
| Time (hh: mm: ss) | 00:31:16 | 51:25:00 | 00:31:29 | 58:48:00 | 00:31:42 | 59:43:00 | 00:48:48 | 68:33:00 |
| Cost* ($) | 16.26 | 1,593.92 | 16.26 | 1,822.80 | 16.38 | 1,851.22 | 25.38 | 2,125.05 |
| Saving** ($) | 1,577.66 | 1,806.54 | 1,834.84 | 2,099.67 | ||||
Median salary for a data analyst in the Kansas City, KS was roughly $62,000 for 2019
Difference of cost of manual and auto methods
Table 02:
specificity - comparison of automated approach versus manual approach
| 2019* | 2018 | 2017 | 2016 | |||||
|---|---|---|---|---|---|---|---|---|
| Method | Auto | Manual | Auto | Manual | Auto | Manual | Auto | Manual |
| Total | 593 | 610 | 809 | 559 | 820 | 598 | 895 | 691 |
| Match | 81% | 75% | 71% | 73% | ||||
January to August
ACKNOWLEDGEMENTS:
We would like to extend our gratitude to KU Cancer Center Leadership for the indispensable guidance and advice they have provided on the design and development of SCiDS.
We would like to extend our gratitude to Drs. Roy Jensen and Matthew S Mayo for the indispensable guidance and advice they provided on the design and development of Publications Search Optimization tool.
FUNDING:
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Cancer Institute (NCI) Cancer Center Support Grant P30 CA168524 and used the Biostatistics and Informatics Shared Resource (BISR).
Footnotes
Declaration of conflicting interests:
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Cancer Center Support Grants (CCSGs) for NCI-designated Cancer Centers (P30): National Institutes of Health (NIH); 2016. [Available from: https://grants.nih.gov/grants/guide/pa-files/PAR-13-386.html.] [Google Scholar]
- 2.Porter AL CS. TECH MINING Exploiting New Technologies for Competitive Advantage. Hoboken, New Jersey: John Wiley & Sons Inc.; 2005. [Google Scholar]
- 3.William M, Michael T, Mahendra Y. Comparative Methods of CCSG Data Collection. Cancer Center Administrators Forum 03/16/2018; Portland OR: Proceedings from Cancer Center Administrators Forum – Information Technology (CCAF – IT); 2018. [Google Scholar]
- 4.Cory A. formstack [Internet]. Cory A, editor: BLASTmedia. 2018. [cited 2019]. Available from: https://www.formstack.com/blog/2018/report-workflow-automation-2018/. [Google Scholar]
- 5.Porter AL, Cunningham SW. Tech mining exploiting new technologies for competitive advantage. Hoboken, N.J.: Wiley; 2005. [Google Scholar]
- 6.Choong MK, Galgani F, Dunn AG, Tsafnat G. Automatic evidence retrieval for systematic reviews. Journal of medical Internet research. 2014;16(10):e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ongenaert M, Dehaspe L. Integrating automated literature searches and text mining in biomarker discovery. Bmc Bioinformatics. 2010;11. [Google Scholar]
- 8.Padilla LA, Desmond RA, Brooks CM, Waterbor JW. Automated Literature Searches for Longitudinal Tracking of Cancer Research Training Program Graduates. Journal of Cancer Education. 2018;33(3):564–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R: A language and environment for statistical computing. Vienna, Austria: R Core Team; 2014. [Available from: http://www.R-project.org/.] [Google Scholar]
- 10.Damiano F. easyPubMed: Search and Retrieve Scientific Publication Records from PubMed 2019. [Available from: https://cran.r-project.org/web/packages/easyPubMed/index.html.] [Google Scholar]
- 11.Requirement for ORCID iDs for Individuals Supported by Research Training, Fellowship, Research Education, and Career Development Awards Beginning in FY 2020 National Institutes of Health (NIH); 2019. [Available from: https://grants.nih.gov/grants/guide/notice-files/NOT-OD-19-109.html.] [Google Scholar]
- 12.Reed C. Build vs. Buy. Cancer Center Administrators Forum; Portland OR: Proceedings from Cancer Center Administrators Forum - Information Technology (CCAF - IT); 2018. [Google Scholar]