Abstract
Longitudinal cancer monitoring is crucial to clinical implementation of precision medicine. There is growing evidence indicating important functions of extracellular vesicles (EVs) in tumor progression and metastasis, including matrix remodeling via transporting matrix metalloproteases (MMPs). However, the clinical relevance of EVs remains largely undetermined, partially owing to challenges in EV analysis. Distinct from existing technologies mostly focused on characterizing molecular constituents of EVs, here we report a nanoengineered lab-on-a-chip system that enables integrative functional and molecular phenotyping of tumor-associated EVs. A generalized, high-resolution colloidal inkjet printing method was developed to allow robust and scalable manufacturing of three-dimensional (3D) nanopatterned devices. With this nanochip platform, we demonstrated integrative analysis of the expression and proteolytic activity of MMP14 on EVs to detect in vitro cell invasiveness and monitor in vivo tumor metastasis, using cancer cell lines and mouse models. Analysis of clinical plasma specimen showed that our technology could be used for cancer detection including accurate classification of age-matched controls and patients with ductal carcinoma in situ, invasive ductal carcinoma, or locally metastatic breast cancer in a training cohort (n = 30, 96.7% accuracy) and an independent validation cohort (n = 70, 92.9% accuracy). With clinical validation, our technology could provide a useful liquid biopsy tool to improve cancer diagnostics and real-time surveillance of tumor evolution in patients to inform personalized therapy.
INTRODUCTION
Although early-stage cancer generally has a favorable prognosis, most cases are diagnosed with local or distant metastases that lead to ~90% of deaths from cancer (1). Poor survival of advanced cancer has been related to the lack of effective therapies and emergent drug resistance, attributed to molecular heterogeneity of tumors (2) and divergent clonal evolution during disease development and treatment (3, 4). Such diverse tumor dynamics pose a major challenge to clinical management of advanced cancer, which requires real-time assessment of disease states to inform decision-making and to optimize treatment. Although widely used in the clinic, radiographic imaging often fails to detect changes in tumor burden. Genomics centered on gene alterations faces challenges in capturing the real-time status of malignancy, such as invasive/metastatic phenotypes (5). Moreover, longitudinal surveillance of tumor evolution is crucial to clinical implementation of precision medicine; however, conventional tissue biopsy is invasive, constrained to a localized snapshot of the tumor, and often unrepeatable. Thus, new tools that complement current methods for accurate tracking of tumor dynamics are urgently needed to improve disease stratification, prognostic prediction, and early detection of metastasis for optimal treatment.
Liquid biopsy offers an attractive alternative for cancer diagnosis and treatment (6-8). Extracellular vesicles (EVs), including exosomes, are rapidly emerging as a new paradigm of liquid biopsy, owing to the distinct properties of these nanovesicles that are (i) enriched with selectively sorted original cell contents, (ii) actively released from live cells instead of shed from apoptotic or damaged cells, and (iii) relatively stable in bodily fluids (9, 10). EVs have been extensively identified as important cell communication mediators and implicated in tumor development and metastasis via various processes, such as epithelial-mesenchymal transition (EMT) (11), extracellular matrix (ECM) remodeling (12, 13), and formation of premetastatic niche (14-16). Relevant to this study, EVs derived from tumors and stromal cells in tumor microenvironments were found to carry matrix metalloproteinases (MMPs) (17-19), key regulators of the ECM. In particular, tumor-derived EVs, including exosomes, were shown to carry functionally active MMP14 that not only degrades type 1 collagen and gelatin but also promotes the expression and activation of other MMPs to enhance the remodeling of ECM (18, 19). Despite these provocative findings, very limited progress has been reported in exploring the clinical value of MMP-mediated functions of EVs in tumor progression and metastasis.
Clinical study of EVs is largely hindered by several key practical challenges, including the lack of standardized methods for efficient and unbiased EV isolation, general unavailability of ultrasensitive and robust biosensing systems for rapid analysis of large clinical cohorts, large sample consumption and assay cost, and poorly defined markers to distinguish tumor-specific EVs from host cell EVs (20). Microfluidics have been extensively applied to address these challenges owing to its inherent advantages in sample consumption and analytical performance (21-26). We recently developed a microfluidic colloidal self-assembly (μCSA) strategy for three-dimensional (3D) nanoengineering of microfluidic biosensors, which immensely improved the sensitivity for small EV (sEV) detection via overcoming the fundamental limits in mass transfer, surface reaction, and boundary effects (23). Compared to the existing microfluidic technologies, this work presents distinct innovations in device engineering and EV marker studies, focusing on improving the translatability for clinical applications. First, distinct from the μCSA-based strategy, we developed a generalized, high-resolution colloidal inkjet printing method that obviates the need for surface pretreatment and is more amenable to mass production of 3D nanoengineered chips for large-scale clinical studies. Second, despite well-demonstrated applications to molecular analysis of EVs, microfluidic technology has not been explored to assess the functional activities of EVs as cancer signatures. We devised a nanoengineered lab-on-a-chip system for multiparametric analysis of EV concentration in circulation, subtype, and enzymolytic activity (EV-CLUE) with ultrahigh sensitivity that required minuscule sample input, enabling longitudinal monitoring of in vivo tumor growth in mice. Third, although EV-mediated transport of MMP14 has been implicated in tumor invasion and metastasis, the clinical value of MMP14 as an EV marker remains largely unexplored. We systematically investigated the MMP14 expression and activity phenotypes of EVs for detecting tumor invasion and metastasis, using cell lines, mouse models, and clinical plasma specimen. We show that our nanochip-based assay could improve the diagnostic performance of EV MMP markers compared to standard assays. If validated by large-scale clinical studies, our technology may provide a useful liquid biopsy tool for longitudinal surveillance of tumor evolution in patients to improve cancer management and precision medicine.
RESULTS
3D nanopatterning of EV-CLUE chips by colloidal inkjet printing
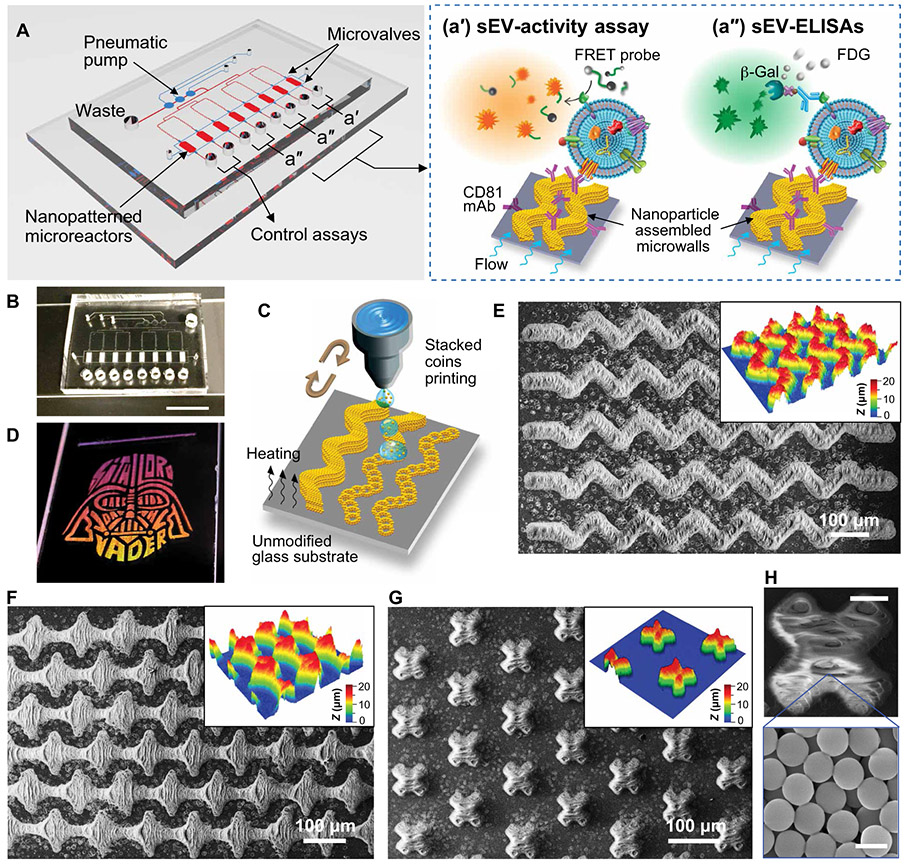
The EV-CLUE chip is a polydimethylsiloxane (PDMS)/glass hybrid device composed of a pneumatic control circuit and an array of eight parallel microchambers patterned with 3D nanostructured microelements (Fig. 1A). The microchambers are flanked by two valves to form the enclosed microreactors for enzymatic detection. Three sEV assays are implemented on the integrated microchip in parallel. We have developed a fluorogenic activity assay that immuno-captures sEVs and specifically measures the proteolytic activity of sEV-carried MMP14 using a fluorescence resonance energy transfer (FRET) peptide probe (Fig. 1, A and a’). This peptide substrate for MMP14 is labeled with a fluorophore and a quencher that can be enzymatically cleaved by MMP14 to generate fluorescence signal. Two sEV enzyme-linked immunosorbent assays (sEV-ELISAs) (22, 23) were also established on chip to quantify the MMP14 protein present on sEVs and the total sEV abundance by probing CD63 and CD9 (Fig. 1, A and a’’). The eight-channel design permits simultaneous analysis of two samples, along with two negative control assays using the phosphate-buffered saline (PBS) blank to determine the background for the activity and expression assays, respectively. The chip fabrication process is similar to that reported in our previous work (22, 23) and detailed in Supplementary Materials and Methods. Figure 1B displays a completed microdevice, highlighting the array of printed sinusoidal micropatterns embedded inside the 30-μm-high assay chambers.
Fig. 1. Colloidal inkjet printing creates the 3D nanopatterned EV-CLUE chip.
(A) Design of the EV-CLUE chip composed of a pneumatic control circuit and an array of eight parallel assay reactors patterned with 3D nanostructured colloidal microelements to enhance immunocapture of circulating sEVs. The pneumatically operated chip integrates three parallel sEV assays: (a’) MMP14 proteolytic activity assay using a specific FRET peptide probe and (a”) ELISA quantitation of sEV MMP14 protein expression and the total sEV concentration determined by CD63 and CD9 in parallel. The eight-channel design allows parallel analysis of two samples, combined with the control assays with PBS blank to determine the background signal for both expression and activity assays. (B) Digital photo of an EV-CLUE chip showing the microreactor array embedded with the printed 3D nanostructures. Scale bar, 1 cm. (C) Schematic of the “stacked coins” colloidal inkjet printing approach for fabrication of 3D self-assembled microelements on an untreated hydrophilic glass substrate. (D) Demonstration of a large-area, complex colloidal crystal pattern printed on a standard microscope slide (2.54 cm wide) with a five-cycle printing of 1-μm silica colloids. The printed nanomaterial patterns display angle-dependent structural colors under white light illumination. (E to G) SEM images and corresponding optical profilometry plots of various micropatterns composed of an array of (E) sinusoidal strips, (F) diamonds, and (G) X shapes, respectively, deposited by 15-cycle inkjet printing of 1-μm silica colloids. (H) SEM image of a micropattern deposited by the high-resolution stacked coins colloidal printing. The magnified image highlights the nanoporous morphology of the self-assembled nanoparticle microstructures. Scale bars, 20 μm (top) and 1 μm (bottom).
In contrast to our μCSA strategy, here we have developed a general inkjet printing method for scalable manufacturing of 3D nanopatterned microchips. Colloidal inkjet printing is an attractive nanomaterial patterning technique (27) that controls the geometry and quality of patterns by exploiting surface modifications to delicately adjust the interactions between surface wetting and evaporation-driven CSA (28-31). This strategy limits the ability to print complex colloidal structures due to the challenges in controlling interfacial interactions during drying. We tried to optimize the protocol of conventional printing (30, 31) but failed to print an array of sinusoidal stripes (fig. S1). Compared to hydrophobic glass surfaces, low surface tension on hydrophilic surfaces is more favorable to printing continuous structures but can cause uncontrolled spread of the printed droplets, leading to poor printing resolution and severely distorted geometries. Evaporation of a drop of colloidal suspension on a hydrophilic surface produces a donut pattern due to the “coffee ring” effect. Adding formamide in the solvent can reduce the coffee-ring effect to produce more uniform assembly of colloids, but it also decreases surface tension, resulting in worse printing resolution and pattern quality.
Distinct from the conventional strategies, we established a “stacked coins” printing approach that exploits the coffee-ring phenomenon to print continuous 3D colloidal patterns on unmodified glass surface (Fig. 1C). We adjusted the droplet volume, substrate temperature, and jetting delay period such that colloidal ring patterns can be deposited individually and overlapped with each other, like offset stacked coins. Multicycle, repeated printing stacks the layers of packed colloids to create a 3D structure of designed geometries. Since drop spacing is a crucial factor that affects the final morphology of the printed structures, we investigated a range of 5 to 20 μm for printing with a 5% (w/w) solution of 1-μm silica colloids; the drop spacing of 10 μm appeared to yield the best printing quality (fig. S2). Using the optimized protocol, we demonstrated a five-cycle printing of 1-μm silica colloids to print a centimeter-scale graphic design on a plain glass slide (Fig. 1D and fig. S3). The pattern exhibited either uniform iridescent structural colors or smooth spectra across the entire pattern due to Bragg diffraction, indicating the high quality of the printed nanomaterial structures (32).
We further tested the method for 15-cycle printing of micropatterns, such as arrays of sinusoidal strips, diamonds, and X shapes. As visualized by SEM and optical profilometry, our method produced 3D self-assembled colloidal patterns of designed geometries (Fig. 1, E to G). Surface profiling by 3D profilometry determined the pattern heights to vary from ~14 to 22 μm. Such topological roughness is expected for inkjet printing and can be further optimized by tuning printing parameters. Nonetheless, rough morphology is favorable for this particular application because it confers greatly increased surface area to enhance EV immunocapture (22, 23). Testing X shapes consisting of 40-μm-wide bars, we estimated the geometric resolution of our printing method as ~20 μm (Fig. 1, G and H). High-magnification SEM of the printed micropatterns verified the highly nanoporous structure of self-assembled silica nanoparticles (Fig. 1H, inset). Moreover, compared to the microchannel-confined CSA method, open-surface printing greatly improves the scalability and the success rate of device fabrication because it negates manual removal of the patterning chip, which can mechanically damage the deposited micropatterns. Overall, these results suggest that our inkjet printing technology provides a general approach capable of high-resolution printing of large-area, complex patterns on hydrophilic surfaces without any chemical pretreatment.
Integrative molecular and functional phenotyping of sEVs by EV-CLUE chip
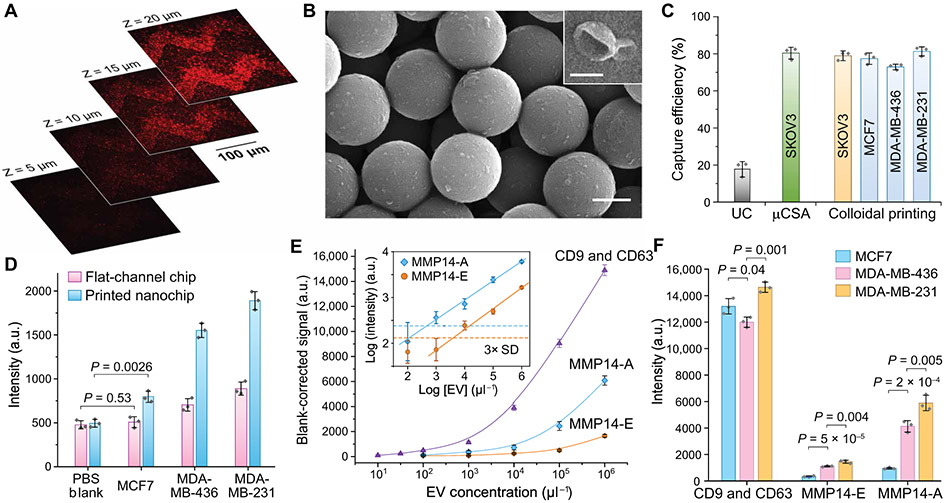
We first characterized sEV immunocapture by the EV-CLUE chip using a well-characterized colon cancer COLO-1 cell–derived EV standard (33). Similar to the ExoProfile chip that we established previously (22), the EV-CLUE system adapts the same sinusoidal patterns, which were printed inside the assay microchambers using the 15-cycle printing protocol. Fluorescently stained COLO-1 EVs were spiked at 106 μl−1 in 10-fold diluted healthy human plasma and injected into the chip coated with anti-CD81 monoclonal antibody (mAb). The confocal fluorescence microscopy images acquired at different depths demonstrated that COLO-1 sEVs were captured on both external and interior surfaces of the micropatterns (Fig. 2A) with a decreasing density gradient inward, indicating flow penetration through the 3D nanoporous structures. SEM imaging visualized high-density capture of sEVs on the mAb-modified silica nanoparticles and the typical spherical and cup-shaped morphologies of captured sEVs (Fig. 2B). The size range of chip-captured EVs was estimated to be 40 to 160 nm from the SEM images, smaller than that of the ultracentrifugation (UC)–purified EVs (~50 to 350 nm) measured by nanoparticle tracking analysis (NTA). These observations for the printed nanostructures are consistent with the sEV capture performance of our μCSA-patterned devices (22, 23).
Fig. 2. Characterization of the EV-CLUE chip for immunoisolation and integrative molecular and activity phenotyping of sEVs.
(A) 2D confocal images at variable depth and (B) representative SEM images of colon cancer COLO1 cell–derived sEVs captured on and inside the self-assembled nanoporous silica micropatterns printed on the substrate. Dil dye-stained COLO1 EVs (106 μl−1) were assayed for confocal fluorescence microscopy. Scale bars, 500 nm (B) and 100 nm (B, inset). (C) Comparison of sEV capture efficiency for standard UC and the nanochips fabricated by the μCSA and colloidal inkjet printing methods. Fluorescently stained EVs of cancer cell lines were spiked in healthy human plasma at 106 μl−1. (D) Comparison of the flat-channel and nanopatterned chips for specific detection of MMP14 in UC-purified vesicles (106 μl−1) from breast cancer cell lines. Two-tailed Student’s t test was used for two-sample comparison, P < 0.05. (E) Calibration of the EV-CLUE chip by measuring the total sEV concentration (determined by CD9 and CD63), MMP14 expression (MMP14-E), and MMP14 proteolytic activity (MMP14-A) of MDA-MB-231 EVs. Inset: Determination of LODs for the MMP14 activity assay from 3 SDs of the backgrounds (dashed lines). (F) Integrative multiparameter analysis of purified EVs (106 μl−1) from three breast cancer cell lines with different invasiveness. Statistical difference was determined by one-way ANOVA with post hoc Tukey’s pairwise multiple comparisons test, P < 0.05. Anti-CD81 capture mAb was used in all cases. All data were presented as mean values with error bars of 1 SD (n = 3).
We evaluated the capture efficiency of EV-CLUE nanochips in comparison with the gold standard UC and the μCSA-engineered chips. EVs were purified from the conditioned culture media of cancer cell lines and characterized by NTA to prepare standards of know quantities (fig. S4). Recent studies have reported abundant expression of tetraspanin proteins (CD9, CD63, and CD81) in sEVs derived from a wide range of cell lines and biofluids and enrichment of some endosome-related proteins in the anti-tetraspanin immuno-captured sEVs versus other types of large EVs and sEV subpopulations (10, 34). Thus, tetraspanin-targeting immunocapture has been proposed for specific isolation of endosome-derived exosomes, despite the possible presence of other tetraspanin-bearing sEV subtypes (10). Using an established depletion method (23), we assessed the anti-CD81 mAb-coated nanochips for capture of cancer cell–derived EVs spiked in healthy human plasma (106 μl−1). The observed capture efficiencies were 78.2 ± 2.6% for SKOV3, 77.4 ± 3.1% for MCF7, 72.9 ± 1.5% for MDA-MB-436, and 81.3 ± 3.4% for MDA-MB-231 (Fig. 2C). Such relatively consistent capture efficiency across various cell lines and cancer types permits downstream quantitative detection of captured sEVs and their molecular constituents. The capture performance of the printed chip was comparable to that of the μCSA-engineered device (80.3 ± 3.2% for SKOV3) and much higher than that of standard UC isolation (17.9 ± 3.9%). sEV isolation using the bovine serum albumin–coated control chips resulted in low non-specific binding (<6.7%), indicating the effectiveness of our assay protocol to suppress the matrix effects on the nanopatterned chip (fig. S5).
On the basis of the nanochip immunocapture, we attempted to develop sandwich MMP ELISA and proteolytic activity assays for molecular and functional phenotyping of tumor-derived sEVs. Here, we targeted MMP14 because it has been credited as a central regulator of cell invasion via degradation of major components of ECM (fibrillar collagens, fibronectin, and vitronectin) and processing of intra- and extracellular proteins (soluble pro-MMPs, cytokines, and growth factors) to promote matrix remodeling and invasive behavior of tumor cells (35). As a test case, three breast cancer cell lines were used: weakly invasive MCF7 cells as the control and two triple-negative breast cancer (TNBC) cell lines, MDA-MB-436 and MDA-MB-231, with metastatic capability. Among three tetraspanins commonly used for sEV capture, CD81 yielded the highest detection sensitivity for these cell lines (fig. S6). Thus, our assays were configured to capture overall sEVs by anti-CD81 mAb and to measure the expression and activity of sEV-bound MMP14 with specific mAb and peptide probes, respectively. Optimized from our previously developed protocol (22, 23), the nanochip sEV-MMP14 ELISA permitted sensitive detection of the low-abundance MMP14+ subpopulation in MCF7-derived EVs, which was otherwise undetectable on a conventional flat-channel chip (Fig. 2D). Higher MMP14 expression was detected in EVs from metastatic MDA-MB-436 and MDA-MB-231 compared to that of MCF7 cells. The printing-based 3D nanostructuring of microfluidic chips improved detection sensitivity of sEV-ELISA. Overexpression of MMP14 in these metastatic breast cancer cell lines has been well documented (36-38). The chip detection of MMP14 was further verified by standard Western blot and microplate enzymatic activity assays (fig. S7).
The sandwich activity assay was then developed by combining sEV immunocapture with the detection of enzymatic activity of MMP14 using a FRET peptide substrate (Fig. 1A). Using a standard microplate assay kit, a commercially available fluorogenic probe was selected and verified for its specificity to MMP14 protein against three soluble and membrane MMPs commonly associated with breast cancer (18, 37, 39-41): MMP9, MMP15, and MMP16 (fig. S8, A and B). Chemical activation of recombinant pro-MMP14 protein by 4-aminophenylmercuric acetate was required to gain enzymatic activity (fig. S8B). In contrast, similar MMP14 activity was detected for EVs of breast cancer cells with and without chemical activation (fig. S8C), indicating that sEV MMP14 was activated. Our findings are in line with the previous reports that EVs transport proteolytically active MMP14 to promote tumor invasion and metastasis in malignancies, including breast cancer (19, 42). Thus, we conducted the MMP14 activity assay without chemical treatment to measure the native activity of circulating sEVs. We further optimized the enzymatic reaction time for the on-chip fluorogenic activity assay to afford a maximal signal/noise ratio (fig. S8D).
Using the optimized assays, the analytical performance of the EV-CLUE technology was systematically calibrated. In addition to the MMP14 assays, a tetraspanin ELISA was assessed for quantifying the total sEV concentration by measuring the combined expression of CD9 and CD63. To this end, we titrated a serial dilution of UC-purified EVs from MDA-MB-231 cells and observed a linear curve of the CD9 and CD63 expression as a function of the total EV concentrations measured by NTA (Fig. 2E). The curve starts to level off when the EV concentration decreases, yielding a low limit of detection (LOD) of ~16 EVs μl−1 (calculated by dividing three SDs of the background by the slope of the calibration plot). These cell line results support the use of the CD9 and CD63 assay for quantitative detection of the total sEV concentration, which we validated further using human plasma samples. Compared with the combined CD9 and CD63 expression, the calibration curve for the sEV MMP14 expression (MMP14-E) indicated that the MMP14+ sEVs account for a small fraction of the overall EV population and were detectable above a calculated EV concentration of ~5 × 103 μl−1 (Fig. 2E, inset). The MMP14 activity (MMP14-A) assay was observed to produce higher signals than the protein quantification, yielding a 10-fold lower LOD of ~5 × 102 EVs μl−1. The EV-CLUE chip was then assessed for integrative molecular and functional phenotyping of tumor-derived sEVs using three breast cancer cell lines. Using equal EV inputs, the sEV MMP14 expression and activity assays differentiated the metastatic capabilities of the cells, with higher detection signals conferred by the activity assay (Fig. 2F). CD9 and CD63 expression provided the highest analytical sensitivity for sEV detection but did not correlate with metastatic phenotypes. These findings highlight the necessity of highly sensitive detection of low-abundance, clinically relevant EV subtypes and suggest that activity analysis could provide a sensitive means to probe pathological phenotypes of tumor-derived sEVs.
Detection of in vitro invasiveness of isogenic cell lines
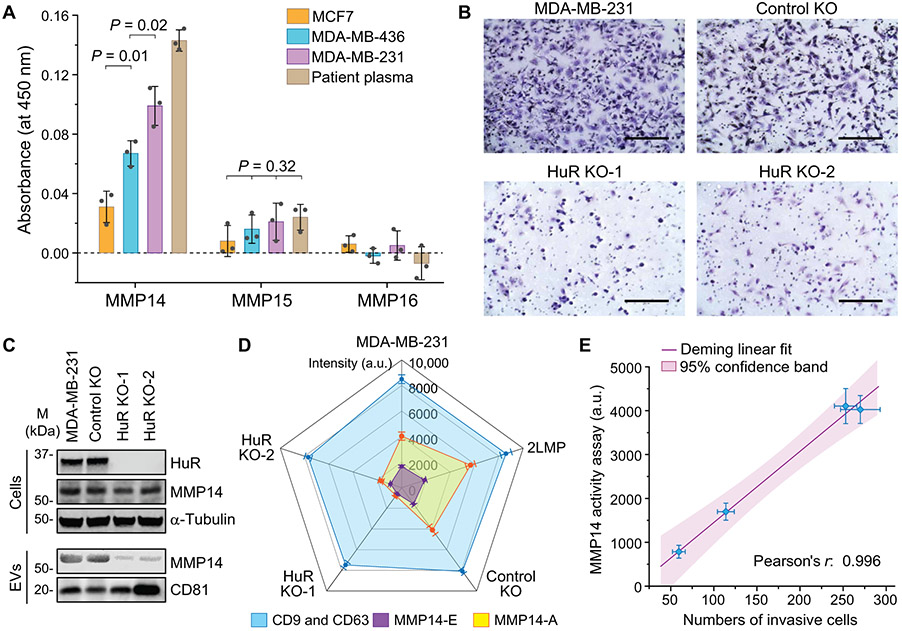
As a proof of concept of potential clinical application, we adapted the EV-CLUE technology for noninvasive measurement of tumor progression and metastasis using breast cancer as a disease model. Studies have identified multiple MMPs in breast cancer–derived EVs, among which MMP14 was the most frequently detected membrane-type MMP (18, 19, 43, 44). Here, we attempted to quantify MMP14 and two major membrane MMPs, MMP15 and MMP16, which have been detected in breast cancer cell lines and tissues (40), but not in EVs. Using standard microplate ELISA kits, MMP14 was readily detected in samples of ~109 EVs purified from three cell lines and the plasma of a patient with metastatic breast cancer (Fig. 3A). The sEV MMP14 expression was observed to differentiate the variable metastatic potential of the cells and plasma sample, validating the chip-based measurements performed with less than 1/100 of the sample quantities. MMP15 expression was very low and could not be used to detect metastatic cells, whereas MMP16 was essentially undetectable (Fig. 3A). On the basis of our results and those of others, our subsequent studies have been focused on only MMP14.
Fig. 3. Functional phenotyping of sEVs by EV-CLUE detects tumor cell invasiveness.
(A)Measurement of the expression of MMP14, MMP15, and MMP16 on breast cancer-derived EVs. One hundred microliters of purified EVs (107 μl−1) from three cell lines and 20 μl of patient plasma (diluted 1:5 in PBS) were assayed using the commercial microplate ELISA kits. Statistical analysis was conducted with one-way ANOVA with post hoc Tukey’s test, P < 0.05. (B) Microscopic images of Matrigel invasion assays for the parental MDA-MB-231 cell line, an isogenic control, and two HuR KO clones. Scale bars, 200 μm. (C) Western blot analysis of HuR and MMP14 proteins in the isogenic MDA-MB-231 cell lines and their EVs purified by UC. Cell lysate (30 μg) and ~1010 EVs from each cell line were assayed with α-tubulin and CD81 as the loading controls, respectively. (D) Radar plot of the nanochip analyses of the total sEV (CD9 and CD63), MMP14-E, and MMP14-A of sEVs isolated from culture media of MDA-MB-231, 2LMP (lung metastatic subline of MDA-MB-231), control, and HuR KO clones. Ten microliters of purified EVs (106 μl−1) was used in each assay on a chip. Signal intensity was subtracted by the corresponding background. (E) Correlation of the measured sEV MMP14 proteolytic activity with the number of invasive cells counted in the Matrigel assays. The linear fitting was performed using the Deming regression model at the 95% confidence level. In all cases, anti-CD81 mAb was used for sEV capture, and each sample was measured in triplicate. All data were presented as means ± SD (n = 3).
The EV-CLUE chip was assessed for integrative molecular and functional phenotyping of tumor-derived sEVs using isogenic cell lines. We established two human antigen R (HuR) CRISPR knockout (KO) clones from MDA-MB-231. HuR is an RNA-binding protein known to promote tumorigenesis and invasion (45). As verified by Matrigel invasion assays, the invasiveness of MDA-MB-231 cells was largely reduced by knocking out HuR (Fig. 3B). Small interfering RNA knockdown of cellular MMP14 expression (fig. S9) was seen to suppress the invasiveness of metastatic MDA-MB-231 cells, as supported by other reported studies (46-48). However, Western blot analysis revealed that HuR KO did not impair the expression of MMP14 protein in cells, but rather attenuated the amount of MMP14 protein carried by the secreted EVs (Fig. 3C). The findings suggest that EV-transported MMP14 plays a role in cell invasion. Therefore, although the mechanism of HuR-mediated regulation of EV transport of MMP14 remains to be elucidated, this HuR KO model provides a useful approach to validate our technology and to specifically assess EV MMP14 as a marker of cell invasion.
Using the EV-CLUE chip, we conducted multiplexed molecular and functional phenotyping of EVs isolated from these isogenic cell lines in comparison with a lung metastatic subline of MDA-MB-231 (2LMP). In contrast to the total sEV abundance, the measured MMP14 phenotypes of sEVs reflected the invasiveness of the parental and KO cell lines (Fig. 3D). Measurements of MMP14 activity conferred a two- to threefold increase in sensitivity over protein quantification. Quantitative comparison by regression analysis revealed a strong linear correlation between the sEV MMP14 proteolytic activity measured by the EV-CLUE chip and the number of invading cells counted in the Matrigel invasion assays (Pearson’s r = 0.996; Fig. 3E). To assess the adaptability of our technology to other malignancies, we also tested a set of isogenic pancreatic cancer MIA PaCa2 cell lines and observed consistent performance of the EV-CLUE chip for detecting in vitro cell invasiveness (fig. S10). Overall, our findings support the potential utility of sEV MMP14 as a marker of tumor invasion and metastasis.
Noninvasive monitoring of tumor evolution in vivo using mouse models
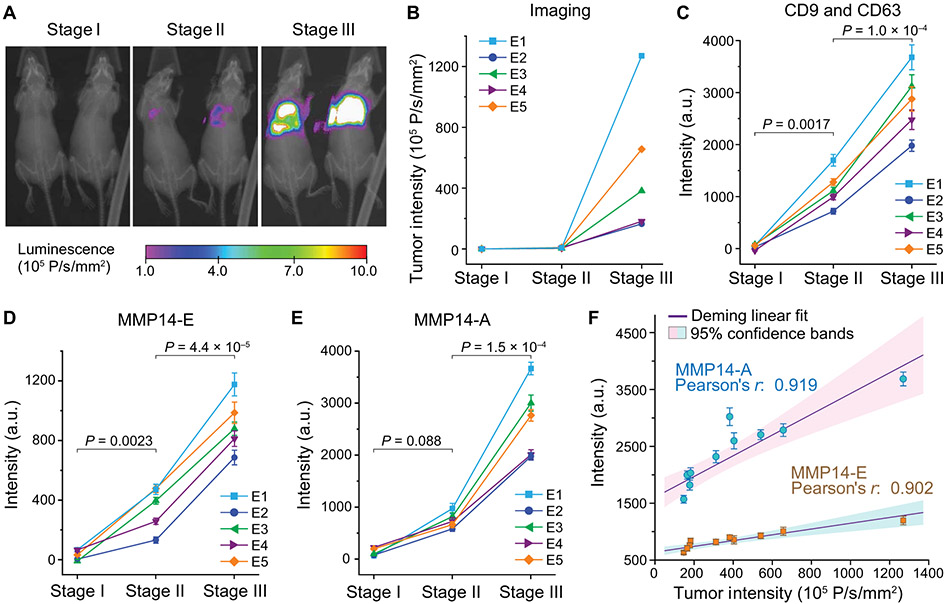
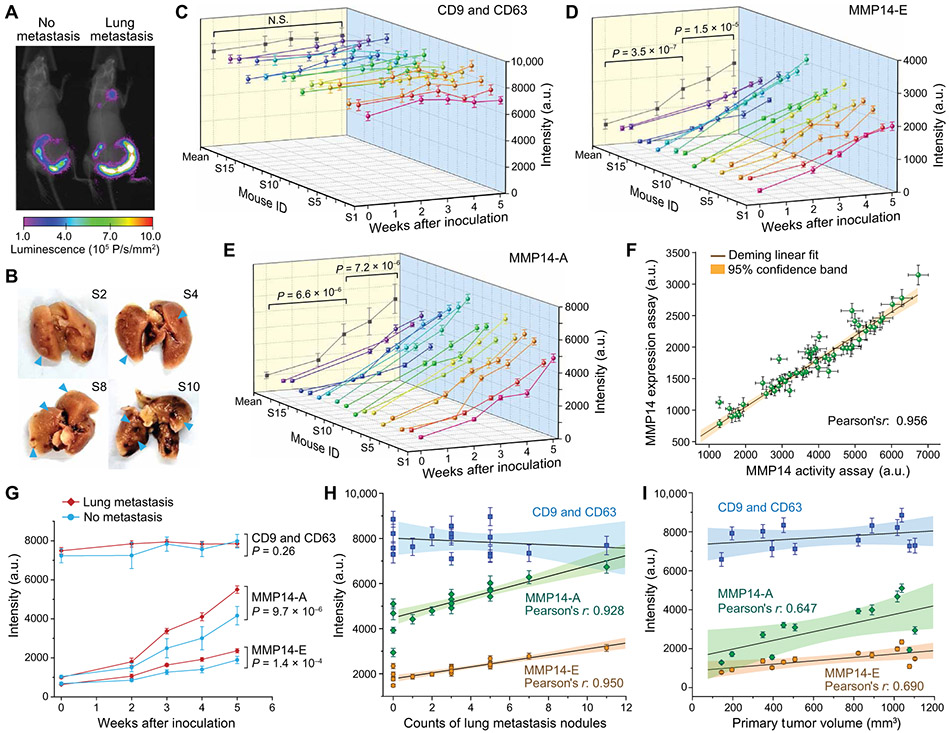
We first assessed the feasibility of using the EV-CLUE technology for longitudinal monitoring of metastatic burden using a mouse model of human breast cancer metastasis. Experimental metastasis assays typically reduce variability and improve the statistical power of analyses by bypassing early steps in the metastatic cascade, therefore providing a robust in vivo model for technology assessment. We injected 106 2LMP-Luc cells (luciferase-expressing lung metastatic subline of MDA-MB-231) into the lateral tail veins of athymic nude female mice and monitored the development and growth of tumors, primarily in the lungs, by imaging twice a week. Figure 4 (A and B) presents the representative images and tumor intensity plots acquired for the same mice at three stages: (i) before inoculation, (ii) initial detection of early metastasis, and (iii) nearly moribund with extensive lung metastases. Blood (~50 μl) was repeatedly collected from each mouse at each of the three stages to prepare plasma for microfluidic analysis. We identified and validated the antibodies for specific analysis of human-derived sEVs in blood plasma of the xenografted mice (table S1 and fig. S11). With these optimized assays, we demonstrated multiparametric analysis of human cancer-derived sEVs directly in 6 μl of mouse plasma per run, which enabled longitudinal monitoring of progressive tumor development in individual mice (n = 5; Fig. 4, C to E, and data file S1). To quantitatively evaluate sEV MMP14 as a marker of tumor burden, we compared the sEV phenotypes with the tumor intensity measured for 10 xenografted mice at stage III of metastasis (Fig. 4F). Both sEV MMP14-E and MMP14-A were found to correlate with the tumor intensity at stage III, and the activity analysis provided a higher sensitivity than the expression analysis.
Fig. 4. Integrative sEV phenotyping for monitoring tumor development in mice using an experimental metastasis model of human breast cancer.
(A and B) An experimental metastasis model in athymic nude mice was established by injecting 106 2LMP-Luc cells into the tail veins of 4-week-old female nude mice. Progression of the lung tumors was monitored by bioluminescent imaging. (A) Representative images and (B) corresponding tumor intensity plots were acquired for each mouse at three stages: (i) before inoculation, (ii) initial detection of early metastasis, and (iii) nearly moribund with extensive lung metastases. (C to E) Multiplexed total sEV (CD9 and CD63), MMP14-E, and MMP14-A analyses of circulating sEVs in mouse plasma using EV-CLUE. Blood (~50 μl) was collected from tail vein at each stage. Plasma (6 μl) was diluted in PBS by five times and analyzed on chip using the anti-human mAbs validated for specific detection of human tumor xenograft-derived sEVs. Each sample was assayed in triplicate to determine the mean and SD (error bars). P values were determined by one-way repeated measures ANOVA with post hoc Tukey’s pairwise multiple comparisons test, P < 0.05. (F) Correlation of the sEV MMP14 expression and activity measured for 10 xenografted mice at stage III with the tumor sizes measured by bioluminescent imaging. All data were presented as means ± SD (n = 3). Deming linear fitting was performed at the 95% confidence level.
We further extended the in vivo mouse studies to a spontaneous metastasis model that recapitulates all of the steps in the pathogenesis of metastasis, to closely mimic the clinical reality. In this orthotopic model, 0.5 × 106 mouse 4T1-Luc breast cancer cells were injected into the mammary fat pad of 4-week-old female BALB/c mice (n = 16). Primary tumor growth was assessed by caliper measurements of the mammary xenografts to calculate tumor volume using the modified ellipsoidal formula (fig. S12) (49), while the development of metastases was monitored by bioluminescence imaging (Fig. 5A). Meanwhile, ~50 μl of blood from the tail vein of each mouse was collected before and after inoculation. When the mice were euthanized at the end of week 5, 12 were found to have lung metastases (Fig. 5B). The EV-CLUE technology enabled multiplexed analysis of the longitudinal plasma samples collected from each mouse (data file S1 and fig. S13). In contrast to the experimental model, the total sEV abundance did not appear to be a potent indicator of tumor development in the spontaneous model (Fig. 5C and fig. S14A). This discrepancy is attributed to the fact that the experimental metastasis assay probed human xenograft-derived sEVs against the background of mouse sEVs, whereas in the spontaneous model, the total sEV assay also detected many wide-type sEVs that masked the dynamics of tumor-derived vesicles. Both sEV MMP14-E and MMP14-A were observed to increase along with tumor growth and metastasis in individual mice (Fig. 5, D and E). Our method detected significant increases in the population means of the MMP14 markers over 1-week tumor development (P = 0.003 for MMP14-E and P = 6.8 × 10−4 for MMP14-A in week 5; fig. S14, B and C). Although the single-mouse longitudinal sEV analysis revealed increased sEV MMP14-E and MMP14-A in several mice (week 4; fig. S13), the population means showed no significant difference (week 4, MMP14-E: P = 0.18; MMP14-A: P = 0.26; fig. S14, B and C), owing to the large interindividual heterogeneity observed. Regression analysis of the data for all 16 mice resulted in a strong linear correlation between the sEV MMP14-E and MMP14-A measurements (Pearson’s r = 0.956, Fig. 5F). These results, combined with the in vitro cell analysis and the experimental metastasis assays, validate the sensitive and specific molecular and functional profiling of tumor-derived sEVs using our technology.
Fig. 5. Longitudinal monitoring of tumor evolution in a spontaneous breast cancer metastasis model.
(A)Representative bioluminescence images of mice without and with lung metastasis developed at week 5. A spontaneous metastasis model in BALB/c mice was established by injecting 0.5 × 106 4T1-Luc cells into the mammary fat pad of 4-week-old female BALB/c mice, and the tumor burden was monitored by bioluminescence imaging. (B) Representative digital photos of lungs isolated from the mice euthanized 5 weeks after tumor inoculation showing metastatic nodules (blue arrowheads). (C to E) Time-lapse, multiplexed measurements of the total concentration (CD9 and CD63), MMP14-E, and MMP14-A of circulating sEVs in individual mice (n = 16) using the EV-CLUE technology. Repeated tail vein collection of ~50 μl of blood from the same mouse was conducted at the indicated time points before and after inoculation. Plasma (6 μl) prepared from the blood samples was diluted by five times and assayed directly on different chips in triplicate to determine the mean and SD (error bars). The averaged results of all samples collected at individual time points were projected on the X-Z plane (means ± SEM). P values were determined by one-way repeated measures ANOVA at P < 0.05. (F) Correlation between the sEV MMP14-E and MMP14-A measured for all plasma samples in (C) to (E). (G) Time-lapse curves of the measurements (means ± SEM) of three sEV markers in the mice that developed primary tumor only (n = 4) or with lung metastasis (n = 12). P values were determined by two-way ANOVA at the 95% confidence level. (H) Correlation of the sEV markers measured for 16 mice at week 5 with the count of lung metastasis nodules. (I) Correlation of the sEV markers with the primary tumor volume measured from weeks 2 to 5 for four mice that developed only primary tumors. The data in (H) and (I) were presented as means ± SD (n = 3 technical repeats). Linear Deming fitting in (F), (H), and (I) was performed at the 95% confidence level.
Next, we compared the longitudinal sEV measurements between subgroups of mice that developed primary tumor (n = 4) or primary tumor with lung metastasis (n = 12). In contrast to the total sEV concentration (P = 0.26), both sEV MMP14-E (P = 1.4 × 10−4) and MMP14-A (P = 9.7 × 10−6) exhibited significantly faster overall rates of increase in the mice developing metastases than those with only primary tumors (Fig. 5G). Statistical comparisons at individual time points also showed that monitoring the change in sEV MMP14 activity provided better statistical power to differentiate the two mouse subgroups than monitoring MMP14 expression (fig. S15), suggesting the potential of the functional activity of sEVs for detection of tumor metastasis. To further assess the correlation between the sEV phenotypes with metastasis, we compared the sEV results with the number of lung metastasis nodules measured at week 5 (Fig. 5H) and the volume of primary tumor measured from weeks 2 to 5 for the four mice, which developed only primary tumors (Fig. 5I). Consistently, the sEV MMP14 markers exhibited significantly stronger correlation with the lung metastases (Pearson’s r = 0.950 for MMP14-E and r = 0.928 for MMP14-A) than the mammary xenografts (Pearson’s r = 0.690 for MMP14-E and r = 0.647 for MMP14-A). Spontaneous metastasis model studies support the feasibility of sEV MMP14 phenotype analysis as a potent biomarker of aggressiveness and metastatic potential of tumors and the superior analytical performance of the nanoengineered chip. We also demonstrated that our EV-CLUE technology enables minimally invasive, real-time monitoring of the dynamics of tumor development in individual mice.
Clinical analysis of breast cancer patient cohorts
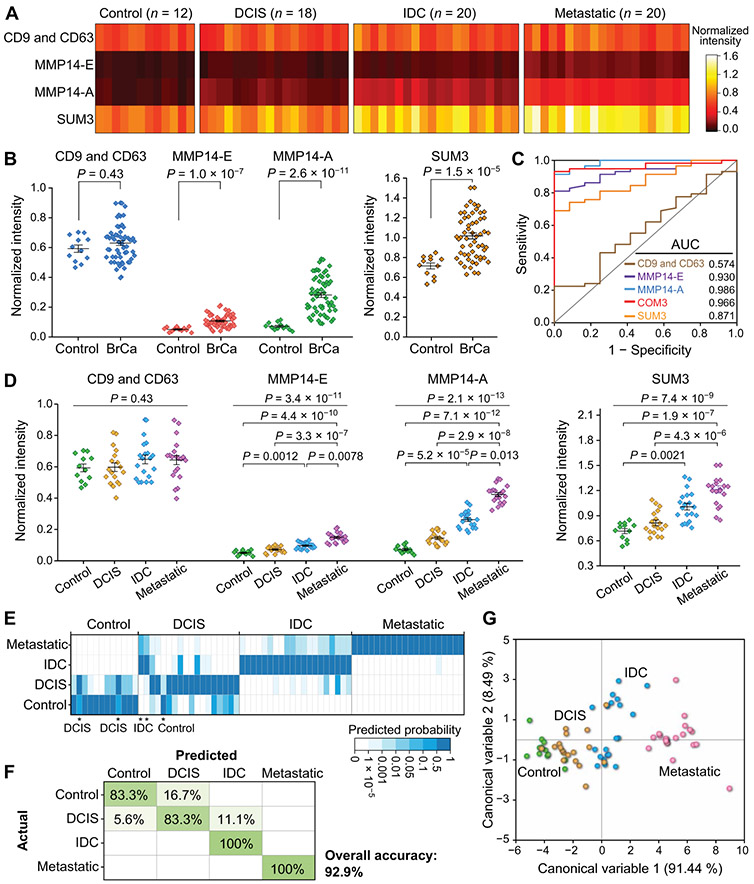
We next assessed the EV-CLUE technology for clinical application to human malignancies, using plasma samples collected from a training cohort of patients with stage 0 to III breast cancer (n = 22) and age-matched noncancer controls (n = 8). The patient cases involved three groups of distinct clinical stages: preinvasive ductal carcinoma in situ (DCIS; n = 8), nonmetastatic invasive ductal carcinoma (IDC; n = 7), and locally advanced IDC with lymph node metastases (n = 7), each of which consists of diverse histological and molecular subtypes, including TNBC, to represent tumor heterogeneity (table S2). Figure 6A summarizes the measurements of individual sEV markers for each subject (data file S2) and the SUM3 signature defined by the unweighted sum of three markers (50, 51). The detection of sEV MMP14 was verified by Western blot analysis of UC-purified EVs, which detected high expression of CD81 in EVs from controls and patients and low-abundance sEV MMP14 in patients, with a notably elevated abundance in the metastatic cases (Fig. 6B). As shown in Fig. 6C, the total sEV concentration was not significantly different between the cancer and control groups (two-tailed Mann-Whitney U test, P = 0.63), whereas sEV MMP14 markers differentiated the two groups (P = 1.1 × 10−4 for MMP14-E and P = 4.1 × 10−6 for MMP14-A), despite large interindividual variations. It is noted that multiparametric combination with the functional activity marker improved the performance of molecular phenotyping of sEVs for cancer detection [P = 1.0 × 10−5 for SUM2 (MMP14-E + MMP14-A) versus P = 1.0 × 10−5 for MMP14-E and P = 0.005 for SUM3 versus P = 0.27 for SUM1 (CD9 and CD63 + MMP14-E); Fig. 6C and fig. S16].
Fig. 6. Integrative sEV phenotyping of human breast cancer in a training cohort.
(A) Heat map of the molecular and activity profiles of plasma sEVs from a training set involving age-matched cancer-free controls (n = 8) and patients with preinvasive DCIS (n = 8), localized IDC (n = 7), or locally metastatic IDC (n = 7). Signals were averaged over triplicate measurements of each sample and normalized against the 99th percentile of all signals after the background subtraction. The color intensity displays the mean of the triplicate measurements of each sample on different chips. SUM3 signature denotes the unweighted sum of three markers. (B) Western blot analysis of MMP14 in plasma EVs purified by UC from a control and patients with breast cancer (BrCa) of various stages. EVs (~1010) from each sample was assayed with CD81 as the loading control. (C) Scatterplots of the sEV markers and SUM3 signature for detecting BrCa against the control. Nonparametric, two-tailed Mann-Whitney U test was used for two-group comparison. (D) ROC curves for LDA of three sEV markers, a three-marker panel (COM3), and the SUM3 signature for breast cancer diagnosis. (E) Scatterplots of the sEV markers and SUM3 signature for differentiating individual groups at progressing disease stages. Kruskal-Wallis one-way ANOVA with post hoc Dunn’s test for pairwise multiple comparisons was used to determine the overall and each group pair’s P value. (F to H) Classification of the training cohort by discriminant analysis of the three-marker panel COM3 was summarized in (F) the heat map of classification probabilities, (G) confusion matrix, and (H) canonical score plot of the first two canonical variables that together capture 99.93% of the variance. The middle line and error bar in (C) and (E) represent the mean and 1 SEM, respectively. All statistical analyses were performed at 95% confidence level.
We quantitatively evaluated the diagnostic metrics of the biomarkers individually and in combinations using a multivariate receiver operating characteristic (ROC) curve analysis strategy based on linear discriminant analysis (LDA) (51, 52). The training set data were processed by LDA to create a discriminant function model for classification of two sample groups. The predicted probabilities yielded from this binary classification procedure were used as the single test variable for ROC analyses. As summarized in Fig. 6D and table S3, among three markers, sEV MMP14-A showed the best diagnostic performance for the training cohort with 0.977 [95% confidence interval (CI), 0.845 to 1] area under the curve (AUC), 95.5% sensitivity (95% CI, 77.2 to 99.9%), 100% specificity (95% CI, 63.1 to 100%), and 96.7% accuracy (95% CI, 82.8 to 99.9%). Evaluation of combinations of the markers showed that multivariate detection with the marker panels afforded comparable or better diagnostic power than univariate detection using single SUM signatures [COM3 (0.977 AUC, 100% sensitivity, 87.5% specificity, 96.7% accuracy) versus SUM3 (0.830 AUC, 81.8% sensitivity, 87.5% specificity, 83.3% accuracy)]. While providing about the same performance as sEV MMP14-A to detect cancer against control, the three-marker panel COM3 improved the classification of patients at variable disease stages.
Figure 6E depicts the assessment of our method for detecting the controls and three subgroups of patients: preinvasive DCIS, nonmetastatic IDC, and locally metastatic IDC. We observed an overall significant increase in sEV MMP14-E [Kruskal-Wallis one-way analysis of variance (ANOVA), P = 4.9 × 10−5] and MMP14-A (P = 1.4 × 10−5) and the SUM3 signature (P = 4.3 × 10−4) with progressive disease stages. Post hoc Dunn’s pairwise multiple comparisons test revealed significant differences between three group pairs: control versus IDC (MMP14-E, P = 0.04; MMP14-A, P = 0.0024), control versus metastatic IDC (MMP14-E, P = 3.2 × 10−5; MMP14-A, P = 2.8 × 10−5), and DCIS versus metastatic IDC (MMP14-E, P = 0.0084; MMP14-A, P = 0.014; Fig. 6E). Moreover, the sEV MMP14 markers were able to discriminate the combined group of invasive and locally advanced IDC cases from the control or preinvasive DCIS group (fig. S17). To further explore the capacity of our method for diagnosis and stratification of breast cancer, we conducted discriminant analysis of the sEV phenotypes individually and in combination for classification of the training cohort. The quadratic method was chosen to generate the discriminant models as the equality test of within-group covariance matrices failed. The classification results obtained with the three-marker panel COM3 are visualized in Fig. 6 (F to H) and results are summarized in fig. S18. It was seen that the discriminant analysis of COM3 correctly classified individual subjects into the four groups except for one DCIS case misidentified as a control (Fig. 6F). The classification results were quantitatively presented as a confusion matrix (Fig. 6G), showing an overall accuracy of 96.7% (95% CI, 82.8 to 99.9%), which was better than that of sEV MMP14-A alone (86.7%; fig. S18). Such improvement demonstrates the advantage of the multiparametric sEV marker panel versus single markers for multiclass diagnostics. To further characterize the effectiveness of our method to discriminate subgroups, we plotted the scores of each subject for the first two canonical variables computed from the discriminant analysis (Fig. 6H). It was clearly visualized that the training samples were classified into four groups with notably better separation among the patient groups at progressing disease stages. A correlation circle was also created by projecting the input variables (sEV markers) in the factors space (fig. S19), showing strong positive correlation of the sEV markers to the first canonical variable that highlights their contribution to accurate disease classification. The correlation circle also revealed a weak but positive correlation of the CD9 and CD63 expression to the canonical variables, which explains its role in improving the accuracy of the multiclass diagnostics when combined with the sEV MMP14 markers (fig. S18).
The EV-CLUE technology was further applied to an independent validation cohort of plasma samples from age-matched cancer-free controls (n = 12) and patients with breast cancer (n = 58; 18 DCIS, 20 nonmetastatic IDC, and 20 locally advanced breast cancer with lymph node metastases; Fig. 7A). Correlation analyses of the sEV MMP14-E and MMP14-A data found no significant difference between the training and validation cohorts (P = 0.46) using one-way analysis of covariance (ANCOVA) and yielded a high Pearson coefficient of 0.956 for two cohorts combined (fig. S20), indicating the adaptability of our assays to specific and reliable analysis of clinical plasma samples. Patients with different histological and molecular subtypes (table S2) were distinguished from controls by probing sEV MMP14-E and MMP14-A (P = 1.0 × 10−7 and P = 2.6 × 10−11, respectively, two-tailed Mann-Whitney U test; Fig. 7B). The validation set data were fed into the discriminant function model established with the training cohort to test its validity for clinical diagnosis. The postprobabilities predicted for the validation cohort without a known a priori disease state were used to conduct ROC analysis (Fig. 7C and table S3). Consistent diagnostic performance of three sEV parameters was observed between the training and validation cohorts in terms of AUC (0.926 versus 0.930 for MMP14-E and 0.977 versus 0.986 for MMP14-A) and accuracy (86.7% versus 85.7% for MMP14-E and 96.7% versus 92.9% for MMP14-A).
Fig. 7. Validation of integrative functional sEV phenotyping for noninvasive diagnosis and monitoring progression and metastasis of BrCa.
(A) Heat map of three sEV markers and their SUMs measured from plasma sEVs from an independent validation cohort of age-matched cancer-free controls (n = 12) and patients with DCIS (n = 18), nonmetastatic IDC (n = 20), and locally metastatic cases (n = 20). The assays and data processing were the same as those for the training cohort (Fig. 6A). (B and C) Evaluation of the sEV markers individually and in combinations for breast cancer diagnosis. Two-tailed Mann-Whitney U test was used for two-group comparisons in (B). (D) Evaluation of the sEV markers and SUM3 signature for differentiating individual groups at progressing disease stages. Kruskal-Wallis one-way ANOVA with post hoc Dunn’s test for pairwise multiple comparisons was used to determine the overall and each group pair’s P value. (E to G) Multivariate classification to assess the combined three sEV markers for identifying the control and patient groups with preinvasive, invasive, and metastatic BrCa, as presented by (E) the heat map of classification probabilities, (F) confusion matrix, and (G) canonical score plot of the first two canonical variables derived from the discriminant analysis of the training cohort. The middle line and error bar in (B) and (D) represent the mean and 1 SEM, respectively. All statistical analyses were performed at 95% confidence level.
Using different statistical approaches, the validation cohort data were then evaluated for detection of the progression stages of breast cancer. Compared to the training cohort, a considerable improvement in differentiating the four groups of subjects in the validation cohort was observed with the Kruskal-Wallis one-way ANOVA followed by the post hoc Dunn’s multiple comparisons test (Fig. 7D). In particular, as opposed to the training cohort, here we detected a significant increase in sEV MMP14-E (P = 0.0078) and MMP14-A (P = 0.013) in the metastatic group versus the localized IDC group. We also observed enhanced statistic distinction between the combined malignant group and the control or DCIS group in the validation cohort, when probing the MMP14+ sEVs (fig. S21). Such improvement could be attributed to the larger sample size of the validation cohort that reduces the sampling variances. Classification analysis of the validation cohort data was then performed with the discriminant function model derived from the training cohort to test its reliability for multiclass diagnosis of breast cancer. The classification accuracy for the training and validation cohorts was consistent (86.7% versus 81.4% for MMP14-A; fig. S18). Among the biomarker combinations tested here, discriminant analysis of COM3 yielded the best classification performance (Fig. 7, E to G). It is noted that all IDC and metastatic IDC cases in the validation cohort were correctly detected, and only two noninvasive DCIS cases were misclassified as invasive, leading to an overall accuracy of 92.9% (95% CI, 84.1 to 97.6%; Fig. 7, E and F). This observation was also reflected in the canonical score plot, which displays separation among the four groups in the validation cohort in the 2D discriminant factor space defined by the training samples (Fig. 7G).
The nanochip-based integrative phenotyping of circulating sEVs was systematically validated by the measurements of the same samples with a variety of gold standard approaches. EVs were isolated by UC from a subset of control and breast cancer samples randomly selected from the training and validation cohorts (n = 10 for each subgroup) and characterized by the NTA and Bradford assays. The NTA results displayed variable mean diameters of ~100 to 160 nm with the major size distribution ranging from ~60 to 400 nm (fig. S22). To assess the CD9 and CD63 assay for quantifying the total EV concentration in human plasma, we plotted the nanochip signals against the EV abundance counted by NTA for these 40 plasma samples (fig. S23). Regression analysis resulted in a strong linear correlation between the two methods (Pearson’s r = 0.931). This plasma analysis, together with the cell line studies, demonstrates our tetraspanin-based assay as a means to quantitatively estimate the abundance of circulating EVs. It should be noted that the applicability of this method may be limited by the heterogeneous tetraspanin expression on EVs and thus needs to be validated in individual cases. We compared the EV numbers and sizes measured by NTA across the control and patient groups, which showed no significant difference (P = 0.53 and P = 0.47, respectively; fig. S24, A, and B). The total EV protein expression measured by the Bradford assay detected the difference between the control group and each of the patient groups, but not among the patient groups (fig. S24C). Compared to these measurements of general EV properties, the nanochip method targeting tumor-associated markers on sEVs greatly augmented diagnostic performance.
To further demonstrate the advantageous performance of our technology, we attempted to compare it to standard microplate ELISA for targeted analysis of sEV MMP14 marker. Sixty samples (n = 15 for each subgroup) from the two cohorts were measured by a commercial microplate EV ELISA kit. This standard assay yielded a similar sEV MMP14-E pattern to the nanochip analysis but with much lower signal intensity (fig. S25A). Comparing the data obtained with the two methods, a strong linear correlation (Pearson’s r = 0.991) was revealed at the high-concentration range, and the lower concentrations were only detectable with the nanochip method (fig. S25B). ROC analysis showed that the nanochip analysis of sEV MMP14-E improved diagnosis of the patients with breast cancer compared to the standard ELISA (AUC, 0.934 versus 0.800; fig. S25C). Moreover, our technology was able to differentiate the individual groups of control and DCIS combined, IDC, and metastatic cases, whereas the standard ELISA only detected the metastatic group (fig. S25D). These comparative results further support the ability of our technology to enhance the diagnostic performance while reducing the sample consumption and assay time by a factor of >5, owing to its improved detection sensitivity. Last, to examine the potential of plasma-borne sEVs as liquid biopsy of solid tumors, breast tissues from four of the patients were assayed with standard hematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining (fig. S26). The expression of MMP14 was barely to weakly detectable in the tumor tissues from the two DCIS cases. For the other two patients with IDC and metastatic IDC, respectively, there was an apparent increase in staining intensity with weak to moderate MMP14 expression observed in the carcinoma compartment of the primary tumors. For comparison, we stained the tumor-adjacent normal breast tissue available from the same IDC patient, which displayed the absence of detectable MMP14 expression. These IHC assay results appear to support the correlation between the biomarker profiles of circulating sEVs and solid tumors and, thus, the potential applications of sEVs as a noninvasive surrogate biopsy of tumors. Collectively, the studies of clinical cohorts suggest the potential adaptability of our integrative molecular and functional sEV phenotyping technology to improve postdiagnosis surveillance of cancer status for early detection of tumor invasion or metastasis.
DISCUSSION
Nanoengineering of microdevices augments the sensitivity, accuracy, and speed of bioanalysis via constructing multiscale systems to combine the advantages of micro- and nanoscale flow dynamics and biochemical reactions (25, 53). This is usually accomplished by top-down nanofabrication using standard nanolithography or other sophisticated physical/chemical methods, as well as by bottom-up surface patterning with nanomaterials. These methods often produce 2D nanofeatures with respect to the dimensions of microscale sensors. Moreover, a key barrier toward practical nanomanufacturing arises from the limited scalability and standardization of these methods, which often involve expensive, sophisticated facilities, and labor-intensive fabrication procedures. Our pilot studies to address these limitations yielded a microchannel-based μCSA strategy for bottom-up 3D nanoengineering of microelements to improve the sensitivity of protein profiling of sEVs (22, 23). While enabling simple and high-quality 3D nanopatterning, this prototyping method uses manual processes, which can result in high cost, batch-to-batch variation in yield, and limited compatibility with mass production.
Here, we expanded the CSA-based multiscale engineering strategy by developing a general, high-resolution colloidal inkjet printing method using an industrial-grade material printer. Inkjet printing is cost-effective, flexible, and scalable and, thus, has been extensively developed for scientific and industrial applications (27, 54, 55), including surface patterning of nanomaterials (28-31). However, these techniques have three major limitations that need to be overcome for our applications. First, they are often limited to low-resolution printing of 2D colloidal patterns with a thickness of a few micrometers. Increasing ink concentration and jetting droplet volume can print thicker materials. However, concentrated particles can easily clog small inkjet nozzles required for high-resolution printing, and the use of large jetting droplets also limits the printing resolution. Second, largely uncontrolled solvent evaporation results in uneven distribution of nanoparticles during CSA and even broken, irregular patterns, making it challenging to print large-scale, continuous features. Last, to better control the geometry and quality of printed patterns, current techniques rely on delicately tuning surface modifications and ink composition to adjust the interactions between surface wetting and evaporation-driven CSA (30, 31). This strategy suffers from difficulties in controlling interfacial interactions during drying and limits the applications to biosensing, which often demand variable surface chemistry. To overcome these limitations, our method uses a multilayer “stacked coins” printing strategy that enables precise printing of complex 3D nanostructured patterns on plain glass surface with a ~20-μm geometric resolution. Such resolution is sufficient to print most existing micromixing architectures in microfluidic devices (56) to leverage biosensing performance. In addition, its ability to directly print on unmodified glass surfaces simplifies device manufacturing and broadens the adaptability to biosensing applications, which often demand variable surface chemistries. Compared to our prior manual μCSA method, this printing-based process using a commercial-grade printer substantially enhanced the production scale, success rate, and robustness of device fabrication. This aids our efforts toward developing industry-compatible manufacturing of 3D nanoengineered bioassay devices.
Building on the advance in chip fabrication, our study aimed to develop new bioanalytical capabilities to facilitate the progress of EV biology and clinical biomarker development. In contrast to existing microfluidic technologies that focus solely on molecular characterization of EVs, here we report a 3D nanoengineered lab-on-a-chip system that integrates an ultrasensitive proteolytic activity assay with quantitative protein immunoassays to define the biofunctional signatures of circulating EVs associated with tumor invasion and metastasis. Using different cell lines, mouse models, and clinical samples, we demonstrated that compared to the protein immunoassay, the activity assay augments the sensitivity for detection of sEV MMP14, enhancing its performance for cancer diagnosis and monitoring. Small sample consumption is another practical advantage of our EV-CLUE system. Mouse models are indispensable tools for cancer research and drug development. Current EV research involving longitudinal studies of mouse models often requires sacrificing a group of mice at each time point to collect sufficient sample quantity for standard bioassays. This approach is expensive, and interindividual heterogeneity can confound data analysis or lead to misleading observations. Our EV-CLUE system enables periodical measurements of small volumes of blood collected minimally invasively from individual mice to monitor tumor growth and metastasis in vivo. As a specific example, we demonstrated the blood test-based surveillance of human cancer cell line xenograft growing in each mouse through examining both total abundance and the MMP14+ subtype of human-derived sEVs in mouse plasma. With a spontaneous mouse metastasis model, we showed that our method enabled single-mouse longitudinal sEV analysis to detect large interindividual heterogeneity and to capture distinct trajectories of tumor development in individuals that could otherwise be masked by the ensemble measurement of different populations. Overall, these studies support our technology as a valuable tool to benefit animal studies of human diseases, especially for challenging or expensive patient-derived xenografts (PDXs) and transgenic models.
Enabled by the technical advances, this work presents a systematic assessment of clinical value of sEV MMP14 for tumor surveillance, using breast cancer as the disease model. EV-mediated transport of proteases is a newly discovered mechanism underlying tumor invasion and metastasis (44). Increasing evidence has suggested that EVs are a major route for cellular secretion of membrane-type metalloproteases, predominantly MMP14, into the extracellular space, which mediates diverse proteolytic activities of cells (18, 19, 43, 44). It was shown that knockdown of MMP14 expression, inhibition of MMP14 proteolytic activity, or inhibition of exosome biogenesis resulted in largely decreased activity of cancer cells to degrade ECM (42, 57). Consistently, we detected considerable expression of activated MMP14 on the purified EVs of breast cancer cell lines, which agrees with the previous observation of EV-bound MMP14 activating pro-MMPs and degrading matrices (19, 42). Furthermore, MMP14 knockdown and HuR KO cells support the possible contributions of sEV MMP14 to cell invasion and its potential as a biomarker of tumor progression and metastasis.
The molecular mechanisms governing activation and EV-mediated trafficking of MMP14 remain largely unknown. This process is known to involve endocytic internalization of membrane MMPs, conversion into functionally active forms, recycling back to plasma membrane, packing into intraluminal vesicles, and secretion as EVs (19). Growing evidence has shown that the traffic of MMP14 is controlled by diverse regulatory pathways and molecular machineries that may be differently programmed in tumor cells to drive ECM degradation and cell invasion depending on their pathological conditions and oncogenic stimuli. For instance, the vesicular soluble N-ethylmaleimide–sensitive factor attachment protein receptor (v-SNARE) family is a key component in the core machinery controlling intracellular trafficking and membrane fusion. The late endosomal v-SNARE, vesicle-associated membrane protein-7, along with a subset of Rab guanosine triphosphatases (Rab27a and Rab5a), was found to form regulatory circuitries for spatially confined delivery of MMP14, which is crucial to the formation and function of invadopodia, membrane structures formed by invasive cells to protrude into ECM (58, 59). Providing key docking sites for multivesicular bodies, invadopodia enhances secretion of MMP14-enriched exosomes that not only supports invadopodia formation but also promotes pericellular proteolysis of ECM to drive tumor progression and metastasis (42). In addition to the exosome biogenesis-associated routes, another v-SNARE, VAMP-3, has been recognized as a regulator for specified sorting and delivery of endosomal MMP14 into shedding microvesicles at plasma membrane to support cancer cell invasion (60). VAMP3-mediated MMP14 exocytosis was found to be regulated by WD repeat and FYVE domain containing 2 (WDFY2), a protein involved in the early endocytic pathway (61). WDFY2 KO leads to enhanced secretion of MMP14+ vesicles and promoted ECM degradation and cell invasion. Differential dysregulation of these regulators, such as Rab5 overexpression and WDFY2 loss, is often implicated in human cancers, reinforcing the diverse and dynamic nature of the regulatory circuitries of MMP14 trafficking during oncogenic transformation. In our studies, it appeared that MMP14 was not a direct target of HuR; however, the KO of HuR down-regulated EV trafficking of MMP14. Such HuR-regulated EV transport of MMP14 might constitute a component of the HuR-dependent regulatory network that promotes tumor development and metastasis. The mechanism underlying the HuR-mediated regulation of EV MMP14 is beyond the scope of this work and is still under investigation. Our in vitro results resonate with the existing evidences that suggest that MMP14-enriched EVs serve as a functional indicator of tumor invasion and metastasis. This is further supported by our in vivo studies of mouse models of breast cancer metastasis.
Previous mechanistic studies have shown the biomedical significance of MMP14-mediated function of EVs, mostly using cell lines and animal models; however, clinical value remains largely undetermined. We assessed the feasibility of probing sEV MMP14 from clinical plasma samples for diagnosis and staging of breast cancer, the most prevalent malignancy and the leading cause of cancer-related death in women (6). The 5-year survival rate for women with localized breast cancer (stage 0 to IIa) is 99% and drops down to 85 and 27% for women with regional lymph node metastases (stage IIb and III) or distant metastases (stage IV), respectively (62). About 25% of cases are diagnosed with in situ breast cancer, of which ~81% are DCIS. Without timely diagnosis and treatment, 20 to 50% of these in situ cases progress to invasive breast cancer with distant metastases (63, 64). However, molecular mechanisms underlying early-stage progression of breast cancer remain elusive. Prior studies have suggested that the genetic alternations driving potential invasion may be present at early stage and showed that transcriptomic profiles were virtually indistinguishable among the distinct stages of progression (65, 66). Because there are no effective markers to predict the risk or detect progression to invasive lesions, it remains challenging to accurately stratify DCIS lesions to improve treatment and eliminate unnecessary overtreatments. Thus, more informative and robust markers are pressingly needed to improve clinical management of breast cancer.
Using two independent cohorts, we showed that the plasma concentration and proteolytic activity of MMP14+ sEVs can be used to distinguish the affected patient group (stage 0 to III) from the control group. Our activity assay yielded the best diagnostic performance, and sEV MMP14 expression and activity appeared to correlate with progressive disease stages, supporting the possibility for longitudinal surveillance of breast carcinomas. We also demonstrated the feasibility of these markers for differentiating noncancer controls or in situ carcinoma from invasive and locally advanced cases, which is the first major stratification of breast cancer types to guide clinical treatment. To further enhance the performance of our method for breast cancer diagnosis and stratification, discriminant analysis of the training cohort (n = 30) was performed to derive a discriminant function model, which was then tested for classifying the patients in the validation cohort (n = 70) without a known a priori disease state. Combining three sEV parameters tested (total concentration, MMP14-E, and MMP14-A), a high overall accuracy of 92.9% (95% CI, 84.1 to 97.6%) was obtained for classifying all four groups in the validation cohort. Overall, our clinical specimen analysis, combined with the cell line and mouse model studies, demonstrated the feasibility of the MMP14-targeted molecular and functional sEV phenotyping for predicting invasive potential, detecting early progression or metastasis with high sensitivity and specificity, and informing individualized treatment of breast cancer.
The current study represents a feasibility study to evaluate the potential clinical utilities of the EV-CLUE system based on sEV MMP14 in cancer diagnosis and monitoring. This technology may be limited in detecting precancerous and pre-EMT conditions, due to the low abundance of tumor-derived EVs available in blood and the limited diagnostic performance of single biomarkers. Our integrated and expandable multichannel device could be multiplexed to analyze multiple functional proteases implicated in tumor invasion and metastasis (44), such as tissue inhibitors of metalloproteinases, to develop sensitive and specific liquid biopsy-based biomarker panels. Blood derivatives (plasma and serum) have been the most commonly used specimen in the studies of EV biomarkers, due in part to their wide clinical availability (20). As the goal of our technology development is to enhance system robustness and clinical adaptability, current device was designed to analyze plasma rather than whole blood, which simplifies the chip design, fabrication, and operation procedures. We envision that our chip can be expanded for whole-blood analysis due to its inherent amenability to multifunctional integration and the availability of microfluidic modules for plasma extraction (67) or direct EV isolation (68) from whole blood. The widely used tail vein injection and orthotopic mammary fat pad models provide a relevant means to assess and validate our technology to monitor tumor invasion and metastasis, despite their biological/clinical limitations. More in-depth studies, such as mechanistic and biomarker studies focused on DCIS-derived local tumor progression, will benefit from other mouse models of breast cancer that could more closely recapitulate the underlying biology, including the intraductal injection and PDX models. Moving our technology toward clinical applications will require rigorous validation with much larger cohorts and longitudinal studies of the development of high-risk in situ breast carcinomas into invasive or metastatic lesions. The amenability to robust and scalable device manufacturing of the inkjet printing technique developed here would greatly facilitate such large-scale translational and clinical studies.
MATERIALS AND METHODS
Study design
This study sought to investigate EV-transported MMP14 as a potential biomarker for monitoring tumor progression and metastasis using a 3D nanopatterned EV-CLUE lab-on-a-chip system. We developed a high-resolution colloidal inkjet printing technique for robust and scalable 3D nanopatterning to augment the potential translatability of our technology. The EV-CLUE chip was designed to integrate the EV ELISA and MMP14 activity assays in parallel for multiparametric analysis of the total concentration and MMP14 expression and activity of EVs in blood plasma. The nanochip-based assays were assessed and optimized to measure in vitro cell invasiveness using cancer cell lines and the isogenic breast and pancreatic cancer cell clones created by CRISPR-Cas9 editing. With the experimental and spontaneous mouse models of breast cancer metastasis, the EV-CLUE chip was validated for longitudinal monitoring of in vivo tumor growth and metastasis in single mice. For the animal studies, on-chip analyses were blinded to the independently prepared plasma samples. Last, we assessed the feasibility of our technology and the sEV MMP14 markers for clinical detection and staging of breast cancer using plasma samples from two independent training (n = 30) and validation cohorts (n = 70) composed of age-matched cancer-free controls and patients with DCIS, nonmetastatic IDC, or locally metastatic breast cancer. The sample size of the small training cohort was computed to meet the desired statistic errors (type I, 0.05; type II, 0.2) with an AUC of 0.8 and a control/patient ratio of 0.5, as described before (23). A machine learning–based diagnostic was combined with the chip-based functional EV analysis to enhance the clinical sensitivity and specificity for patient classification. Furthermore, extensive comparative and correlation studies with standard analytical methods were performed to validate the EV-CLUE measurements. Three technical replicates were conducted for all measurements.
Statistical analysis
Mean, SD, SEM, and LOD were calculated with standard formulas. Deming linear fitting was performed at the 95% confidence level to determine the Pearson correlation coefficient between different variables. To determine whether parametric or nonparametric tests will be used, we conducted normality test and found that the assumption of normality cannot be rejected for the cell line and mouse model data but is rejected for the human studies, at P < 0.05. Hence, for cell line experiments, two-tailed Student’s t test was performed for two-group comparison and one-way ANOVA with post hoc Tukey’s pairwise multiple comparisons test for multigroup comparison. For mouse models, longitudinal tumor growth was conducted with one-way repeated measures ANOVA with post hoc Tukey’s test. The mouse groups with or without lung metastasis were assessed by two-way ANOVA followed by Tukey’s test. For human specimen, group difference was assessed using nonparametric, two-tailed Mann-Whitney U test for two groups or Kruskal-Wallis one-way ANOVA with Dunn’s pairwise multiple comparisons test for multiple groups. An LDA-based ROC analysis was adopted to evaluate the diagnostic metrics of biomarkers. The training set data were first processed by LDA to classify the subjects into the control and cancer groups, and the resultant discriminant function model was used for binary classification of the validation cohort without a known a priori disease state. The predicted probabilities yielded for both training and validation cohorts were used to conduct ROC curves. Optimal cutoff points were selected using the maximum Youden’s index to determine the sensitivity, specificity, and accuracy of disease diagnosis. For multigroup classification, quadratic discriminant analysis was conducted as the equality test of within-group covariance matrices failed. The training cohort was first analyzed to generate the discriminant function model, which was used to classify the patients in the validation cohort; 95% CIs for AUC were determined using the DeLong method and exact CIs for sensitivity, specificity, and accuracy were calculated based on the binomial distribution. All statistical analyses were performed at a 95% confidence level using Excel 2018, OriginPro 2019, and GraphPad Prism 8.
Supplementary Material
Fig. S1. Investigation of surface wettability and ink composition for colloidal inkjet printing.
Fig. S2. Optimization of drop spacing for the stacked coins printing.
Fig. S3. High-quality colloidal inkjet printing.
Fig. S4. NTA plots of EVs isolated from breast cancer cell lines and patient plasma by UC.
Fig. S5. Specificity of sEV immunoisolation with the EV-CLUE chips.
Fig. S6. Optimization of capture antibodies for immunodetection of sEV-MMP14.
Fig. S7. Verification of MMP14 expression on EVs derived from breast cancer cells.
Fig. S8. Optimization of the sEV MMP14 activity assay.
Fig. S9. Knockdown of MMP14 reduces MDA-MB-231 cell invasion.
Fig. S10. Detection of the invasiveness of pancreatic cancer cells.
Fig. S11. Validation of antibodies for sEV analysis in the mouse models.
Fig. S12. Growth of subcutaneously xenografted tumors in 16 mice.
Fig. S13. Time-lapse, multiparametric measurements of circulating sEVs in 16 mice using the EV-CLUE technology.
Fig. S14. Box-whiskers charts with overlapping data points of time-lapse analysis of circulating sEVs in individual mice.
Fig. S15. Comparison of MMP14 expression and activity of plasma sEVs in the mice developing primary tumors with or without lung metastasis.
Fig. S16. Scatterplots of the SUM signatures for detecting breast cancer from the control.
Fig. S17. Detection of the combined group of invasive and metastatic IDC from the control and DCIS groups in the training cohort.
Fig. S18. Confusion matrices from discriminant classification of the patient cohorts.
Fig. S19. Correlation circle derived from discriminant analysis of the training cohort.
Fig. S20. Correlation between the sEV MMP14 expression and activity measured for all samples of the training and validation cohorts.
Fig. S21. Detection of the combined group of advanced cases from the control and DCIS groups in the validation cohort.
Fig. S22. NTA plots of EVs isolated from patient plasma.
Fig. S23. Correlation between the measurements of EVs in patient plasma with the EV-CLUE chip and NTA.
Fig. S24. Characterization of UC-purified EVs from the sample set used in fig. S22.
Fig. S25. Comparison of the EV-CLUE nanochip and standard microplate ELISA for analysis of breast cancer samples.
Fig. S26. Representative images of H&E and IHC staining of MMP14 in patient-matched primary tumor tissues.
Table S1. Antibodies and ELISA kits used in this research.
Table S2. Summary of two cohorts of women controls and patients.
Table S3. Statistical analyses of diagnostic performance of sEV markers for the training and validation cohorts.
Data file S2. Original data for the on-chip measurements of patient samples in Figs. 6 and 7.
Acknowledgments:
We thank the microfabrication core facility at the KU COBRE Center for Molecular Analysis of Disease Pathways (NIH P20GM103638) for device fabrication and A. K. Godwin and R. Madan at KUMC and KU Cancer Center’s Biospecimen Repository Core Facility funded in part by NCI P30 CA168524 for providing deidentified clinical samples and histological analysis. We thank D. Dixon for helpful discussions and suggestions.
Funding: This study was supported in part by NIH grants R21 CA207816, R21 EB024101, and R33 CA214333 (to Y. Zeng); R01 CA191785 (to L.X.); and R01 CA243445 (to Y. Zeng and L.X.) and Department of Defense Breast Cancer Research Program Breakthrough Level II grant BC151845 (to L.X.).
Footnotes
Competing interests: P.Z. and Y. Zeng are co-inventors on a U.S. provisional patent application based on this work (no. 62/984,541; title: Methods for inkjet printing objects for microfluidic devices). Y. Zeng holds equity interest in Clara Biotech Inc. and serves on its scientific advisory board. All other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Chaffer CL, Weinberg RA, A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, Brewer DS, Kallio HML, Högnäs G, Annala M, Kivinummi K, Goody V, Latimer C, O’Meara S, Dawson KJ, Isaacs W, Emmert-Buck MR, Nykter M, Foster C, Kote-Jarai Z, Easton D, Whitaker HC; ICGC Prostate Group, Neal DE, Cooper CS, Eeles RA, Visakorpi T, Campbell PJ, McDermott U, Wedge DC, Bova GS, The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagogo-Jack I, Shaw AT, Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol 15, 81–94 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Burrell RA, McGranahan N, Bartek J, Swanton C, The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Arnedos M, Vicier C, Loi S, Lefebvre C, Michiels S, Bonnefoi H, Andre F, Precision medicine for metastatic breast cancer—Limitations and solutions. Nat. Rev. Clin. Oncol 12, 693–704 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N, Analysis of circulating tumor DNA to monitor metastatic breast cancer. New Engl. J. Med 368, 1199–1209 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Siravegna G, Marsoni S, Siena S, Bardelli A, Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol 14, 531–548 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Rossi G, Ignatiadis M, Promises and pitfalls of using liquid biopsy for precision medicine. Cancer Res. 79, 2798–2804 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ, Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol 15, 617–638 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ, Reassessment of exosome composition. Cell 177, 428–445.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Syn N, Wang L, Sethi G, Thiery J-P, Goh B-C, Exosome-mediated metastasis: From epithelial–;mesenchymal transition to escape from immunosurveillance. Trends Pharmacol. Sci 37, 606–617 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL, Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542–1556 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Genschmer KR, Russell DW, Lal C, Szul T, Bratcher PE, Noerager BD, Abdul Roda M, Xu X, Rezonzew G, Viera L, Dobosh BS, Margaroli C, Abdalla TH, King RW, McNicholas CM, Wells JM, Dransfield MT, Tirouvanziam R, Gaggar A, Blalock JE, Activated PMN exosomes: Pathogenic entities causing matrix destruction and disease in the lung. Cell 176, 113–126.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, Garcia-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D, Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol 17, 816–826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D, Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med 18, 883–891 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Mark MT, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D, Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolo V, D’Ascenzo S, Violini S, Pompucci L, Festuccia C, Ginestra A, Vittorelli ML, Canevari S, Pavan A, Matrix-degrading proteinases are shed in membrane vesicles by ovarian cancer cells in vivo and in vitro. Clin. Exp. Metastasis 17, 131–140 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Taraboletti G, D’Ascenzo S, Borsotti P, Giavazzi R, Pavan A, Dolo V, Shedding of the matrix metalloproteinases MMP-2, MMP-9, and MT1-MMP as membrane vesicle-associated components by endothelial cells. Am. J. Pathol 160, 673–680 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakulinen J, Sankkila L, Sugiyama N, Lehti K, Keski-Oja J, Secretion of active membrane type 1 matrix metalloproteinase (MMP-14) into extracellular space in microvesicular exosomes. J. Cell. Biochem 105, 1211–1218 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach J-M, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Zavec AB, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MÁ, Brigstock DR, Brisson A, Broekman MLD, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DRF, Caruso S, Chamley LW, Chang Y-T, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FAW, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Chaudhuri AD, de Candia P, De Santana EF, De Wever O, del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TAP, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DCI, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG-E, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano S.-i., Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ, Kornek M, Kosanović MM, Kovács ÁF, Krämer-Albers E-M, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee M-S, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li ITS, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz ÁM, Lötvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SLN, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG, Meehan KL, Mertens I, Minciacchi VR, Möller A, Jørgensen MM, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-t Hoen ENM, Hooten NN, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Østergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BCH, Pink RC, Pisetsky DS, von Strandmann EP, Polakovicova I, Poon IKH, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KMA, Rughetti A, Russell AE, Saá P, Sahoo S, Salas-Huenuleo E, Sánchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schøyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PRM, Silva AM, Skowronek A, Snyder OL, Soares RP, Sódar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BWM, van der Grein SG, Van Deun J, van Herwijnen MJC, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ, Veit TD, Vella LJ, Velot É, Verweij FJ, Vestad B, Viñas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MHM, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yáñez-Mó M, Yin H, Yuana Y, Zappulli V, Zarubova J, Žėkas V, Zhang J.-y., Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK, Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H, New technologies for analysis of extracellular vesicles. Chem. Rev 118, 1917–1950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P, Zhou X, Zeng Y, Multiplexed immunophenotyping of circulating exosomes on nano-engineered ExoProfile chip towards early diagnosis of cancer. Chem. Sci 10, 5495–5504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK, Zeng Y, Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng 3, 438–451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao H, Zhou X, Zeng Y, Microfluidic exponential rolling circle amplification for sensitive microRNA detection directly from biological samples. Sensors Actuators B Chem. 279, 447–457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reátegui E, van der Vos KE, Lai CP, Zeinali M, Atai NA, Aldikacti B, Floyd FP Jr., Khankhel AH, Thapar V, Hochberg FH, Sequist LV, Nahed BV, Carter BS, Toner M, Balaj L, Ting DT, Breakefield XO, Stott SL, Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat. Commun 9, 175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H, Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med 18, 1835–1840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Li W, Qin M, Zhou H, Zhang X, Li F, Song Y, Printable functional chips based on nanoparticle assembly. Small 13, 1503339 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Ding H, Zhu C, Tian L, Liu C, Fu G, Shang L, Gu Z, Structural color patterns by electrohydrodynamic jet printed photonic crystals. ACS Appl. Mater. Interfaces 9, 11933–11941 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Park J, Moon J, Shin H, Wang D, Park M, Direct-write fabrication of colloidal photonic crystal microarrays by ink-jet printing. J. Colloid Interface Sci 298, 713–719 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Sowade E, Blaudeck T, Baumann RR, Inkjet printing of colloidal nanospheres: Engineering the evaporation-driven self-assembly process to form defined layer morphologies. Nanoscale Res. Lett 10, 362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L, Dong Z, Kuang M, Li Y, Li F, Jiang L, Song Y, Printing patterned fine 3D structures by manipulating the three phase contact line. Adv. Funct. Mater 25, 2237–2242 (2015). [Google Scholar]
- 32.Liu P, Bai L, Yang J, Gu H, Zhong Q, Xie Z, Gu Z, Self-assembled colloidal arrays for structural color. Nanoscale Adv. 1, 1672–1685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P, He M, Zeng Y, Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip 16, 3033–3042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C, Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U.S.A 113, E968–E977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itoh Y, Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 44-46, 207–223 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Shen Q, Lee ES, Pitts RL, Wu MH, Yuan SY, Tissue inhibitor of metalloproteinase-2 regulates matrix metalloproteinase-2–mediated endothelial barrier dysfunction and breast cancer cell transmigration through lung microvascular endothelial cells. Mol. Cancer Res 8, 939–951 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Köhrmann A, Kammerer U, Kapp M, Dietl J, Anacker J, Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: New findings and review of the literature. BMC Cancer 9, 188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devy L, Huang L, Naa L, Yanamandra N, Pieters H, Frans N, Chang E, Tao Q, Vanhove H, Lejeune A, van Gool R, Sexton DJ, Kuang G, Rank D, Hogan S, Pazmany C, Ma YL, Schoonbroodt S, Nixon AE, Ladner RC, Hoet R, Henderikx P, TenHoor C, Rabbani SA, Valentino ML, Wood CR, Dransfield DT, Selective inhibition of matrix metalloproteinase-14 blocks tumor growth, invasion, and angiogenesis. Cancer Res. 69, 1517–1526 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Ren F, Tang R, Zhang X, Madushi WM, Luo D, Dang Y, Li Z, Wei K, Chen G, Overexpression of MMP family members functions as prognostic biomarker for breast cancer patients: A systematic review and meta-analysis. PLOS ONE 10, e0135544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGowan PM, Duffy MJ, Matrix metalloproteinase expression and outcome in patients with breast cancer: Analysis of a published database. Ann. Oncol 19, 1566–1572 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Kruger S, Elmageed ZYA, Hawke DH, Wörner PM, Jansen DA, Abdel-Mageed AB, Alt DU, Izadpanah R, Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer 14, 44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM, Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 5, 1159–1168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gangoda L, Liem M, Ang C-S, Keerthikumar S, Adda CG, Parker BS, Mathivanan S, Proteomic profiling of exosomes secreted by breast cancer cells with varying metastatic potential. Proteomics 17, 1600370 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Sanderson RD, Bandari SK, Vlodavsky I, Proteases and glycosidases on the surface of exosomes: Newly discovered mechanisms for extracellular remodeling. Matrix Biol. 75-76, 160–169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B, Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int. J. Mol. Sci 14, 10015–10041 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrari R, Martin G, Tagit O, Guichard A, Cambi A, Voituriez R, Vassilopoulos S, Chavrier P, MT1-MMP directs force-producing proteolytic contacts that drive tumor cell invasion. Nat. Commun 10, 4886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kraning-Rush CM, Carey SP, Lampi MC, Reinhart-King CA, Microfabricated collagen tracks facilitate single cell metastatic invasion in 3D. Integr. Biol 5, 606–616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ota I, Li X-Y, Hu Y, Weiss SJ, Induction of a MT1-MMP and MT2-MMP-dependent basement membrane transmigration program in cancer cells by Snail1. Proc. Natl. Acad. Sci. U.S.A 106, 20318–20323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faustino-Rocha A, Oliveira PA, Pinho-Oliveira J, Teixeira-Guedes C, Soares-Maia R, da Costa RG, Colaço B, Pires MJ, Colaço J, Ferreira R, Ginja M, Estimation of rat mammary tumor volume using caliper and ultrasonography measurements. Lab. Anim 42, 217–224 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Yang KS, Im H, Hong S, Pergolini I, Del Castillo AF, Wang R, Clardy S, Huang C-H, Pille C, Ferrone S, Yang R, Castro CM, Lee H, Del Castillo CF, Weissleder R, Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci. Transl. Med 9, eaal3226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C, Zhao J, Tian F, Cai L, Zhang W, Feng Q, Chang J, Wan F, Yang Y, Dai B, Cong Y, Ding A, Sun J, Tan W, Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers. Nat. Biomed. Eng 3, 183–193 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Ko J, Bhagwat N, Yee SS, Ortiz N, Sahmoud A, Black T, Aiello NM, McKenzie L, O’Hara M, Redlinger C, Romeo J, Carpenter EL, Stanger BZ, Issadore D, Combining machine learning and nanofluidic technology to diagnose pancreatic cancer using exosomes. ACS Nano 11, 11182–11193 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH, Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nat. Nanotechnol 9, 969–980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daly R, Harrington TS, Martin GD, Hutchings IM, Inkjet printing for pharmaceutics—A review of research and manufacturing. Int. J. Pharm 494, 554–567 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Fukuda K, Someya T, Recent progress in the development of printed thin-film transistors and circuits with high-resolution printing technology. Adv. Mater 29, 1602736 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Cai G, Xue L, Zhang H, Lin J, A review on micromixers. Micromachines 8, 274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC, Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: Defining the stages of invadopodia formation and function. Cancer Res. 66, 3034–3043 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Steffen A, Le Dez G, Poincloux R, Recchi C, Nassoy P, Rottner K, Galli T, Chavrier P, MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr. Biol 18, 926–931 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Linder S, Scita G, RABGTPases in MT1-MMP trafficking and cell invasion: Physiology versus pathology. Small GTPases 6, 145–152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clancy JW, Sedgwick A, Rosse C, Muralidharan-Chari V, Raposo G, Method M, Chavrier P, D’Souza-Schorey C, Regulated delivery of molecular cargo to invasive tumour-derived microvesicles. Nat. Commun 6, 6919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sneeggen M, Pedersen NM, Campsteijn C, Haugsten EM, Stenmark H, Schink KO, WDFY2 restrains matrix metalloproteinase secretion and cell invasion by controlling VAMP3-dependent recycling. Nat. Commun 10, 2850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, SEER Cancer Statistics Review, 1975–2016. National Cancer Institute. Bethesda, MD, https://seer.cancer.gov/csr/1975_2016/, based on November 2018 SEER data submission, posted to the SEER web site, April 2019. [Google Scholar]
- 63.Independent UK Panel on Breast Cancer Screening, The benefits and harms of breast cancer screening: An independent review. Lancet 380, 1778–1786 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Sanders ME, Schuyler PA, Simpson JF, Page DL, Dupont WD, Continued observation of the natural history of low-grade ductal carcinoma in situ reaffirms proclivity for local recurrence even after more than 30 years of follow-up. Mod. Pathol 28, 662–669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma X-J, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou Y-X, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC, Gene expression profiles of human breast cancer progression. Proc. Natl. Acad. Sci. U.S.A 100, 5974–5979 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doebar SC, Sieuwerts AM, de Weerd V, Stoop H, Martens JWM, van Deurzen HM, Gene expression differences between ductal carcinoma in situ with and without progression to invasive breast cancer. Am. J. Pathol 187, 1648–1655 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Yeh E-C, Fu C-C, Hu L, Thakur R, Feng J, Lee LP, Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv 3, e1501645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu M, Ouyang Y, Wang Z, Zhang R, Huang P-H, Chen C, Li H, Li P, Quinn D, Dao M, Suresh S, Sadovsky Y, Huang TJ, Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. U.S.A 114, 10584–10589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang T, Zhang M, Dreher DD, Zeng Y, Ultrasensitive microfluidic solid-phase ELISA using an actuatable microwell-patterned PDMS chip. Lab Chip 13, 4190–4197 (2013). [DOI] [PubMed] [Google Scholar]
- 70.He M, Crow J, Roth M, Zeng Y, Godwin AK, Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 14, 3773–3780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Investigation of surface wettability and ink composition for colloidal inkjet printing.
Fig. S2. Optimization of drop spacing for the stacked coins printing.
Fig. S3. High-quality colloidal inkjet printing.
Fig. S4. NTA plots of EVs isolated from breast cancer cell lines and patient plasma by UC.
Fig. S5. Specificity of sEV immunoisolation with the EV-CLUE chips.
Fig. S6. Optimization of capture antibodies for immunodetection of sEV-MMP14.
Fig. S7. Verification of MMP14 expression on EVs derived from breast cancer cells.
Fig. S8. Optimization of the sEV MMP14 activity assay.
Fig. S9. Knockdown of MMP14 reduces MDA-MB-231 cell invasion.
Fig. S10. Detection of the invasiveness of pancreatic cancer cells.
Fig. S11. Validation of antibodies for sEV analysis in the mouse models.
Fig. S12. Growth of subcutaneously xenografted tumors in 16 mice.
Fig. S13. Time-lapse, multiparametric measurements of circulating sEVs in 16 mice using the EV-CLUE technology.
Fig. S14. Box-whiskers charts with overlapping data points of time-lapse analysis of circulating sEVs in individual mice.
Fig. S15. Comparison of MMP14 expression and activity of plasma sEVs in the mice developing primary tumors with or without lung metastasis.
Fig. S16. Scatterplots of the SUM signatures for detecting breast cancer from the control.
Fig. S17. Detection of the combined group of invasive and metastatic IDC from the control and DCIS groups in the training cohort.
Fig. S18. Confusion matrices from discriminant classification of the patient cohorts.
Fig. S19. Correlation circle derived from discriminant analysis of the training cohort.
Fig. S20. Correlation between the sEV MMP14 expression and activity measured for all samples of the training and validation cohorts.
Fig. S21. Detection of the combined group of advanced cases from the control and DCIS groups in the validation cohort.
Fig. S22. NTA plots of EVs isolated from patient plasma.
Fig. S23. Correlation between the measurements of EVs in patient plasma with the EV-CLUE chip and NTA.
Fig. S24. Characterization of UC-purified EVs from the sample set used in fig. S22.
Fig. S25. Comparison of the EV-CLUE nanochip and standard microplate ELISA for analysis of breast cancer samples.
Fig. S26. Representative images of H&E and IHC staining of MMP14 in patient-matched primary tumor tissues.
Table S1. Antibodies and ELISA kits used in this research.
Table S2. Summary of two cohorts of women controls and patients.
Table S3. Statistical analyses of diagnostic performance of sEV markers for the training and validation cohorts.
Data file S2. Original data for the on-chip measurements of patient samples in Figs. 6 and 7.