Abstract
The left ventricular (LV) ejection fraction (EF) is key to prognosis in dilated cardiomyopathy (DCM). Circulating microRNAs have emerged as reliable biomarkers for heart diseases, included DCM. Clinicians need improved tools for greater clarification of DCM EF categorization, to identify high-risk patients. Thus, we investigated whether microRNA profiles can categorize DCM patients based on their EF. 179-differentially expressed circulating microRNAs were screened in two groups: (1) non-idiopathic DCM; (2) idiopathic DCM. Then, 26 microRNAs were identified and validated in the plasma of ischemic-DCM (n = 60), idiopathic-DCM (n = 55) and healthy individuals (n = 44). We identified fourteen microRNAs associated with echocardiographic variables that differentiated idiopathic DCM according to the EF degree. A predictive model of a three-microRNA (miR-130b-3p, miR-150-5p and miR-210-3p) combined with clinical variables (left bundle branch block, left ventricle end-systolic dimension, lower systolic blood pressure and smoking habit) was obtained for idiopathic DCM with a severely reduced-EF. The receiver operating characteristic curve analysis supported the discriminative potential of the diagnosis. Bioinformatics analysis revealed that miR-150-5p and miR-210-3p target genes might interact with each other with a high connectivity degree. In conclusion, our results revealed a three-microRNA signature combined with clinical variables that highly discriminate idiopathic DCM categorization. This is a potential novel prognostic biomarker with high clinical value.
Subject terms: Biomarkers, Cardiology, Diseases, Medical research, Molecular medicine
Introduction
Dilated cardiomyopathy (DCM) is a heterogeneous heart disease characterized by the presence of left ventricular (LV) dilatation and systolic dysfunction in the absence of abnormal loading conditions or coronary artery disease sufficient to cause global systolic impairment1,2. This entity encompasses different phenotypes, clinical features and etiologies with a common final pathway. The prevalence of DCM is estimated at more than one in 250 individuals and it has increased by 27% in the last ten years. Hence, DCM is a health burden worldwide that imposes huge economic healthcare concerns due to high hospitalization, morbidity and mortality rates3.
The latest guidelines have established that the systolic ejection fraction (EF) discerns between heart failure (HF) categories4. Left ventricular ejection fraction (LVEF) has been strongly associated with adverse cardiac outcome in DCM5–7. Severely depressed EF, below 35%, has been shown in randomized clinical trials as a sudden death risk factor8. Nevertheless, the need for expert staff and inaccessible or excessive radiation imaging techniques impede clinical follow-up. Furthermore, highly time-consuming imaging techniques restrict access to the LV systolic function in the DCM population. Circulating levels of B-type natriuretic peptide or related molecules are very useful for clinical tailoring. However, they are not highly specific, as they are not good as systolic function markers4. The lack of solid biomarkers and the limitations of imaging cardiovascular tests to categorize LVEF in DCM urge the investigation of a new, reliable tool.
MicroRNAs (miRNAs) are evolutionary conserved non-coding RNA molecules of 19–25 nucleotides that play crucial roles in modulating the expression of most proteins at post-transcriptional level9. Their properties make them the most widely studied extracellular RNAs as diagnostic, prognostic and therapeutic markers in the cardiovascular field10–12. Several studies have described the relationship between DCM and miRNA profiles13–15. Therefore, a deeper knowledge of miRNA and the LVEF subcategories in the DCM population is needed to identify high-risk patients.
In the current study, we evaluated plasma miRNAs as potential biomarkers to discriminate severe from moderate reduced EF in a DCM population.
Results
Demographics and clinical manifestations in the patients
The subject characteristics are shown in Table 1. The mean age for control, idiopathic, and ischemic DCM patients was 39.1 ± 12.8, 67.3 ± 9.2, and 68.2 ± 8.4 years old. Most of the patients enrolled were men. As expected, LVEF and LVESD differed significantly between DCM patients and control subjects (p < 0.0001, for both comparisons). DCM group exhibited an increase in left atrium diameter (LA) and volume, sphericity index, and E/e’ ratio, compared with the control group. Regarding DCMMOD group, ischemic patients showed significantly reduced heart rate, LVEF and right ventricle diameter compared with idiopathic patients (p = 0.014, p = 0.020 and p = 0.024, respectively). As to the DCMSEV group, 10 ischemic patients had diabetes mellitus type 2, while only 4 idiopathic patients showed this comorbidity (p = 0.01). In both cases, sphericity index was significantly reduced in ischemic patients (p = 0.047 for DCMMOD group and p = 0.03 for DCMSEV group).
Table 1.
Baseline characteristics of of the study groups.
| Variables | DCMMOD | p | DCMSEV | p | Healthy control | ||
|---|---|---|---|---|---|---|---|
| Etiology | Idiopathic | Ischemic | Idiopathic | Ischemic | |||
| n | 40 | 46 | 15 | 14 | 44 | ||
| Demographics | |||||||
| Age (years) | 67.6 ± 9.4 | 68.0 ± 8.3 | NS | 66.5 ± 8.4 | 68.7 ± 8.4 | NS | 39.1 ± 12.8 |
| Sex (male, %) | 70.0 | 76.1 | NS | 60.0 | 92.9 | 0.045 | 52.3 |
| SBP (mm Hg) | 124.1 ± 21.0 | 125.0 ± 14.5 | NS | 113.8 ± 17.5 | 118.7 ± 11.3 | NS | 114.4 ± 8.6 |
| DBP (mm Hg) | 71.7 ± 6.8 | 72.2 ± 11.1 | NS | 73.1 ± 10.6 | 70.8 ± 9.8 | NS | 74.4 ± 8.1 |
| Heart rate (b.p.m) | 71.0 ± 13.3 | 65.1 ± 12.9 | 0.014 | 67.1 ± 5.4 | 68.0 ± 11.5 | NS | 63.7 ± 11.4 |
| LBBB (%) | 15 | 8.7 | NS | 33.3 | 14.3 | NS | 0 |
| Body mass index (kg/m2) | 29.4 ± 5.6 | 29.1 ± 4.4 | NS | 28.0 ± 5.3 | 27.5 ± 3.7 | NS | 24.0 ± 3.5 |
| Diabetes Mellitus (%) | 40 | 46.7 | NS | 26.7 | 71.4 | 0.019 | 0 |
| Smoking habit (%) | 40 | 47.8 | NS | 73 | 69.2 | NS | 0 |
| II-III NYHA functional class (%) | 12.5 | 10.9 | NS | 20 | 35.7 | NS | 0 |
| Medications | |||||||
| ACE I (%) | 42.5 | 41.3 | NS | 60 | 35.7 | NS | 0 |
| Antiplatelet (%) | 37.5 | 30.4 | NS | 26.7 | 42.9 | NS | 0 |
| ARA II (%) | 33.3 | 41.3 | NS | 55 | 71.4 | 0.047 | 0 |
| Beta-blocker (%) | 65 | 71.7 | NS | 100 | 100 | NS | 0 |
| Statin (%) | 22.5 | 32.6 | NS | 33.3 | 28.6 | NS | 29.5 |
| Echocardiography | |||||||
| LVEF (%) | 39.0 ± 4.5 | 36.7 ± 4.0 | 0.020 | 24.1 ± 4.4 | 25.7 ± 3.6 | NS | 66.2 ± 5.2 |
| LVEDD (mm) | 63.1 ± 4.1 | 63.3 ± 6.2 | NS | 60.5 ± 5.4 | 60.4 ± 5.8 | NS | 46.2 ± 4.9 |
| LVESD (mm) | 45.8 ± 14.0 | 46.1 ± 13.5 | NS | 54.6 ± 5.4 | 42.9 ± 18.6 | NS | 28.4 ± 5.8 |
| Right ventricle (mm) | 37 ± 8.5 | 33.4 ± 7.3 | 0.024 | 37.9 ± 11.7 | 34.9 ± 7.2 | NS | 27.2 ± 4.0 |
| MAPSE | 11 ± 2.3 | 10.9 ± 2.1 | NS | 1 ± 3.2 | 9.5 ± 2.0 | NS | 18.9 ± 2.2 |
| TAPSE | 20 ± 4.7 | 19.8 ± 4.7 | NS | 19. 2 ± 6.3 | 19.5 ± 3.6 | NS | 21.4 ± 3.3 |
| Aortic root (mm) | 33.9 ± 5.0 | 33.2 ± 4.8 | NS | 30. 3 ± 6.2 | 32.2 ± 3.5 | NS | – |
| E/e’ ratio | 15.2 ± 6.8 | 15.1 ± 6.5 | NS | 15.2 ± 8.1 | 16.9 ± 8.7 | NS | 7.5 ± 1.7 |
| Left Atrium (mm) | 46.7 ± 9.2 | 44.3 ± 7.7 | NS | 41.1 ± 8.2 | 45.1 ± 7.3 | NS | 35.8 ± 8.3 |
| Sphericity index | 0.8 ± 0.3 | 0.7 ± 0.1 | 0.047 | 0.9 ± 0.27 | 0.7 ± 0.1 | 0.032 | 0.5 ± 0.1 |
Data presented as mean ± SD for continuous variables and as percentage for categorical variables. NS, no significant. Abbreviations: ACE I, angiotensin-converting enzyme; ARA II, angiotensin II receptor antagonist; b.p.m, beats per minute; DBP, diastolic blood pressure; DCMMOD, dilated cardiomyopathy with ejection fraction ≥ 31–49%; DCMSEV, dilated cardiomyopathy with ejection fraction ≤ 30%; E/e’, ratio of mitral early diastolic flow velocity over tissue Doppler lateral mitral annular lengthening velocity; LBBB, left bundle branch block; LVEDD, left ventricle end-diastolic diameter; LVEF, left ventricle ejection fraction; LVESD, left ventricle end-systolic diameter; MAPSE, mitral annular plane systolic excursion; NYHA, New York Heart Association; SBP, systolic blood pressure; TAPSE, tricuspid annular plane systolic excursion.
Screening study
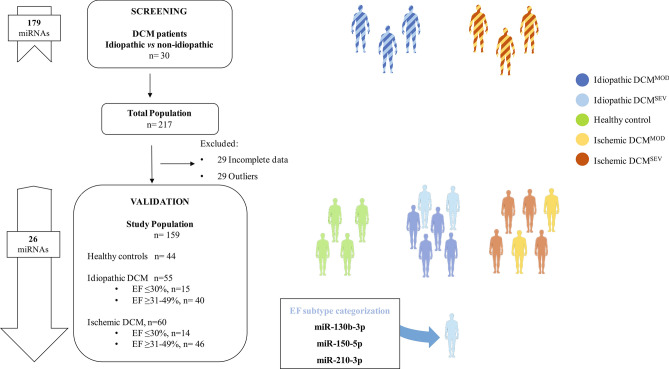
The study design is shown in Fig. 1. We first screened 179-differentially expressed circulating miRNAs in 30 idiopathic and non-idiopathic DCM age- and sex-matched patients (see Table S1 and Fig. S1 in Supplementary Material). In total, 26 miRNAs were significantly expressed according to the selection criteria at high levels and statistical significance (median Cq < 32 and detected in at least 80% of all samples and p < 0.05). The candidates selected for further analysis were let-7 g-5p, let-7a-5p, miR-1-3p, miR-16-5p, miR-16–2-3p, miR-19b-3p, miR-25-3p, miR-29a-3p, miR-30b-5p, miR-30e-3p, miR-130b-3p, miR-133a-3p, miR-133b, miR-142-3p, miR-145-5p, miR-150-5p, miR-192-5p, miR-199a-3p, miR-210-3p, miR-215-5p, miR-324-3p, miR-363-3p, miR-454-3p, miR-532-5p, miR-629-5p and miR-660-5p (Table 2).
Figure 1.
Flowchart of the study design strategy. This figure illustrates the experimental workflow of the study including screening, validation and EF status of DCM patients. Abbreviations: DCM, dilated cardiomyopathy; DCMMOD, dilated cardiomyopathy with ejection fraction ≥ 31–49%; DCMSEV, dilated cardiomyopathy with ejection fraction ≤ 30%; EF, ejection fraction.
Table 2.
Peripheral microRNA levels in the Idiopathic and Ischemic DCM vs. healthy control.
| microRNA | Healthy control | Idiopathic DCM | Ischemic DCM | |||||
|---|---|---|---|---|---|---|---|---|
| n | median (Q1-Q3) | n | median (Q1-Q3) | p value | n | median (Q1-Q3) | p value | |
| let-7a-5p | 44 | 6.543 (6.308–6.832) | 55 | 6.511 (6.308–7.207) | 0.6636 | 60 | 7.155 (6.507–7.626) | 0.0001 |
| let-7 g-5p | 44 | 6.268 (6.095–6.477) | 55 | 6.303 (6.095–6.939) | 0.3767 | 60 | 6.555 (6.147–7.034) | 0.0236 |
| miR-1-3p | 44 | 4.101 (3.878–4.460) | 55 | 4.238 (3.878–5.059) | 0.0611 | 60 | 4.491 (3.552–5.038) | 0.1142 |
| miR-16-5p | 44 | 7.634 (7.379–7.900) | 55 | 7.658 (7.379–8.304) | 0.0046 | 60 | 8.025 (7.714–8.300) | 0.0017 |
| miR-16–2-3p | 44 | 4.967 (4.706–5.281) | 55 | 7.035 (6.773–5.609) | 0.0043 | 60 | 5.363 (5.003–5.573) | 0.0026 |
| miR-19b-3p | 44 | 7.014 (6.773–7.230) | 55 | 6.542 (6.199–7.637) | 0.008 | 60 | 7.517 (7.103–7.751) | 0.0006 |
| miR-25-3p | 44 | 6.487 (6.199–6.751) | 55 | 6.230 (6.052–7.178) | 0.0011 | 60 | 6.917 (6.620–7.127) | 0.0017 |
| miR-29a-3p | 44 | 5.480 (5.297–5.643) | 55 | 4.951 (4.730–6.233) | 0.0001 | 60 | 5.951 (5.630–6.304) | 0.0001 |
| miR-30b-5p | 44 | 6.197 (6.052–6.431) | 55 | 4.913 (4.745–6.739) | 0.9142 | 60 | 6.525 (6.032–6.992) | 0.0229 |
| miR-30e-3p | 44 | 4.923 (4.745–5.165) | 55 | 5.072 (4.858–5.559) | 0.145 | 60 | 5.346 (4.793–5.722) | 0.0127 |
| miR-130b-3p | 44 | 5.058 (4.858–5.275) | 55 | 4.483 (4.167–5.849) | 0.0001 | 60 | 5.610 (5.056–5.904) | 0.0002 |
| miR-133a-3p | 44 | 4.451 (4.167–4.741) | 55 | 4.164 (3.843–5.412) | 0.0001 | 60 | 4.873 (4.273–5.439) | 0.0042 |
| miR-133b | 44 | 4.129 (3.843–4.437) | 55 | 6.950 (6.690–5.028) | 0.0019 | 60 | 4.491 (3.909–5.105) | 0.0026 |
| miR-142-3p | 44 | 6.932 (6.690–7.217) | 55 | 5.454 (5.279–7.588) | 0.9753 | 60 | 7.425 (7.007–8.022) | 0.0001 |
| miR-145-5p | 44 | 5.423 (5.279–5.629) | 55 | 5.504 (5.313–6.044) | 0.2254 | 60 | 5.867 (5.473–6.385) | 0.0001 |
| miR-150-5p | 44 | 5.520 (5.313–5.696) | 55 | 4.970 (4.665–6.133) | 0.1293 | 60 | 5.803 (5.377–6.053) | 0.0634 |
| miR-192-5p | 44 | 4.928 (4.665–5.152) | 55 | 6.231 (6.110–5.607) | 0.0708 | 60 | 5.293 (4.893–5.486) | 0.0075 |
| miR-199a-3p | 44 | 6.218 (6.110–6.390) | 55 | 4.512 (4.314–6.967) | 0.0007 | 60 | 6.628 (6.018–7.044) | 0.0047 |
| miR-210-3p | 44 | 4.514 (4.314–4.705) | 55 | 4.809 (4.544–5.317) | 0.0001 | 60 | 5.039 (4.683–5.478) | 0.0001 |
| miR-215-3p | 44 | 4.770 (4.544–4.997) | 55 | 5.536 (5.299–5.397) | 0.0479 | 60 | 5.101 (4.720–5.397) | 0.0098 |
| miR-324-3p | 44 | 4.892 (4.728–5.139) | 55 | 5.318 (4.969–5.554) | 0.0079 | 60 | 5.397 (4.957–5.689) | 0.0002 |
| miR-363-3p | 44 | 5.260 (4.969–5.555) | 55 | 3.845 (3.281–5.946) | 0.002 | 60 | 5.750 (5.389–5.893) | 0.0006 |
| miR-454-3p | 44 | 3.759 (3.281–4.098) | 55 | 4.704 (4.454–4.437) | 0.7829 | 60 | 4.130 (3.298–4.699) | 0.0061 |
| miR-532-5p | 44 | 4.668 (4.454–4.854) | 55 | 5.535 (5.297–5.344) | 0.083 | 60 | 5.207 (4.782–5.398) | 0.0002 |
| miR-629-5p | 44 | 4.332 (4.104–4.680) | 55 | 5.281 (4.854–5.72) | < 0.0001 | 60 | 5.182 (4.769–5.813) | < 0.0001 |
| miR-660-5p | 44 | 4.936 (4.730–5.162) | 55 | 5.436 (5.104–5.639) | 0.0001 | 60 | 5.454 (5.092–5.614) | 0.0001 |
Data presented as median (Q1-Q3). Samples size is indicated. Abbreviations: DCM. dilated cardiomyopathy.
Validation study and association with a population that has a reduced ejection fraction and DCM
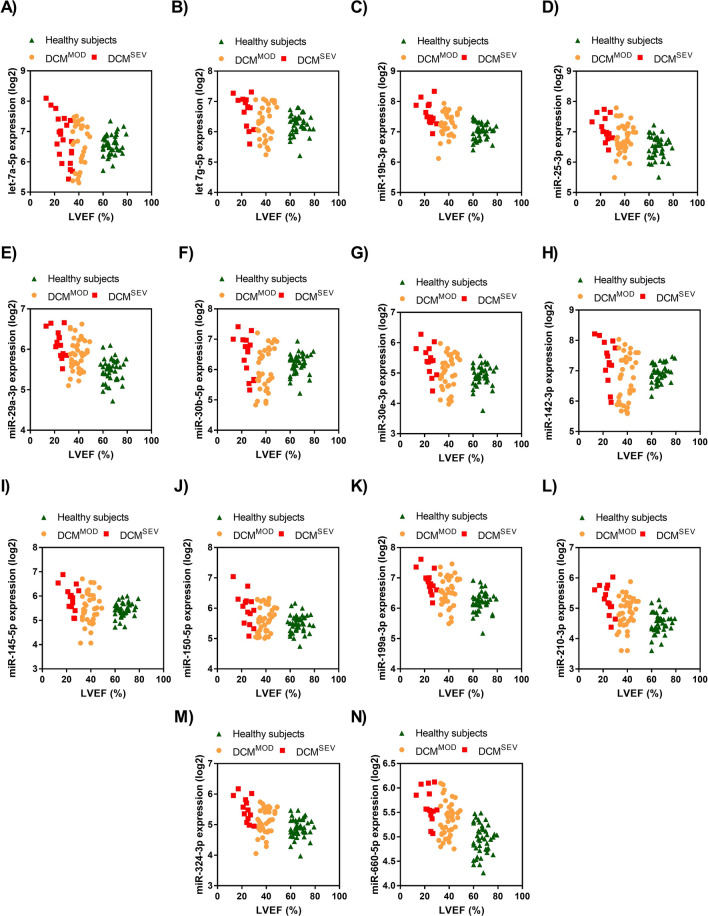
The ischemic and idiopathic DCM populations are presented as DCMMOD and DCMSEV groups based on their LVEF, ≤ 30% and > 31–49%, respectively. We investigated the potential value of the miRNA candidates to discriminate between the DCMSEV vs. the DCMMOD population and their association with echocardiographic parameters (Table 3 and Table S2). Only the idiopathic DCMSEV group showed an association between LVEF and fourteen miRNA candidates (Table 4). These miRNAs were let-7a-5p, let 7 g-5p, miR-19b-3p, miR-25-3p, miR-29a-3p, miR-30b-5p, miR-30e-3p, miR-142-3p, miR-145-5p, miR-150-5p, miR-199a-3p, miR-210-3p, , miR-324-3p and miR-660-5p. No association was found in the idiopathic DCMMOD cohort with LVEF (Table 4). Moreover, the correlation between other echocardiographic variables and individual miRNAs was also identified. All these miRNAs significantly correlated with LVEF in DCMSEV patients. In addition, these miRNAs in the DCMMOD group were significantly correlated with LV end-systolic dimension (LVESD) and the left atrium dimension (LA).
Table 3.
Peripheral microRNA levels for LVEF categorization in idiopathic and ischemic DCM.
| microRNA | Idiopathic DCM | Ischemic DCM | ||||
|---|---|---|---|---|---|---|
| DCMSEV | DCMMOD | p value | DCMSEV | DCMMOD | p value | |
| median (Q1-Q3) | median (Q1-Q3) | median (Q1-Q3) | median (Q1-Q3) | |||
| let-7a-5p | 6.460 (5.732–7.080) | 6.909 (6.059–7.226) | 0.009 | 7.206 (6.547–7.594) | 7.125 (6.342–7.707) | 0.893 |
| let-7 g-5p | 6.272 (5.894–6.482) | 6.668 (6.000–7.010) | 0.003 | 6.712 (5.940–6.993) | 6.548 (6.138–7.054) | 0.997 |
| miR-1-3p | 4.772 (4.493–5.241) | 4.398 (2.779–4.950) | 0.003 | 4.732 (2.779–5.128) | 4.432 (3.242–4.989) | 0.995 |
| miR-16-5p | 8.127 (7.898–8.513) | 7.900 (7.619–8.221) | 0.552 | 7.930 (7.425–8.251) | 8.040 (7.681–8.301) | 0.997 |
| miR-16–2-3p | 5.386 (5.132–5.694) | 5.233 (5.016–5.559) | 0.651 | 5.338 (4.857–5.550) | 5.381 (4.994–5.588) | 0.999 |
| miR-19b-3p | 7.218 (6.949–7.581) | 7.449 (7.112–7.637) | 0.044 | 7.486 (6.814–7.671) | 7.539 (7.100–7.775) | 0.997 |
| miR-25-3p | 6.697 (6.293–7.524) | 6.938 (6.638–7.175) | 0.007 | 6.858 (6.320–7.047) | 6.919 (6.577–7.153) | 0.996 |
| miR-29a-3p | 5.746 (5.576–6.095) | 5.973 (5.718–6.258) | 0.0326 | 6.077 (5.176–6.289) | 5.949 (5.635–6.329) | 0.825 |
| miR-30b-5p | 5.880 (5.498–6.397) | 6.488 (5.634–6.862) | 0.011 | 6.684 (5.846–6.923) | 6.468 (5.982–7.027) | 0.946 |
| miR-30e-3p | 4.926 (4.563–5.320) | 5.371 (4.697–5.622) | 0.001 | 5.507 (4.551–5.699) | 5.186 (4.779–5.781) | 0.952 |
| miR-130b-3p | 5.653 (5.371–5.923) | 5.493 (5.236–5.846) | 0.956 | 5.625 (4.955–5.872) | 5.542 (5.048–5.920) | 0.834 |
| miR-133a-3p | 5.197 (4.906–5.412) | 4.895 (4.429–5.433) | 0.365 | 4.890 (3.212–5.439) | 4.851 (4.269–5.429) | 0.968 |
| miR-133b | 4.784 (4.459–5.196) | 4.434 (3.899–4.994) | 0.193 | 4.689 (3.204–5.061) | 4.485 (3.890–5.111) | 0.760 |
| miR-142-3p | 6.770 (5.961–7.348) | 7.233 (6.286–7.600) | 0.0005 | 7.749 (6.859–8.046) | 7.384 (6.741–8.023) | 0.340 |
| miR-145-5p | 5.389 (5.113–5.758) | 5.654 (5.089–6.126) | 0.005 | 6.005 (5.466–6.175) | 5.761 (5.221–6.408) | 0.998 |
| miR-150-5p | 5.539 (5.143–6.016) | 5.790 (5.418–6.156) | 0.004 | 5.446 (5.031–5.806) | 5.852 (5.451–6.066) | 0.916 |
| miR-192-5p | 5.192 (5.107–5.614) | 5.119 (4.857–5.579) | 0.438 | 5.179 (4.712–5.523) | 5.294 (4.827–5.508) | 0.757 |
| miR-199a-3p | 6.527 (6.240–6.733) | 6.715 (6.367–7.000) | 0.036 | 6.906 (5.961–7.026) | 6.544 (6.104–7.081) | 0.962 |
| miR-210-3p | 4.781 (4.340–5.179) | 5.058 (4.655–5.364) | 0.003 | 5.023 (4.382–5.479) | 5.040 (4.679–5.479) | 0.797 |
| miR-215-3p | 5.158 (4.784–5.616) | 4.993 (4.662–5.389) | 0.773 | 5.084 (4.449–5.356) | 5.110 (4.670–5.401) | 0.874 |
| miR-296a-3p | 6.099 (5.849–6.409) | 5.883 (5.590–6.168) | 0.032 | 6.077 (5.176–6.289) | 5.949 (5.635–6.329) | 0.825 |
| miR-324-3p | 5.040 (4.827–5.385) | 5.360 (4.960–5.570) | 0.001 | 5.454 (4.695–5.689) | 5.371 (4.945–5.697) | 0.858 |
| miR-363-3p | 5.698 (5.512–6.065) | 5.576 (5.255–5.928) | 0.778 | 5.757 (5.094–5.908) | 5.712 (5.349–5.896) | 0.999 |
| miR-454-3p | 3.069 (3.069–4.437) | 3.069 (3.069–4.438) | 0.926 | 4.442 (3.654–4.635) | 4.051 (3.069–4.758) | 0.418 |
| miR-532-5p | 5.152 (4.748–5.622) | 4.917 (4.459–5.315) | 0.285 | 5.207 (4.468–5.407) | 5.192 (4.741–5.403) | 0.985 |
| miR-629-5p | 4.902 (4.651–5.566) | 5.538 (5.112–5.903) | 0.123 | 5.465 (5.306–5.918) | 5.852 (5.451–5.065) | 0.979 |
| miR-660-5p | 5.184 (5.049–5.899) | 5.465 (5.196–5.570) | 0.034 | 5.454 (4.608–5.697) | 5.4504 (5.050–5.624) | 0.992 |
Data presented as median (Q1-Q3). Samples size is indicated. Abbreviations: DCM. dilated cardiomyopathy; DCMMOD, dilated cardiomyopathy with ejection fraction ≥ 31–49%; DCMSEV, dilated cardiomyopathy with ejection fraction ≤ 30%.
Table 4.
Correlation between the echocardiographic parameter and individual microRNAs for LVEF categorization in idiopathic DCM.
| microRNA | LVEF (%) | LVESD (mm) | Left atrial dimension (mm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DCMSEV | DCMMOD | DCMSEV | DCMMOD | DCMSEV | DCMMOD | |||||||
| Pearson r | p | Pearson r | p | Pearson r | p | Pearson r | p | Pearson r | p | Pearson r | p | |
| Let-7a-5p | − 0.677 | 0.006 | 0.140 | 0.388 | − 0.103 | 0.726 | − 0.489 | 0.002 | 0.277 | 0.318 | − 0.362 | 0.022 |
| Let-7 g-5p | − 0.609 | 0.016 | 0.181 | 0.269 | − 0.286 | 0.321 | − 0.408 | 0.011 | 0.368 | 0.269 | − 0.532 | < 0.001 |
| miR-1-3p | − 0.190 | 0.498 | 0.204 | 0.206 | 0.046 | 0.875 | − 0.218 | 0.183 | 0.242 | 0.385 | − 0.433 | 0.005 |
| miR-16-5p | − 0.190 | 0.498 | 0.204 | 0.206 | 0.046 | 0.875 | − 0.218 | 0.183 | 0.242 | 0.385 | − 0.433 | 0.005 |
| miR-16–2-3p | − 0.438 | 0.102 | 0.130 | 0.424 | 0.044 | 0.882 | − 0.559 | < 0.001 | 0.219 | 0.432 | − 0.262 | 0.102 |
| miR-19b-3p | − 0.527 | 0.043 | 0.193 | 0.233 | 0.111 | 0.705 | − 0.444 | 0.005 | 0.276 | 0.319 | − 0.306 | 0.054 |
| miR-25-3p | − 0.538 | 0.039 | 0.106 | 0.515 | 0.077 | 0.795 | − 0.544 | < 0.001 | 0.155 | 0.580 | − 0.246 | 0.127 |
| miR-29a-3p | − 0.641 | 0.010 | 0.168 | 0.300 | 0.180 | 0.538 | − 0.261 | 0.109 | 0.337 | 0.219 | − 0.336 | 0.034 |
| miR-30b-5p | − 0.603 | 0.017 | 0.179 | 0.269 | − 0.312 | 0.278 | − 0.376 | 0.018 | 0.313 | 0.256 | − 0.436 | 0.005 |
| miR-30e-3p | − 0.629 | 0.012 | 0.234 | 0.145 | − 0.130 | 0.658 | − 0.335 | 0.037 | 0.385 | 0.157 | − 0.347 | 0.028 |
| miR-130b-3p | − 0.394 | 0.146 | 0.094 | 0.564 | 0.109 | 0.710 | − 0.296 | 0.067 | 0.554 | 0.032 | − 0.269 | 0.093 |
| miR-133a-3p | − 0.455 | 0.088 | 0.092 | 0.574 | 0.160 | 0.584 | − 0.359 | 0.025 | 0.208 | 0.456 | − 0.348 | 0.028 |
| miR-133b | − 0.361 | 0.186 | 0.179 | 0.268 | 0.028 | 0.924 | − 0.194 | 0.236 | 0.140 | 0.619 | − 0.444 | 0.004 |
| miR-142-3p | − 0.569 | 0.027 | 0.210 | 0.193 | − 0.080 | 0.787 | − 0.417 | 0.008 | 0.359 | 0.189 | − 0.349 | 0.027 |
| miR-145-5p | − 0.614 | 0.015 | 0.130 | 0.423 | 0.109 | 0.711 | − 0.425 | 0.007 | 0.324 | 0.239 | − 0.382 | 0.015 |
| miR-150-5p | − 0.540 | 0.038 | 0.178 | 0.272 | − 0.148 | 0.614 | − 0.357 | 0.025 | 0.327 | 0.234 | − 0.419 | 0.007 |
| miR-192-5p | − 0.464 | 0.082 | 0.237 | 0.141 | 0.152 | 0.603 | − 0.536 | < 0.001 | 0.191 | 0.495 | − 0.252 | 0.117 |
| miR-199a-3p | − 0.692 | 0.004 | 0.184 | 0.257 | 0.065 | 0.826 | − 0.198 | 0.228 | 0.396 | 0.144 | − 0.401 | 0.010 |
| miR-210-3p | − 0.555 | 0.032 | 0.188 | 0.244 | 0.056 | 0.848 | − 0.405 | 0.011 | 0.285 | 0.304 | − 0.287 | 0.073 |
| miR-215-3p | − 0.425 | 0.114 | 0.201 | 0.213 | − 0.003 | 0.993 | − 0.583 | 0.000 | 0.109 | 0.699 | − 0.301 | 0.059 |
| miR-324-3p | − 0.667 | 0.007 | 0.183 | 0.257 | − 0.053 | 0.857 | − 0.448 | 0.004 | 0.270 | 0.330 | − 0.293 | 0.066 |
| miR-363-3p | − 0.463 | 0.082 | 0.075 | 0.644 | 0.070 | 0.811 | − 0.582 | < 0.001 | 0.125 | 0.657 | − 0.189 | 0.244 |
| miR-454-3p | − 0.692 | 0.004 | 0.085 | 0.603 | − 0.290 | 0.315 | − 0.293 | 0.070 | 0.250 | 0.368 | − 0.416 | 0.008 |
| miR-532-5p | − 0.480 | 0.070 | 0.283 | 0.077 | − 0.058 | 0.844 | − 0.487 | 0.002 | 0.217 | 0.438 | − 0.308 | 0.053 |
| miR-629-5p | − 0.383 | 0.159 | − 0.057 | 0.729 | 0.017 | 0.954 | − 0.419 | 0.008 | 0.151 | 0.592 | − 0.057 | 0.728 |
| miR-660-5p | − 0.500 | 0.058 | 0.102 | 0.530 | 0.313 | 0.276 | − 0.538 | < 0.001 | 0.231 | 0.407 | − 0.118 | 0.467 |
Abbreviations: DCMMOD, dilated cardiomyopathy with ejection fraction ≥ 31–49%; DCMSEV, dilated cardiomyopathy with ejection fraction ≤ 30%; LVEF, left ventricle ejection fraction; LVESD, left ventricle end-systolic diameter. Coefficient significant at p < 0.05.
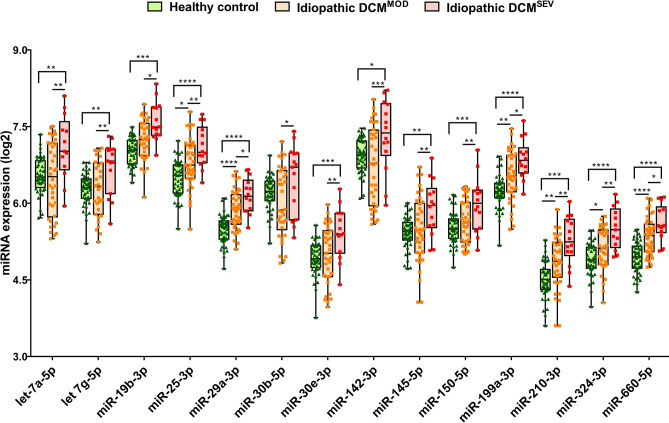
A graphical illustration of the potential miRNA expression levels to discriminate between DCMSEV and DCMMOD in the idiopathic group is represented in Figs. 2 and 3. In all cases, the average miRNA levels were significantly higher in the idiopathic DCM group than in healthy subjects. Moreover, we found a significant increase in these circulating miRNA levels in plasma from DCMSEV compared with DCMMOD idiopathic patients.
Figure 2.
Boxplots of miRNA expression levels, normalized to cel-miR-39-3p, in healthy subjects, idiopathic DCMMOD and DCMSEV. The analysis was carried out using qPCR. Data are presented in log2. Data represent the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: DCMMOD, dilated cardiomyopathy with ejection fraction ≥ 31–49%; DCMSEV, dilated cardiomyopathy with ejection fraction ≤ 30%.
Figure 3.
Correlation between the LVEF parameter and individual microRNAs for LVEF categorization in idiopathic DCM. (A-N) LVEF was independently negatively related with let-7a-5p (A), let 7 g-5p (B), miR-19b-3p (C), miR-25-3p (D), miR-29a-3p (E), miR-30b-5p (F), miR-30e-3p (G), miR-142-3p (H), miR-145-5p (I), miR-150-5p (J), miR-199a-3p (K), miR-210-3p (L), , miR-324-3p (M) and miR-660-5p (N) levels. Abbreviations: DCMMOD, dilated cardiomyopathy with ejection fraction ≥ 31–49%; DCMSEV, dilated cardiomyopathy with ejection fraction ≤ 30%.
Combining miRNAs for DCM detection and subtype categorization
The ability of these miRNAs to discriminate between DCM with moderate and severe EF impairment was assessed using AUC-ROC. All individual miRNAs show an AUC ≥ 0.7, except for miR-30b-5p, miR-210-3p and miR-324-3p that were 0.69, 0.68 and 0.69, respectively. The highest AUC values achieved by single miRNAs was obtained for miR-145-5p with an AUC of 0.78 (95% CI: 0.65–0.91; p = 0.0029). The AUC values obtained for let-7a-5p, let 7 g-5p, miR-19b-3p, miR-25-3p, miR-29a-3p, miR-30e-3p, miR-142-3p, miR-150-5p, miR-199a-3p, and miR-660-5p when DCMSEV were compared to DCMMOD in the idiopathic group, as well as sensitivity for a whole range of specificities, are shown in Table 5.
Table 5.
Comparisons of single microRNAs as predictors of idiopathic DCMSEV from DCMMOD and their diagnostic value.
| miRNA | AUC (95% CI) | Sensitivity (%) | Specificity (%) | p |
|---|---|---|---|---|
| Let-7a-5p | 0.73 (0.57–0.88) |
81.58 68.42 63.16 |
38.46 46.15 61.54 |
0.0155 |
| Let-7 g-5p | 0.71 (0.55–0.86) |
75.68 72.97 64.86 |
46.67 60 66.67 |
0.0213 |
| miR-19b-3p | 0.77 (0.62–0.92) |
76.67 66.67 63.33 |
50 57.14 78.57 |
0.0050 |
| miR-25-3p | 0.70 (0.55–0.86) |
78.95 68.42 60.53 |
35.71 57.14 64.29 |
0.0259 |
| miR-29a-3p | 0.70 (0.55–0.86) |
72.5 70 52.5 |
50 57.14 71.43 |
0.0244 |
| miR-30b-5p | 0.69 (0.52–0.86) |
82.35 73.53 55.88 |
46.67 53.33 60 |
0.0354 |
| miR-30e-3p | 0.72 (0.57–0.87) |
78.38 72.97 70.27 |
35.71 42.86 64.29 |
0.0161 |
| miR-142-3p | 0.76 (0.62–0.91) |
75.86 72.41 65.52 |
46.67 60 73.33 |
0.0046 |
| miR-145-5p | 0.78 (0.65–0.91) |
78.95 73.68 65.79 |
46.15 61.54 69.23 |
0.0029 |
| miR-150-5p | 0.73 (0.58–0.89) |
84.62 79.49 69.23 |
38.46 46.15 69.23 |
0.0119 |
| miR-199a-3p | 0.72 (0.58–0.87) |
71.05 68.42 63.16 |
57.14 64.29 71.43 |
0.0145 |
| miR-210-3p | 0.68 (0.52–0.85) |
81.58 68.42 63.16 |
40 53.33 66.67 |
0.0382 |
| miR-324-3p | 0.69 (0.52–0.86) |
85.71 77.14 60 |
35.71 42.86 64.29 |
0.0417 |
| miR-660-5p | 0.73 (0.58–0.87) |
75 70 60 |
57.14 64.29 71.43 |
0.0129 |
Abbreviations: AUC, area under the curve; CI, confidence interval; DCMSEV, dilated cardiomyopathy with ejection fraction ≤ 30%; miRNA, microRNA; ROC, receiver operating characteristic.
A predictive model to determinate the association of microRNAs with a very reduced ejection fraction in idiopathic DCM
Statistical modelling was used to determine the association of the fourteen miRNA candidates with the patient characteristics. A multivariate model was built up with three predictive circulating miRNAs, miR-130b-3p, miR-150-5p and miR-210-3p. The multivariate logistic regression analyses revealed that circulating miR-130b-3p, miR-150-5p and miR-210-3p levels were associated with left bundle branch block (LBBB), LVESD, lower systolic blood pressure (SBP) and smoking habit in our idiopathic DCMSEV group. Based on this analysis, LBBB (OR: 18.13; 95% CI: 1.28- 257.21; p = 0.014), LVESD (OR: 1.24; 95% CI: 1.01–1.53; p = 0.01), SBP (OR: 0.97; 95% CI: 0.91–1.03; p = 0.329), smoking habit (OR: 01; 95% CI: 1.44–630.02; p = 0.007), plasma miR-130b-3p (OR: 0; 95% CI: 0.1–1.57; p = 0.01), miR-150-5p (OR: 36.06; 95% CI: 1.64–794.78; p = 0.006) and miR-210-3p (OR: 687.53; 95% CI: 0.9–527,268.21; p < 0.001) were independent influencing factors for idiopathic DCMSEV (Table 6). In addition, the Akaike Information Criterion (AIC) of our model showed a value of 35.406. The AIC for a model just with a combination of miR-150-5p and miR-210-3p levels showed a higher value (AIC: 61.393). This lower value indicates that our model has a better fit.
Table 6.
Multivariate logistic regression analysis for identification of independent predictors of idiopathic DCMSEV patients.
| Variable | OR | 95% CI | p |
|---|---|---|---|
| LBBB (%) | 18.13 | 1.28–257.21 | 0.014 |
| LVESD (mm) | 1.24 | 1.01–1.53 | 0.01 |
| SBP (mm Hg) | 0.97 | 0.91–1.03 | 0.329 |
| Smoking habit | 30.1 | 1.44–630.02 | 0.007 |
| miR-130b-3p | 0 | 0.1–1.57 | 0.01 |
| miR-150-5p | 36.06 | 1.64–794.78 | 0.006 |
| miR-210-3p | 687.53 | 0.9–527,268.21 | < 0.001 |
AIC value: 35.406.
The model included LBBB (%), LVESD (mm), SBP (mm Hg), smoking habit and the levels of miR-130b-3p,miR-150-5p and miR-210-3p. Abbreviations: AIC, Aikaike Information Criterion; CI, confidence interval; LBBB, left bundle branch block; LVESD, left ventricular end-systolic diameter; OR, odds ratio; SBP, systolic blood pressure.
The potential usefulness of our model for identifying the idiopathic DCMSEV population was obtained with a ROC curve analysis. As shown in Fig. 4, the AUC was 0.96 (95% CI: 0.884–1.00; p < 0.001).
Figure 4.
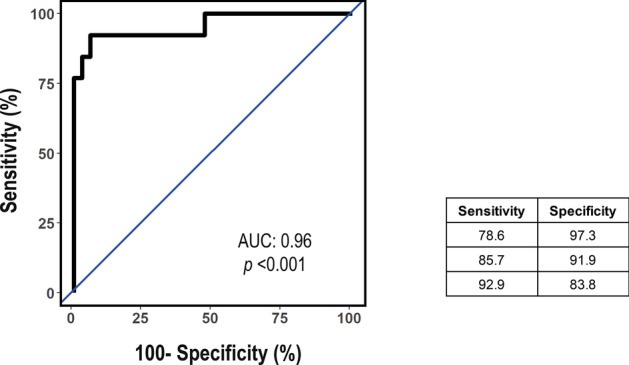
ROC analysis for the multivariate logistic regression model predicting idiopathic DCMSEV patients. The mean AUC for the averaged ROC curves are presented. The data are presented as the AUC and 95% CI and corresponding sensitivity for a range of specificities. The AUC is 0.96 (95% CI: 0.884–1.00; p < 0.001). Abbreviations: AUC, area under the curve; CI, confidence interval; DCMSEV, dilated cardiomyopathy with ejection fraction ≤ 30%; ROC, receiver operating characteristic.
miRNA-gene network analysis
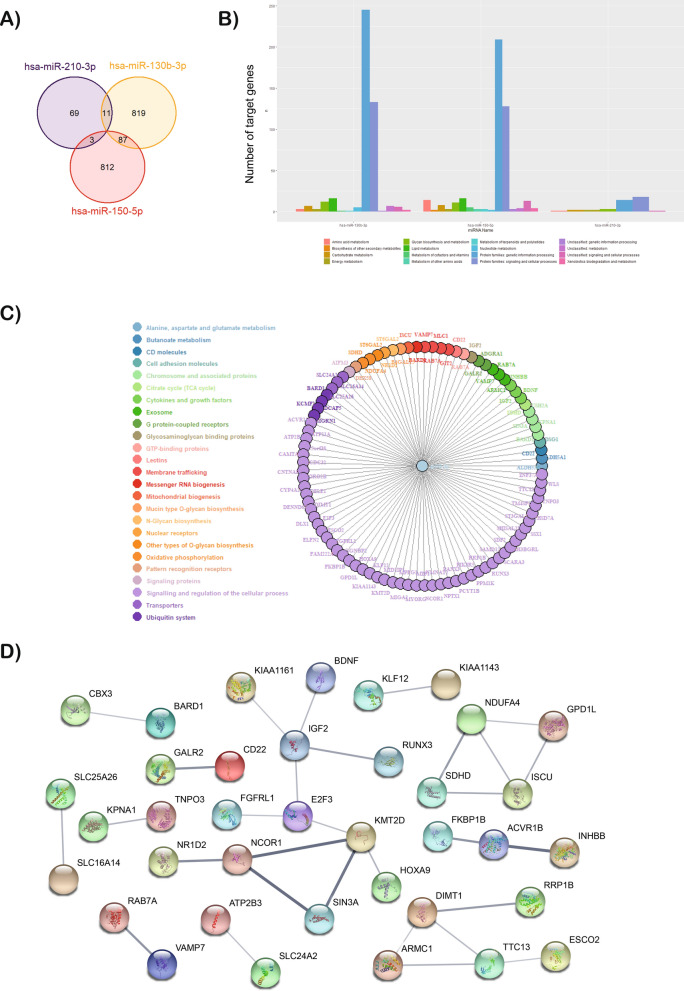
The hypothetical functions of three miRNAs were assessed with the miRDB database to reach the target mRNAs. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database identified the biological and molecular processes. The miR-130b-3p, miR-150-5p and miR-210-3p were associated with 917, 902 and 83 mRNAs in the miRDB database, respectively (Fig. 5A). The analysis revealed that most of these genes are involved in signalling and regulation of the cellular and metabolic process and genetic information processing (Figs. 5B–C). For instance, the genes are deeply involved in the transcription pathway since these miRNAs regulate the expression of the RNA Polymerase II, transcription factors or alternative splicing. In addition, other target genes are implicated in the nitrogen compound metabolic process or in response to oxygen levels, among others (extended in Fig. S3).
Figure 5.
The miRNA-gene network for miRNAs associated with genes related to idiopathic DCMSEV. (A) Venn diagram showing overlap of gene targets of miR-130b-3p, miR-150-5p and miR-210-3p. (B) GO and KEGG analysis of miR-130b-3p, miR-150-5p and miR-210-3p target genes; the x-axis indicates KEGG pathway categories and the y-axis indicates the target numbers of genes. (C) miRNA-gene network for miR-210-3p. The network was built using 83 targeted genes and predicted interactions from the miRDB database. (D) PPI analysis performed using the STRING database for the target genes of miR-210-3p. The disconnected nodules are hidden. Bold lines indicate stronger association. Abbreviations: DCMSEV, dilated cardiomyopathy with ejection fraction ≤ 30%.
On the other hand, the protein–protein interaction networks (PPI) analysis networks related to miR-210-3p were analysed using the search tool for the retrieval of interacting genes (STRING) (Fig. 5D). Our in-silico model showed that ISCU and other mitochondrial activity-related proteins, as well as SDHD and NDUFA4, are targets of miR-210-3p. Moreover, the analysis revealed that genes targeted by miR-150 and miR-210 showed a significant PPI enrichment (p = 0.00459) (Figure S4 and Table S3). These results demonstrated that target genes of miR-210-3p and miR-150-5p may interact with each other.
Discussion
We analysed the circulating miRNA signature of the ischemic and idiopathic DCM population stratified in severely and moderately reduced EF, ≤ 30% and 31–49% respectively. Our study reports, for the first time, a signature of three circulating miRNAs (miR-130b-3p, miR-150-5p and miR-210-3p) that allow us to discriminate between idiopathic DCMMOD and DCMSEV subjects. Furthermore, this circulating miRNA panel in plasma defines idiopathic DCMSEV from ischemic etiology.
LVEF is the cornerstone of DCM outcome. Optimal pharmacological and non-pharmacological decisions, mostly based on LVEF, have improved DCM prognosis drastically16. Major arrhythmogenic episodes, sudden death events and mortality are related to DCM with lower EF ≤ 35%7. The impact of stratifying the severity of systolic impairment is crucial in DCM as shown6,7,17, but access to the EF has several limitations. The need for expert professionals, intra- and inter-observational differences or time-consuming imaging techniques in transthoracic echocardiography or cardiac magnetic resonance, as well as inaccessible or excessive radiation imaging techniques, make it difficult to monitor the patient. The lack of a specific circulating biomarker to stratify LVEF and the limitations of cardiac imaging tests lead to the need to investigate a novel tool for this at-risk population.
Some studies have struggled to relate preserved and impaired LVEF to HF categorization18–20. The dysregulation of miRNAs related to DCM diagnosis and its etiology has scarcely been characterized nor categorization of the EF has not been reported in this population14,15,21. Some groups have worked on the reverse remodeling of LV in DCM or the signalling network22. To the best of our knowledge, no study has focused on LVEF in the DCM population and circulating miRNA.
In the present study, fourteen plasma miRNAs showed discriminative power to define DCMSEV vs. DCMMOD idiopathic patients based on their correlation to some echocardiographic variables. The idiopathic DCMSEV group showed an association between LVEF with these miRNAs. In the case of DCMMOD patients, the miRNAs were associated with LVESD, and LA. Remarkably, it has been suggested that pressure overload in earlier stages of DCM and elevated LV filling pressures lead to a hemodynamic imbalance that justifies the increased LA and LVESD in this cohort23,24.. Interestingly, some of these fourteen miRNAs have been related to HF and DCM patients with an EF < 30%, compared with healthy controls. Circulating levels of let-7a-5p were significantly upregulated in HF patients25. Myocardial and circulating miR-30 levels were significantly increased in DCM patients22. We present a novel, reliable fingerprint of circulating miRNAs to discriminate between idiopathic DCMMOD and DCMSEV subjects.
A multiparametric model was performed as a potential tool for the diagnosis of idiopathic DCMSEV. A panel comprised of a combination of three circulating miRNAs (miR-130b-3p, miR-150-5p and miR-210-3p) with four clinical variables (LBBB, LVESD, SBP and smoking habit) showed a high-yield diagnostic accuracy with an AUC of 0.96. Significantly, miR-150-5p and miR-210-3p had a positive value while miR-130b-3p had a negative value for a diagnosis of idiopathic DCM.SEV.
A total of 1902 genes were predicted to be targeted by these three miRNAs. Although they have been described as key hypoxia-related miRNA, our in-silico analysis revealed that they are more than silent players in hypoxia. Hence, these miRNAs may influence DCMSEV devolvement, promoting heart injury. MiR-210-3p and miR-150-5p share targeted genes linked within the cellular proliferation, differentiation oxidative stress and apoptosis26–29. The miR-210 expression has been extensively studied in several cardiovascular-related diseases with controversial results30–33. Its overexpression reduces mitochondrial reactive oxygen species production, and promotes cardiomyocyte proliferation, cell survival and angiogenesis post-myocardial infarction30,31. In disagreement, Sun et al. confirmed that miR-210 induced oxidative stress and repressed mitochondrial function34. Our in-silico model showed that several mitochondrial activity-related proteins, are targets of miR-210-3p. Downregulation of these proteins may influence mitochondrial activity. Hence, our data are in agreement with previous studies that demonstrate impaired mitochondrial function in idiopathic DCM35,36. On the other hand, miR‐150 levels have been described as a responsive protector in ischemic heart disease37–39 and DCM40. MiR-150 protects the heart from ischemic injury by inhibiting the inflammatory response and repressing pro-apoptotic gene expression39. Otherwise, a different role has been defined for miR-150. Its levels were upregulated in the infarct area compared with other cardiac zones, altering the LV remodeling37. Furthermore, the downregulation of miR-150 targeted genes intensifies cardiac injury by deregulation of cell cycle-dependent intracellular Ca2+ concentration, and oxidative stress increase29; accompanied by an increase in reactive nitrogen disease, DNA damage and cardiomyocyte death41. MiR-130b-3p is negatively associated in our model. Instead of having a cytoprotective role, miR-130b-3p promotes cardiomyocytes injury42. Although these three miRNAs have been studied in several cardiovascular-related diseases, their role is still unknown regarding idiopathic DCM.
Using predictive tools, we report an additive value to our model, including some clinical variables such as LBBB, LVESD, SBP and smoking habit. LBBB, SBP and LVESD are a risk factor marker of HF5,43. Interestingly, LBBB and lower EF have widely shown a better response to non-pharmacological treatment, such as an implantable defibrillator44–46. Both clinical markers are related to neurohormonal activation in HF leading to severe myocardial damage43. Our circulating levels of miRNAs may reflect a dysregulation of the cardiomyocyte protecting processes arising from severe impairment of the LVEF in the DCMSEV population.
We have described a circulating miRNA signature as a useful and non-invasive tool for LVEF classification in idiopathic DCM patients. In addition, we have defined a parametric model, a miRNA panel and four clinical parameters that categorize DCM based on LV systolic impairment with the highest diagnostic values reported to date. The bioinformatics analysis may elucidate the underlying molecular and cellular mechanisms of idiopathic DCM. Our results present a step-forward personalized therapeutic strategy and management of this entity.
There are several limitations to the current study. The study sample was small, but the patients who were included were strictly DCM subjects. Recently, it has been reported that sex differences might play a role in the prognosis of DCM47,48. In our case, larger sample size is needed to validate these data. Therefore, before these novel biomarkers can be routinely used in clinical practice, the data should be replicated and extended to larger populations. Secondly, data on natriuretic peptides, troponin, or reverse remodeling were not available in all patients to compare to our results. Our sample was of patients in a chronic situation, who were not hospitalized but were prospectively followed in the outpatient clinic. Future studies should include these data. The dysregulation of miRNAs related to DCM diagnosis and its etiology has scarcely been characterized and characterization of the EF has not been reported in this population. In the bioinformatic analysis, we only identified the genes targeted by miR-210-3p and miR-150-5p, since both are positively related to our logistic regression model. MiR-130b-3p was not included in the PPI analysis since it is negatively related to our logistic model. In addition, the analysis of 1902 genes did not show clear results due to a large number of interactions and data overload. Finally, we have no evidence that these circulating miRNAs are directly secreted into the extracellular space from the heart. Thus, dedicated experiments on human heart samples are necessary to verify these results and bioinformatic predictions.
Conclusion
In conclusion, we identified a miRNAs fingerprint that is differentially expressed in the idiopathic DCMSEV population. This signature arises as a potential clinical biomarker to discriminate DCM etiology and stratify its risk, based on the LVEF. The clinical usefulness of this miRNA panel as a diagnostic tool could lead to tailored treatment improving the DCM population outcome. In addition, the bioinformatics analysis results may be useful in elucidating the underlying mechanisms of idiopathic DCM.
Methods
Study population and design
This was a prospective cross-sectional study. Inclusion criteria were patients with clinical features of DCM1, a LV end-diastolic diameter larger than 56 mm and a LVEF below 50%. Exclusion criteria were genetic DCM or any cardiovascular, life-limiting systemic condition or an infectious or tumoral condition that could influence the DCM definition or miRNA results. Only patients older than 18 years old were enrolled. A total of 159 consecutive subjects were included in the study: (1) 55 idiopathic DCM patients, (2) 60 ischemic DCM and (3) 44 healthy controls who had been referred to the Cardiology Department of the University Hospital Puerta del Mar, Cádiz, Spain. The study design is shown in Fig. 1. LVEF was classified into two categories based on prior studies: ≤ 30%, severely reduced EF (DCMSEV) and 31–49% moderately reduced EF (DCMMOD)49,50. A transthoracic echocardiography protocol was performed as described previously13,15. Echocardiographic methods for the measurement of EF were the apical biplane method of disks (modified Simpson’s rule) and when this was not possible, Teicholz51. Detailed anthropometric, clinical and pharmacological information was obtained from each subject including family history, symptoms of HF, an electrocardiogram, a 24-h Holter electrocardiogram and, when appropriate, cardiac magnetic resonance. Our institution’s ethics committee (Comité de Ética de la Investigación de Cádiz) approved the study protocol. The study was performed in full compliance with the Helsinki II Declaration. All participants provided written informed consent.
Blood collection
Ten millilitres of peripheral blood were collected in K2-ethylenediaminetetraacetic acid tubes (BD) after 10 h overnight fasting and were immediately centrifuged (1500 xg, 15 min, 4 °C). The blood was processed within 4 h after isolation. The upper layer containing plasma was divided into aliquots and stored at -80ºC until further analysis.
RNA isolation
Total RNA was isolated using a miRNeasy Serum/Plasma Kit (Qiagen), according to the manufacturer’s instructions. A total of 200 µL of plasma aliquots were used to obtain enough final volume. To normalize extracellular miRNAs, a mix of synthetic Caenorhabditis Elegans miR-39-3p (cel-miR-39-3p) (1.6 × 108 copies/µL) (Qiagen) and UniSp6 (Qiagen) was added to each sample. RNA was stored at -80 °C.
MicroRNA real-time reverse transcriptase-polymerase chain reaction
RNA was reverse transcribed using the miRCURY LNA RT Kit (Qiagen) as previously described15. The reverse transcription reaction was performed under the following conditions: 60 min at 42 ºC, heat-inactivated for 5 min at 95 ºC and immediately cooled to 4 ºC. Then cDNA was stored at − 20 ºC. Quantitative real-time PCR (qPCR) was performed according to the protocol of miRCURY LNA SYBR Green PCR Kit (Qiagen). CFX Connect PCR System (BioRad) was used to perform the qPCR, at 95 °C for 2 min, followed by 40 cycles of 95 °C for 10 s, and 56 °C for 60 s, followed by melting curve analysis. qPCR amplification curves were evaluated with CFX Manager™ software (BioRad). The specificity of the amplification was corroborated by melting curve analysis.
The miRCURY LNA miRNA Serum/Plasma Focus PCR Panel (Qiagen) was used for the screening study. This panel contains 179 miRNA primer sets expressed in human serum/plasma. Then, custom 96-wells Pick-&-Mix microRNA PCR plates (Qiagen) were used to validate each miRNA candidate. To account for the variability between the plates, the interplate calibrator UniSp3 was analysed. Cqs above 35 cycles were censored at the minimum level observed for each miRNA. cel-miR-39-3p levels were stable across all samples. Relative quantification was performed using the 2 − ΔCq method, where ΔCq = CqmiRNA − Cqcel-miR-39-3p miRNA levels were log-transformed before being used in the statistical analyses. We have previously used this approach in the field of circulating non-coding RNAs13,15.
miRNA-gene network analysis
The miRNAs obtained were tested using the miRDB database (http://mirdb.org/) to predict the targeted genes52. A database analysis to identify the biological function was performed using GO enrichment analysis (http://geneontology.org/)53 and the KEGG (https://www.kegg.jp/kegg/) software54. R software (www.r-project.org) was assessed to build up miRNA-mRNA55. The STRING database (http://www.string-db.org/) was used to analyse the PPI networks56.
Statistical analysis
Continuous variables are shown as mean ± standard deviation. Categorical variables are expressed as frequency and percentage of patients (%). Intergroup comparisons of miRNAs levels were performed using non-parametric Mann–Whitney and Kruskal–Wallis rank tests for continuous variables. The Pearson correlation coefficient was used for correlations between echocardiographic and clinical parameters vs. log2 miRNAs. An analysis of differences between groups was performed using analysis of variance. Receiver operating characteristic (ROC) curves that characterize the diagnostic performance of candidate miRNAs and logistic regression models were plotted to determine the area under the curve (AUC) and the specificity and sensitivity of the optimal cut-offs. ROC curves were generated by plotting sensitivity against 100-specificity. Data were presented as the AUC and 95% confidence intervals. The relationships between miRNAs and LVEF status were assessed using logistic regression. Multiple logistic regression modelling was applied to perform the miRNA panel. The regression coefficients of each miRNA that was significantly associated with the outcome were applied to estimate the miRNA panel value. The Wilcoxon test and iterating combinations between our miRNA candidates, as well as echocardiographic and clinical covariates, were used to construct several models. The changes in p-values of their variables were evaluated by the Wald test and a likelihood ratio. The statistical software package R (www.r-project.org) was used for all analyses55.
Supplementary Information
Acknowledgements
This study was supported by the Fundación Pública Andaluza Progreso y Salud para la Financiación, co-financed by the European Regional Development Fund (ERDF) (PI-0048-2017), Plan Propio de INIBICA (PI-INBICA2019-13), Fundación Pública Andaluza Progreso y Salud (PI 0136-2018). The study was also supported by a grant from the Spanish Society of Cardiology (SEC) for Basic Research (0011-2019). We would like to thank Galan Pacheco for statistical support.
Abbreviations
- AUC
Area under the curve
- AIC
Akaike Information Criterion
- CI
Confidence interval
- EF
Ejection fraction
- DCM
Dilated cardiomyopathy
- GO
Gene Ontology
- HF
Heart failure
- ISCU
Iron-sulphur cluster homolog
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LA
Left atrial dimension
- LBBB
Left bundle branch block
- LV
Left ventricle
- LVEF
Left ventricle ejection fraction
- LVESD
LV end-systolic dimension
- miRNA
MicroRNA
- OR
Odd ratio
- PPI
Protein–protein interaction networks
- qPCR
Quantitative real-time PCR
- ROC
Receiver-operator characteristic
- SBP
Systolic blood pressure
- STRING
Search Tool for the Retrieval of Interacting Genes
Author contributions
M.C-D., T.B., A.M.and R.T. conceived the experiments, M.Q-F., M.R., O.C., A.M. and R.T. recruited the subjects, M.C-D., T.B., J.C-D.and R.T. conducted the experiments, M.C-D., T.B., J.C-D.and R.T. analyzed the results. M.C-D., T.B., A.M. and R.T. wrote the manuscript. All authors reviewed the manuscript.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maria Calderon-Dominguez, Email: mariacalderond@gmail.com.
Rocio Toro, Email: rocio.toro@uca.es.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-87086-1.
References
- 1.Elliott P, Andersson B, Arbustini E, et al. Classification of the cardiomyopathies: A position statement from the european society of cardiology working group on myocardial and pericardial diseases. Eur. Heart. J. 2008;29(2):270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 2.Merlo M, Cannatà A, Gobbo M, Stolfo D, Elliott PM, Sinagra G. Evolving concepts in dilated cardiomyopathy. Eur. J. Heart Fail. 2017 doi: 10.1002/ejhf.1103. [DOI] [PubMed] [Google Scholar]
- 3.Schultheiss HP, Fairweather DL, Caforio ALP, et al. Dilated cardiomyopathy. Nat. Rev. Dis. Prim. 2019 doi: 10.1038/s41572-019-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branca L, Sbolli M, Metra M, Fudim M. Heart failure with mid-range ejection fraction: pro and cons of the new classification of Heart Failure by European Society of Cardiology guidelines. ESC Hear Fail. 2020;7(2):381–399. doi: 10.1002/ehf2.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witt CM, Wu G, Yang D, Hodge DO, Roger VL, Cha YM. Outcomes with left bundle branch block and mildly to moderately reduced left ventricular function. JACC Hear Fail. 2016;4(11):897–903. doi: 10.1016/j.jchf.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Gentile P, Merlo M, Cannatà A, et al. Dilated cardiomyopathy with mid-range ejection fraction at diagnosis: characterization and natural history. J. Am. Heart Assoc. 2019;8(17):e010705. doi: 10.1161/JAHA.118.010705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angaran P, Dorian P, Ha ACT, et al. Association of left ventricular ejection fraction with mortality and hospitalizations. J. Am. Soc. Echocardiogr. 2020;S0894–7317(20):30009–30012. doi: 10.1016/j.echo.2019.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Rusnak J, Behnes M, Weiß C, et al. Impact of left ventricular ejection fraction on recurrent ventricular tachyarrhythmias in recipients of implantable cardioverter defibrillators. Cardiology. 2020 doi: 10.1159/000504876. [DOI] [PubMed] [Google Scholar]
- 9.Calderon-Dominguez M, Belmonte T, Quezada-Feijoo M, et al. Emerging role of microRNAs in dilated cardiomyopathy: evidence regarding etiology. Transl. Res. 2019;215:86–101. doi: 10.1016/j.trsl.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Stepien E, Costa MC, Kurc S, Drozdz A, Cortez-Dias N, Enguita FJ. The circulating non-coding RNA landscape for biomarker research: Lessons and prospects from cardiovascular diseases review-article. Acta Pharmacol Sin. 2018;39(7):1085–1099. doi: 10.1038/aps.2018.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colpaert RMW, Calore M. MicroRNAs in cardiac diseases. Cells. 2019;8(7):737. doi: 10.3390/cells8070737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu D, Thum T. RNA-based diagnostic and therapeutic strategies for cardiovascular disease. Nat. Rev. Cardiol. 2019;16(11):661–674. doi: 10.1038/s41569-019-0218-x. [DOI] [PubMed] [Google Scholar]
- 13.Toro R, Blasco-Turrión S, Morales-Ponce FJ, et al. Plasma microRNAs as biomarkers for Lamin A/C-related dilated cardiomyopathy. J. Mol. Med. 2018;96(8):845–856. doi: 10.1007/s00109-018-1666-1. [DOI] [PubMed] [Google Scholar]
- 14.Zaragoza C, Saura M, Hernández I, et al. Differential expression of circulating miRNAs as a novel tool to assess BAG3-associated familial dilated cardiomyopathy. Biosci Rep. 2019;39(3):BSR20180934. doi: 10.1042/BSR20180934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belmonte T, Mangas A, Calderon-Dominguez M, et al. Peripheral microRNA panels to guide the diagnosis of familial cardiomyopathy. Transl Res. 2020;218:1–15. doi: 10.1016/j.trsl.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Al-Khatib SM, Sanders GD, Carlson M, et al. Preventing tomorrow’s sudden cardiac death today. Dissemination of effective therapies for sudden cardiac death prevention. Am. Heart J. 2008;156(4):613–622. doi: 10.1016/j.ahj.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 17.Wu T, Chen Y, Du Y, et al. Circulating exosomal miR-92b-5p is a promising diagnostic biomarker of heart failure with reduced ejection fraction patients hospitalized for acute heart failure. J. Thorac Dis. 2018;10(11):6211–6220. doi: 10.21037/jtd.2018.10.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson CJ, Gupta SK, O’Connell E, et al. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur. J. Heart Fail. 2015;17(4):405–415. doi: 10.1002/ejhf.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen F, Yang J, Li Y, Wang H. Circulating microRNAs as novel biomarkers for heart failure. Hell J. Cardiol. 2018;59(4):209–214. doi: 10.1016/j.hjc.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Wong LL, Zou R, Zhou L, et al. Combining circulating MicroRNA and NT-proBNP to detect and categorize heart failure subtypes. J. Am. Coll Cardiol. 2019;73(11):1300–1313. doi: 10.1016/j.jacc.2018.11.060. [DOI] [PubMed] [Google Scholar]
- 21.Domínguez F, Cuenca S, Bilińska Z, et al. Dilated cardiomyopathy due to BLC2-associated athanogene 3 (BAG3) mutations. J. Am. Coll Cardiol. 2018;72(20):2471–2481. doi: 10.1016/j.jacc.2018.08.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dziewięcka E, Totoń-Żurańska J, Wołkow P, et al. Relations between circulating and myocardial fibrosis-linked microRNAs with left ventricular reverse remodeling in dilated cardiomyopathy. Adv. Clin. Exp. Med. 2020;29(3):285–293. doi: 10.17219/acem/115088. [DOI] [PubMed] [Google Scholar]
- 23.Ponikowski P, Voors AA, Anker SD, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 2016;37(27):2129–2200m. doi: 10.1093/eurheartj/ehw128. [DOI] [PubMed] [Google Scholar]
- 24.Merlo M, Gentile P, Artico J, et al. Arrhythmic risk stratification in patients with dilated cardiomyopathy and intermediate left ventricular dysfunction. J. Cardiovasc. Med. 2019;20(5):343–350. doi: 10.2459/JCM.0000000000000792. [DOI] [PubMed] [Google Scholar]
- 25.Marques FZ, Vizi D, Khammy O, Mariani JA, Kaye DM. The transcardiac gradient of cardio-microRNAs in the failing heart. Eur. J. Heart Fail. 2016;18(8):1000–1008. doi: 10.1002/ejhf.517. [DOI] [PubMed] [Google Scholar]
- 26.Priest JR, Osoegawa K, Mohammed N, et al. De Novo and rare variants at multiple loci support the oligogenic origins of atrioventricular septal heart defects. PLoS Genet. 2016;12(4):1–25. doi: 10.1371/journal.pgen.1005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan R, Wen XY, Wu J, et al. Knockdown of ZNF403 inhibits cell proliferation and induces G2/M arrest by modulating cell-cycle mediators. Mol. Cell Biochem. 2012;365(1–2):211–222. doi: 10.1007/s11010-012-1262-6. [DOI] [PubMed] [Google Scholar]
- 28.King JC, Moskowitz IPG, Burgon PG, et al. E2F3 plays an essential role in cardiac development and function. Cell Cycle. 2008;7(23):3775–3780. doi: 10.4161/cc.7.23.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Kong M, Jiang D, Qian J, Duan Q, Dong A. MicroRNA-150 aggravates H2O2-induced cardiac myocyte injury by down-regulating c-myb gene. Acta Biochim Biophys Sin (Shanghai). 2013;45(9):734–741. doi: 10.1093/abbs/gmt067. [DOI] [PubMed] [Google Scholar]
- 30.Mutharasan RK, Nagpal V, Ichikawa Y, Ardehali H. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am. J. Physiol. Hear Circ. Physiol. 2011 doi: 10.1152/ajpheart.01080.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arif M, Pandey R, Alam P, et al. MicroRNA-210-mediated proliferation, survival, and angiogenesis promote cardiac repair post myocardial infarction in rodents. J. Mol. Med. 2017;95(12):1369–1385. doi: 10.1007/s00109-017-1591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bian WS, Shi PX, Mi XF, et al. MIR-210 protects cardiomyocytes from OGD/R injury by inhibiting E2F3. Eur. Rev. Med. Pharmacol. Sci. 2018;22(3):743–749. doi: 10.26355/eurrev_201802_14305. [DOI] [PubMed] [Google Scholar]
- 33.Qiao XR, Wang L, Liu M, Tian Y, Chen T. MiR-210-3p attenuates lipid accumulation and inflammation in atherosclerosis by repressing IGF2. Biosci. Biotechnol. Biochem. 2020;84(2):321–329. doi: 10.1080/09168451.2019.1685370. [DOI] [PubMed] [Google Scholar]
- 34.Sun W, Zhao L, Song X, et al. MicroRNA-210 Modulates the Cellular Energy Metabolism Shift during H2O2 -Induced Oxidative Stress by Repressing ISCU in H9c2 Cardiomyocytes. Cell Physiol. Biochem. 2017;43(1):383–394. doi: 10.1159/000480417. [DOI] [PubMed] [Google Scholar]
- 35.Marin-Garcia J, Goldenthal MJ, Pierpont MEM, Ananthakrishnan R. Impaired mitochondrial function in idiopathic dilated cardiomyopathy: biochemical and molecular analysis. J. Card Fail. 1995;1(4):285–291. doi: 10.1016/1071-9164(95)90003-9. [DOI] [PubMed] [Google Scholar]
- 36.Jarreta D, Orús J, Barrientos A, et al. Mitochondrial function in heart muscle from patients with idiopathic dilated cardiomyopathy. Cardiovasc Res. 2000;45(4):860–865. doi: 10.1016/S0008-6363(99)00388-0. [DOI] [PubMed] [Google Scholar]
- 37.Devaux Y, Vausort M, McCann GP, et al. MicroRNA-150: A novel marker of left ventricular remodeling after acute myocardial infarction. Circ. Cardiovasc. Genet. 2013;6(3):290–298. doi: 10.1161/CIRCGENETICS.113.000077. [DOI] [PubMed] [Google Scholar]
- 38.Lin X, Zhang S, Huo Z. Serum circulating miR-150 is a predictor of post-acute myocardial infarction heart failure. Int Heart J. 2019;60(2):280–286. doi: 10.1536/ihj.18-306. [DOI] [PubMed] [Google Scholar]
- 39.Tang YY, Wang Y, Park KM, et al. MicroRNA-150 protects the mouse heart from ischaemic injury by regulating cell death. Cardiovasc Res. 2015;106(3):387–397. doi: 10.1093/cvr/cvv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scrutinio D, Conserva F, Passantino A, Iacoviello M, Lagioia R, Gesualdo L. Circulating microRNA-150-5p as a novel biomarker for advanced heart failure: a genome-wide prospective study. J. Hear Lung Transplant. 2017;36(6):616–624. doi: 10.1016/j.healun.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 41.Feridooni T, Hotchkiss A, Remley-Carr S, Saga Y, Pasumarthi KBS. Cardiomyocyte specific ablation of p53 is not sufficient to block doxorubicin induced cardiac fibrosis and associated cytoskeletal changes. PLoS ONE. 2011;6:7. doi: 10.1371/journal.pone.0022801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu X, Wang Y, Pang L, Huang J, Sun X, Chen X. miR-130 aggravates acute myocardial infarction-induced myocardial injury by targeting PPAR-γ. J. Cell Biochem. 2018;119(9):7235–7244. doi: 10.1002/jcb.26903. [DOI] [PubMed] [Google Scholar]
- 43.Lund LH, Jurga J, Edner M, et al. Prevalence, correlates, and prognostic significance of QRS prolongation in heart failure with reduced and preserved ejection fraction. Eur. Heart J. 2013;34(7):529–539. doi: 10.1093/eurheartj/ehs305. [DOI] [PubMed] [Google Scholar]
- 44.Daubert C, Gold MR, Abraham WT, et al. Prevention of disease progression by cardiac resynchronization therapy in patients with asymptomatic or mildly symptomatic left ventricular dysfunction: insights from the European cohort of the REVERSE (Resynchronization Reverses Remodeling in Systolic Le. J. Am. Coll Cardiol. 2009;54(20):1837–1846. doi: 10.1016/j.jacc.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 45.Al-Khatib SM, Stevenson WG, Ackerman MJ, et al. 2017 AHA/ACC/HRS Guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death. Circulation. 2018;138(13):e272–e391. doi: 10.1161/CIR.0000000000000549. [DOI] [PubMed] [Google Scholar]
- 46.Sanders GD, Hlatky MA, Owens DK. Cost-effectiveness of implantable cardioverter-defibrillators. N. Engl. J. Med. 2005;353(14):1471–1480. doi: 10.1056/NEJMsa051989. [DOI] [PubMed] [Google Scholar]
- 47.Cannatà A, Fabris E, Merlo M, et al. Sex differences in the long-term prognosis of dilated cardiomyopathy. Can. J. Cardiol. 2020;36(1):37–44. doi: 10.1016/j.cjca.2019.05.031. [DOI] [PubMed] [Google Scholar]
- 48.Yogasundaram H, Qi A, Nguyen Q, Oudit GY. Battle of the sexes: differential prognosis by sex in dilated cardiomyopathy. Can. J. Cardiol. 2020;36(1):7–10. doi: 10.1016/j.cjca.2019.06.031. [DOI] [PubMed] [Google Scholar]
- 49.Bänsch D, Antz M, Boczor S, et al. Primary prevention of sudden cardiac death in idiopathic dilated cardiomyopathy: the Cardiomyopathy Trial (CAT) Circulation. 2002;105(12):1453–1458. doi: 10.1161/01.CIR.0000012350.99718.AD. [DOI] [PubMed] [Google Scholar]
- 50.Dewan P, Rørth R, Jhund PS, et al. Differential impact of heart failure with reduced ejection fraction on men and women. J. Am. Coll Cardiol. 2019;73(1):29–40. doi: 10.1016/j.jacc.2018.09.081. [DOI] [PubMed] [Google Scholar]
- 51.Galderisi M, Cosyns B, Edvardsen T, et al. Standardization of adult transthoracic echocardiography reporting in agreement with recent chamber quantification, diastolic function, and heart valve disease recommendations: An expert consensus document of the European Association of Cardiovascular Imag. Eur. Heart J. Cardiovasc Imaging. 2017;18(12):1301–1310. doi: 10.1093/ehjci/jex244. [DOI] [PubMed] [Google Scholar]
- 52.Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Team RC. R: A Language and Environment for Statistical Computing. https://www.r-project.org
- 56.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.