FIG. 1.
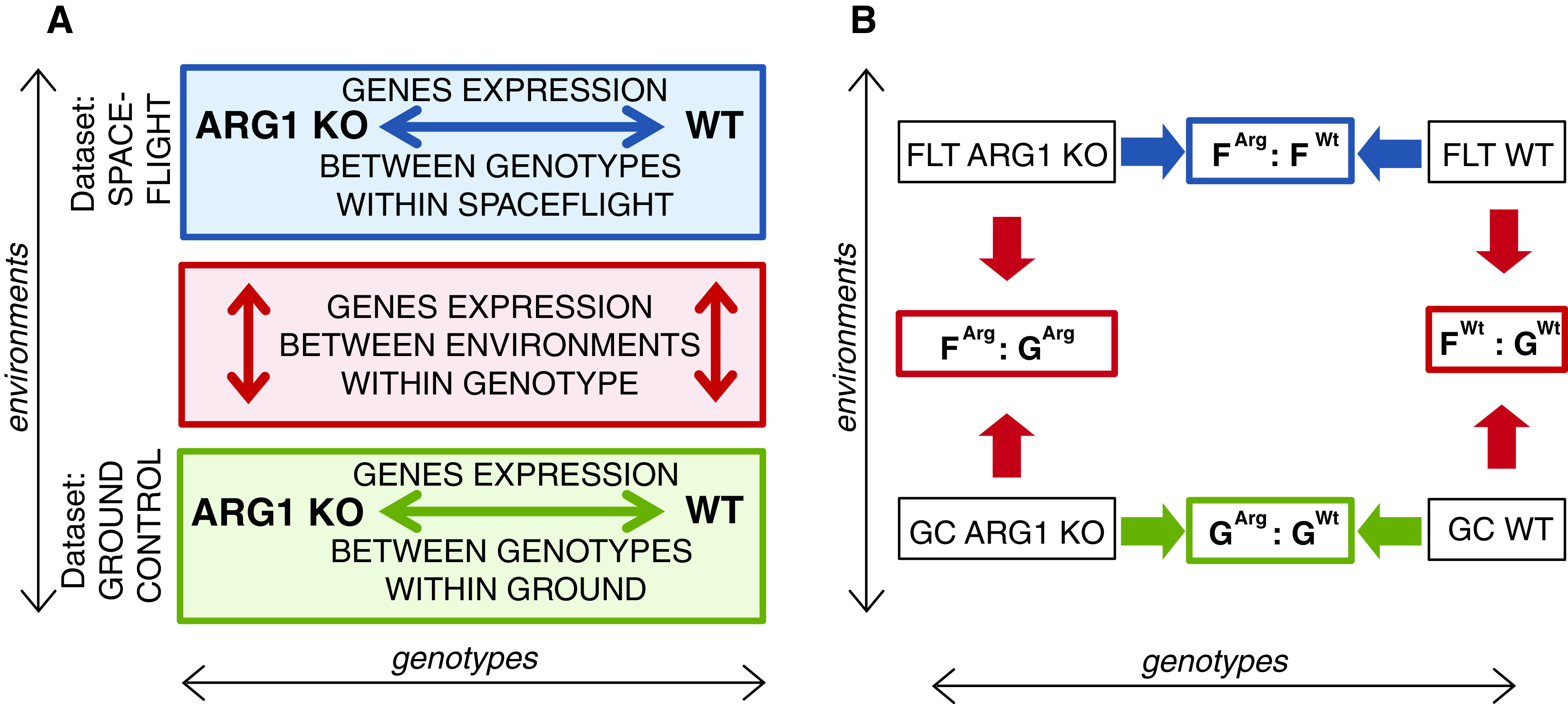
Graphical presentation of the two approaches used in the microarray data analysis. (A) ARG1 KO and WT mark the gene expression profiles for respective cell samples. Solid arrows represent the direction of comparison of the gene expression profiles. The red box and arrows indicate the first approach for data analysis—differentially expressed genes were identified between cells grown in the two environments: spaceflight and the comparable ground controls. The green box and arrow indicate the first part of the second approach for data analysis—differentially expressed genes were identified between wild-type (WT) and arg1 mutant (ARG1 KO) genotypes on the ground. The blue box and arrow indicate the second part of the second approach for data analysis—differentially expressed genes were identified between wild-type (WT) and arg1 mutant (ARG1 KO) genotypes in spaceflight. (B) Microarray data comparison groups used to obtain the significantly differentially expressed genes between the samples.
