Abstract
Approximately 70% of advanced breast cancer patients will develop bone metastases, which accounts for ∼90% of cancer-related mortality. Breast cancer circulating tumor cells (CTCs) establish metastatic tumors in the bone after a close interaction with local bone marrow cells including pericytes and osteoblasts, both related to resident mesenchymal stem/stromal cells (BM-MSCs) progenitors. In vitro recapitulation of the critical cellular players of the bone microenvironment and infiltrating CTCs could provide new insights into their cross-talk during the metastatic cascade, helping in the development of novel therapeutic strategies. Human BM-MSCs were isolated and fractionated according to CD146 presence. CD146+ cells were utilized as pericyte-like cells (PLCs) given the high expression of the marker in perivascular cells, while CD146− cells were induced into an osteogenic phenotype generating osteoblast-like cells (OLCs). Transwell migration assays were performed to establish whether primary breast cancer cells (3384T) were attracted to OLC. Furthermore, proliferation of 3384T breast cancer cells was assessed in the presence of PLC- and OLC-derived conditioned media. Additionally, conditioned media cultures as well as transwell co-cultures of each OLCs and PLCs were performed with 3384T breast cancer cells for gene expression interrogation assessing their induced transcriptional changes with an emphasis on metastatic potential. PLC as well as their conditioned media increased motility and invasion potential of 3384T breast cancer cells, while OLC induced a dormant phenotype, downregulating invasiveness markers related with migration and proliferation. Altogether, these results indicate that PLC distinctively drive 3384T cancer cells to an invasive and migratory phenotype, while OLC induce a quiescence state, thus recapitulating the different phases of the in vivo bone metastatic process. These data show that phenotypic responses from metastasizing cancer cells are influenced by neighboring cells at the bone metastatic niche during the establishment of secondary metastatic tumors.
Keywords: Cancer, bone, metastatic cascade, tumor, organs on chips, pericytes
Impact statement
Accurate in vitro human models of the bone extravasation site are critical for the understanding of breast cancer metastasis and the development of precision medicine. We recapitulated the endosteal niche with the exclusive use of primary human cells to elucidate the effect of perivascular cells and osteoblasts on breast cancer metastasis. Thus, we were able to drive cancer cells to a more invasive, migratory phenotype using perivascular-like cells, while osteoblast-like cells drove them to quiescence, recapitulating the metastatic niche and the initial phases of the bone metastatic process. These data support the thesis that phenotypic response from cancer cells can be controlled by neighboring cells and will support the engineering of an organ-on-chip approach which allows the elucidation of a compounded effect of more than one cell type involved in the metastasis of breast cancer cells into bone and provide a platform for their mechanistic interrogation and therapy development.
Introduction
Metastasis is the central event in cancer and accounts for 90% of cancer-related mortality.1 In the case of advanced breast cancer, approximately 70% of patients will develop bone metastases. A deeper understanding of how clinical metastasis is initiated, particularly in key sites like bone, is crucial for the development of precision treatment strategies. Since probing the bone microenvironment in healthy humans or cancer patients is fundamentally complex, development of accurate in vitro human models of the perivascular, primary central marrow, and endosteal niches of the bone is critically important.
Bone metastatic invasion constitutes a multi-step process, starting with extravasation of circulating tumor cells (CTCs) into the bone marrow close to the endosteal surface, followed by their close interaction with the local environment which determines the fate of the distant tumor before it becomes clinically detectable.2 Throughout these steps, invading CTCs chemically and physically interact with different cellular players, all related with local progenitors of mesenchymal phenotype (mesenchymal stem/stromal cells, MSCs). A fraction of MSCs localize at the perivascular space surrounding bone marrow sinusoids exhibiting a pericytic phenotype,2–4 controlling the extravasation of CTC via secretion of the chemoattractant stromal cell-derived factor 1 (Sdf-1) and capturing them through CD146-dependent anchoring capabilities.3–5 On the other hand, MSCs serve as progenitors of bone-forming osteoblasts,3,5 which participate in the regulation of various biological phenomena determining the future of the extravasated cancer cells. These include chemoresistance6–8 and their entrance into a state of either dormancy (i.e. quiescence) or growth following proliferation and angiogenesis induction (i.e. angiogenic switch).
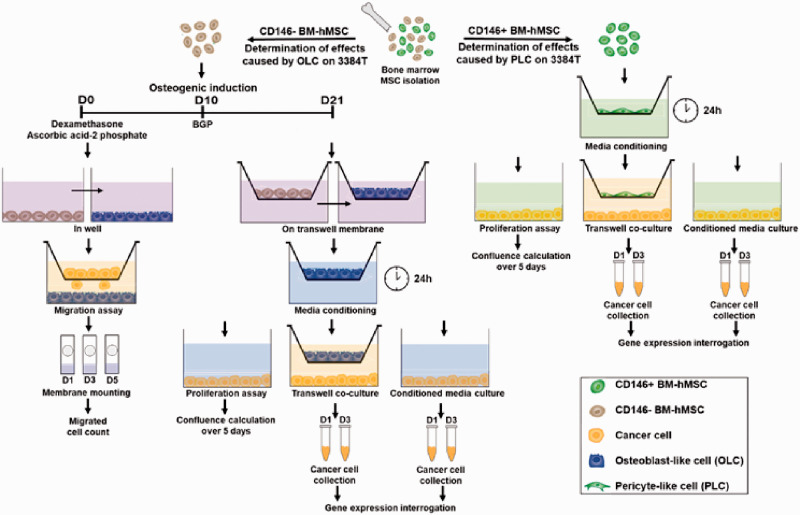
In order to study these complex cellular interactions, several models have been utilized;3,9–15 however, most of these studies use murine cells in combination with human tumor cells and immortalized cell lines. While valuable information has been extracted from these studies, it is important to consider that metastatic molecular mechanisms have variations between species.16–19 Therefore, our work aims to contribute to the understanding of the underlying mechanisms of breast cancer cell metastasis to bone, by focusing on the influential effects that both pericytes and osteoblasts have on invading breast cancer CTCs, using human primary cells exclusively. In this study, we exploited a dual role MSCs can have within the bone marrow, reproduced using in vitro assays: first, as perisinusoidal pericytes interacting with invading CTCs; and second as progenitors of bone-forming osteoblasts. Human bone marrow-derived mesenchymal stem cells (hBM-hMSCs) were fractionated based on their surface expression of CD146, using CD146+ as pericyte-like cells (PLCs), whereas CD146− were induced towards the osteoblastic lineage to produce osteoblast-like cells (OLCs, Figure 1).20–23 In doing so, along with transwell co-cultures and conditioned-media cultures we recapitulated the critical cellular components of the endosteal niche,24 which has been shown to play a critical role during metastatic invasion of CTCs and subsequent induction of quiescence of extravasated cancer cells.25
Materials and methods
hBM-MSCs culture and characterization
hBM-MSCs were obtained from consent-signed, de-identified healthy female patients (N = 2, BMC2101 and BMC2149). All hBM-MSCs were cultured as adhesion-dependent cells in complete media containing low glucose Dulbecco’s Modified Eagle Medium (LG-DMEM; #12430054, Gibco, Grand Island, NY, USA) and 10% fetal bovine serum (FBS; 89510194, VWR, Radnor, PA, USA) and maintained in a humidified incubator at 37°C and 5% CO2. Cells were detached using TrypLE (#12604013, Gibco, Grand Island, NY, USA) and were expanded to passage 4–5, at which point cells were plated for experiments or stored at −80°C until further use.
All hBM-MSCs (passage 3) were characterized by the minimum criteria including phenotypic expression. For morphologic assessments and growth kinetics, hBM-MSCs were plated at low confluence in complete media, and plates were transferred into an IncuCyte® ZOOM System (Essen BioScience, Inc., Ann Arbor, MI, USA) for five days for live image analysis of growth kinetics.
Flow cytometric analysis of phenotype was performed on all hBM-MSCs at passage 3. Briefly, cell suspensions were stained with anti-human antibodies against positive markers CD90, CD73, CD105, CD44 (Invitrogen, Waltham, MA, USA), negative markers HLA-DR, CD34, CD45, and CD31 (Miltenyi, Auburn, CA, USA), and corresponding isotype controls along with Ghost Dye™ 780 Red Viability dye (#13-0865, Tonbo Biosciences, San Diego, CA, USA) for 20 min at 4°C. Stained cells were washed twice and transferred into the wells of a 96-well plate that was loaded into a CytoFLEX S Flow Cytometer Platform using CytoExpert software (Beckman Coulter, Brea, CA, USA), as well as unstained controls. Acquisition of 20,000 events was achieved. Flow cytometric analysis of the phenotypic expressions of each marker was performed by gating strategy of scatter, singlets, and live cells.
hBM-MSC fractionation
hBM-MSCs were fractionated by magnet-activated cell sorting (MACS) based on the presence of CD146 using CELLection Biotin Binder kit (#1153310; Invitrogen, Waltham, MA, USA). Briefly, cells were washed with a buffer consisting of Dulbecco’s phosphate-buffered saline (DPBS; #14190144, Gibco, Grand Island, NY, USA) with 0.1% bovine serum albumin (BSA; A2153-50G, Sigma-Aldrich, St. Louis, MO, USA) and 2 mmol/L ethylenediaminetetraacetic acid (EDTA; #E9884, Sigma-Aldrich, St. Louis, MO, USA), followed by a 20-min incubation at 4°C in the presence of 100 µg biotinylated CD146 antibody (#ab77928, Abcam, Cambridge, UK ). Cells were then washed and resuspended in 1 mL of buffer with 30 µL of pre-washed Dynabeads and incubated at 4°C for 20 min while rocking. Cell suspension was then increased to 5 mL by adding buffer. The tube was placed in a magnetic stand for 2 min. While in the stand, the supernatant which contained CD146− cells was collected and transferred to a new 15 mL tube and the cells left in the first tube were resuspended in 5 mL of buffer. Both tubes were placed in a magnetic stand to repeat the process three times to obtain pure populations of cells. CD146− cells were used for osteogenic induction and CD146+ cells were used as PLCs.
Osteogenic induction
CD146- hBM-MSCs from each donor (N = 2) from passages 4–5 were plated at 26,000 cells/cm2 in LG-DMEM supplemented with 10% FBS. Media was changed every two to three days. Once confluency was reached, media was switched to serum-containing LG-DMEM supplemented with 1% (v/v) each of 10−5 mol/L dexamethasone (D2915, Sigma-Aldrich, St. Louis, MO, USA) and 12 mmol/L ascorbic acid-2 phosphate (A8960, Sigma-Aldrich, St. Louis, MO, USA). Media was changed every three to four days until day 10 at which point 1% 200 mmol/L β-glycerophosphate (G9422, Sigma-Aldrich, St. Louis, MO, USA) was added to the media until day 21 of culture. Control hBM-MSCs continued to receive serum-containing LG-DMEM only. Differentiations were performed on transwell membranes with 3 µm pores for transwell co-cultures, on fibronectin-coated glass coverslips for immunofluorescent analysis, and on culture plates for migration assays and bone matrix calcium quantification.
Immunofluorescent staining
OLCs and control hBM-MSCs from each donor (N = 2) cultured on glass cover slips were rinsed with PBS (10010023, Gibco, Grand Island, NY, USA) and fixed with 4% paraformaldehyde (PFA; 15710, Electron Microscopy Sciences, Hatfield, PA, USA) for 15 min at room temperature followed by three PBS washes. They were then permeabilized with 0.25% Triton X-100 (1001976232, Sigma Aldrich, St. Louis, MO, USA) and blocked for 1 h with 5% normal goat serum (50062Z, Molecular Probes, Eugene, OR, USA) and 1% BSA. Samples were then incubated with Phalloidin (A12379, Invitrogen, Waltham, MA, USA) and DAPI (D3571, Molecular Probes, Eugene, OR, USA) diluted in 1% blocking buffer for 1 h at 37°C, then washed again three times. ProLong Platinum (P36970, Invitrogen, Waltham, MA, USA) was used as mounting medium and a fluorescence microscope was used to obtain images.
Bone matrix calcium quantification
Samples designated as OLCs or control hBM-MSCs from each donor (n = 3 per condition for each of the 2 donors) were rinsed with distilled water and fixed with 4% PFA for 15 min at room temperature. Wells were then rinsed again with distilled water and covered with 1% alizarin red (A5533, Sigma-Aldrich, St. Louis, MO, USA) solution for 1 h at room temperature. Afterwards, wells were washed for 15 min with distilled water until water came out clear. In order to elute the stain, 10% cetylpyridinium chloride (190177, MP Biomedicals, Solon, OK, USA) was added to each well for 1 h at room temperature. Elutions from each sample were plated in triplicate and absorbance at 584 nm was obtained using a plate reader.
Breast cancer cell culture and characterization
Triple negative primary human breast cancer tumor cells (cell identity 3384T, procured from the Live Tumor Culture Core, University of Miami) from passage 125 were cultured in BCMI-L media (Live Tumor Culture Core, University of Miami). Media was changed every two to three days and cells were characterized by phenotypic expressions of breast cancer-related markers. Briefly, 3384T were prepared as cell suspensions and stained with anti-human antibodies against E-cadherin, Ep-CAM (Molecular Probes, Eugene, OR, USA), vimentin, CD49f, N-cadherin, CXCR4 (Invitrogen, Waltham, MA, USA), and CD146 (Miltenyi, Bergisch Gladbach, Germany). Cells were subsequently washed twice and transferred into the wells of a 96-well plate for flow cytometry acquisition and analysis as described above.
Migration assay
3384T breast cancer cells were labeled with fluorescent membrane dye PKH26 (PKH26GL, Sigma Aldrich, St. Louis, MO, USA) according to manufacturer’s instructions, and seeded on ThinCert membranes for 24-well plates with 8 µm pores at 7500 cells/cm2 and cultured for 24 h before being placed in wells containing BCMI-L or wells with OLCs derived from each donor (N = 2) in BCMI-L media. Membranes were fixed and mounted on days 1, 3, and 5. Briefly, a cotton swab was used to wipe the apical side of the ThinCerts to remove any cells that had not migrated. Then the membranes were washed three times with PBS and fixed with 4% PFA for 10 min followed by another three washes with PBS. Membranes were then mounted using Prolong Platinum with DAPI. Three cell counts were performed manually by different individuals who counted three random fields of view at 20X in each of three membranes per condition per donor (n = 3 for control, experimental groups were pooled together to yield n = 6). Calculations included data obtained from both donors (n = 3 for each donor). Cells were required to express both PKH26 and DAPI to be included in the count.
Breast cancer cell proliferation
Proliferation of 3384T breast cancer cells was assessed in BCMI-L media, and in conditioned media collected from OLCs or PLCs (conditions labeled as 3384T + OLCCM and 3384T + PLCCM, respectively). For preparation of the conditioned media, OLCs and PLCs were cultured in BCMI-L media from each donor (N = 2) from passage 4–5 for 24 h. The 3384T breast cancer cells at passage 125 were plated in triplicate for each donor at low confluence and plates were transferred into an IncuCyte® ZOOM System for five days for time-lapse imaging and analysis of proliferation every 6 h using IncuCyte ZOOM’s Confluence Processing analysis tool. Data presented is the average of that obtained from both donors (n = 3 for control, n = 6 for experimental groups).
Transwell co-cultures
3384T breast cancer cells from passage 123 were seeded at 7500 cells/cm2. After 48 h transwell inserts were placed on the wells with either OLCs or PLCs (conditions labeled as 3384T + OLCTW and 3384T + PLCTW, respectively) from each donor (N = 2) from passage 4–5 (seeded at 7500 cells/cm2 48 h prior) which were cultured in BCMI-L media for 24 h (Day 0). The 3384T breast cancer controls were maintained in the wells without a transwell insert. On days 1 and 3, 3384T + OLCTW, 3384T + PLCTW, and 3384T were collected by incubation with 0.25% trypsin (J63688, Alfa Aesar, Haverhill, MA, USA) for 3 min at 37°C, neutralization with serum-containing DMEM and then centrifugation at 500g for 4 min. Supernatants were aspirated and RNAlater (AM7020, Invitrogen, Waltham, MA, USA) was added to each sample for long-term storage at −20°C until RNA extraction.
Conditioned media cultures
The 3384T breast cancer cells from passage 123 were seeded at 7500 cells/cm2. After 48 h media was changed to BCMI-L (3384T), PLC-conditioned BCMI-L (3384T + PLCCM), or OLC-conditioned BCMI-L (3384T + OLCCM) from each of two donors (Day 0). On days 1 and 3, 3384T, 3384T + PLCCM, and 3384T + OLCCM were collected and processed as described above.
Gene expression analyses
RNA was extracted from samples of each condition (3384T control, 3384T + OLCTW, 3384T + PLCTW, 3384T + OLCCM, and 3384T + PLCCM) using RNeasy Plus Mini Kit (74136, Qiagen, Germantown, MD, USA) and then used to synthesize cDNA using VILO Superscript cDNA Synthesis Kit (11754050, Invitrogen, Waltham, MA, USA) per manufacturers’ instructions. The resulting cDNA (1 µg) was used as template for qualitative real-time polymerase chain reaction (qRT-PCR) using SYBR GreenER kit (11762100, Invitrogen, Waltham, MA, USA) per manufacturer’s instructions to analyze mRNA expression levels of FGF13 (fibroblast growth factor 13) linked to cancer cell migration and proliferation, CD9 (motility-related protein 1) a marker of cancer cell motility, CD146 (melanoma cell adhesion molecule) which enables the binding of CXCR4 to Sdf-1 secreted by osteoblasts, CX43 (connexin 43) a breast cancer tumor suppressor, CXCR4 (C-X-C motif chemokine receptor 4) involved in migration and proliferation as well as being the ligand to Sdf-1 secreted by osteoblasts, CXCR2 (C-X-C motif chemokine receptor 2) linked to proliferation and formation of the metastatic niche, COL I (collagen I) involved in tumor progression, COL IV (collagen IV) which protects breast cancer cells from apoptosis, COL VI (collagen VI) a marker of tumor progression, CDH1 (E-cadherin) which is involved in invasion and cell differentiation in cancer cells, CDH2 (N-cadherin) linked to migration and proliferation of cancer cells, CDH11 (cadherin-11) involved in cancer cell migration and invasion, VIM (vimentin) a marker of cancer cell migration, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a housekeeping control. The primers used in this investigation are listed in Supplementary Table S1. Samples were performed in triplicate and all Ct values were normalized using GAPDH as a housekeeping gene and fold expressions were calculated compared to 3384T control group using the ΔΔCt method.
Additionally, gene expression profiles of day 3 3384T + OLCTW and 3384T + PLCTW were obtained using RT2 Profiler PCR Arrays specified for cancer stem cells (PAHS-176Z, Qiagen, Germantown, MD, USA). According to the manufacturer’s instructions, qRT-PCR was performed on each sample by combining 1 µg of cDNA with associated SYBR Green Mastermix (330503, Qiagen, Germantown, MD, USA) and Invitrogen™ UltraPure™ Distilled Water (10977015, Invitrogen, Waltham, MA, USA) which was then loaded into the wells of a profiler plate and loaded into a CFX Connect™ Real-Time PCR Detection System thermocycler (BioRad, Hercules, CA, USA). All Ct values were uploaded into Qiagen’s Gene Analysis Center for normalization using all included housekeeping genes. Relative gene expressions were normalized to 3384T control group and represented as clustergrams for all genes.
All gene expression analyses include n = 2 for controls and n = 4 for experimental groups, two from each donor, where n indicates biological replicates.
Statistical analyses
Results are represented as mean ± standard error mean for all analyses using Prism v8 software (GraphPad, San Diego, CA, USA). Data were analyzed using a two-tailed unpaired t-test, with statistical significance defined as P ≤ 0.05. Additionally, outliers from the migration assay were identified and removed using ROUT with Q = 1%.
Results
hBM-MSC characterization
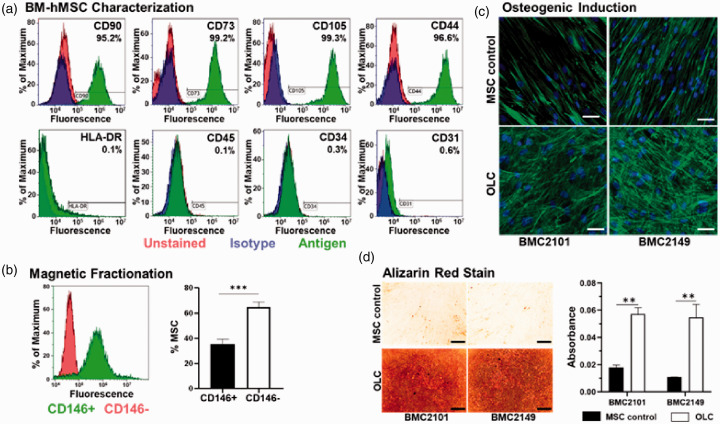
To confirm mesenchymal phenotype, hBM-MSCs from the two healthy young female donors were immune-typified using flow cytometry. The combination of both cell populations was positive (>95%) for CD90, CD73, CD44, and CD105, as well as negative (<5%) for CD34, CD45, CD31, and HLA-DR (Figure 2(a)), confirming their MSC phenotypes.
Figure 2.
Phenotype of primary hBM-MSCs and osteogenic induction. (a) Surface antigen expression of markers (CD90, CD73, CD105, and CD44) demonstrated the basal expression of primary hBM-MSCs. Additional, non-mesenchymal, markers (HLA-DR, CD45, CD34, and CD31) were negative. (b) Magnetic fractionation based on the presence (green) or absence (red) of CD146; and quantification of CD146+ cells and CD146− cells post-fractionation (results from both patients were pooled, n = 6). (c) hBM-MSCs from two young healthy donors underwent osteogenic induction to obtain OLCs. OLCs obtained presented more cuboidal, spread-out morphology compared to the spindle-like hBM-MSCs. (d) Significantly higher levels of calcium in OLCs matrix as revealed by alizarin red stain (n = 3 for each donor, scale bars represent 300 µm). ** indicates P < 0.01, *** indicates P < 0.001. (A color version of this figure is available in the online journal.)
Figure 1.
Experimental design. hBM-MSCs were isolated from human bone marrow and fractionated according to CD146 presence. CD146− cells were induced into an osteogenic phenotype (OLCs) and CD146+ cells were determined to be pericyte-like cells (PLCs). Transwell migration assays were performed to determine if 3384T were attracted to OLCs. Additionally, proliferation assays were performed where 3384T breast cancer cells were cultured for five days in OLCCM and PLCCM. Finally, transwell co-cultures of each OLCTW and PLCTW were performed with 3384T breast cancer cells as well as cultures of 3384T breast cancer cells in OLCCM and PLCCM. The 3384T cells were collected on day 1 and day 3 for gene expression interrogation. (A color version of this figure is available in the online journal.)
PLC and OLC attainment
hBM-MSCs were fractionated by MACS to obtain two subpopulations: CD146+ and CD146−. CD146+ hBM-MSCs were defined as PLCs while CD146− hBM-MSCs were used to induce osteogenesis to obtain OLCs. Post-fractionation cell counts from both donors show CD146+ cells accounted for 35.27% of all MSCs while CD146− cells made up the remaining 64.7%. Flow cytometry results (Figure 2(b)) indicated that 95.98% of cells in the PLC population expressed CD146 and no CD146− cells were detected. Immunofluorescent staining was performed to confirm osteoblast-like morphology and revealed that, after osteogenic induction, OLCs present a more spread, cuboidal phenotype when compared to control hBM-MSCs which maintain spindle-like morphology (Figure 2(c)). Alizarin red staining, used to determine the capacity of OLCs to function properly, showed significantly more calcium deposition in OLC ECM than in that of the control hBM-MSCs (Figure 2(d)), suggesting that the OLCs obtained were functional ECM-depositing osteoblastic cells.
Breast cancer cell characterization
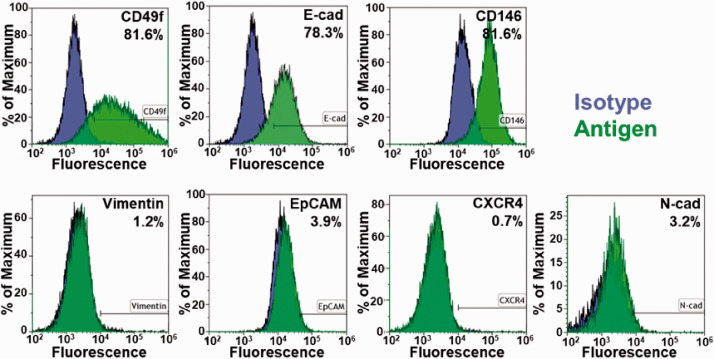
Given that these are primary human cancer cells, surface antigen expression of markers was analyzed to determine the basal expression of the 3384T breast cancer cells (Figure 3). EpCAM (3.93%), N-cadherin (3.22%), CXCR4 (0.69%), and vimentin (1.22%) were minimally expressed while E-cadherin (78.27%), CD146 (81.55%), and CD49f (81.62%) were highly expressed. The results indicate that 3384T breast cancer cells are highly aggressive, with migratory potential, and considerable chemoresistance.26–30
Figure 3.
Phenotype of primary breast cancer cells (3384T). Surface antigen expression of markers related to epithelial-to-mesenchymal transition (EpCAM, E-cadherin, N-cadherin, vimentin) demonstrated the basal expression of primary breast cancer cells. Additional markers of interest were used to investigate the mechanisms implicated in BCC aggressiveness (CD146, CD49f), migration (CXCR4, CD146), and chemoresistance (CD49f). (A color version of this figure is available in the online journal.)
3384T breast cancer cell migration
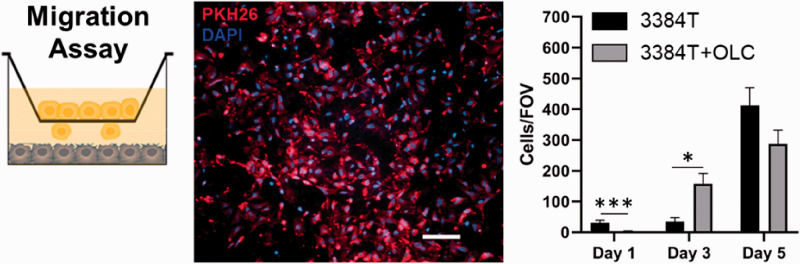
Given the osteotropic nature of breast cancer CTCs, we performed a migration assay to determine whether OLCs would draw 3384T breast cancer cells. The cell counts (n = 3 for control, n = 6 for 3384T cultured in presence of OLCs) show that on day 1, 3384T breast cancer cells cultured in BCMI-L media alone migrated significantly more than those in the presence of OLCs, while on day 3, 3384T cultured with OLCs presented increased migration when compared to controls cultured in BCMI-L media alone (Figure 4). Overall, no correlation could be found, suggesting the migratory effects may be time dependent.
Figure 4.
Migration assay showed time-dependent effect. The 3384T stained with PKH26 (red) migrated through ThinCert pores and membranes were fixed, stained with DAPI (blue) and mounted on days 1, 3, and 5. Cells presenting both stains were counted (n = 3 for control, n = 6 for experimental group). Scale bar represents 100 µm. (A color version of this figure is available in the online journal.)
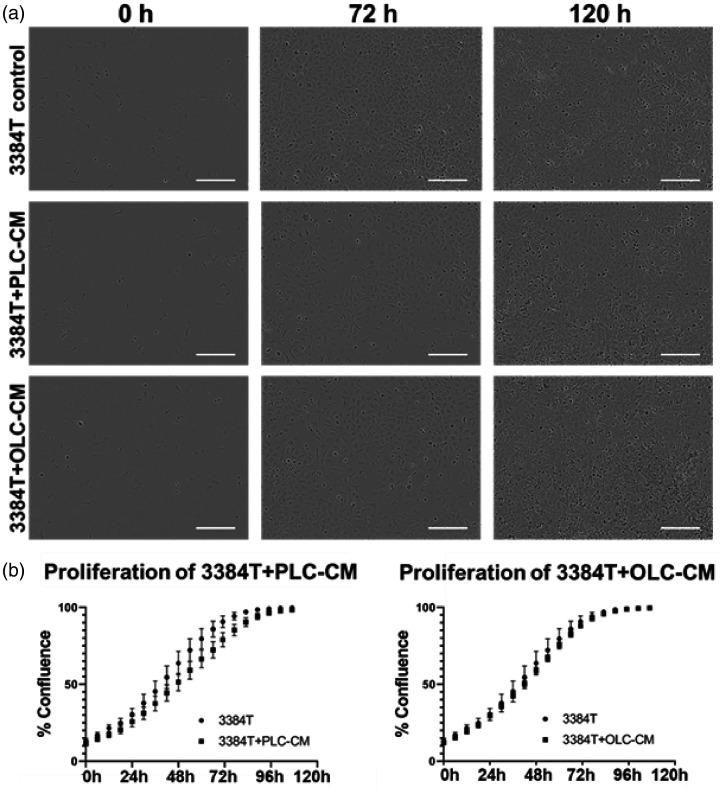
Breast cancer cell proliferation
3384T breast cancer cells (n = 3), 3384T + OLCCM (n = 6), and 3384T + PLCCM (n = 6) were used to determine proliferative changes as a result of the conditioned media in an effort to establish whether OLCs or PLCs would initiate a tumorigenic-like phenotype in 3384T breast cancer cells (Figure 5). Although not statistically significant, proliferation rates suggest that 3384T breast cancer cells proliferated slower in the presence of PLCCM compared to the control. On the other hand, cells incubated with OLCCM did not appear to have altered proliferation patterns (Figure 5(b)). In both cases, the biggest difference between groups was found around the 54 h mark.
Figure 5.
Proliferation assay. Assessment of growth kinetic effects in 3384T breast cancer cells cultured in PLCCM and OLCCM. (a) Representative images obtained from IncuCyte® ZOOM System at 0, 72, and 120 h for each condition (scale bars represent 50 µm). (b) Growth kinetics of 3384T, 3384T + OLCCM, and 3384T + PLCCM in the course of five days (n = 3 for control, n = 6 for experimental groups).
Gene expression responses
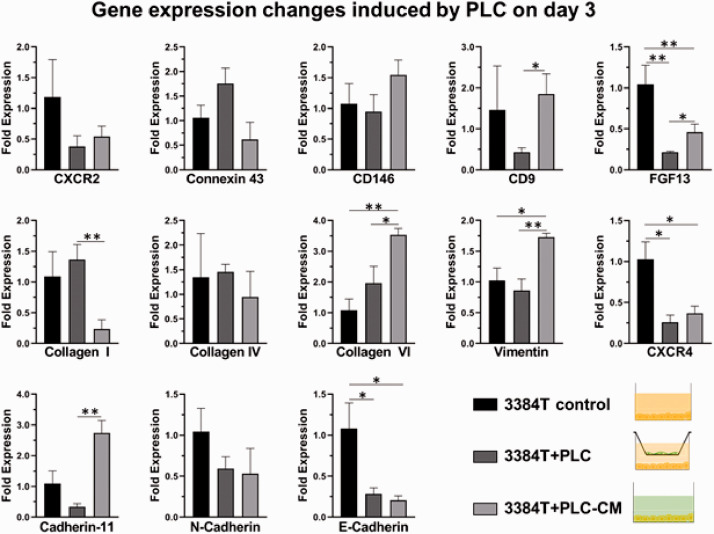
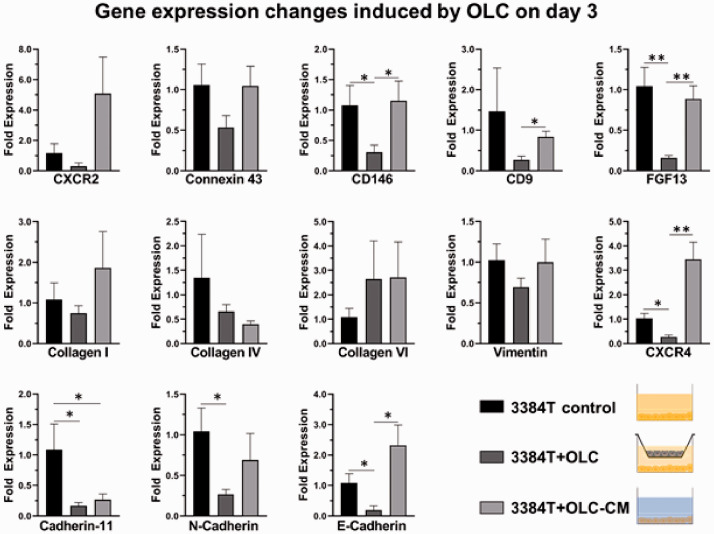
Osteoblasts and pericytes have a key role in the extravasation and metastasis of breast cancer cells. In order to specifically determine each of their roles, genetic expression changes were used to determine the effects of OLCs and PLCs, as well as their respective conditioned media, on 3384T breast cancer cells. RT-qPCR data from day 1 (Supplementary Figures S1 and S2) did not suggest clear effects, while data derived from day 3 cultures showed significant changes. In the case of 3384T + PLCTW and 3384T + PLCCM, the following results were observed (Figure 6): both conditions present low expression of E-cadherin, CXCR2, CXCR4, and FGF13; suggesting de-differentiation of the cancer cells and increased invasion. On the other hand, they present high expression of collagen VI, linked to tumor progression. Furthermore, 3384T + PLCTW presented low expression of CD9 and increased expression of Collagen I, while 3384T + PLCCM underwent upregulation of Vimentin and Cadherin-11; all of which have a key role in migration and invasion. Additionally, 3384T + OLCTW and 3384T +OLCCM showed decreased expression of Cadherin-11 and N-cadherin in 3384T breast cancer cells which relates to decreased migration and invasion, as well as increased expression of Collagen VI, a tumor progression marker. 3384T + OLCTW also underwent downregulation of CD9, FGF13, CD146, CXCR4, and CX43, indicating decreased migration and increased tumor potential, while culturing 3384T + OLCCM increased expression of CXCR2 which is linked to tumor niche formation (Figure 7).
Figure 6.
PLCs induced genetic expression shifts after three days. RT-qPCR results revealed altered gene expression levels in 3384T breast cancer cells cultured for three days with PLCTW and PLCCM (3384T control n = 2, all others n = 4). * indicates P < 0.05, ** indicates P < 0.01. (A color version of this figure is available in the online journal.)
Figure 7.
OLCs induced genetic expression shifts after three days. RT-qPCR results revealed altered gene expression levels in 3384T breast cancer cells cultured for three days with OLCTW and OLCCM (3384 T control n = 2, all others n = 4). * indicates P < 0.05, ** indicates P < 0.01. (A color version of this figure is available in the online journal.)
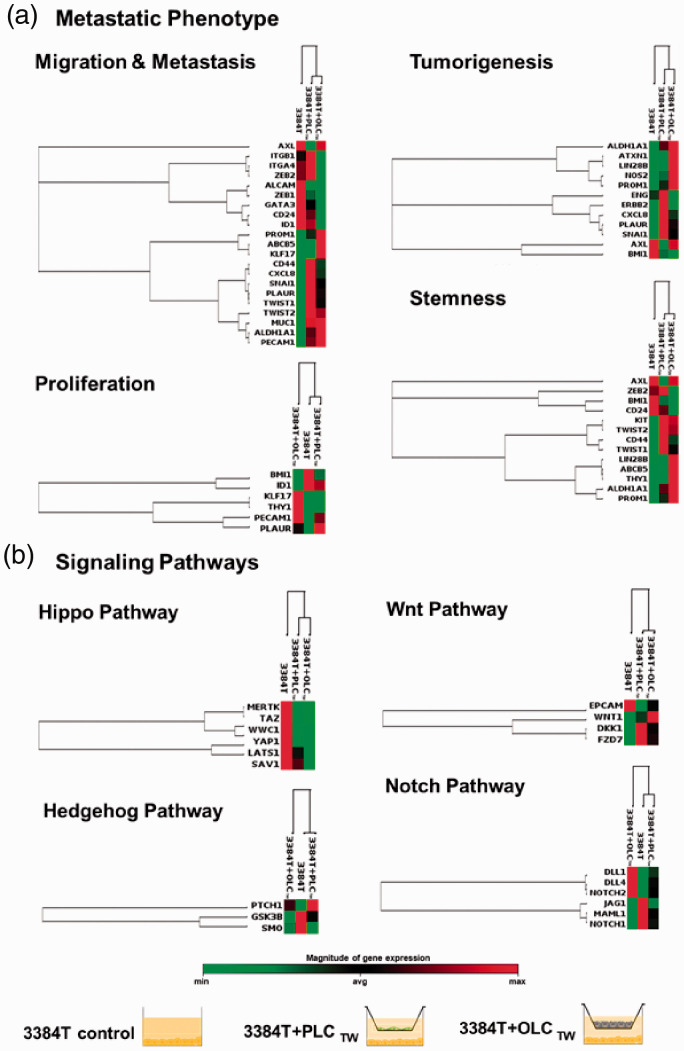
Additionally, RT2 Profiler PCR Arrays specified for cancer stem cells were used to determine gene expression profiles of day 3 3384T + OLCTW and 3384T + PLCTW. Genes were categorized according to function and the data were arranged using unsupervised hierarchal clustering. In the panels related to metastatic behavior (Figure 8(a)) 3384T + PLCTW presented substantial expression of genes related to increased migration (ITGA4, ZEB2, ALCAM, GATA3, PROM1, ABCB5, CXCL8, SNAI1, TWIST1, TWIST2, MUC1, ALDH1A1, PECAM1); as well as a downregulation of CD24 and upregulation of CD44, also linked to invasion and breast cancer stem cells.31–33 ITGB1 and PLAUR were upregulated, which are markers of blood vessel remodeling.34–37 The majority of tumorigenesis markers (ALDH1A1, PROM1, NOS2, ERBB2, CXCL8, PLAUR, SNAI1, and ENG) were considerably upregulated. While expression of AXL and LIN28B, cancer stem cell markers, was downregulated; ZEB2, KIT, TWIST1, TWIST2, ABCB5, ALDH1A1, and PROM1 were upregulated. Lastly, with regards to proliferation markers, there was downregulation of BMI1, IDM1, as well as upregulation of PECAM1 which is a proliferation suppressor.38
Figure 8.
Transcription profile changes. Unsupervised hierarchical clustering of genes shows altered expression of genes associated with (a) migration and metastasis, proliferation, tumorigenesis, and stemness, as well as of genes associated with (b) signal transduction in the Hippo, Hedgehog, Wnt, and Notch pathways. The three culture conditions (3384T, 3384T + PLCTW, 3384T + OLCTW at day 3) are represented on the top of each cluster (3384T control n = 2, all others n = 4). (A color version of this figure is available in the online journal.)
The 3384T + OLCTW underwent marked expression changes when compared to the control (Figure 8(a)). It presented mixed expression alterations in the migration and metastasis panel, most likely because these genes are also involved in processes related to the establishment of a secondary tumor and stemness.37–50 The majority of the changes seen in the proliferation panel (upregulation of KLF7, THY1, PECA1, and PLAUR as well as downregulation of BMI1 and ID1) are linked to decrease in proliferation. Although the tumorigenesis panel shows two downregulated genes (ENG, BMI1), the rest were upregulated. Finally, 3384T + OLCTW presented upregulation of the majority of markers involved in stemness (except for ZEB2, BMI1, and CD24) and downregulation of CD24 coupled with upregulation of CD44, which is an important indicator of cancer stem cells.
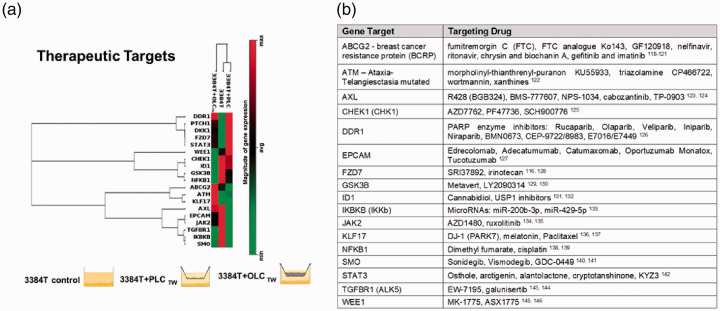
Regarding signal transduction effects, 3384T + OLCTW and 3384T + PLCTW presented a downregulation of Hippo and Hedgehog pathways as well as the Wnt pathway (Figure 8(b)) through inhibition by DKK1. On the other hand, the Notch pathway had a combination of upregulated and downregulated ligands and receptors (Figure 8(b)). Additionally, RT2 Profiler PCR Arrays also yielded possible therapeutic targets (Figure 9(a)) based on specific altered gene expressions including the upregulation of DDR1, PTCH1, DKK1, FZD7, STAT3, and ATM, as well as the downregulation of CHEK1, ID1, GSK3B, NFKB1, ABCG2, AXL, EPCAM, JAK2, TGFBR1, IKBKB, and SMO in 3384T cultured in the presence of both OLCs and PLCs. KLF17 was upregulated in 3384T + PLCTW and WEE1 was downregulated by PLCs while upregulated by OLCs.
Figure 9.
Transcription profile of genes involved in critical signaling pathways and drug targets. (a) Unsupervised hierarchical clustering of genes shows altered gene expression of cancer therapy drug targets. The three culture conditions (3384T, 3384T + PCLTW, 3384T + OCLTW at day 3) are represented on the top of each cluster (3384 T control n = 2, all others n = 4). (b) Candidate drugs that have been evaluated in pre-clinical to clinical trials which target genes with altered expression in transwell co-cultures or conditioned media conditions. (A color version of this figure is available in the online journal.)
Discussion
Investigation of the paracrine interactions between the endosteal niche cellular components and invading breast cancer CTCs is paramount to understanding the various metastatic cascade steps, and secondarily to design novel therapies. Herein, we show that two important cell populations that are part of the endosteal niche, perisinusoidal pericytes and osteoblasts, represented by PLCs and OLCs, respectively, distinctively participate inducing phenotypic changes in invading CTCs. The exclusive use of human primary cells allowed in vitro studies that more closely resemble human physiology than previously reported. Specifically, we used primary hBM-MSCs to derive PLCs and OLCs as well as primary breast cancer tumor cells which according to surface antigen expression were highly aggressive cells with migratory potential and considerable potential chemoresistance.26–30 PLCs and OLCs induced distinct phenotypes in primary breast cancer cells 3384T. PLCs as well as PLCCM increased motility and invasion potential in 3384T breast cancer cells while OLCs appeared to induce a dormant phenotype in 3384T breast cancer cells, decreasing markers of migration and proliferation. These phenotypic changes mirror the behavior or CTCs in vivo as they initially migrate through the blood vessel wall while exposed to perivascular cells pre-extravasation and then enter a quiescent stage once in the bone parenchyma post-extravasation while in close contact with osteoblasts. Additionally, both OLCs and PLCs downregulated genes in the Hippo pathway, shielding cancer cells from apoptosis, while upregulating cancer stem cell and tumorigenesis markers.
Effects of PLCs on 3384T breast cancer cells
3384T + PLCTW experienced a considerable change in gene expression (ITGA4, ZEB2, ALCAM, GATA3, PROM1, ABCB5, CXCL8, SNAI1, TWIST1, TWIST2, MUC1, ALDH1A1, PECAM1, E-cadherin, CD9, and collagen I) related to increased migration, invasion, metastasis, and aggressiveness.28,29,38,42,51–71 Additionally, these cells have downregulation of CD24 and upregulation of CD44 which is also linked to invasion and distant metastasis.32,33 Interestingly, collagen VI, ITGB1, and PLAUR were upregulated, indicating an increase in markers associated with vessel remodeling during the extravasation process.34–37 These findings correlate with the metastatic stage where CTCs interact with PLCs. During extravasation, CTCs migrate through the blood vessel wall directly affecting its composition and causing vascular leakiness72 in order to invade the bone.
While there was some decreased expression of a few tumorigenesis markers, including ATXN1, LIN28B, AXL, and BMI1,40,73–75 in 3384T + PLCTW, most of them (ALDH1A1, PROM1, NOS2, ERBB2, CXCL8, PLAUR, SNAI1, and ENG) were greatly upregulated, which indicates increased tumorigenic phenotype, neovascularization, and tissue remodeling.37,44,46,47,50,63,76,77 This phenotypic change suggests that cells in contact with PLC are being primed to form distant metastasis.
When looking into stemness markers in 3384T + PLCTW, even though two of the cancer stem cell markers (AXL and LIN28B)40,78 were downregulated; ZEB2, KIT, TWIST1, TWIST2, ABCB5, ALDH1A1, and PROM1, involved in self-renewal, de-differentiation, and the establishment of cancer stem cells,45,48–50,79–81 were all significantly upregulated. At the same time, there was E-cadherin downregulation which is linked to loss of differentiation.28,29 Additionally, these cells presented an upregulation of CD44 and downregulation of CD24. CD44+/CD24− have been shown to correlate with cancer-initiating cells,39 and more specifically, with breast cancer stem cells.31 Altogether this expression shift indicates that 3384T + PLCTW have increased stem properties in line with initiating and maintaining a secondary tumor once they reach the endosteal niche.
The 3384T + PLCTW presented an overall decrease in proliferation, according to marker expression. There was downregulation of BMI1, IDM1, CXCR2, and CXCR4, all linked to cell proliferation,82–86 as well as upregulation of PECAM1 which is a proliferation suppressor.38 While PLAUR, which is also associated with increased proliferation, was upregulated, it is a critical marker for migration and matrix remodeling as well, which are key aspects of extravasation.37 Previous studies87–90 have shown that tumor cells present decreased expression of genes related to proliferation when increasing expression of motility and invasion genes. Additionally, findings indicate that 3384T present with dedifferentiated phenotype (most notably due to the downregulation of E-cadherin28,29) which has been determined to be associated with increased invasion and migration.51,52
In order to isolate the effect of PLC on 3384T, conditioned media experiments were performed. The 3384T + PLCCM presented low expression of E-cadherin, linked to loss of differentiation and increased invasion.28,29 The 3384T + PLCCM presented upregulated expression of Collagen VI, involved in blood vessel remodeling, to a higher degree than 3384T + PLCTW. While 3384T + PLCCM did not experience transcriptional changes in CD9, or Collagen I like 3384T-PLCTW did, they did present increased expression of Vimentin and Cadherin-11, which are linked to migration and invasion to bone.28,91–95 Overall, these differences from 3384T + PLCTW suggest that although PLC cytokines activate metastatic genotypes in cancer cells, in the presence of cancer cells, PLCs attenuate their invasion potential. In addition, and similarly to 3384T + PLCTW, CXCR2 and CXCR4 were downregulated in 3384T + PLCCM, indicating a decrease in cell proliferation.82–84 This was consistent with the slight decrease observed in the proliferation assay which presented the largest decrease between 54 and 72 h, coinciding with the time point at which these RT-qPCR and RT2 samples were obtained (Figure 4).
Effect of OLC on 3384T breast cancer cells
3384T + OLCTW presented overall upregulation of tumorigenesis and cancer stem cell markers, suggesting that OLCs modify the cancer cells to initiate a tumor, establish the metastatic site by remodeling the surrounding ECM, and initiating angiogenic and neovascularization processes. In addition to the stemness and tumorigenesis markers previously mentioned, 3384T + OLC presented an upregulation of Collagen VI linked to tumor progression and inhibition of cancer cell apoptosis.34,96
The 3384T-OLCTW transcription level from the profiler panel related to migration and metastasis (Figure 8(a)) appears to not have a clear trend; however, a closer look reveals that the upregulated markers related to increased migration are also critical in the establishment of a secondary tumor and are linked to processes such as colonization, adaptation to a foreign environment, tumorigenesis, recapitulation of the primary tumor, skeletal metastasis initiation, and stemness.37–50 This could be the reason why our representative migration assay demonstrated no clear trend and the migration of 3384T, which reinforces the complexity of migratory pathways where some mobility markers were upregulated while others were downregulated.
Most expression changes seen in the proliferation panel (Figure 8(a)) yield a decrease in proliferation except for PLAUR, which is related to matrix remodeling.37 In addition, there was a downregulation in expression of Cadherin-11, N-cadherin, FGF13, CD146, and CXCR4, which are all associated to migration, invasion, and proliferation.27,84,97–103 CD9 was downregulated suggesting increased malignancy,55,56 even though there have been reports of increased CD9 in osteotropic breast cancer cells.104 Thus, OLCs appear to induce a quiescent phenotype in 3384T breast cancer cells, decreasing markers of migration and proliferation. The adoption of a quiescent phenotype agrees with the literature as osteoblasts play a critical role in the induction of dormancy in post-extravasation cancer cells.105
To isolate the effect of OLCs on 3384T breast cancer cells, conditioned media experiments were performed. The most important differences that were found between 3384T + OLCTW and 3384T + OLCCM relate to migration and proliferation markers which were downregulated in 3384T + OLCTW but to a lesser degree or not at all in 3384T + OLCCM (CD146, CD9, FGF13, N-cadherin). This difference could explain why the proliferation assay did not result in reduced proliferation of 3384T + OLCCM, as well as the fact that the biggest difference found in the proliferation assay took place between 42 and 54 h. Nevertheless, there was an upregulation of CXCR2, which is involved in the establishment and maintenance of the metastatic niche most likely by promoting angiogenesis.106,107 Collagen I and CX43, both of which promote survival of cancer cells, were upregulated when compared to 3384T + OLCTW. Additionally, increased expression of E-cadherin in 3384T + OLCCM when compared to the other two groups could indicate a reversal to an epithelial phenotype. Lastly, we speculate that the significant upregulation of CXCR4 could be a response to Sdf-1 secreted by OLCs. Altogether, the differences in 3384T + OLCCM suggest, much like in 3384T + PLCCM, that in the absence of osteoblast-like cancer cells present a more aggressive genotype and that the presence of OLCs attenuate their metastatic potential.
PLC and OLC modulate signaling pathways in 3384T breast cancer cells
PLCs and OLCs induced important changes in the key cell signaling pathways Hippo, Hedgehog, Wnt, and Notch. Under both co-culture conditions, 3384T breast cancer cells show decreased Hippo pathway gene expression which in turn protects cancer cells from apoptosis.108 Both 3384T + PLC and 3384T + OLC presented inactivated Hedgehog as seen by the downregulation of GSK3β109 as well as the increase in PTCH1 which downregulates SMO, thereby decreasing cancer cell proliferation.110,111 PLCs and OLCs also had a similar effect on Wnt, given that DKK1 is Wnt negative regulator112 and the increase of WNT1 degrades β-catenin which also downregulates EPCAM resulting in decreased cell proliferation.113,114 Finally, changes in Notch ligand and receptor expression as well as the signal transducer MAML1 were observed in the Notch pathway. Some of these changes have been previously reported to have specific effects on the pathway modulating properties related to metastasis. DLL4 upregulation has been linked to vascular remodeling and angiogenesis,115,116 NOTCH2 upregulation is associated with cancer stem cell phenotype,115 while decreased expression of JAG1and NOTCH1 inhibits proliferation.117,118 Therefore, the overall effect that PLCs and OLCs had on 3384T essential cell pathways was one of protection of cancer cells, decreased proliferation and increased tumorigenesis, falling in line with what was discussed above.
Clinical relevance
Clinically relevant cancer therapeutic targets were also dramatically altered by PLCs and OLCs (Figure 9(a)). These genes have been previously identified, and are currently under pre-clinical and clinical investigation with therapeutic inhibitors117 (Figure 9(b)). Discovering preliminary concordance between the genes regulated in our study, and the genes that are being targeted by candidate drugs in pre-clinical to clinical development provides initial validation that our PLC and OLC transwell co-culture set up could be employed to discover more novel targets. In the future, these interactions can be sequentially studied using organ-on-chip technology which can integrate both PLCs and OLCs in the same system in a more physiologically relevant manner in order to better understand the compounded effect of these critical cell populations as well as additional ones such as endothelial cells or cancer-associated fibroblasts. Additionally, microfluidics could allow for cancer cells to be circulated in a tissue-engineered endosteal niche, much like CTCs do in vivo, in order to track them throughout the metastatic process, exposing them to individual critical cell populations sequentially or at the same time. This would enable the elucidation of cell–cell and paracrine interactions that drive the establishment of the metastatic niche and eventually, the stimuli that drive the colonizing cells to proliferation and formation of the secondary tumor.
Conclusions
In this study, we recreated the endosteal niche using primary human cells in order to understand the effect of two of its most important cell populations (perivascular cells and osteoblasts) on breast cancer metastasis and the formation of the secondary tumor in bone. Overall, the results indicate that primary hBM-MSC-derived perivascular-like cells drive 3384T cancer cells to a more invasive, migratory phenotype, while osteoblast-like cells drive them to quiescence and the establishment of the metastatic niche, recapitulating cancer cell behavior in vivo. These data provide a baseline to support the thesis that phenotypic response from cancer cells can be controlled by neighboring cells, which could explain why CTCs choose specific sites to extravasate. While this study elucidated the individual effects of each endosteal cell population, an organ-on-chip approach would allow the interrogation of a compounded effect with two or more different cell types that play a part in the extravasation of breast cancer cells into, as well as their establishment of a secondary site in, bone. This approach would also allow for the sequential exposure of cancer cells to different critical cell populations as it occurs in vivo (first they are exposed to pericytes, then to osteoblasts) enabling a high fidelity recapitulation of the metastatic niche in order to gain insight into the metastatic process as well as uncovering new therapy targets.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220971599 for Human-derived osteoblast-like cells and pericyte-like cells induce distinct metastatic phenotypes in primary breast cancer cells by Vera Mayo, Annie C Bowles, Laura E Wubker, Ismael Ortiz, Albert M Cordoves, Richard J Cote, Diego Correa and Ashutosh Agarwal in Experimental Biology and Medicine
Footnotes
Authors’ contributions: VM, AA, DC, and RJC conceived and designed study; VM, ACB, LEW, IO, and AMC performed experiments; VM and ACB analyzed and interpreted data; DC provided critical cells; VM wrote the manuscript; RJC, DC, and AA provided critical revisions. All authors read and approved the final manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was partially supported by the National Cancer Institute of the National Institutes of Health under Award Number U01CA233363 (AA and RJC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ORCID iDs: Ismael Ortiz https://orcid.org/0000-0002-5214-567X
Diego Correa https://orcid.org/0000-0002-9004-4300
Ashutosh Agarwal https://orcid.org/0000-0002-2397-9886
SupplementAL MATERIAL: Supplemental material for this article is available online.
REFERENCES
- 1.Spano D, Heck C, De Antonellis P, Christofori G, Zollo M. Molecular networks that regulate cancer metastasis. Seminars in Cancer Biology 2012; 22:234–49 [DOI] [PubMed] [Google Scholar]
- 2.Crisan M, Yap S, Casteilla L, Chen C-W, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 2008; 3:301–13 [DOI] [PubMed] [Google Scholar]
- 3.Correa D, Somoza RA, Lin P, Schiemann WP, Caplan AI. Mesenchymal stem cells regulate melanoma cancer cells extravasation to bone and liver at their perivascular niche. Int J Cancer 2016; 138:417–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Correa D. Mesenchymal stem cells during tumor formation and dissemination. Curr Stem Cell Rep 2016; 2:174–82 [Google Scholar]
- 5.Caplan AI, Correa D. PDGF in bone formation and regeneration: new insights into a novel mechanism involving MSCs. J Orthop Res 2011; 29:1795–803 [DOI] [PubMed] [Google Scholar]
- 6.Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 2009; 9:274–84 [DOI] [PubMed] [Google Scholar]
- 7.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006; 8:315–7 [DOI] [PubMed] [Google Scholar]
- 8.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003; 3:537–49 [DOI] [PubMed] [Google Scholar]
- 9.Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, Moretti M, Kamm RD. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 2014; 35:2454–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao S, Ha L, Cheng G, Wan Y, Xia Y, Sosnoski DM, Mastro AM, Zheng SY. A spontaneous 3D bone-on-a-chip for bone metastasis study of breast cancer cells. Small 2018; 14:1702787. [DOI] [PubMed] [Google Scholar]
- 11.Torisawa Y-S, Spina CS, Mammoto T, Mammoto A, Weaver JC, Tat T, Collins JJ, Ingber DE. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat Methods 2014; 11:663. [DOI] [PubMed] [Google Scholar]
- 12.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc Natl Acad Sci U S A 2015; 112:214–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marturano-Kruik A, Nava MM, Yeager K, Chramiec A, Hao L, Robinson S, Guo E, Raimondi MT, Vunjak-Novakovic G. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc Natl Acad Sci U S A 2018; 115:1256–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samatov TR, Shkurnikov MU, Tonevitskaya SA, Tonevitsky AG. Modelling the metastatic cascade by in vitro microfluidic platforms. Prog Histochem Cytochem 2015; 49:21–9 [DOI] [PubMed] [Google Scholar]
- 15.Arrigoni C, De Luca P, Gilardi M, Previdi S, Broggini M, Moretti M. Direct but not indirect co-culture with osteogenically differentiated human bone marrow stromal cells increases RANKL/OPG ratio in human breast cancer cells generating bone metastases. Mol Cancer 2014; 13:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czekanska E, Stoddart M, Richards R, Hayes J. In search of an osteoblast cell model for in vitro research. Eur Cell Mater 2012; 24:1–17 [DOI] [PubMed] [Google Scholar]
- 17.Lefley D, Howard F, Arshad F, Bradbury S, Brown H, Tulotta C, Eyre R, Alférez D, Wilkinson JM, Holen I. Development of clinically relevant in vivo metastasis models using human bone discs and breast cancer patient-derived xenografts. Breast Cancer Res 2019; 21:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxena M, Christofori G. Rebuilding cancer metastasis in the mouse. Mol Oncol 2013; 7:283–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Jong M, Maina T. Of mice and humans: are they the same? – implications in cancer translational research. J Nucl Med 2010; 51:5010–4 [DOI] [PubMed] [Google Scholar]
- 20.Caplan AI. Mesenchymal stem cells: cell–based reconstructive therapy in orthopedics. Tissue Eng 2005; 11:1198–211 [DOI] [PubMed] [Google Scholar]
- 21.Caplan AI. Mesenchymal stem cells. J Orthop Res 1991; 9:641–50 [DOI] [PubMed] [Google Scholar]
- 22.Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 2007; 131:324–36 [DOI] [PubMed] [Google Scholar]
- 23.Kolbe M, Xiang Z, Dohle E, Tonak M, Kirkpatrick CJ, Fuchs S. Paracrine effects influenced by cell culture medium and consequences on microvessel-like structures in cocultures of mesenchymal stem cells and outgrowth endothelial cells. Tissue Eng Part A 2011; 17:2199–212 [DOI] [PubMed] [Google Scholar]
- 24.Tormin A, Li O, Brune JC, Walsh S, Schütz B, Ehinger M, Ditzel N, Kassem M, Scheding S. CD146 expression on primary nonhematopoietic bone marrow stem cells is correlated with in situ localization. Blood 2011; 117:5067–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiozawa Y, Pedersen EA, Havens AM, Jung Y, Mishra A, Joseph J, Kim JK, Patel LR, Ying C, Ziegler AM. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J Clin Invest 2011; 121:1298–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye F, Qiu Y, Li L, Yang L, Cheng F, Zhang H, Wei B, Zhang Z, Sun L, Bu H. The presence of EpCAM−/CD49f+ cells in breast cancer is associated with a poor clinical outcome. J Breast Cancer 2015; 18:242–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Yan X. CD146, a multi-functional molecule beyond adhesion. Cancer Lett 2013; 330:150–62 [DOI] [PubMed] [Google Scholar]
- 28.Pishvaian MJ, Feltes CM, Thompson P, Bussemakers MJ, Schalken JA, Byers SW. Cadherin-11 is expressed in invasive breast cancer cell lines. Cancer Res 1999; 59:947–52 [PubMed] [Google Scholar]
- 29.Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res 2001; 3:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Miragaya J, González-Suárez E. Tumor-initiating CD49f cells are a hallmark of chemoresistant triple negative breast cancer. Mol Cell Oncol 2017; 4:e1338208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res 2008; 10:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R, Badve S, Nakshatri H. CD44+/CD24-breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res 2006; 8:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24−/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res 2005; 11:1154–9 [PubMed] [Google Scholar]
- 34.Chen P, Cescon M, Bonaldo P. Collagen VI in cancer and its biological mechanisms. Trends Mol Med 2013; 19:410–7 [DOI] [PubMed] [Google Scholar]
- 35.Brakebusch C, Fässler R. β 1 integrin function in vivo: adhesion, migration and more. Cancer Metastasis Rev 2005; 24:403–11 [DOI] [PubMed] [Google Scholar]
- 36.Kurozumi A, Goto Y, Matsushita R, Fukumoto I, Kato M, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T. Tumor-suppressive micro RNA-223 inhibits cancer cell migration and invasion by targeting ITGA 3/ITGB 1 signaling in prostate cancer. Cancer Sci 2016; 107:84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayanaswamy PB, Baral TK, Haller H, Dumler I, Acharya K, Kiyan Y. Transcriptomic pathway analysis of urokinase receptor silenced breast cancer cells: a microarray study. Oncotarget 2017; 8:101572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Righi L, Deaglio S, Pecchioni C, Gregorini A, Horenstein AL, Bussolati G, Sapino A, Malavasi F. Role of CD31/platelet endothelial cell adhesion molecule-1 expression in in vitro and in vivo growth and differentiation of human breast cancer cells. Am J Pathol 2003; 162:1163–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003; 100:3983–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gjerdrum C, Tiron C, Høiby T, Stefansson I, Haugen H, Sandal T, Collett K, Li S, McCormack E, Gjertsen BT. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci 2010; 107:1124–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holland SJ, Pan A, Franci C, Hu Y, Chang B, Li W, Duan M, Torneros A, Yu J, Heckrodt TJ. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res 2010; 70:1544–54 [DOI] [PubMed] [Google Scholar]
- 42.Davies SR, Dent C, Watkins G, King JA, Mokbel K, Jiang WG. Expression of the cell to cell adhesion molecule, ALCAM, in breast cancer patients and the potential link with skeletal metastasis. Oncol Rep 2008; 19:555–61 [PubMed] [Google Scholar]
- 43.Dydensborg A, Rose A, Wilson B, Grote D, Paquet M, Giguere V, Siegel P, Bouchard M. GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene 2009; 28:2634–42 [DOI] [PubMed] [Google Scholar]
- 44.Swaminathan SK, Roger E, Toti U, Niu L, Ohlfest JR, Panyam J. CD133-targeted paclitaxel delivery inhibits local tumor recurrence in a mouse model of breast cancer. J Controlled Release 2013; 171:280–7 [DOI] [PubMed] [Google Scholar]
- 45.Wilson BJ, Saab KR, Ma J, Schatton T, Pütz P, Zhan Q, Murphy GF, Gasser M, Waaga-Gasser AM, Frank NY. ABCB5 maintains melanoma-initiating cells through a proinflammatory cytokine signaling circuit. Cancer Res 2014; 74:4196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T, Chen Y, Han X, Wu K. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev 2016; 31:61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soini Y, Tuhkanen H, Sironen R, Virtanen I, Kataja V, Auvinen P, Mannermaa A, Kosma V-M. Transcription factors zeb1, twist and snai1 in breast carcinoma. BMC Cancer 2011; 11:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foubert E, De Craene B, Berx G. Key signalling nodes in mammary gland development and cancer. The Snail1-Twist1 conspiracy in malignant breast cancer progression. Breast Cancer Res 2010; 12:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang X, Cai Y, Liu J, Wang Z, Wu Q, Zhang Z, Yang C, Yuan L, Ouyang G. Twist2 contributes to breast cancer progression by promoting an epithelial–mesenchymal transition and cancer stem-like cell self-renewal. Oncogene 2011; 30:4707–20 [DOI] [PubMed] [Google Scholar]
- 50.Wang R, Yang L, Li S, Ye D, Yang L, Liu Q, Zhao Z, Cai Q, Tan J, Li X. Quercetin inhibits breast cancer stem cells via downregulation of aldehyde dehydrogenase 1A1 (ALDH1A1), chemokine receptor type 4 (CXCR4), mucin 1 (MUC1), and epithelial cell adhesion molecule (EpCAM). Med Sci Monit 2018; 24:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gabbert H, Wagner R, Moll R, Gerharz C-D. Tumor dedifferentiation: an important step in tumor invasion. Clin Exp Metastasis 1985; 3:257–79 [DOI] [PubMed] [Google Scholar]
- 52.Kaihara T, Kusaka T, Nishi M, Kawamata H, Imura J, Kitajima K, Itoh-Minami R, Aoyama N, Kasuga M, Oda Y. Dedifferentiation and decreased expression of adhesion molecules, E-cadherin and ZO-1, in colorectal cancer are closely related to liver metastasis. J Exp Clin Cancer Res 2003; 22:117–23 [PubMed] [Google Scholar]
- 53.Orr FW, Lee J, Duivenvoorden WC, Singh G. Pathophysiologic interactions in skeletal metastasis. Cancer 2000; 88:2912–8 [DOI] [PubMed] [Google Scholar]
- 54.Ono M, Handa K, Withers DA, Hakomori S-I. Motility inhibition and apoptosis are induced by metastasis-suppressing gene product CD82 and its analogue CD9, with concurrent glycosylation. Cancer Res 1999; 59:2335–9 [PubMed] [Google Scholar]
- 55.Ikeyama S, Koyama M, Yamaoko M, Sasada R, Miyake M. Suppression of cell motility and metastasis by transfection with human motility-related protein (MRP-1/CD9) DNA. J Exp Med 1993; 177:1231–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shanafelt TD, Geyer SM, Bone ND, Tschumper RC, Witzig TE, Nowakowski GS, Zent CS, Call TG, LaPlant B, Dewald GW. CD49d expression is an independent predictor of overall survival in patients with chronic lymphocytic leukaemia: a prognostic parameter with therapeutic potential. Br J Haematol 2008; 140:537–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waldschmidt JM, Simon A, Wider D, Müller SJ, Follo M, Ihorst G, Decker S, Lorenz J, Chatterjee M, Azab AK. CXCL 12 and CXCR 7 are relevant targets to reverse cell adhesion-mediated drug resistance in multiple myeloma. Br J Haematol 2017; 179:36–49 [DOI] [PubMed] [Google Scholar]
- 58.Qi S, Song Y, Peng Y, Wang H, Long H, Yu X, Li Z, Fang L, Wu A, Luo W. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. PloS One 2012; 7:e38842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tachezy M, Zander H, Gebauer F, Marx A, Kaifi JT, Izbicki JR, Bockhorn M. Activated leukocyte cell adhesion molecule (CD166) – its prognostic power for colorectal cancer patients. J Surg Res 2012; 177:e15–20 [DOI] [PubMed] [Google Scholar]
- 60.Jezierska A, Olszewski WP, Pietruszkiewicz J, Olszewski W, Matysiak W, Motyl T. Activated leukocyte cell adhesion molecule (ALCAM) is associated with suppression of breast cancer cells invasion. Med Sci Monit 2006; 12:BR245–56 [PubMed] [Google Scholar]
- 61.Mehra R, Varambally S, Ding L, Shen R, Sabel MS, Ghosh D, Chinnaiyan AM, Kleer CG. Identification of GATA3 as a breast cancer prognostic marker by global gene expression Meta-analysis. Cancer Res 2005; 65:11259–64 [DOI] [PubMed] [Google Scholar]
- 62.Yan W, Cao QJ, Arenas RB, Bentley B, Shao R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial-mesenchymal transition. J Biol Chem 2010; 285:14042–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nadal R, Ortega FG, Salido M, Lorente JA, Rodríguez-Rivera M, Delgado-Rodríguez M, Macià M, Fernández A, Corominas JM, García-Puche JL. CD133 expression in circulating tumor cells from breast cancer patients: potential role in resistance to chemotherapy. Int J Cancer 2013; 133:2398–407 [DOI] [PubMed] [Google Scholar]
- 64.Yao J, Yao X, Tian T, Fu X, Wang W, Li S, Shi T, Suo A, Ruan Z, Guo H. ABCB5‐ZEB1 axis promotes invasion and metastasis in breast cancer cells. Oncol Res 2017; 25:305–16 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Gales D, Clark C, Manne U, Samuel T. The chemokine CXCL8 in carcinogenesis and drug response. ISRN Oncol 2013; 2013:2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olmeda D, Moreno-Bueno G, Flores JM, Fabra A, Portillo F, Cano A. SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer Res 2007; 67:11721–31 [DOI] [PubMed] [Google Scholar]
- 67.Hong J, Zhou J, Fu J, He T, Qin J, Wang L, Liao L, Xu J. Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Res 2011; 71:3980–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Wang W, Wang W, Yang R, Wang T, Su T, Weng D, Tao T, Li W, Ma D. Correlation of TWIST2 up-regulation and epithelial–mesenchymal transition during tumorigenesis and progression of cervical carcinoma. Gynecol Oncol 2012; 124:112–8 [DOI] [PubMed] [Google Scholar]
- 69.McGuckin MA, Walsh MD, Hohn BG, Ward BG, Wright RG. Prognostic significance of MUC1 epithelial mucin expression in breast cancer. Hum Pathol 1995; 26:432–9 [DOI] [PubMed] [Google Scholar]
- 70.Rakha EA, Boyce RW, El-Rehim DA, Kurien T, Green AR, Paish EC, Robertson JF, Ellis IO. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod Pathol 2005; 18:1295–304 [DOI] [PubMed] [Google Scholar]
- 71.Croker AK, Rodriguez-Torres M, Xia Y, Pardhan S, Leong HS, Lewis JD, Allan AL. Differential functional roles of ALDH1A1 and ALDH1A3 in mediating metastatic behavior and therapy resistance of human breast cancer cells. IJMS 2017; 18:2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang Y. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer 2017; 17:302–17 [DOI] [PubMed] [Google Scholar]
- 73.Grange C, Lanzardo S, Cavallo F, Camussi G, Bussolati B. Sca-1 identifies the tumor-initiating cells in mammary tumors of BALB-neuT transgenic mice. Neoplasia 2008; 10:1433–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou J, Ng S-B, Chng W-J. LIN28/LIN28B: an emerging oncogenic driver in cancer stem cells. Int J Biochem Cell Biol 2013; 45:973–8 [DOI] [PubMed] [Google Scholar]
- 75.Chiba T, Miyagi S, Saraya A, Aoki R, Seki A, Morita Y, Yonemitsu Y, Yokosuka O, Taniguchi H, Nakauchi H. The polycomb gene product BMI1 contributes to the maintenance of tumor-initiating side population cells in hepatocellular carcinoma. Cancer Res 2008; 68:7742–9 [DOI] [PubMed] [Google Scholar]
- 76.Ambs S, Glynn SA. Candidate pathways linking inducible nitric oxide synthase to a basal-like transcription pattern and tumor progression in human breast cancer. Cell Cycle 2011; 10:619–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ursini-Siegel J, Schade B, Cardiff RD, Muller WJ. Insights from transgenic mouse models of ERBB2-induced breast cancer. Nat Rev Cancer 2007; 7:389–97 [DOI] [PubMed] [Google Scholar]
- 78.Asiedu MK, Beauchamp-Perez FD, Ingle JN, Behrens MD, Radisky DC, Knutson KL. AXL induces epithelial-to-mesenchymal transition and regulates the function of breast cancer stem cells. Oncogene 2014; 33:1316–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Zhou Y, Yang Z, Chen B, Huang W, Liu Y, Zhang Y. MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial–mesenchymal transition in breast cancer. Tumour Biol 2017; 39:1010428317690998. [DOI] [PubMed] [Google Scholar]
- 80.Regan J, Kendrick H, Magnay F, Vafaizadeh V, Groner B, Smalley M. c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene 2012; 31:869–83 [DOI] [PubMed] [Google Scholar]
- 81.Gopisetty G, Xu J, Sampath D, Colman H, Puduvalli V. Epigenetic regulation of CD133/PROM1 expression in glioma stem cells by Sp1/myc and promoter methylation. Oncogene 2013; 32:3119–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sharma B, Nawandar DM, Nannuru KC, Varney ML, Singh RK. Targeting CXCR2 enhances chemotherapeutic response, inhibits mammary tumor growth, angiogenesis, and lung metastasis. Mol Cancer Ther 2013; 12:799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balkwill F. Chemokine biology in cancer. Semin Immunol 2003;49–55 [DOI] [PubMed] [Google Scholar]
- 84.Smith MC, Luker KE, Garbow JR, Prior JL, Jackson E, Piwnica-Worms D, Luker GD. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res 2004; 64:8604–12 [DOI] [PubMed] [Google Scholar]
- 85.Proctor E, Waghray M, Lee CJ, Heidt DG, Yalamanchili M, Li C, Bednar F, Simeone DM. Bmi1 enhances tumorigenicity and cancer stem cell function in pancreatic adenocarcinoma. PloS One 2013; 8:e55820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tobin NP, Sims AH, Lundgren KL, Lehn S, Landberg G. Cyclin D1, Id1 and EMT in breast cancer. BMC Cancer 2011; 11:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res 2004; 64:8585–94 [DOI] [PubMed] [Google Scholar]
- 88.Wang W, Wyckoff JB, Goswami S, Wang Y, Sidani M, Segall JE, Condeelis JS. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res 2007; 67:3505–11 [DOI] [PubMed] [Google Scholar]
- 89.Gil-Henn H, Patsialou A, Wang Y, Warren MS, Condeelis JS, Koleske AJ. Arg/Abl2 promotes invasion and attenuates proliferation of breast cancer in vivo. Oncogene 2013; 32:2622–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang W, Goswami S, Sahai E, Wyckoff JB, Segall JE, Condeelis JS. Tumor cells caught in the act of invading: their strategy for enhanced cell motility. Trends Cell Biol 2005; 15:138–45 [DOI] [PubMed] [Google Scholar]
- 91.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol 1999; 147:631–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gilles C, Polette M, Mestdagt M, Nawrocki-Raby B, Ruggeri P, Birembaut P, Foidart J-M. Transactivation of vimentin by β-catenin in human breast cancer cells. Cancer Res 2003; 63:2658–64 [PubMed] [Google Scholar]
- 93.Kallergi G, Papadaki MA, Politaki E, Mavroudis D, Georgoulias V, Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res 2011; 13:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci 2011; 68:3033–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamashita N, Tokunaga E, Kitao H, Hisamatsu Y, Taketani K, Akiyoshi S, Okada S, Aishima S, Morita M, Maehara Y. Vimentin as a poor prognostic factor for triple-negative breast cancer. J Cancer Res Clin Oncol 2013; 139:739–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sethi T, Rintoul RC, Moore SM, MacKinnon AC, Salter D, Choo C, Chilvers ER, Dransfield I, Donnelly SC, Strieter R. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: a mechanism for small cell lung cancer growth and drug resistance in vivo. Nat Med 1999; 5:662–8 [DOI] [PubMed] [Google Scholar]
- 97.Manfredi JJ. Tumor suppression by p53 involves inhibiting an enabler, FGF13. Proc Natl Acad Sci U S A 2017; 114:632–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hollern DP, Swiatnicki MR, Rennhack JP, Misek SA, Matson BC, McAuliff A, Gallo KA, Caron KM, Andrechek ER. E2F1 drives breast cancer metastasis by regulating the target gene FGF13 and altering cell migration. Sci Rep 2019; 9:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garcia S, Dalès J-P, Charafe-Jauffret E, Carpentier-Meunier S, Andrac-Meyer L, Jacquemier J, Andonian C, Lavaut M-N, Allasia C, Bonnier P. Poor prognosis in breast carcinomas correlates with increased expression of targetable CD146 and c-Met and with proteomic basal-like phenotype. Hum Pathol 2007; 38:830–41 [DOI] [PubMed] [Google Scholar]
- 100.Zeng Q, Li W, Lu D, Wu Z, Duan H, Luo Y, Feng J, Yang D, Fu L, Yan X. CD146, an epithelial-mesenchymal transition inducer, is associated with triple-negative breast cancer. Proc Natl Acad Sci U S A 2012; 109:1127–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zabouo G, Imbert A-M, Jacquemier J, Finetti P, Moreau T, Esterni B, Birnbaum D, Bertucci F, Chabannon C. CD146 expression is associated with a poor prognosis in human breast tumors and with enhanced motility in breast cancer cell lines. Breast Cancer Res 2009; 11:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Imbert A-M, Garulli C, Choquet E, Koubi M, Aurrand-Lions M, Chabannon C. CD146 expression in human breast cancer cell lines induces phenotypic and functional changes observed in epithelial to mesenchymal transition. PloS One 2012; 7:e43752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mariotti A, Perotti A, Sessa C, Rüegg C. N-cadherin as a therapeutic target in cancer. Expert Opin Investig Drugs 2007; 16:451–65 [DOI] [PubMed] [Google Scholar]
- 104.Kischel P, Bellahcene A, Deux B, Lamour V, Dobson R, De Pauw E, Clezardin P., Castronovo V. Overexpression of CD9 in human breast cancer cells promotes the development of bone metastases. Anticancer Res 2012; 32:5211–20 [PubMed] [Google Scholar]
- 105.Yu-Lee L-Y, Yu G, Lee Y-C, Lin S-C, Pan J, Pan T, Yu K-J, Liu B, Creighton CJ, Rodriguez-Canales J. Osteoblast-secreted factors mediate dormancy of metastatic prostate cancer in the bone via activation of the TGFβRIII–p38MAPK–pS249/T252RB pathway. Cancer Res 2018; 78:2911–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nannuru KC, Sharma B, Varney ML, Singh RK. Role of chemokine receptor CXCR2 expression in mammary tumor growth, angiogenesis and metastasis. J Carcinog 2011; 10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Romero-Moreno R, Curtis KJ, Coughlin TR, Miranda-Vergara MC, Dutta S, Natarajan A, Facchine BA, Jackson KM, Nystrom L, Li J. The CXCL5/CXCR2 axis is sufficient to promote breast cancer colonization during bone metastasis. Nat Commun 2019; 10:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development 2010; 137:4135–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Science AAftAo. GSK3 inhibits hedgehog signals. Sci Signaling 2002; 2002:tw135 [Google Scholar]
- 110.Della Corte CM, Bellevicine C, Vicidomini G, Vitagliano D, Malapelle U, Accardo M, Fabozzi A, Fiorelli A, Fasano M, Papaccio F. SMO gene amplification and activation of the hedgehog pathway as novel mechanisms of resistance to anti-epidermal growth factor receptor drugs in human lung cancer. Clin Cancer Res 2015; 21:4686–97 [DOI] [PubMed] [Google Scholar]
- 111.Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The role of the Hedgehog signaling pathway in cancer: a comprehensive review. Bosn J Basic Med Sci 2018; 18:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Niida A, Hiroko T, Kasai M, Furukawa Y, Nakamura Y, Suzuki Y, Sugano S, Akiyama T. DKK1, a negative regulator of wnt signaling, is a target of the β-catenin/TCF pathway. Oncogene 2004; 23:8520–6 [DOI] [PubMed] [Google Scholar]
- 113.Forget M, Turcotte S, Beauseigle D, Godin-Ethier J, Pelletier S, Martin J, Tanguay S, Lapointe R. The wnt pathway regulator DKK1 is preferentially expressed in hormone-resistant breast tumours and in some common cancer types. Br J Cancer 2007; 96:646–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by wnt–β-catenin signaling in hepatocellular carcinoma. Cancer Res 2007; 67:10831–9 [DOI] [PubMed] [Google Scholar]
- 115.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development 2011; 138:3593–612 [DOI] [PubMed] [Google Scholar]
- 116.Ridgway J, Zhang G, Wu Y, Stawicki S, Liang W-C, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature 2006; 444:1083–7 [DOI] [PubMed] [Google Scholar]
- 117.Zhang W, Lu W, Ananthan S, Suto MJ, Li Y. Discovery of novel frizzled-7 inhibitors by targeting the receptor’s transmembrane domain. Oncotarget 2017; 8:91459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I. Expression of notch-1 and its ligands, delta-like-1 and jagged-1, is critical for glioma cell survival and proliferation. Cancer Res 2005; 65:2353–63 [DOI] [PubMed] [Google Scholar]
- 119.Ni Z, Bikadi Z, F, Rosenberg M, Mao Q. Structure and function of the human breast cancer resistance protein (BCRP/ABCG2). Curr Drug Metab 2010; 11:603–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert Opin Drug Metab Toxicol 2008; 4:1–15 [DOI] [PubMed] [Google Scholar]
- 121.Mao Q. Role of the breast cancer resistance protein (ABCG2) in drug transport. AAPS J 2005; 7:E118–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ahmed-Belkacem A, Pozza A, Macalou S, Boumendjel A, Di Pietro A. Inhibitors of cancer cell multidrug resistance mediated by breast cancer resistance protein (BCRP/ABCG2). Anticancer Drugs 2006; 17:239–43 [DOI] [PubMed] [Google Scholar]
- 123.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov 2009; 8:547–66 [DOI] [PubMed] [Google Scholar]
- 124.Chen F, Song Q, Yu Q. Axl inhibitor R428 induces apoptosis of cancer cells by blocking lysosomal acidification and recycling independent of Axl inhibition. Am J Cancer Res 2018; 8:1466. [PMC free article] [PubMed] [Google Scholar]
- 125.Shen Y, Chen X, He J, Liao D, Zu X. Axl inhibitors as novel cancer therapeutic agents. Life Sci 2018; 198:99–111 [DOI] [PubMed] [Google Scholar]
- 126.Ma CX, Janetka JW. Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med 2011; 17:88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cerrato A, Morra F, Celetti A. Use of poly ADP-ribose polymerase [PARP] inhibitors in cancer cells bearing DDR defects: the rationale for their inclusion in the clinic. J Exp Clin Cancer Res 2016; 35:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Eyvazi S, Farajnia S, Dastmalchi S, Kanipour F, Zarredar H, Bandehpour M. Antibody based EpCAM targeted therapy of cancer, review and update. Curr Cancer Drug Targets 2018; 18:857–68 [DOI] [PubMed] [Google Scholar]
- 129.Li X-X, Zheng H-T, Peng J-J, Huang L-Y, Shi D-B, Liang L, Cai S-J. RNA-seq reveals determinants for irinotecan sensitivity/resistance in colorectal cancer cell lines. Int J Clin Exp Pathol 2014; 7:2729. [PMC free article] [PubMed] [Google Scholar]
- 130.Edderkaoui M, Chheda C, Soufi B, Zayou F, Hu RW, Ramanujan VK, Pan X, Boros LG, Tajbakhsh J, Madhav A. An inhibitor of GSK3B and HDACs kills pancreatic cancer cells and slows pancreatic tumor growth and metastasis in mice. Gastroenterology 2018; 155:1985–98 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kunnimalaiyaan S, Schwartz VK, Jackson IA, Gamblin TC, Kunnimalaiyaan M. Antiproliferative and apoptotic effect of LY2090314, a GSK-3 inhibitor, in neuroblastoma in vitro. BMC Cancer 2018; 18:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McAllister SD, Christian RT, Horowitz MP, Garcia A, Desprez P-Y. Cannabidiol as a novel inhibitor of id-1 gene expression in aggressive breast cancer cells. Mol Cancer Ther 2007; 6:2921–7 [DOI] [PubMed] [Google Scholar]
- 133.Mistry H, Hsieh G, Buhrlage SJ, Huang M, Park E, Cuny GD, Galinsky I, Stone RM, Gray NS, D'Andrea AD. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol Cancer Ther 2013; 12:2651–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li D, Wang H, Song H, Xu H, Zhao B, Wu C, Hu J, Wu T, Xie D, Zhao J. The microRNAs miR-200b-3p and miR-429-5p target the LIMK1/CFL1 pathway to inhibit growth and motility of breast cancer cells. Oncotarget 2017; 8:85276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell 2009; 16:487–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim JW, Gautam J, Kim JE, Kim J, Kang KW. Inhibition of tumor growth and angiogenesis of tamoxifen-resistant breast cancer cells by ruxolitinib, a selective JAK2 inhibitor. Oncol Lett 2019; 17:3981–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ismail I, Kang H, Lee H, Kim J, Hong S. DJ-1 upregulates breast cancer cell invasion by repressing KLF17 expression. Br J Cancer 2014; 110:1298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.El-Sokkary GH, Ismail IA, Saber SH. Melatonin inhibits breast cancer cell invasion through modulating DJ-1/KLF17/ID-1 signaling pathway. J Cell Biochem 2019; 120:3945–57 [DOI] [PubMed] [Google Scholar]
- 139.Kastrati I, Siklos MI, Calderon-Gierszal EL, El-Shennawy L, Georgieva G, Thayer EN, Thatcher GR, Frasor J. Dimethyl fumarate inhibits the nuclear factor κB pathway in breast cancer cells by covalent modification of p65 protein. J Biol Chem 2016; 291:3639–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Godwin P, Baird A-M, Heavey S, Barr M, O'Byrne K, Gately KA. Targeting nuclear factor-kappa B to overcome resistance to chemotherapy. Front Oncol 2013; 3:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Benvenuto M, Masuelli L, De Smaele E, Fantini M, Mattera R, Cucchi D, Bonanno E, Di Stefano E, Frajese GV, Orlandi A. In vitro and in vivo inhibition of breast cancer cell growth by targeting the hedgehog/GLI pathway with SMO (GDC-0449) or GLI (GANT-61) inhibitors. Oncotarget 2016; 7:9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li Y, Song Q, Day BW. Phase I and phase II sonidegib and vismodegib clinical trials for the treatment of paediatric and adult MB patients: a systemic review and Meta-analysis. Acta Neuropathol Commun 2019; 7:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Qin J-J, YL, Zhang J, Zhang W-D. STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J Exp Clin Cancer Res 2019; 38:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.de Gramont A, Faivre S, Raymond E. Novel TGF-β inhibitors ready for prime time in onco-immunology. Oncoimmunology 2017; 6:e1257453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Park C-Y, Son J-Y, Jin CH, Nam J-S, Kim D-K, Sheen YY. EW-7195, a novel inhibitor of ALK5 kinase inhibits EMT and breast cancer metastasis to lung. Eur J Cancer 2011; 47:2642–53 [DOI] [PubMed] [Google Scholar]
- 146.Do K, Doroshow JH, Kummar S. Wee1 kinase as a target for cancer therapy. Cell Cycle 2013; 12:3348–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Matheson CJ, Backos DS, Reigan P. Targeting WEE1 kinase in cancer. Trends Pharmacol Sci 2016; 37:872–81 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220971599 for Human-derived osteoblast-like cells and pericyte-like cells induce distinct metastatic phenotypes in primary breast cancer cells by Vera Mayo, Annie C Bowles, Laura E Wubker, Ismael Ortiz, Albert M Cordoves, Richard J Cote, Diego Correa and Ashutosh Agarwal in Experimental Biology and Medicine