Abstract
The population of catfish, Clarias batrachus has substantially diminished in various countries and studies show that another related species Clarias gariepinus is replacing it. The better adaptability and survivability of C. gariepinus over C. batrachus could be attributed to the metabolic differences between these two species, which is primarily regulated by mitochondrial activities. To understand the reasons behind this phenomenon, we performed in silico analyses to decipher the differences between the proteins encoded by the mitochondrial genome of these two related species. Our analysis revealed that out of thirteen, twelve proteins encoded by the mitochondrial genome of these two species have substantial variations between them. We characterised these variations by analysing their effect on secondary structure, intrinsic disorder predisposition, and functional impact on protein and stability parameters. Our data show that most of the parameters are changing between these two closely related species. Altogether, we demonstrate the molecular insights into the mitochondrial genome-encoded proteins of these two species and predict their effect on protein function and stability that might be helping C. gariepinus to gain survivability better than the C. batrachus.
Keywords: Clarias batrachus, Clarias gariepinus, Mitochondrial genome, Variations, Protein stability
Highlights
-
•
Analysis of proteins encoded by mitogenome of C. batrachus and C. gariepinus identified several variations.
-
•
The most frequent variations among these two related species were observed for NADH dehydrogenase subunit 3.
-
•
The variations contributed to alteration in secondary structure and intrinsic protein disorder.
-
•
These variations in mitogenome might help C. gariepinus to better adapt to the environmental conditions.
1. Introduction
The popularity of Clarias batrachus (Linnaeus, 1758) has been attributed to its taste, high protein and low fat content, and its medicinal values [1,2]. Among Asian countries, the most prominent species of Clarias (family Clariidae) used for aquaculture are C. batrachus in India, C. fuscus (Taiwan and Hawaii) and C. macrocephalus (South-East Asia) [3]. Recently, the population of C. batrachus significantly declined due to the anthropogenic activities and the introduction of morphologically similar exotic catfish, C. gariepinus (Burchell,1822) [3,4]. Subsequently, C. gariepinus has further increased the vulnerability of C. batrachus [5,6] that has led to listing of C. batrachus in ‘‘Critically Endangered’’ category of the IUCN (IUCN, 2007) red list of threatened species [4]. Similar observations were also reported by Na-Nakorn and Brummett [7] who emphasized that the wild type population must be conserved from C. gariepinus, which may endanger the purity and viability of wild populations.
Since, C. batrachus is slowly moving toward the extinction; therefore, it is rational to understand the genetic makeup of this species so that proper conservational strategies can be implemented. A study by Khedkar et al. [8] revealed the lack of genetic diversity in C. batrachus populations found in major riverine system of India that can contribute to inbreeding and disease susceptibility. A comparative study on C. batrachus and C. gariepinus from India revealed up to 99% substitution by C. gariepinus in fish markets [3]. At genetic level the C. gariepinus and C. batrachus have distinct characteristics. Karyotyping of these two species revealed that the C. gariepinus has 56 chromosomes (2n = 28) while C. batrachus has 104 chromosomes (2n = 52) [9]. Recently, C. batrachus genome has been sequenced and its annotation revealed the presence of 22,914 genes [10]. Similar study on C. gariepinus is required to better compare the overall genomic variations present between these two species.
The better adaptation of the fish populations has been shown to be associated with positive selection pressure [11,12]. The molecular mechanisms of these adaptations are not properly understood; however, few studies have linked to the key processes of physiology, metabolism, better antioxidant and DNA repair mechanisms [[13], [14], [15]]. One of the most promising players required for better adaptation is the enhancement of energy metabolism. At cellular levels, the mitochondria regulate energy metabolism events and play an indispensable role in the species diversity and differentiation [16]. Therefore, mitochondrial genome (mitogenome) has been widely studied to understand the genetic diversity of different populations in fish species. Furthermore, the small genetic material of the mitogenome enables researchers to comprehensibly analyse the relatively rapid substitution rate as well as the maternal inheritance [17,18]. The variations in mitochondrial DNA (mtDNA) are influenced by environmental conditions affecting metabolic processes, which eventually led to implications in fitness, such as, caviomorph rodents related to anoxic subterranean environments [19], monkeys to altitude [20], killer whales (Orcinus orca) [21] and Pacific salmon (Oncorhynchus sp.) [22] to latitudinal clines. Altogether, studies on the regulation of mtDNA variations are keys for understanding the selection events.
This study aimed to understand the molecular determinants of better survivability and adaptability of C. gariepinus over C. batrachus. Here, we analysed the variations present between the proteins encoded by the mitochondrial genome of C. gariepinus and C. batrachus to dissect the possible mechanisms of C. gariepinus adaptability. Our data revealed that there are many variations present in C. gariepinus protein that alters protein disorder parameters, secondary structure, and stability that might interfere in protein functioning. Recently, various algorithms have been developed to predict several aspects of protein structure, such as the prediction of secondary-structure [23], state of protein disorder, solvent accessibility of its amino acid residues [24], alteration in stability and function due to an amino acid substitution, sites for protein interactions [25], etc. Among these, predicting protein disorder has gained considerable interest in the last decade [26]. In eukaryotic proteins, the intrinsically disordered regions (IDRs) are abundant and are associated with various cellular functions [27,28]. IDRs are part of the protein that do not possess distinct three-dimensional structure, but are nevertheless functional [29]. Here, we used several bioinformatics tools to characterise the variations among the mitochondrial proteins of C. gariepinus and C. batrachus.
2. Material and methods
2.1. Sequence retrieval
As of 30th August 2020, the NCBI-genome-database has seven complete mitochondrial genome sequences of C. batrachus and C. gariepinus as shown in Table 1. Among these, three sequences were reported from both India (KC572134.1, NC_023923.1, and KM259918.1) and China (KY767672.1, KT001082.1, and NC_027661.1), while one from Hungary (KT809508.1). All three Indian sequences were for C. batrachus and the Hungarian sequence was for C. gariepinus. Of the three sequences reported from China, two were for C. gariepinus (KT001082.1 and NC_027661.1) and one was for C. batrachus (KY767672.1). Since, only China has reported both sequences of C. batrachus and C. gariepinus; therefore, we decided to perform a comparative study of the proteins encoded by the mitochondrial genome of these two species from China. Further, the two C. gariepinus sequence reported from China was similar (KT001082.1 and NC_027661.1), there were no variations present between them; therefore, we used one of the C. gariepinus sequence (NC_027661.1) for further analysis. We extracted the proteins encoded by the mitochondrial genome of KY767672.1 (C. batrachus) and NC_027661.1 (C. gariepinus) and used for analysis (Table 2). The protein identifier accession numbers of the proteins encoded by the mitochondrial genome of above two mentioned species are listed in Table 2.
Table 1.
List of complete mitochondrial genome sequences of C. batrachus and C. gariepinus reported worldwide till August 2020.
| S. No. | Species | Accession Number |
Submission date | Reported from | Authors | Institute |
|---|---|---|---|---|---|---|
| 1 | Clarias batrachus | KY767672 | 14-MAR-2017 | China | Ma,A. et al., | College of Fisheries, Henan normal University, Henan, 453007, China |
| 2 | Clarias gariepinus | NC_027661 | 01-JUN-2015 | China | Han,C. et al., | College of Life Science, Sun Yat-Sen University, Guangdong 510000, China |
| 3 | Clarias gariepinus | KT001082 | 01-JUN-2015 | China | Han,C. et al., | College of Life Science, Sun Yat-Sen University, Guangdong 510000, China |
| 4 | Clarias gariepinus | KT809508 | 22-SEP-2015 | Hungary | Kovacs, B. et al., | Department of Aquaculture, Szent Istvan University, Godollo 2100, Hungary |
| 5 | Clarias batrachus | KM259918 | 01-AUG-2014 | India | Kushwaha,B. et al., | Molecular Biology and Biotechnology, National Bureau of Fish Genetic Resources, UP, 226002, India |
| 6 | Clarias batrachus | KC572134 | 01-FEB-2013 | India | Mohindra,V. et al., | Fish Conservation Division, National Bureau of Fish Genetic Resources, UP, 226002, India |
| 7 | Clarias batrachus | NC_023923 | 01-FEB-2013 | India | Mohindra,V. et al., | Fish Conservation Division, National Bureau of Fish Genetic Resources, UP 226002, India |
Table 2.
List of protein Accession Number used in this study.
| S. No. | Protein | Clarias batrachus (Accession number) | Clarias gariepinus (Accession number) |
|---|---|---|---|
| 1 | NADH dehydrogenase subunit 1 | AUX80750.1 | YP_009160664.1 |
| 2 | NADH dehydrogenase subunit 2 | AUX80751.1 | YP_009160665.1 |
| 3 | cytochrome c oxidase subunit 1 | AUX80752.1 | YP_009160666.1 |
| 4 | cytochrome c oxidase subunit 2 | AUX80753.1 | YP_009160667.1 |
| 5 | ATP synthase F0 subunit 8 | AUX80754.1 | YP_009160668.1 |
| 6 | ATP synthase F0 subunit 6 | AUX80755.1 | YP_009160669.1 |
| 7 | cytochrome c oxidase subunit 3 | AUX80756.1 | YP_009160670.1 |
| 8 | NADH dehydrogenase subunit 3 | AUX80757.1 | YP_009160671.1 |
| 9 | NADH dehydrogenase subunit 4L | AUX80758.1 | YP_009160672.1 |
| 10 | NADH dehydrogenase subunit 4 | AUX80759.1 | YP_009160673.1 |
| 11 | NADH dehydrogenase subunit 5 | AUX80760.1 | YP_009160674.1 |
| 12 | NADH dehydrogenase subunit 6 | AUX80761.1 | YP_009160675.1 |
| 13 | cytochrome b | AUX80762.1 | YP_009160676.1 |
2.2. Multiple sequence alignments
The CLUSTAL Omega webserver developed at EMBL’s European Bioinformatics Institute, UK [30] was used for multiple sequence alignment with each mitochondrial genome encoded protein from C. batrachus and C. gariepinus.
2.3. Secondary structure predictions
To obtain a basic idea about the probable secondary structure, we compared the protein from C. batrachus and C. gariepinus using CFSSP (Chou and Fasman secondary structure prediction) [30] tool. This webserver predicts the most appropriate secondary structure, including, α-helix, β-sheet, and turns from the input peptide sequence.
2.4. Evaluating the protein intrinsic disorders
The intrinsic disorder distribution of each individual residues of the target polypeptide chain was evaluated by PONDR-VSL2 webserver [31]. The per-residue disorder predisposition scores have been designated on a scale from 0 to 1. The value of ‘0’ depicts fully ordered residues and ‘1’ represents fully disordered residues. Value ‘0.5’ is considered threshold above which is considered disordered while, below ‘0.5’ is considered ordered.
2.5. Prediction of impact of variation on protein function and stability
To predict the impact of variations on the function of protein, PROVEAN (Protein Variation Effect Analyzer) was used [32]. This tool predicts the implication of variation on protein function. The predefined threshold score of PROVEAN is -2.5. The variation is considered ‘deleterious’, if the score is equal to or below -2.5. Similarly, the variation is considered ‘neutral’, if the score is above the threshold. To analyse the effect of variations on protein stability I-mutant suite webserver was used [33]. I-mutant suite predicts the difference in free energy (ΔΔG) between the wild-type and mutant sequence. The positive value represents an increase in stability, while negative ΔΔG represents destabilisation.
3. Results
3.1. Identification of variations between the proteins encoded by the mitochondrial genome of Clarias batrachus and Clarias gariepinus
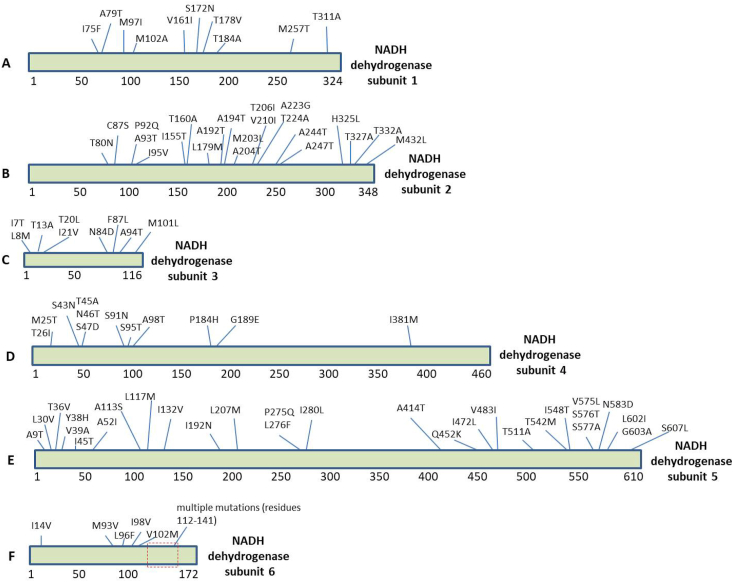
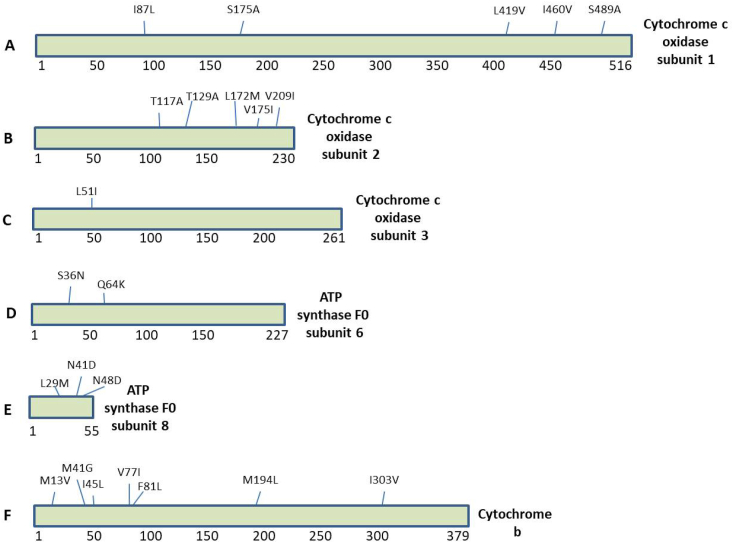
We downloaded (NCBI-genome database) the two mitochondrial sequences reported from China as discussed in methods and extracted the thirteen polypeptide sequences encoded by its mitogenome (Table 2). All these protein sequences obtained from C. batrachus and C. gariepinus were aligned using CLUSTAL Omega tool to check for similarities or variations. The C. batrachus mitochondrial proteins were used as wild-type sequences for this analysis. Using this algorithm, we identified the variations present between C.batrachus and C. gariepinus listed in Table 3. The most frequent variations were observed for NADH dehydrogenase subunit 3 that has 7.75% residues mutated (Table 3). Interestingly, we did not observe any change in amino acids for NADH dehydrogenase subunit 4L that means this protein is 100% conserved between these two species. The most number of amino acid variations at 29 sites was observed for NADH dehydrogenase subunit 5 (Table 3). Further, the location of the variation site for each protein is shown as schematics. Fig. 1 (A-F) shows the variations observed in NADH dehydrogenase subunits (subunit 1, 2, 3, 4, 5 and 6). The Fig. 2 (A-F) shows the variations observed for Cytochrome C Oxidase subunits (subunit 1, 2 and 3), ATP synthase F0 subunits (subunit 6 and 8) and Cytochrome b.
Table 3.
Summary of variations observed between C. batrachus and C. gariepinus. The data was obtained by comparing the mitochondrial protein sequences of C. gariepinus and C. batrachus.
| S. No | Mitochondrial genome encoded Proteins | Total Protein length | Number of point mutations observed in C. gariepinus (C. batrachus used as wild type) | % mutant residues in C. gariepinus |
|---|---|---|---|---|
| 1 | NADH dehydrogenase subunit 1 | 324 | 10 | 3.08 |
| 2 | NADH dehydrogenase subunit 2 | 348 | 22 | 6.32 |
| 3 | Cytochrome c oxidase subunit I | 516 | 5 | 0.96 |
| 4 | Cytochrome c oxidase subunit II | 230 | 5 | 2.12 |
| 5 | ATP synthase F0 subunit 8 | 55 | 3 | 5.45 |
| 6 | ATP synthase F0 subunit 6 | 227 | 2 | 0.88 |
| 7 | Cytochrome c oxidase subunit III | 261 | 1 | 0.38 |
| 8 | NADH dehydrogenase subunit 3 | 116 | 9 | 7.75 |
| 9 | NADH dehydrogenase subunit 4L | 98 | No mutation | 0 |
| 10 | NADH dehydrogenase subunit 4 | 460 | 11 | 2.39 |
| 11 | NADH dehydrogenase subunit 5 | 608 | 29 | 4.7 |
| 12 | NADH dehydrogenase subunit 6 | 172 | Five point mutations and multiple mutations from residue 112-141 | – |
| 13 | Cytochrome b | 379 | 7 | 1.84 |
Fig. 1.
The sequence alignment of the NADH Dehydrogenase subunits encoded by mitochondrial genome of C. batrachus and C. gariepinus reported from China. (A–F) The amino acid substitutions are highlighted in the schematics of individual proteins.
Fig. 2.
The sequence alignment of the Cytochrome C Oxidase subunits, ATP Synthase F0 subunits and Cytochrome b encoded by mitochondrial genome of C. batrachus and C. gariepinus reported from China. (A–F) The amino acid substitutions are highlighted in the schematics of individual proteins.
3.2. The secondary structures of proteins are altered due to the variations
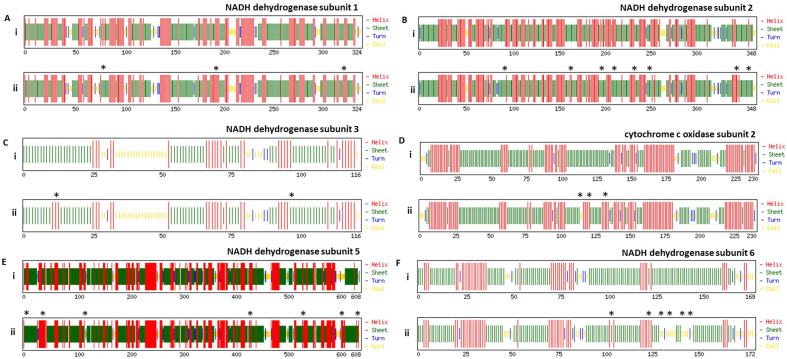
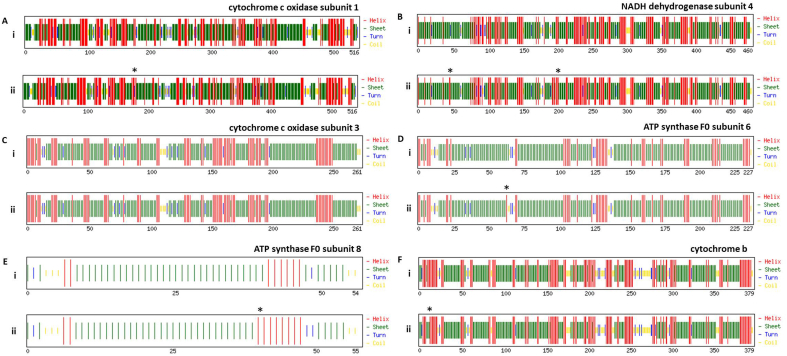
Next, we studied the effect of these variations on the secondary structure of individual proteins. We used CFSSP webserver to predict the secondary structure of the polypeptide sequences, as explained in the methods section. Fig. 3 demonstrates the secondary structures of NADH dehydrogenase subunits (subunit 1, 2, 3, 5 and 6). The location of variations in secondary structure has been marked by an asterisk in each panel. The detailed analysis revealed that the NADH dehydrogenase subunit 1 has acquired changes in secondary structure at three positions (Fig. 3A, marked by asterisk). Among those, one position is leading to gain in the beta-sheet structure while rest two positions are leading to gain in alpha-helix (Fig. 3A, compare panel i and ii, location is marked by an asterisk). Similar analysis was performed for other subunits as well and our data show that NADH dehydrogenase subunit 2, 3, 5 and 6 have variations in secondary structures at 8, 2, 7 and 5 locations, respectively (Fig. 3B, C, E and F). The Cytochrome C Oxidase subunit 2 showed variation in secondary structure at three locations (Fig. 3D). We performed similar secondary structure predictions with other proteins that include Cytochrome C Oxidase subunits (subunit 1 and 3), ATP synthase F0 subunits (subunit 6 and 8) and Cytochrome b (supplementary fig. 1). Detailed analysis revealed that the change in the secondary structure was observed for Cytochrome C Oxidase subunit 1 at one location only (suppl. fig 1A), Cytochrome C Oxidase subunit 3 did not show any variation (suppl. fig. 1C). Similarly, ATP synthase F0 subunit 6 and 8 shows variation at one location each (suppl. Fig. 1D and E). The Cytochrome b also shows variation at one location only (suppl. fig. 1F). Altogether, our secondary structure prediction revealed alteration in secondary structure due to the variations in amino acid sequences between C. batrachus and C. gariepinus.
Fig. 3.
Effect of amino acid substitutions on the secondary structure of proteins encoded by mitochondrial genome of C. batrachus and C. gariepinus. Panel (i) represents sequence of C. batrachus and panel (ii) represents sequence of C. gariepinus. (A–F) represents the individual proteins and asterisk show the locations of the variation in the secondary structure of C. gariepinus protein.
3.3. The variations alter the intrinsic disorder parameters
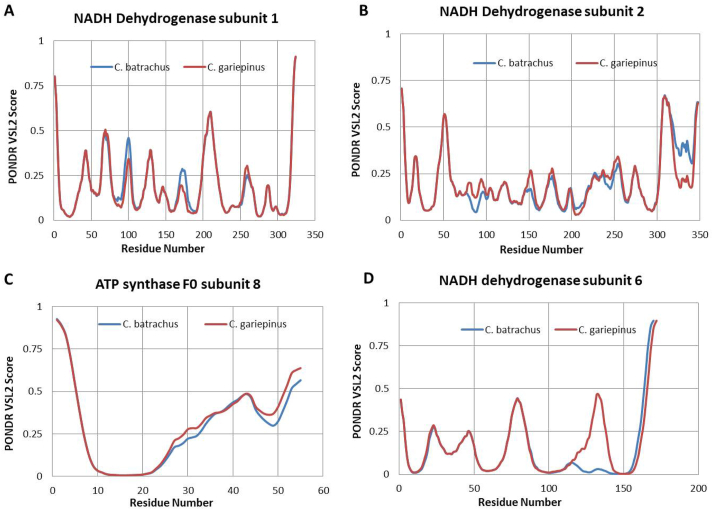
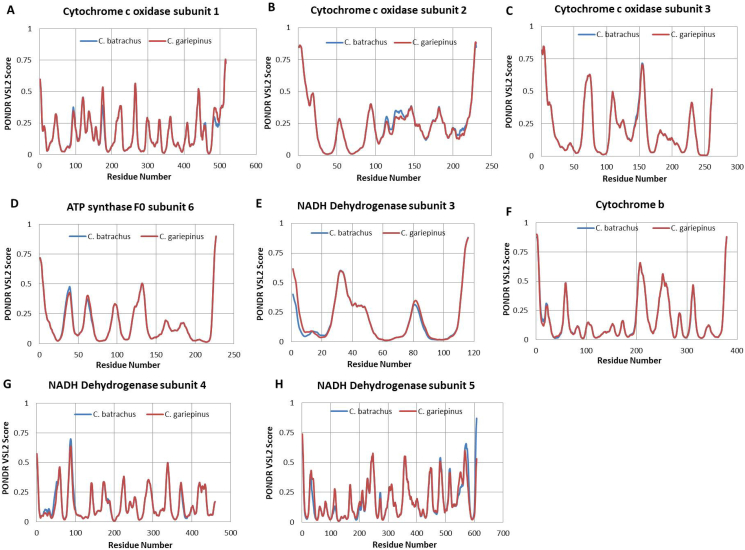
The differences between the proteins encoded by the mitogenome of C. batrachus and C. gariepinus can be additionally demonstrated via the analysis of the per-residue intrinsic disorder predispositions of these proteins. Results of this analysis are shown in Fig. 4 and supplementary fig. 2, which illustrates the intrinsic disorder propensity of the proteins of the two species. The detailed analysis revealed that many amino acid substitutions do not lead to alteration in local disorder propensity; however, some of them cause noticeable alterations in the disorder predisposition (Fig. 4). For instance, NADH dehydrogenase subunit 1 local disorder predisposition in the vicinity of residue 100 and 175 was decreased in the C. gariepinus (Fig. 4A). The NADH dehydrogenase subunit 2 shows the loss of a peak at the end of the protein sequence in C. gariepinus (Fig. 4B). Similar analysis was performed using the other NADH dehydrogenase subunits and minor variations were observed for subunit 3, 4 and 5 (supplementary fig. 2). A new noticeable prominent peak was observed for NADH dehydrogenase subunit 6 of C. gariepinus (Fig. 4D) in the vicinity of residue 120, whereas the C. batrachus lacks it (Fig. 4D). Furthermore, the Cytochrome C Oxidase subunit 1, 2 and ATP synthase F0 subunit 6 show minor variations in disorder propensity (supplementary fig. 2). Cytochrome C Oxidase subunit 3 and Cytochrome b do not show any variation (supplementary fig. 2). The ATP synthase F0 subunit 8 shows an overall increase in protein disorder propensity (Fig. 4C). Altogether, our data indicate a change in intrinsic disorder propensity among few proteins encoded by mtDNA of these two species.
Fig. 4.
Comparisons of the intrinsic disorder predisposition of the C. batrachus and C. gariepinus. A disorder threshold is indicated at score = 0.5, residues/regions with the disorder scores >0.5 are considered as disordered. (A–D) Each panel represents the disorder parameters of individual proteins including NADH dehydrogenase subunit 1, 2, 6 and ATP Synthase F0 subunit 8.
3.4. The variations in amino acid also alter the protein functions and stability parameters
Subsequently, to understand the functional impact of the variations, we performed additional predictions that include protein function prediction (PROVEAN score) and change in stability (ΔΔG). We performed these two analyses for all variations observed in this study as shown in Table 4 and supplementary table 1 (complete list). The functional impact was demonstrated in terms of ‘neutral and deleterious’. Our analysis revealed that most of the variations were having no effect on protein function (neutral). However, few of them were deleterious to protein function (Table 4). The P92Q substitution in NADH dehydrogenase subunit 2 was observed as deleterious (Table 4). Furthermore, T26I of NADH dehydrogenase subunit 4, I192N of NADH dehydrogenase subunit 5 and M41G of Cytochrome b was found to be deleterious (Table 4 and supplementary table 1). Similarly, the stability predictions revealed that most of the variations decrease protein stability ΔΔG more than (-1.5 Kcal/mol) that includes I75F of NADH dehydrogenase subunit 1, I7T of NADH dehydrogenase subunit 3, I192 N and I548T of NADH dehydrogenase subunit 5. However, only one substitution S172N of NADH dehydrogenase subunit 1 (supplementary table 1) led to increase in stability with high reliability index (more than 7). Altogether, our analysis revealed that various amino acid substitutions alter protein functions and stability parameters.
Table 4.
Protein structural analysis was performed by PROVEAN and I-mutant suite. The effect of mutation on protein function and stability was analysed using these two webservers.
| NADH dehydrogenase subunit 1 | |||||
|---|---|---|---|---|---|
| Variant | PROVEAN score | Prediction (cutoff = -2.5) | ΔΔG value (Kcal/mol) | Stability | Reliability index |
| I75F | −0.166 | Neutral | −1.64 | Decrease | 8 |
| M257T |
−1.562 |
Neutral |
−0.98 |
Decrease |
7 |
| NADH dehydrogenase subunit 2 | |||||
| Variant |
PROVEAN score |
Prediction (cutoff = -2.5) |
ΔΔG value (Kcal/mol) |
Stability |
Reliability index |
| P92Q | −6.87 | Deleterious | −0.99 | Decrease | 8 |
| I155T | 1.306 | Neutral | −2 | Decrease | 8 |
| L179M | −1.156 | Neutral | −0.99 | Decrease | 5 |
| A223G | −0.754 | Neutral | −1.17 | Decrease | 7 |
| T224A | −0.061 | Neutral | −1.07 | Decrease | 9 |
| T327A |
0.926 |
Neutral |
−0.91 |
Decrease |
7 |
| NADH dehydrogenase subunit 3 | |||||
| Variant |
PROVEAN score |
Prediction (cutoff = -2.5) |
ΔΔG value (Kcal/mol) |
Stability |
Reliability index |
| I7T |
−1.135 |
Neutral |
−1.74 |
decrease |
2 |
| NADH dehydrogenase subunit 4 | |||||
| Variant |
PROVEAN score |
Prediction (cutoff = -2.5) |
ΔΔG value (Kcal/mol) |
Stability |
Reliability index |
| T45A | −1.572 | Neutral | −1.34 | decrease | 8 |
| P184H | −1.243 | Neutral | −1.32 | decrease | 7 |
| I381M |
2.144 |
Neutral |
−1.46 |
decrease |
8 |
| NADH dehydrogenase subunit 5 | |||||
| Variant |
PROVEAN score |
Prediction (cutoff = -2.5) |
ΔΔG value (Kcal/mol) |
Stability |
Reliability index |
| L30V | −0.729 | Neutral | −1.29 | decrease | 7 |
| Y38H | 2.038 | Neutral | −1.28 | increase | 1 |
| V39A | −1.142 | Neutral | −1.46 | decrease | 9 |
| I45T | 1.663 | Neutral | −2.2 | decrease | 8 |
| A113S | 1.653 | Neutral | −0.93 | decrease | 9 |
| L117M | 0.532 | Neutral | −1.37 | decrease | 7 |
| I192N | −4.605 | Deleterious | −1.73 | decrease | 7 |
| L207M | −1.057 | Neutral | −1.12 | decrease | 7 |
| P275Q | 1.906 | Neutral | −1.02 | decrease | 7 |
| I472L | −1.473 | Neutral | −0.9 | decrease | 7 |
| I548T | 1.504 | Neutral | −1.66 | decrease | 2 |
| V575L | 1.36 | Neutral | −1.08 | decrease | 2 |
| S577A |
−1.158 |
Neutral |
−1.03 |
decrease |
7 |
| NADH dehydrogenase subunit 6 | |||||
| Variant |
PROVEAN score |
Prediction (cutoff = -2.5) |
ΔΔG value (Kcal/mol) |
Stability |
Reliability index |
| L96F | −1.391 | Neutral | −1.02 | decrease | 5 |
| V102M |
−0.471 |
Neutral |
−1.11 |
decrease |
6 |
| Cytochrome c oxidase subunit 1 | |||||
| Variant |
PROVEAN score |
Prediction (cutoff = -2.5) |
ΔΔG value (Kcal/mol) |
Stability |
Reliability index |
| L419V | 0.253 | Neutral | −1.15 | decrease | 6 |
| I460V |
−0.294 |
Neutral |
−0.97 |
decrease |
7 |
| Cytochrome c oxidase subunit 2 | |||||
| Variant |
PROVEAN score |
Prediction (cutoff = -2.5) |
ΔΔG value (Kcal/mol) |
Stability |
Reliability index |
| T117A | −0.316 | Neutral | −1.08 | decrease | 5 |
| L172M |
0.575 |
Neutral |
−1.35 |
decrease |
7 |
| ATP synthase F0 subunit 8 | |||||
| Variant |
PROVEAN score |
Prediction (cutoff = -2.5) |
ΔΔG value (Kcal/mol) |
Effect/stability |
Reliability index |
| L29M |
−0.39 |
Neutral |
−1.08 |
decrease |
8 |
| Cytochrome b | |||||
| Variant |
PROVEAN score |
Prediction (cutoff = -2.5) |
ΔΔG value (Kcal/mol) |
Stability |
Reliability index |
| M41G | −3.169 | Deleterious | −1.06 | decrease | 8 |
| F81L | 2.572 | Neutral | −1.19 | decrease | 5 |
4. Discussion
All 13 proteins encoded by mtDNA are components of the electron transport chain and play a critical role in the organismal energy metabolism [34]. Therefore, mitochondrial genes are key molecules that connect the mechanisms of the evolution of the energy consumption [35]. Even in higher eukaryotes, the mtDNA polymorphisms have been linked to altered mitochondrial matrix pH and intracellular calcium dynamics [36] and contribute to the generation of reactive oxygen species (ROS) in murine cells [37]. Further, rare functional variants in mtDNA enhances aerobic performance of human athletes [38].
Here, we have comparatively studied the proteins encoded by the mitochondrial genome of C. batrachus and C. gariepinus to understand the impact of variations on protein structure and function. Our data revealed that the NADH dehydrogenase subunit 3 of C. gariepinus displayed the highest variability with 7.75% residues substituted compared with C. batrachus. Other NADH dehydrogenases also showed variations including subunit 2 (6.32%), subunit 5 (4.7%), subunit 4 (2.39%) and subunit 1 (3.08%) indicating that NADH dehydrogenases exhibit high levels of substitution in C. gariepinus. ATP synthase F0 subunit 8 also showed high variation (5.45%). Cytochrome C oxidase subunits 1, 3 showed a low level of variation (<1%). Ample studies revealed a positive association of proteins involved in oxidative phosphorylation with increased energetic demands (reviewed in Ref. [39]). The variations observed between these two species might be favouring the energy metabolism of C. gariepinus. The rapid rate of mutation in mtDNA makes it possible to produce advantageous or disadvantageous phenotypes in a relatively short time. Variations in protein sequences correlate with adaptation, such as a study examined the rhodopsin sequences from 2056 species of fish identified the occurrence of the same missense mutation and its independent selection toward light environments across these fishes [40]. Earlier studies have linked the variations in the protein encoded by mitogenome might contribute to the phenotypic variation in poultry and livestock. For instance, in cattle, mtDNA variation has been linked to economic traits such as increased production of milk, calving rates, increased weight and many others [[41], [42], [43], [44]]. Our result correlates with these studies and it seems that the NADH dehydrogenases and others displayed the variability in the C. gariepinus that might be associated with their positive selection; however, future experimental data are required to validate our current in silico predictions. Furthermore, the NADH enzymes play a vital role in respiratory-chain activities, thus influencing energy supply [45,46]. Foote et al. [21] identified two positively selected amino acid sites in the mitochondrial genes of killer whales (Orcinus orca) that influences its metabolic performance. Similarly, positive selection of mitochondrial genes were also observed in migratory Pacific salmon [22].
Subsequently, we show that variations between the mitochondrial proteins among these two species translate to an alteration in secondary structure, intrinsic protein disorder, protein stability that can impact protein function. Our data revealed a considerable alteration in protein disorder parameters of NADH dehydrogenase subunit 1, 2, and 6 (Fig. 4) among these two species that can have potential impact on protein function. Several IDRs are involved in folding of their interacting partners with variable degrees, thereby the resultant complexes exhibit structural and functional heterogeneity [47,48]. Proteins with IDRs retain extreme thermal and acid stability as well as remain functional under these adverse conditions. Furthermore, IDRs provide advantages to its carriers, at the molecular, supra-molecular, and organismal levels [49]. Studies on IDRs of human proteome have revealed that a considerable fraction of disease-associated mutations reside within the IDRs and the substitutions of residues have significant functional impact [50,51]. Whether the high variation rate of the proteins observed here could be related to phenotypic variation between C. batrachus and C. gariepinus, needs further investigation. The information reported in this study could be useful for future studies on mitogenome variations between two closely related fish species and their role in better adaptation to the environmental conditions.
CRediT authorship contribution statement
Gyanendra Bahadur Chand: Methodology, Validation, Writing - original draft and & editing.
Sushant Kumar: Methodology, Validation, Visualization, Writing - original draft and & editing.
Gajendra Kumar Azad: Conceptualization, Supervision, Methodology, Validation, Visualization, Writing - original draft and & editing.
Declaration of competing interest
Authors declare no conflict of interests.
Acknowledgements
We acknowledge Patna University, Patna, Bihar (India) for providing necessary infrastructural support. No funding was used to conduct this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.100985.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
figs1.
Effect of amino acid substitutions on the secondary structure of proteins encoded by mitochondrial genome of C. batrachus and C. gariepinus. Panel (i) represents sequence of C. batrachus and panel (ii) represents sequence of C. gariepinus. (A-F) represents the individual proteins and asterisk show the locations of the variation in the secondary structure of C. gariepinus protein.
figs2.
Comparisons of the intrinsic disorder predisposition of the C. batrachus and C. gariepinus. A disorder threshold is indicated at score = 0.5, residues/regions with the disorder scores >0.5 are considered as disordered. (A-H) Each panel represents the disorder parameters of individual protein (mentioned in each panel)
References
- 1.Debnath S. Clarias batrachus, the medicinal fish: an excellent candidate for aquaculture & employment generation. Int. Conf. Asia Agric. Anim. 2011 [Google Scholar]
- 2.Islam R., Mondol L.K., Sheikh L., Rahman S.S., Islam M., Rahman A. Identification of fatty acid profile, lipid characterization and nutritional status of Clarias batrachus. Nutr. Sci. Food Technol. 2013 doi: 10.7243/2054-1848-1-1. [DOI] [Google Scholar]
- 3.Khedkar G.D., Tiknaik A.D., Shinde R.N., Kalyankar A.D., Ron T.B., Haymer D. Mitochondrial DNA; 2016. High Rates of Substitution of the Native Catfish Clarias batrachus by Clarias gariepinus in India. [DOI] [PubMed] [Google Scholar]
- 4.Argungu L.A., Christianus A., Amin S.M.N., Daud S.K., Sirsaj S.S., Rahman M.A. Asian catfish clarias batrachus (linnaeus, 1758) getting critically endangered. Asian J. Anim. Vet. Adv. 2013 doi: 10.3923/ajava.2013.168.176. [DOI] [Google Scholar]
- 5.Ahmad R., Pandey R.B., Anf S.H., Nabi N., Mumtaz J., ul Hasnain A. Polymorphic β and γ lens crystallins demonstrate latitudinal distribution of threatened walking catfish Clarias batrachus (Linn.) populations in North-Western India. J. Biol. Sci. 2012 doi: 10.3923/jbs.2012.98.104. [DOI] [Google Scholar]
- 6.Beata Więcaszek S.K. Morphometric characteristic of asian walking catfish Clarias batrachus. Electron. J. Polish Agric. Univ. 2010 [Google Scholar]
- 7.Na-Nakorn U., Brummett R.E. Use and exchange of aquatic genetic resources for food and aquaculture: Clarias catfish. Rev. Aquacult. 2009 doi: 10.1111/j.1753-5131.2009.01010.x. [DOI] [Google Scholar]
- 8.Khedkar G.D., Reddy A.C.S., Mann P., Ravinder K., Muzumdar K. Clarias batrachus (Linn.1758) population is lacking genetic diversity in India. Mol. Biol. Rep. 2010 doi: 10.1007/s11033-009-9517-3. [DOI] [PubMed] [Google Scholar]
- 9.Maneechot N., Yano C.F., Bertollo L.A.C., Getlekha N., Molina W.F., Ditcharoen S., Tengjaroenkul B., Supiwong W., Tanomtong A., De Bello Cioffi M. Genomic organization of repetitive DNAs highlights chromosomal evolution in the genus Clarias (Clariidae, Siluriformes) Mol. Cytogenet. 2016 doi: 10.1186/s13039-016-0215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N., Bao L., Zhou T., Yuan Z., Liu S., Dunham R., Li Y., Wang K., Xu X., Jin Y., Zeng Q., Gao S., Fu Q., Liu Y., Yang Y., Li Q., Meyer A., Gao D., Liu Z. Genome sequence of walking catfish (Clarias batrachus) provides insights into terrestrial adaptation. BMC Genom. 2018 doi: 10.1186/s12864-018-5355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien K.M. Mitochondrial biogenesis in cold-bodied fishes. J. Exp. Biol. 2011 doi: 10.1242/jeb.046854. [DOI] [PubMed] [Google Scholar]
- 12.Greenway R., Barts N., Henpita C., Brown A.P., Rodriguez L.A., Rodríguez Peña C.M., Arndt S., Lau G.Y., Murphy M.P., Wu L., Lin D., Tobler M., Kelley J.L., Shaw J.H. Convergent evolution of conserved mitochondrial pathways underlies repeated adaptation to extreme environments. Proc. Natl. Acad. Sci. U.S.A. 2020 doi: 10.1073/pnas.2004223117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokolova I. Mitochondrial adaptations to variable environments and their role in animals' stress tolerance. Integr. Comp. Biol. 2018 doi: 10.1093/icb/icy017. [DOI] [PubMed] [Google Scholar]
- 14.White C.R., Alton L.A., Frappell P.B. Metabolic cold adaptation in fishes occurs at the level of whole animal, mitochondria and enzyme. Proc. R. Soc. B Biol. Sci. 2012 doi: 10.1098/rspb.2011.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shtolz N., Mishmar D. The mitochondrial genome–on selective constraints and signatures at the organism, cell, and single mitochondrion levels. Front. Ecol. Evol. 2019 doi: 10.3389/fevo.2019.00342. [DOI] [Google Scholar]
- 16.Stairs C.W., Leger M.M., Roger A.J. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Philos. Trans. R. Soc. B Biol. Sci. 2015 doi: 10.1098/rstb.2014.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison R.G. Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends Ecol. Evol. 1989 doi: 10.1016/0169-5347(89)90006-2. [DOI] [PubMed] [Google Scholar]
- 18.Zardoya R., Meyer A. Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Mol. Biol. Evol. 1996 doi: 10.1093/oxfordjournals.molbev.a025661. [DOI] [PubMed] [Google Scholar]
- 19.Tomasco I.H., Lessa E.P. The evolution of mitochondrial genomes in subterranean caviomorph rodents: adaptation against a background of purifying selection. Mol. Phylogenet. Evol. 2011 doi: 10.1016/j.ympev.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Yu L., Wang X., Ting N., Zhang Y. Mitogenomic analysis of Chinese snub-nosed monkeys: evidence of positive selection in NADH dehydrogenase genes in high-altitude adaptation. Mitochondrion. 2011 doi: 10.1016/j.mito.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Foote A.D., Morin P.A., Durban J.W., Pitman R.L., Wade P., Willerslev E., Gilbert M.T.P., Fonseca R.R.D. Positive selection on the killer whale mitogenome. Biol. Lett. 2011 doi: 10.1098/rsbl.2010.0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garvin M.R., Bielawski J.P., Gharrett A.J. Positive darwinian selection in the piston that powers proton pumps in Complex I of the mitochondria of Pacific salmon. PloS One. 2011 doi: 10.1371/journal.pone.0024127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuff J.A., Barton G.J. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins Struct. Funct. Genet. 2000 doi: 10.1002/1097-0134(20000815)40:3<502::AID-PROT170>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 24.Adamczak R., Porollo A., Meller J. Accurate prediction of solvent accessibility using neural networks-based regression. Proteins Struct. Funct. Genet. 2004 doi: 10.1002/prot.20176. [DOI] [PubMed] [Google Scholar]
- 25.Ofran Y., Rost B., ISIS Interaction sites identified from sequence. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btl303. [DOI] [PubMed] [Google Scholar]
- 26.Uversky V.N. Intrinsically disordered proteins and their “Mysterious” (meta)physics. Front. Physiol. 2019 doi: 10.3389/fphy.2019.00010. [DOI] [Google Scholar]
- 27.Peng Z., Yan J., Fan X., Mizianty M.J., Xue B., Wang K., Hu G., Uversky V.N., Kurgan L. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell. Mol. Life Sci. 2014 doi: 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuxreiter M., Tóth-Petróczy Á., Kraut D.A., Matouschek A.T., Lim R.Y.H., Xue B., Kurgan L., Uversky V.N. Disordered proteinaceous machines. Chem. Rev. 2014 doi: 10.1021/cr4007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T., Kim P.M., Kriwacki R.W., Oldfield C.J., Pappu R.V., Tompa P., Uversky V.N., Wright P.E., Babu M.M. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014 doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obradovic Z., Peng K., Vucetic S., Radivojac P., Dunker A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins Struct. Funct. Genet. 2005 doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 32.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PloS One. 2012 doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005 doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheffler I.E. Molecular genetics of succinate:quinoi oxidoreductase in eukaryotes. Prog. Nucleic Acid Res. Mol. Biol. 1998 doi: 10.1016/S0079-6603(08)60895-8. [DOI] [PubMed] [Google Scholar]
- 35.da Fonseca R.R., Johnson W.E., O’Brien S.J., Ramos M.J., Antunes A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008 doi: 10.1186/1471-2164-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kazuno A.A., Munakata K., Nagai T., Shimozono S., Tanaka M., Yoneda M., Kato N., Miyawaki A., Kato T. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006 doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno-Loshuertos R., Acín-Pérez R., Fernández-Silva P., Movilla N., Pérez-Martos A., De Cordoba S.R., Gallardo M.E., Enríquez J.A. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 2006 doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 38.Kiiskilä J., Moilanen J.S., Kytövuori L., Niemi A.K., Majamaa K. Analysis of functional variants in mitochondrial DNA of Finnish athletes. BMC Genom. 2019 doi: 10.1186/s12864-019-6171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garvin M.R., Bielawski J.P., Sazanov L.A., Gharrett A.J. Review and meta-analysis of natural selection in mitochondrial complex I in metazoans. J. Zool. Syst. Evol. Res. 2015 doi: 10.1111/jzs.12079. [DOI] [Google Scholar]
- 40.Hill J., Enbody E.D., Pettersson M.E., Sprehn C.G., Bekkevold D., Folkvord A., Laikre L., Kleinau G., Scheerer P., Andersson L. Recurrent convergent evolution at amino acid residue 261 in fish rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2019 doi: 10.1073/pnas.1908332116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin Y.H., Chen S.Y., Lai S.J. Polymorphisms of mitochondrial ATPASE 8/6 genes and association with milk production traits in holstein cows. Anim. Biotechnol. 2012 doi: 10.1080/10495398.2012.686468. [DOI] [PubMed] [Google Scholar]
- 42.Fernández A.I., Alves E., Fernández A., De Pedro E., López-García M.A., Ovilo C., Rodríguez M.C., Silió L. Mitochondrial genome polymorphisms associated with longissimus muscle composition in Iberian pigs. J. Anim. Sci. 2008 doi: 10.2527/jas.2007-0568. [DOI] [PubMed] [Google Scholar]
- 43.Zhang B., Chen H., Hua L., Zhang C., Kang X., Wang X., Pan C., Lan X., Lei C. Novel SNPs of the mtDNA ND5 gene and their associations with several growth traits in the Nanyang cattle breed. Biochem. Genet. 2008 doi: 10.1007/s10528-008-9152-z. [DOI] [PubMed] [Google Scholar]
- 44.Sutarno J.M., Cummins J., Greeff A.J., Lymbery Mitochondrial DNA polymorphisms and fertility in beef cattle. Theriogenology. 2002 doi: 10.1016/S0093-691X(02)00664-7. [DOI] [PubMed] [Google Scholar]
- 45.Chomyn A. Mitochondrial genetic control of assembly and function of complex I in mammalian cells. J. Bioenerg. Biomembr. 2001 doi: 10.1023/A:1010791204961. [DOI] [PubMed] [Google Scholar]
- 46.Bai Y., Shakeley R.M., Attardi G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell Biol. 2000 doi: 10.1128/mcb.20.3.805-815.2000. [DOI] [Google Scholar]
- 47.Uversky V.N. Intrinsic disorder-based protein interactions and their modulators. Curr. Pharmaceut. Des. 2013 doi: 10.2174/1381612811319230005. [DOI] [PubMed] [Google Scholar]
- 48.Uversky V.N. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem. Soc. Rev. 2011 doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- 49.Uversky V.N. Paradoxes and wonders of intrinsic disorder: stability of instability. Intrinsically Disord. Proteins. 2017 doi: 10.1080/21690707.2017.1327757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uversky V.N., Oldfield C.J., Dunker A.K. Intrinsically disordered proteins in human diseases: introducing the D 2 concept. Annu. Rev. Biophys. 2008 doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 51.Vacic V., Markwick P.R.L., Oldfield C.J., Zhao X., Haynes C., Uversky V.N., Iakoucheva L.M. Disease-associated mutations disrupt functionally important regions of intrinsic protein disorder. PLoS Comput. Biol. 2012 doi: 10.1371/journal.pcbi.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.