Abstract
Intratumoral heterogeneity is tightly associated with the failure of anticancer treatment modalities including conventional chemotherapy, radiation therapy, and molecularly targeted therapy. Such heterogeneity is generated in an evolutionary manner not only as a result of genetic alterations but also by the presence of cancer stem cells (CSCs). CSCs are proposed to exist at the top of a tumor cell hierarchy and are undifferentiated tumor cells that manifest enhanced tumorigenic and metastatic potential, self-renewal capacity, and therapeutic resistance. Properties that contribute to the robustness of CSCs include the abilities to withstand redox stress, to rapidly repair damaged DNA, to adapt to a hyperinflammatory or hyponutritious tumor microenvironment, and to expel anticancer drugs by the action of ATP-binding cassette transporters as well as plasticity with regard to the transition between dormant CSC and transit-amplifying progenitor cell phenotypes. In addition, CSCs manifest the phenomenon of metabolic reprogramming, which is essential for maintenance of their self-renewal potential and their ability to adapt to changes in the tumor microenvironment. Elucidation of the molecular underpinnings of these biological features of CSCs is key to the development of novel anticancer therapies. In this review, we highlight the pathological relevance of CSCs in terms of their hallmarks and identification, the properties of their niche—both in primary tumors and at potential sites of metastasis—and their resistance to oxidative stress dependent on system xc (−).
Keywords: Cancer stem cell, CD44 variant, Epithelial-to-mesenchymal transition (EMT), Intratumoral heterogeneity, Niche, Plasticity
Abbreviations: ABC, ATP-binding cassette; ALDH, Aldehyde dehydrogenase; BMP, Bone morphogenetic protein; CAF, Cancer-associated fibroblast; CagA, Cytotoxin-associated gene A; CD44v, CD44 variant; CSC, Cancer stem cell; CTC, Circulating tumor cell; DTC, Disseminated tumor cell; ECM, Extracellular matrix; EGF, Epidermal growth factor; E/M, Epithelial/mesenchymal; EMT, Epithelial-to-mesenchymal transition; EpCAM, Epithelial cell adhesion moleculeE; SRP1, Epithelial splicing regulatory protein 1; GSC, Glioma stem cell; GSH, reduced glutathione; HGF, Hepatocyte growth factor; HNSCC, Head and neck squamous cell cancer; IL, Interleukin; MAPK, mitogen-activated protein kinase; MET, mesenchymal-to-epithelial transition; Nrf2, nuclear factor erythroid 2–related factor 2; NSCLC, non–small cell lung cancer; OXPHOS, Oxidative phosphorylation; Prrx1, Paired-related homeodomain transcription factor 1; ROS, Reactive oxygen species; TGF-β, Transforming growth factor–β
Highlights
-
•
Intratumoral heterogeneity driven by CSCs is responsible for therapeutic resistance.
-
•
CTCs survive in the distant organs and achieve colonization, causing metastasis.
-
•
E/M hybrid cancer cells due to partial EMT exhibit the highest metastatic potential.
-
•
The CSC niche regulates stemness in metastatic disease as well as in primary tumor.
-
•
Activation of system xc(-) by CD44 variant in CSCs is a promising therapeutic target.
1. Emerging concept and molecular hallmarks of cancer stem cells
1.1. Hallmarks of cancer stem cells
Cancer stem cells (CSCs) have been defined as a “cellular population within a tumor which exhibits the capacity to self-renew and to cause the heterogeneous lineages of cancer cells that comprise the tumor tissue” [[1], [2], [3]]. Accumulating evidence suggests that CSCs also constitute a small subpopulation of tumor cells that show an enhanced potential for metastatic dissemination and enhanced resistance to anticancer therapies. CSCs possess the property of “robustness,” which refers to several biological characteristics including metabolic reprogramming [4,5], resistance to oxidative stress [4,6], an immunosuppressive phenotype [7,8], the ability to rapidly repair damaged DNA [9,10], plasticity in the transition between dormant and transit-amplifying progenitor cells [11,12], an enhanced ability to expel anticancer agents via ATP-binding cassette (ABC) transporters [13,14], and the capacity to adapt to a hyperinflammatory or hyponutritious microenvironment [15,16]. In particular, metabolic reprogramming is thought to be crucial for CSCs to maintain their self-renewal potential and their ability to adapt to marked changes in the tumor microenvironment [5,17].
For a typical instance, metabolic plasticity has been characterized in glioma stem cells (GSCs) [18]. Examination of the metabolic requirements of GSCs during tumor initiation revealed that both glycolysis and oxidative phosphorylation (OXPHOS) are able to independently sustain the emergence of primary disease [19]. Both aerobic glycolysis and OXPHOS can also maintain tumor propagation by isogenic GSCs, and GSCs that are dependent on OXPHOS are able to switch to a glycolytic phenotype in response to metabolic stress, indicative of the plasticity of GSC metabolism [18]. A promising strategy to overcome this metabolic heterogeneity and plasticity of GSCs might therefore be dual blockade of glycolysis and OXPHOS. Indeed, treatment with both glycolysis inhibitor 2-deoxyglucose (2DG) and anti-diabetic drug metformin has been shown to be effective for suppression of tumor growth and metastasis in various preclinical cancer models [20]. Heterogeneity and plasticity of cancer metabolism thus contribute to therapeutic resistance [5,21]. Another example of this phenomenon is provided by malignant melanoma [22,23]. The oncogenic protein BRAF(V600E) induces down-regulation of oxidative enzymes and mitochondrial number and function and thereby enhances production of lactate in melanoma cells [22]. This metabolic reprogramming triggered by activated BRAF is mediated by suppression of the transcriptional factors MITF and PGC1α, both of which are major regulators of mitochondrial biogenesis and function [22]. However, melanoma cells that express the histone H3 lysine-4 (H3K4) demethylase JARID1B at a high level proliferate slowly and are highly dependent on mitochondrial metabolism, and they become enriched in residual disease after chemotherapy [23]. Combination therapy that targets both the bulk and JARID1Bhigh tumor cells is therefore a potential new treatment approach for melanoma.
The clinical relevance of CSCs is highlighted by demonstrations of their intrinsic resistance to chemotherapy and radiation, with such resistance being largely attributable to their quiescence [24,25]. In addition, the discovery of treatment- or microenvironment-induced phenotypic conversion of differentiated cancer cells into CSCs has revealed such plasticity to be a key challenge to the eradication of malignancy [12,26]. For example, in vitro studies have provided evidence that ovarian tumor cells can acquire CSC characteristics after their exposure to cisplatin [27]. Mechanisms underlying the therapeutic resistance of CSCs include the up-regulation of drug-efflux pumps and antiapoptotic proteins as well as interactions of these cells with the protective niche that they foster and inhabit [13,28]. ABC subfamily B member 5 (ABCB5) is highly expressed in primary glioblastoma multiforme, with its expression level being correlated with that of the CSC marker protein CD133 and with poor patient survival [29]. Blockade of ABCB5 was found to attenuate G2-M cell cycle arrest and to promote cell death induced by temozolomide in glioblastoma cells, suggesting that ABCB5 is a marker and mediator of chemoresistance. Another example has been provided by the study of induced CSCs, which can be established by the introduction of oncogenes such as those for MYC or RAS into somatic stem cells [30]. A syngeneic mouse model of mature B cell lymphoma based on the induced CSC model has been established [30,31], and it was recently shown that malignant lymphoma cells in this model are resistant to Fas-mediated apoptosis as a result of up-regulation of the antiapoptotic protein Livin [32]. The bromodomain proteins BRD2 and BRD4 were found to be responsible for this up-regulation of Livin by binding to upstream regions of the Livin gene that overlap with regions enriched for histone H3 lysine-27 acetylation (H3K27ac).
1.2. CSC marker molecules that determine stemness
Surrogate assays for CSCs include tumorsphere formation in vitro and limiting-dilution tumorigenicity analysis in immunocompromised mice, the latter of which is the gold standard method [2,30]. Cell surface markers—including CD44, CD133, and epithelial cell adhesion molecule (EpCAM)—as well as aldehyde dehydrogenase (ALDH) enzymatic activity can also contribute to the identification of CSCs. CD44 is an integral membrane glycoprotein that serves as a cell surface receptor for extracellular matrix (ECM) components such as hyaluronate and osteopontin [33]. As described in detail below (Section 4), CD44 is overexpressed in CSCs, and alternatively spliced variants of the protein are thought to play a key role in cancer development and progression [34]. EpCAM is a type 1 transmembrane glycoprotein that contributes to the acquisition and maintenance of stemness by coupling with β-catenin and thereby activating the canonical Wnt signaling pathway and up-regulating the expression of stem cell genes such as those for SOX2, MYC, and KLF4 [35,36]. In addition, EpCAM forms a complex with the amino acid transporter LAT1, which mediates the uptake of leucine and thereby activates mammalian target of rapamycin (mTOR) signaling in CSCs [[37], [38], [39]]. Members of the ALDH family of enzymes catalyze the oxidation of endogenous and exogenous aldehyde substrates to their corresponding carboxylic acids [40], and ALDH enzymatic activity is a major functional marker for the identification of both normal and cancer stem cells [41]. ALDH activation has been found to be positively correlated with radiation resistance and tumor recurrence [40].
CSCs were initially identified in acute myeloid leukemia [42,43]. Since this initial discovery, CSCs have been identified by flow cytometry–based prospective analyses in a wide variety of human malignancies including those of the breast (CD44+/CD24–/low/ALDH1high cells), brain (CD15+/CD133+ cells), prostate (CD44+/CD24–/low cells), liver (AFP+/EpCAM+/N-MYChigh cells), where AFP is α-fetoprotein, colon (CD133+/EpCAMhigh/CD44+ cells), ovary (CD44+/CD117+/EpCAM+ cells), and pancreas (CD44+/CD24+/ESA+ cells), where ESA is epithelial specific antigen [[44], [45], [46], [47], [48], [49], [50]]. These cell subpopulations are relatively rare and form secondary tumors that recapitulate the pathology and intratumoral heterogeneity of the original malignancy on transplantation into experimental immune-deficient mice [2,30]. Intratumoral heterogeneity driven by the presence of CSCs is primarily responsible for the failure to achieve a uniform therapeutic effect among cancer cells as a whole [2]. Whereas CD133, also known as prominin 1, has long served as a marker to identify CSCs [51], tumor cells negative for this glycoprotein have also been found to manifest tumorigenic potential, which questions the legitimacy of CD133 as a bona fide CSC marker [52,53]. Although markers such as CD44 and CD133 have been detected in CSCs for a wide range of tumor types, the accurate isolation and identification of CSCs remains a challenge [2,53,54]. The functional characterization of cancer cell subpopulations defined by putative CSC markers is thus crucial for further CSC research.
Increasing evidence suggests that lineage tracing and single-cell analysis are useful for the identification of CSCs [[55], [56], [57], [58], [59]]. Analysis of Lgr5-positive intestinal crypt stem cells in APC mutant mice, which develop intestinal adenoma, revealed that these cells constituted ~5%–10% of the total tumor cell population, a proportion similar to that for such cells in normal crypts [55]. Furthermore, lineage tracing revealed that these Lgr5-positive tumor cells were able to generate all of the other cell types present in the adenoma, indicating that they are multipotent stem cells of the adenoma [55]. Stromal cues derived from the osteopontin-rich CSC niche have also been found to influence cancer cell hierarchy in established colon adenocarcinoma and to have marked effects on the tumor mass as a whole [58]. This finding regarding the molecular determinants of stemness has the potential to shift the focus from targeting of tumor cell subpopulations that express conventional CSC markers toward targeting of the niche. Studies of organoid xenografts and other mouse models have revealed that targeted ablation of Lgr5-positive cancer cells in colorectal adenocarcinoma is sufficient to disrupt tumor growth [60,61]. However, tumor expansion was found to immediately resume after cessation of Lgr5-positive CSC depletion, revealing Lgr5-positive cancer cells to be a major contributor to tumor growth as well as pronounced plasticity within the tumor tissue.
2. EMT and plasticity contribute to the expansion of CSCs
2.1. Orchestration between EMT and MET in the development of metastatic disease
Cancer cells that have undergone epithelial-to-mesenchymal transition (EMT) are highly likely to invade surrounding tissue and to metastasize, and thereby to give rise to the life-threatening manifestations of disease progression [[62], [63], [64]]. The central role played by loss of the adhesion molecule E-cadherin in the EMT program is revealed by the actions of several EMT-inducing transcription factors that facilitate acquisition of a mesenchymal phenotype characterized by the induction of vimentin and N-cadherin [65]. Loss of E-cadherin promotes Wnt signaling and is associated with an increased abundance of the transcription factor Snail in the nucleus [66]. EMT has long been thought to result in an increase in the number of CSCs at the invasive front of tumors and in metastatic foci [[67], [68], [69]]. The EMT program that allows breast cancer cells to disseminate from a primary lesion promotes their self-renewal capacity. Indeed, breast cancer cells that have undergone EMT acquire CSC properties, including the ability to self-renew and tumorigenicity in association with a CD44+/CD24−/low/ALDHhigh phenotype [70]. Several transcription factors that induce EMT in cancer cells have been identified, including FOXC2 in basal-like breast cancer [71,72]; SIP1 (also known as ZEB2) in ovarian, breast, hepatic, and colorectal tumors [[73], [74], [75]]; and Snail, Slug, and Twist in various types of solid tumor [63,76]. For instance, FOXC2 expression is sufficient to promote EMT and the acquisition of CSC properties—including chemoresistance, the capacity for tumor initiation, and metastatic competence—in transformed human mammary epithelial cells [72].
In contrast, mesenchymal-to-epithelial transition (MET) is a key step in the colonization and proliferation of cancer cells at the premetastatic niche [77,78]. MET is thought to result from the interactions of metastatic cancer cells with the premetastatic niche and to promote cell survival and growth coincident with reversion from the mesenchymal phenotype to the parental cancer phenotype [64,79]. This scenario is consistent with Paget's theory that circulating tumor cells (CTCs) (the “seeds”) of certain cancers can survive in the “soil” of distant organs and thereby achieve colonization and give rise to metastatic disease [[78], [79], [80], [81]]. Knockdown of paired-related homeodomain transcription factor 1 (Prrx1) induces MET in and promotes metastatic colonization of the lungs by breast cancer cells [82]. Unlike classical EMT transcription factors such as Snail, Slug, and ZEB1, the loss of Prrx1 is required for metastatic colonization by cancer cells in vivo, with the cells reverting to the epithelial phenotype concomitant with the acquisition of stem cell properties [82,83]. Breast cancer cells overexpressing Prrx1 thus fail to give rise to lung metastases, whereas silencing of Prrx1 promotes efficient lung colonization, suggesting that MET is crucial for pulmonary metastasis [82,84]. These various observations thus suggest that the development of metastatic disease requires orchestration of both EMT and MET.
2.2. Partial EMT and induction of E/M hybrid CTCs
Cancer cells have also been found to undergo partial or incomplete EMT, which is thought to increase their invasive potential, to generate both CSCs and CTCs, and to promote resistance to anticancer drugs [84,85]. Cancer cells thus often manifest an epithelial/mesenchymal (E/M) hybrid phenotype characterized by the simultaneous expression of both epithelial and mesenchymal markers during the processes of invasion and distant metastasis [[84], [85], [86]]. In addition, the coexpression of both epithelial and mesenchymal markers is associated with resistance to chemotherapy and a poor clinical outcome in several tumor types [84,[87], [88], [89]]. Single-cell transcriptomics analysis applied to evaluate the heterogeneity of head and neck squamous cell cancer (HNSCC) cells identified a partial EMT program defined by incomplete activation of EMT transcription factors such as Grhl2 and Ovol2 [90,91]. Knockdown of Grhl2 and Ovol2 in these HNSCC cells impaired collective cell migration, which is a hallmark of partial EMT [91]. The E/M hybrid phenotype is associated with increased cancer stemness, whereas the fully epithelial or mesenchymal phenotypes are associated with loss of both stem cell markers and tumorigenicity [92]. Cancer cells that have undergone partial EMT have been found to be localized at the leading edge of tumors and to show the highest metastatic potential in cooperation with cancer-associated fibroblasts (CAFs) [84,90,93]. CAFs have thus been shown to enhance the invasive potential of E/M hybrid-type tumor cells, which are associated with epithelial-type cancer cell clusters, leading to collective invasion of both epithelial and E/M hybrid tumor cell clusters [[93], [94], [95]]. In the clinical setting, individuals with advanced-stage breast cancer in whom CTCs manifested the E/M hybrid phenotype (defined by expression of cytokeratin and ALDH1 as well as nuclear localization of Twist 1) after chemotherapy had a shorter progression-free survival compared with those whose CTCs did not have this phenotype, and this association was especially prominent in patients with HER2-negative breast cancer [96].
2.3. Plasticity between CSCs and other tumor cells regulated by epigenetic modification
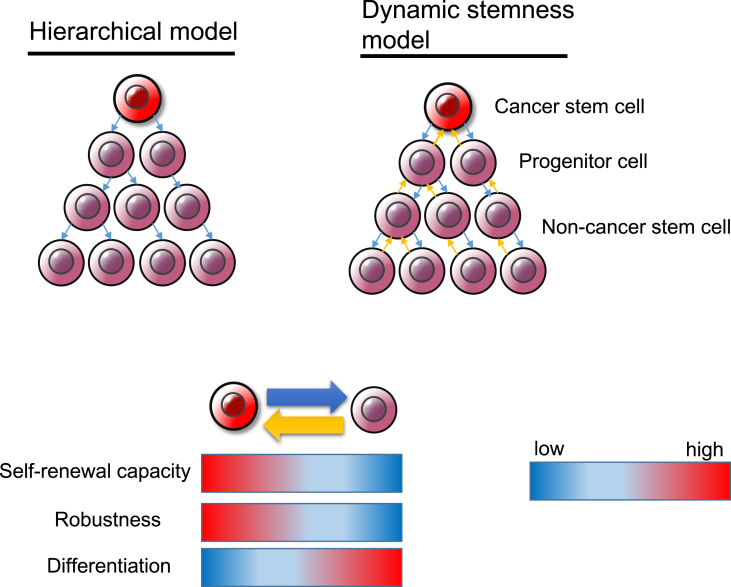
According to the “dynamic stemness model,” CSCs are not static entities but rather undergo dynamic and reversible changes depending on the tumor microenvironment [[97], [98], [99]] (Fig. 1). Although it may seem paradoxical that CSCs, which are located at the top of a hierarchical tumor cell population, undergo reversible phenotypic alterations, epigenetic changes induced by various factors including chronic inflammation, excessive oxidative stress, and hypoxic stimuli enhance the plasticity of the transition between mostly quiescent, symmetrically dividing CSCs and proliferative, asymmetrically dividing cells and can thereby increase the number of non-CSC tumor cells [67,99,100]. Indeed, the dual nature of CSCs allows them to adapt to changes in the tumor microenvironment. However, this plasticity of CSCs hinders their clinical identification in vivo [101,102]. Certain tumor cells follow a phenotypic plasticity model in which phenotypic heterogeneity is driven largely by reversible changes in the tumorigenic cell subpopulation rather than a hierarchical model supported by irreversible epigenetic changes. Tumorigenic malignant melanoma cells show unlimited tumorigenicity in serial transplantation experiments regardless of the expression status of potential CSC markers including ABCB5, CD133, and CD271 [103,104]. In addition, the reversible acquisition of stemlike phenotypes has been observed in glioblastoma in response to transient exposure to nitric oxide (NO) produced by endothelial NO synthase (eNOS) expressed in the tumor vascular endothelium, a perivascular niche [105]. NO thus activates the Notch signaling pathway in malignant glioma cells exposed to platelet-derived growth factor and thereby induces the acquisition of CSC-like characteristics [105,106]. GSCs are therefore unlikely to conform to the hierarchical model based on CSCs as a minor undifferentiated subpopulation with marked tumorigenic potential. The transition between CSCs and non-CSC tumor cells appears to be dependent on both tumor type and the tumor microenvironment. A treatment-induced transient decrease in the extent of cancer heterogeneity is thought to reflect enrichment of CSCs in minimal residual disease [2,5,107]. Long-term treatment of melanoma with vemurafenib, which targets constitutively active mutant BRAF, increases expression of the histone demethylase JARID1B, which was found to be highly expressed in slow-cycling cells with a stemlike phenotype, leading to the development of adaptive resistance [23]. Regulation of the abundance of trimethylated H3K4 by JARID1B constitutes an epigenetic switch at several proto-oncogenes and tumor suppressor genes whose expression is related to the plasticity between CSCs and other tumor cells [23,108] (Fig. 1). Crosstalk between oncogenic signaling pathways and reversible epigenetic alterations may thus give rise to adaptive resistance to anticancer treatment and other types of exogenous stress in the tumor microenvironment.
Fig. 1.
The hierarchical and dynamic stemness models. In the hierarchical model, cancer stem cells (CSCs) give rise to differentiated non-CSCs and thereby contribute to the maintenance and growth of tumor tissue. The coexistence of both CSCs and non-CSC tumor cells is responsible for intratumoral heterogeneity. In contrast, the dynamic stemness model emphasizes the plasticity between CSCs and non-CSCs. According to this model, the transition between undifferentiated CSCs and differentiated non-CSCs is reversible.
3. Role of the CSC niche in maintenance of stemness and the potential for distant metastasis
3.1. Components of the tumor microenvironment that maintain stemness
The niche is a specialized local site in the stromal microenvironment of stem cells that integrates signals reflecting tissue and organismal state and regulates stem cell fate commitment and epithelial cell plasticity during tissue homeostasis and regeneration [109,110]. The niche may be cellular in nature, as appears to be the case for melanocyte stem cells located in the bulge area of hair follicles and Paneth cells located in intestinal crypts, which are thought to form niches for normal hair follicle stem cells and intestinal stem cells, respectively [111,112]. Stemness of CSCs has also been found to be supported by niches whose components include endothelial cells, osteoblasts, and ECM molecules such as hyaluronic acid and osteopontin [113,114]. In addition, CAFs, tumor-associated macrophages, undifferentiated mesenchymal stem cells, and immune cells in the tumor stroma serve as cellular components of niches [115,116]. Such stromal cells contribute to formation and maintenance of the niche by providing growth factors such as epidermal growth factor (EGF), hepatocyte growth factor (HGF), and transforming growth factor-β (TGF-β) as well as proinflammatory cytokines such as tumor necrosis factor-α (TNF-α) and various interleukins including IL-1β and IL-6 [[117], [118], [119]]. An inflammatory microenvironment is beneficial for cancer cells in that it results in activation of the nuclear factor-κB (NF-κB) signaling pathway [[120], [121], [122]]. The cytokine network not only promotes tumor development but also maintains stemness that underlies tumor metastasis and recurrence. Furthermore, both CAFs and tumor-associated macrophages drive stemness of cancer cells as well as neoangiogenesis, remodeling of ECM, and attenuation of the host immune response [123,124]. Evidence suggests that resistance to cisplatin can be triggered by IL-6 and interferon released by stromal cells [125,126]. The CSC niche is thus an essential regulator of stemness not only in primary tumors but also in metastasized disease [110,127].
3.2. Harmonious interplay between CSCs and their niche
The relevance of the interaction between CSCs and their niche is supported by the fact that the loss of the niche microenvironment results in depletion of the CSC population [128,129]. The reliance of CSCs on niche signals appears to be a general phenomenon, having been demonstrated in a wide variety of tumor types. In addition to maintaining the CSC pool and promoting the proliferation of primary cancer cells, the CSC niche plays a role in the reversion of non-CSC tumor cells to CSCs by a process related to EMT and thereby promotes tumor invasion and dissemination (Fig. 1) [130,131]. Studies of breast cancer cells have identified a subpopulation of non-CSCs that are highly plastic and can adopt the CSC state [[132], [133], [134]]. This transition may be the result of a stochastic event or can be driven by the niche [134,135]. Molecular components and stiffness of the tumor stroma associated with the premetastatic niche can induce the stemness phenotype and plasticity in premetastatic lesions by eliciting EMT-related signaling pathways [136,137]. In turn, cells that have undergone EMT secrete ECM proteins to help construct a permissive niche for themselves at potential metastatic sites [138]. Whereas most disseminated tumor cells (DTCs) arriving at new organs manifest EMT-like features associated with a stem cell gene signature and maintain a quiescent state [139], the formation of metastases requires that they adopt a more epithelial phenotype, down-regulate the stem cell gene program related to quiescence, and up-regulate the expression of proliferative genes related to stemness, such as that for c-MYC.
Driver gene mutations allow gastric CSCs to survive and proliferate independently of their native niche constraint [140]. The amplification of genes encoding receptor tyrosine kinases has also been found to render cancer cells independent of niche factors such as EGF, fibroblast growth factor 10 (FGF10), and HGF [141]. In addition, the independence of pancreatic ductal adenocarcinoma with regard to niche factors is mainly acquired as a result of driver mutations, although loss of dependence on Wnt ligands was found to be mediated by epigenetic changes, suggestive of a complex niche-adaptation process during the development of such tumors [142]. Given the loss of dependence of advanced neoplasms on niche factors, it remains to be determined whether therapeutic strategies that target only the niche could provide the “silver bullet’ to eliminate CSCs.
3.3. Self-seeding model and metastatic niche model
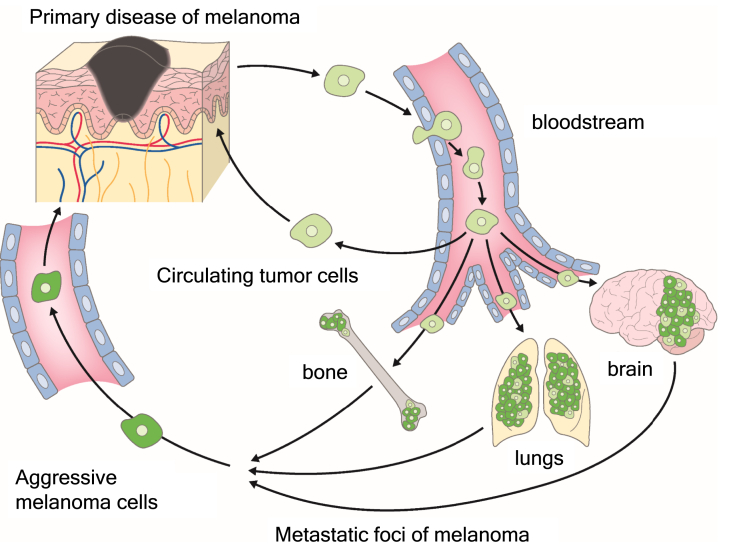
CTCs are described as tumor cells that are shed by primary disease into the vasculature and which then circulate in the bloodstream of cancer patients [[143], [144], [145]]. It has been thought that EpCAM-positive tumor cells circulate in the body even in the primary stages of disease, long before they metastasize to various organs [145]. CTCs that survive in the bloodstream have been found to manifest an E/M hybrid phenotype, to be resistant to anoikis, and to be able to exit the bloodstream efficiently [85,146]. CTCs derived from squamous cell cancer, however, have been found to be EpCAM negative and to be enriched in cells in early E/M hybrid states. Tumor cells with an E/M hybrid phenotype not only exhibited increased lung colonization ability in vivo, but also underwent intravasation into the blood circulation more efficiently [147]. CTCs also have the potential to return to and grow in their tumor of origin, in a process referred to as “tumor self-seeding” [148]. Self-seeding of cancers in mouse models is preferentially mediated by aggressive CTCs including those with a bone, brain, or lung metastatic tropism [148,149] (Fig. 2). Unlike the colonization of distant organs, self-seeding requires little, if any, additional adaptation of CTCs to the recipient tumor microenvironment [81,148]. Self-seeding tends to enrich metastatic cancer cell subpopulations that are more aggressive compared with the bulk cell population of the primary disease. Tumor-derived cytokines such as IL-6 and IL-8 act as CTC attractants, whereas matrix metalloproteinase 1 and fascin-1, a component of the actin cytoskeleton, are implicated as major mediators of CTC infiltration into the primary disease [150].
Fig. 2.
Tumor self-seeding by circulating tumor cells (CTCs). CTCs tend to reinfiltrate an established tumor, enriching it with aggressive cells that have withstood a period of dissemination. This process, known as “tumor self-seeding,” may have consequences for tumor growth and the generation of metastatic cell progeny.
Clusters of CTCs detected in the bloodstream have been shown to comprise oligoclonal cancer cell aggregates rather than to arise from intravascular aggregation of tumor cells, and they are associated with increased metastatic capacity and poorer patient outcome compared with single CTCs [[151], [152], [153]]. CTC clusters present in the blood of patients with breast cancer are strongly positive for mesenchymal markers and weakly positive for epithelial markers, indicative of the role of partial EMT in metastatic dissemination of tumor cells [146].
A subpopulation of CTCs is able to colonize distant organs and to persist as DTCs. In turn, an even smaller fraction of DTCs is able to progress to give rise to distant metastases. The metastatic niche model proposes that DTCs seek out compatible niches able to promote their survival and proliferation, and that the metastatic niche evolves in association with disease progression [131]. The “seed and soil” hypothesis first proposed the necessity of a specific compatible “soil” for the growth of metastases, forming the basis for the concept of the metastatic niche that is now supported by recent experimental studies [79,80,154,155]. Metastatic niches can be formed either on arrival of DTCs in the recipient tissue, or under the influence of secreted factors or exosomes released by the primary tumor prior to the seeding of DTCs [156]. Many DTCs in bone marrow of cancer patients have been found to have undergone EMT [144,157]. Although this is an important finding, given that a subpopulation of DTCs are the precursors of macrometastases, it is not yet possible to predict which specific DTCs will survive and escape dormancy to give rise to such lesions. Bone marrow is a relatively hypoxic environment, raising the possibility that hypoxia-induced EMT processes in local stromal cells and arriving cancer cells promote colonization of the niche followed by a period of dormancy [81,158,159]. Mounting evidence suggests the existence of premetastatic niches in various organs including lymph nodes, lung, liver, bone, and, to a lesser extent, the brain [155]. Metastatic niche formation also occurs through occupancy of preexisting resident stem cell niches by DTCs. For instance, DTCs derived from advanced prostate carcinoma frequently metastasize to bone marrow in association with the production and secretion of parathyroid hormone-related protein (PTHrP) [160,161]. This protein promotes bone remodeling, which likely facilitates the homing of DTCs to bone marrow and their occupation of the osteoblastic niche for normal hematopoietic stem cells [162,163]. In addition, interaction of the osteoblast-derived chemokine CXCL12 (also known as SDF1) with its receptor CXCR4 expressed on the surface of prostate cancer cells drives the homing of the cancer cells to bone marrow [162], suggesting that the CXCL12-CXCR4 axis promotes the survival and proliferation of DTCs. Neutralizing antibodies to CXCR4 have been found to be effective for prevention of prostate carcinoma metastasis in a preclinical model [164,165].
3.4. Dormancy of DTCs which potentially promotes metastasis and chemoresistance
Metastasis development can occur after removal of the primary disease and a long period of time without clinical symptoms. Quiescent DTCs may thus exist undetected for long periods—years or even decades—and may account for prolonged asymptomatic minimal residual disease and therapeutic resistance [158,[166], [167], [168]]. The bone morphogenetic protein (BMP) 4 inhibitor COCO (also known as DAND5) was found to prevent the onset of dormancy in solitary breast cancer cells by activating a self-renewal program and restricting quiescence [169], suggesting that dormant DTCs may need to inhibit BMP signaling in order to escape the quiescent state and become proliferative. TGF-β2 and BMP4 or BMP7 signaling in dormant DTCs derived from HNSCC activates the mitogen-activated protein kinase (MAPK) p38, inhibits the MAPK ERK, and induces expression of the cyclin-dependent kinase inhibitors such as p21 and p27 [170]. Canonical (mediated by SMAD1 or SMAD5) and noncanonical signaling by TGF-β2 results in up-regulation of the transcriptional regulator DEC2 and BMP7 signaling and induces the expression of NDRG1 in prostate cancer cells, which in turn leads to induction of cell cycle inhibitors and cancer cell dormancy [171]. Importantly, systemic inhibition of the TGF-β2 receptor TGF-β-R1 or of p38 MAPK activity was found to awaken dormant DTCs and to drive metastasis at multiple organs including liver, spleen, lung, and bone marrow [170]. These findings thus suggest the importance of inhibition of TGF-β signaling for the activation of quiescent DTCs.
Autophagy is an evolutionarily conserved process for the degradation of cellular macromolecules and organelles [172,173]. Autophagy allows dormant DTCs to maintain metabolic robustness while inhibiting signaling by the PI3K (phosphatidylinositol 3-kinase)–Akt pathway [174]. For instance, ARHI, a RAS homolog and promoter of autophagy that is down-regulated in ~60%–70% of ovarian cancers, was actually found to be up-regulated together with p21 in dormant ovarian cancer cells in vivo [175]. Inhibition of autophagy with chloroquine in ovarian tumor cells that had been rendered dormant by AHRI expression in vivo resulted in attenuation of tumor regrowth. Autophagy is thus thought to promote the survival of cancer cells during dormancy and to support their resistance to therapy.
4. CD44 and system xc(−) promote resistance to redox stress in CSCs
4.1. The CD44v-xCT-GSH axis confers resistance to oxidative stress
The adhesion molecule CD44, which binds to ECM components such as hyaluronic acid and osteopontin, has been identified as a CSC marker [[176], [177], [178]]. Alternative splicing of the primary transcript of the CD44 gene results in the generation of various CD44 isoforms, which are classified as CD44 standard (CD44s) or CD44 variant (CD44v) depending on the absence or presence of sequences encoded by variant exons [179]. The isoforms CD44v3 and CD44v6 enhance the metastatic potential and drug resistance of breast cancer and malignant melanoma cells, respectively [[180], [181], [182]]. CD44v3 interacts with monocarboxylate transporters 1 and 4, both of which are responsible for the transport of lactate in breast cancer cells [182]. CD44v6 interacts with c-Met, a receptor tyrosine kinase that binds HGF [183], and thereby increases the potential of melanoma cells to migrate to distant organs such as the brain [180]. Furthermore, CSCs that express CD44v8-10 are enriched at the invasive front of gastric and breast carcinoma [67,68]. The expression of CD44v8-10 was found to be inversely correlated with that of c-MYC in a manner dependent on the tumor microenvironment [68]. The RNA binding protein ESRP1 (epithelial splicing regulatory protein 1) and the epigenetic modulator HP1γ (heterochromatin protein 1γ) contribute to the alternative splicing of CD44 pre-mRNA [[184], [185], [186], [187]]. In addition, both normal and cancer cells have been shown to change the splicing pattern of CD44 so as to increase the expression of CD44v during the formation and maintenance of organoids or spheroids in three-dimensional culture with ECM components, suggesting that expression of variant isoforms of CD44 is associated with epithelial organization [188,189]. Of note, gene set enrichment analysis (GSEA) showed that the relation between CD44v-positive and -negative cells is similar to that between induced pluripotent stem cells and mouse embryonic fibroblasts [190], suggesting that CD44v might contribute to maintenance of the undifferentiated stemlike phenotype [191,192].
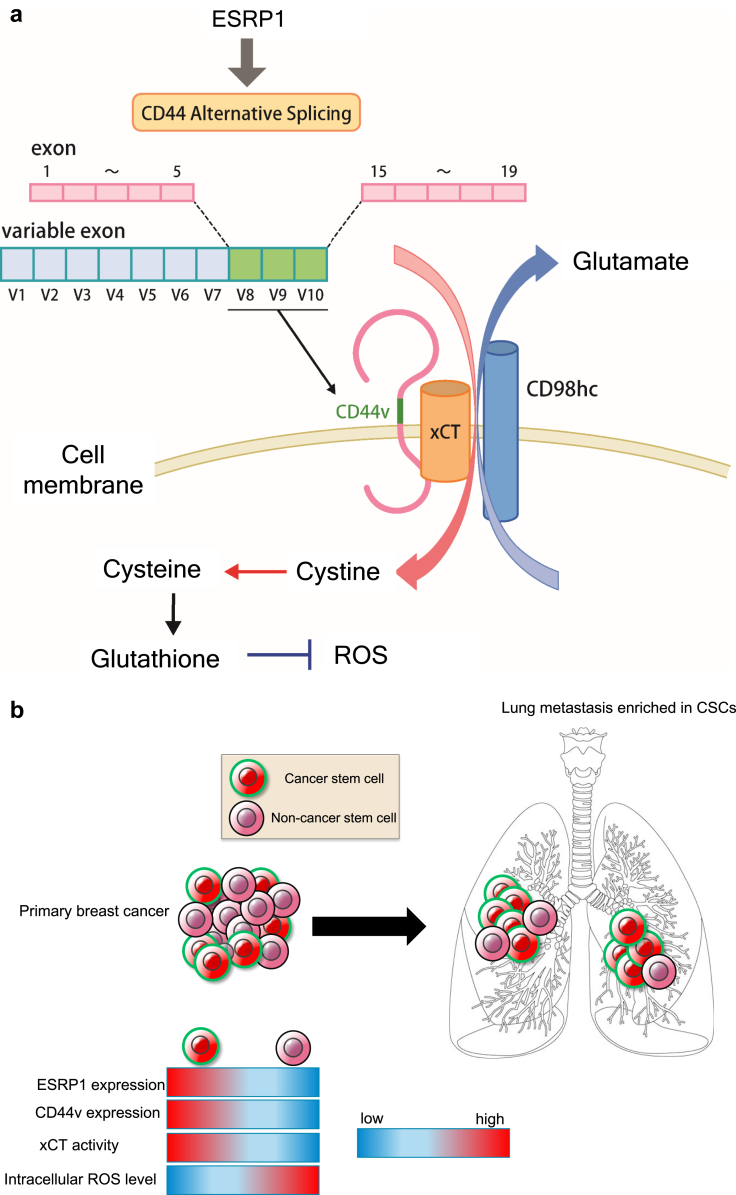
CD44v has also been implicated in resistance to oxidative stress, with CD44v8-10 having been found to interact with and to stabilize xCT (SLC7A11) at the cell surface [6] (Fig. 3a). xCT and CD98 heavy chain (4F2) constitute an antiporter complex known as system xc (−), which mediates the exchange of extracellular cystine for intracellular glutamate [6,39,193]. Cysteine as well as glycine and glutamate are essential substrates for the synthesis of the reduced form of glutathione (GSH). The availability of cysteine is rate limiting for GSH synthesis, however, with the result that the ESRP1-CD44v-xCT-GSH axis is essential for the antioxidant system specific to CSCs [2,6,190]. CD44v8-10 promotes GSH synthesis by facilitating the import of cystine, thereby increasing the intracellular concentration of cysteine. The elimination of reactive oxygen species (ROS) by GSH inhibits the activation of p38 MAPK signaling [6,194] and thereby prevents ROS-induced programmed cell death, senescence, or differentiation of epithelial cancer cells. Furthermore, ferroptosis, an iron-dependent type of regulated necrotic cell death, is related to excessive ROS-induced lipid peroxidation [[195], [196], [197]]. Activation of system xc (−) prevents ferroptosis mediated by glutathione peroxidase 4 (GPX4) [198,199]. The ESRP1-CD44v-xCT-GSH axis in CSCs thus prevents oxidative stress-induced cell death.
Fig. 3.
Regulation of redox stress by the ESRP1-CD44v-xCT-GSH axis. (a) Epithelial splicing regulatory protein 1 (ESRP1) contributes to alternative splicing of CD44 pre-mRNA that generates CD44v8-10 (CD44v). CD44v stabilizes the xCT (SLC7A11) subunit of system xc (−) at the plasma membrane and thereby promotes the intracellular uptake of cystine. Given that cysteine is an essential and rate-limiting factor for synthesis of the reduced form of glutathione (GSH), the interaction between CD44v and xCT protects cells from oxidative stress and the induction of ferroptosis. (b) CD44v-positive cancer stem cells (CSCs) with high levels of GSH are predominantly responsible for colonization of the premetastatic niche for breast cancer cells in the lungs. In contrast to CD44v-negative differentiated tumor cells, CD44v-positive cancer cells are sensitive to sulfasalazine (SSZ).
4.2. CD44v-xCT interaction plays an important role in both carcinogenesis and metastasis
Infection with Helicobacter pylori is responsible for the development of gastric adenocarcinoma [200]. The intracellular accumulation of the protein encoded by cytotoxin-associated gene A (CagA) of the bacterium as a result of impaired autophagy has been observed in CSCs of gastric carcinoma [172,201,202]. Autophagy-dependent degradation of CagA, which is a type IV secretion effector of H. pylori, is activated by depletion of GSH and the consequent increase in oxidative stress and activation of Akt signaling [202]. Activated Akt in CD44v8-10–negative cells induces degradation of the tumor suppressor p53 by the ubiquitin-proteasome system, which in turn results in the degradation of CagA. In contrast, given the resistance to ROS conferred by the CD44v-xCT-GSH axis, gastric CSCs highly expressing CD44v8-10 do not manifest the autophagy-dependent degradation of CagA and contribute to carcinogenesis [202]. These findings thus indicate that redox balance regulated by the CD44v-xCT-GSH axis determines cell fate during the development of gastric cancer in a manner dependent on the extent of CagA accumulation.
Regulation of redox stress is important not only for therapeutic resistance but also for the metastatic potential of cancer cells [190] (Fig. 3b). Highly metastatic murine 4T1 breast cancer cells include a subpopulation positive for CD44v8-10, and 4T1 cells depleted of ESRP1 form significantly fewer nodules and smaller metastatic foci in the lungs after their injection into mammary fat pads than do control 4T1 cells. Although loss of ESRP1 did not affect the expression levels of E-cadherin, ZO-1, N-cadherin, vimentin, or key EMT-related transcriptional factors, it shifted CD44 expression from variant (epithelial) to standard (mesenchymal) isoforms [184,190]. Furthermore, microarray analysis showed that ESRP1-positive cancer cells are undifferentiated relative to ESRP1-negative cells. This finding is consistent with the notion that CD44v8-10–positive tumor cells have a CSC-like phenotype, serving as the cell of origin for metastatic lesions in the experimental model of aggressive breast cancer [190]. ESRP1 likely serves as a determinant of CSC robustness by promoting resistance to oxidative stress. Indeed, tumor-entrained neutrophils were shown to accumulate in the lungs before the arrival of metastatic cells as well as to inhibit lung metastasis in a different murine model of metastatic breast cancer, with this latter effect being mediated by the NADPH-dependent generation of ROS [203,204]. Such an exogenous insult associated with the premetastatic niche may contribute to selective evolutionary pressure to enrich the CSC population.
4.3. Therapeutic targeting of system xc(−) with sulfasalazine
Sulfasalazine, a conventional agent used for the treatment of inflammatory bowel disease and rheumatoid arthritis [205], inhibits the cystine transport activity of xCT. Given that targeting of xCT with sulfasalazine increases the sensitivity of CD44v8-10–positive cancer cells to oxidative stress [6], this drug has been examined in clinical trials for patients with advanced gastric adenocarcinoma or non–small cell lung cancer (NSCLC), both of which express CD44v8-10 [[206], [207], [208]]. This is a typical example of drug repositioning [209,210]. Furthermore, CD44v8-10–positive HNSCC cells have been shown to be susceptible to sulfasalazine, whereas CD44v8-10–negative HNSCC cells are instead sensitive to cetuximab, an antibody to the EGF receptor [211]. The survival and proliferation of CD44v8-10–positive HNSCC cells thus depend on xCT-mediated cystine transport, and inhibition of system xc (−) by sulfasalazine selectively triggers ferroptotic death in these undifferentiated cancer cells. Given the heterogeneity of CD44v expression level among cancer cells in HNSCC, combination therapy with both sulfasalazine and cetuximab may prove effective for tumor eradication [211].
Transcription of the xCT gene is induced by redox stress due to the depletion of cystine or exposure to electrophilic agents, with this effect being mediated through binding of the transcription factor Nrf 2 (nuclear factor erythroid 2–related factor 2) to its response element in the promoter region of the gene [39,212]. The combination of sulfasalazine and auranofin, both of which are disease-modifying antirheumatic drugs (DMARDs), simultaneously inhibits cystine uptake by xCT as well as Nrf2-mediated transcription of the xCT gene [213]. Indeed, translocation of Nrf2 into the nucleus has been shown to increase not only xCT expression but also cystine uptake, indicative of functional activation of system xc (−) [39,214]. Exposure of normal airway epithelial cells to cigarette smoke was recently shown to result in up-regulation of xCT [215], likely as a consequence of transient activation of the Nrf2 signaling pathway [214]. In many cases of NSCLC, the nuclear localization of constitutively active Nrf2 as a result of loss-of-function mutations of the gene encoding the ubiquitin ligase substrate-adapter protein KEAP1 has been shown to prevent the chemotherapy-induced accumulation of ROS [216], consistent with the poor 5-year overall survival rate of patients with NSCLC overexpressing xCT [215]. Also of note, system xc (−) is a key regulator of metabolic reprogramming with overarching effects on glucose metabolism, glutamine dependency, and the balance between reduced and oxidized forms of glutathione in CSCs, all of which are determinants of cancer development and progression.
5. Conclusion
The concept of CSCs in the field of oncology has attracted increasing attention and has led to a growing interest in the molecular machinery underlying the therapeutic resistance attributed to these cells. Resistance to conventional chemotherapy is thus thought to emerge as a result of selective pressure that leads to enrichment of drug-tolerant CSCs. In general, CSCs manifest “robustness” with regard to their ability to repair DNA damage, to withstand oxidative stress, and to adapt to characteristics of the tumor microenvironment such as a deficiency of glucose and growth factors, all of which properties contribute to the maintenance of stemness. Given bona fide markers for the accurate identification of CSCs have yet to be identified, it is important to uncover the function of conventional CSC markers in the maintenance of stemness. Indeed, CSC markers including CD44, CD133, and EpCAM have been found to promote tumor progression, metastasis, and chemoresistance through a variety of signaling pathways. CD44v-positive cancer cells manifest an increased capacity to defend against oxidative stress as a result of enhanced xCT-mediated cystine uptake and consequent GSH synthesis. Furthermore, the favorable tumor microenvironment provided by the CSC niche contributes to intratumoral heterogeneity with regard to genetic alterations and epigenetic modifications. The development of molecularly targeted drugs to eliminate CSCs is thus being pursued as a “silver bullet” for eradication of cancer composed of heterogeneous cell populations.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by a KAKENHI grant from Japan Society for the Promotion of Science (20H00518, to H.S.).
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
References
- 1.Clarke M.F., Dick J.E., Dirks P.B., Eaves C.J., Jamieson C.H., Jones D.L. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Canc Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida G.J., Saya H. Therapeutic strategies targeting cancer stem cells. Canc Sci. 2016;107:5–11. doi: 10.1111/cas.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clara J.A., Monge C., Yang Y., Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells - a clinical update. Nat Rev Clin Oncol. 2020;17:204–232. doi: 10.1038/s41571-019-0293-2. [DOI] [PubMed] [Google Scholar]
- 4.Wu Z., Wei D., Gao W., Xu Y., Hu Z., Ma Z. TPO-induced metabolic reprogramming drives liver metastasis of colorectal cancer CD110+ tumor-initiating cells. Cell Stem Cell. 2015;17:47–59. doi: 10.1016/j.stem.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida G.J. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J Exp Clin Canc Res. 2015;34:111. doi: 10.1186/s13046-015-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishimoto T., Nagano O., Yae T., Tamada M., Motohara T., Oshima H. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Canc Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 7.Codony-Servat J., Rosell R. Cancer stem cells and immunoresistance: clinical implications and solutions. Transl Lung Cancer Res. 2015;4:689–703. doi: 10.3978/j.issn.2218-6751.2015.12.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Q., Long W., Xing C., Chu J., Luo M., Wang H.Y. Cancer stem cells and immunosuppressive microenvironment in glioma. Front Immunol. 2018;9:2924. doi: 10.3389/fimmu.2018.02924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maugeri-Sacca M., Bartucci M., De Maria R. DNA damage repair pathways in cancer stem cells. Mol Canc Therapeut. 2012;11:1627–1636. doi: 10.1158/1535-7163.MCT-11-1040. [DOI] [PubMed] [Google Scholar]
- 10.Skvortsov S., Debbage P., Lukas P., Skvortsova I. Crosstalk between DNA repair and cancer stem cell (CSC) associated intracellular pathways. Semin Canc Biol. 2015;31:36–42. doi: 10.1016/j.semcancer.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Weinberg R., Fisher D.E., Rich J. Dynamic and transient cancer stem cells nurture melanoma. Nat Med. 2010;16:758. doi: 10.1038/nm0710-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thankamony A.P., Saxena K., Murali R., Jolly M.K., Nair R. Cancer stem cell plasticity - a deadly deal. Front Mol Biosci. 2020;7:79. doi: 10.3389/fmolb.2020.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean M. ABC transporters, drug resistance, and cancer stem cells. J Mammary Gland Biol Neoplasia. 2009;14:3–9. doi: 10.1007/s10911-009-9109-9. [DOI] [PubMed] [Google Scholar]
- 14.Begicevic R.R., Falasca M. ABC transporters in cancer stem cells: beyond chemoresistance. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18112362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishimoto T., Oshima H., Oshima M., Kai K., Torii R., Masuko T. CD44+ slow-cycling tumor cell expansion is triggered by cooperative actions of Wnt and prostaglandin E2 in gastric tumorigenesis. Canc Sci. 2010;101:673–678. doi: 10.1111/j.1349-7006.2009.01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida G.J., Saya H. EpCAM expression in the prostate cancer makes the difference in the response to growth factors. Biochem Biophys Res Commun. 2014;443:239–245. doi: 10.1016/j.bbrc.2013.11.093. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y.A., Wang C.Y., Hsieh Y.T., Chen Y.J., Wei Y.H. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle. 2015;14:86–98. doi: 10.4161/15384101.2014.974419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibao S., Minami N., Koike N., Fukui N., Yoshida K., Saya H. Metabolic heterogeneity and plasticity of glioma stem cells in a mouse glioblastoma model. Neuro Oncol. 2018;20:343–354. doi: 10.1093/neuonc/nox170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saga I., Shibao S., Okubo J., Osuka S., Kobayashi Y., Yamada S. Integrated analysis identifies different metabolic signatures for tumor-initiating cells in a murine glioblastoma model. Neuro Oncol. 2014;16:1048–1056. doi: 10.1093/neuonc/nou096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong J.H., Park E.S., Liang J., Dennison J.B., Tsavachidou D., Nguyen-Charles C. Dual inhibition of tumor energy pathway by 2-deoxyglucose and metformin is effective against a broad spectrum of preclinical cancer models. Mol Canc Therapeut. 2011;10:2350–2362. doi: 10.1158/1535-7163.MCT-11-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loponte S., Lovisa S., Deem A.K., Carugo A., Viale A. The many facets of tumor heterogeneity: is metabolism lagging behind? Cancers. 2019;11 doi: 10.3390/cancers11101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haq R., Shoag J., Andreu-Perez P., Yokoyama S., Edelman H., Rowe G.C. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Canc Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roesch A., Vultur A., Bogeski I., Wang H., Zimmermann K.M., Speicher D. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Canc Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore N., Lyle S. Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J Oncol. 2011;2011 doi: 10.1155/2011/396076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W., Dong J., Haiech J., Kilhoffer M.C., Zeniou M. Cancer stem cell quiescence and plasticity as major challenges in cancer therapy. Stem Cell Int. 2016;2016:1740936. doi: 10.1155/2016/1740936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das P.K., Pillai S., Rakib M.A., Khanam J.A., Gopalan V., Lam A.K.Y. Plasticity of cancer stem cell: origin and role in disease progression and therapy resistance. Stem Cell Rev Rep. 2020;16:397–412. doi: 10.1007/s12015-019-09942-y. [DOI] [PubMed] [Google Scholar]
- 27.Wiechert A., Saygin C., Thiagarajan P.S., Rao V.S., Hale J.S., Gupta N. Cisplatin induces stemness in ovarian cancer. Oncotarget. 2016;7:30511–30522. doi: 10.18632/oncotarget.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y.H., Scadden D.T. Harnessing the apoptotic programs in cancer stem-like cells. EMBO Rep. 2015;16:1084–1098. doi: 10.15252/embr.201439675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C.A.A., Banerjee P., Wilson B.J., Wu S., Guo Q., Berg G. Targeting the ABC transporter ABCB5 sensitizes glioblastoma to temozolomide-induced apoptosis through a cell-cycle checkpoint regulation mechanism. J Biol Chem. 2020;295:7774–7788. doi: 10.1074/jbc.RA120.013778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugihara E., Saya H. Complexity of cancer stem cells. Int J Canc. 2013;132:1249–1259. doi: 10.1002/ijc.27961. [DOI] [PubMed] [Google Scholar]
- 31.Sugihara E., Shimizu T., Kojima K., Onishi N., Kai K., Ishizawa J. Ink4a and Arf are crucial factors in the determination of the cell of origin and the therapeutic sensitivity of Myc-induced mouse lymphoid tumor. Oncogene. 2012;31:2849–2861. doi: 10.1038/onc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugihara E., Hashimoto N., Osuka S., Shimizu T., Ueno S., Okazaki S. The inhibitor of apoptosis protein Livin confers resistance to Fas-mediated immune cytotoxicity in refractory lymphoma. Canc Res. 2020 doi: 10.1158/0008-5472.CAN-19-3993. [DOI] [PubMed] [Google Scholar]
- 33.Chen C., Zhao S., Karnad A., Freeman J.W. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Zuo X., Xie K., Wei D. The role of CD44 and cancer stem cells. Methods Mol Biol. 2018;1692:31–42. doi: 10.1007/978-1-4939-7401-6_3. [DOI] [PubMed] [Google Scholar]
- 35.Imrich S., Hachmeister M., Gires O. EpCAM and its potential role in tumor-initiating cells. Cell Adh Migr. 2012;6:30–38. doi: 10.4161/cam.18953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oishi N., Yamashita T., Kaneko S. Molecular biology of liver cancer stem cells. Liver Cancer. 2014;3:71–84. doi: 10.1159/000343863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D., Hemler M.E. Metabolic activation-related CD147-CD98 complex. Mol Cell Proteomics. 2005;4:1061–1071. doi: 10.1074/mcp.M400207-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia P., Xu X.Y. PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. Am J Cancer Res. 2015;5:1602–1609. [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida G.J. The harmonious interplay of amino acid and monocarboxylate transporters induces the robustness of cancer cells. Metabolites. 2021:11. doi: 10.3390/metabo11010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark D.W., Palle K. Aldehyde dehydrogenases in cancer stem cells: potential as therapeutic targets. Ann Transl Med. 2016;4:518. doi: 10.21037/atm.2016.11.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vassalli G. Aldehyde dehydrogenases: not just markers, but functional regulators of stem cells. Stem Cell Int. 2019;2019:3904645. doi: 10.1155/2019/3904645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabbath K.D., Ball E.D., Larcom P., Davis R.B., Griffin J.D. Heterogeneity of clonogenic cells in acute myeloblastic leukemia. J Clin Invest. 1985;75:746–753. doi: 10.1172/JCI111756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffin J.D., Lowenberg B. Clonogenic cells in acute myeloblastic leukemia. Blood. 1986;68:1185–1195. [PubMed] [Google Scholar]
- 44.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J. Identification of a cancer stem cell in human brain tumors. Canc Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 46.Patrawala L., Calhoun T., Schneider-Broussard R., Li H., Bhatia B., Tang S. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 47.Ricci-Vitiani L., Lombardi D.G., Pilozzi E., Biffoni M., Todaro M., Peschle C. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 48.Li C., Heidt D.G., Dalerba P., Burant C.F., Zhang L., Adsay V. Identification of pancreatic cancer stem cells. Canc Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 49.Burgos-Ojeda D., Rueda B.R., Buckanovich R.J. Ovarian cancer stem cell markers: prognostic and therapeutic implications. Canc Lett. 2012;322:1–7. doi: 10.1016/j.canlet.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida G.J. Beyond the warburg effect: N-myc contributes to metabolic reprogramming in cancer cells. Front Oncol. 2020;10:791. doi: 10.3389/fonc.2020.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanai N., Alvarez-Buylla A., Berger M.S. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 52.LaBarge M.A., Bissell M.J. Is CD133 a marker of metastatic colon cancer stem cells? J Clin Invest. 2008;118:2021–2024. doi: 10.1172/JCI36046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irollo E., Pirozzi G. CD133: to be or not to be, is this the real question? Am J Transl Res. 2013;5:563–581. [PMC free article] [PubMed] [Google Scholar]
- 54.Feng L., Huang S., An G., Wang G., Gu S., Zhao X. Identification of new cancer stem cell markers and signaling pathways in HER2positive breast cancer by transcriptome sequencing. Int J Oncol. 2019;55:1003–1018. doi: 10.3892/ijo.2019.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schepers A.G., Snippert H.J., Stange D.E., van den Born M., van Es J.H., van de Wetering M. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 56.Driessens G., Beck B., Caauwe A., Simons B.D., Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J., Li Y., Yu T.S., McKay R.M., Burns D.K., Kernie S.G. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lenos K.J., Miedema D.M., Lodestijn S.C., Nijman L.E., van den Bosch T., Romero Ros X. Stem cell functionality is microenvironmentally defined during tumour expansion and therapy response in colon cancer. Nat Cell Biol. 2018;20:1193–1202. doi: 10.1038/s41556-018-0179-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fendler A., Bauer D., Busch J., Jung K., Wulf-Goldenberg A., Kunz S. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat Commun. 2020;11:929. doi: 10.1038/s41467-020-14700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimokawa M., Ohta Y., Nishikori S., Matano M., Takano A., Fujii M. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature. 2017;545:187–192. doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- 61.de Sousa e Melo F., Kurtova A.V., Harnoss J.M., Kljavin N., Hoeck J.D., Hung J. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 62.Heerboth S., Housman G., Leary M., Longacre M., Byler S., Lapinska K. EMT and tumor metastasis. Clin Transl Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato R., Semba T., Saya H., Arima Y. Concise review: stem cells and epithelial-mesenchymal transition in cancer: biological implications and therapeutic targets. Stem Cell. 2016;34:1997–2007. doi: 10.1002/stem.2406. [DOI] [PubMed] [Google Scholar]
- 64.Bakir B., Chiarella A.M., Pitarresi J.R., Rustgi A.K. EMT, MET, plasticity, and tumor metastasis. Trends Cell Biol. 2020 Oct;30(10):764–776. doi: 10.1016/j.tcb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blanco M.J., Moreno-Bueno G., Sarrio D., Locascio A., Cano A., Palacios J. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 2002;21:3241–3246. doi: 10.1038/sj.onc.1205416. [DOI] [PubMed] [Google Scholar]
- 67.Yoshida G.J. The heterogeneity of cancer stem-like cells at the invasive front. Canc Cell Int. 2017;17:23. doi: 10.1186/s12935-017-0393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida G.J., Saya H. Inversed relationship between CD44 variant and c-Myc due to oxidative stress-induced canonical Wnt activation. Biochem Biophys Res Commun. 2014;443:622–627. doi: 10.1016/j.bbrc.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 69.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morel A.P., Lievre M., Thomas C., Hinkal G., Ansieau S., Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mani S.A., Yang J., Brooks M., Schwaninger G., Zhou A., Miura N. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104:10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hollier B.G., Tinnirello A.A., Werden S.J., Evans K.W., Taube J.H., Sarkar T.R. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Canc Res. 2013;73:1981–1992. doi: 10.1158/0008-5472.CAN-12-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vandewalle C., Comijn J., De Craene B., Vermassen P., Bruyneel E., Andersen H. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prislei S., Martinelli E., Zannoni G.F., Petrillo M., Filippetti F., Mariani M. Role and prognostic significance of the epithelial-mesenchymal transition factor ZEB2 in ovarian cancer. Oncotarget. 2015;6:18966–18979. doi: 10.18632/oncotarget.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kahlert C., Lahes S., Radhakrishnan P., Dutta S., Mogler C., Herpel E. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clin Canc Res. 2011;17:7654–7663. doi: 10.1158/1078-0432.CCR-10-2816. [DOI] [PubMed] [Google Scholar]
- 76.Acloque H., Adams M.S., Fishwick K., Bronner-Fraser M., Nieto M.A. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gunasinghe N.P., Wells A., Thompson E.W., Hugo H.J. Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Canc Metastasis Rev. 2012;31:469–478. doi: 10.1007/s10555-012-9377-5. [DOI] [PubMed] [Google Scholar]
- 78.Yao D., Dai C., Peng S. Mechanism of the mesenchymal-epithelial transition and its relationship with metastatic tumor formation. Mol Canc Res. 2011;9:1608–1620. doi: 10.1158/1541-7786.MCR-10-0568. [DOI] [PubMed] [Google Scholar]
- 79.Langley R.R., Fidler I.J. The seed and soil hypothesis revisited--the role of tumor-stroma interactions in metastasis to different organs. Int J Canc. 2011;128:2527–2535. doi: 10.1002/ijc.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fidler I.J., Poste G. The "seed and soil" hypothesis revisited. Lancet Oncol. 2008;9:808. doi: 10.1016/S1470-2045(08)70201-8. [DOI] [PubMed] [Google Scholar]
- 81.Massague J., Obenauf A.C. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ocana O.H., Corcoles R., Fabra A., Moreno-Bueno G., Acloque H., Vega S. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Canc Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 83.Takano S., Reichert M., Bakir B., Das K.K., Nishida T., Miyazaki M. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev. 2016;30:233–247. doi: 10.1101/gad.263327.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 85.Saitoh M. Involvement of partial EMT in cancer progression. J Biochem. 2018;164:257–264. doi: 10.1093/jb/mvy047. [DOI] [PubMed] [Google Scholar]
- 86.Shibue T., Weinberg R.A. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.George J.T., Jolly M.K., Xu S., Somarelli J.A., Levine H. Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Canc Res. 2017;77:6415–6428. doi: 10.1158/0008-5472.CAN-16-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamashita N., Tokunaga E., Iimori M., Inoue Y., Tanaka K., Kitao H. Epithelial paradox: clinical significance of coexpression of E-cadherin and vimentin with regard to invasion and metastasis of breast cancer. Clin Breast Canc. 2018;18:e1003–e1009. doi: 10.1016/j.clbc.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 89.Fustaino V., Presutti D., Colombo T., Cardinali B., Papoff G., Brandi R. Characterization of epithelial-mesenchymal transition intermediate/hybrid phenotypes associated to resistance to EGFR inhibitors in non-small cell lung cancer cell lines. Oncotarget. 2017;8:103340–103363. doi: 10.18632/oncotarget.21132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624 e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jolly M.K., Tripathi S.C., Jia D., Mooney S.M., Celiktas M., Hanash S.M. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7:27067–27084. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Strauss R., Li Z.Y., Liu Y., Beyer I., Persson J., Sova P. Analysis of epithelial and mesenchymal markers in ovarian cancer reveals phenotypic heterogeneity and plasticity. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoshida G.J. Regulation of heterogeneous cancer-associated fibroblasts: the molecular pathology of activated signaling pathways. J Exp Clin Canc Res. 2020;39:112. doi: 10.1186/s13046-020-01611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pearson G.W. Control of invasion by epithelial-to-mesenchymal transition programs during metastasis. J Clin Med. 2019;8 doi: 10.3390/jcm8050646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang C., Cao M., Liu Y., He Y., Du Y., Zhang G. Inducible formation of leader cells driven by CD44 switching gives rise to collective invasion and metastases in luminal breast carcinomas. Oncogene. 2019;38:7113–7132. doi: 10.1038/s41388-019-0899-y. [DOI] [PubMed] [Google Scholar]
- 96.Papadaki M.A., Stoupis G., Theodoropoulos P.A., Mavroudis D., Georgoulias V., Agelaki S. Circulating tumor cells with stemness and epithelial-to-mesenchymal transition features are chemoresistant and predictive of poor outcome in metastatic breast cancer. Mol Canc Therapeut. 2019;18:437–447. doi: 10.1158/1535-7163.MCT-18-0584. [DOI] [PubMed] [Google Scholar]
- 97.Hatina J. The dynamics of cancer stem cells. Neoplasma. 2012;59:700–707. doi: 10.4149/neo_2012_092. [DOI] [PubMed] [Google Scholar]
- 98.Islam F., Qiao B., Smith R.A., Gopalan V., Lam A.K. Cancer stem cell: fundamental experimental pathological concepts and updates. Exp Mol Pathol. 2015;98:184–191. doi: 10.1016/j.yexmp.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 99.Cabrera M.C., Hollingsworth R.E., Hurt E.M. Cancer stem cell plasticity and tumor hierarchy. World J Stem Cell. 2015;7:27–36. doi: 10.4252/wjsc.v7.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Csermely P., Hodsagi J., Korcsmaros T., Modos D., Perez-Lopez A.R., Szalay K. Cancer stem cells display extremely large evolvability: alternating plastic and rigid networks as a potential Mechanism: network models, novel therapeutic target strategies, and the contributions of hypoxia, inflammation and cellular senescence. Semin Canc Biol. 2014 doi: 10.1016/j.semcancer.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 101.Harris J.F., Best M.W. Dynamic heterogeneity: metastatic variants to liver are generated spontaneously in mouse embryonal carcinoma cells. Clin Exp Metastasis. 1988;6:451–462. doi: 10.1007/BF01784376. [DOI] [PubMed] [Google Scholar]
- 102.Roesch A., Fukunaga-Kalabis M., Schmidt E.C., Zabierowski S.E., Brafford P.A., Vultur A. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Quintana E., Shackleton M., Foster H.R., Fullen D.R., Sabel M.S., Johnson T.M. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Canc Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Quintana E., Shackleton M., Sabel M.S., Fullen D.R., Johnson T.M., Morrison S.J. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Charles N., Ozawa T., Squatrito M., Bleau A.M., Brennan C.W., Hambardzumyan D. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010;6:141–152. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stockhausen M.T., Kristoffersen K., Poulsen H.S. The functional role of Notch signaling in human gliomas. Neuro Oncol. 2010;12:199–211. doi: 10.1093/neuonc/nop022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ghiaur G., Gerber J., Jones R.J. Concise review: cancer stem cells and minimal residual disease. Stem Cell. 2012;30:89–93. doi: 10.1002/stem.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 109.Scadden D.T. Nice neighborhood: emerging concepts of the stem cell niche. Cell. 2014;157:41–50. doi: 10.1016/j.cell.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li L., Neaves W.B. Normal stem cells and cancer stem cells: the niche matters. Canc Res. 2006;66:4553–4557. doi: 10.1158/0008-5472.CAN-05-3986. [DOI] [PubMed] [Google Scholar]
- 111.Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rabbani P., Takeo M., Chou W., Myung P., Bosenberg M., Chin L. Coordinated activation of Wnt in epithelial and melanocyte stem cells initiates pigmented hair regeneration. Cell. 2011;145:941–955. doi: 10.1016/j.cell.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guerrouahen B.S., Al-Hijji I., Tabrizi A.R. Osteoblastic and vascular endothelial niches, their control on normal hematopoietic stem cells, and their consequences on the development of leukemia. Stem Cell Int. 2011;2011:375857. doi: 10.4061/2011/375857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gattazzo F., Urciuolo A., Bonaldo P. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim Biophys Acta. 2014;1840:2506–2519. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cully M. Tumour microenvironment: fibroblast subtype provides niche for cancer stem cells. Nat Rev Canc. 2018;18:136. doi: 10.1038/nrc.2018.18. [DOI] [PubMed] [Google Scholar]
- 116.Prager B.C., Xie Q., Bao S., Rich J.N. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019;24:41–53. doi: 10.1016/j.stem.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Veirman K., Rao L., De Bruyne E., Menu E., Van Valckenborgh E., Van Riet I. Cancer associated fibroblasts and tumor growth: focus on multiple myeloma. Cancers. 2014;6:1363–1381. doi: 10.3390/cancers6031363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jinushi M., Baghdadi M., Chiba S., Yoshiyama H. Regulation of cancer stem cell activities by tumor-associated macrophages. Am J Cancer Res. 2012;2:529–539. [PMC free article] [PubMed] [Google Scholar]
- 119.Kfoury Y., Scadden D.T. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16:239–253. doi: 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 120.Hoesel B., Schmid J.A. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Canc. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rinkenbaugh A.L., Baldwin A.S. The NF-kappaB pathway and cancer stem cells. Cells. 2016;5 doi: 10.3390/cells5020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yoshida G.J., Azuma A., Miura Y., Orimo A. Activated fibroblast program orchestrates tumor initiation and progression; molecular mechanisms and the associated therapeutic strategies. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20092256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wei X., Yang S., Pu X., He S., Yang Z., Sheng X. Tumor-associated macrophages increase the proportion of cancer stem cells in lymphoma by secreting pleiotrophin. Am J Transl Res. 2019;11:6393–6402. [PMC free article] [PubMed] [Google Scholar]
- 125.Jinushi M., Chiba S., Yoshiyama H., Masutomi K., Kinoshita I., Dosaka-Akita H. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc Natl Acad Sci U S A. 2011;108:12425–12430. doi: 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Salvagno C., Ciampricotti M., Tuit S., Hau C.S., van Weverwijk A., Coffelt S.B. Therapeutic targeting of macrophages enhances chemotherapy efficacy by unleashing type I interferon response. Nat Cell Biol. 2019;21:511–521. doi: 10.1038/s41556-019-0298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Plaks V., Kong N., Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Borovski T., De Sousa E.M.F., Vermeulen L., Medema J.P. Cancer stem cell niche: the place to be. Canc Res. 2011;71:634–639. doi: 10.1158/0008-5472.CAN-10-3220. [DOI] [PubMed] [Google Scholar]
- 129.Zhao Y., Dong Q., Li J., Zhang K., Qin J., Zhao J. Targeting cancer stem cells and their niche: perspectives for future therapeutic targets and strategies. Semin Canc Biol. 2018;53:139–155. doi: 10.1016/j.semcancer.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 130.Weidenfeld K., Barkan D. EMT and stemness in tumor dormancy and outgrowth: are they intertwined processes? Front Oncol. 2018;8:381. doi: 10.3389/fonc.2018.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ingangi V., Minopoli M., Ragone C., Motti M.L., Carriero M.V. Role of microenvironment on the fate of disseminating cancer stem cells. Front Oncol. 2019;9:82. doi: 10.3389/fonc.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chaffer C.L., Brueckmann I., Scheel C., Kaestli A.J., Wiggins P.A., Rodrigues L.O. Normal and neoplastic nonsystem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chaffer C.L., Marjanovic N.D., Lee T., Bell G., Kleer C.G., Reinhardt F. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gupta P.B., Fillmore C.M., Jiang G., Shapira S.D., Tao K., Kuperwasser C. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 135.Giancotti F.G. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lopez-Novoa J.M., Nieto M.A. Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol Med. 2009;1:303–314. doi: 10.1002/emmm.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wei S.C., Fattet L., Tsai J.H., Guo Y., Pai V.H., Majeski H.E. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat Cell Biol. 2015;17:678–688. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 139.Lawson D.A., Bhakta N.R., Kessenbrock K., Prummel K.D., Yu Y., Takai K. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Date S., Sato T. Mini-gut organoids: reconstitution of the stem cell niche. Annu Rev Cell Dev Biol. 2015;31:269–289. doi: 10.1146/annurev-cellbio-100814-125218. [DOI] [PubMed] [Google Scholar]
- 141.Nanki K., Toshimitsu K., Takano A., Fujii M., Shimokawa M., Ohta Y. Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell. 2018;174:856–869 e17. doi: 10.1016/j.cell.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 142.Seino T., Kawasaki S., Shimokawa M., Tamagawa H., Toshimitsu K., Fujii M. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell. 2018;22:454–467 e6. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 143.Yap T.A., Lorente D., Omlin A., Olmos D., de Bono J.S. Circulating tumor cells: a multifunctional biomarker. Clin Canc Res. 2014;20:2553–2568. doi: 10.1158/1078-0432.CCR-13-2664. [DOI] [PubMed] [Google Scholar]
- 144.Williams E.D., Gao D., Redfern A., Thompson E.W. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat Rev Canc. 2019;19:716–732. doi: 10.1038/s41568-019-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Raimondi C., Naso G., Gradilone A., Gianni W., Cortesi E., Gazzaniga P. Circulating tumor cells in cancer therapy: are we off target? Curr Cancer Drug Targets. 2010;10:509–518. doi: 10.2174/156800910791517163. [DOI] [PubMed] [Google Scholar]
- 146.Yu M., Bardia A., Wittner B.S., Stott S.L., Smas M.E., Ting D.T. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 148.Norton L., Massague J. Is cancer a disease of self-seeding? Nat Med. 2006;12:875–878. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
- 149.Comen E., Norton L. Self-seeding in cancer. Recent Results Canc Res. 2012;195:13–23. doi: 10.1007/978-3-642-28160-0_2. [DOI] [PubMed] [Google Scholar]
- 150.Kim M.Y., Oskarsson T., Acharyya S., Nguyen D.X., Zhang X.H., Norton L. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aceto N., Bardia A., Miyamoto D.T., Donaldson M.C., Wittner B.S., Spencer J.A. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Friedl P., Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 153.Cheung K.J., Gabrielson E., Werb Z., Ewald A.J. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155:1639–1651. doi: 10.1016/j.cell.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Akhtar M., Haider A., Rashid S., Al-Nabet A. Paget's "seed and soil" theory of cancer metastasis: an idea whose time has come. Adv Anat Pathol. 2019;26:69–74. doi: 10.1097/PAP.0000000000000219. [DOI] [PubMed] [Google Scholar]
- 155.Peinado H., Zhang H., Matei I.R., Costa-Silva B., Hoshino A., Rodrigues G. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Canc. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 156.Dasgupta A., Lim A.R., Ghajar C.M. Circulating and disseminated tumor cells: harbingers or initiators of metastasis? Mol Oncol. 2017;11:40–61. doi: 10.1002/1878-0261.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Alix-Panabieres C., Mader S., Pantel K. Epithelial-mesenchymal plasticity in circulating tumor cells. J Mol Med (Berl) 2017;95:133–142. doi: 10.1007/s00109-016-1500-6. [DOI] [PubMed] [Google Scholar]
- 158.Sosa M.S., Bragado P., Aguirre-Ghiso J.A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Canc. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hiraga T. Hypoxic microenvironment and metastatic bone disease. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19113523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bryden A.A., Islam S., Freemont A.J., Shanks J.H., George N.J., Clarke N.W. Parathyroid hormone-related peptide: expression in prostate cancer bone metastases. Prostate Cancer Prostatic Dis. 2002;5:59–62. doi: 10.1038/sj.pcan.4500553. [DOI] [PubMed] [Google Scholar]
- 161.Asadi F., Kukreja S. Parathyroid hormone-related protein in prostate cancer. Crit Rev Eukaryot Gene Expr. 2005;15:15–28. doi: 10.1615/critreveukaryotgeneexpr.v15.i1.20. [DOI] [PubMed] [Google Scholar]
- 162.Shiozawa Y., Pedersen E.A., Havens A.M., Jung Y., Mishra A., Joseph J. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J Clin Invest. 2011;121:1298–1312. doi: 10.1172/JCI43414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yoneda T., Hiraga T. Crosstalk between cancer cells and bone microenvironment in bone metastasis. Biochem Biophys Res Commun. 2005;328:679–687. doi: 10.1016/j.bbrc.2004.11.070. [DOI] [PubMed] [Google Scholar]
- 164.Sun Y.X., Schneider A., Jung Y., Wang J., Dai J., Cook K. Skeletal localization and neutralization of the SDF-1(CXCL12)/CXCR4 axis blocks prostate cancer metastasis and growth in osseous sites in vivo. J Bone Miner Res. 2005;20:318–329. doi: 10.1359/JBMR.041109. [DOI] [PubMed] [Google Scholar]
- 165.Gravina G.L., Mancini A., Muzi P., Ventura L., Biordi L., Ricevuto E. CXCR4 pharmacological inhibition reduces bone and soft tissue metastatic burden by affecting tumor growth and tumorigenic potential in prostate cancer preclinical models. Prostate. 2015;75:1227–1246. doi: 10.1002/pros.23007. [DOI] [PubMed] [Google Scholar]
- 166.Gomis R.R., Gawrzak S. Tumor cell dormancy. Mol Oncol. 2017;11:62–78. doi: 10.1016/j.molonc.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Aguirre-Ghiso J.A. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Canc. 2007;7:834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Goss P.E., Chambers A.F. Does tumour dormancy offer a therapeutic target? Nat Rev Canc. 2010;10:871–877. doi: 10.1038/nrc2933. [DOI] [PubMed] [Google Scholar]
- 169.Gao H., Chakraborty G., Lee-Lim A.P., Mo Q., Decker M., Vonica A. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell. 2012;150:764–779. doi: 10.1016/j.cell.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bragado P., Estrada Y., Parikh F., Krause S., Capobianco C., Farina H.G. TGF-beta 2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nat Cell Biol. 2013;15:1351–1361. doi: 10.1038/ncb2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kobayashi A., Okuda H., Xing F., Pandey P.R., Watabe M., Hirota S. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J Exp Med. 2011;208:2641–2655. doi: 10.1084/jem.20110840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yoshida G.J. Therapeutic strategies of drug repositioning targeting autophagy to induce cancer cell death: from pathophysiology to treatment. J Hematol Oncol. 2017;10:67. doi: 10.1186/s13045-017-0436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yorimitsu T., Klionsky D.J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schewe D.M., Aguirre-Ghiso J.A. ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc Natl Acad Sci U S A. 2008;105:10519–10524. doi: 10.1073/pnas.0800939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lu Z., Luo R.Z., Lu Y., Zhang X., Yu Q., Khare S. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–3929. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Prince M.E., Sivanandan R., Kaczorowski A., Wolf G.T., Kaplan M.J., Dalerba P. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Collins A.T., Berry P.A., Hyde C., Stower M.J., Maitland N.J. Prospective identification of tumorigenic prostate cancer stem cells. Canc Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 178.Goodison S., Urquidi V., Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189–196. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lynch K.W. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- 180.Marzese D.M., Liu M., Huynh J.L., Hirose H., Donovan N.C., Huynh K.T. Brain metastasis is predetermined in early stages of cutaneous melanoma by CD44v6 expression through epigenetic regulation of the spliceosome. Pigment Cell Melanoma Res. 2015;28:82–93. doi: 10.1111/pcmr.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hu J., Li G., Zhang P., Zhuang X., Hu G. A CD44v(+) subpopulation of breast cancer stem-like cells with enhanced lung metastasis capacity. Cell Death Dis. 2017;8:e2679. doi: 10.1038/cddis.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Slomiany M.G., Grass G.D., Robertson A.D., Yang X.Y., Maria B.L., Beeson C. Hyaluronan, CD44, and emmprin regulate lactate efflux and membrane localization of monocarboxylate transporters in human breast carcinoma cells. Canc Res. 2009;69:1293–1301. doi: 10.1158/0008-5472.CAN-08-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schmidt D.S., Klingbeil P., Schnolzer M., Zoller M. CD44 variant isoforms associate with tetraspanins and EpCAM. Exp Cell Res. 2004;297:329–347. doi: 10.1016/j.yexcr.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 184.Brown R.L., Reinke L.M., Damerow M.S., Perez D., Chodosh L.A., Yang J. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Horiguchi K., Sakamoto K., Koinuma D., Semba K., Inoue A., Inoue S. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene. 2012;31:3190–3201. doi: 10.1038/onc.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Saint-Andre V., Batsche E., Rachez C., Muchardt C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 187.Zhou H.L., Luo G., Wise J.A., Lou H. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res. 2014;42:701–713. doi: 10.1093/nar/gkt875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Olsson E., Honeth G., Bendahl P.O., Saal L.H., Gruvberger-Saal S., Ringner M. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Canc. 2011;11:418. doi: 10.1186/1471-2407-11-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yoshida G.J., Saya H., Zouboulis C.C. Three-dimensional culture of sebaceous gland cells revealing the role of prostaglandin E2-induced activation of canonical Wnt signaling. Biochem Biophys Res Commun. 2013;438:640–646. doi: 10.1016/j.bbrc.2013.07.129. [DOI] [PubMed] [Google Scholar]
- 190.Yae T., Tsuchihashi K., Ishimoto T., Motohara T., Yoshikawa M., Yoshida G.J. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883. doi: 10.1038/ncomms1892. [DOI] [PubMed] [Google Scholar]
- 191.Quintanilla R.H., Jr., Asprer J.S., Vaz C., Tanavde V., Lakshmipathy U. CD44 is a negative cell surface marker for pluripotent stem cell identification during human fibroblast reprogramming. PLoS One. 2014;9 doi: 10.1371/journal.pone.0085419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Song Z., Ji Q., Zhao H., Nie Y., He Z., Chen Y. Generation of CD44 gene-deficient mouse derived induced pluripotent stem cells: CD44 gene-deficient iPSCs. In Vitro Cell Dev Biol Anim. 2014;50:874–882. doi: 10.1007/s11626-014-9786-6. [DOI] [PubMed] [Google Scholar]
- 193.Bassi M.T., Gasol E., Manzoni M., Pineda M., Riboni M., Martin R. Identification and characterisation of human xCT that co-expresses, with 4F2 heavy chain, the amino acid transport activity system xc. Pflügers Archiv. 2001;442:286–296. doi: 10.1007/s004240100537. [DOI] [PubMed] [Google Scholar]
- 194.Ito K., Hirao A., Arai F., Takubo K., Matsuoka S., Miyamoto K. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 195.Cao J.Y., Dixon S.J. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73:2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dixon S.J., Patel D.N., Welsch M., Skouta R., Lee E.D., Hayano M. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3 doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sato M., Kusumi R., Hamashima S., Kobayashi S., Sasaki S., Komiyama Y. The ferroptosis inducer erastin irreversibly inhibits system xc- and synergizes with cisplatin to increase cisplatin's cytotoxicity in cancer cells. Sci Rep. 2018;8:968. doi: 10.1038/s41598-018-19213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hatakeyama M. The role of Helicobacter pylori CagA in gastric carcinogenesis. Int J Hematol. 2006;84:301–308. doi: 10.1532/IJH97.06166. [DOI] [PubMed] [Google Scholar]
- 201.Yong X., Tang B., Li B.S., Xie R., Hu C.J., Luo G. Helicobacter pylori virulence factor CagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun Signal. 2015;13:30. doi: 10.1186/s12964-015-0111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tsugawa H., Suzuki H., Saya H., Hatakeyama M., Hirayama T., Hirata K. Reactive oxygen species-induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem-like cells. Cell Host Microbe. 2012;12:764–777. doi: 10.1016/j.chom.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 203.Granot Z., Henke E., Comen E.A., King T.A., Norton L., Benezra R. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Canc Cell. 2011;20:300–314. doi: 10.1016/j.ccr.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sceneay J., Smyth M.J., Moller A. The pre-metastatic niche: finding common ground. Canc Metastasis Rev. 2013;32:449–464. doi: 10.1007/s10555-013-9420-1. [DOI] [PubMed] [Google Scholar]
- 205.Plosker G.L., Croom K.F. Sulfasalazine: a review of its use in the management of rheumatoid arthritis. Drugs. 2005;65:1825–1849. doi: 10.2165/00003495-200565130-00008. [DOI] [PubMed] [Google Scholar]
- 206.Shitara K., Doi T., Nagano O., Imamura C.K., Ozeki T., Ishii Y. Dose-escalation study for the targeting of CD44v(+) cancer stem cells by sulfasalazine in patients with advanced gastric cancer (EPOC1205) Gastric Cancer. 2017;20:341–349. doi: 10.1007/s10120-016-0610-8. [DOI] [PubMed] [Google Scholar]
- 207.Shitara K., Doi T., Nagano O., Fukutani M., Hasegawa H., Nomura S. Phase 1 study of sulfasalazine and cisplatin for patients with CD44v-positive gastric cancer refractory to cisplatin (EPOC1407) Gastric Cancer. 2017;20:1004–1009. doi: 10.1007/s10120-017-0720-y. [DOI] [PubMed] [Google Scholar]
- 208.Otsubo K., Nosaki K., Imamura C.K., Ogata H., Fujita A., Sakata S. Phase I study of salazosulfapyridine in combination with cisplatin and pemetrexed for advanced non-small-cell lung cancer. Canc Sci. 2017;108:1843–1849. doi: 10.1111/cas.13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Xue H., Li J., Xie H., Wang Y. Review of drug repositioning approaches and resources. Int J Biol Sci. 2018;14:1232–1244. doi: 10.7150/ijbs.24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 211.Yoshikawa M., Tsuchihashi K., Ishimoto T., Yae T., Motohara T., Sugihara E. xCT inhibition depletes CD44v-expressing tumor cells that are resistant to EGFR-targeted therapy in head and neck squamous cell carcinoma. Canc Res. 2013;73:1855–1866. doi: 10.1158/0008-5472.CAN-12-3609-T. [DOI] [PubMed] [Google Scholar]
- 212.Sasaki H., Sato H., Kuriyama-Matsumura K., Sato K., Maebara K., Wang H. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J Biol Chem. 2002;277:44765–44771. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]
- 213.Harris I.S., Treloar A.E., Inoue S., Sasaki M., Gorrini C., Lee K.C. Cancer Cell; 2015. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. [DOI] [PubMed] [Google Scholar]
- 214.Habib E., Linher-Melville K., Lin H.X., Singh G. Expression of xCT and activity of system xc(-) are regulated by NRF2 in human breast cancer cells in response to oxidative stress. Redox Biol. 2015;5:33–42. doi: 10.1016/j.redox.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ji X., Qian J., Rahman S.M.J., Siska P.J., Zou Y., Harris B.K. xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene. 2018 Sep;37(36):5007–5019. doi: 10.1038/s41388-018-0307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bauer A.K., Hill T., 3rd, Alexander C.M. The involvement of NRF2 in lung cancer. Oxid Med Cell Longev. 2013;2013:746432. doi: 10.1155/2013/746432. [DOI] [PMC free article] [PubMed] [Google Scholar]