Abstract
β-thalassemia, an autosomal recessive blood disorder that reduces the production of hemoglobin, is majorly caused by the point mutation of the HBB gene resulting in reduced or absent β-globin chains of the hemoglobin tetramer. Animal models recapitulating both the phenotype and genotype of human disease are valuable in the exploration of pathophysiology and for in vivo evaluation of novel therapeutic treatments. The docile temperament, short vital cycles, and low cost of rabbits make them an attractive animal model. However, β-thalassemia rabbit models are currently unavailable. Here, using CRISPR/Cas9-mediated genome editing, we point mutated the rabbit β-globin gene HBB2 with high efficiency and generated a β-thalassemia rabbit model. Hematological and histological analyses demonstrated that the genotypic mosaic F0 displayed a mild phenotype of anemia, and the heterozygous F1 exhibited typical characteristics of β-thalassemia. Whole-blood transcriptome analysis revealed that the gene expression was altered in HBB2-targeted when compared with WT rabbits. And the highly expressed genes in HBB2-targeted rabbits were enriched in lipid and iron metabolism, innate immunity, and hematopoietic processes. In conclusion, using CRISPR-mediated HBB2 knockout, we have created a β-thalassemia rabbit model that accurately recapitulates the human disease phenotype. We believe this tool will be valuable in advancing the investigation of pathogenesis and novel therapeutic targets of β-thalassemia and associated complications.
Keywords: genetic disease, animal model, CRISPR/Cas, hemoglobin, gene therapy, β-thalassemia, rabbit
Abbreviations: ESC, embryonic stem cell; Hb, hemoglobin concentration; HCT, hematocrit; MCH, mean corpuscular hemoglobin; MCV, mean corpuscular volume; RBC, red blood cell; RDW, red cell distribution width; RET, reticulocyte; SSA, single-stranded annealing
Thalassemia is a heterogeneous group of inherited blood disorders characterized by reduced hemoglobin synthesis (1). β-thalassemia major is its severe form and caused by mutations in the HBB gene and reduction or absence of β-globin chain synthesis. As a result, the α-globin chains are free and unpaired; they aggregate and precipitate within red cells and consequently cause ineffective erythropoiesis and severe anemia (2), with complications such as immune defects (3), metabolic disorders, and endocrine dysfunctions (4). To date, over 200 different HBB gene mutations have been identified that cause β-thalassemia. Most of those mutations are single nucleotide substitutions, deletions, or insertions of oligonucleotides leading to the frameshift (5). At present, the most effective treatment for thalassemia is allogenic bone marrow transplantation, but fully matched donors are rare (6). Despite chronic blood transfusion and iron chelation therapy remarkably improving the duration and quality of life of patients with thalassemia in industrialized countries, the transfusion-related complications, including high rates of alloimmunization and blood-borne infectious sequelae, become a major source of morbidity (7). Besides, the high-cost treatment is not a feasible option in many developing countries where thalassemia is a common disease. Therefore, the need for the development of new curative approaches for thalassemia is urgent.
Given the complex clinical and molecular mechanisms of β-thalassemia, animal models are essential for understanding its pathophysiological and hemoglobinopathy mechanisms and validating future therapeutic strategies in vivo. So far, mouse was the only animal used for mimicking human thalassemia. A group of thalassemia mouse models have been generated; these mouse thalassemia models display many features of the β-hemoglobinopathies and provide enormous insight into the mechanisms of gene regulation (8). Spontaneous and insertional disruption Hbb-b1 gene deletion mice have been characterized (9, 10, 11). The heterozygotes show very mild thalassemia comparable with human patients. While homozygotes are severely anemic and lethal. The transgenic mouse models containing mutant human β-globin genomic fragments were also reported (12). The mouse models show thalassemia phenotype in heterozygous β-globin double knockout (KO) background (13), which could not make the same gene mutation as that in human, and are not suitable for preclinical testing of gene therapy.
Notably, the rabbit is another standard experimental model for the study of various human diseases. Compared with mice, rabbits have unique features of physiology, anatomy, and genetics and more similarities with humans (14). Although transgenic rabbits have been developed and contribute greatly to the study of lipid metabolism (15), cardiovascular (16) and infectious diseases (17), the lack of embryonic stem cells (ESCs) hampers the gene targeting of rabbit and consequently limits the various applications of rabbit model. The advent of customized nucleases, such as ZFN (18), TALEN (19), and CRISPR (20) technologies, has changed the circumstance of gene editing. In combination with embryo microinjection, these customized nucleases have been efficiently used for generation of various gene editing rabbits with different applications. Remarkably, the comparative genomic analysis revealed that the β-globin gene cluster of rabbits is close to that of humans and contains three functional globin genes: the embryonic gene ε-globin gene (HBE), fetal γ-globin gene (HBG), and adult β-globin gene (HBB2), implying that the rabbit can be a better model to study human β-thalassemia. As a proof of concept, herein we successfully generated the first β-thalassemia rabbit model using CRISPR/Cas9-mediated insertion/deletion(indel) mutagenesis, and typical phenotypes of β-thalassemia were characterized in the HBB2-targeted rabbit models. The β-thalassemia rabbit models also harbored the various abnormalities of lipid and iron metabolism and innate immunity, in addition to hematopoietic defects.
Results
Optimization of the concentration of Cas9 mRNA and gRNA in rabbit embryos
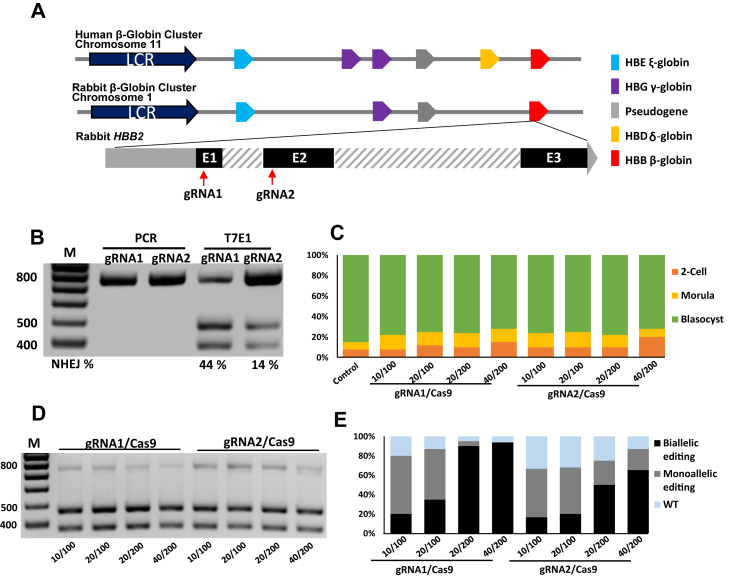
Two gRNAs targeting the exon I and exon II of the rabbit HBB2 gene were designed and synthesized (Fig. 1A) respectively. Single-stranded annealing (SSA) assay was performed to test the cleavage efficiency of the two gRNAs, Cas9/gRNA-expressing and SSA EGFP reporter vectors were constructed and transfected into HEK293 cells, flow cytometry was applied to analyze the recovery of EGFP gene, and the results indicated that gRNA1 exhibited higher cleavage activity (Fig. S1). Then we validate the targeting efficiency of the two gRNAs on endogenous HBB2 gene by transfecting Cas9/gRNA-expressing plasmids into rabbit fetal fibroblast cells. The T7E1 assay showed that gRNA1 was more effective, which was consistent with the SSA assay result (Fig. 1B). To optimize the concentration of Cas9 mRNA and gRNA, capped polyadenylated Cas9 mRNA and gRNA were produced by in vitro transcription and microinjected into one-cell-stage embryos with different concentrations, the injected embryos were cultured for 5 days to the blastocyst stage for testing the targeting efficiency. In comparison with the controls, the microinjection of Cas9 mRNA and gRNA showed no significant impact on the development of embryos (Fig. 1C). The T7E1 assay showed an efficient gene mutation in the injected blastocysts, even at the lowest concentration of Cas9 mRNA and gRNA (100/10 ng/μl) (Fig. 1D). Single-embryo sequencing was applied to identify mutations in the injected embryos, the results indicated that gRNA1 was more effective and the higher concentration of Cas9 mRNA and gRNA led to more biallelic mutagenesis efficiency (Fig. 1E).
Figure 1.
CRISPR/Cas9-mediated gene targeting of rabbit HBB2.A, genomic structure of the β-globin gene cluster and schematic diagram of the Cas9/gRNA targeting sites in rabbit HBB2. Comparative genomics analysis showed that the β-globin gene families are highly conserved between rabbit and human, which contain only one adult β-globin gene (highlighted in red box); Two gRNAs were designed targeting exon1 and exon2 of rabbit HBB2, gRNA1 and gRNA2 are highlighted in red arrows. B, mutation detection of Cas9/gRNA1 and Cas9/gRNA2 in rabbit fetal fibroblasts by T7E1 cleavage assay. C, development of different stage of rabbit embryos with no injection (control group) and coinjected with Cas9/gRNA1 and Cas9/gRNA2 in vitro. D, mutation detection of Cas9/gRNA1 and Cas9/gRNA2 with different concentration in rabbit embryos by T7E1 cleavage assay. E, mutagenic efficiency of embryo injection of Cas9/gRNA detected by Sanger sequencing.
Generation of HBB2-targeted rabbits via embryo injection
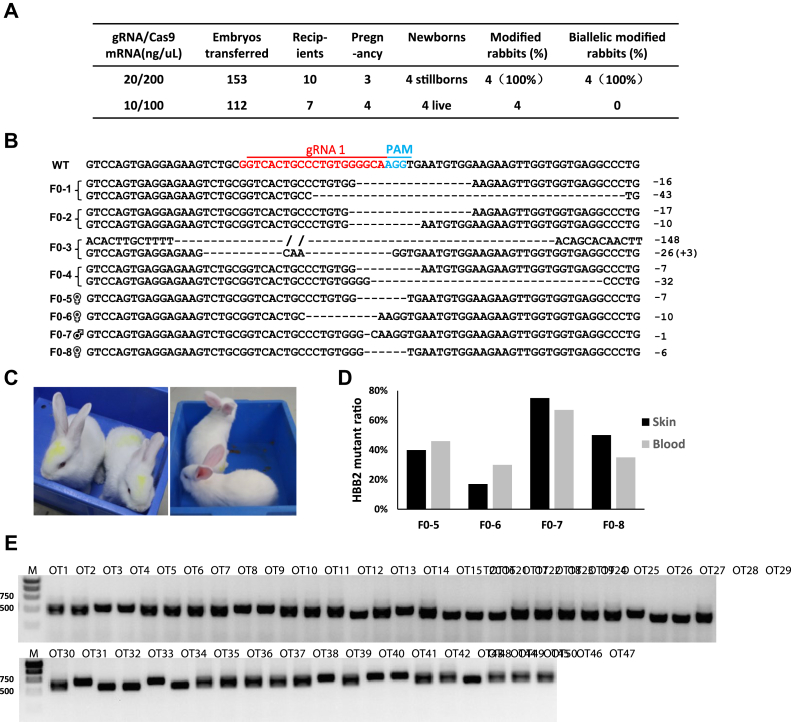
Given the higher effectivity of gRNA1, we initially applied the microinjection of a high concentration of Cas9 mRNA and gRNA1 (200/20 ng/μl) into the cytoplasm of one-cell rabbit embryos. A total of 153 injected embryos were transferred into the oviducts of ten surrogate rabbits (Fig. 2A). After 1-month gestation, only three of the surrogate rabbits developed to term and delivered four stillborn fetuses. Genotyping by PCR and sequencing indicated that all these four stillborn fetuses showed biallelic mutagenesis (Fig. 2B). The fact that human patients with the homozygous forms of β0-thalassemia are lethal in the untreated state and the mice homozygous for the Hbb-b1 and Hbb-b2 double KOs are stillborn or die within hours after birth, implying that the biallelic mutation of the rabbit adult β-globin gene was lethal.
Figure 2.
Generation of HBB2-targeted rabbit via embryo injection of Cas9/gRNA.A, summary of embryo injection and transplantation. B, genotype of HBB2-targeted rabbits detected by T-A cloning and Sanger sequencing. C, photographs of HBB2-targeted rabbits F0-5, F0-6, F0-7, and F0-8 at 1 month old. D, HBB2 mutant proration in the skin and blood of the genotypic mosaic F0 rabbits. E. T7E1 assays of the predicated off-target sites (Fig. S2).
Therefore, to achieve the heterozygous HBB2-targeted rabbits, we then applied a low concentration of Cas9 mRNA and gRNA1 (100/10 ng/μl) for embryo microinjection. A total of 112 injected embryos were transferred into seven surrogates (Fig. 2A). Two of them successfully developed to term and gave birth to four rabbit pups (F0, one male, three females) after 30 days’ gestation (Fig. 2C). Genomic DNA was extracted from the ear tissues and blood of the newborns for genotyping. Through T-A cloning and Sanger sequencing, HBB2 gene mutations were detected in all four newborns carrying indels at the expected target locus (Fig. 2B). All of the four rabbits exhibited genotypic mosaicism, which contained both wild-type HBB2 allele and various prorations of the mutant allele (F0-5, −7 bp; F0-6, −10 bp; F0-7, −1 bp; F0-8, −6 bp). The mutant prorations of the skin and blood were also detected and showed to be heterogeneous (Fig. 2D).
In consideration of the failed generation of homozygous HBB2-targeted rabbits and high frequency of off-target mutagenesis reported in various animals and cell lines, we sought to detect the off-target effect of CRISPR/Cas9. The potential off-target sites were predicted using CRISPOR (21). The 48 highest-scored potential off-target sites of the HBB2 gRNA1 were selected for T7E1 cleavage assay (Fig. S2) and no cleavage was detected (Fig. 2E), indicating the high fidelity of the CRISPR/Cas9-mediated gene editing.
Germline transmission and phenotype of the HBB2-targeted rabbits
Given the genotypic mosaicism of the HBB2-targeted founders, germline transmission of the mutant HBB2 allele is crucial for the establishment of β-thalassemia rabbit model. Initially, we mated the male founder F0-7 with the other three female founders at the age of 8 months. Unfortunately, the F0-5 rabbit died of childbirth, F0-6 gave birth of two stillborns, and F0-8 was not successfully pregnant. Then, we mated the male founder F0-7 with three female WT rabbits and obtained a total of 15 offspring (F1). Genotyping and T7E1 assay indicated that five of them carried the same mutant HBB2 allele (−1 bp) with that of the founder F0-7(Fig. 3A).
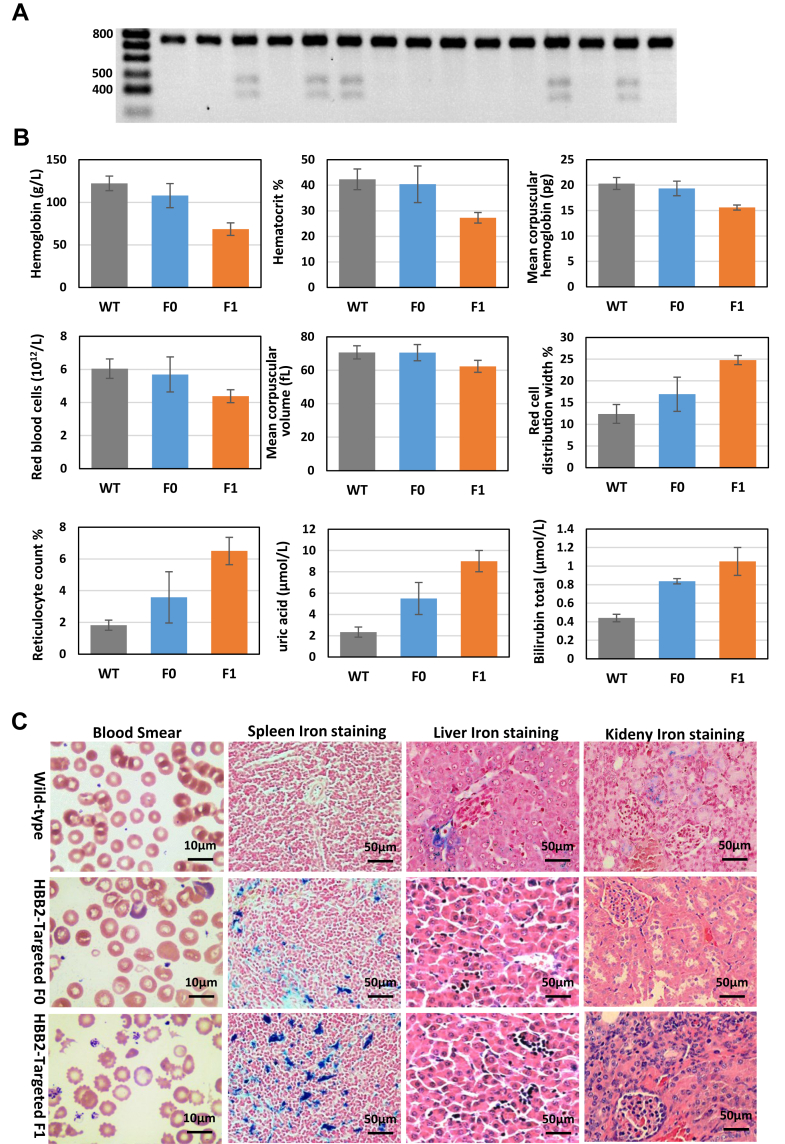
Figure 3.
Heritability and phenotype of HBB2-targeted rabbits.A, T7E1 cleavage assay for mutant HBB2 detection in F1 HBB2-targeted rabbits. B, hematologic features of HBB2-targeted rabbits. Hematological values are expressed as means+ SD. Hb, hemoglobin concentration; HCT, hematocrit; MCH, mean corpuscular hemoglobin; MCV, mean corpuscular volume; RBC, red blood cells; RDW, red cell distribution width; Retic, reticulocyte count. C, peripheral blood smears of WT and HBB2-targeted rabbits. Scale bars: 10 μm. Iron staining of the spleen, liver, and kidney in WT and HBB2-targeted rabbits. Scale bars: 50 μm.
The blood was collected from the WT, F0, and F1 HBB2-targeted rabbits for examination of the hematological characteristic. In comparison with the WT rabbits, both F0 and F1 showed decreased hemoglobin concentration (Hb), hematocrit (HCT), mean corpuscular hemoglobin (MCH), red blood cell (RBC), mean corpuscular volume (MCV), and RBC counts, which accompanied by increased red cell distribution width (RDW), reticulocytes (RET), uric acid, and total bilirubin (Fig. 3B). All these changes were similar to the relative parameters of human patients with β-thalassemia. The morphology of the red cells was directly observed under microscopy and blood smear treatment (Fig. 3C, Fig. S3). Frequent acanthocytes were found in the blood of both F0 and F1 heterozygous rabbits. The blood smear results showed that the HBB2-targeted rabbits contained marked macrocytic and microcytic hypochromic red cells, target cells, schistocytes, and burr cells. Pappenheimer bodies, which were derived from precipitated alpha-globin chains and granules of iron, also presented in the RBCs. All these characteristics indicated that the HBB2-targeted rabbits exhibited typical pathological features of β-thalassemia and the phenotypes of F1 heterozygous rabbits were apparently more severe than that of F0 rabbits.
Furthermore, histological analyses were performed in the spleen, liver, and kidney from WT and HBB2-targeted rabbits. The result showed that the spleens of the HBB2-targeted rabbits display a massive expansion of red pulps and dramatic pooling of sinusoidal erythrocytes (Fig. S3). Blue-purple staining showed marked iron deposits in the spleen, liver, and kidney of the HBB2-targeted rabbits; it is similar to that of human patients in which the abnormal stimulation of extramedullary erythropoiesis induces spontaneous iron overload (Fig. 3C).
Whole-blood transcriptome profiles
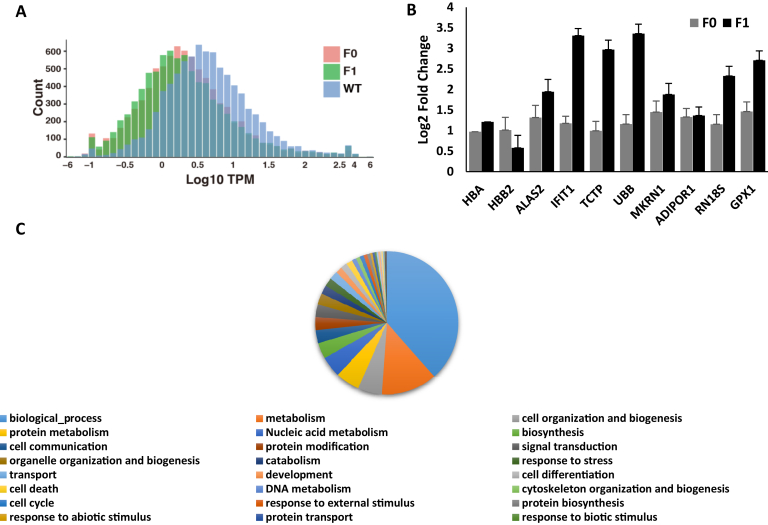
To further investigate the transcriptional differences between the WT and HBB2-targeted rabbits, total RNA was extracted from the peripheral blood of two F0, two F1, and four WT rabbits to synthesize the cDNA libraries and was used for RNA sequencing. A total of 8236 genes was detected, the majority of genes were fewer than ten copies, and only a small proportion of genes were highly expressed. In comparison with WT, HBB2-targeted rabbits harbored more low-expressing genes in the blood (Fig. 4A); however, most of the highly expressed genes were overexpressed in HBB2-targeted rabbits (Fig. 4B). We found that HBA was highly expressed in the blood of F0 and F1rabbits, whereas HBB2 was overexpressed in F0 rabbits, the expression levels of HBA and HBB2 resulted in the different anemic features of F0 and F1 rabbits. GO classification indicated that the highly expressed genes were involved in biological process, metabolism, cell organization, and biogenesis (Fig. 4C).
Figure 4.
The whole-blood transcriptome profiles of WT and HBB2-targeted rabbits.A, the expression level of genes in WT and HBB2-targeted F0 and F1 rabbits. B, logs-fold change of the top ten highly expressed genes in HBB2-targeted F0 and F1 rabbits. C, go classification of the genes expressed in the blood of HBB2-targeted rabbits.
Differential gene expression patterns of the HBB2-targeted rabbits
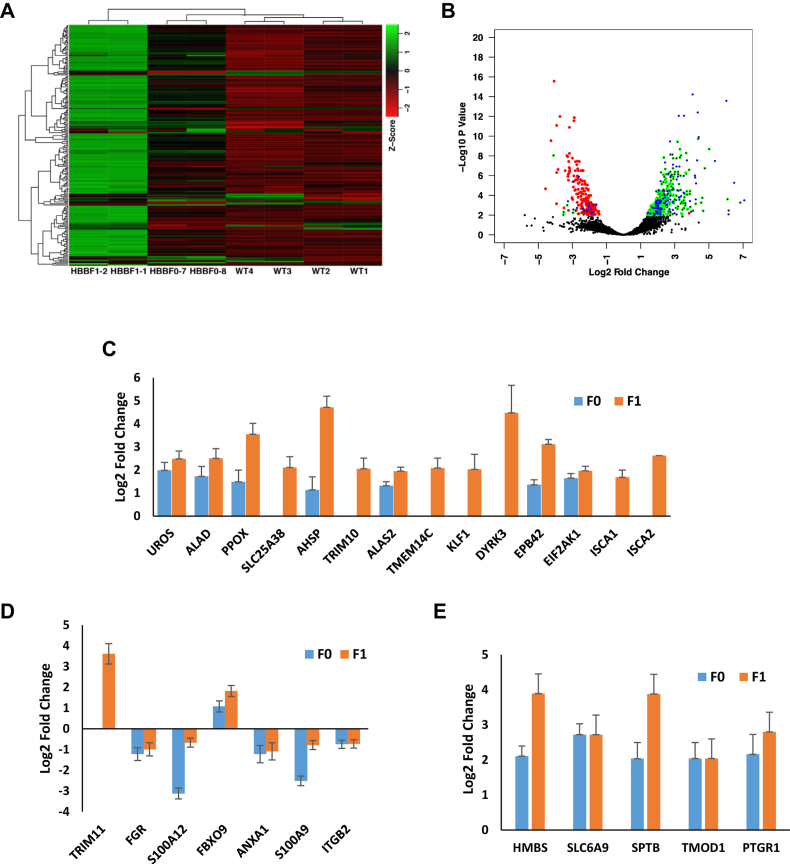
We then applied DESeq2 (22) to investigate the differential gene expression patterns of WT and HBB2-targeted rabbits. Gene set enrichment and principal component analysis indicated that the WT and HBB2-targeted rabbits featured distinctly different blood transcriptome patterns (Fig. 5A, Fig. S4A). In comparison with WT (p < 0.01), 130 genes were overexpressed in HBB2-targeted rabbits, 19 and 299 genes were highly expressed exclusively in F0 and F1, respectively. Seven of the lower-expressed genes were identified in HBB2-targeted rabbits, 58 and 10 genes were found in F0 and F1, respectively (Fig. 5B, Fig. S4B). Further gene interaction network analyses by GeneMANIA showed that the spectrin alpha chain, erythrocytic 1 (SPTA1) gene was highly coexpressed with hemoglobin genes (Fig. S4C), which indicated that the HBB2-targeted F0 rabbits possessed great erythrocytic repopulating ability.
Figure 5.
Differential gene expression pattern in the blood of HBB2-targeted rabbits.A, the cluster heat map of the top200 genes with the highest variance in WT and HBB2-targeted rabbits. B, volcano plots of differential gene expression level. The padj <0.05 (FDR < 0.1), HBB2-targeted-F0 (red dots), HBB2-targeted F1 (blue dots). C, the differential expression of genes related to hematopoiesis. D, the differential expression of genes related to innate immune. E, the differential expression of genes related to various blood disorders.
GO enrichment analysis (p < 0.05) showed that the highly expressed genes in HBB2-targeted rabbits were enriched in lipid metabolism and mitophagy, such as “GO:0033211 adiponectin-activated signaling pathway,” “GO:0044539 long-chain fatty acid import,” and “GO:0000422 mitophagy.” Furthermore, the genes highly expressed in F1 were mostly encircled in hematopoietic processes, such as “GO:0030218 erythrocyte differentiation,” “GO:0006783 heme biosynthetic process,” and “GO:0016226 iron sulfur cluster assembly” (Fig. S4D). We identified that 14 genes enriched in the hematopoiesis GO term were significantly overexpressed in the blood of F1 (Fig. 5C).
Thalassemia is associated with several immune deficiencies, in addition to anemia. A recent study demonstrated that innate immune cells, neutrophils in particular, are defective in human patients with β-thalassemia (23). We found that nine genes (TRIM11, FGR, S100A9, S100A12, FBXO9, TRIM10, ANXA1, S100A9, and ITGB2) related to innate immune response and neutrophil chemotaxis were expressed differently in the HBB2-targeted rabbits (Fig. 5D). A close examination of these differentially expressed genes after filtering at an adjusted p-value <0.01 (FDR < 0.1) and log2-fold-change >2. We also identified that five genes (HMBS (24), SLC6A9 (25, 26), SPTB (27), TMOD1 (28), and PTGR1 (29)) associated with various blood disorders were overexpressed in the HBB2-targeted rabbits (Fig. 5E). Gene interaction network analysis indicated that these genes interact with each other intimately (Fig. S4E).
Discussion
Animal models are crucial for understanding the mechanisms of hemoglobinopathies and validating future therapeutic strategies. To date, the only available β-thalassemia animal models are mouse models generated by KO β-globin genes or transgenic human mutant β-globin genes (8). Although these mouse models have improved our understanding of hemoglobinopathy, the gaps between mice and humans limit the knowledge from preclinical studies to the clinic. To overcome this limitation, here we successfully established a HBB2-targeted rabbit models by coinjection of Cas9 mRNA and gRNA into rabbit zygotes, which accurately matched the mutation pattern of human thalassemia patients. The hematological and histological analyses indicated that both the HBB2-targeted F0 and F1 rabbits exhibited typical characteristics of β-thalassemia clinical features. Moreover, the blood transcriptome analysis indicated that in addition to hematopoiesis, the HBB2-targeted rabbits exhibited innate immunity and lipid metabolism abnormalities, which are major concerns in patients with β-thalassemia (30).
Rabbits have been extensively used in the research of cardiovascular and metabolic diseases. For genotypic aspect, the rabbit β-globin gene cluster contains only one adult β-globin gene that is similar to human. Hence, rabbits are supposed to be a more suitable model for thalassemia disease than mice. However, due to the unavailability of pluripotent stem cells with ability to result in germline chimera and the low efficiency of somatic nuclear transfer in rabbits, which are the two classical approaches for generating gene targeted animals, the HBB2-targeted rabbits have not been established yet. Recent advances in genome editing have greatly prompted the generation of gene-targeted rabbits (20). By utilizing NHEJ to repair the double-strand breaks induced by Cas9/gRNA, we successfully achieved gene-edited rabbits harboring indel mutation in the HBB2 gene, which mimicked the genotype of most β-thalassemia patients (2). Compared with the previously reported mouse models, which were generated by large DNA fragment deletion, our established HBB2-targeted rabbit models are applicable for preclinical testing of gene correction in vivo.
For technological aspect, as reported in mice (31) and other animals (32), the efficiency of Cas9/gRNA-mediated gene targeting is associated with the concentration of injected Cas9 mRNA and gRNA, especially for the biallelic targeting efficiency. In particular, targeting the lethal gene is of vital importance, as in this study. We initially failed to obtain HBB2-targeted rabbits by embryo injection of a high concentration of Cas9 mRNA and gRNA (200/20 ng/μl). To avoid biallelic targeting, which is lethal for rabbits, we then used a lower concentration of Cas9 mRNA and gRNA (100/10 ng/μl) and successfully generated four HBB2-targeted chimeric rabbits with a variety of indel mutations from1 bp to 10 bp. By mating the F0 chimeric rabbits with wild-type ones, we achieved five F1 HBB2-targeted rabbits with deletion of 1 bp. Although the off-target effect (33) of CRISPR is a great concern and has been frequently reported (34), we did not detect off-target mutagenesis in the HBB2-targeted rabbits by silico predictions.
For phenotype aspect, examination of peripheral blood smears demonstrated marked anisocytosis, poikilocytosis, and target cells were presented in both the HBB2-targeted F0 and F1 rabbits, which was similar to the red cell morphology in β-thalassemia patients. Hematological analyses indicated that the heterozygous F1 rabbits exhibited typical characteristics of β-thalassemia, and the chimeric F0 rabbits showed a mild phenotype of anemia, which is majorly due to the rescue of wild-type genotypic hematopoietic stem cells. However, histological analyses suggested that unlike most untransfused β-thalassemia patients, the HBB2-targeted rabbits develop spontaneous iron deposits in the spleen at early stage, which may be useful to study the pathophysiology of iron overload in different organs and develop pharmacological-based therapies.
For transcriptome aspect, blood transcriptome analysis indicated that the HBB2-targeted rabbits featured different transcriptional patterns from WT. HBA was highly expressed in both F0 and F1 rabbits, whereas HBB2 was overexpressed in F0 rabbits. We identified 14 of the genes overexpressed in the blood of F1 related to hematopoiesis, in which seven genes showed no difference in F0 and WT. Furthermore, gene interaction analysis showed that SPTA1, an erythroid specific marker (35), was coexpressed with other hemoglobin genes in F0, this might imply that the normal hematopoietic stem cells in the chimeric rabbits possessed greater differentiation tendency toward RBCs to make up for the function loss of HBB2 gene, and therefore F0 rabbits exhibited milder symptoms than F1 counterparts. This also reminds us that β-thalassemia could be a suitable disease candidate of gene therapy through correcting the mutant HBB2 gene, where the correction of small ratio of cells is sufficient to relieve the symptoms. Patients with thalassemia suffer from various complications such as immune abnormalities, metabolic and endocrinologic disorders. Here, we found that the genes differently expressed in HBB2-targeted rabbits were significantly enriched in lipid and iron metabolism, protein translation, innate immunity, and hematopoietic processes, the genes related to hematopoiesis and immunity have also been identified in patients with various blood disorders. Taken together, here we described the first HBB2-targeted rabbits, which accurately recapitulated the typical genotypic and phenotypic features of β-thalassemia patients. This model will advance the further study of pathological mechanisms and development of new therapeutic strategies for β-thalassemia and related complications.
Experimental procedures
Animals
The rabbits used in the experiments were the New Zealand rabbit strain and obtained from the Center for Laboratory Animal Sciences of Southern Medical University. All experiments were performed according to the guidelines for animal experiments of the Department of Science and Technology of Guangdong Province (China) and approved by the Animal Research Ethics Committee of the Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences.
gRNA design and RNA synthesis
Two sgRNAs targeting the rabbit HBB2 exons were designed (sgRNA1 targeting ExonI and sgRNA2 targeting ExonII) according to the following website: http://crispr.mit.edu/. The sequences of the sgRNAs were as follows: sgRNA1 (GGTCACTGCCCTGTGGGGCAAGG) and sgRNA2 (GGTTCTTCGAGTCCTTTGGGG). Two complementary DNA oligos of gRNA were synthesized for each target site. The two DNA oligos (10 μM) were denatured at 95 °C for 10 min and annealed at room temperature before they were cloned between two BbsI sites of the pUC57-T7 vector (Addgene ID: 51306). Subsequent sequencing analysis was performed to select the correct gRNA that contained the target site sequence. The template of gRNA for in vitro transcription was PCR products obtained from gRNA vectors by using the primer pair (T7-F: 5′-GAAATTAATACGACTCACTATA-3′; T7-R: 5′-AAAAAAAGCACCGACTCGGTGCCAC-3′) with a high-fidelity enzyme (Takara). The gRNAs were transcribed using a T7 High Yield RNA Synthesis Kit (NEB) and purified using an miRNeasy Mini Kit (Qiagen). The Cas9 expression vector was linearized using NotI and transcribed using the mMESSAGE mMACHINE SP6 Kit (Ambion) to produce capped Cas9 RNA. The concentration and quality of the synthesized Cas9 mRNA and gRNA were measured using NanoDrop 2000 and agarose gel (1%) electrophoresis, respectively.
Microinjection and embryo transfer
The female New Zealand White rabbits (6–8 months old) were superovulated with FSH (100 IU). Approximately 3 days later, the rabbits were mated and then injected with 100 IU human chorionic gonadotrophin. The day after mating, the donor rabbits were sacrificed, and embryos were collected from the oviducts. Various concentrations of Cas9/gRNA mRNA were microinjected into the pronuclear (PN)-stage embryos. Approximately 35 to 40 injected embryos were transferred into the oviduct of a recipient rabbit. Surplus embryos were cultured to blastocyst stage for in vitro embryo development testing, and evaluation of gene targeting efficiency in Cas9/gRNA injected embryos was conducted.
Gene mutation detection in embryos and newborns by using PCR
The injected embryos were collected at the blastocyst stage. DNA of each embryo was extracted using a lysis buffer (1% NP40 plus 60 ng/μl protein K) for T7E1 assay and sequencing to detect the targeting efficiency. The genomic DNA of newborn rabbits was extracted using a DNA extraction kit (Takara) according to the manufacturer’s instructions. The PCR primer sequences used for mutation detection were as follows: rHBBF (GGGTCTGGGAGATACATAGAAGGAA) and rHBBR (CTTCCCATTCTAAACAACACCCTGA). The PCR products were gel purified and cloned into the pMD18-T vector (Takara). At least 10 T-A clones were used for sequencing to obtain the detailed information of gene mutation.
Off-target assay
Potential off-target cleavage sites of sgRNA1 were predicted using CRISPOR online software (http://crispor.tefor.net/). The top 48 HBB2-sgRNA1 potential off-target sites were identified. Approximately 500 to 700 bp genomic fragments containing the potential off-target site were amplified using PCR. The PCR products were confirmed using T7E1 enzyme digestion and sequenced to identify any mutations.
Hematological analysis of blood
Approximately 2 ml of blood samples was collected from the auricular vein of WT and HBB2-targeted rabbits. The 2 μl of blood samples was used for peripheral blood smears according to the manufacturer’s instructions (Giemsa Stain Kit, ZBS9-BA-4017). The blood smears were stained with Wright–Giemsa and examined under a Nikon microscope. Hemoglobin electrophoresis test was performed following the instructions of the CAPILLARYS HEMOGLOBIN(E) kit. A total of 18 μl of blood of each sample and five volumes of diluted reagent were added to the well of the cuvette and mixed thoroughly. The hemoglobin profiles of the samples were measured using a CAPILLARYS 2 FLEX-PIERCING instrument (Sebia). The rest of the blood was used for hematological analyses. The Hb, HCT, MCH, RBCs, MCV, RBC counts, RDW, and RET for each sample were measured by following the manufacturer’s instructions (Sysmex XN-9000). Uric acid and total bilirubin for each sample were determined using the Roche cobas 8000 analyzer series.
Histological analysis
The tissues of the liver, lung, heart, spleen, and kidney from the WT and HBB2-targeted rabbits (euthanized at 6 months of age) were fixed in 4% paraformaldehyde (pH 7.4) for 48 h, then embedded in paraffin wax, and sectioned for slides. Hematoxylin and eosin staining and Perls staining (Leagene DJ0001) were performed separately using standard protocols. The samples were then viewed under a Nikon microscope.
Whole-blood transcriptome analysis
Blood samples were collected using auricular venipuncture from four 6-month-old wild-type rabbits. HBB2-targeted founders F0-7, F0-8, and the offsprings F1-1 and F1-2. Total RNA was isolated using the RNAprep Pure Hi-Blood Kit (TIANGEN). cDNA libraries were prepared from high-quality RNA by using an Illumina TruSeq RNA sample prep kit following the manufacturer’s instruction (Illumina). Individual RNA-seq libraries were sequenced in triplicate at 100 bp/sequence paired-end reads by using an Illumina HiSeq 2500 sequencer (Illumina, TruSeq PE Cluster Kit v3, cBot, and TruSeq BS Kit v3). The clean reads were quantified using Salmon. The cDNA reference file came from Ensembl. Transcripts per million normalized counts were used for the transcriptome profile analysis. Differential expression analysis was performed using DEseq2.
Data availability
All data are contained within the article and the Supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
Y. Y. had the idea; X. L., L. L., and X. S. designed the experiments; X. L. and X. K analyzed the data; Y. Y., S. H., B. C. conducted most of the experiments; Y. X., B. S., Q. Z., H. W., Z. O., and Y. X. contributed to the experiments or provided critical advice; X. S. and L. L. provided funding; Y. Y. wrote the article, and X. L. and L. L. approved the final version for submission.
Funding and additional information
This work was supported by the National Key Research and Development Program of China, Stem Cell and Translational Research [grant numbers 2019YFA0111500, 2019YFA0110804]; the National Natural Science Foundation of China [grant numbers 31801124, 82071801, 81671121, 31872800]; the Guangzhou Science and Technology Project [grant numbers 201803010048, 201904010024]; the Guangzhou City Science and Technology Key Topics Project [201904020025]; the Guangdong Basic and Applied Basic Research Foundation [2019A1515010755]; the Health Science and technology project of Guangzhou [20191A011087]; the Natural Science Foundation of Guangdong Province [2020A0505100062]; Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory [2018GZR0201002].
Edited by Qi-Qun Tang
Contributor Information
Xiaoping Li, Email: lixiaoping@mail.sysu.edu.cn.
Liangxue Lai, Email: lai_liangxue@gibh.ac.cn.
Xiaofang Sun, Email: xiaofangsun@gzhmu.edu.cn.
Supporting information
References
- 1.Cohen A.R., Galanello R., Pennell D.J., Cunningham M.J., Vichinsky E. Thalassemia. Hematology Am. Soc. Hematol. Educ. Program. 2004:14–34. doi: 10.1182/asheducation-2004.1.14. [DOI] [PubMed] [Google Scholar]
- 2.Cao A., Galanello R. Beta-thalassemia. Genet. Med. 2010;12:61–76. doi: 10.1097/GIM.0b013e3181cd68ed. [DOI] [PubMed] [Google Scholar]
- 3.Farmakis D., Giakoumis A., Polymeropoulos E., Aessopos A. Pathogenetic aspects of immune deficiency associated with beta-thalassemia. Med. Sci. Monit. 2003;9:RA19–RA22. [PubMed] [Google Scholar]
- 4.Shamshirsaz A.A., Bekheirnia M.R., Kamgar M., Pourzahedgilani N., Bouzari N., Habibzadeh M., Hashemi R., Shamshirsaz A.A., Aghakhani S., Homayoun H., Larijani B. Metabolic and endocrinologic complications in beta-thalassemia major: A multicenter study in Tehran. BMC Endocr. Disord. 2003;3:4. doi: 10.1186/1472-6823-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danjou F., Anni F., Galanello R. Beta-thalassemia: From genotype to phenotype. Haematologica. 2011;96:1573–1575. doi: 10.3324/haematol.2011.055962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baronciani D., Angelucci E., Potschger U., Gaziev J., Yesilipek A., Zecca M., Orofino M.G., Giardini C., Al-Ahmari A., Marktel S., de la Fuente J., Ghavamzadeh A., Hussein A.A., Targhetta C., Pilo F. Hemopoietic stem cell transplantation in thalassemia: A report from the European Society for Blood and Bone Marrow Transplantation Hemoglobinopathy Registry, 2000-2010. Bone Marrow Transplant. 2016;51:536–541. doi: 10.1038/bmt.2015.293. [DOI] [PubMed] [Google Scholar]
- 7.Vichinsky E., Neumayr L., Trimble S., Giardina P.J., Cohen A.R., Coates T., Boudreaux J., Neufeld E.J., Kenney K., Grant A., Thompson A.A., the CDC Thalassemia Investigators Transfusion complications in thalassemia patients: A report from the Centers for Disease Control and Prevention (CME) Transfusion. 2014;54:972–981. doi: 10.1111/trf.12348. quiz 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McColl B., Vadolas J. Animal models of beta-hemoglobinopathies: Utility and limitations. J. Blood Med. 2016;7:263–274. doi: 10.2147/JBM.S87955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skow L.C., Burkhart B.A., Johnson F.M., Popp R.A., Popp D.M., Goldberg S.Z., Anderson W.F., Barnett L.B., Lewis S.E. A mouse model for beta-thalassemia. Cell. 1983;34:1043–1052. doi: 10.1016/0092-8674(83)90562-7. [DOI] [PubMed] [Google Scholar]
- 10.Shehee W.R., Oliver P., Smithies O. Lethal thalassemia after insertional disruption of the mouse major adult beta-globin gene. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3177–3181. doi: 10.1073/pnas.90.8.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang B., Kirby S., Lewis J., Detloff P.J., Maeda N., Smithies O. A mouse model for beta 0-thalassemia. Proc. Natl. Acad. Sci. U. S. A. 1995;92:11608–11612. doi: 10.1073/pnas.92.25.11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Opazo J.C., Hoffmann F.G., Storz J.F. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12950–12955. doi: 10.1073/pnas.0804392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamsai D., Zaibak F., Khongnium W., Vadolas J., Voullaire L., Fowler K.J., Gazeas S., Fucharoen S., Williamson R., Ioannou P.A. A humanized mouse model for a common beta0-thalassemia mutation. Genomics. 2005;85:453–461. doi: 10.1016/j.ygeno.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Shiomi M. Rabbit as a model for the study of human diseases. In: Houdebine L.-M., Fan J., editors. Rabbit Biotechnology. Springer Netherlands; Dordrecht: 2009. pp. 49–63. [Google Scholar]
- 15.Araki M., Fan J., Challah M., Bensadoun A., Yamada N., Honda K., Watanabe T. Transgenic rabbits expressing human lipoprotein lipase. Cytotechnology. 2000;33:93–99. doi: 10.1023/A:1008115429679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng X. Transgenic rabbit models for studying human cardiovascular diseases. Comp. Med. 2012;62:472–479. [PMC free article] [PubMed] [Google Scholar]
- 17.Peng X., Knouse J.A., Hernon K.M. Rabbit models for studying human infectious diseases. Comp. Med. 2015;65:499–507. [PMC free article] [PubMed] [Google Scholar]
- 18.Flisikowska T., Thorey I.S., Offner S., Ros F., Lifke V., Zeitler B., Rottmann O., Vincent A., Zhang L., Jenkins S., Niersbach H., Kind A.J., Gregory P.D., Schnieke A.E., Platzer J. Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song J., Zhong J., Guo X., Chen Y., Zou Q., Huang J., Li X., Zhang Q., Jiang Z., Tang C., Yang H., Liu T., Li P., Pei D., Lai L. Generation of RAG 1- and 2-deficient rabbits by embryo microinjection of TALENs. Cell Res. 2013;23:1059–1062. doi: 10.1038/cr.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Q., Zhang Q., Yang H., Zou Q., Tang C., Fan N., Lai L. Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen. (Lond.) 2014;3:12. doi: 10.1186/2045-9769-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canver M.C., Haeussler M., Bauer D.E., Orkin S.H., Sanjana N.E., Shalem O., Yuan G.-C., Zhang F., Concordet J.-P., Pinello L. Integrated computational guide design, execution, and analysis of arrayed and pooled CRISPR genome editing experiments. bioRxiv. 2017 doi: 10.1101/125245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siwaponanan P., Siegers J.Y., Ghazali R., Ng T., McColl B., Ng G.Z., Sutton P., Wang N., Ooi I., Thiengtavor C., Fucharoen S., Chaichompoo P., Svasti S., Wijburg O., Vadolas J. Reduced PU.1 expression underlies aberrant neutrophil maturation and function in beta-thalassemia mice and patients. Blood. 2017;129:3087–3099. doi: 10.1182/blood-2016-07-730135. [DOI] [PubMed] [Google Scholar]
- 24.Martinez di Montemuros F., Di Pierro E., Biolcati G., Rocchi E., Bissolotti E., Tavazzi D., Fiorelli G., Cappellini M.D. Acute intermittent porphyria: Heterogeneity of mutations in the hydroxymethylbilane synthase gene in Italy. Blood Cells Mol. Dis. 2001;27:961–970. doi: 10.1006/bcmd.2001.0466. [DOI] [PubMed] [Google Scholar]
- 25.Magor G.W., Tallack M.R., Gillinder K.R., Bell C.C., McCallum N., Williams B., Perkins A.C. KLF1-null neonates display hydrops fetalis and a deranged erythroid transcriptome. Blood. 2015;125:2405–2417. doi: 10.1182/blood-2014-08-590968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., Zhang W., Ma S.F., Desai A.A., Saraf S., Miasniakova G., Sergueeva A., Ammosova T., Xu M., Nekhai S., Abbasi T., Casanova N.G., Steinberg M.H., Baldwin C.T., Sebastiani P. Hypoxic response contributes to altered gene expression and precapillary pulmonary hypertension in patients with sickle cell disease. Circulation. 2014;129:1650–1658. doi: 10.1161/CIRCULATIONAHA.113.005296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallagher P.G., Forget B.G. Hematologically important mutations: Spectrin and ankyrin variants in hereditary spherocytosis. Blood Cells Mol. Dis. 1998;24:539–543. doi: 10.1006/bcmd.1998.0217. [DOI] [PubMed] [Google Scholar]
- 28.Moyer J.D., Nowak R.B., Kim N.E., Larkin S.K., Peters L.L., Hartwig J., Kuypers F.A., Fowler V.M. Tropomodulin 1-null mice have a mild spherocytic elliptocytosis with appearance of tropomodulin 3 in red blood cells and disruption of the membrane skeleton. Blood. 2010;116:2590–2599. doi: 10.1182/blood-2010-02-268458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crosslin D.R., McDavid A., Weston N., Zheng X., Hart E., de Andrade M., Kullo I.J., McCarty C.A., Doheny K.F., Pugh E., Kho A., Hayes M.G., Ritchie M.D., Saip A., Crawford D.C. Genetic variation associated with circulating monocyte count in the eMERGE network. Hum. Mol. Genet. 2013;22:2119–2127. doi: 10.1093/hmg/ddt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borgna-Pignatti C., Gamberini M.R. Complications of thalassemia major and their treatment. Expert Rev. Hematol. 2011;4:353–366. doi: 10.1586/ehm.11.29. [DOI] [PubMed] [Google Scholar]
- 31.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo X., Zhang T., Hu Z., Zhang Y., Shi Z., Wang Q., Cui Y., Wang F., Zhao H., Chen Y. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–714. doi: 10.1242/dev.099853. [DOI] [PubMed] [Google Scholar]
- 33.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4 doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolgast L.R., Cannizzarro L.A., Ramesh K.H., Xue X., Wang D., Bhattacharyya P.K., Gong J.Z., McMahon C., Albanese J.M., Sunkara J.L., Ratech H. Spectrin isoforms: Differential expression in normal hematopoiesis and alterations in neoplastic bone marrow disorders. Am. J. Clin. Pathol. 2011;136:300–308. doi: 10.1309/AJCPSA5RNM9IGFJF. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and the Supporting information.