Abstract
Background and Aims
TRIM21 is a ubiquitin E3 ligase that is implicated in numerous biological processes including immune response, cell metabolism, redox homeostasis, and cancer development. We recently reported that TRIM21 can negatively regulate the p62-Keap1-Nrf2 antioxidant pathway by ubiquitylating p62 and prevents its oligomerization and protein sequestration function. As redox homeostasis plays a pivotal role in many cancers including liver cancer, we sought to determine the role of TRIM21 in hepatocarcinogenesis.
Methods
We examined the correlation between TRIM21 expression and the disease using publicly available data sets and 49 cases of HCC clinical samples. We used TRIM21 genetic knockout mice to determine how TRIM21 ablation impact HCC induced by the carcinogen DEN plus phenobarbital (PB). We explored the mechanism that loss of TRIM21 protects cells from DEN-induced oxidative damage and cell death.
Results
There is a positive correlation between TRIM21 expression and HCC. Consistently, TRIM21-knockout mice are resistant to DEN-induced hepatocarcinogenesis. This is accompanied by decreased cell death and tissue damage upon DEN treatment, hence reduced hepatic tissue repair response and compensatory proliferation. Cells deficient in TRIM21 display enhanced p62 sequestration of Keap1 and are protected from DEN-induced ROS induction and cell death. Reconstitution of wild-type but not the E3 ligase-dead and the p62 binding-deficient mutant TRIM21 impedes the protection from DEN-induced oxidative damage and cell death in TRIM21-deficient cells.
Conclusions
Increased TRIM21 expression is associated with human HCC. Genetic ablation of TRIM21 leads to protection against oxidative hepatic damage and decreased hepatocarcinogenesis, suggesting TRIM21 as a preventive and therapeutic target.
Keywords: TRIM21, Hepatocellular Carcinoma, p62, Nrf2, Diethylnitrosamine
Abbreviations used in this paper: 8-oxo-dG, 8-oxo-2-deoxyguanosine; α-SMA, α-smooth muscle actin; AFP, α-fetoprotein; cDNA, complementary DNA; DEN, diethylnitrosamine; DMEM, Dulbecco’s modified Eagle’s medium; FBS, fetal bovine serum; HCC, hepatocellular carcinoma; IHC, immunohistochemistry; Keap1, Kelch-like ECH-associated protein 1; KO, knockout; LD, ligase dead; MEF, mouse embryonic fibroblast; mRNA, messenger RNA; Nrf2, nuclear factor E2-related factor 2; PB, phenobarbital; PBS, phosphate-buffered saline; PBST, phosphate-buffered saline with 0.02% Tween 20; PCNA, proliferation cell nuclear antigen; PCR, polymerase chain reaction; PI, propidium iodide; RNA-seq, RNA sequencing; ROS, reactive oxygen species; SDS, sodium dodecyl sulfate; TCGA, The Cancer Genome Atlas; TRIM21, tripartite motif-containing protein 21; WB, western blotting; WT, wild-type
Summary.
The ubiquitin E3 ligase Tripartite motif-containing protein 21 (TRIM21) is highly expressed in hepatocellular carcinoma (HCC). Genetic ablation of TRIM21 suppresses diethylnitrosamine (DEN)-induced hepatocarcinogenesis by activating the p62-Keap1-Nrf2 antioxidative pathway.
A major risk factor in hepatocellular carcinoma (HCC) is dysregulated redox homeostasis that is associated with various oncogenic events such as hepatitis B and C viral infection, obesity-induced nonalcoholic fatty liver disease, and alcohol-associated fatty liver diseases.1, 2, 3 Increased reactive oxygen species (ROS) can lead to oncogenic events such as elevated mitogenic signaling, genomic instability, increased metastatic potential, and altered tumor microenvironment. On the one hand, it is generally thought that reducing oxidative stress can be cancer preventive by lowering the chance of accumulating oncogenic mutations. On the other hand, it has been increasingly recognized that chronic and excessive antioxidant response can be oncogenic by promoting cancer cell survival and reducing anticancer immune response. Therefore, the precise role of ROS in cancer is highly context-dependent, vastly varies among tissue types, and is largely affected by the intensity, timing, and duration of the oxidative stress.4
The Kelch-like ECH-associated protein 1 (Keap1)–nuclear factor E2-related factor 2 (Nrf2)–ARE (antioxidant response element) pathway plays a major role in regulating cellular redox balance. Keap1 negatively regulates the antioxidant response by interacting with and promoting proteasomal degradation of the antioxidant transcription factor Nrf2.5,6 In response to oxidative stress, the suppression of Keap1 on Nrf2 is relieved by 2 means: the canonical way that is through Keap1 oxidation and failure of promoting Nrf2 proteasomal degradation, and the noncanonical way via Keap1’s sequestration in SQSTM1/p62-mediated protein aggregation. In the noncanonical pathway, in response to oxidative stress, p62 dimerizes via the hydrogen bond between lysine7 (K7) and aspartate 69 (D69) within its PB1 domain, which facilitates the subsequent p62 aggregation and protein sequestration function. As Keap1 interacts with p62 via its KIR domain, it is then sequestered in the p62-mediated protein aggregates. This Keap1 sequestration frees up Nrf2 and facilitates Nrf2 nuclear translocation and transcriptional activation of its targets including many antioxidative genes. The p62-Keap1-Nrf2 antioxidant pathway was found to be closely associated with a wide spectrum of cancers, including HCC.7, 8, 9, 10, 11, 12 Ironically, similar to the elusive picture as to how cellular redox balance is involved in oncogenesis, the precise role of the p62-Keap1-Nrf2 pathway in cancer especially in HCC remains elusive and controversial.9,13, 14, 15, 16, 17
We recently reported that a RING finger domain-containing ubiquitin E3 ligase tripartite motif-containing protein 21 (TRIM21) can directly interact with and ubiquitylate p62 at K7 via K63-linkage. This K7 ubiquitylation of p62 abolishes the K7-D69 hydrogen bond formation and hence p62 oligomerization, aggregation, and sequestration functions.18 TRIM21-mediated p62 K7 ubiquitylation leads to the failure of Keap1 sequestration and suppressed antioxidant response. Conversely, TRIM21-deficient cells display increased p62 oligomerization, protein aggregation, Keap1 sequestration, Nrf2 activation, and antioxidant response.18 Therefore, TRIM21 functions as a negative regulator of cellular antioxidant response. In light of the elusive role of redox balance and the p62-Keap1-Nrf2 pathway in cancer, we set out to study the expression of TRIM21 in human HCC samples, and test how TRIM21 may impact HCC using a mouse model that is treated with the carcinogen DEN.
Results
TRIM21 Expression Is Elevated in Human HCC
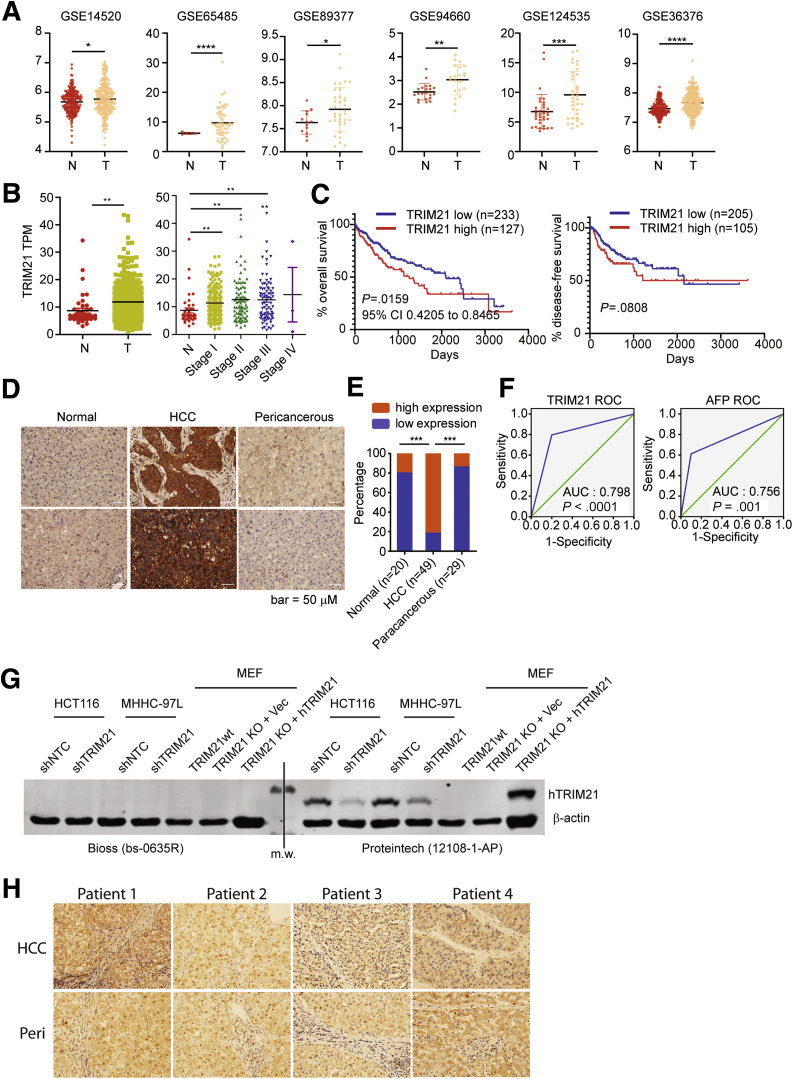
We first examined the expression of TRIM21 in human HCC using publicly available datasets. In the Gene Expression Omnibus Series,19 6 of 18 studies showed elevated TRIM21 expression in HCC comparing with normal liver tissues (Figure 1A). The Cancer Genome Atlas (TCGA) also showed that TRIM21 transcript level is elevated in tumor tissues and positively correlates with disease progression (Figure 1B), and that the expression level of TRIM21 correlates with a worse overall survival although not with disease-free survival (Figure 1C). These observations are opposite to that was reported in GSE14520, in which increased TRIM21 correlates with better survival20 and in other studies in the TCGA. To closely examine the role of TRIM21 in HCC, we performed TRIM21 immunohistochemistry (IHC) using cohorts of 49 HCC samples, 29 paracancerous tissues, and 20 hepatic hemangioma tissues as normal liver sample controls. TRIM21 protein level was markedly higher in HCC tumor tissues than that in nonmalignant liver tissues from peripheral hepatic hemangioma or in pericancerous tissues (Figure 1D and E). Among the 49 patients, 35 (71.43%) were infected with hepatitis B virus and 36 (73.47%) patients were infected with cirrhosis, suggesting a potential etiological connection between TRIM21 expression and these HCC risk factors. We also compared the diagnostic capacity of TRIM21 with the commonly used HCC biomarker α-fetoprotein (AFP). TRIM21 showed higher area under the receiver-operating characteristic curve (= 0.798) than AFP (= 0.756) (Figure 1F). The diagnostic performance was calculated and showed the sensitivity (79.6% vs 61.2%), specificity (80.0% vs 90.0%), positive predictive value (90.7% vs 93.8%), negative predictive value (61.5% vs 48.6%), and accuracy (79.7% vs 69.6%) for TRIM21 (TRIM21 high expression) vs AFP (AFP ≥15 ng/mL), respectively. Taken together, these analyses indicate that TRIM21 expression is elevated in HCC and correlates with a poorer prognosis.
Figure 1.
Increased TRIM21 expression is associated with HCC. (A) In the Gene Expression Omnibus Series, 6 studies showed elevated TRIM21 expression in HCC compared with normal liver tissues. (B) TCGA shows increased TRIM21 TPM in liver cancer comparing with normal tissues. (C)TRIM21 high expression indicates poor prognosis with decreasing overall survival according to TCGA data analysis, while the disease-free survival is trending lower yet not significant. (D) Liver tissue sections were stained for TRIM21 by immunohistochemistry (IHC). Representative TRIM21 staining on normal liver tissues from peripheral hepatic hemangioma (left panel; n = 20), on tumor tissues from HCC (middle panel; n = 49), and on paracancerous tissues 2 cm away from the HCC (right panel; n = 29). (E) TRIM21 expression is increased in HCC tissues compared with the paracancerous tissues and normal liver tissues. Relative intensity of staining in each section was assessed blindly and assigned a value ranging from 0 to 4 according to intensity of staining: score 0, negative staining, no positive cells; score 1, weak staining, the ratio of positive cells to total cells ≤25%; score 2, moderately positive, the ratio of positive cells to total cells ≤50%; score 3, moderately positive, the ratio of positive cells to total cells ≤75%; score 4, strongly positive, the ratio of positive cells to total cells>75%. Low expression defines the score from 0 to 2 and high expression is the score 3–4. (F) Receiver-operating characteristic (ROC) curves of the ability of TRIM21 or AFP high expression to the diagnosis of HCC. (G) TRIM21 was silenced in HCT116 and MHHC-97L human cells, or reconstituted with vector or HA-hTRIM21 in TRIM21–/– MEFs. Cells were lysed in RIPA buffer (1% SDS) and subjected to WB. The TRIM21 antibody obtained from Bioss (bs-0635R) or Proteintech (12018-1-AP) was used to detect human TRIM21. Note that the Bioss antibody fails to recognize human TRIM21, and neither antibody recognizes murine TRIM21. (H) TRIM21 antibody (Bioss bs-0635R) was used for IHC on HCC tumor tissues and paracancerous tissues collected 2 cm away from the HCC. Note that the Bioss antibody fails to detect difference in IHC staining in paracancerous and HCC tissues. ∗P < .05. ∗∗P < .01. ∗∗∗P < .001.
These results are opposite to a previous study that showed high level of TRIM21 in normal liver tissue but decreased TRIM21 in HCC tumor tissues.20 To reconcile the seemingly drastically different results between ours and the previous report, we acquired the same TRIM21 antibody as described in Ding et al.20 To our surprise, this antibody failed to recognize both endogenous and ectopically expressed TRIM21 by immunoblotting (Figure 1G) and did not detect high signal in IHC of both peri-cancerous and HCC tissues (Figure 1H). Therefore, while we cannot fully explain why the results in Ding et al20 are drastically different from ours, we suspect that the discrepancy may be caused, at least in part, by the different resources and validity of reagents. Taken together, our previous analyses indicate that TRIM21 expression is elevated in HCC and correlates with a poorer prognosis.
TRIM21-Deficiency Inhibits HCC Development in the Diethylnitrosamine/Phenobarbital Model
We next determined the role of TRIM21 in HCC development. We chose to use the chemical carcinogen diethylnitrosamine (DEN)/phenobarbital (PB)-induced HCC model, which has been characterized to have histology and gene expression similar to human HCC.21, 22, 23 DEN is bioactivated in the liver by cytochrome P450 to cause oxidative damage and genotoxic lesions and subsequent tissue repair response. PB promotes the progression from DEN-induced early dysplastic lesions to fully malignant tumors. TRIM21 wild-type (WT) and knockout (KO) male mice were given a single intraperitoneal injection of DEN at day 14 after birth. All WT mice developed HCC 10 months later. Strikingly, TRIM21–/– mice showed markedly less HCC development than the WT mice, while the heterozygote TRIM21+/– mice showed an intermediate phenotype (Figure 2A–D). Close examination of the tumor tissues showed that solid type HCC that was poorly differentiated (including tumor giant cell) in WT, moderately differentiated in TRIM21+/–, and well differentiated in TRIM21–/– mice, respectively (Figure 2E). While both WT and KO tumors expressed high levels of a HCC marker HSP70, the WT tumor cells proliferated much faster than the KO tumor cells (Figure 2F). Tissue fibrotic response was also markedly decreased in TRIM21–/– mice, as indicated by the expression of α-smooth muscle actin (α-SMA) and Sirius Red staining (Figure 2G). These data indicate that TRIM21 deficiency confers resistance to DEN/PB-induced HCC development in mice.
Figure 2.
TRIM21 ablation leads to decreased HCC induced by DEN/PB. (A) TRIM21+/+ (n = 8), TRIM21+/– (n = 15), and TRIM21–/– (n = 14) 14-day-old male mice were intraperitoneal injected with DEN (5 mg/kg), then fed with 0.05% PB in drinking water 7 days later. Livers from DEN/PB-treated mice were collected after 10 months. Representative images are shown. (B) Liver weight and body weight were measured 10 months after DEN/PB treatment. Ratio of liver weight to body 8 was plotted. (C) Total tumor number was counted and plotted. Each dot represents 1 mouse. (D) Tumors with >0.3 cm diameter was counted and plotted. (E) Liver tissue sections were processed for hematoxylin and eosin staining. TRIM21+/+ (left panel), TRIM21+/– (middle panel), and TRIM21–/– (right panel) liver tissue showed solid type hepatocellular carcinoma, with poorly differentiated (including tumor giant cell), moderately differentiated, and well differentiated, respectively. (F) Liver tissues sections were stained for HSP70 and PCNA. PCNA-positive cells were counted and the means of 3 randomly selected fields are shown. (G) Liver tissue sections were processed for Sirius Red staining and α-SMA/DAPI staining by immunofluorescence. Representative images are shown. ∗P < .05. ∗∗P < .01. ∗∗∗P < .001. IOD, integrated optical density.
TRIM21–/– Mice Are Protected From DEN-Induced Liver Damage
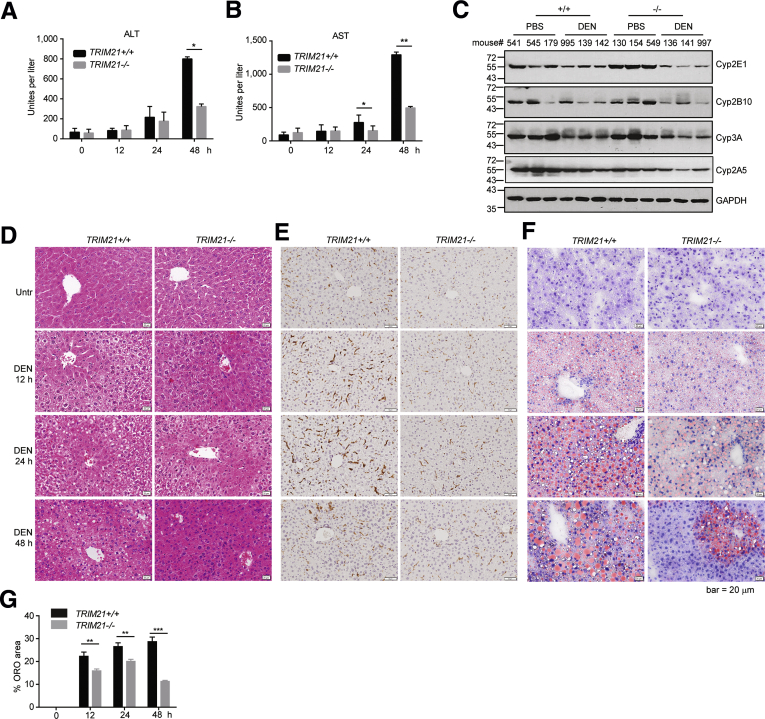
DEN is a genotoxic agent that induces oxidative DNA damage, cell death, and dysplastic lesions.24,25 The resulting chromatin instability, activating oncogenic mutations, and compensatory proliferation of the surviving hepatocytes contribute to DEN-induced carcinogenesis.26,27 Therefore, we examined whether TRIM21–/– mice were protected from DEN-induced acute liver damage. WT and TRIM21 KO mice were treated with 1 intraperitoneal injection of DEN for 12, 24, and 48 hours. TRIM21–/– mice were protected from acute liver damage caused by DEN treatment as indicated by the serological levels of alanine aminotransferase (Figure 3A) and aspartate aminotransferase (Figure 3B). Both the untreated WT and TRIM21–/– livers showed similar levels of the DEN-metabolizing enzymes, suggesting that the decreased acute liver damage in TRIM21–/– mice was not due to decreased expression of the DEN-metabolizing enzymes (Figure 3C). Hematoxylin and eosin staining showed that in WT mice, DEN treatment for 12 and 24 hours induced perivenular necrosis, with hepatocytes showing microvesicular change and infiltration of scattered neutrophils, lymphocytes, and monocytes (Figure 3D and E), with cells around the portal tracts spared. These changes were resolving at 48 hours (Figure 3D and E). The extent of liver damage was drastically less severe in TRIM21 KO mice at all time points (Figure 3D). Consistently, DEN treatment led to more profound infiltration of F4/80-positive monocytes in WT than in KO livers (Figure 3E). Moreover, acute DEN treatment is known to induce accumulation of lipid droplets via oxidative inhibition of β-oxidation enzymes.28 This DEN-induced accumulation of lipid droplets was significantly suppressed in TRIM21–/– livers at 12- and 24-hour time points, which was further resolved at 48 hours (Figure 3F and G). These results indicate that TRIM21–/– mice are protected from acute oxidative hepatocyte damage induced by DEN treatment.
Figure 3.
TRIM21–/–livers are protected from DEN-induced liver damage. (A, B) Eight-week-old TRIM21+/+ and TRIM21–/– male mice were treated with 200-mg/kg DEN via intraperitoneal injection. Cardiac blood was collected. (A) Alanine aminotransferase and (B) aspartate aminotransferase were measured before and after 12, 24, and 48 hours treatment. (C) Liver tissues (control and 48 hours DEN treatment) were lysed in RIPA buffer and subjected to WB for enzymes related to DEN metabolism. (D) Hematoxylin and eosin staining of formalin-fixed, paraffin-embedded liver sections. (E) F-4/80 staining indicates more profound monocyte infiltration in WT livers upon DEN treatment. (F) Oil Red O (ORO) staining of frozen liver sections with hematoxylin counterstaining. Red indicates lipid, purple indicates nuclei. Representative images are shown. (G) Quantitation of the area occupied by lipid inclusions within the frozen liver sections after staining of sections with ORO. Data are presented as mean ± SEM of 3 replicates. ∗∗P < .01. ∗∗∗P < .001.
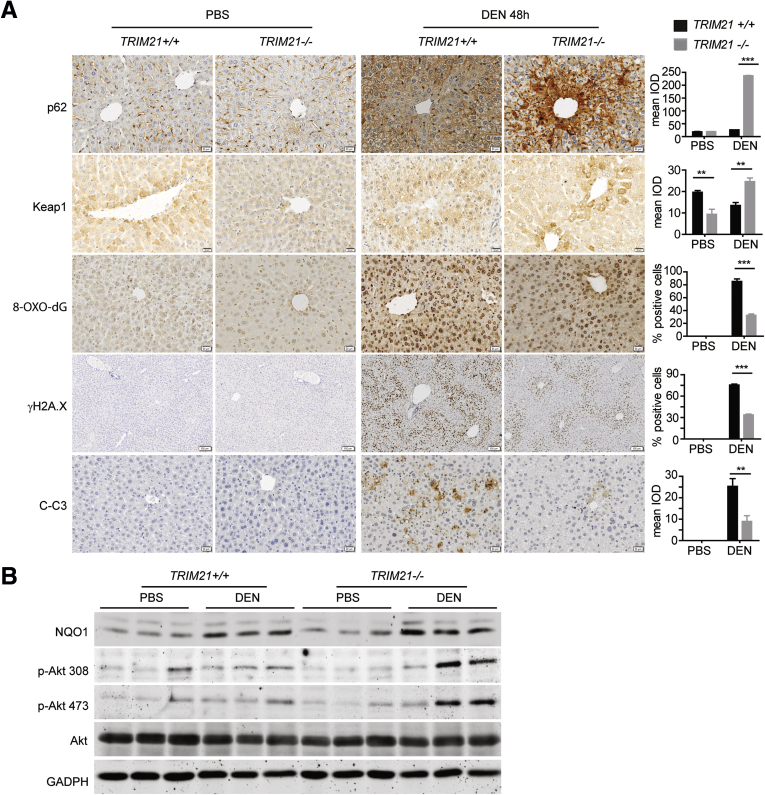
We showed previously that TRIM21 is a negative regulator of the p62-Keap1-Nrf2 antioxidant pathway, and that TRIM21-deficient mice are protected from oxidative damage in arsenic-induced liver damage and aortic transverse operation–induced heart failure.18 Our previous results suggest that TRIM21 deficiency confers protection from DEN-induced oxidative hepatocyte damage. We then looked at whether the p62-Keap1-Nrf2 pathway is involved in DEN-induced liver damage. Indeed, consistent with our previous finding that TRIM21 negatively regulates p62 oligomerization and protein aggregation, TRIM21 KO livers showed increased p62 and Keap1 aggregation upon DEN treatment, as indicated by increased integrated optical density of both proteins (Figure 4A), as well as increased NQO1 expression indicative of elevated Nrf2 activity (Figure 4B). These were accompanied by decreased H2A.X phosphorylation (a DNA damaging marker), 8-oxo-2-deoxyguanosine (8-oxo-dG) (an oxidative derivative of deoxyguanosine), and cleaved caspase-3 (Figure 4A). Interestingly, Akt activation as indicated by phosphorylated Akt was higher in DEN-treated TRIM21 KO livers (Figure 4B), suggesting that Akt may play a cytoprotective role in the hepatocytes exposed to DEN, although this increased phospho-Akt may also lead to predisposition of increased tumorigenesis. Collectively, these results indicate that TRIM21 KO livers have increased p62-Keap1-Nrf2 antioxidative activity and are protected from DEN-induced oxidative damage.
Figure 4.
TRIM21–/–livers are protected from DEN-induced liver damage. (A) Eight-week-old TRIM21+/+ and TRIM21–/– mice were treated with 200-mg/kg DEN via intraperitoneal injection. Livers were collected 48 hours later and processed for IHC analysis. Mean integrated optical density was determined for p62, Keap1, and cleaved caspase-3 (C-C3) because they are predominantly localized in the cytosol. Cells with positive nuclear staining of 8-oxo-dG and γH2A.X staining were counted and indicated as the percentage to total cells. Representative images are shown. Data are presented as mean ± SEM of 3 replicates. (B) Liver tissues were lysed in RIPA buffer and subjected to WB for indicated proteins. ∗∗P < .01. ∗∗∗P < .001.
TRIM21 KO Mouse Livers Show Enhanced Antioxidant Capacity and Decreased Compensatory Proliferation
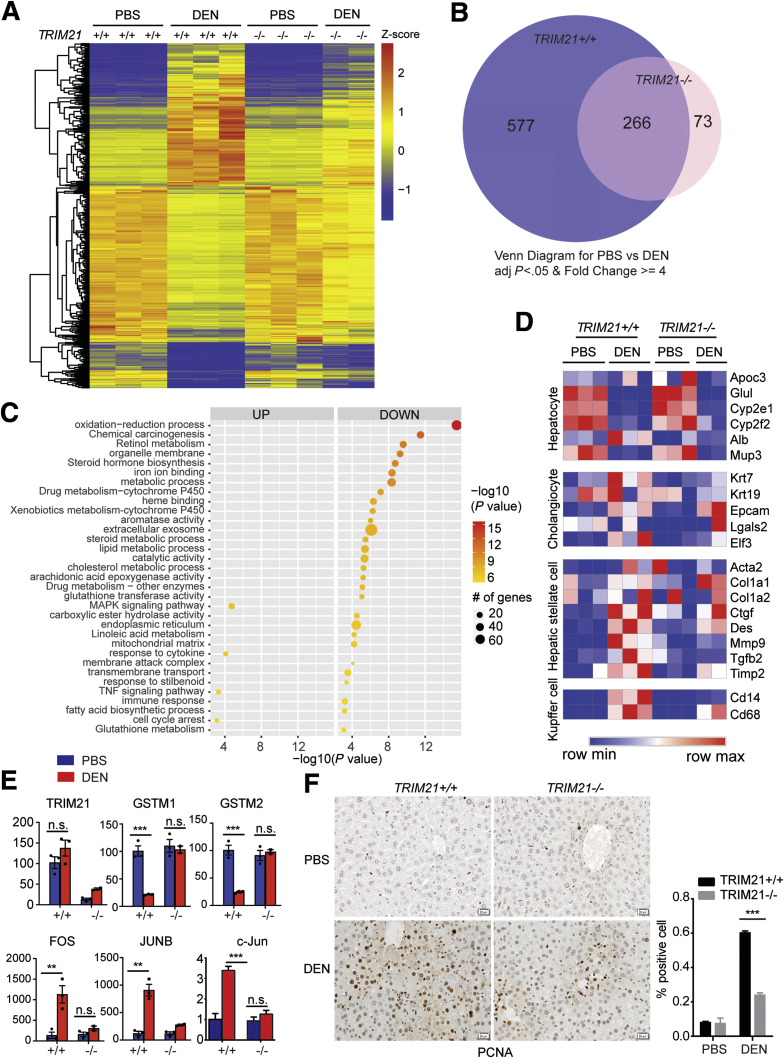
To further determine the effect of TRIM21 ablation on the liver tissue response to DEN treatment, we performed RNA sequencing (RNA-seq) analysis of liver tissues of TRIM21 WT or KO mice treated with DEN for 48 hours. Differentially expressed genes were identified by a cutoff of 4-fold and P < .05 (Figure 5A). Upon DEN treatment, markedly more genes in TRIM21 WT (n = 843) were altered than in TRIM21 KO (n = 339), with 266 genes in common (Figure 5B; Supplementary Table S1). Gene Ontology analysis revealed that the 577 genes uniquely altered in TRIM21 WT livers in response to acute DEN treatment were associated with redox regulation and chemical carcinogenesis (Figure 5C). In contrast, no significant enrichment was found in the 73 genes specifically altered in TRIM21 KO livers. The enrichment associated with chemical carcinogenesis was not significant in the 266 commonly altered genes. While DEN treatment did not significantly alter hepatocyte and cholangiocyte markers in WT and KO livers, it significantly induced the expression of makers of hepatic stellate cells and Kupffer cells (Figure 5D), consistent with our observations that DEN-induced fibrosis was lower in the KO livers than in the WT livers (Figure 2G). In addition, no significant difference in cytochrome P450 (Cyp2e1 and Cyp2f2) expression was observed between TRIM21 WT vs KO livers, before and after DEN treatment (Figure 5D), consistent with our observation by immunoblotting (Figure 3C). Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) analysis confirmed the significantly decreased expression of antioxidant genes GSTM1 and GSTM2 and increased expression of proliferation-related protogenes c-FOS, c-JUN, and JUNB in TRIM21 WT but not KO livers (Figure 5E). Consistently, IHC analysis demonstrated that the proliferation cell nuclear antigen (PCNA) was highly induced upon DEN treatment in TRIM21 WT but not in KO livers (Figure 5F). Taken together, these results indicate that TRIM21 KO livers have better antioxidant capacity and decreased compensatory hepatic proliferation compared with TRIM21 WT livers.
Figure 5.
DEN-induced hepatocyte proliferation is suppressed in TRIM21–/–liver. (A) Eight-week-old TRIM21+/+ and TRIM21–/– male mice were treated with PBS or 200-mg/kg DEN by intraperitoneal injection. Liver tissues were collected after 48 hours for RNA-seq. Hierarchical clustering and heatmap illustration of differentially expressed genes under indicated conditions. Color key indicates Z-scores after normalization. (B) Venn diagram showing overlap of genes differentially expressed in PBS-treated vs DEN-treated TRIM21+/+ and TRIM21–/– mice. Analysis was restricted to genes, which showed a 4-fold difference and yielded P < .05. (C) Dot plots showing the enriched terms from Gene Ontology analysis using differentially expressed genes identified from DEN vs PBS treatment in TRIM21 WT mice (the 577 unique genes altered by >4-fold in TRIM21-WT mice as shown in B). Note that genes involved in redox regulation, chemical carcinogenesis, and retinol metabolism are identified. (D) Heatmap shows that DEN-induced expressions of hepatic stellate cell and Kupffer cell markers were markedly decreased in TRIM21 KO livers, while no obvious difference was detected in hepatocyte markers. (E) Quantitative reverse-transcription PCR for the DEN-treated TRIM21 WT and KO liver samples, showing decreased antioxidant response and increased proliferation signals DEN-treated WT mice (n = 3 per group). (F) IHC analysis of PCNA expression in livers of PBS- and DEN-treated TRIM21 WT and KO mice. PCNA-nuclear positive cells were counted and the ratio of positive cells was calculated. Representative images are shown. Data are presented as means ± SEM of 3 mice in each group. ∗∗∗P < .001. FPKM, fragments per kilobase of transcript per million; n.s., nonsignificant.
TRIM21-Deficient Cells Are Protected From DEN-Induced Oxidative Damage via p62-Mediated Sequestration of Keap1
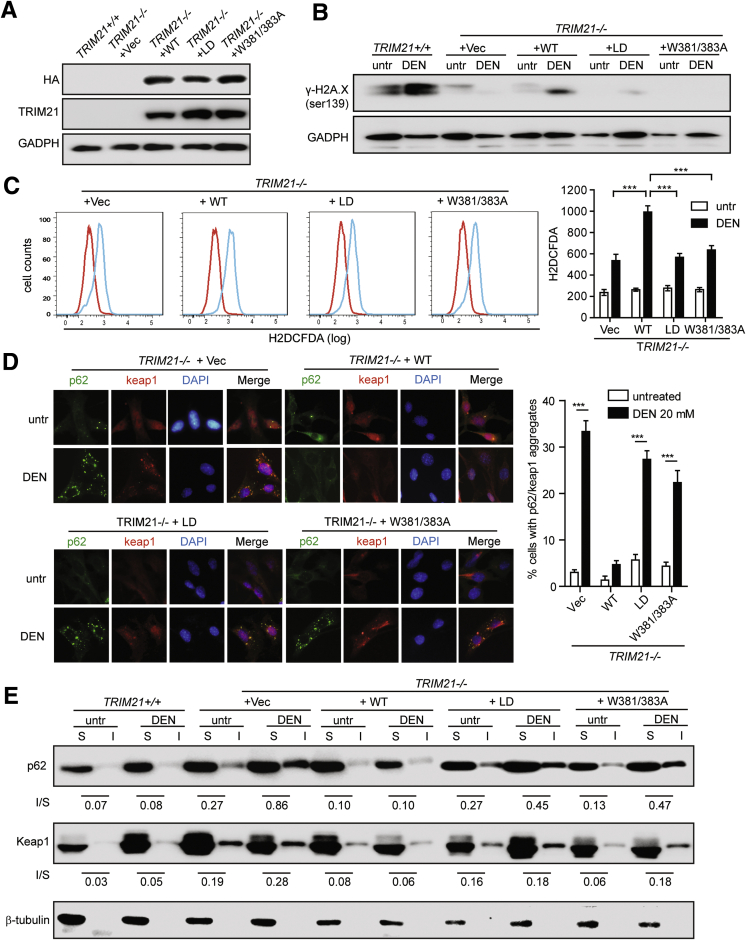
We showed previously that TRIM21 interacts with and ubiquitylates p62 and suppresses p62 oligomerization and Keap1 sequestration, which negatively regulates cellular antioxidant response.18 To determine whether the protection of TRIM21 KO livers from DEN treatment is via this mechanism, we utilized cultured TRIM21 KO mouse embryonic fibroblast (MEFs) and the human cancer cell line SMMC-7721 for acute DEN treatment. Although DEN is commonly used for in vivo studies, it has also been shown to possess cytotoxicity in cultured cells via similar mechanisms as in vivo.29, 30, 31 TRIM21-KO MEFs were reconstituted with HA-tagged WT, the ubiquitin E3 ligase dead (LD), and p62 binding-deficient mutant (W381/383A) mutants of TRIM21 (Figure 6A). DEN treatment induced DNA damage as indicated by H2A.X phosphorylation (γH2A.X) in TRIM21 WT MEFs, which was largely suppressed in TRIM21 KO MEFs (Figure 6B). Reconstitution of WT, but not the E3 LD and the p62 interaction–deficient (W381/382A) mutants, resumed the DEN-induced DNA damage (Figure 6B). Consistently, WT TRIM21 reconstituted cells displayed increased ROS indicated by H2DCFDA staining, while the LD and W381/383A mutants showed similar level of ROS as the KO MEFs (Figure 6C). Immunofluorescence analysis showed that DEN induced the formation of protein aggregates that were positive for p62 and Keap1 in TRIM21 KO MEFs and those reconstituted with the LD and W381/383A mutants but not with WT TRIM21 (Figure 6D). Cellular fractionation of detergent (1% Triton X-100) soluble and insoluble fractions also showed that DEN treatment led to increased insoluble p62 and Keap1 in TRIM21 KO MEFs, which was reduced by the reconstitution of WT but not the LD and W381/383 mutants of TRIM21 (Figure 6E). These results indicate that TRIM21 negatively regulates DEN-induced p62 aggregation and Keap1 sequestration. Ablation of TRIM21 leads to decreased oxidative stress.
Figure 6.
TRIM21 silencing protects cells from DEN-induced oxidative damage.TRIM21–/– MEFs reconstituted with vector, HA-TRIM21 WT, HA-TRIM21 LD, and HA-TRIM21 W381/383A mutant. (A) Cells were lysed in RIPA buffer with 1% SDS and subjected to WB. (B–E) Cells were treated with DEN (20 mM) for 6 hours. (B) Cells were harvested and probed for indicated proteins by WB. (C) Cells were stained with H2DCFDA and analyzed by flow cytometry. Quantification of relative H2DCFDA intensity (geometry mean of 3 repeats) is shown on the right. ∗∗∗P < .001. (D) Cells were subjected to IF with Keap1 and p62 antibodies and observed under deconvolution microscope. Cells with p62/Keap1 aggregates were counted blindly. Data shown are the averages plus SD of at least 3 countings with over 200 cells. ∗∗∗P < .001. (E) Cells were separated into Triton X-100 (1%) soluble and insoluble fractions, and 30-μg soluble proteins and corresponding volume of insoluble proteins were used for WB. The numbers below Western blot panels indicate the ratios of absolute levels of insoluble and soluble proteins, respectively.
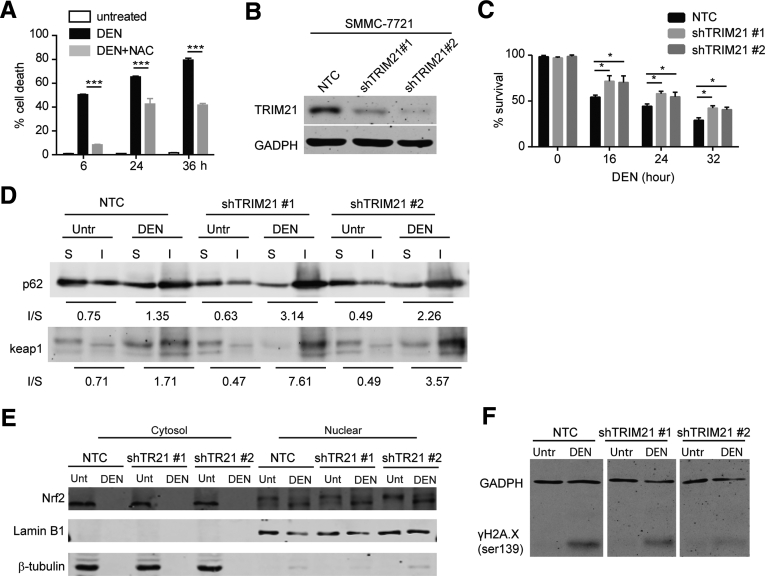
Similarly, in the human SMMC-7721 cancer cell line, acute DEN treatment led to significant amount of cell death, which was alleviated by the ROS scavenger N-acetylcysteine (Figure 7A). This indicates that DEN-induced cell death is mediated by increased oxidative stress (Figure 7A). We then used 2 independent short hairpin RNA of TRIM21 to silence TRIM21 (Figure 7B). Upon DEN treatment in vitro, the knockdown cells showed increased cell viability (Figure 7C). Consistent with increased p62 sequestration of Keap1 in TRIM21-deficient cells upon other oxidative stress,18 DEN treatment led to increased p62 and Keap1 in the detergent-insoluble fraction in the TRIM21-silenced cells (Figure 7D). This was correlated with increased Nrf2 in the nuclear fraction (Figure 7E), and decreased DNA damage as indicated by H2A.X phosphorylation (γH2A.X) (Figure 7F). Taken together, the previous results demonstrate that ablation of TRIM21 leads to increased p62 oligomerization and Keap1 sequestration, which protects hepatocytes from DEN-induced genotoxic damage and cell death.
Figure 7.
Silencing of TRIM21 protects cells from DEN-induced genotoxic damage and cell death. (A) SMMC-7721 cells were treated with DEN (50 mM) alone or together with N-acetylcysteine (5 mM) for indicated time. Cell viability was analyzed by PI exclusion. ∗∗∗P < .001. (B) TRIM21 was silenced in SMMC-7721 cells using 2 independent short hairpin RNAs. Cells was lysed in RIPA buffer with 1% SDS and subjected to WB. (C) SMMC-7721 with nontargeted control, shTRIM21 #1 and shTRIM21 #2 were treated with DEN (50 mM) for indicated periods of time. Cell viability was determined by trypan blue exclusion. Shown are the mean plus SD of 3 countings. ∗P < .05. (D) SMMC-7721 with nontargeted control, shTRIM21 #1, and shTRIM21 #2 were treated with DEN (50 mM) for 6 hours. Cells were lysed in immunoprecipitation lysis buffer containing 1% Triton X-100. The insoluble fraction was dissolved in RIPA buffer containing 1% SDS. Both Triton X-100 soluble and insoluble fractions were subjected to WB, and 30-μg soluble proteins and corresponding volume of insoluble proteins were used for WB. The numbers below Western blot panels indicate the ratios of absolute levels of insoluble and soluble proteins, respectively. (E) SMMC-7721 cells were treated with DEN (50 mM) for 6 hours. Cells were separated into cytoplasmic and nuclear fractions, and 20-μg cytosolic proteins and corresponding volume of nuclear fractions were used for WB. (F) SMMC-7721 cells were treated with DEN (50 mM) for 6 hours. Cells was lysed in RIPA buffer (1% SDS) and subjected to WB.
Discussion
In this study, we report that TRIM21 expression is elevated in human HCC by examining clinical samples and publicly available datasets. We also show that TRIM21 KO mice are resistant to hepatocarcinogenesis induced by carcinogen DEN. The decreased HCC development is correlated with decreased ROS production, DNA damage, and cell death in TRIM21-deficient mice and cells upon DEN treatment. As DEN is known to induce HCC by causing genomic instability, cellular damage, and subsequent compensatory proliferation of the remaining survived cells, our data indicate that ablation of TRIM21 plays a protective role in maintaining hepatocyte homeostasis and survival and hence reduces carcinogenesis.
Redox regulation plays a critical role in organismal homeostasis. While certain level of ROS is beneficial by serving as signaling molecules and by promoting antipathogen responses, elevated ROS can cause oxidative damage to macromolecules thus genomic instability, cellular damage, and ultimately cell death. The role of ROS in cancer has been complex and is dependent on tissue types and temporal stages of tumor initiation and development. Antioxidant response on one hand can prevent tumor initiation by maintaining cellular redox hemostasis and suppressing oxidative damage, on the other hand can enhance survival and growth of established tumor cells hence facilitate tumor progression and therapy-resistance.32, 33, 34 We have previously shown that TRIM21, a RING finger-containing E3 ubiquitin ligase, negatively regulates the p62-Keap1-Nrf2 antioxidant pathway.18 Therefore, it is foreseeable that loss of TRIM21 should be equivalent to gain of function of p62 and Nrf2 and of loss of function of Keap1, in their capacity in regulating redox balance and cellular homeostasis. Our previous study showed that TRIM21 KO hearts are protected from cardiac failure induced by pressure overload and livers protected from arsenic-induced liver damage,18 and the current study shows that TRIM21-deficient cells and liver are protected from DEN-induced oxidative damage. Indeed, numerous studies on the p62-Keap1-Nrf2 axis have demonstrated consistent results in protection against cellular and tissue damages. Activation of Nrf2 attenuated myocardial ischemia and reperfusion injury35 and oxidative liver damage.36, 37, 38, 39, 40
In cancer, the p62-Keap1-Nrf2 axis plays a paradoxical role.13,41 While some studies showed that activation of Nrf2 promotes tumorigenesis,42, 43, 44 others showed it functions as a tumor-suppressor by maintaining redox homeostasis.45,46 KEAP1-inactivating mutations and Nrf2-activating mutations are prevalent in a wide spectrum of cancers, suggesting that NRF2 functions as an oncoprotein. Increased p62 in hepatocytes promotes HCC,9 whereas increased p62 in hepatic stellate cells suppresses liver inflammation, fibrosis, and HCC.16 Therefore, the role of the p62-Keap1-Nrf2 axis in cancer is highly context dependent and may vary depending on different stages of tumor initiation and progression. NRF2 activation may inhibit chemically induced tumor initiation but accelerates tumor progression at tumor progression stages by increasing the antioxidant capacity for cancer cells against the harsh tumor microenvironment. Similarly, the role of TRIM21 in cancer is complex. While genetic deletion of TRIM21 or reduced TRIM21 expression has been found to associate with the development and poor prognosis in non-small cell lung cancer and diffuse large B cell lymphoma,47, 48, 49 overexpression of TRIM21 has been reported to promote tumor aggressiveness and therapy resistance in glioma, nasopharyngeal carcinoma, colon and pancreatic cancer.50, 51, 52 In HCC, while one study shows that TRIM21 is a favorable molecule whose downregulation promotes cancer,20 several Gene Expression Omnibus Series studies, the TCGA dataset, and our own analysis using clinical samples indicate an opposite effect of TRIM21 (ie, its elevated expression correlates with HCC incidence and poor outcome) (Figure 1). It is possible that TRIM21 plays different and even opposite roles in HCC owing to the multifaceted causes and risk factors of this highly heterologous disease. Moreover, it is important to note that while our current study shows that TRIM21 inhibition can decrease HCC development by protecting hepatocytes against oxidative DNA damage, TRIM21’s other functions such as preventing guanosine 5'-monophosphate synthases (GMPs) from stabilizing p53 hence promoting on cogenesis53 and its role in immune checkpoint regulation may also contribute to its oncogenic activity. Nonetheless, our study uncovers an important role of TRIM21 in HCC development and suggests that it may serve as a prognostic marker and therapeutic target in HCC as it functions as an important regulator of the redox homeostasis.
Materials and Methods
Mouse Experiments
TRIM21 KO mice with C57Bl/6J background were kind gift from Dr Keiko Ozato.54 To induce HCC, 14-day-old male mice were injected via intraperitoneal with DEN (5 mg/kg; Sigma-Aldrich, St. Louis, MO; N-0756), then fed with 0.05% PB in drinking water 7 days later. This protocol has been shown to induce HCC in 100% male mice with C57BL/6 background in 20–40 weeks.22,23 All mouse experiments were done in compliance with the Institutional Animal Care and Use Committee guidelines at Rutgers University. For acute DEN genotoxicity, 8-week-old male TRIM21 WT and KO mice were treated with 1 dose of 200 mg/kg DEN by intraperitoneal injection. Mice were weighted and liver tissues harvested 12, 24, or 48 hours posttreatment.
Data and Code Availability
The datasets generated are available in TCGA (https://portal.gdc.cancer.gov/) and Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds). The RNA-seq data of DEN-treated mice have been uploaded and are available at Gene Expression Omnibus (GSE164369).
Quantification and Statistical Analysis
Immunoblotting images were quantified by densitometry using ImageJ 2.1.0/1.53C (National Institutes of Health, Bethesda, MD), and protein expressions are expressed as relative band intensities. All statistical analyses were performed with more than 3 independent biological replicates for cell culture studies or with the indicated numbers of animals for mouse studies. Independent 2-sample and 1-sample t tests were used to make comparisons between 2 groups and to evaluate whether fold changes are different from 1, respectively. The analyses were mainly carried out using GraphPad Prism (GraphPad Software, San Diego, CA) or Microsoft Excel (Microsoft Corporation, Redmond, WA).
Plasmids
Human TRIM21 complementary DNA (cDNA) was amplified using primers 5′-CCCAAGCTTACCATG TACCCATAC GATGTTCCAGATTACGCG ATGGCTTCAG CAGCACGC-3′ and 5′-CG GAATTCTCAATAGTCAGTGGATC CTTG-3′ and were cloned into the retroviral LPC vector. TRIM21 LD and W381/383A mutants were generated following standard site-directed mutagenesis procedure. The primers used for TRIM21 mutagenesis were: for LD C16A, 5′-GGAGGTCACAGCCCCTATCTGCCTGGACCCCTTC G-3′ and 5′- GCAGATAGGGGCTGTGACCTCCTCCCACATC-3′; for W381/383A, 5′-CTGGACAATTG CGTTGGC GAACAAACAAAAATATGAGGCTG-3′ and 5′-TTTTGTTTGTTCGCCAACGC AATTGTCCA GAAGCCACTCTTG-3′.
Cell Death Assay
Cell viability was quantified by either PI exclusion or Trypan blue staining. For PI exclusion, cells were collected and resuspended in culture medium with PI (1 μg/mL). Cell viability was determined by flow cytometry (BD Biosciences, Franklin Lakes, NJ; FACSCanto II) and analyzed using FlowJo 7.6.1 software (FlowJo, Ashland, OR). For Trypan blue staining, 0.5 mL of cells (1×105 cells per mL) were mixed with 0.1 mL of 0.4% Trypan blue and incubated at room temperature for 3 minutes. Cells were counted under a phase-contrast light microscope.
ROS Detection
A total of 5 × 105 cells were plated into 6-cm petri dish. After overnight recovery, cells were untreated or treated with 20-mM DEN. six hours after treatment, cells were incubated with H2DCFDA (MCE HY-D0940, 5 μM final concentration) for 30 minutes in dark. Cells were resuspended in phosphate-buffered saline (PBS) containing 10% fetal bovine serum (FBS) and then analyzed by flow cytometry (FACSCanto II) using FlowJo 7.6.1 software.
Cell Culture, Transfection, and Retroviral Infection
HEK293T cells and MEFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 100 units/mL penicillin, and 100-μg/mL streptomycin (Invitrogen, Carlsbad, CA). Lipofectamine 2000 (Invitrogen) was used for transfection. TRIM21–/– MEFs were infected with the retroviral linear predictive coding vector to generate reconstruction with HA-TRIM21 WT, HA-TRIM21 LD, and HA-TRIM21 W381/383A, respectively. One day after infection, the virus-containing medium was replaced with fresh medium. Repeat viral infection every 12 hours for 3 times. Cells were cultured for at least 5 days to reach sufficient protein expression level that was verified by immunoblotting with HA-Tag or TRIM21 antibodies.
SMMC-7721 and MHHC-97L cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 100 units/mL penicillin, and 100-μg/mL streptomycin (Invitrogen). Lipofectamine 2000 (Invitrogen) was used for transfection. Cells were infected with the retroviral nontargeted control, shTRIM21#1, and shTRIM21#2, respectively. One day after infection, the virus-containing medium was replaced with fresh medium. Viral infection was repeated every 12 hours for 3 times. Cells were cultured for at least 5 days before verifying TRIM21 silencing by immunoblotting.
Antibodies
The following antibodies were used: AKT (Cell Signaling, Danvers, MA; #4695; 1:1000 for western blotting [WB]), Phospho-AKT308 (Cell Signaling; #2965; 1:1000 for WB), Phospho-AKT473 (Cell Signaling; #4508; 1:1000 for WB), Cytochrome p450 2E1 (Abcam, Cambridge, United Kingdom; ab28146; 1:2000 for WB), Cytochrome p450 2B10 (Santa Cruz, Dallas, TX; sc-53242; 1:2000 for WB), Cytochrome p450 2D1 (1:2000 for WB),55 Cytochrome p450 3A (1:2000 for WB),56 NQO1 (Abcam; ab28947; 1:1000 for WB), Nrf2 (Abcam; ab62352; 1:1000 for WB), GADPH (Cell Signaling; #97166; 1:5000 for WB), β-actin (Cell Signaling; #4970; 1:1000 for WB), β-tubulin (Cell Signaling; #86298; 1:1000 for WB), Lamin B1 (Cell Signaling; #12586, 1:1000 for WB), p62 (Novus Biologicals, Littleton, CO; C11, H00008878-MOL; 1:1000 for WB and IF), Keap1 (Proteintech, Rosemont, IL; 10503-2-AP; 1:1000 for WB and IF), Phospho-histone H2A.X (Ser139) (Cell Signaling; #9718; 1:1000 for WB and 1:100 for IHC), Ki-67 (Cell Signaling; #12202; 1:100 for IHC), PCNA (Cell Signaling; #13110; 1:100 for IHC), 8-oxo-dG (R&D Systems, Minneapolis, MN; 4354-MC-050; 1:100 for IHC), cleaved caspase-3 (Cell Signaling; #9661; 1:300 for IHC), HA-Tag (Cell Signaling; #3742; 1:1000 for WB), TRIM21 (Abcam; ab207728; 1:1000 for WB; Proteintech; 12108-1-AP; 1:1000 for WB, 1:100 for IHC), F-4/80 (Cell Signaling; #70076; 1:200 for IHC), HSP70 (Abcam; ab2787; 1:200 for IHC).
Triton X-100 Soluble/Insoluble Fractionation and Nuclear/Cytosolic Fractionation
For Triton X-100 soluble/insoluble fractionation, liver tissues or cells (1 × 106) were lysed in RIPA buffer containing 1% Triton X-100. Cell and tissue lysates were centrifuged at 400 g for 10 minutes at 4°C. After centrifugation, pellets were separated from supernatants, resuspended in a lysis buffer containing 1% sodium dodecyl sulfate (SDS), softly sonicated 3 times (6 seconds each time) on ice, and boiled in SDS sample buffer (insoluble fraction). Supernatants were also boiled in SDS sample buffer (soluble fraction). Both the Triton X-100 soluble and insoluble fractions were subjected to WB. A total of 30-μg soluble proteins and corresponding volume of insoluble proteins were used for WB. Nuclear/cytosolic fractionation was performed using the Nuclear Extraction Kit (Abcam; ab113474)
Immunofluorescence
Cells were fixed in 4% paraformaldehyde in PBS for 15 minutes at room temperature. After 3 washes with PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 15 minutes. After a brief wash with PBS with 0.02% Tween 20 (PBST) and 1.5% FBS, cells were incubated with primary antibodies in PBST with 1.5% FBS for 1 hours at room temperature. Cells were washed 3 times with PBST, followed by incubation with fluorophore-conjugated secondary antibodies in PBST with 1.5% FBS for 1 hour in dark at room temperature. Cells were then washed 3 times with PBST. After 2 washes with dH2O, the cells were mounted with ProLong Gold antifade reagent with DAPI (Invitrogen; 2047396). Slides were observed and imaged using inverted fluorescence microscope (Olympus, Tokyo, Japan; IX71) and analyzed using Image-Pro Plus software (version 5.1.0.20; Media Cybernetics, Rockville, MD).
RNA Extraction, Complementary DNA Synthesis, and PCR
After treatment, total RNA of the cells was isolated with RNeasy kit (Qiagen, Hilden, Germany). Reverse transcription was carried out with 2 μg of total RNA using the Superscript III First Strand Synthesis system (Invitrogen). The synthesized cDNA was used for real-time quantitative PCR with the PerfeCTa SYBR Green Super mix (Quanta Bioscience, Beverly, MA; 95055) on the StepOnePlus (Applied Biosystems, Foster City, CA). The following primers were used for quantitative reverse-transcription PCR: PCNA: forward 5′-TTTGAGGCACGCCTGATCC-3′; reverse 5′-GGAGACGTGAGACGAGTCCAT-3′:
c-Jun: forward 5′-CCTTCTACGACGATGCCCTC-3′; reverse 5′-GGTTCAAGGTCATGCTCTGTTT-3′; TRIM21 forward 5′-GGATCATCCTGGAAAGGAGTGG-3′ and reverse 5′-GTGCTTGTTAGGTCTGGGGC-3′ ; GSTM1: forward 5′-TGTTTGAGCCCAAGTGCCTGGA-3′ and reverse 5′-TAGGTGTTGCGATGTAGCGGCT-3′; GSTM2: forward 5′-AGAGCAATGCCATCCTGCGCTA-3′ and reverse 5′-GTGTCCATAGCCTGGTTCTCCA-3′; FOS: forward 5′- GGGAATGGTGAAGACCGTGTCA-3′ and reverse 5′-GCAGCCATCTTATTCCGTTCCC-3′; JUNB: forward 5′-GACCTGCACAAGATGAACCACG-3′ and reverse 5′ ACTGCTGAGGTTGGTGTAGACG-3′.
RNA-Seq and Data Analysis
RNA-seq was performed by the University of Texas Health San Antonio Genome Sequencing Facility. Approximately 500-ng total RNA was used for RNA-seq library preparation following the Illumina TruSeq stranded messenger RNA (mRNA) sample preparation guide (Illumina, San Diego, CA). The first step in the workflow involves purifying the poly-A–containing mRNA molecules using poly-T oligo–attached magnetic beads. Following purification, mRNA is fragmented into small pieces using divalent cautions under elevated temperature. The cleaved RNA fragments are copied into first strand cDNA using reverse transcriptase and random primers. This is followed by second strand cDNA synthesis using DNA polymerase I and RNase H. Strand specificity is achieved by replacing dTTP with dUTP in the Second Strand Marking Mix. These cDNA fragments then go through an end repair process, the addition of a single “A” base, and then ligation of the adapters. The products are then purified and enriched with PCR to create the final RNA-seq library. After RNA-seq libraries were subjected to quantification process, pooled for cBot amplification and subsequent 50-bp single-read sequencing run with Illumina HiSeq 3000 platform. After the sequencing run, demultiplexing with Bcl2fastq2 was employed to generate the fastq file for each sample. An average of 37 million reads was obtained for this set of samples.
The raw reads were aligned to the reference Mus musculus genome (UCSC mm9) with TopHat2.57 Genes were annotated and quantified by HTSeq,58 and DEseq was used to find differentially expressed genes.59 Significant differentially expressed genes were identified with Benjamini and Hochberg–adjusted P value <.05 and fold change ≥4 when the gene has read abundance with reads per kilobase per million reads ≥1 within at least 1 treatment (samples). Gene Ontology analysis was performed using the DAVID online tool (http://david.abcc.ncifcrf.gov/).60 Plots were generated and the related statistics were performed using R version 3.4.1 (R Foundation for Statistical Computing, Vienna, Austria).
Histology, IHC, and Immunohistofluorescence
Liver tissues were fixed and processed for hematoxylin and eosin and IHC staining. In brief, paraffin-embedded liver sections were deparaffinized, rehydrated, and microwave heated for 10 minutes in 0.01-mol/L citrate buffer (pH 6.0) for antigen retrieval. A total of 3% hydrogen peroxide was applied to block endogenous peroxidase activity. After 30 minutes of blocking with 5% goat serum, primary antibodies (TRIM21, γH2A.X, p62, Keap1, PCNA, cleaved caspase 3, 8-oxo-dG, F-4/80, or control IgG) were applied and incubated overnight at 4°C. After wash, slides were incubated with biotinylated secondary antibody and the streptavidin-biotin complex (Vector Laboratories, Burlingame, CA) were applied, each for 60 minutes at room temperature with an interval washing. After rinsing with PBS, slides were immersed for 5 minutes in the coloring substrate DAB (Vector Labs, Burlingame, CA; cat SK-4100) 0.4 mg/mL with 0.003% hydrogen peroxide, then rinsed with distilled water, counterstained with hematoxylin, dehydrated, and coverslipped.
For immunohistofluorescence analysis, liver optimum cutting temperature compound (OCT) cryosections were fixed in cold acetone for 15 minutes and immunostained with anti-α-SMA (1:100; Abcam; cat ab124964) overnight at 4°C. After wash with PBS for 3 times, cryosections were incubated with goat anti-rabbit IgG-Alexa488 (1:500; Invitrogen, Grand Island, NY) and donkey anti-guinea pig IgG-Alexa651 (1:500; Millipore, Temecula, CA) for 1 hour at room temperature. Images were visualized and captured using the All-in-One BZ-X700 fluorescence microscope (Keyence, Elmwood Park, NJ).
For Sirius Red staining, paraffin-embedded liver sections were deparaffinized, rehydrated, and stained with hematoxylin for 10 minutes, followed by staining in Picrosirius red (Direct Red 80 [Sigma Aldrich; 365548], Picric acid [Sigma-Aldrich; 197378]) for 1 hour. After washing in 2 changes of 0.5% acidified water, slides were dehydrated and coverslipped.
For Oil Red O staining, liver frozen sections were fixed in cold propylene glycol for 2 minutes, then incubated slide in Oil Red O solution (Oil Red O stain kit; ab150678) for 8 minutes. Slides were differentiated in 85% propylene glycol for 1 minutes, rinsed in 2 changes of distilled water, incubated in hematoxylin for 2 minutes, and softly rinsed in tap water and 2 changes of distilled water. A coverslip was mounted using an aqueous mounting medium. Images were visualized and captured using the Virtual/Digital Slice microscope (VS120; Olympus).
Access to Data
All authors had access to all data and have reviewed and approved the final manuscript.
Acknowledgments
The authors thank Dr Keiko Ozato (National Institutes of Health) for the TRIM21 KO mouse strain. The authors thank Drs Chung S. Yang and Tony Ah-Ng Kong for insightful discussions.
CRediT Authorship Contributions
Fang Wang (Data curation: Equal; Methodology: Equal; Validation: Equal; Writing – original draft: Supporting)
Ye Zhang (Data curation: Equal; Formal analysis: Equal; Methodology: Equal; Validation: Equal; Writing – original draft: Equal)
Jianliang Shen (Data curation: Supporting; Methodology: Supporting; Validation: Supporting)
Bin Yang (Data curation: Supporting)
Weiwei Dai (Data curation: Supporting)
Junrong Yan (Formal analysis: Supporting)
Sara Maimouni (Data curation: Supporting)
Heineken Q. Daguplo (Data curation: Supporting)
Sara Coppola (Data curation: Supporting)
Yingtang Gao (Data curation: Supporting)
Yijun Wang (Project administration: Supporting)
Zhi Du (Project administration: Supporting)
Kesong Peng (Data curation: Supporting; Formal analysis: Supporting)
Hui Liu (Data curation: Supporting)
Qin Zhang (Data curation: Supporting)
Fei Tang (Data curation: Supporting)
Peng Wang (Data curation: Supporting)
Shenglan Gao (Data curation: Supporting)
Yongbo Wang (Formal analysis: Supporting)
Wen-Xing Ding (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting)
Grace Guo (Formal analysis: Supporting; Methodology: Supporting)
Fengmei Wang (Project administration: Supporting)
Wei-Xing Zong (Conceptualization: Lead; Funding acquisition: Lead; Writing – original draft: Lead)
Footnotes
Conflicts of Interest The authors disclose no conflicts.
Funding The study was supported by National Institutes of Health grants R01CA224550 (to Wei-Xing Zong) and R21ES029258 (to Grace Guo). Shenglan Gao was supported by the Shanghai Pujiang Program 19PJ1401900 and the National Natural Science Foundation 31970714. The University of Texas Health San Antonio Genome Sequencing Facility is supported by National Institutes of Health-National Cancer Institute P30 CA054174 (University of Texas Health San Antonio Cancer Center), National Institutes of Health Shared Instrument grant 1S10OD021805-01 (S10 grant), and Cancer Prevention and Research Institute of Texas Core Facility Award (RP160732).
Supplementary Material
References
- 1.Choi J. Oxidative stress, endogenous antioxidants, alcohol, and hepatitis C: pathogenic interactions and therapeutic considerations. Free Radic Biol Med. 2012;52:1135–1150. doi: 10.1016/j.freeradbiomed.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Gambino R., Musso G., Cassader M. Redox balance in the pathogenesis of nonalcoholic fatty liver disease: mechanisms and therapeutic opportunities. Antioxid Redox Signal. 2011;15:1325–1365. doi: 10.1089/ars.2009.3058. [DOI] [PubMed] [Google Scholar]
- 3.Ivanov A.V., Valuev-Elliston V.T., Tyurina D.A., Ivanova O.N., Kochetkov S.N., Bartosch B., Isaguliants M.G. Oxidative stress, a trigger of hepatitis C and B virus-induced liver carcinogenesis. Oncotarget. 2017;8:3895–3932. doi: 10.18632/oncotarget.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dodson M., de la Vega M.R., Cholanians A.B., Schmidlin C.J., Chapman E., Zhang D.D. Modulating NRF2 in disease: timing is everything. Annu Rev Pharmacol Toxicol. 2019;59:555–575. doi: 10.1146/annurev-pharmtox-010818-021856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 6.Lau A., Wang X.J., Zhao F., Villeneuve N.F., Wu T., Jiang T., Sun Z., White E., Zhang D.D. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao L., Chandra P.K., Moroz K., Zhang X., Thung S.N., Wu T., Dash S. Impaired autophagy response in human hepatocellular carcinoma. Exp Mol Pathol. 2014;96:149–154. doi: 10.1016/j.yexmp.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inami Y., Waguri S., Sakamoto A., Kouno T., Nakada K., Hino O., Watanabe S., Ando J., Iwadate M., Yamamoto M., Lee M.S., Tanaka K., Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umemura A., He F., Taniguchi K., Nakagawa H., Yamachika S., Font-Burgada J., Zhong Z., Subramaniam S., Raghunandan S., Duran A., Linares J.F., Reina-Campos M., Umemura S., Valasek M.A., Seki E., Yamaguchi K., Koike K., Itoh Y., Diaz-Meco M.T., Moscat J., Karin M. p62, Upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell. 2016;29:935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin G.Z., Dong H., Yu W.L., Li Y., Lu X.Y., Yu H., Xian Z.H., Dong W., Liu Y.K., Cong W.M., Wu M.C. A novel panel of biomarkers in distinction of small well-differentiated HCC from dysplastic nodules and outcome values. BMC Cancer. 2013;13:161. doi: 10.1186/1471-2407-13-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zatloukal K., Stumptner C., Fuchsbichler A., Heid H., Schnoelzer M., Kenner L., Kleinert R., Prinz M., Aguzzi A., Denk H. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito T., Ichimura Y., Taguchi K., Suzuki T., Mizushima T., Takagi K., Hirose Y., Nagahashi M., Iso T., Fukutomi T., Ohishi M., Endo K., Uemura T., Nishito Y., Okuda S., Obata M., Kouno T., Imamura R., Tada Y., Obata R., Yasuda D., Takahashi K., Fujimura T., Pi J., Lee M.S., Ueno T., Ohe T., Mashino T., Wakai T., Kojima H., Okabe T., Nagano T., Motohashi H., Waguri S., Soga T., Yamamoto M., Tanaka K., Komatsu M. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun. 2016;7:12030. doi: 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniguchi K., Yamachika S., He F., Karin M. p62/SQSTM1-Dr. Jekyll and Mr. Hyde that prevents oxidative stress but promotes liver cancer. FEBS Lett. 2016;590:2375–2397. doi: 10.1002/1873-3468.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 16.Duran A., Hernandez E.D., Reina-Campos M., Castilla E.A., Subramaniam S., Raghunandan S., Roberts L.R., Kisseleva T., Karin M., Diaz-Meco M.T., Moscat J. p62/SQSTM1 by binding to vitamin D receptor inhibits hepatic stellate cell activity, fibrosis, and liver cancer. Cancer Cell. 2016;30:595–609. doi: 10.1016/j.ccell.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ngo H.K.C., Kim D.H., Cha Y.N., Na H.K., Surh Y.J. Nrf2 mutagenic activation drives hepatocarcinogenesis. Cancer Res. 2017;77:4797–4808. doi: 10.1158/0008-5472.CAN-16-3538. [DOI] [PubMed] [Google Scholar]
- 18.Pan J.A., Sun Y., Jiang Y.P., Bott A.J., Jaber N., Dou Z., Yang B., Chen J.S., Catanzaro J.M., Du C., Ding W.X., Diaz-Meco M.T., Moscat J., Ozato K., Lin R.Z., Zong W.X. TRIM21 Ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol Cell. 2016;61:720–733. doi: 10.1016/j.molcel.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao L., Xiong D.D., He R.Q., Yang X., Lai Z.F., Liu L.M., Huang Z.G., Wu H.Y., Yang L.H., Ma J., Li S.H., Lin P., Yang H., Luo D.Z., Dang Y.W., Chen G. MIR22HG as a tumor suppressive lncRNA in HCC: a comprehensive analysis integrating RT-qPCR, mRNA-seq, and microarrays. Onco Targets Ther. 2019;12:9827–9848. doi: 10.2147/OTT.S227541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Q., He D., He K., Zhang Q., Tang M., Dai J., Lv H., Wang X., Xiang G., Yu H. Downregulation of TRIM21 contributes to hepatocellular carcinoma carcinogenesis and indicates poor prognosis of cancers. Tumour Biol. 2015 doi: 10.1007/s13277-015-3572-2. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa H. Recent advances in mouse models of obesity- and nonalcoholic steatohepatitis-associated hepatocarcinogenesis. World J Hepatol. 2015;7:2110–2118. doi: 10.4254/wjh.v7.i17.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heindryckx F., Colle I., Van Vlierberghe H. Experimental mouse models for hepatocellular carcinoma research. Int J Exp Pathol. 2009;90:367–386. doi: 10.1111/j.1365-2613.2009.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakiri L., Wagner E.F. Mouse models for liver cancer. Mol Oncol. 2013;7:206–223. doi: 10.1016/j.molonc.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verna L., Whysner J., Williams G.M. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol Ther. 1996;71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 25.Aleksic K., Lackner C., Geigl J.B., Schwarz M., Auer M., Ulz P., Fischer M., Trajanoski Z., Otte M., Speicher M.R. Evolution of genomic instability in diethylnitrosamine-induced hepatocarcinogenesis in mice. Hepatology. 2011;53:895–904. doi: 10.1002/hep.24133. [DOI] [PubMed] [Google Scholar]
- 26.Naugler W.E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A.M., Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 27.Connor F., Rayner T.F., Aitken S.J., Feig C., Lukk M., Santoyo-Lopez J., Odom D.T. Mutational landscape of a chemically-induced mouse model of liver cancer. J Hepatol. 2018;69:840–850. doi: 10.1016/j.jhep.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa H., Hayata Y., Kawamura S., Yamada T., Fujiwara N., Koike K. Lipid metabolic reprogramming in hepatocellular carcinoma. Cancers (Basel) 2018;10 doi: 10.3390/cancers10110447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong Q., Shi G., Zhang Q., Zhang Y., Levy D., Zhong S. Role of phosphorylated histone H3 serine 10 in DEN-induced deregulation of Pol III genes and cell proliferation and transformation. Carcinogenesis. 2013;34:2460–2469. doi: 10.1093/carcin/bgt219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wree A., Johnson C.D., Font-Burgada J., Eguchi A., Povero D., Karin M., Feldstein A.E. Hepatocyte-specific Bid depletion reduces tumor development by suppressing inflammation-related compensatory proliferation. Cell Death Differ. 2015;22:1985–1994. doi: 10.1038/cdd.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson M.E., Lahiri S., Chow J.D., Byrne F.L., Hargett S.R., Breen D.S., Olzomer E.M., Wu L.E., Cooney G.J., Turner N., James D.E., Slack-Davis J.K., Lackner C., Caldwell S.H., Hoehn K.L. Inhibition of hepatic lipogenesis enhances liver tumorigenesis by increasing antioxidant defence and promoting cell survival. Nat Commun. 2017;8:14689. doi: 10.1038/ncomms14689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentric G., Maillet V., Paradis V., Couton D., L'Hermitte A., Panasyuk G., Fromenty B., Celton-Morizur S., Desdouets C. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest. 2015;125:981–992. doi: 10.1172/JCI73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakurai T., He G., Matsuzawa A., Yu G.Y., Maeda S., Hardiman G., Karin M. Hepatocyte necrosis induced by oxidative stress and IL–1 alpha release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell. 2008;14:156–165. doi: 10.1016/j.ccr.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haque E., Karim M.R., Salam Teeli A., Smiech M., Leszczynski P., Winiarczyk D., Parvanov E.D., Atanasov A.G., Taniguchi H. Molecular mechanisms underlying hepatocellular carcinoma induction by aberrant NRF2 activation-mediated transcription networks: interaction of NRF2-KEAP1 controls the fate of hepatocarcinogenesis. Int J Mol Sci. 2020:21. doi: 10.3390/ijms21155378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bubb K.J., Kok C., Tang O., Rasko N.B., Birgisdottir A.B., Hansen T., Ritchie R., Bhindi R., Reisman S.A., Meyer C., Ward K., Karimi Galougahi K., Figtree G.A. The NRF2 activator DH404 attenuates adverse ventricular remodeling post-myocardial infarction by modifying redox signalling. Free Radic Biol Med. 2017;108:585–594. doi: 10.1016/j.freeradbiomed.2017.04.027. [DOI] [PubMed] [Google Scholar]
- 36.Enomoto A., Itoh K., Nagayoshi E., Haruta J., Kimura T., O'Connor T., Harada T., Yamamoto M. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 37.Ni H.M., Boggess N., McGill M.R., Lebofsky M., Borude P., Apte U., Jaeschke H., Ding W.X. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127:438–450. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bae S.H., Sung S.H., Oh S.Y., Lim J.M., Lee S.K., Park Y.N., Lee H.E., Kang D., Rhee S.G. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013;17:73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Wang C., Niu Q., Ma R., Song G., Hu Y., Xu S., Li Y., Wang H., Li S., Ding Y. The variable regulatory effect of arsenic on Nrf2 signaling pathway in mouse: a systematic review and meta-analysis. Biol Trace Elem Res. 2019;190:362–383. doi: 10.1007/s12011-018-1549-x. [DOI] [PubMed] [Google Scholar]
- 40.Taguchi K., Kensler T.W. Nrf2 in liver toxicology. Arch Pharm Res. 2020;43:337–349. doi: 10.1007/s12272-019-01192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartolini D., Dallaglio K., Torquato P., Piroddi M., Galli F. Nrf2-p62 autophagy pathway and its response to oxidative stress in hepatocellular carcinoma. Transl Res. 2018;193:54–71. doi: 10.1016/j.trsl.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Mathew R., Karp C.M., Beaudoin B., Vuong N., Chen G., Chen H.Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R.S., Karantza-Wadsworth V., White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., Scrimieri F., Winter J.M., Hruban R.H., Iacobuzio-Donahue C., Kern S.E., Blair I.A., Tuveson D.A. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni H.M., Woolbright B.L., Williams J., Copple B., Cui W., Luyendyk J.P., Jaeschke H., Ding W.X. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu C., Huang M.T., Shen G., Yuan X., Lin W., Khor T.O., Conney A.H., Kong A.N. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 46.Khor T.O., Huang M.T., Kwon K.H., Chan J.Y., Reddy B.S., Kong A.N. Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res. 2006;66:11580–11584. doi: 10.1158/0008-5472.CAN-06-3562. [DOI] [PubMed] [Google Scholar]
- 47.Zhao B., Bepler G. Transcript map and complete genomic sequence for the 310 kb region of minimal allele loss on chromosome segment 11p15.5 in non-small-cell lung cancer. Oncogene. 2001;20:8154–8164. doi: 10.1038/sj.onc.1205027. [DOI] [PubMed] [Google Scholar]
- 48.Brauner S., Zhou W., Backlin C., Green T.M., Folkersen L., Ivanchenko M., Lofstrom B., Xu-Monette Z.Y., Young K.H., Moller Pedersen L., Boe Moller M., Sundstrom C., Enblad G., Baecklund E., Wahren-Herlenius M. Reduced expression of TRIM21/Ro52 predicts poor prognosis in diffuse large B-cell lymphoma patients with and without rheumatic disease. J Intern Med. 2015 doi: 10.1111/joim.12375. [DOI] [PubMed] [Google Scholar]
- 49.Jin Y., Zhao X., Zhang Q., Zhang Y., Fu X., Hu X., Wan Y. Cancer-associated mutation abolishes the impact of TRIM21 on the invasion of breast cancer cells. Int J Biol Macromol. 2020;142:782–789. doi: 10.1016/j.ijbiomac.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Z., Wang Y., Yun D., Huang Q., Meng D., Li Q., Zhang P., Wang C., Chen H., Lu D. TRIM21 overexpression promotes tumor progression by regulating cell proliferation, cell migration and cell senescence in human glioma. Am J Cancer Res. 2020;10:114–130. [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang P., Li X., He Q., Zhang L., Song K., Yang X., He Q., Wang Y., Hong X., Ma J., Liu N. TRIM21-SERPINB5 aids GMPS repression to protect nasopharyngeal carcinoma cells from radiation-induced apoptosis. J Biomed Sci. 2020;27:30. doi: 10.1186/s12929-020-0625-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen J.Q., Irby R.B. TRIM21 is a novel regulator of Par-4 in colon and pancreatic cancer cells. Cancer Biol Ther. 2017;18:16–25. doi: 10.1080/15384047.2016.1252880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddy B.A., van der Knaap J.A., Bot A.G., Mohd-Sarip A., Dekkers D.H., Timmermans M.A., Martens J.W., Demmers J.A., Verrijzer C.P. Nucleotide biosynthetic enzyme GMP synthase is a TRIM21-controlled relay of p53 stabilization. Mol Cell. 2014;53:458–470. doi: 10.1016/j.molcel.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 54.Yoshimi R., Chang T.H., Wang H., Atsumi T., Morse H.C., 3rd, Ozato K. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J Immunol. 2009;182:7527–7538. doi: 10.4049/jimmunol.0804121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu J., Zhang Q.Y., Genter M.B., Lipinskas T.W., Negishi M., Nebert D.W., Ding X. Purification and characterization of heterologously expressed mouse CYP2A5 and CYP2G1: role in metabolic activation of acetaminophen and 2,6-dichlorobenzonitrile in mouse olfactory mucosal microsomes. J Pharmacol Exp Ther. 1998;285:1287–1295. [PubMed] [Google Scholar]
- 56.Ma X., Shah Y., Cheung C., Guo G.L., Feigenbaum L., Krausz K.W., Idle J.R., Gonzalez F.J. The PREgnane X receptor gene-humanized mouse: a model for investigating drug-drug interactions mediated by cytochromes P450 3A. Drug Metab Dispos. 2007;35:194–200. doi: 10.1124/dmd.106.012831. [DOI] [PubMed] [Google Scholar]
- 57.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated are available in TCGA (https://portal.gdc.cancer.gov/) and Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds). The RNA-seq data of DEN-treated mice have been uploaded and are available at Gene Expression Omnibus (GSE164369).