Abstract
Background & Aims
A coordinated stress and regenerative response is important after hepatocyte damage. Here, we investigate the phenotypes that result from genetic abrogation of individual components of the checkpoint kinase 2/transformation-related protein 53 (p53)/cyclin-dependent kinase inhibitor 1A (p21) pathway in a murine model of metabolic liver injury.
Methods
Nitisinone was reduced or withdrawn in Fah-/- mice lacking Chk2, p53, or p21, and survival, tumor development, liver injury, and regeneration were analyzed. Partial hepatectomies were performed and mice were challenged with the Fas antibody Jo2.
Results
In a model of metabolic liver injury, loss of p53, but not Chk2, impairs the oxidative stress response and aggravates liver damage, indicative of a direct p53-dependent protective effect on hepatocytes. Cell-cycle control during chronic liver injury critically depends on the presence of both p53 and its downstream effector p21. In p53-deficient hepatocytes, unchecked proliferation occurs despite a strong induction of p21, showing a complex interdependency between p21 and p53. The increased regenerative potential in the absence of p53 cannot fully compensate the surplus injury and is not sufficient to promote survival. Despite the distinct phenotypes associated with the loss of individual components of the DNA damage response, gene expression patterns are dominated by the severity of liver injury, but reflect distinct effects of p53 on proliferation and the anti-oxidative stress response.
Conclusions
Characteristic phenotypes result from the genetic abrogation of individual components of the DNA damage-response cascade in a liver injury model. The extent to which loss of gene function can be compensated, or affects injury and proliferation, is related to the level at which the cascade is interrupted. Accession numbers of repository for expression microarray data: GSE156983, GSE156263, GSE156852, and GSE156252.
Keywords: DNA Damage, Oxidative Stress Response, CHK2, HCC
Abbreviations used in this paper: aCGH, array comparative genomic hybridization; ATM, ataxia telangiectasia mutated; BCL2, B-cell lymphoma 2; CDK, cyclin-dependent kinase; CHK, checkpoint kinase; FAA, fumarylacetoacetate; FAH, fumarylacetoacetate hydrolase; GSEA, gene set enrichment analysis; GSH, glutathione; GSSG, glutathione disulfide; IP, immunoprecipitation; NRF2, nuclear factor erythroid 2-related factor 2; NTBC, nitisinone; p16, cyclin-dependent kinase inhibitor 2A; p21, cyclin-dependent kinase inhibitor 1A; p53, transformation-related protein 53; RB, retinoblastoma; TUNEL, terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling
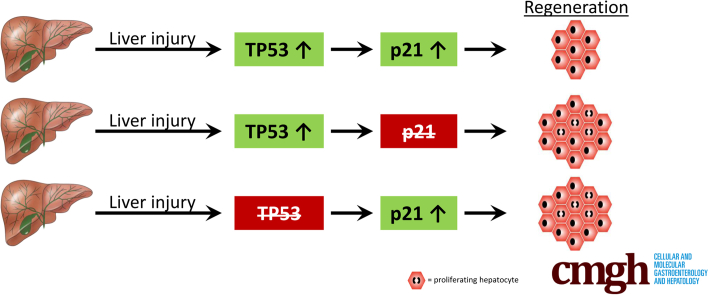
Graphical abstract
Summary.
Functional annotation of individual components of the DNA damage response pathway in the liver shows distinct genotype–phenotype correlations and unveils their complex interplay in response to liver injury.
Exposure to extrinsic factors that directly or indirectly induce DNA damage activates a cascade of response mechanisms in the injured liver. Induction of survival pathways that loosen the breaks on proliferative control in hibernating hepatocytes is critical to enable efficient regeneration and facilitate survival.1, 2, 3, 4 At the same time, maintaining coordinated guidance through the cell cycle is necessary to ensure that survival does not happen at the expense of malignant transformation, which is especially challenging in the face of chronic liver injury. Activation of transformation-related protein 53 (p53) serves as the central mediator of cell-cycle control and as a major barrier against malignant transformation.5
Inactivation of DNA damage response checkpoints can influence tumor development. For instance, patients with mutations in ataxia telangiectasia mutated (ATM) are prone to carcinogenesis. Paradoxically, loss of ATM delays carcinogen-induced hepatocarcinogenesis in mice, which has been attributed to a strong compensatory activation of ataxia telangiectasia and Rad3 related, checkpoint kinase (CHK)1, and p53.6 After double-strand breaks, ATM phosphorylates and activates CHK2, a serine/threonine protein kinase with a variety of substrates, including p53 and cyclin-dependent kinase inhibitor 1A (p21).7 CHK2 mutations have been identified in human cancers and loss of CHK2 predisposes transgenic mice to tumorigenesis, although they are not prone to spontaneous cancer development.8 The role of CHK2 in chronic liver injury and hepatocarcinogenesis is currently unknown.
After activation, p53 orchestrates various biological processes such as cell-cycle arrest, apoptosis, and DNA repair. Extensive damage may trigger apoptosis to eradicate the affected cells or lead to senescence via p21 and cyclin-dependent kinase inhibitor 2A (p16). After milder damage, p53 can transcriptionally activate cell-cycle regulators and facilitate DNA repair and survival. In the murine liver, loss of p53 alone leads to formation of hepatocellular carcinomas with a long latency of 14–20 months, but dramatically accelerates carcinogen- and oncogene-induced tumor development.2,9,10 Although the capacity of p53 to suppress liver tumor development is well established, increasing evidence supports the context-dependent role of p53 in acute liver diseases. On the one hand, there is evidence—albeit controversial—that p53 protects hepatocytes from acetaminophen-induced liver injury by regulating drug-metabolizing enzymes and initiating liver regeneration.11,12 On the other hand, activation of p53 has been implicated in the progression of alcoholic and nonalcoholic liver injury by increasing oxidative stress and apoptosis.13,14
p21 is one of the main effectors of p53, with considerable tumor–suppressive potential that mediates cell-cycle arrest and senescence in response to DNA damage.15 However, data from preclinical model systems challenged the view of p21 as an exclusive tumor suppressor.16 In this regard, we previously showed that the degree of liver injury and the strength of p21 activation determine its effects on hepatocyte proliferation and hepatocarcinogenesis.17 Deletion of p21 led to continuous hepatocyte proliferation in mice with severe injury, allowing animals to survive, but also resulted in rapid tumor development. In contrast, liver proliferation was reduced and hepatocarcinogenesis was delayed in p21-deficient mice with moderate injury.
The aim of this study was to delineate the role of p53 in relation to its downstream target p21 and its upstream regulator CHK2 during chronic liver disease. We used a metabolic mouse model of hereditary tyrosinemia type 1, which is an autosomal-recessive human disease caused by the genetic inactivation of the fumarylacetoacetate hydrolase (FAH). FAH is expressed mainly in the liver and in the kidneys, where it catalyzes the last step in the tyrosine catabolism pathway. Accumulation of toxic metabolites, such as fumarylacetoacetate (FAA), causes liver injury and severely increases susceptibility to liver cancer development. The drug nitisinone (also known as NTBC) blocks the pathway upstream of the formation of FAA and is used to treat patients with hereditary tyrosinemia type 1. The murine model of Fah deficiency recapitulates the phenotypic manifestations of human liver disease.18, 19, 20
Results
Loss of p53 Does Not Rescue Survival Despite Unchecked Proliferation in Mice With Severe Liver Injury
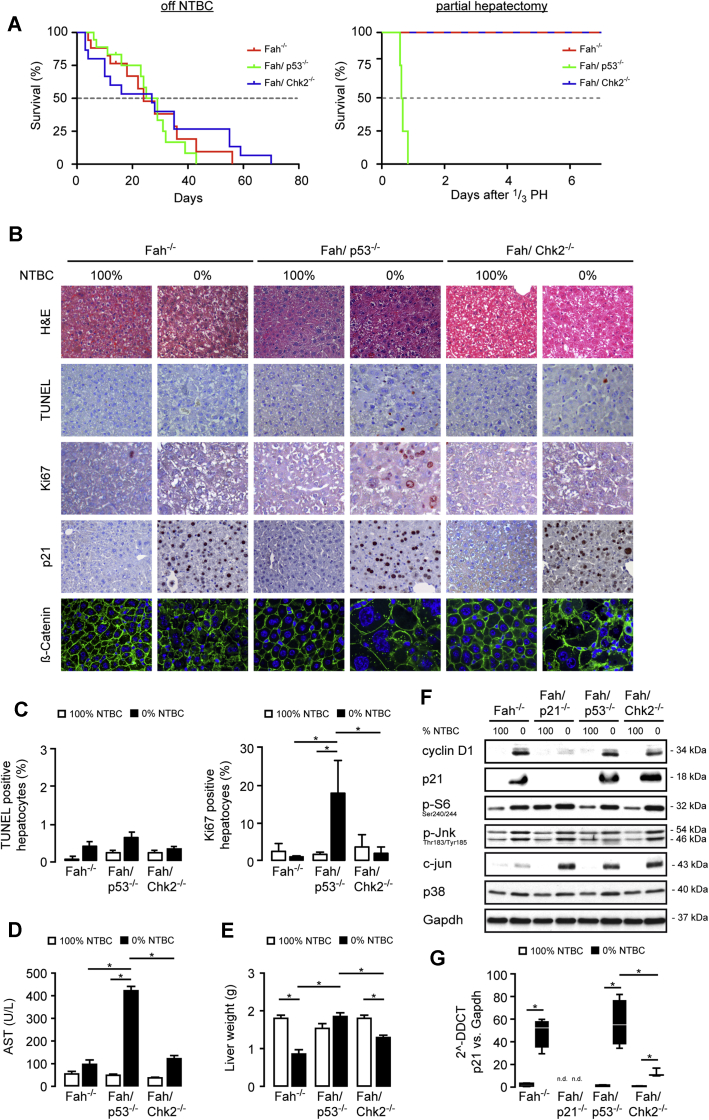
To study the role of p53 and CHK2 in acute and chronic liver injury, we crossed Fah-/- mice with the Chk2-/- strain (denoted Fah/Chk2-/-) or with animals with liver-specific loss of p53 (AlfpCre; p53fl/fl, denoted as Fah/p53-/-). Both double-knockout strains, as well as Fah-/- controls, did not show any overt morphologic or biochemical phenotype within the observational time frame of 15 months when supplemented with 100% NTBC. We previously showed that after NTBC withdrawal, Fah-/- mice die of acute liver failure within 2–4 months, whereas Fah/p21-/- mice survive for more than 6 months.3,17 To assess the consequences of loss of p53 or CHK2 on acute liver injury in Fah-/- mice, we withdrew NTBC from 8-week-old Fah-/-, Fah/p53-/-, and Fah/Chk2-/- mice. All mice eventually died of liver failure, with a median overall survival independent of the presence of p53 or CHK2 (Fah-/-: median overall survival, 24 d; n = 17; Fah/p53-/-: median overall survival, 27 d; n = 17; Fah/Chk2-/- mice: median overall survival, 26.5 d; n = 26) (Figure 1A, left). Characteristic of hepatic injury, liver histology on day 14 after NTBC withdrawal showed multiple small foci of necroinflammation and dysplastic hepatocytes (H&E and β-catenin staining) (Figure 1B), and a few scattered terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL)-positive hepatocytes in Fah-/- as well as in double-knockout mice (Figure 1B and C). Although loss of p53 did not affect overall survival, increased transaminase levels indicated that liver injury was most pronounced in the absence of p53 compared with Fah-/- and Fah/Chk2-/- mice (Figure 1D). The injury phenotype of p53-deficient mice was unveiled further when we performed one-third partial hepatectomies in mice taken off NTBC. Although Fah-/- and Fah/Chk2-/- mice tolerated this additional damaging insult, none of the p53-deficient animals survived beyond day 1 (Figure 1A, right).
Figure 1.
p53 does not affect survival but is required for p21-mediated cell-cycle arrest in Fah-deficient mice with severe liver injury. (A) Kaplan–Meier survival curves of Fah-/- (n = 17), Fah/p53-/- (n = 17), and Fah/Chk2-/- (n = 26) mice on 0% NTBC (left) and Fah-/- (n = 8), Fah/p53-/- (n = 4), and Fah/Chk2-/- (n = 8) mice after one-third partial hepatectomy (14 days off NTBC, right). (B–F) Eight-week-old Fah-/-, Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- mice on 100% NTBC or 14 days after NTBC withdrawal. (B) Representative H&E, TUNEL, Ki67, and p21 immunohistochemistry, as well as immunofluorescence staining for ß-catenin. Fah/p53-/- livers show numerous Ki67-positive hepatocytes despite strong activation of p21. (C) Left: The relative percentage of TUNEL-positive hepatocytes was not significantly different between the individual genotypes. Means ± SEM across multiple mice (n = 4–6). Right: The relative percentage of Ki67-positive hepatocytes was increased in p53-deficient mice after NTBC withdrawal. Means ± SEM across multiple mice (n = 4–6). (D) NTBC withdrawal lead to markedly increased plasma levels of aspartate aminotransferase (AST) in the absence of p53. (E) p53-deficient mice retain their liver weight after 14 days of NTBC withdrawal. Means ± SEM (n = 6). (F) Immunoblots on total liver lysates from pooled samples (n = 4) to detect expression levels of the indicated cell-cycle–related proteins. (G) p21 levels were analyzed by quantitative polymerase chain reaction from Fah-/-, Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- mice treated with either 100% or 14 days with 0% NTBC. DDCT, delta-delta Ct; Gapdh, glyceraldehyde-3-phosphate dehydrogenase. ∗P ≤ .05.
We previously showed that the near-complete block in proliferation in the hepatocyte compartment of Fah-/- mice off NTBC, which is fully reversible after restarting NTBC treatment,21 is caused by a strong induction of p21.17 Consequently, genetic loss of p21 resulted in continuous hepatocyte proliferation and a significantly prolonged survival of Fah/p21-/- mice taken off NTBC.3 Although p53 is one of the master regulators of p21, NTBC withdrawal led to a significant up-regulation of p21 messenger RNA and protein expression in both Fah/p53-/- and Fah/Chk2-/- mice, indicating that p21 was induced independently of p53 and CHK2 (Figure 1B, F, and G). Surprisingly, although only few Ki67-positive cells were detectable in Fah/Chk2-/- mice on 0% NTBC similar to Fah-/- mice, a large fraction of proliferating hepatocytes was present in Fah/p53-/- mice despite the strong induction of p21 (Figure 1B and C).
To delineate whether proliferation-related pathways show a distinct activation profile in the proliferative vs the non-proliferative genotypes, we generated liver lysates from mice on NTBC and 14 days off NTBC from all groups, including Fah/p21-/- mice, and performed immunoblotting for cyclin D1, c-Jun N-terminal kinase/Jun signaling, mechanistic target of rapamycin kinase mediators (ribosomal protein S6 phosphorylation), and p38 (Figure 1F). Overall, although this basic characterization coherently showed activated signaling in the 0% NTBC livers compared with the respective 100% NTBC controls, we did not observe any consistent differences between the distinct genotypes that could differentiate between the groups with and without hepatocyte proliferation.
In agreement with the increased liver proliferation phenotype, Fah/p53-/- mice were able to maintain their liver weight independent of NTBC withdrawal, whereas the lack of a proliferative response in Fah-/- and Fah/Chk2-/- mice was accompanied by a significantly reduced liver weight 2 months after NTBC withdrawal (Figure 1E).
Taken together, we previously showed that severe liver injury induced by NTBC withdrawal leads to activation of p21, and genetic loss of p21 enables a strong proliferative response and prolongs survival.3,17 Here, we show that in the absence of p53, FAA-induced liver injury is aggravated, and is accompanied by increased hepatocyte proliferation, similar to our observation in Fah/p21-/- mice. The pronounced expression of p21 in the Fah/p53-/- mice indicates that p21 requires intact p53 to induce the profound cell-cycle arrest characteristic of Fah-/- mice with liver injury. Despite the strong proliferative response and in contrast to the Fah/p21-/- phenotype, Fah/p53-/- mice show the same high mortality as Fah-/- mice off NTBC. Partial hepatectomy as an additional insult to the liver distinguished the p53-deficient from the Chk2-deficient and Fah-/- control mice. This suggests that the overall potential of the p53-deficient liver to compensate for acute liver injury is reduced, but that if the injury does not exceed a certain threshold, compensatory proliferation can partially rescue and protect Fah/p53-/- mice from a further increase in early lethality. Unlike loss of p53, Chk2 deficiency neither aggravates FAA-induced liver injury nor alters the proliferative arrest observed in Fah-/- mice.
Apoptosis Resistance of Fah-/- Hepatocytes Is Not Fully Reversed by Loss of Either p53, Chk2, or p21
Fah-/- mice develop a profound resistance against FAS-induced apoptosis after NTBC withdrawal,22,23 which might serve as a physiologic protective mechanism in a situation of imminent liver failure. Apoptosis resistance, however, is reduced in Fah/p21-/- mice, implicating that p21 plays a role in the regulation of FAS-induced apoptosis in injured hepatocytes.3 This observation raises the question whether protection from apoptosis is strictly dependent on the presence of p21, or whether CHK2 and p53 might exert similar effects on the apoptosis sensitivity of FAH-deficient hepatocytes.
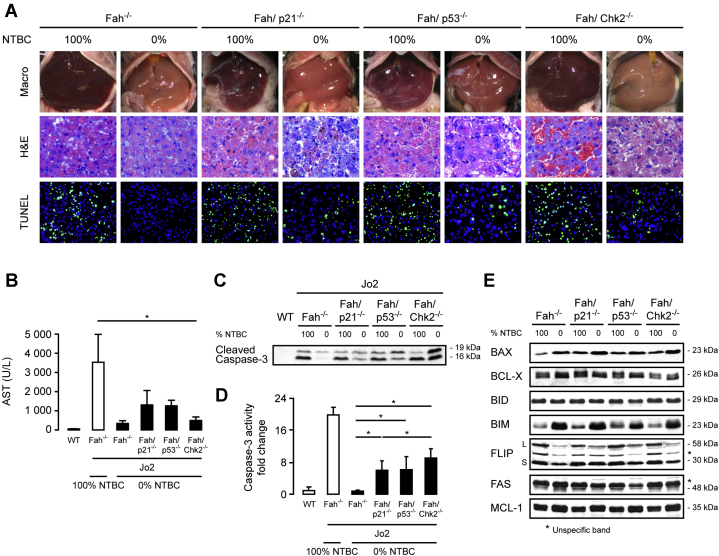
To address this question, Fah-/-, Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- mice on 100% NTBC treatment or 14 days after NTBC withdrawal were challenged with the anti-FAS monoclonal antibody Jo2. After Jo2-mediated activation of the apoptotic machinery, microscopic examination of liver sections from Fah-/- mice on 100% NTBC showed pronounced hepatocyte destruction with massive hemorrhage and multiple TUNEL-positive hepatocytes, which was markedly suppressed in Fah-/- mice on 0% NTBC, in line with our previous report (Figure 2A and Sherr and Roberts24). In all double-knockout mice, including a Fah/p21-/- cohort, surrogate markers of liver damage (transaminase levels) and apoptosis (TUNEL-positive hepatocytes, cleaved caspase-3) were increased (Figure 2A–D) when compared with Fah-/- mice off NTBC, but the animals did not present with the fulminant liver failure phenotype observed in Jo2-challenged Fah mice under 100% NTBC supplementation in the absence of concomitant injury. At this point, it is still enigmatic how apoptosis resistance is mediated in Fah-deficient hepatocytes in the context of liver injury.25 Notably, loss of p21, p53, and CHK2 did not significantly alter the expression levels of pro-apoptotic or anti-apoptotic proteins such as B-cell lymphoma 2 (BCL2)-associated X protein, B-cell lymphoma-extra large, Bcl-2 homology 3 interacting domain death agonist, BCL2-like 11 (apoptosis facilitator), FADD-homologous ICE/CED-3-like protease-like inhibitory protein, myeloid cell leukemia 1, and FAS receptor in healthy or injured hepatocytes compared with Fah-/- controls (Figure 2E).
Figure 2.
Apoptosis sensitivity was partially restored in Chk2, p53, and p21-deficient Fah-/-hepatocytes. (A–E) Eight-week-old Fah-/-, Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- mice on 100% NTBC treatment or 14 days after NTBC withdrawal were challenged with the anti-Fas antibody Jo2. (A) Representative macroscopic images of Fas-challenged animals, as well as H&E and TUNEL staining. Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- mice show morphologic signs of aggravated liver damage and increased numbers of TUNEL-positive hepatocytes. (B) Plasma levels of aspartate aminotransferase (AST) as a surrogate marker for liver injury are increased in Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- mice compared with Fah-/- mice off NTBC after Jo2 treatment, but do not reach the levels observed in Fah-/- mice on full NTBC supplementation (n = 3–5 per group, means ± SEM). Control: wild-type (WT) mice. (C and D) Jo2 treatment leads to increased cleaved caspase-3 expression (C, immunoblot on whole liver lysates) and (D) activity in Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- off NTBC compared with Fah-/- controls (n = 3–6). Activity values were normalized against WT. Means ± SEM are shown. (E) Immunoblots to detect the levels of several pro-apoptotic and anti-apoptotic proteins in pooled liver lysates from Fah-/-, Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- mice on 100% NTBC and 14 days after NTBC withdrawal (n = 3–4). ∗P ≤ .05. BAX, BCL2-associated X protein; BCL-X, B-cell lymphoma-extra large; BID, BH3 interacting domain death agonist; BIM, BCL2-like 11; FLIP, CASP8 and FADD-like apoptosis regulator; MCL-1, myeloid cell leukemia sequence 1.
Together, these data indicate that the mechanisms that can protect hepatocytes from apoptosis in the context of liver injury remain at least partially intact in the absence of p53, p21, or CHK2. However, loss of either p53, p21, or CHK2 results in a shift toward an apoptosis phenotype.
p53 Is Essential for Survival of Fah-Deficient Mice With Moderate Chronic Liver Injury
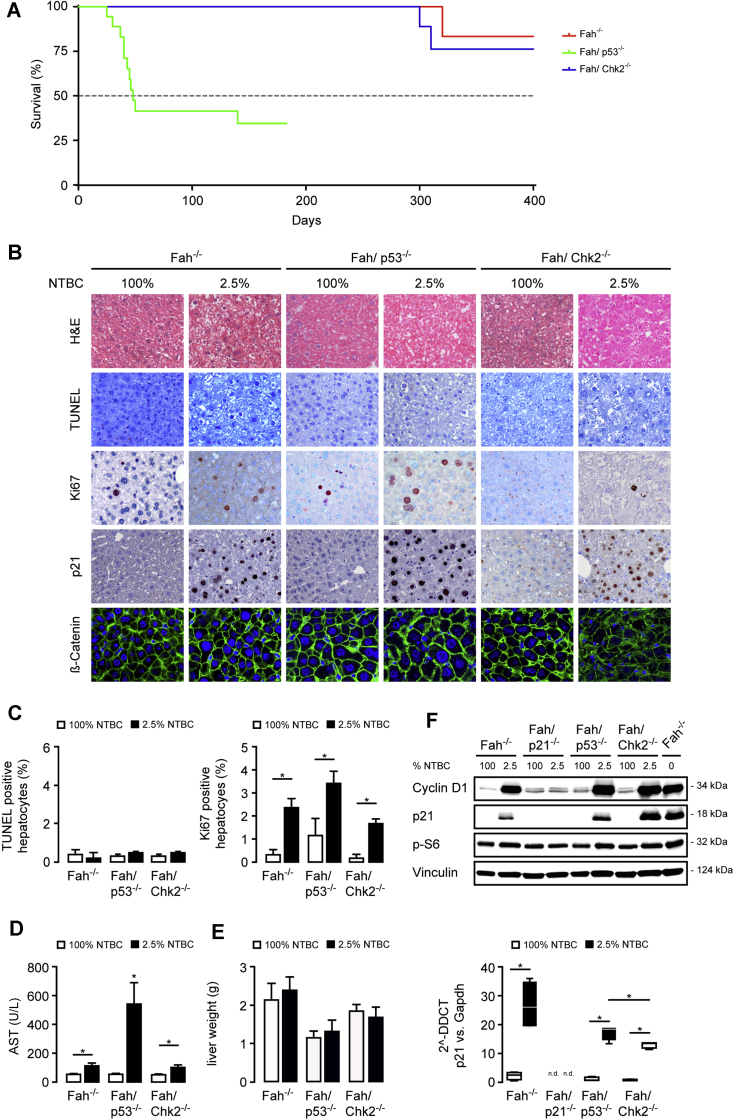
The reaction of the liver to injury is highly context-dependent and influenced by the severity of the damaging insult. To analyze the role of p53 and CHK2 in an experimental setting of moderate long-term injury and to define their involvement in liver cancer formation, mice were exposed to a reduced treatment regimen of NTBC (2.5% of the normal dose) for up to 12 months. Fah-/- and Fah/Chk2-/- mice survived the low-dose NTBC treatment (Figure 3A). In contrast, the majority of Fah/p53-/- mice died, with a median overall survival of 48 days after NTBC reduction with some long-term survivors, indicating that the p53-regulated stress response is critically important for the survival of Fah-deficient mice during chronic liver injury (Figure 3A). Histologically, livers from Fah-/- and Fah/Chk2-/- mice on low-dose NTBC showed multiple abnormal hepatic features with mild acinar inflammation and prominent hepatocyte size variations (Figure 3B, H&E and β-catenin staining). In all groups, TUNEL-positive hepatocytes were scarce, further highlighting that apoptosis is not a predominant mechanism of cell death in FAA-induced acute and chronic liver injury (Figure 3B and C). Similar to the respective cohort off NTBC, aggravated biochemical and histologic liver injury also was evident in Fah/p53-/- mice on 2.5% NTBC (Figure 3B and D). In contrast to the profound cell-cycle arrest observed in Fah-deficient mice on 0% NTBC, several proliferating hepatocytes were found in livers of Fah-/- mice with moderate liver injury despite increased p21 levels (Ki67 staining) (Figure 3B and C). Further supporting the proliferative phenotype, cyclin D levels were increased in Fah-/-, Fah/p53-/-, and Fah/Chk2-/- mice (Figure 3F and Marhenke et al26), and there was no significant difference in liver weight between mice on and off NTBC (Figure 3E).
Figure 3.
Loss of p53 aggravates liver injury and increases mortality in mice with moderate liver injury. (A) Kaplan–Meier survival curves of Fah-/- (n = 15), Fah/p53-/- (n = 15), and Fah/Chk2-/- (n = 15) mice on 2.5% NTBC treatment. (B–E) Fah-/-, Fah/p53-/-, and Fah/Chk2-/- mice 4 weeks on 2.5% NTBC compared with the respective 100% NTBC controls. (B) Representative H&E, immunohistochemistry, and immunofluorescence images as indicated. (C) Left: The relative percentage of TUNEL-positive hepatocytes was not significantly different between the genotypes. Means ± SEM across multiple mice (n = 4–6). Right: The relative percentage of Ki67-positive hepatocytes was increased after NTBC reduction. Means ± SEM across multiple mice (n = 4–6). (D) NTBC reduction lead to markedly increased plasma levels of aspartate aminotransferase (AST) in the absence of p53. (E) Liver weight was retained in all mice after 14 days of NTBC reduction. Means ± SEM (n = 4–6). (F) Upper: Immunoblots on total liver lysates from pooled samples (n = 4) for key cell-cycle–related proteins. Lower: p21 levels were analyzed by quantitative polymerase chain reaction from Fah-/-, Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- mice treated with either 100% or 4 weeks on 2.5% NTBC. ∗P ≤ .05. DDCT, delta-delta Ct; Gapdh, glyceraldehyde-3-phosphate dehydrogenase.
In contrast to the impaired regenerative response in Fah/p21-/- mice with moderate liver injury,17 the percentage of proliferating hepatocytes was similar in Fah-/- mice with or without loss of p53-/- or Chk2-/-. However, the increased hepatocyte proliferation was not sufficient to fully compensate the more severe liver injury and prevent the increased mortality in the majority of Fah/p53-/- mice, whereas the less pronounced injury in Fah/Chk2-/- mice allowed long-term survival similar to Fah-/- mice.
Loss of p53 Significantly Accelerates Tumor Development in Mice With Moderate Chronic Liver Injury
Liver tumor development can be critically influenced by predisposing genetic events, the degree of liver injury, and the proliferation rate, further complicated by the fact that these parameters frequently are interdependent. For instance, in Fah/p21-/- mice, tumor development correlated directly with the proliferative response and loss of p21 either accelerated or delayed tumorigenesis, depending on the severity of liver injury.17
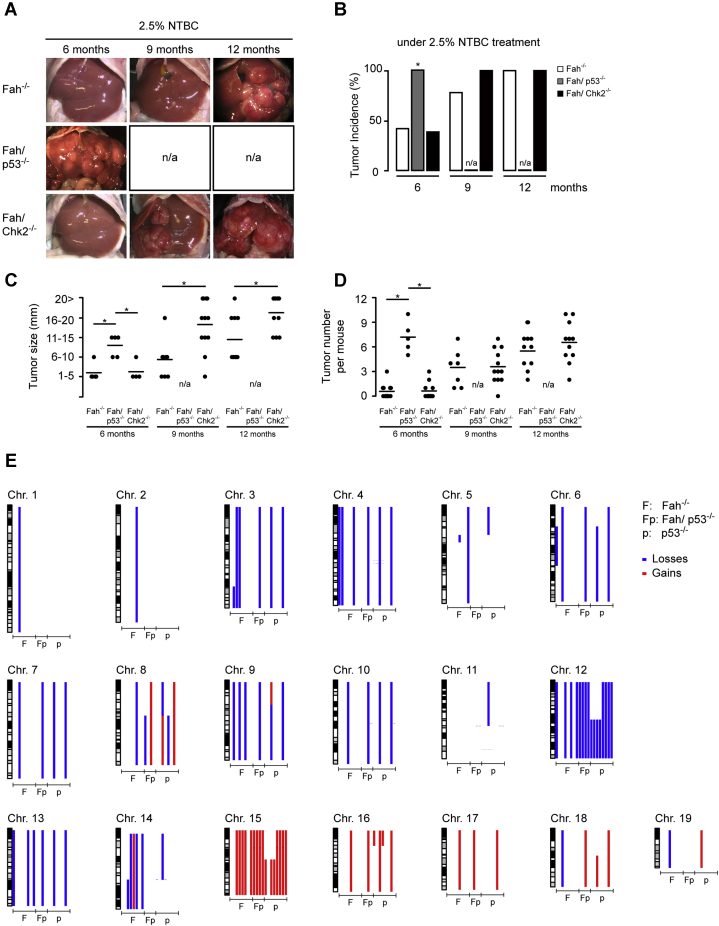
Liver tumors were evident by macroscopic and histologic examination in less than half of Fah-/- mice (n = 10) after 6 months with a steady increase over time, reaching 76% after 9 months (n = 20) and 100% after 12 months (n = 20) (Figure 4A and B). In contrast to our observations in Fah/p21-/- mice, surviving Fah/p53-/- on 2.5% NTBC showed accelerated tumor development: the surviving Fah/p53-/- mice exhibited several large tumor foci with full penetrance and had to be killed at 6 months (n = 6) (Figure 4A, B, and D). Tumor incidence in Fah/Chk2-/- and Fah-/- mice was similar (n = 13) (Figure 4B), indicating that loss of CHK2 neither substantially accelerates tumorigenesis-like p53 deficiency nor delays tumorigenesis as previously observed in p21-deficient mice.17 Of note, the tumor size was increased in Fah/Chk2-/- mice compared with Fah-/- mice after 9 (n = 12; P ≤ .05) and 12 months (n = 11; P ≤ .05) (Figure 4C).
Figure 4.
Loss of p53 accelerates tumor development but does not increase chromosomal instability in FAA-induced tumors. (A–D) Fah-/-, Fah/p53-/-, and Fah/Chk2-/- mice received either 100% or 2.5% NTBC for 6, 9, and 12 months. (A and B) Loss of p53 accelerated liver tumor formation in Fah-deficient mice. (C and D) Size and tumor numbers at the indicated time points. Each dot represents the maximum value measured per mouse, lines indicate the mean value. (E) aCGH on liver tumors from Fah-/- (F) and Fah/p53-/- (Fp) mice on 2.5% NTBC shows numerous chromosomal aberrations. Tumors from p53-/- mice were included as controls (p). ∗P ≤ .05. n/a, not available.
Next, we addressed whether loss of p53 in the FAH context alters the chromosomal profile of resulting tumors. Array comparative genomic hybridization (aCGH) was performed on liver tumors dissected from Fah-/- and Fah/p53-/- mice, and we included a control group of tumors that arose in p53-/- mice. Chromosomal gains and losses were evident in all liver tumors examined (Fah-/-, n = 8; Fah/p53-/-, n = 4; p53-/-, n = 8) (Figure 4E). A recurring chromosomal event across all genotypes was a copy number gain of chromosome 15, overlapping the c-Myc locus, which also is amplified frequently in human hepatocellular carcinoma (syntenic region on chromosome 8). In addition, chromosomal losses on chromosome 12 occurred in most p53-deficient tumors. Overall, aCGH did not show a striking increase in genomic instability in p53-deficient vs p53-proficient tumors.
In summary, loss of p53 not only dramatically increases liver injury and mortality of Fah-deficient mice, but also significantly accelerates tumor development in the liver, thereby confirming the strong tumor-suppressive role of p53. In contrast to the accelerated tumor development in p53-deficient mice and the delayed tumorigenesis in p21-deficient mice, loss of CHK2 did not significantly modulate tumor occurrence in the Fah model.
Gene Expression Profiles Are Dominated by the Severity of Liver Injury, But Also Reflect p53-Dependent Effects on Proliferation and Oxidative Stress Response
Thus far, our focused approaches did not show phenotype-specific differences in signaling pathway activation (Figures 1F and 3F). To better understand how loss of p53 modulates the cellular stress response, and to identify gene expression profiles that are associated with liver injury, regeneration, survival, and tumor development, we performed microarray analysis on reverse-transcribed RNA extracted from Fah-/- and Fah/p53-/- mice with moderate (3 months on 2.5% NTBC, n = 4) or severe liver injury (0% NTBC for 2 weeks, n = 4), and their respective controls (100% NTBC, n = 4). In addition, based on our observation that loss of p53 under 0% NTBC was not sufficient to recapitulate the prolonged overall survival of Fah-deficient mice lacking p21 expression, we included samples from the Fah/p21-/- cohort (n = 4) to directly compare how loss of p21 vs p53 affects the adaptive response in Fah-/- mice with severe liver injury.
Gene set enrichment analysis (GSEA) using the 50 hallmark gene sets derived from the Molecular Signature Database was performed to identify relevant biological processes.27 First, transcriptional profiles from mice with moderate and severe liver injury were compared with profiles from the corresponding healthy controls. Several significantly enriched gene sets were identified, and the majority of these gene sets were allocated to 4 main classes: p53/DNA damage response, oxidative stress response, inflammation, and proliferation (Table 1). Overall, these gene sets thematically recapitulate our previous observations on the role of p21,3,17 mechanistic target of rapamycin kinase,19 and nuclear factor erythroid 2-related factor 2 (NRF2)20 in the Fah-/- model.
Table 1.
Classification of Hallmark Gene Sets
| Classes | Hallmark gene sets | Genes, n |
|---|---|---|
| p53/DNA damage response | 282 | |
| HALLMARK_p53_PATHWAY | ||
| HALLMARK_DNA_REPAIR | ||
| Oxidative stress | 614 | |
| HALLMARK_XENOBIOTIC_METABOLISM | ||
| HALLMARK_REACTIVE_OXIGEN_SPECIES_PATHWAY | ||
| HALLMARK_HEME_METABOLISM | ||
| HALLMARK_BILE_ACID_METABOLISM | ||
| Inflammation | 508 | |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | ||
| HALLMARK_INFLAMMATORY_RESPONSE | ||
| HALLMARK_ALLOGRAFT_REJECTION | ||
| HALLMARK_TNFa SIGNALING VIA NFKB | ||
| HALLMARK_IL6, JAK, STAT3 SIGNALING | ||
| Proliferation | 791 | |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | ||
| HALLMARK_MTORC1_SIGNALING | ||
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | ||
| HALLMARK_G2M_CHECKPOINT | ||
| HALLMARK_MYC_TARGETS_V2 | ||
| HALLMARK_MYC_TARGETS_V1 | ||
| HALLMARK_E2F_TARGETS | ||
| HALLMARK_MITOTIC_SPINDLE |
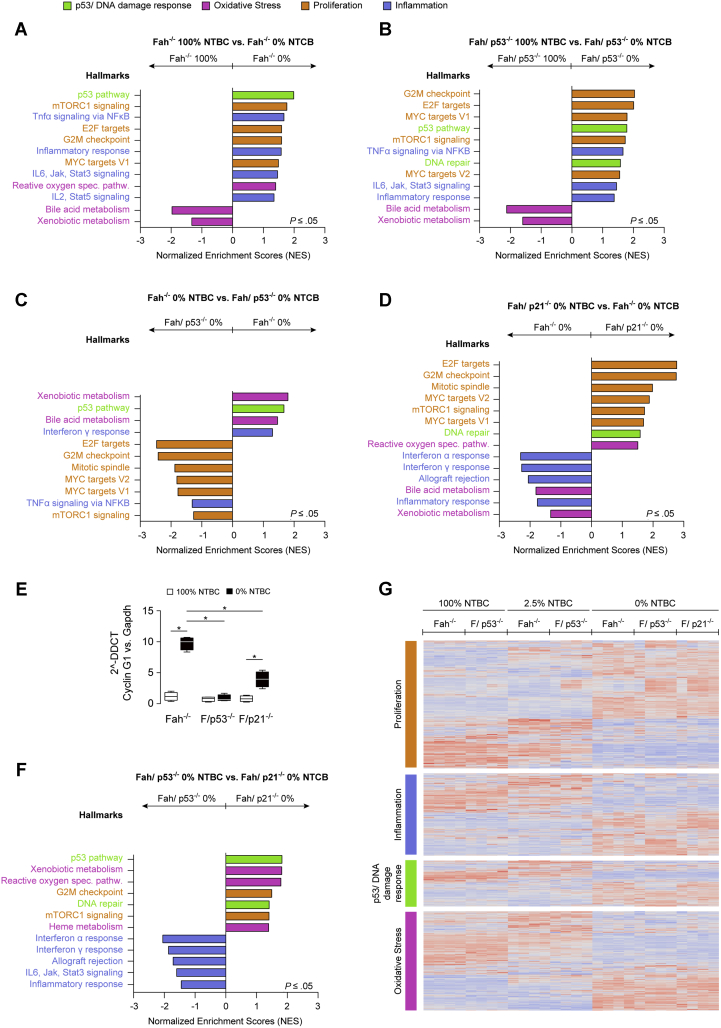
As expected, the p53/DNA damage response class segregated with liver injury in p53-proficient mice when compared with their 100% counterparts (Figure 5A, green). Of note, this class also was enriched in p53-deficient mice compared with healthy double-knockout mice (Figure 5B), indicating that a subset of genes in the p53 pathway gene set can be regulated independently of p53 during liver injury. On the contrary, when comparing different genotypes, the p53/DNA damage response class was enriched in p53-proficient mice compared with p53-deficient mice, which validates the dependency of these gene sets on the presence of intact p53 (Figure 5C).
Figure 5.
Gene expression profiling in p53-proficient and p53-deficient mice with moderate and severe liver injury. (A–D and F) Normalized Enrichment Scores (NES) of hallmark gene sets stratified into the categories p53/DNA damage response, oxidative stress, proliferation, and inflammation. (E) Quantitative polymerase chain reaction (PCR) analysis shows that p53 deficient Fah-/- mice off NTBC lack the decisive transcriptional up-regulation of cyclin G1. (G) Gene expression patterns are prominently influenced by the type of liver injury and less affected by the different genotypes. DDCT, delta-delta Ct; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; G2M, G2/M-phase; IL, interleukin; Jak, Janus kinase; mTorc1, mTOR complex 1; MYC, myelocytomatosis oncogene; NFKB, nuclear factor κB; Stat3, signal transducer and activator of transcription 3; TNF, tumor necrosis factor.
Gene sets assigned to the proliferation class were enriched significantly in severe injury conditions (Figure 5A and C, orange) in both p53-proficient and p53-deficient mice compared with the respective healthy controls. When comparing mice with liver injury, proliferation gene sets were enriched in the absence of p53 (Figure 5C) and p21 (Figure 5D). Next, we specifically analyzed the proliferation class for differentially regulated genes that segregated with the p53 status. Among the 791 genes included in the proliferation class, 190 genes were significantly differentially regulated between Fah-/- and Fah/p53-/- mice, but only 3 genes (epidermal growth factor receptor, cyclin G1, asparagine synthetase) were more than 2-fold up-regulated in Fah -/- mice compared with Fah/p53-/- mice. Of these 3 genes, cyclin G1 has been described previously as a p53-dependent cell-cycle inhibitor in hepatocytes, and increased levels of cyclin G1 were confirmed in Fah-/- mice with severe liver injury, although they were significantly lower in Fah/p53-/- and Fah/p21-/- mice (Figure 5E), indicating that cyclin G1 might contribute to the cell-cycle arrest in Fah-/- mice with severe liver injury.
Oxidative stress response–related gene sets include a broad array of genes that are either induced or suppressed to counteract increased oxidative stress after NTBC reduction. In agreement with our previous findings, gene sets included in the category of oxidative stress response were differently regulated in Fah-/- mice with severe liver injury (Figure 5A). Aggravated liver injury and high mortality of Fah/p53-/- mice was accompanied by an impaired oxidative stress response after NTBC withdrawal compared with the p53-proficient Fah-/- mice (Figure 5C). The oxidative stress response class also was differentially regulated between Fah/p53-/- and Fah/p21-/- mice, which likely contributes to the more severe liver injury and high mortality of Fah/p53-/- mice compared with Fah/p21-/- mice taken off NTBC despite the high hepatocyte proliferation rate in both genotypes (Figure 5F).
Despite the experimental evidence that Fah-/- mice respond differently to injury depending on the presence or absence of p53 or p21, it should be noted that the global gene expression patterns, and also the regulation within the hallmark gene sets, were influenced most prominently by the degree of liver injury and less affected by the genotype of the respective genetic groups (Figure 5G).
Together, the comprehensive GSEA confirmed the strong activation of the p53 pathway and an oxidative stress response in the Fah model. The gene set class proliferation was induced in Fah/p53-/- and Fah/p21-/- mice. Moreover, increased liver injury and high mortality of Fah/p53-/- mice occurred despite a compensatory induction of p53-related genes and was accompanied by an impaired oxidative stress response after NTBC reduction and withdrawal.
p21 Fails to Inhibit Retinoblastoma Phosphorylation in Fah/p53-/- Mice
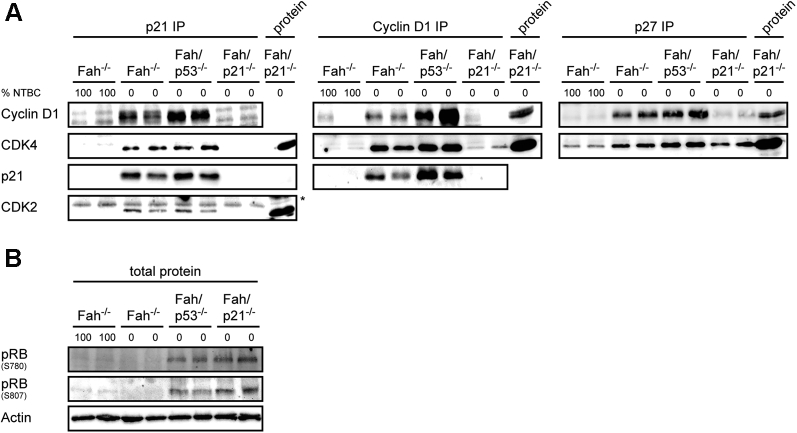
The gene expression analysis confirmed a strong induction of proliferation-related genes in Fah/p53-/- mice with severe liver injury occurred despite the p21 expression (Figure 1F). D-type cyclins and the kinases cyclin-dependent kinase (CDK)4 and CDK6 regulate G0–G1–S progression by contributing to the phosphorylation and subsequent inactivation of the retinoblastoma (RB) gene product. Assembly of active cyclin D–CDK complexes in response to mitogenic signals is regulated negatively by the cyclin-dependent kinase inhibitors p21 and p27. On the other hand, a noncatalytic function of cyclin D–dependent kinases is the sequestration of p21 and Cyclin-dependent kinase inhibitor 1B (p27) during the G1 phase to relieve cyclin E–CDK2 from their constraint, thereby facilitating its activation later in the G1 phase.24 To understand whether the unexpected finding that p21 failed to inhibit RB phosphorylation and cell-cycle progression in Fah/p53-/- mice results from an impaired cyclin/CDK/cyclin-dependent kinase inhibitor complex formation, we performed immunoprecipitation (IP) of the individual components of the complex. IP with an antibody against p21 showed that p21 associated with cyclin D1, CDK4, and also with CDK2 in Fah/p53-/- and in Fah -/- mice (Figure 6A). The interaction of cyclin D1/CDK4 and p21 was confirmed by IP of cyclin D1. Similarly, IP with an anti-p27 antibody showed an interaction of p27 with cyclin D1 and CDK4 in both Fah/p53-/- and in Fah -/- mice. In line with the strong regenerative response, RB was phosphorylated only in Fah/p53-/- mice and Fah/p21-/- mice, but not in Fah -/- mice with severe liver injury (Figure 6B).
Figure 6.
Formation of cyclin/CDK/cyclin-dependent kinase inhibitor (CKI) complexes during liver injury. (A) Extracts from liver tissue were subjected to immunoprecipitation and Western analysis. Association of p21 with cyclin D1, CDK4, and CDK2; association of cyclin D1 with CDK4 and p21; association of p27 with cyclin D1 and CDK4. (B) Immunoblots on total liver lysates from 2 samples of each group for phospho-RB (pRB) were performed.
Together, these data indicate that the interaction of p21 with cyclin D1, CDK4, and CDK2, and the interaction of p27 with cyclin D1 and CDK4, is not impaired in Fah/p53-/- mice. In contrast to Fah-/- mice, this interaction, however, fails to inhibit RB phosphorylation and cell-cycle progression of hepatocytes during liver injury.
Fah/p53-/- Mice Show an Impaired Oxidative Stress Response
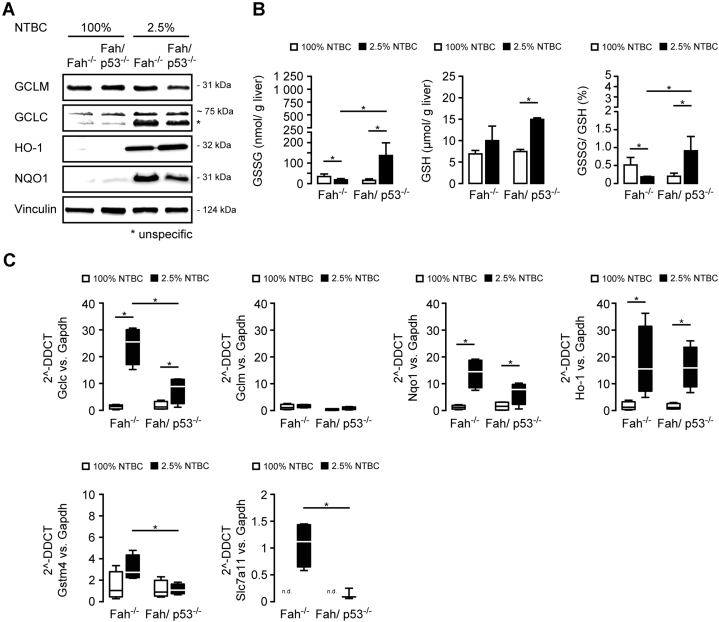
The gene expression analysis suggests that an impaired oxidative stress response contributes to the high mortality of Fah/p53-/- mice. FAA is a highly electrophilic compound that induces DNA damage and oxidative stress in vitro and in vivo.28 In line with these studies, we previously showed that the NRF2-mediated anti-oxidative stress response is critically important for the survival of Fah-deficient mice,20 and a strong induction of NRF2 target genes such as reduced nicotinamide adenine dinucleotide phosphate dehydrogenase, quinone 1 and heme oxygenase 1 was evident in Fah-/- and Fah/p53-/- mice (Figure 7A and C).
Figure 7.
Fah/p53-/-mice show an impaired oxidative stress response. (A–E) Fah-/- and Fah/p53-/- mice were treated with 100% or 2.5% NTBC for 4 weeks. (A) Immunoblot on pooled liver samples (n = 4) for key regulators of the oxidative stress response. (B) GSSG/GSH ratio shows an impaired oxidative stress response in Fah/p53-/- mice. (C) Quantitative polymerase chain reaction (PCR) confirms significantly lower levels of selected members of the anti-oxidative stress response (Gclc, Gstm4, and Slc7a11) in p53-deficient Fah-/- mice. Medians in box-and-whisker plots (Tukey test). ∗P ≤ .05. DDCT, delta-delta Ct; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; Gclc, glutamate-cysteine ligase, catalytic subunit; Gclm, glutamate-cysteine ligase, modifier subunit; HO-1, heme oxygenase 1; n.d., not detectable; NQO1, NAD(P)H dehydrogenase, quinone 1.
Accumulation of electrophilic compounds is counterbalanced by changes in antioxidants such as glutathione (GSH). Previously, it has been shown that acute FAA accumulation leads to a depletion of GSH in vivo,20 which enhances FAA-induced cell death and also amplifies its mutagenicity.29 The biosynthesis of glutathione is dependent on the availability of the cystine/glutamate transporter (solute carrier family 7 [cationic amino acid transporter, y+ system], member 11), a member of heterodimeric sodium-dependent amino acid transporter that imports cystine and exports glutamate, and of a glutamate-cysteine ligase, which consists of a heavy catalytic subunit (glutamate cysteine ligase, catalytic subunit) and a light regulatory subunit (glutamate cysteine ligase, modifier subunit). Electrophilic compounds and products of oxidative stress subsequently are detoxified by conjugation with glutathione by enzymes such as glutathione S-transferase mu4. In agreement with our previous observation,20 Gclc, Gstm4, and Slc7a11 were strongly induced in Fah-/- mice after NTBC reduction, whereas they were significantly lower in p53-deficient livers (Figure 7A and C). In line with the compensatory increase in GSH production upon liver injury, GSH levels were increased in both groups on 2.5% NTBC (Figure 7B). However, glutathione disulfide (GSSG) as the oxidized form of GSH was increased exclusively in liver lysates of Fah/p53-/- mice, leading to an increased GSSG:GSH ratio suggestive of a net shift toward insufficient compensation (Figure 7B).
Together, accumulation of FAA leads to a strong oxidative stress response in the liver, which is impaired in Fah/p53-/- mice. Notably, loss of p53 could not be compensated by other regulators of the oxidative stress response such as NRF2, suggesting that a coordinated expression of cytoprotective p53 target genes is critically important to protect Fah-deficient mice against FAA-induced liver injury.
Discussion
p53 is embedded in an intricate network of upstream regulators and downstream effectors that, in a concerted action, determine how a cell responds to damage. This response is highly flexible and context-dependent, and despite extensive efforts, a universal set of p53 target genes has not yet been defined.5 In the Fah model, loss of p53 severely aggravated liver injury and increased mortality. Overall, the phenotype of Fah/p53-/- mice was reminiscent of our previous observations in Fah-/- mice with targeted deletion of Nrf2.20 Both p53 and NRF2 play a fundamental role in maintaining the redox homeostasis of hepatocytes by controlling the expression of key components of the glutathione and thioredoxin antioxidant system, as well as enzymes involved in reduced nicotinamide adenine dinucleotide phosphate regeneration and xenobiotic detoxification. Compared with Fah-/- controls, the GSSG:GSH ratio was increased significantly in Fah/p53-/- mice as a consequence of the insufficient ability of hepatocytes to compensate the FAA-induced oxidative stress. Nevertheless, the antioxidant N-acetylcysteine alone was not sufficient to relieve the increased liver injury in Fah/p53-/- mice (data not shown) or in Fah/Nrf2-/- mice.20 Thus, our data suggest that both a coordinate p53- and NRF2-regulated anti-oxidative stress response is required to protect hepatocytes from injury.
The liver has a remarkable ability to regenerate, and both moderate and severe liver damage upon NTBC reduction was associated with increased proliferation in the absence of p53. However, the proliferative response in p53-deficient mice on low-dose NTBC was not sufficient to prevent early mortality. In contrast, mice with severe acute injury (0% NTBC) were capable of mounting a compensatory proliferative response that rescued them from accelerated death. However, the delivery of an additional insult to the p53-deficient liver through partial hepatectomy tipped the delicate balance toward liver failure.
Several previous studies have delineated the critical involvement of p21 as one of the pivotal p53 target genes in liver regeneration, and we have shown that p21 induces a profound cell-cycle arrest in Fah-deficient mice with severe liver injury.17 Unexpectedly, the strong induction of p21 is not dependent on p53 or on its upstream regulator CHK2. Moreover, the p53-independent p21 induction is not able to prevent hepatocyte proliferation, indicating that p21 is necessary but not sufficient to inhibit cell-cycle progression in the liver.
Activation of established pathways that regulate liver regeneration was evident in all mice with liver injury, but did not correlate with the proliferative capabilities of the different cohorts. Of note, the physical interaction of p21 with cyclin D1, CDK4, and CDK2 was not impaired in Fah/p53-/- mice, but failed to inhibit phosphorylation of RB. On the molecular level, NTBC withdrawal led to a strong induction of genes associated with proliferation, irrespective of the genotype-dependent phenotypes. Among these genes, transcriptional regulation of cyclin G1 correlated with the proliferative response in Fah-/- and Fah/p53-/- mice. Cyclin G1 was identified previously as a p53 target gene that not only regulates stabilization of p53 by promoting protein degradation through Mouse double minute 2 homolog phosphorylation, but also acts as a cell-cycle checkpoint in hepatocytes.30, 31, 32 Thus, cell-cycle progression in the absence of p53 might be facilitated in part by low levels of cyclin G1, but additional studies are necessary to unravel the molecular mechanism why p21 fails to inhibit hepatocyte proliferation in the absence of p53 during chronic liver injury.
p53-/p21-mediated cell-cycle arrest in Fah mice is accompanied by a resistance to apoptosis, reminiscent of a senescence phenotype.33,34 Notably, although loss of either p21 or p53 restored hepatocyte proliferation, apoptosis resistance was relieved only partially in Fah/p21-/-, Fah/p53-/-, and Fah/Chk2-/- mice challenged with the FAS antibody Jo2. This suggests that the apoptosis resistance of Fah-/- mice cannot be fully attributed to p53-/p21-mediated senescence.
One potential limitation of our study could be the choice of a conditional liver-specific p53 knockout in contrast to the constitutive and ubiquitous loss of gene expression in p21-/- and Chk2-/- knockout mice. The conditional p53 allele was chosen because of the early tumor incidence in p53-/- mice (mainly lymphomas and sarcomas), whereas p21-/- and Chk2-/- knockout mice do not have an overt phenotype. Moreover, tyrosine metabolism occurs only in hepatocytes and proximal renal tubular cells, leading to FAA-induced injury specifically in these 2 organs. We therefore do not envision that loss of p21 or CHK2 in nonparenchymal cells has a major impact on the observed phenotypes.
In summary, we show that, in a model of metabolic liver injury, loss of p53, but not of Chk2, impairs the oxidative stress response and aggravates liver damage, indicating that, beyond its role as a tumor suppressor, intact p53 exerts an immediate protective effect on hepatocytes.
The increased regenerative potential in the absence of p53 is only partially capable of compensating the surplus injury and not sufficient to promote survival. Our work highlights that, although gene expression patterns are dominated by the severity of the damaging insult, characteristic phenotypes are created through genetic abrogation of individual components of the multilevel cascade that controls cell-cycle progression. The extent to which loss of gene function can be compensated, or affects liver injury and proliferation, is related to the level at which the cascade is interrupted. Although loss of the upstream regulator Chk2 is dispensable in the model, cell-cycle control during chronic liver injury critically depends on the presence of both p53 and its downstream effector p21. Our work shows a complex interdependency between p21 and p53: neither the p53-independent induction of p21 nor activation of p53 in the absence of p21 is sufficient to prevent hepatocyte proliferation. Although loss of either p53 or p21 can elicit a strong proliferative response, only loss of p53 leads to increased liver injury and mortality, which cannot be compensated by increased liver regeneration.
Thus, damaging insults to the liver activate complex and interwoven response programs, whose net result is determined by the susceptibility to the damaging agent and the ability to regenerate. The type and the degree of injury, but also the underlying genetic landscape of the organism, are key modifiers of the injury response and can produce distinct, or even opposite, phenotypical outcomes.
Materials and Methods
Animal Experiments
All animal experiments were approved by local authorities (Lower Saxony State Office for Consumer Protection and Food Safety). The B6;129-Fahtm1Mgo (Fah-/-)35 mice were crossed to the C57BL/6J-Tg(Alb1-cre)7Gsc-FVB.129-Trp53tm1Brn (p53fl/fl/Alfp-Cre+), C57BL/6-Cdkn1atm1Ty1/J (p21-/-), and C57BL/6-Chk2tm1Mak (Chk2-/-) strain for several generations to generate Fahtm1Mgo-C57BL/6J-Tg(Alb1-cre)7Gsc-FVB.129-Trp53tm1Brn (Fah-/-/p53fl/fl/Alfp-Cre, ie, Fah/p53-/-), B6;129-Fahtm1Mgo-C57BL/6-Cdkn1atm1Ty1/J (Fah/p21-/-), and C57BL/6-Fahtm1Mgo-C57BL/6- Chk2tm1Mak (Fah/Chk2-/-) double-knockout mice. Drinking water was supplemented with 7.5 mg/L NTBC18 unless otherwise indicated, and mice had access to food (Altromin1324 M; Altromin, Lage, Germany) and water ad libitum. Mice were housed in individually ventilated cages and all experiments were performed during the daytime of a 14-/10-hour day/night cycle. Kaplan–Meier survival curves were generated after NTBC reduction (2.5%) or withdrawal (0%) as previously published.21 Tumor incidence (including size and number) were evaluated for all genotypes at the same time, data from Fah-/- (included in Figure 4) and Fah/p21-/- mice were reported previously.17
DNA Microarray Hybridization and Analysis
The quality and integrity of the total RNA were analyzed on an Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). A total of 500 ng total RNA was applied for Cy3-labeling reaction using the 1-color Quick Amp Labeling protocol (Agilent Technologies). Labeled complementary RNA was hybridized to Agilent's murine 4 × 44 k microarrays for 16 hours at 68°C and scanned using the Agilent DNA Microarray Scanner. Expression values were calculated by the software package Feature Extraction 10.5.1.1 (Agilent Technologies). Statistical analysis of the expression data was performed using the Gene Spring Software package (Agilent Technologies). Raw data were analyzed further using R package Limma. Raw data were log2-transformed and quantile-normalized.36 All clustering was performed using the Bioconductor package ComplexHeatmap.37 GSEA were conducted using the R version of the GSEA software provided by github of the Broad Institutes (https://github.com/GSEA-MSigDB/GSEA_R).38
Monoclonal Antibody Fas Injection and Caspase-3 Activity Assay
Ten-week-old mice on 100% NTBC or 14 days after NTBC withdrawal received an intraperitoneal injection with a monoclonal anti-Fas antibody (Jo2 purified anti-mouse CD95, 0.6 μg/g body weight diluted in 200 μL phosphate-buffered saline). Six hours after the injection, mice were killed and their livers were explanted. Caspase-3 activity was assayed using a synthetic tetrapeptide fluorogenic substrate (Ac-DEVD-AMC; BD Bioscience, San Jose, CA) in the presence or absence of the pan-caspase inhibitor (Ac-DEVD-CHO; BD Pharmingen), according to the manufacturer’s recommendations.
One-Third Partial Hepatectomy
Mice were anesthetized with an intraperitoneal injection of Ketanest/Rompun (Bayer, Leverkusen, Germany). An abdominal midline incision was performed and the liver was exposed. After ligation, the left liver lobe was resected. The abdominal wall was sutured and mice recovered from anesthesia on a warming pad.
Histology and Immunostaining
Liver tissue was fixed in 3.5%–3.7% formaldehyde, processed, and paraffin-embedded. Sections (5 μm) were stained with H&E or processed further for immunohistochemistry. Ki67 (ab16667, 1:200; Abcam, Cambridge, UK), p21 (ab188224, 1:500; Abcam), and ß-catenin (ab6302, 1:100; Abcam) staining were performed using standard immunohistochemistry protocols. TUNEL staining was performed according to the manufacturer's protocols (Roche Diagnostics GmbH, Mannheim, Germany).
GSH/GSSG Measurements
Total glutathione (GSH + GSSG) and GSSG were measured as described.1
Immunoblotting
Frozen liver tissue was homogenized using an Ultra-Turrax (IKA, Staufen, Germany) for 10 seconds in AMP-activated protein kinase lysis buffer (50 mmol/L HEPES, 50 mmol/L KCl, 50 mmol/L Natrium fluorid, 5 mmol/L Sodium pyrophosphate decahydrate, 1 mmol/L EDTA, 1 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 5 mmol/L ß-glycerophosphate, 1 mmol/L dithiothreitol, 1 mmol/L sodium orthovanadate, and 1% [v/v] Nonidet P40) containing Complete Protease Inhibitor mixture (Roche, Germany) and centrifuged for 10 minutes at 16,000 × g. All antibodies were used at 1:1000 dilutions unless otherwise indicated.
C-jun (9165), cleaved caspase-3 (9664), Flip (3210), p38 (9212), phospho-Jnk (Thr193/Tyr185) (9251), phospho-ribosomal protein S6 (Ser240/244) (5364, 1:500, bovine serum albumin), and vinculin (13901, 1:2000) were from Cell Signaling (Leiden, Netherland).
BCL2-associated X protein (sc-493), FAS (sc-716), and glyceraldehyde-3-phosphate dehydrogenase (sc-32233) were from Santa Cruz Biotechnology Inc. (Heidelberg, Germany). BCL2-like 1 (610211; BD Biosciences), Bcl-2 homology 3 interacting domain death agonist at 1:2000 (AF860; Bio-Techne GmbH, formerly R&D Systems, Wiesbaden-Nordenstadt, Germany), BCL2-like 11 (apoptosis facilitator) (B7929; Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany), CASP8 and FADD-like apoptosis regulator (AAP-440; Stressgen), heme oxygenase 1 (ADI-SPA-896F, 1:500; Enzo Life Sciences GmbH, Lörrach, Germany), and myeloid cell leukemia sequence 1 (Mcl-1) (600-401-394; Rockland Immunochemicals Inc, Limerick, PA) also were used.
Cyclin D1 (ab134175, 1:500), glutamate cysteine ligase, catalytic subunit (ab53179, 1:500), glutamate cysteine ligase, modifier subunit (ab153967, 1:500), reduced nicotinamide adenine dinucleotide phosphate dehydrogenase, quinone 1 (ab80588, 1:500), p21 (ab188224, 1:500) phospho-RB S780 (ab184702), and phospho-RB S807 (ab184796) were from Abcam.
Horseradish-peroxidase–conjugated secondary antibodies were purchased from Santa Cruz Biotechnologies (sc-2357) and Cell Signaling (7074).
IP
For IP we used the Pierce Classic IP Kit (26146, Fisher Scientific GmbH, Part of Thermo Fisher Scientific, Schwerte, Germany) according to the manufacturer’s protocol. Frozen liver tissue (50 mg) was homogenized using an Ultra-Turrax (IKA) for 10 seconds in the lysis buffer contained in the kit, to which we added Complete Protease Inhibitor mixture (Roche) and phosphatase inhibitors (2.5 mmol/L sodium pyrophosphate, 1 mmol/L ß-glycerophosphate, and 1 mmol/L sodium orthovanadate). For immune complex formation we used 5 μg of a purified antibody and 500 μg precleared liver lysate. The following antibodies from Abcam (UK) were used: CDK2 (ab32147), CDK4 (ab199728), cyclin D1 (ab134175) p21 (ab188224), and p27 (ab190851). For antibody detection, the Clean-Blot IP Detection Reagent from Thermo Scientific (21230) was used.
Transaminase Levels
Blood was collected in lithium heparin tubes (LH1.3; SARSTEDT AG & Co. KG (Nümbrecht, Germany), Olympus Deutschland GmbH (Hamburg, Germany) and transaminase activity was measured using an Olympus AU 400 System (Olympus, Germany).
RNA Isolation and Quantitative Polymerase Chain Reaction
Total RNA was extracted from liver tissue (n = 4) using the NucleoSpin RNA II kit (Macherey-Nagel GmbH & Co KG, Düren, Germany) and complementary DNA was synthesized using the Transcriptor High Fidelity Complementary DNA Kit (Roche). The relative quantification of gene expression was performed by TaqMan Gene Expression Assay in a QuantStudio 12K Flex Real-Time Polymerase Chain Reaction System (Fisher Scientific) according to the manufacturer’s protocol. Glyceraldehyde-3-phosphate dehydrogenase served as an internal control. The following TaqMan (Life Technologies GmbH Darmstadt, Germany) probes were used: Mm00432359_m1 (Ccnd1), Mm00432448_m1 (p21), and Mm99999915_g1 (glyceraldehyde-3-phosphate dehydrogenase).
aCGH
Agilent oligonucleotide aCGH for genomic DNA analysis (SurePrint G3 Mouse CGH Microarray Kit 4 × 180K; Agilent Technologies, Santa Clara, CA) was performed on frozen tumor tissue. Isolated genomic DNA (1 μg) was digested and labeled by random priming with cyanine 5-deoxyuridine triphosphate (tumor DNA) or cyanine 3-deoxyuridine triphosphate (reference DNA) using the SureTag DNA Labeling Kit (Agilent Technologies) according to the manufacturer’s protocol. As reference, spleen genomic DNA was used. Microarray slides were scanned using a G2565CA microarray scanner (Agilent Technologies), and raw data were extracted using the Feature Extraction software (v.10.7.3.1; Agilent Technologies) for further analyses with the Genomic Workbench software (v.7.0.4.0; Agilent Technologies). The extracted files were analyzed with the statistical algorithm ADM-2.
Statistical Analysis
Data are represented as means ± SEM or as medians in box-and-whisker plots (Tukey test). Data were analyzed by analysis of variance, followed by the Student t test to determine significance. P values were considered statistically significant when P ≤ .05.
Acknowledgments
The authors thank Eric Jende and Meriame Nassiri for their expert technical assistance.
CRediT Authorship Contributions
Laura Elisa Buitago-Molina, PhD (Conceptualization: Equal; Formal analysis: Equal; Investigation: Equal; Writing – review & editing: Equal)
Silke Marhenke, PhD (Conceptualization: Equal; Formal analysis: Equal; Investigation: Equal; Writing – review & editing: Equal)
Diana Becker (Formal analysis: Equal; Investigation: Equal)
Robert Geffers, PhD (Formal analysis: Equal; Investigation: Equal)
Timo Itzel (Formal analysis: Equal)
Andreas Teufel, MD/ PhD (Formal analysis: Equal)
Hartmut Jaeschke, PhD (Formal analysis: Equal; Investigation: Supporting)
André Lechel, PhD (Formal analysis: Equal; Investigation: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Kristian Unger, PhD (Formal analysis: Equal)
Jens U. Marquardt, MD (Formal analysis: Supporting; Writing – review & editing: Supporting)
Michael Saborowski, MD (Conceptualization: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting)
Anna Saborowski, MD (Conceptualization: Equal; Formal analysis: Equal; Funding acquisition: Supporting; Project administration: Equal; Writing – original draft: Lead)
Arndt Vogel, MD (Conceptualization: Lead; Formal analysis: Equal; Funding acquisition: Lead; Project administration: Lead; Supervision: Lead; Writing – original draft: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was funded by the Deutsche Forschungsgemeinschaft (German Research Foundation) grants SFB/TR 209–314905040 (A.S., A.V., M.S.), grant BU2722/2-3 (L.E.B-M) and Vo959/9-1 (A.V.) and supported by Else Kröner-Fresenius Foundation grant 2015_A225 (A.S.).
References
- 1.Plentz R.R., Park Y.N., Lechel A., Kim H., Nellessen F., Langkopf B.H., Wilkens L., Destro A., Fiamengo B., Manns M.P., Roncalli M., Rudolph K.L. Telomere shortening and inactivation of cell cycle checkpoints characterize human hepatocarcinogenesis. Hepatology. 2007;45:968–976. doi: 10.1002/hep.21552. [DOI] [PubMed] [Google Scholar]
- 2.Katz S.F., Lechel A., Obenauf A.C., Begus-Nahrmann Y., Kraus J.M., Hoffmann E.M., Duda J., Eshraghi P., Hartmann D., Liss B., Schirmacher P., Kestler H.A., Speicher M.R., Rudolph K.L. Disruption of Trp53 in livers of mice induces formation of carcinomas with bilineal differentiation. Gastroenterology. 2012;142:1229–1239 e3. doi: 10.1053/j.gastro.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Willenbring H., Sharma A.D., Vogel A., Lee A.Y., Rothfuss A., Wang Z., Finegold M., Grompe M. Loss of p21 permits carcinogenesis from chronically damaged liver and kidney epithelial cells despite unchecked apoptosis. Cancer Cell. 2008;14:59–67. doi: 10.1016/j.ccr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boege Y., Malehmir M., Healy M.E., Bettermann K., Lorentzen A., Vucur M., Ahuja A.K., Bohm F., Mertens J.C., Shimizu Y., Frick L., Remouchamps C., Mutreja K., Kahne T., Sundaravinayagam D., Wolf M.J., Rehrauer H., Koppe C., Speicher T., Padrissa-Altes S., Maire R., Schattenberg J.M., Jeong J.S., Liu L., Zwirner S., Boger R., Huser N., Davis R.J., Mullhaupt B., Moch H., Schulze-Bergkamen H., Clavien P.A., Werner S., Borsig L., Luther S.A., Jost P.J., Weinlich R., Unger K., Behrens A., Hillert L., Dillon C., Di Virgilio M., Wallach D., Dejardin E., Zender L., Naumann M., Walczak H., Green D.R., Lopes M., Lavrik I., Luedde T., Heikenwalder M., Weber A. A dual role of caspase-8 in triggering and sensing proliferation-associated DNA damage, a key determinant of liver cancer development. Cancer Cell. 2017;32:342–359 e10. doi: 10.1016/j.ccell.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kastenhuber E.R., Lowe S.W. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teoh N., Pyakurel P., Dan Y.Y., Swisshelm K., Hou J., Mitchell C., Fausto N., Gu Y., Farrell G. Induction of p53 renders ATM-deficient mice refractory to hepatocarcinogenesis. Gastroenterology. 2010;138:1155–1165. doi: 10.1053/j.gastro.2009.11.008. e1–2. [DOI] [PubMed] [Google Scholar]
- 7.Hirao A., Kong Y.Y., Matsuoka S., Wakeham A., Ruland J., Yoshida H., Liu D., Elledge S.J., Mak T.W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 8.Hirao A., Cheung A., Duncan G., Girard P.M., Elia A.J., Wakeham A., Okada H., Sarkissian T., Wong J.A., Sakai T., De Stanchina E., Bristow R.G., Suda T., Lowe S.W., Jeggo P.A., Elledge S.J., Mak T.W. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol. 2002;22:6521–6532. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zender L., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Mu D., Lucito R., Powers S., Lowe S.W. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClendon A.K., Dean J.L., Ertel A., Fu Z., Rivadeneira D.B., Reed C.A., Bourgo R.J., Witkiewicz A., Addya S., Mayhew C.N., Grimes H.L., Fortina P., Knudsen E.S. RB and p53 cooperate to prevent liver tumorigenesis in response to tissue damage. Gastroenterology. 2011;141:1439–1450. doi: 10.1053/j.gastro.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borude P., Bhushan B., Gunewardena S., Akakpo J., Jaeschke H., Apte U. Pleiotropic role of p53 in injury and liver regeneration after acetaminophen overdose. Am J Pathol. 2018;188:1406–1418. doi: 10.1016/j.ajpath.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J., Wen Y., Zhou Y., Jiang Y., Chen Y., Zhang H., Guan L., Yao X., Huang M., Bi H. p53 attenuates acetaminophen-induced hepatotoxicity by regulating drug-metabolizing enzymes and transporter expression. Cell Death Dis. 2018;9:536. doi: 10.1038/s41419-018-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomita K., Teratani T., Suzuki T., Oshikawa T., Yokoyama H., Shimamura K., Nishiyama K., Mataki N., Irie R., Minamino T., Okada Y., Kurihara C., Ebinuma H., Saito H., Shimizu I., Yoshida Y., Hokari R., Sugiyama K., Hatsuse K., Yamamoto J., Kanai T., Miura S., Hibi T. p53/p66Shc-mediated signaling contributes to the progression of non-alcoholic steatohepatitis in humans and mice. J Hepatol. 2012;57:837–843. doi: 10.1016/j.jhep.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Derdak Z., Villegas K.A., Harb R., Wu A.M., Sousa A., Wands J.R. Inhibition of p53 attenuates steatosis and liver injury in a mouse model of non-alcoholic fatty liver disease. J Hepatol. 2013;58:785–791. doi: 10.1016/j.jhep.2012.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhury A.R., Ju Z., Djojosubroto M.W., Schienke A., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., Lee H.W., von Zglinicki T., Ganser A., Schirmacher P., Nakauchi H., Rudolph K.L. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- 16.De la Cueva E., Garcia-Cao I., Herranz M., Lopez P., Garcia-Palencia P., Flores J.M., Serrano M., Fernandez-Piqueras J., Martin-Caballero J. Tumorigenic activity of p21Waf1/Cip1 in thymic lymphoma. Oncogene. 2006;25:4128–4132. doi: 10.1038/sj.onc.1209432. [DOI] [PubMed] [Google Scholar]
- 17.Buitrago-Molina L.E., Marhenke S., Longerich T., Sharma A.D., Boukouris A.E., Geffers R., Guigas B., Manns M.P., Vogel A. The degree of liver injury determines the role of p21 in liver regeneration and hepatocarcinogenesis in mice. Hepatology. 2013;58:1143–1152. doi: 10.1002/hep.26412. [DOI] [PubMed] [Google Scholar]
- 18.Grompe M., Lindstedt S., al-Dhalimy M., Kennaway N.G., Papaconstantinou J., Torres-Ramos C.A., Ou C.N., Finegold M. Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1995;10:453–460. doi: 10.1038/ng0895-453. [DOI] [PubMed] [Google Scholar]
- 19.Buitrago-Molina L.E., Pothiraju D., Lamle J., Marhenke S., Kossatz U., Breuhahn K., Manns M.P., Malek N., Vogel A. Rapamycin delays tumor development in murine livers by inhibiting proliferation of hepatocytes with DNA damage. Hepatology. 2009;50:500–509. doi: 10.1002/hep.23014. [DOI] [PubMed] [Google Scholar]
- 20.Marhenke S., Lamle J., Buitrago-Molina L.E., Canon J.M., Geffers R., Finegold M., Sporn M., Yamamoto M., Manns M.P., Grompe M., Vogel A. Activation of nuclear factor E2-related factor 2 in hereditary tyrosinemia type 1 and its role in survival and tumor development. Hepatology. 2008;48:487–496. doi: 10.1002/hep.22391. [DOI] [PubMed] [Google Scholar]
- 21.Endig J., Buitrago-Molina L.E., Marhenke S., Reisinger F., Saborowski A., Schutt J., Limbourg F., Konecke C., Schreder A., Michael A., Misslitz A.C., Healy M.E., Geffers R., Clavel T., Haller D., Unger K., Finegold M., Weber A., Manns M.P., Longerich T., Heikenwalder M., Vogel A. Dual role of the adaptive immune system in liver injury and hepatocellular carcinoma development. Cancer Cell. 2016;30:308–323. doi: 10.1016/j.ccell.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Vogel A., van Den Berg I.E., Al-Dhalimy M., Groopman J., Ou C.N., Ryabinina O., Iordanov M.S., Finegold M., Grompe M. Chronic liver disease in murine hereditary tyrosinemia type 1 induces resistance to cell death. Hepatology. 2004;39:433–443. doi: 10.1002/hep.20077. [DOI] [PubMed] [Google Scholar]
- 23.Vogel A., Aslan J.E., Willenbring H., Klein C., Finegold M., Mount H., Thomas G., Grompe M. Sustained phosphorylation of Bid is a marker for resistance to Fas-induced apoptosis during chronic liver diseases. Gastroenterology. 2006;130:104–119. doi: 10.1053/j.gastro.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherr C.J., Roberts J.M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 25.Schungel S., Buitrago-Molina L.E., Nalapareddy P., Lebofsky M., Manns M.P., Jaeschke H., Gross A., Vogel A. The strength of the Fas ligand signal determines whether hepatocytes act as type 1 or type 2 cells in murine livers. Hepatology. 2009;50:1558–1566. doi: 10.1002/hep.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marhenke S., Buitrago-Molina L.E., Endig J., Orlik J., Schweitzer N., Klett S., Longerich T., Geffers R., Sanchez Munoz A., Dorrell C., Katz S.F., Lechel A., Weng H., Krech T., Lehmann U., Dooley S., Rudolph K.L., Manns M.P., Vogel A. p21 promotes sustained liver regeneration and hepatocarcinogenesis in chronic cholestatic liver injury. Gut. 2014;63:1501–1512. doi: 10.1136/gutjnl-2013-304829. [DOI] [PubMed] [Google Scholar]
- 27.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorquera R., Tanguay R.M. Fumarylacetoacetate, the metabolite accumulating in hereditary tyrosinemia, activates the ERK pathway and induces mitotic abnormalities and genomic instability. Hum Mol Genet. 2001;10:1741–1752. doi: 10.1093/hmg/10.17.1741. [DOI] [PubMed] [Google Scholar]
- 29.Jorquera R., Tanguay R.M. The mutagenicity of the tyrosine metabolite, fumarylacetoacetate, is enhanced by glutathione depletion. Biochem Biophys Res Commun. 1997;232:42–48. doi: 10.1006/bbrc.1997.6220. [DOI] [PubMed] [Google Scholar]
- 30.Gordon E.M., Ravicz J.R., Liu S., Chawla S.P., Hall F.L. Cell cycle checkpoint control: the cyclin G1/Mdm2/p53 axis emerges as a strategic target for broad-spectrum cancer gene therapy - a review of molecular mechanisms for oncologists. Mol Clin Oncol. 2018;9:115–134. doi: 10.3892/mco.2018.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen M.R., Factor V.M., Thorgeirsson S.S. Regulation of cyclin G1 during murine hepatic regeneration following Dipin-induced DNA damage. Hepatology. 1998;28:537–546. doi: 10.1002/hep.510280235. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L., Samuels T., Winckler S., Korgaonkar C., Tompkins V., Horne M.C., Quelle D.E. Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways. Mol Cancer Res. 2003;1:195–206. [PubMed] [Google Scholar]
- 33.Chen J.J., Yang T., Song S.H., Liu Q.G., Sun Y., Zhao L.H., Fu Z.R., Wang M.J., Hu Y.P., Chen F. Senescence suppressed proliferation of host hepatocytes is precondition for liver repopulation. Biochem Biophys Res Commun. 2019;516:591–598. doi: 10.1016/j.bbrc.2019.06.103. [DOI] [PubMed] [Google Scholar]
- 34.Wang C., Chen W.J., Wu Y.F., You P., Zheng S.Y., Liu C.C., Xiang D., Wang M.J., Cai Y.C., Zhao Q.H., Borjigin U., Liu W., Xiong W.J., Wangensteen K.J., Wang X., Liu Z.M., He Z.Y. The extent of liver injury determines hepatocyte fate toward senescence or cancer. Cell Death Dis. 2018;9:575. doi: 10.1038/s41419-018-0622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grompe M., Al-Dhalimy M., Finegold M., Ou C.N., Burlingame T., Kennaway N.G., Soriano P. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 1993;7:2298–2307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- 36.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. article 3. [DOI] [PubMed] [Google Scholar]
- 37.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102 doi: 10.1073/pnas.0506580102. 15545–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]